Ufmylation is the post-translational modification of proteins through the addition of UFM1. Nahorksi et al. identify mutations in UFM1 and in UFC1, which encodes an enzyme required for ufmylation, in individuals with severe early-onset encephalopathy with progressive microcephaly. The findings suggest an essential role for ufmylation in human brain development.
Keywords: ufmylation, UFM1, UFC1, encephalopathy, epilepsy
Abstract
The post-translational modification of proteins through the addition of UFM1, also known as ufmylation, plays a critical developmental role as revealed by studies in animal models. The recent finding that biallelic mutations in UBA5 (the E1-like enzyme for ufmylation) cause severe early-onset encephalopathy with progressive microcephaly implicates ufmylation in human brain development. More recently, a homozygous UFM1 variant was proposed as a candidate aetiology of severe early-onset encephalopathy with progressive microcephaly. Here, we establish a locus for severe early-onset encephalopathy with progressive microcephaly based on two families, and map the phenotype to a novel homozygous UFM1 mutation. This mutation has a significantly diminished capacity to form thioester intermediates with UBA5 and with UFC1 (the E2-like enzyme for ufmylation), with resulting impaired ufmylation of cellular proteins. Remarkably, in four additional families where eight children have severe early-onset encephalopathy with progressive microcephaly, we identified two biallelic UFC1 mutations, which impair UFM1-UFC1 intermediate formation with resulting widespread reduction of cellular ufmylation, a pattern similar to that observed with UFM1 mutation. The striking resemblance between UFM1- and UFC1-related clinical phenotype and biochemical derangements strongly argues for an essential role for ufmylation in human brain development. The hypomorphic nature of UFM1 and UFC1 mutations and the conspicuous depletion of biallelic null mutations in the components of this pathway in human genome databases suggest that it is necessary for embryonic survival, which is consistent with the embryonic lethal nature of knockout models for the orthologous genes.
Introduction
Post-translational modification greatly expands the functional repertoire of proteins beyond the confines of their coding sequence, and serves a diverse array of regulatory roles in protein turnover, localization and interactions (Hitosugi and Chen, 2014; Tompa et al., 2014). Ubiquitination is a well-established post-translational modification involving a series of enzymatic reactions that add ubiquitin to its target protein (Hershko and Ciechanover, 1998). These reactions involve a few E1 activating enzymes, dozens of E2 conjugating enzymes and hundreds of E3 ligating enzymes. More than a dozen ubiquitin-like (UBL) proteins have been found to similarly modify their target proteins (Schulman and Harper, 2009). UBL are classified into those that are activated and conjugated to substrates (type I) e.g. SUMO, NEDD8, ATG8, ATG12, URM1, UFM1, FAT10, and ISG15, and those that do not undergo conjugation (type II) (Cappadocia and Lima, 2018).
UFM1 (ubiquitin-fold modifier 1), a 9.1-kDa protein with a tertiary structure similar to ubiquitin, is among the most recently identified type I UBL proteins (Komatsu et al., 2004). The process of covalently attaching UFM1 to its target proteins, termed ufmylation, requires E1 activating enzymes (UBA5), E2 conjugating enzymes (UFC1), and an E3 ligase (UFL1) (Daniel and Liebau, 2014). UFM1 is activated by UBA5, forming a high-energy thioester bond. Activated UFM1 is then transferred to UFC1, in a similar thioester linkage (Komatsu et al., 2004). Ufmylation is completed by the catalysis of UFM1 transfer to the substrate protein through the action of the E3 enzyme UFL1 (Tatsumi et al., 2010). The physiological context of ufmylation remains incompletely understood. The literature on the relevance of ufmylation to human health was originally limited to cancer and, to a much lesser extent, complex diseases such as diabetes, ischaemic heart diseases and alcoholic liver disease (Liu et al., 2014; Yoo et al., 2015; Wei and Xu, 2016). As with other biochemical processes, however, Mendelian diseases involving deficiency of the individual components of ufmylation offered a unique opportunity to reveal the extent to which ufmylation influences human physiology. We and others have reported hypomorphic mutations in UBA5 in children with severe infantile onset epileptic encephalopathy (Colin et al., 2016; Duan et al., 2016; Muona et al., 2016). Additionally, Hamilton et al. (2017) reported that a homozygous mutation in UFM1 caused a severe early-onset encephalopathy with progressive microcephaly, although the mechanism remained unclear. In this report, we provide evidence that it is the process of ufmylation that is essential for normal nervous system development and function, based on the identification of UFM1 and UFC1 biallelic mutations, which we show lead to a remarkably similar neurological phenotype and accompanying impairment in ufmylation.
Materials and methods
Human subjects
Patients and available relatives were recruited with informed consent as part of IRB-approved research protocols (RAC# 2080006 and 2121053; StV 11/09). Clinical data, including laboratory and imaging studies, were collected from all participants. Blood was collected in EDTA tubes for DNA extraction and in Na-heparin tubes for the establishment of lymphoblastoid cell lines.
Autozygome analysis
Genomewide single nucleotide polymorphism (SNP) genotyping using Axiom SNP Chip (Affymetrix) from all available patients and relatives was pursued to determine the candidate autozygome as described before (Alkuraya, 2010, 2012). Runs of homozygosity >2 Mb were considered surrogates of autozygosity given the consanguineous nature of the study families as determined by AutoSNPa. Homozygosity mapping was performed on all available family members using HomozygosityMapper (http://www.homozygositymapper.org/).
Exome sequencing and variant filtering
Exome capture was performed using TruSeq Exome Enrichment kit (Illumina) following the manufacturer’s protocol. Samples were prepared as an Illumina sequencing library, and in the second step, the sequencing libraries were enriched for the desired target using the Illumina Exome Enrichment protocol. The captured libraries were sequenced using Illumina HiSeq 2000 Sequencer. The reads were mapped against UCSC hg19 (http://genome.ucsc.edu/) by BWA (http://bio-bwa.sourceforge.net/). The SNPs and indels were detected by SAMtools (http://samtools.sourceforge.net/). Variants from whole exome sequencing (WES) were filtered such that only novel (or very low frequency <0.1%), coding/splicing, homozygous variants that are within the candidate autozygome (autozygous intervals exclusive to the affected individuals) and are predicted to be pathogenic were considered as likely causal variants (Alkuraya, 2013, 2016). Frequency of variants was determined using publicly available variant databases (1000 Genomes, Exome Variant Server and ExAC) as well as a database of 2369 in-house ethnically-matched exomes. Pathogenicity is likely if the mutation is loss-of-function (splicing/truncating) or, in the case of missense/in-frame indels, removes a highly conserved amino acid and is predicted to be pathogenic by the three in silico prediction modules PolyPhen, SIFT and CADD.
