Abstract
In virtually all eukaryotic organisms, linker DNA between nucleosomes is associated with a histone termed linker histone or histone H1. In Saccharomyces cerevisiae, HHO1 encodes a putative linker histone with very significant homology to histone H1. The encoded protein is expressed in the nucleus, but has not been shown to affect global chromatin structure, nor has its deletion shown any detectable phenotype. In vitro chromatin assembly experiments with recombinant HHO1p have shown that it is able to complex with dinuncleosomes in a similar manner to histone H1. Here we report that while disruption of HHO1 has little affect on RNA levels of most cellular transcripts, there are numerous exceptions. Measurement of HHO1p concentration in the wild-type cell showed a stoichiometry of about one HHO1p molecule per 37 nucleosomes. Localization of HHO1p in the chromatin, using an immunoprecipitation technique, showed preferential HHO1p binding to rDNA sequences. These results suggest that HHO1p may play a similar role to linker histones, but at restricted locations in the chromatin.
INTRODUCTION
Eukaryotic DNA is packaged in the nucleus in a very precise way that enables it to contend with numerous topological and physical constraints. The underlying structure of DNA compaction is extremely well conserved in eukaryotes ranging from yeast to man (reviewed in 1). The fundamental packing unit, known as the nucleosome (2,3), consists of two full superhelical turns of DNA wrapped around a core containing two each of the four conserved core histones H2A, H2B, H3 and H4. Digestion of the nucleosome with micrococcal nuclease yields a core particle containing 146 bp of DNA. Each nucleosome is separated from the next by a region of linker DNA, which is associated with a single molecule of a fifth, less conserved histone usually referred to as the linker histone or H1. The linker DNA can vary in length from close to 0 to 80 nucleotide pairs, depending on the species.
The linker histone has been shown to be located in the interior of the folded structure (4) where the DNA enters and leaves the nucleosome. By binding to the DNA at these positions, the linker histones fix the entry/exit angle of the DNA and thus, together with other constraints, such as length and rigidity of the linker DNA, contribute to the formation of higher order chromatin structure. In vitro they show a strong preference for binding of superhelical/supercoiled DNA, compared with linear or relaxed circular DNA.
Curiously, much of the work attempting to identify a function for the nearly ubiquitous histone H1 has not been very rewarding, though the molecule has been extensively studied. It has been proposed that H1 is involved in transcription control (see for example 5) and in setting the linker distance between nucleosomes (6–8), though conflicting evidence has been presented for both proposed functions. There have been a few successful attempts to disrupt the genes encoding H1 and/or the H1 isoforms. Surprisingly, elimination of all of these gene products in Tetrahymena (9) and of histone H1(zero) in mice (10) did not result in dramatic phenotypes. In tobacco plants, overexpressing a gene encoding histone H1 from Arabidopsis thaliana caused a decreased level of their own H1 (11). The total histone H1 level increased 2.3–2.8 times. While the resulting drastic change in H1:DNA stoichiometry had a limited effect on basal cellular functions, it did affect plant morphology and flowering. These results indicate that, at least in plants, the regulatory function of H1 with respect to transcription may be limited to a class of genes responsible for specific developmental programs.
In Tetrahymena, linker histone is required for activated expression of at least one gene called CyP (encoding a cysteine protease) that is strongly induced by starvation (12). H1 is not required for repression of CyP in growing cells, but it is required for activated expression upon starvation (13). Another gene, ngoA, also induced by starvation, requires H1 for normal basal repression in growing cells, but does not require H1 for activated expression in starved cells. Importantly, phosphorylation of H1 mimicked its depletion (14). Most recently, it was shown that the effect of phosphorylation in Tetrahymena is due to the overall charge of the small domain where phosphorylation occurs (15).
In Xenopus it was shown that different variants of H1 are necessary for selective transcriptional silencing of regulatory genes that are required for mesodermal differentiation (16). In addition, TFIIIA transcription of 5S ribosomal RNA (rRNA) genes is regulated by histone H1 in Xenopus. An increase in histone H1 content in the cell specifically restricts 5S rRNA gene transcription, while a decrease in histone H1 within chromatin facilitates activation of the oocyte 5S rRNA genes (17). A number of different isoforms of this histone have been described in various cell types [e.g. H5 and H1(zero)] (18) which appear to be differentially expressed in a number of tissues at varying stages of differentiation (5,19,20).
In chicken tissue culture cells, deletion of all but one of the H1 genes caused changes in the pattern of proteins analyzed on 2-dimensional gels, though cell growth was unaffected (21). Overexpression of H1 variants in mouse tissue culture cells specifically increased (rather than repressed) basal and induced expression from the MMTV promoter (22). Most recently, it was shown that histone H1 is essential for a long lifespan in the fungus Ascobolus immersus (23). Taken together, these results indicate that H1 function in vivo may be gene specific and that it may not be a general structural component of chromatin.