In silico modelling
3D experimental structures were retrieved from the Protein Data Bank (PDB) and analysed using PYMOL (www.pymol.org).
Pull-down assay
Recombinant proteins were purified with glutathione S-transferase (GST) affinity purification system according to the manufacturer’s protocol (GE Healthcare). The GST moiety of all purified proteins without GST-UBA5 was removed on column by PreScission protease (GE Healthcare). GST-UBA5 bound to Glutathione Sepharose® 4B (GE Healthcare) was incubated for 20 min at 4°C with indicated purified proteins in pull-down assay buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.05% Nonidet P-40). The pulled-down protein complexes were extensively washed with pull-down assay buffer, and the obtained samples were subjected to NuPAGE® (4–12% acrylamide gradient) and Coomassie brilliant blue staining.
Cell culture
HEK293T cells were grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% foetal bovine serum (FBS), 5 U/ml penicillin, and 50 µg/ml streptomycin. Lymphoblasts were cultured in RPMI 1640 supplemented by 10% FBS, 5 U/ml penicillin, and 50 µg/ml streptomycin. To generate UFM1- and UFC1-knockout cells, each UFM1 and UFC1 guide RNA designed using the CRISPR Design tool (http://crispr.mit.edu/) was subcloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene #42230), a human codon-optimized SpCas9 and chimeric guide RNA expression plasmid. HEK293T cells were co-transfected with the pX330 and pEGFP-C1 (#6084-1, Clontech Laboratories) vectors, and cultured for 2 days. Thereafter, the GFP-positive cells were sorted and expanded. Loss of UFM1 and of UFC1 was confirmed by heteroduplex mobility assay followed by immunoblot analysis with anti-UFM1 and anti-UFC1 antibodies, respectively.
Immunoblot analysis
Cells were lysed with ice-cold TNE buffer (10 mM Tris-Cl, pH 7.5, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, and protease inhibitors). The samples were separated using the NuPAGE® system (Invitrogen) on 12% Bis-Tris gels in NuPAGE® MOPS SDS Running Buffer, and transferred to polyvinylidene difluoride (PVDF) membranes. Antibodies against FLAG (Medical & Biological Laboratories Co., Ltd., M185-3L), UFC (Abcam, ab189251) and UFM1 (Abcam, ab109305) were purchased from the indicated suppliers. Anti–UBA5 and UFM1 polyclonal antibodies were described previously (Komatsu et al., 2004). The immunoreactive bands were detected by LAS-4000 (GE Healthcare UK Ltd.). The quantitative densitometric analyses of FLAG-UBA5-MYC-UFM1, FLAG-UFC1-MYC-UFM1, endogenous UBA5-UFM1 intermediate, and endogenous UFC1-UFM1 intermediate relative to free FLAG-UBA5, FLAG-UFC1, endogenous UBA5, and endogenous UFC1 were carried out using Multi Gauge Version 3.2 Image software (Fuji Film, Tokyo, Japan). Statistical analysis was performed using an unpaired t-test (Welch test). The data represent the means ± standard error (SE) of three separate experiments.
In vitro thioester formation assay
In vitro thioester formation assay was conducted as previously reported (Komatsu et al., 2004). Briefly, recombinant GST-UFM1ΔC2, GST-UFM1ΔC2R81C, GST- UFM1ΔGly83, GST-UBA5, GST-UFC1, GST-UFC1R23Q and GST-UFC1T106I were produced in Escherichia coli and recombinant proteins were purified by chromatography on Glutathione Sepharose® 4B (GE Healthcare). After digestion of GST by PreScission Protease (GE Healthcare), the recombinant proteins were dialyzed against 50 mM BisTris (pH 6.5), 100 mM NaCl, 10 mM MgCl2, and 0.1 mM DTT (reaction buffer). Thioester formation reactions contained reaction buffer with 0.8 µg UFM1ΔC2, UFM1ΔC2R81C or UFM1ΔGly83 and some of the following: 5 mM ATP, 0.08 (for UFC1-UFM1 thioester formation assay) or 0.8 µg (for UBA5-UFM1 thioester formation assay) UBA5 and 0.8 µg UFC1, UFC1R23Q or UFC1T106I. Reactions were incubated for 5 min at 25°C and stopped by the addition of NuPAGE® LDS Sample Buffer lacking reducing agent, followed by a 10-min incubation at 37°C, NuPAGE® (4–12% acrylamide gradient) and Coomassie brilliant blue staining. Data shown are representative of three separate experiments.
Assessment of apoptosis in response to endoplasmic reticulum stress
HeLa or SH-SY5Y cells were plated into six-well plates at 7.5 × 105 cells/ml. After 18 h they were transfected with either 2 µg GFP, 1 µg UFM1WT/1 µg GFP or 1 µg UFM1R81C/1 µg GFP using FuGENE® HD or X-tremeGENE™ HP, respectively according to the manufacturer’s protocols. Media was changed after 24 h and tunicamycin added after 48 h. For endoplasmic reticulum stress marker analysis, cells were incubated with 5 µg/ml tunicamycin for 8 h. mRNA was extracted from cells using the RNeasy® Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. The concentration of the mRNA was calculated and 1 µg converted to cDNA using the qScript™ cDNA synthesis kit (Quanta Biosciences). Levels of HSPA5, DDIT3 and GAPDH were assayed using pre-designed TaqMan® Gene Expression Assays (ThermoFisher Scientific). The level of gene expression was normalized to that of GAPDH for each sample assessed. For assessment of apoptosis in response to endoplasmic reticulum stress, cells were incubated in different concentrations of tunicamycin for 48 h. They were then, trypsinized and stained with Annexin V, Alexa Fluor® 647 conjugate (ThermoFisher Scientific) and DAPI to assess early and late stages of apoptosis, respectively. Cells were analysed on a BD LSR Fortessa™ flow cytometer, selecting only those expressing GFP. Data were analysed using FlowJo software, and cells expressing either DAPI and/or Annexin V were considered apoptotic.