In yeast, biochemical efforts to definitively identify histone H1 were unsuccessful. However, as the sequence of the entire yeast genome became known (24), Landsman (25) asked whether an H1 homolog exists in yeast. Using the conserved globular domain of histone H1 as a basis for searching the Saccharomyces database, he identified an open reading frame (ORF) encoding a putative protein that has very significant homology to the globular domains of many H1 proteins. The yeast putative H1, termed HHO1p, has an N-terminal peptide that is lysine-rich, followed by a well-conserved globular domain, followed by a short peptide with similar composition to the C-terminal H1 tails, followed by another globular domain. No other linker histone molecules are known to contain two globular domains.
Ushinsky et al. (26) disrupted HHO1. They found no observable phenotype, but found that HHO1p tagged with green fluorescence protein and expressed from a plasmid localizes to the nucleus.
Patterton et al. (27) performed a comprehensive survey of potential phenotypes that might result from an HHO1 deletion, but they too were unable to detect a phenotype particular to hho1Δ. They tested basal transcription levels in a minimal PHO5 promoter and showed no difference between wild-type and hho1Δ cells. They also showed that HHO1p is not required for telomeric repression, nor is it required for efficient sporulation. However, they showed that purified recombinant HHO1p, like histone H1, was able to form a stable ternary complex with a reconstituted core dinucleosome in vitro at a molar ratio of one. They also reported that the reconstituted nucleosomes showed a kinetic pause at ∼168 bp after micrococcal nuclease digestion of chromatin, as do nucleosomes associated with histone H1. In vivo bulk chromatin structure was unaffected by deletion of HHO1.
Puig et al. (28) asked whether structural changes could be noted at a specific locus in the chromatin following HHO1 disruption. They chose a particular locus in the genome which includes POT1 and YIL161w that is located between two strongly micrococcal nuclease hypersensitive sites and is flanked by an array of 13 strictly positioned nucleosomes. They too were unable to detect a difference in the nucleosome spacing between wild-type and hho1Δ strains.
Despite all the above data that could indicate that HHO1p does not have a significant function in the cell, Spellman et al. (29), using gene array technology, reported that the only genes transcribed during S phase in yeast are HHO1 and those genes encoding the core histones (HTA1, HTA2 encoding H2A, HTB1, HTB2 encoding H2B, HHT1, HHT2, encoding H3 and HHF1, HHF2 encoding H4). As HHO1 transcription is coordinately regulated with the core histone genes, it is likely that HHO1p somehow functions in a coordinated fashion with the core histones.
In this study, we further addressed the assignment of HHO1p as the H1 linker histone in Saccharomyces cerevisiae. We show that in wild-type cells, HHO1 is both transcribed and translated and that the protein co-purifies with the core histones. We measured its relative stoichiometry to the core histones in the cell, finding that it is found in far fewer copies in the cell than nucleosomes. Using a DNA array technique we show that HHO1 disruption does have a transcription effect on a subset of genes. Using chromatin immunoprecipitation, we directly probed where HHO1p might be located in the chromatin and showed that it is preferentially concentrated at the repeated sequences that encode rRNA.
MATERIALS AND METHODS
Yeast strains and media
Strain CY26 (MATα, ura3-52, leu2-Δ1, his3-Δ200, trp1-Δ1, lys2-801, ade2-101) was considered wild-type with respect to HHO1. Standard yeast YPD and selective dropout media were used throughout.
hho1Δ strain construction
The complete HHO1 ORF was amplified by PCR from a chromosomal DNA template from strain CY26 using primers IF1 and IF2 (Table 1). Following digestion with XbaI and EcoRI, the PCR product was ligated into a similarly digested pBluescript II SK+ vector. An internal 527 nt Eco47III–BclI fragment of HHO1 was then replaced with an Eco47III–BamH1 fragment containing the HIS3 gene. CY26 was transformed using the LiAcet procedure (30) with a linear EcoRI–NotI fragment from the above construct that contained 99 nt upstream of the HHO1 coding sequence, the first 149 nt of the HHO1 ORF, HIS3, a stop codon, the last 95 nt of the HHO1 ORF and 203 nt downstream of the HHO1 ORF. Transformants were selected on medium lacking histidine. The disruption was verified by PCR and restriction analysis.
Table 1. Primers used in PCR reactions.
| Name |
Sequence |
Comments |
| IF1 | GGTCTAGAACAAATAGAGAAGGGAGAG | For hho1Δ construction. XbaI site underlined |
| IF2 | GGAATTCTTAAAGAGGAGGAGCAAC | For hho1Δ construction. EcoRI site underlined |
| IF3 | AATACGGTAATCAGATGAGC | For p25 construction |
| IF4 | ATTGAACCATATGCGTGGAGAGTTTGACCTTC | For p25 construction. NdeI site underlined |
| IF5 | TATTGCTATCACCATTGACATTCTCGTTTGGATATTCACTTTTTATCGTAATATATGTGTACTTTGC | For construction of chromosomal HHO1p–2HA |
| IF6 | CCCCTCCGGCATTATTAAAC | For construction of chromosomal HHO1p–2HA |
| IF7 | AAGAATTCATGGCACCCAAGAAATCC | pGEX-HHO1 construction. EcoRI site underlined |
| IF8 | ACCGCTCGAGGGATATTCACTTTTTACGTG | pGEX-HHO1 construction. XhoI site underlined |
| IF9 | AGGACGTCATAGAGGGTGAGAATC | nt 201–224 of 25S rDNA transcript |
| IF10 | TTGACTTACGTCGCAGTCCTCAGT | nt 585–608 of 25S rDNA transcript |
| IF11 | AATAGCCGGTCGCAAGACTGTGATT | nt 350–374 of large 35S rDNA transcript (precedes 18S rDNA) |
| 1F12 | CCACCTATTCCCTCTTGCTAGAAG | nt 611–634 of large 35S rDNA transcript (precedes 18S rDNA) |
| IF13 | CTAGTAACAAGGCTAAGATATCAG | CUP1 |
| IF14 | GTAAGCCGATCCCATTACCGACAT | CUP1 |
| IF17 | TGACAACATCAACAACGAAG | HSP26 |
| IF18 | TAGGGAAACCGAAACCAG | HSP26 |
| IF19 | GTGTGCAGATTTACTTGACC | FET3 |
| IF20 | ACCTTTCATCCCGTCTTC | FET3 |
| IF21 | TATCGGGCTCAACAGTTTATT | RPL25 |
| IF22 | GGCCTTCTTGATTTGGTATTT | RPL25 |
| IF23 | AGAGAGTGAAGACGAGGGTG | CDC28 |
| IF24 | TTGGAAGTAGGGGTGGATG | CDC28 |
| IF25 | CTTTACAAAGCGAATCGTCTT | SPS100 |
| IF26 | CCATGATGCCACGGTA | SPS100 |
| IF27 | GGTTCAATCGAGTGCTACACTTAT | FRE1 |
| IF28 | GCTTGAACATTGGGATGCTAACTT | FRE1 |
| IF29 | GCATCCAAGACAGTAGCATCGAGT | CTR1 |
| IF30 | CATCCCTGAAGAGCTACTGTTATC | CTR1 |
| IF31 | GGCCTCTGCTATAAATATCAACGA | SAG1 |
| IF32 | AGCCTGTTGCGAAACATAGCATTT | SAG1 |
| IF33 | TGGCTGCTAATGATGACATCAATG | LYS9 |
| IF34 | GTGGTCGATACCTGGATCCAACCC | LYS9 |
HHO1–2HAp strain construction
The full-length HHO1 ORF was amplified by PCR from strain CY26 chromosomal DNA using primers IF3 and IF4. An 867 nt ClaI–NdeI fragment containing sequences upstream of the HHO1 ORF and the entire HHO1 ORF without a stop codon was ligated to pJL36 (31) that had been digested with Bsu15I and NdeI. The result was the HHO1 ORF fused in-frame to two tandem hemagglutinin (2HA) sequences. The resulting plasmid (termed p25) served as a template in PCR using primers IF5 and IF6. The PCR product contained the terminal 49 nt of the HHO1 ORF, 2HA, plasmid sequence containing TRP1 and the 47 nt immediately downstream of the HHO1 ORF. This fragment was used to transform CY26, selecting for tryptophan auxotrophy. Transformants were verified for proper chromosomal integration by PCR and restriction analysis. The resultant construct replaces the HHO1 ORF with HHO1–2HA at the original chromosomal location.
Protein extraction and western analysis
Total yeast proteins were obtained as follows. Log phase yeast were disrupted at 4°C in 33 mM Tris–HCl, pH 7.5, 12% glycerol, 1.5 mM MgCl2, 0.1 mM EDTA, 1 mM PMSF, 0.2 µM pepstatin A, 1.2 µM leupeptin, 0.5 mM DTT, 0.3 M KCl by vortexing for several minutes with glass beads (0.5 mm diameter). Lysate was recovered by centrifugation.
Histones were extracted from isolated nuclei. Nuclei were isolated by Zymolyase treatment of whole cells in a buffer containing 1.08 M sorbitol, 20 mM KPO4, 0.5 mM CaCl2, 5 mM DTT and 1 mM PMSF. Cells were pelleted, resuspended in a lysis buffer containing 18% Ficoll 400, 20 mM KPO4, pH 6.8, 0.5 mM CaCl2, 3 mM DTT, 1 mM PMSF, 1 mM AEBSF, 100 mM Na butyrate and 1 µM trichostatin A and homogenized with a Teflon pestle. Homogenate was layered onto Percoll gradients formed from 32.5% Percoll and spun for 20 min at 14 250 r.p.m. in a Beckman JA-17 rotor. The nuclei were removed from the gradient, pelleted and resuspended in 0.25 M sorbitol, 10 mM Tris–HCl, pH 6.8, 1% NP-40, 125 mM NaCl, 3 mM DTT, 1 mM PMSF, 100 mM sodium butyrate, 1 µM trichostatin A. Following washing several times, the nuclei were suspended in water. HCl was added to 0.25 N and histones were extracted by incubation for 2 h on ice. The crude histone preparation was precipitated with TCA, washed with acetone and dissolved in 8 M urea.