Results
Identification of novel severe early infantile encephalopathy phenotypes
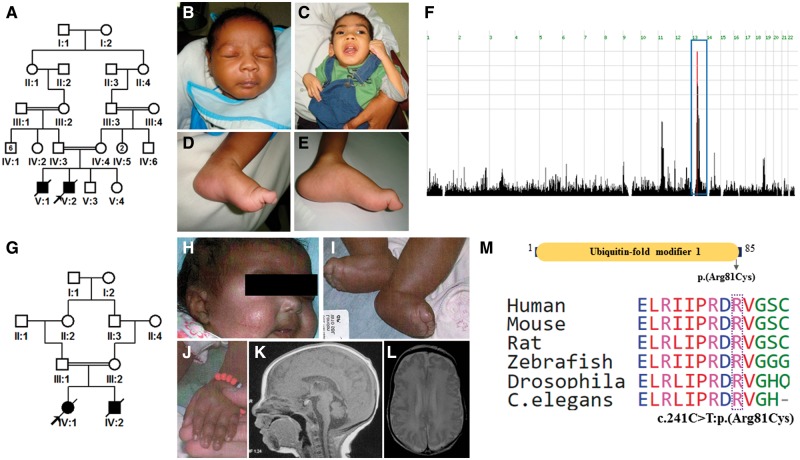
Two Sudanese families were recruited independently by C.G.W. and F.S.A., each with two children presenting with the core phenotype of profound global developmental delay, failure to thrive, progressive microcephaly and refractive epilepsy. Salient clinical features include subtle facial dysmorphism, severe axial hypotonia and appendicular hypertonia. Available brain imaging revealed dysmyelination and volume loss. Hypsarrhythmia was documented on EEG. The severity of the presentation is evidenced by the premature death of two of the four patients at ages 9 months and 8.5 years, respectively (Table 1, Fig. 1 and Supplementary Tables 1 and 2).
Table 1.
Summary of the clinical features of patients with UFM1 and UFC1 mutations
| ID | 12DG0178 | 12DG1577 | 14DG0050 | 16DG1614 | MDL-17-3196 | MDL-17-3892 | 17DG0828 | ID76366 | UK1 | UK2 | 10DG0945 | 10DG0946 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | UFC1 | UFC1 | UFC1 | UFC1 | UFC1 | UFC1 | UFC1 | UFC1 | UFM1 | UFM1 | UFM1 | UFM1 |
| Mutation | c.317C>T p.(Thr106Ile) | c.317C>T p.(Thr106Ile) | c.317C>T p.(Thr106Ile) | c.317C>T p.(Thr106Ile) | c.317C>T p.(Thr106Ile) | c.317C>T p.(Thr106Ile) | c.317C>T p.(Thr106Ile) | c.68G>A p.(Arg23Gln) | c.241C>T p.(Arg81Cys) | c.241C>T p.(Arg81Cys) | c.241C>T p.(Arg81Cys) | c.241C>T p.(Arg81Cys) |
| Age | 16 years | 23 years | 3 years | 5 years | 5 years | 31 months | 8 years | 4 years | 13 months | 13 months | 2 years | 1 year |
| Gender | F | F | F | F | F | F | F | M | F | M | M | M |
| Microcephalya | + | + | - | + | + | + | + | + | + | + | + | + |
| Short Stature | + | + | - | + | + | + | + | + | + | + | + | + |
| Underweight | + | + | + | + | + | + | + | + | + | + | + | + |
| GDD | + | + | + | + | + | + | + | + | + | + | + | + |
| Seizures | + | - | - | + | + | - | - | + | + | + | + | + |
GDD = global developmental delay; F = female; M = male.
aMicrocephaly was always secondary i.e. postnatal.
Figure 1.
UFM1-related clinical phenotype. (A) Pedigree of Family 1 with UFM1 mutation. (B and C) Images from Family 1 exhibiting lack of major facial dysmorphism. (D and E) Pes cavus as a result of abnormal tone in Patients 10DG0945 and 10DG0946, respectively (Family 1). (F) Genome-wide homozygosity mapping revealed a single critical locus. Blue box indicates haplotype (Chr13:36292810-42302880) identical by descent. (G) Pedigree of Family 2 with UFM1 mutation. (H) Facial image of Patient UK1 (Family 2) showing full cheeks. (I and J) Peripheral oedema of Patients UK1 and UK2 (Family 2). (K and L) Brain MRI of Patient UK1 showing cerebellar hypoplasia and thin corpus callosum and frontal cortical polymicrogyria. (M) Schematic diagram of UFM1 protein representing ubiquitin-fold modifier 1 domain with the mutation indicated. The Arg81 at the mutation site is highly conserved from humans to C. elegans.
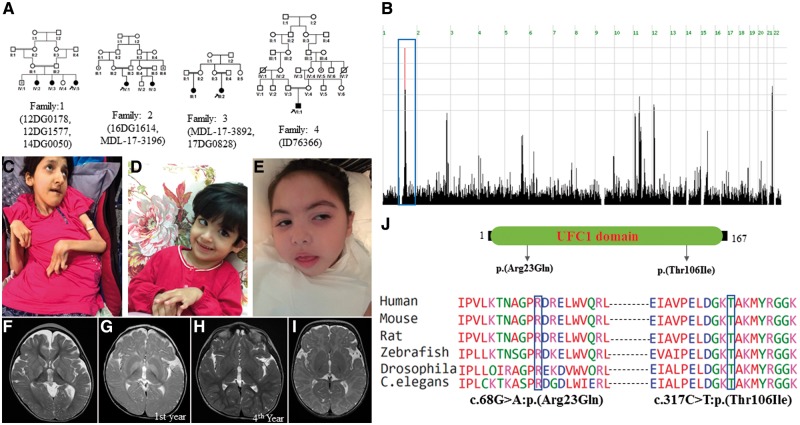
Independently, we encountered a highly similar phenotype in three Saudi families and one Swiss family with eight affected members (one of which was previously described, albeit briefly) (Anazi et al., 2016). They all presented with severe early infantile encephalopathy, progressive microcephaly, axial hypotonia, appendicular hypertonia and refractory epilepsy. Although brain MRI findings were largely non-specific, some had evidence of basal ganglia involvement (Table 1, Fig. 2 and Supplementary Tables 1 and 2). Phenotypic comparison of our patients with UFM1 and UFC1 mutations to previously reported patients with UFM1 and UBA5 mutations can be found in Table 2.
Figure 2.
UFC1-related clinical phenotype. (A) Pedigrees of the families with UFC1 mutation. (B) Genome-wide homozygosity plot highlighting a single critical locus from Families 1–3, who harbour the p.(Thr106Ile) mutation. Blue box indicates haplotype (Chr1:158957700-162094700) identical by descent. (C) Clinical images showing multiple joint contractures and emaciation (Patient 17DG0828). (D and E) Clinical images of younger patients suggest a progressive nature of emaciation (Patients 14DG0050 and 16DG1614, respectively). (F) Brain MRI of Patient 16DG1614 showing bilateral hyperintense signals in the basal ganglia. (G and H) Brain MRI of Patient MDL-17-3196 showing bilateral hyperintense signals in the basal ganglia in the first year that resolved in a repeated brain MRI in the fourth year of age. (I) Brain MRI of Patient ID76366 showing markedly delayed myelination. (J) Schematic diagram of UFC1 protein showing UFC1 domain and mutation site. Mutations Arg23Gln and Thr106Ile are highly conserved from humans to C. elegans.