RNA purification and northern blotting
Yeast were grown in YPD medium at 30°C to an OD600 of 1.0. Total yeast RNA was purified using the hot acid/phenol procedure (32). Aliquots of RNAs were resuspended in water, fractionated through a 1% formaldehyde–agarose gel and transferred to a nitrocellulose membrane (Magna; MSI). Northern hybridization was carried out according to the method of Sambrook et al. (33). The presence of HHO1 mRNA was detected by hybridization with a 32P-labeled internal Eco47III–XmnI fragment of HHO1. Other RNA species were detected by hybridization to probes amplified from gene fragments obtained by PCR (see Table 1).
DNA array
Strain CY26 was made HIS+ by transformation with an Eco47III–BamHI fragment containing the full-length HIS3 gene. This strain was fully isogenic to strain CY26 hho1::HIS3 except for deletion of the HHO1 gene. For purification of total RNA strains CY26HIS+ and CY26hho1::HIS3 were grown in YPD medium at 30°C. Total RNA was purified from log phase cells. Poly(A)+ RNA was purified using the Oligotex mRNA Midi Kit (Qiagen). Hybridization to a DNA array containing all S.cerevisiae ORFs with fluorescently labeled cDNA made from a template of each of these mRNA preparations was performed by V.Iyer in P.Brown’s laboratory at Stanford University (34,35).
Nucleosome repeat length determination and resistance to micrococcal nuclease
Chromatin preparation and micrococcal nuclease treatments were carried out as described (36). The average length of the nucleosome repeat was calculated as in Godde et al. (37).
Measurement of HHO1p:nucleosome stoichiometry
To directly measure the number of HHO1p molecules per cell we replaced the native HHO1 gene with a HA-tagged version. In parallel, recombinant glutathione S-transferase (GST)–HHO1–2HAp was prepared from bacterial extracts. Serial dilutions of known amounts of the GST–HHO1–2HAp protein were compared by western analysis to HA-tagged HHO1p in known amounts of yeast extract using an anti-HA antibody (Babco).
Construction of the HHO1–2HAp producing strain is described above. A bacterial strain producing GST–HHO1–2HAp was constructed as follows. HHO1 was PCR amplified from a wild-type strain using primers IF7 and IF8. The PCR fragment was digested with EcoRI and XhoI and inserted into the EcoRI and XhoI sites of pGEX-4T-1 (Amersham Pharmacia). The resulting plasmid, pGEX-HHO1, contains an in-frame fusion of GST to HHO1p. To produce the GST–HHO1–2HAp fusion, a HindIII–NotI fragment of pGEX-HHO1 containing the HHO1 C-terminus was exchanged with a HindIII–NotI fragment from plasmid p25 containing the C-terminal portion of HHO1 fused to the 2HA tag. Strain BL21(DE3) was transformed with pGEX-HHO1-HA. GST and the GST–HHO1–2HAp protein fusion were purified following the Amersham Pharmacia protocol.
Chromatin immunoprecipitation
Immunoprecipitation of DNA was carried out essentially as described (38). Briefly, 50 ml of yeast encoding HHO1–2HAp were grown in YPD medium to an OD600 of ∼1.0. Formaldehyde was added to the culture medium to a final concentration of 1% and mixed slowly for 15 min on a nutator at room temperature. The reaction was quenched with glycine to a final concentration of 125 mM. Following washing, cells were resuspended in 400 µl of lysis buffer containing 50 mM HEPES–KOH, pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.1% SDS and Complete protease inhibitors (Roche) according to the manufacturer’s instructions. Cells were disrupted with glass beads (0.5 mm diameter) by vigorous vortexing for 45 min at 4°C. The lysate was sonicated (W-375; Heatsystems Ultrasonic) on ice using a fine probe with short pulses for a total of ∼20 s, taking care that the lysate did not become warm. The lysate was spun for 15 min in a microfuge. Aliquots of the supernatant were saved for preparing total genomic DNA. The size class of the sheared DNA was ∼1000 bp. The remainder was mixed with anti-HA monoclonal antibody (Babco) and slowly mixed overnight. The amount of antibody used was carefully chosen by titration of the antibody, such that the amount routinely used was about twice the amount needed to achieve maximal DNA precipitation. The solution was mixed with 50 µl of a 50% suspension of protein A–Sepharose beads (Amersham Pharmacia) for 1 h at 4°C on a nutator, spun briefly in a microfuge and extensively washed with 10 mM Tris–HCl pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% Na deoxycholate, 1 mM EDTA and finally with 10 mM Tris–HCl, pH 8.0, 1 mM EDTA. Crosslinking of the DNA was reversed in both the immunoprecipitated DNA and the total genomic DNA samples by addition of 100 µl of 50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1% SDS and heating at 65°C overnight. DNA was isolated by adding 2 µg glycogen as carrier followed by proteinase K digestion, phenol/chloroform extraction and ethanol precipitation. Quantitative limiting multiplex PCR analysis (24 cycles) was performed as described (39) using an α-32P-radiolabeled nucleotide triphosphate. Control amplifications of total cellular DNA contained ∼1 ng template per amplification (extrapolated following serial dilutions). Immunoprecipitated DNA concentrations were too low to be measured. PCR products were separated on 6% polyacrylamide gels, visualized by autoradiography and quantified using a Fujifilm BAS-1500 phosphorimager, with TINA analysis software (v.2.09c). Specificity in immunoprecipitation of specific bands was noted when their intensity significantly increased relative to a normalized internal control.
Southern hybridization to detect possible enrichment of repeated DNA sequences in immunoprecipitated DNA
DNA was isolated from log phase yeast as described (30). Following restriction enzyme digestion it was separated by agarose gel electrophoresis and stained with ethidium bromide. Repeated DNA bands were identified by comparison with published data (40). DNA in the gel was Southern blotted to a nylon (Magna; MSI) membrane using standard techniques.
To identify repeated DNA sequences that might be specifically bound to HHO1p, immunoprecipitated DNA was used as probe (radioactively labeled with a HexaLabel DNA Labeling Kit; MBI) in a hybridization experiment with the DNA in the blotted gel. Prehybridization was carried out for 5 h at 60°C in 7% SDS, 1.5× SSPE, 10% PEG (mol wt 10 000), 100 µg/ml salmon sperm (41). Following overnight hybridization in the same buffer, the membrane was washed three times at 60°C with 0.1× SSC, 0.1% SDS and applied to film for autoradiography.
RESULTS
HHO1 is both transcribed and translated in the cell
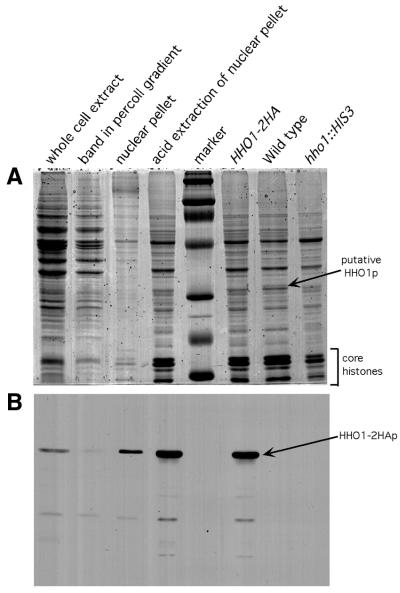
As it had been reported that disruption of HHO1 had no observable phenotype, we asked whether HHO1p is produced in the wild-type cell. To that end, we replaced the chromosomal HHO1 gene at its chromosomal location with a construct that produced a HHO1–2HA fusion protein from the native HHO1 promoter. We then performed a western blot on whole cell extracts using an anti-HA antibody. As can be seen in Figure 1B, HHO1p is easily detected in these cells. The measured molecular weight is ∼35 kDa. Though the actual molecular weight is ∼30 kDa with the two HA tags, this aberrant mobility is in agreement with previous observations (27).
Figure 1.
Histone preparations from yeast nuclei. (A) Coomassie staining of fractions from various stages of purification. Lane 1, whole cell extract; lane 2, band in Percoll gradient; lane 3, nuclear pellet; lane 4, acid extraction of nuclear pellet; lane 5, marker (mol. wt 220, 97.4, 66, 46, 30 21.5 and 14.3 kDa, respectively); lane 6, HHO1–2HA; lane 7, wild-type; lane 8, hho1::HIS3. The putative HHO1p band is present in the wild-type but not in hho1::HIS3 and has a higher molecular weight in the HHO1–2HA lane. (B) Western analysis of (A) with anti-HA antibody.
HHO1p is located in the nucleus and co-purifies with core histones
As HHO1p has been described as the putative yeast histone H1, we asked whether HHO1p co-purifies with core histones extracted from isolated nuclei. To that end, we acid extracted nuclei isolated from a strain in which the native HHO1p gene was replaced with a construct that encodes HHO1–2HAp fusion protein. As controls, we did the same for nuclei isolated from a wild-type strain and an isogenic strain lacking HHO1p. We then performed SDS–PAGE on samples isolated from all phases of the purification process. The gel was western blotted and probed using an anti-HA antibody. As can be seen in Figure 1, the HHO1–2HAp protein was quantitatively recovered in the acid-soluble fraction that had been extracted from nuclei.
Coomassie staining of the acid-soluble proteins extracted from the wild-type strain run on SDS–PAGE showed a band (marked in the figure) migrating slightly faster than the 2HA-tagged band identified in the western blot. The band was absent in extracts made from the hho1Δ strain. We concluded, therefore, that the band represents HHO1p. It is evident from the gel that the HHO1p levels in the cell are considerably lower than the core histone levels (see below).