Table 2.
Comparison of phenotype between this study patients with UFM1 and UFC1 mutations and those previously reported with mutations in UFM1 and UBA5
| Comparison between UFM1-UBA5-UFC1 pathway reported cases | ||||
|---|---|---|---|---|
| Current cohort | UFM1-related cases | UBA5-related cases | ||
| Gene | UFC1 | UFM1 | UFM1(PubMed: 28931644) | UBA5 |
| (PubMed: 27545681, 27545674, 28965491) | ||||
| Mutation | c.317C>T; c.68G>A | c.241C>T | c.-273_-271delTCA | c.1111G>A |
| c.904C>T | ||||
| c.971_972insC | ||||
| c.778G>A | ||||
| c.1165G>T | ||||
| c.169A>G | ||||
| c.503G>A | ||||
| c.164G>A | ||||
| c.684G > A | ||||
| Number of cases | 8 | 4 | 16 | 19 |
| Failure to thrive | 100% (8/8) | 100% (4/4) | 63% (10/16) | 89% (8/9) |
| Short stature | 88% (7/8) | 100% (4/4) | 75% (12/16) | 77% (10/13) |
| Microcephaly | 88% (7/8) | 100% (4/4) | 100% (16/16) | 100% (18/18) |
| Global developmental delay | 100% (8/8) | 100% (4/4) | 100% (16/16) | 100% (19/19) |
| Seizures | 50% (4/8) | 100% (4/4) | 75% (12/16) | 84% (16/19) |
| Brain MRI | ||||
| Basal ganglia abnormality | 33% (2/6) | 0% (0/4) | 100% (16/16) | 0% (0/17) |
| Delayed CNS myelination | 17% (1/6) | 75% (3/4) | 100% (16/16) | 24% (4/17) |
| Cerebellar hypoplasia | 0% (0/6) | 75% (3/4) | 81% (13/16) | 24% (4/17) |
| Mortality | 0% (0/8) | 100% (4/4) | 56% (9/16) | 24% (4/19) |
UFM1 and UFC1 define novel loci for severe infantile encephalopathy with progressive microcephaly
Although the two Sudanese families are not known to be related, they originate from the same village in Sudan. Indeed, autozygome analysis revealed a single shared autozygous interval (Chr13:36292810-42302880) between the affected members with the same ancestral haplotype, strongly supporting a recessive founder mutation. Exome sequencing of the index in each of the two families revealed the same sole novel homozygous variant within this interval: UFM1: NM_016617.3:c.241C>T:p.(Arg81Cys) (Fig. 1).
Similarly, genotyping of the Saudi families with a similar encephalopathy phenotype revealed that they all share the same ancestral haplotype (Chr1:158957700-162094700), again supporting a recessive founder mutation (Fig. 2). Indeed, exome sequencing on four of the seven patients revealed a single novel homozygous variant within the critical locus: UFC1: NM_016406.3:c.317C>T:p.(Thr106Ile), which segregated with the phenotype in all four families as revealed by targeted Sanger sequencing. Exome sequencing on a Swiss patient revealed a novel homozygous variant in UFC1: NM_016406.3:c.68G>A:p.(Arg23Gln) inherited from the heterozygous parents. Mutations at Thr106 and Arg23 involve highly conserved residues (Fig. 2).
Hypomorphic effect of UFM1 mutation on the UFM1-system
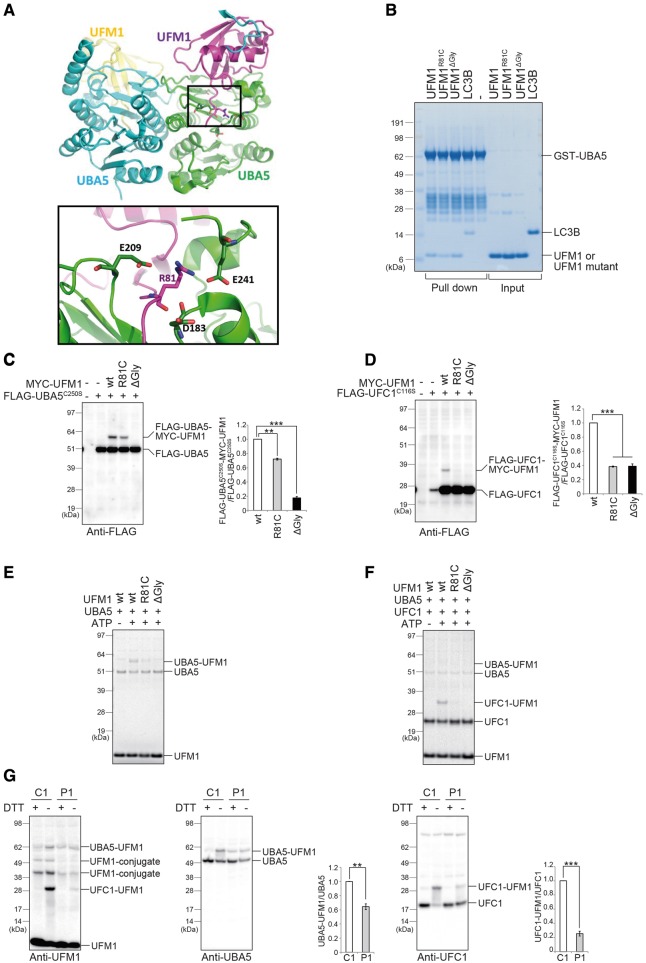
The UFM1 variant is predicted pathogenic by PolyPhen (0.538/possibly damaging), SIFT (0/ deleterious) and CADD (35), as are the UFC1 variants c.317C>T:p.(Thr106Ile) [PolyPhen (0.998/probably damaging), SIFT (0/deleterious), CADD (32)] and c.68G>A:p.(Arg23Gln) [PolyPhen (0.901/probably damaging), SIFT (0.02/deleterious) and CADD (35)]. We therefore carried out experiments to assess if and in which way these mutations affect ufmylation. The C-terminal tail of UFM1 (residues 79–83) is essential for its adenylation by the E1 enzyme UBA5 and subsequent thioester formation with UBA5 (Oweis et al., 2016). The crystallographic structure of UBA5 in the complex with UFM1 (PDB ID 5IAA) shows that the tail region, encompassing Arg81, directly binds to UBA5 (Fig. 3A). Since Arg81 interacts with the negatively charged residues Glu241, Glu209 and Asp183 of UBA5 (Fig. 3A). The substitution of Arg81 with a shorter and hydrophobic Cys is expected to markedly reduce the strength of the interaction between UFM1 and UBA5. Accordingly, an in vitro pull-down assay revealed that the mutant has a lower binding affinity to UBA5 than wild-type UFM1 (Fig. 3B). Next, we investigated whether UBA5 is able to activate UFM1R81C. To do this, we used a UBA5-mutant whose active site, cysteine (Cys250) was substituted with a serine (termed UBA5C250S). When the cysteine residue at the active site of E1 and E2 enzymes is replaced by a serine, an O-ester bond instead of a thioester bond is formed with its respective modifier proteins, which become stable even under reducing conditions (Komatsu et al., 2004). To exclude an effect of endogenous UFM1, we generated UFM1-deficient HEK293T cells (Supplementary Fig. 1A). As expected, when we co-expressed FLAG-UBA5C250S and wild-type MYC-UFM1 into UFM1-deficient HEK293T cells, MYC-UFM1 formed a stable intermediate with FLAG-UBA5C250S (Fig. 3C). Such intermediate formation was completely abrogated when UBA5C250S was co-expressed with MYC-UFM1ΔGly83, whose Gly83 essential for the adenylation and activation through UBA5 is deleted (Fig. 3C). Though we still observed the intermediate in the case of MYC-UFM1R81C, its level was only ∼75% of that of wild-type UFM1 (Fig. 3C).