Disruption of HHO1 results in increased transcription from its promoter
Previous investigators have shown that disruption of HHO1 had no observable phenotype (26,27). We therefore disrupted HHO1 by replacing most of the coding region with HIS3 (see Materials and Methods). This construct left the promoter and the first 149 of the 777 nt of the HHO1 coding region intact. As expected, we could not detect any mutant phenotype.
HHO1 disruption was reported to mildly affect CYC1 expression (26). We sought to determine whether RNA levels of the HHO1 gene are affected by its disruption. As the disrupted gene can still be transcribed, we compared the in vivo levels of the disrupted and wild-type transcripts. As can be seen in Figure 2 (upper left), HHO1 disruption results in a substantial increase in the levels of its own transcript, indicating a possible feedback mechanism in the regulation of HHO1 transcript levels in the cell. As the transcript has been modified it has a different size from the original.
Figure 2.

Northern analysis of whole cell RNA from strains that were wild-type for HHO1 (WT) or were hho1Δ (Δ). Each filter was probed with a radiolabeled PCR fragment containing sequences from the gene listed above each panel. Below the panel is an ethidium stain of the 18S and 28S rRNAs in each sample showing that equal amounts of RNA were loaded in each lane.
Disruption of HHO1 has little affect on the RNA levels of most transcripts in the cell
Though HHO1 disruption did not have a detectable phenotype (27), our results (above) and those of others (26) did show that the presence of HHO1 affected the RNA levels of several transcripts. We sought, therefore, to determine how the absence of HHO1p in the cell would affect the levels of all other transcripts. To this end, poly(A)+ RNA from wild-type and isogenic hho1Δ strains was used to make cDNA. This cDNA was hybridized to a microarray of DNA containing every ORF in yeast (as in 42). The results indicated that while the transcription of very few genes was affected by the HHO1 disruption, a few transcripts were more abundant in the hho1Δ strain and a few transcripts were more abundant in the wild-type strain. The results for a few sample genes were confirmed using northern analysis (Fig. 2).
The average nucleosome distance and sensitivity to micrococcal nuclease is the same in wild-type and hho1Δ strains
As HHO1p has significant sequence homology to histone H1 in other organisms, it has been considered to be the putative yeast histone H1. Transcription during the S phase of the cell cycle in yeast is unique to core histones and HHO1, strongly supporting this contention. As histone H1 has been reported to affect nucleosome spacing and sensitivity to micrococcal nuclease (43–45) and recombinant HHO1p was shown to bind dinucleosomes, we compared the average nucleosome spacing and sensitivity to micrococcal nuclease in wild-type and hho1Δ cells using micrococcal nuclease. As earlier reported (28), we saw no difference in nucleosome spacing between the strains (data not shown). We calculated the nucleosome repeat to be ∼161 nt, which compares well with the published 165 nt (6). The sensitivity was measured as the concentration of micrococcal nuclease necessary to achieve identical nuclease digestion patterns. No difference was found between the wild-type and hho1Δ strains.
HHO1p is not stoichiometric with core histones in vivo
In higher organisms, histone H1 is thought to be associated with all or most nucleosomes. We reasoned that this might not be true for HHO1p since cells lacking HHO1p did not show a difference in nucleosome spacing. In addition, Coomassie staining of the isolated histones showed considerably less HHO1p than each of the core histones.
To directly measure the levels of HHO1p molecules in the cell, we made a recombinant GST–HHO1–2HAp construct that we could isolate to homogeneity from bacteria and directly measure its concentration. We then loaded known amounts of GST–HHO1–2HAp on a polyacrylamide gel together with known amounts of protein extracted from a yeast strain producing HHO1p–2HA. A western blot of this gel can be seen in Figure 3. By extrapolating from the intensities of the GST–HHO1–2HAp bands and comparing them to the HHO1–2HAp bands, we calculated a level of about 2000 copies of HHO1p per cell. This is ∼37-fold fewer molecules than the estimated number of nucleosomes in the cell and represents about one molecule of HHO1p per three genes. This number necessarily means that HHO1p cannot be responsible for packing of the majority of the nucleosomes and cannot directly affect transcription of most genes.
Figure 3.
Quantitation of HHO1p in the cell. Western analysis was performed comparing known amounts of recombinant GST–HHO1p–2HA (58 kDa) with unknown quantities of HHO1p–2HA (30 kDa). Lanes 1–7 contain 37.5, 18.7, 9.4, 4.7, 2.3, 1.2 and 0.6 ng GST–HHO1p–2HA, respectively. Lanes 8–14 contain whole cell lysate from 13.2 × 107, 6.6 × 107, 3.3 × 107, 1.6 × 107, 0.8 × 107 and 0.4 × 107 cells, respectively.