Figure 3.
Hypomorphic effect of the UFM1 mutation on the UFM system. (A) Molecular basis for the effect of the UFM1R81C mutation. Top: Crystal structure of the heterodimeric complex between UFM1 (magenta and yellow) and UBA5 (cyan and green), taken from PDB id 5IAA. Bottom: Magnification of the boxed region. The view is tilted horizontally by ∼30° compared to the top panel. For clarity, only side chains of Arg81 and of Arg81-interacting residues are shown. (B) In vitro pull-down assay. Pull-down assay with GST-UBA5 and UFM1, UFM1 mutants or LC3B. GST-UBA5 conjugated with Glutathione Sepharose® 4B was incubated with purified recombinant UFM1, UFM1 mutants or LC3B. LC3B is known to interact with UBA5. The pulled-down complexes were subjected to NuPAGE® (4–12% acrylamide gradient) and Coomassie brilliant blue staining. GST-UBA5, LC3B, UFM1 and UFM1 mutants are indicated. (C and D) Immunoblot assay. Indicated constructs (0.1 µg for UBA5C250S, 0.5 µg for UFC1C116S, and 2 µg for UFM1 or mutants) were expressed in UFM1-deficient HEK293T cells. Twenty-four hours after transfection, the cell lysates were subjected to immunoblot analysis with anti-FLAG antibody. Bar graphs indicate the quantitative densitometric analyses of FLAG-UBA5-MYC-UFM1 and FLAG-UFC1-MYC-UFM1 intermediates relative to free FLAG-UBA5 and FLAG-UFC1, respectively. Statistical analyses were performed using the unpaired t-test (Welch test). The data represent the means ± SE of three separate experiments. **P < 0.01 and ***P < 0.001. (E and F) In vitro thioester formation assay of UFM1 by UBA5 (E) and of UFM1 by UFC1 (F). The assay was conducted as described in the ‘Materials and methods’ section. Data shown are representative of three separate experiments. (G) Immunoblot analysis in case (P1: 10DG0945, Individual V1 in Fig. 1A) and control (C1: a healthy Sudanese young female) lymphoblasts. Reducing (DTT plus) and non-reducing (DTT minus) samples were prepared from lymphoblasts and subjected to immunoblot analysis for UFM1 (left), UBA5 (middle), and UFC1 (right). We used a hand-made anti-UFM1 antibody in this experiment since the commercial antibody did not recognize UFM1R81C. Bar graphs indicate the quantitative densitometric analyses of UBA5-UFM1 and UFC1-UFM1 intermediates relative to free UBA5 and UFC1, respectively. Statistical analysis was performed using the unpaired t-test (Welch test). The data represent the means ± SE of three separate experiments. **P < 0.01 and ***P < 0.001.
The defective activation of UFM1R81C by the UBA5 E1 enzyme is expected to also hamper the subsequent step, namely the formation of the intermediate with its cognate E2-like enzyme UFC1. Therefore, we investigated whether UFM1R81C forms an intermediate with UFC1 in cells. When wild-type MYC-UFM1 was expressed together with FLAG-UFC1C116S in which the active site, cysteine (Cys116), was substituted with serine, we clearly detected the intermediate of UFC1C116S with MYC-UFM1 (Fig. 3D). Conversely, the formation of the UFM1-UFC1 intermediate was almost abolished when MYC-UFM1R81C or MYC-UFM1ΔGly83 were expressed together with UFC1C116S (Fig. 3D). In good agreement with those analyses, an in vitro thioester formation assay revealed that while wild-type UFM1 forms an intermediate with UBA5, UFM1R81C slightly had the ability to form the intermediate (Fig. 3E). We also observed that UFM1R81C is hardly transferred to UFC1 (Fig. 3F). In the next series of experiments, we tested the effect of the UFM1R81C mutation on the intermediate formation with endogenous UBA5 and UFC1 as well as UFM1-conjugation, using lymphoblasts derived from an affected individual. Expression of free UFM1 protein in lymphoblasts from an affected individual was comparable to control cells (Fig. 3G, left panel). By immunoblot analysis with non-reducing samples, we detected the intermediates of UFM1-UBA5 and of UFM1-UFC1. As predicted, the level of UFM1-UBA5 intermediates was significantly lower in lymphoblasts of affected individual compared to control cells (Fig. 3G, middle panel). Likewise, the formation of the UFM1-UFC1 intermediate markedly declined (Fig. 3G, right panel). We also found that the level of two UFM1-conjugates with cellular proteins in patient-derived lymphoblasts was significantly reduced compared to control lymphoblasts (Fig. 3G, left panel). Taken together, our data show that UFM1R81C has a hypomorphic effect on the UFM1-system and provide a plausible mechanistic explanation.
Hypomorphic effect of UFC1 mutations on the UFM1-system
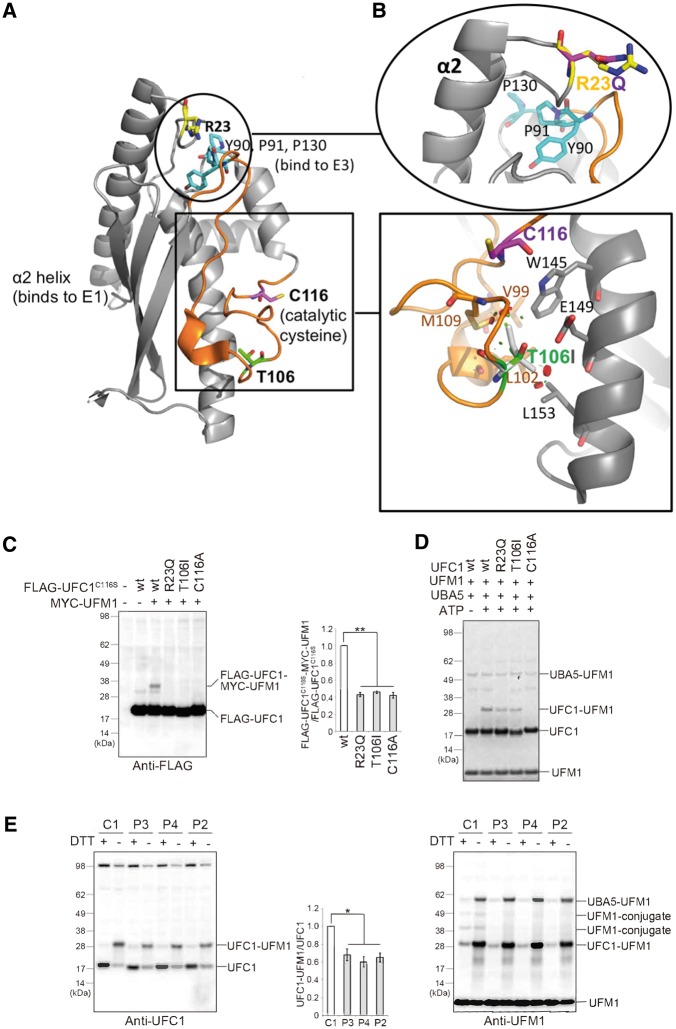
The UFC1 structure consists of the catalytic core domain conserved in all E2-like enzymes and an additional N-terminal helix (Mizushima et al., 2007). Thr106 on the UFC1 structure is on the opposite site of the proposed UBA5 binding site (involving helix α2) (Liu et al., 2009), suggesting that mutation of this residue does not affect the interaction with UBA5. Indeed, an in vitro pull-down assay showed that the Thr106Ile mutation does not have any effect on binding to UBA5 (Supplementary Fig. 2). Thr106 is located in a coiled region close to the catalytic Cys116, and within the site mapped to be required for interactions with UFM1 (Liu et al., 2009) (orange segment in Fig. 4A). In the crystal structure of unliganded UFC1, Thr106 interacts with the hydrophobic core of the protein and is mostly excluded from the solvent (Fig. 4B). Replacing the polar threonine with the slightly larger and completely hydrophobic isoleucine is expected to change the anchoring, stereochemistry and dynamics of this region, affecting the intermediate formation of UFM1 with UFC1. Similarly, Arg23 is close (∼10 Å) to the residues Tyr90, Pro91 and Pro130 that form a substructure and is potentially involved in E3-binding (Liu et al., 2009) (Fig. 4A). Arg23 is also near to the helix α2, which is involved in binding to E1 (Mizushima et al., 2007). Consequently, the substitution of the positively charged Arg23 with a shorter and uncharged glutamine may affect binding of UFC1 to E1 or E3 (Fig. 4B). Therefore, we tested whether the UFC1T106I and UFC1R23Q variants impact on the ufmylation. To do this, HEK293T cells deleting UFC1 were generated (Supplementary Fig. 1B), and MYC-UFM1 together with FLAG-UFC1C116S, FLAG-UFC1T106I/C116S or FLAG-UFC1R23Q/C116S was expressed in the UFC1-deficient HEK293T cells. While an UFC1-UFM1 intermediate was clearly detected in FLAG-UFC1C116S-expressing UFC1-deficient HEK293T cells, only a faint intermediate formation was observed in the case of FLAG-UFC1T106I/C116S and FLAG-UFC1R23Q/C116S (Fig. 4C). Our in vitro thioester formation assay also showed that while wild-type UFC1 formed an intermediate with UFM1, UFC1T106I and UFC1R23Q substantially reduced this ability (Fig. 4D). In lymphoblasts isolated from three affected individuals with UFC1T106I, the intermediate formation with endogenous UFM1 was significantly suppressed in comparison with that in control lymphoblasts (Fig. 4E, left panel). Similar to the case of UFM1R81C, the UFM1-conjugate formation with cellular proteins was impaired in patient-derived lymphoblasts (Fig. 4E, right panel). Taken together, we concluded that although the mutations UFM1R81C and UFC1T106I UFC1R23Q affect different proteins, they all impact ufmylation. Hence our analysis explains the similar patient phenotype by showing that these mutations cause phenotypically similar hypomorphic effects on the UFM1-system.
Figure 4.
Hypomorphic effect of UFC1 mutants on the UFM system. (A) Localization of the key binding sites on UFC1. Binding sites for E1 and E3 are indicated. Boxed region shows the active site, and the coiled region for binding site to UFM1 is coloured in orange. The mutated residues Arg23 and Thr106 are highlighted in yellow and green, respectively. Encircled region shows the proximity of R23 with regions involved in binding to E1 (helix α2) and E3 (Tyr90, Pro91, Pro130; cyan). (B) Bottom: Magnification of Thr106. Residues involved in hydrophobic or polar interactions with Thr106 are shown as stick models. Colouring as in A, except for the mutant Ile106, which is shown as light grey stick model, with positions of minor clashes indicated as discs. Top: Magnification of Arg23. Colouring as in A, except for the mutant Gln23, which is shown as magenta stick model. (C) Immunoblot assay. Indicated constructs (0.5 μg for UFC1C116S, UFC1C116S/T106I or UFC1C116S/R23Q and 2 μg for UFM1) were expressed in UFC1-deficient HEK293T cells. Twenty-four hours after transfection, the cell lysates were subjected to immunoblot analysis with indicated antibodies. Bar graphs indicate the quantitative densitometric analyses of FLAG-UFC1-MYC-UFM1 intermediates relative to free FLAG-UFC1. Statistical analyses were performed using the unpaired t-test (Welch test). The data represent the means ± SE of three separate experiments. *P < 0.05. (D) In vitro thioester formation assay of UFM1 by UFC1. The assay was conducted as described in the ‘Materials and methods’ section. Data shown are representative of three separate experiments. (E) Immunoblot analysis in case (P2, P3, P4: V:1) and control (C1: a healthy Sudanese young females) lymphoblasts. Reducing (DTT plus) and non-reducing (DTT minus) samples were prepared from lymphoblasts and subjected to immunoblot analysis for UFC1 (left), and UFM1 (right). Bar graph indicates the quantitative densitometric analysis of UFC1-UFM1 intermediates relative to free UFC1. Statistical analysis was performed using the unpaired t-test (Welch test). The data represent the means ± SE of three separate experiments. **P < 0.01.