Chromatin immunoprecipitation of DNA bound to HHO1–2HAp does not generally show preferential binding of HHO1p to differentially expressed genes
Both northern analysis and microarray data indicated that a small subset of genes may be differentially transcribed in a strain lacking HHO1p. Since we determined that there are fewer HHO1p molecules than genes in the cell, we asked whether we could detect differential binding of HHO1p to the differentially expressed genes. Using a chromatin immunoprecipitation technique followed by limiting multiplex PCR (38), we looked at both promoter and coding regions of numerous genes that were differentially expressed in the hho1Δ strain. To date, we have been unable to detect any specificity in HHO1p binding to these regions. An example of such a multiplex PCR reaction is shown in Figure 4C (right).
Figure 4.
Immunoprecipitation of HHO1p–2HA-associated chromatin. (A) Ethidium stained agarose gel of restriction digested whole cell DNA. M, λ Eco130I + MluI digested DNA marker (sizes in bp are to left of figure). Lane 1, EcoRI; lane 2, HindIII; lane 3, BglII; lane 4, XhoI. Repeated DNA sequences can be seen as bands on the gel. (B) Southern blot of left panel using immunoprecipitated DNA as probe. All the bands that appear were identified in Foley et al. (41) as encoding rRNA. No other repeated sequences were detected. (C) (Left) rDNA and CUP1 sequences (primers IF9–IF14) were multiplex PCR amplified from total genomic and immunoprecipitated DNA as described in Materials and Methods. The sizes of the amplicons are listed in parentheses. Lane 1, total genomic DNA; lane 2, immunoprecipated DNA. Duplicate lanes are duplicate PCR reactions. Band intensities are quantified in Table 2. (Right) LYS9, CTR1, SAG1, FET3 and FRE1 (primers IF27–IF34) were PCR amplified as in the left panel. All these genes are single copy in the genome. Neither rDNA nor CUP1 were included in the right panel as they are in multiple copies in the genome and they overwhelm the PCR reaction when amplified in the same tube. While none of the DNA fragments in the right panel is specifically immunoprecipitated one relative to the other, it is evident in the left panel that only the rDNA sequences are preferentially immunoprecipitated. About 10-fold more DNA template was used in the multiplex PCR reaction for the single copy genes (right) as for the multicopy genes (left).
HHO1–2HAp does show preferential binding to genes encoding rRNA
The above data do not preclude the possibility that HHO1p does bind specific regions in the chromatin that were not tested. We therefore asked whether we could detect such sequences directly without probing specific genes. To that end, we used the DNA that had been immunoprecipitated with the anti-HA antibody in the immunoprecipitation technique as a probe in a Southern blot with total genomic DNA following restriction digestion. Orlando et al. used a similar technique to identify DNA specifically immunoprecipated from Drosophila extracts (46). Thus our experiment, at a minimum, was expected to detect repeated sequences that might specifically be bound to HHO1p. As can be seen in Figure 4A, restriction digestion with several enzymes yielded specific bands which have been attributed to specific repeated sequences in the genome (40). When the DNA in the agarose gel in Figure 4A was blotted to nitrocellulose and probed with DNA that had been immunoprecipitated with anti-HA, only a subset of the bands was visible (Fig. 4B). All those bands could be attributed to genes encoding rRNA (40). No other bands were visible, even on extended exposure, though many other bands representing repeated sequences could clearly be seen on the ethidium stained gel before transfer. This experiment was repeated several times and the results were the same.
To confirm these results, we probed the immunoprecipitated DNA using a limiting multiplex PCR reaction as done in the experiment with differentially expressed genes. We used two different sets of primers for rDNA amplification, each coding for a portion of the 35S rRNA precursor, and one set for CUP1 amplification. CUP1, which is also repeated in the genome, was used as internal control. We quantified the radioactivity in each of the amplified DNA bands (Table 2) and normalized each of the rDNA bands to the CUP1 amplicon in the same lane. The results clearly show (Fig. 4C and Table 2) that the immunoprecipitated rDNA sequences (Fig. 4C, left, lanes marked 2) were preferentially immunoprecipitated by antibody bound to HHO1–2HAp when compared to the control CUP1 DNA (Fig. 4C, left, lanes marked 1). rDNA sequences near the start of the large 35S rDNA repeat just upstream of the sequence encoding 18S rRNA (285 bp in the PCR) were 1.7-fold more abundant in the immunoprecipitated DNA than in total genomic DNA, while rDNA sequences within the region encoding 25S rRNA (408 bp in the PCR) were enriched 2.5-fold. As mentioned above, we were unable to specifically immunoprecipitate DNA from other parts of the genome (example in Fig. 4C, right). These experiments serve as controls for the results with the rDNA, as they show the consistency and repeatability of the technique.