Mutation in UFM1 does not affect endoplasmic reticulum stress-mediated apoptosis
Studies have reported that the pathogenesis of ufmylation-related encephalopathy is associated with endoplasmic reticulum stress-mediated apoptosis. Thus we tested UFM1 mutation (p.R81C) effect on endoplasmic reticulum stress. We found no significant impact of UFM1 wild-type or mutant expression on the induction of endoplasmic reticulum stress markers CHOP (DDIT3) and BIP (HSPA5) in either HeLa or SH-SY5Y cells, after 8 h treatment with 5 µg/ml tunicamycin (Supplementary Fig. 3). Neither did we note appreciable difference in endoplasmic reticulum stress-induced apoptosis in cells expressing UFM1 wild-type or UFM1 p.R81C, after 48 h incubation in tunicamycin at various concentrations.
Discussion
Our understanding of the pathogenesis of infantile encephalopathy at the molecular level has greatly expanded in recent years due in large part to the rapidly growing use of genomics to diagnose and classify this highly heterogeneous group of disorders. The remarkable diversity and breadth of implicated molecular pathways are consistent with the highly complex nature of the brain (Hamdan et al., 2017). Indeed, the observation that the majority of these conditions are not associated with significant systemic findings, further supports the notion that brain vulnerability is inherent to its complexity. This is especially remarkable when one considers the fundamental and ubiquitous nature of the many biological processes that are impaired in the various genetic forms of infantile encephalopathy, including post-translational modification (Alazami et al., 2015; Anazi et al., 2016, 2017).
Ufmylation is a highly conserved post-translational modification with each of its components having a corresponding orthologue in all multicellular organisms. In this study, we show that this pathway is critically required for normal brain development and function in humans consistent with suggestive data from model organisms. For example, the fruitfly models of UBA5 and UFM1 deficiency display increased mortality, locomotive defects, and abnormal neuromuscular junctions (Duan et al., 2016). Similarly, deficiency of the UBA5 orthologue in Caenorhabditis elegans results in increased susceptibility to induced seizures and pharynx grinder paralysis as well as abnormal sensorial behaviour (Colin et al., 2016). Impaired motility and seizure-like activity were also observed in zebrafish uba5 morphants (Colin et al., 2016). More relevant to the phenotype of the patients we described in this study is our recently published brain-specific conditional knockout of Ufm1 under the nestin promoter (Muona et al., 2016), which allowed us to directly observe the brain pathology associated with impaired ufmylation in vivo. Although these mice appeared normal at birth, they uniformly died in the first day after birth. Histopathological examination revealed that their brains were microcephalic with evidence of increased apoptosis, consistent with the proposed role of ufmylation in neuronal development and survival (Muona et al., 2016).
Similar to our previous work on UBA5-related severe infantile encephalopathy, we show that mutations in UFM1 itself as well as in UFC1, encoding the sole E2 conjugating enzyme for UFM lead to widespread impairment of ufmylation. Our results show a reduction rather than abrogation of ufmylation with the activity of UFM1 and UFC1 mutants at 60–75% of their wild-type counterparts. A recently reported promoter mutation in UFM1 in the context of infantile encephalopathy with or without epilepsy is similarly predicted to only reduce but not eliminate transcription of an otherwise normal transcript (Hamilton et al., 2017). The clinical phenotype of previously reported patients with UBA5 (Colin et al., 2016; Duan et al., 2016; Muona et al., 2016) and UFM1 (Hamilton et al., 2017) mutations are similar to our UFM1 and UFC1 patients, particularly regarding failure to thrive, short stature, microcephaly, GDD, seizures, basal ganglia abnormality, delayed CNS myelination, and cerebellar hypoplasia (Table 2). These observations strongly argue for a minimum threshold required for embryonic viability, and this would be consistent with the mouse embryonic lethal phenotype observed in the complete knockout of Uba5, Ufbp1, Ufl1 or Ufm1 (M.K., unpublished data) (Tatsumi et al., 2011; Cai et al., 2015; Zhang et al., 2015).
The pathogenesis of ufmylation-related encephalopathy remains unclear e.g. is the disease process a progressive apoptosis of selected neuron classes, or a neuronal dysfunction that becomes increasingly evident during development? The known localization of the ufmylation cascade and target proteins to the luminal side of the endoplasmic reticulum and the proposed role in regulating the unfolded protein response (UPR) and endoplasmic reticulum stress-mediated apoptosis, suggest a potential mechanistic link (Lemaire et al., 2011; Zhang et al., 2015; Ishimura et al., 2017). Expanded endoplasmic reticulum network and increased endoplasmic reticulum volume in response to tunicamycin, both markers of endoplasmic reticulum stress, have indeed been observed in fibroblasts from patients with UBA5 mutations (Colin et al., 2016). Long-term endoplasmic reticulum stress driven by mutation of specific disease-related genes is known to cause various adult-onset neurodegenerative phenotypes by overwhelming the UPR and inducing apoptosis (Hetz and Saxena, 2017). While we found no evidence of UFM1 wild-type or p.R81C overexpression affecting endoplasmic reticulum stress induction or apoptosis in response to tunicamycin (Supplementary Fig. 3), further work should serve to investigate the mechanisms by which defects in ufmylation lead to early-onset encephalopathy in humans. However, it may be that the ufmylation cascade, rather than a global effect on endoplasmic reticulum or UPR, could disrupt neuron function through effects on neural-essential proteins; the neuronal cell adhesion molecule (NCAM) interacts and co-localizes with UFC1, and CDK5 activity is controlled by CDK5RAP3, which aggregates with UFL1 and UFSP2 at the endoplasmic reticulum membrane (Homrich et al., 2014).
In conclusion, our study suggests that impaired ufmylation leads to a recognizable syndrome of severe infantile encephalopathy and progressive microcephaly with or without epilepsy. Further studies are needed to discern the exact pathomechanism of ufmylation-related neurodevelopmental disorder, which may lead to possible therapies especially when one considers the hypomorphic nature of the observed mutations.
Supplementary Material
Acknowledgements
We thank the study families for their enthusiastic participation. We also thank the Sequencing and Genotyping Core Facilities at KFSHRC for their technical support.
Funding
M.N. is supported by the Wellcome Trust. R.I. is supported by Grant-in-Aid for JSPS Research Fellows (JP16J07037). C.G.W. acknowledges support from the NIHR Cambridge Biomedical Research Campus. M.K. is supported by Grant-in-Aid for Scientific Research on Innovative Areas (JP25111006 and 15K21749 to M.K.), a Japan Society for the Promotion of Science (an A3 foresight program, to M.K.), and the Takeda Science Foundation (to M.K.). F.S.A. is supported by King Salman Center for Disability Research and King Abdulaziz City for Science and Technology (13-BIO1113-20, and Saudi Human Genome Program). S.T.A. and F.J.G.V. are supported by funding from King Abdullah University of Science and Technology (KAUST). A.R. is supported by radiz—Rare Disease Initiative Zürich, Clinical Research Priority Program for Rare Diseases of the University of Zurich.
Supplementary material
Supplementary material is available at Brain online.
References
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 2015; 10: 148–61. [DOI] [PubMed] [Google Scholar]
- Alkuraya FS. Autozygome decoded. Genet Med 2010; 12: 765–71. [DOI] [PubMed] [Google Scholar]
- Alkuraya FS. Discovery of rare homozygous mutations from studies of consanguineous pedigrees. Curr Protoc Hum Genet 2012; Chapter 6: Unit 6.12. [DOI] [PubMed] [Google Scholar]
- Alkuraya FS. The application of next-generation sequencing in the autozygosity mapping of human recessive diseases. Hum Genet 2013; 132: 1197–211. [DOI] [PubMed] [Google Scholar]
- Alkuraya FS. Discovery of mutations for Mendelian disorders. Hum Genet 2016; 135: 615–23. [DOI] [PubMed] [Google Scholar]
- Anazi S, Maddirevula S, Faqeih E, Alsedairy H, Alzahrani F, Shamseldin H, et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol Psychiatry 2016; 22: 615–24. [DOI] [PubMed] [Google Scholar]
- Anazi S, Maddirevula S, Salpietro V, Asi YT, Alsahli S, Alhashem A, et al. Expanding the genetic heterogeneity of intellectual disability. Hum Genet 2017; 136: 1419–29. [DOI] [PubMed] [Google Scholar]
- Cai Y, Pi W, Sivaprakasam S, Zhu X, Zhang M, Chen J, et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet 2015; 11: e1005643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev 2018; 118: 889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E, Daniel J, Ziegler A, Wakim J, Scrivo A, Haack TB, et al. Biallelic variants in UBA5 reveal that disruption of the UFM1 cascade can result in early-onset encephalopathy. Am J Hum Genet 2016; 99: 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J, Liebau E. The ufm1 cascade. Cells 2014; 3: 627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Shi Y, Yu L, Zhang G, Li J, Lin Y, et al. UBA5 mutations cause a new form of autosomal recessive cerebellar ataxia. PLoS One 2016; 11: e0149039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet 2017; 101: 664–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EM, Bertini E, Kalaydjieva L, Morar B, Dojčáková D, Liu J, et al. UFM1 founder mutation in the Roma population causes recessive variant of H-ABC. Neurology 2017; 89: 1821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998; 67: 425–79. [DOI] [PubMed] [Google Scholar]
- Hetz C, Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 2017; 13: 477–91. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Chen J. Post-translational modifications and the Warburg effect. Oncogene 2014; 33: 4279–85. [DOI] [PubMed] [Google Scholar]
- Homrich M, Wobst H, Laurini C, Sabrowski J, Schmitz B, Diestel S. Cytoplasmic domain of NCAM140 interacts with ubiquitin-fold modifier-conjugating enzyme-1 (Ufc1). Exp Cell Res 2014; 324: 192–9. [DOI] [PubMed] [Google Scholar]
- Ishimura R, Obata M, Kageyama S, Daniel J, Tanaka K, Komatsu M. A novel approach to assess the ubiquitin-fold modifier 1-system in cells. FEBS Lett 2017; 591: 196–204. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Chiba T, Tatsumi K, Iemura Si, Tanida I, Okazaki N, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 2004; 23: 1977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K, Moura RF, Granvik M, Igoillo-Esteve M, Hohmeier HE, Hendrickx N, et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS One 2011; 6: e18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Forouhar F, Eletsky A, Atreya HS, Aramini JM, Xiao R, et al. NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway. J Struct Funct Genomics 2009; 10: 127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Li J, Tillman B, French B, French S. Ufmylation and FATylation pathways are downregulated in human alcoholic and nonalcoholic steatohepatitis, and mice fed DDC, where Mallory–Denk bodies (MDBs) form. Exp Mol Pathol 2014; 97: 81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T, Tatsumi K, Ozaki Y, Kawakami T, Suzuki A, Ogasahara K, et al. Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem Biophys Res Commun 2007; 362: 1079–84. [DOI] [PubMed] [Google Scholar]
- Muona M, Ishimura R, Laari A, Ichimura Y, Linnankivi T, Keski-Filppula R, et al. Biallelic variants in UBA5 link dysfunctional UFM1 ubiquitin-like modifier pathway to severe infantile-onset encephalopathy. Am J Hum Genet 2016; 99: 683–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oweis W, Padala P, Hassouna F, Cohen-Kfir E, Gibbs DR, Todd EA, et al. Trans-binding mechanism of ubiquitin-like protein activation revealed by a UBA5-UFM1 complex. Cell Rep 2016; 16: 3113–20. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol 2009; 10: 319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Sou YS, Tada N, Nakamura E, Iemura S, Natsume T, et al. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem 2010; 285: 5417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Yamamoto-Mukai H, Shimizu R, Waguri S, Sou YS, Sakamoto A, et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat Commun 2011; 2: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Davey NE, Gibson TJ, Babu MM. A million peptide motifs for the molecular biologist. Mol Cell 2014; 55: 161–9. [DOI] [PubMed] [Google Scholar]
- Wei Y, Xu X. UFMylation: a unique & fashionable modification for life. Genomics Proteomics Bioinformatics 2016; 14: 140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HM, Park JH, Jeon YJ, Chung CH. Ubiquitin-fold modifier 1 acts as a positive regulator of breast cancer. Front Endocrinol 2015; 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhu X, Zhang Y, Cai Y, Chen J, Sivaprakasam S, et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ 2015; 22: 1922–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.