Table 2. Quantitation of band intensities in Figure 4C.
| |
Lane 1 left |
Lane 1 right |
Lane 2 left |
Lane 2 right |
| rDNA (408) | 1322 | 1333 | 4680 | 4791 |
| rDNA (285) | 3765 | 4182 | 9685 | 9690 |
| CUP1 (177) | 256 | 223 | 346 | 325 |
We next asked whether we could detect differences in the steady-state levels of rRNA in wild-type and hho1Δ strains. We preformed a northern blot using total cellular RNA from both strains. We probed it with a sequence (YER32W) we knew not to be differentially expressed in the two strains from the DNA array data and then with a sequence encoding rRNA (data not shown). Identical amounts of rRNA were found in the two stains when normalized to YER32W.
DISCUSSION
Previous work had been unable to establish a phenotype for hho1Δ cells. Though it was shown that HHO1 is transcribed (29), none of the work to date has addressed the question of the existence or amount of HHO1p in the cell when transcribed from its native chromosomal location. To address these issues, we asked whether we could detect HHO1p in cellular extracts. To this end, we replaced the HHO1 gene with an epitope-tagged version, transcribed from its native promoter. Northern blot experiments showed that the levels of HHO1 transcript did not change following addition of the HA tag (data not shown). We showed in a western blot that these cells produce HHO1–2HAp and that it co-purified with histones isolated from nuclei. In extracts enriched for histones made from the nuclei of HHO1 and hho1Δ strains, we were able to identify a single protein at the appropriate molecular weight on Coomassie stained gels that was present in wild-type extracts but was absent in extracts made from the HHO1 disruption. We observed that this band had a higher mobility when it was tagged with two copies of HA, supporting its identification as HHO1p in the wild-type cells.
We noted that the Coomassie staining of this band was considerably less intense than staining of the core histones (see Fig. 1A). Our analysis indicated that there are about 2000 HHO1p molecules per nucleus. This is about 37-fold fewer than the presumed number of nucleosomes. These figures may not be completely accurate, as specific degradation of HHO1–2HAp during preparation of the extract would lower the estimate of HHO1–2HAp in the cell. It should be noted that our extraction procedures were carried out in the presence of protease inhibitors and that almost no degradation products were seen on the Coomassie stained polyacrylamide gels nor in the western blots. However, even if our estimate of HHO1p content in the cell is somewhat inaccurate, the results indicate that there are far fewer HHO1p molecules than nucleosomes in the cell. Thus, HHO1p is unlikely to be directly responsible for general chromatin packing in yeast. It is not surprising, therefore, that our data and data from other laboratories show no difference in average nucleosome spacing between wild-type and hho1Δ cells.
DNA array experiments comparing expression of all ORFs in wild-type and hho1Δ strains showed that the levels of a limited number of transcripts are affected by the absence of HHO1p in the cell, as expected by the HHO1p:nucleosome stoichiometry. These data may explain the lack of an observed phenotype in hho1Δ cells.
We used the array information as a guide to try to locate the relatively low numbers of HHO1p molecules in the chromatin using a chromatin immunoprecipitation technique. While these attempts have so far been unsuccessful, possibly indicating that the effect of HHO1 disruption on transcription may be indirect, HHO1p molecules were preferentially found at the repeated genes encoding rRNA. Other repeated sequences that were observed in the ethidium stained gel of total DNA did not hybridize to the immunoprecipitated probe, so we believe that the data indicate preferential binding of HHO1p to rDNA. While a northern experiment indicated no difference in steady-state rRNA levels in wild-type and hho1Δ strains, these data do not preclude the possibility that rDNA transcription rates could be different in the two strains as a result of different chromatin structure. It was found that both rDNA amplicons were specifically immunoprecipitated, though to different degrees. It will be interesting, therefore, to better map HHO1p on the rDNA chromatin. As this region is repeated, our results cannot distinguish binding of many HHO1p molecules to a subset of the repeats from increased uniform binding to all the repeats.
Our results suggest that while yeast HHO1p is not involved in global chromatin compaction, as is histone H1 in other eukaryotes, it may serve a similar function at the local level. It is particularly interesting that HHO1p is preferentially found at the rDNA locus, since this region apparently has a particular chromatin structure (39,47). It will be very interesting to determine how a HHO1 disruption affects the rate of polymerase I transcription in this region and whether its chromatin structure is affected. It is not surprising that the steady-state rRNA levels are not different in wild-type and hho1Δ strains as rRNA levels are carefully controlled in the cell (48).
We believe it is possible that specificity of HHO1p binding to particular regions of the chromatin may be a function of local chromatin structure, rather than sequence specificity. It has been noted, for example, that histone H1 preferentially binds supercoiled DNA (49,50). HHO1p may bind these and other regions, helping to stabilize chromatin structure, and thus influence accessibility to RNA polymerase. It will be interesting to check these predictions at the locus encoding rRNA. In the light of recent evidence showing the effect of histone H1 phosphorylation on transcription of specific genes (14,15,51), it will also be important to probe the phosphorylation state of HHO1p at specific positions in the chromatin and in the nucleus in general.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Vishy Iyar in Patrick Brown’s laboratory for performing the DNA array experiments and making that data available to us. We thank Michael Grunstein for sharing protocols and reagents necessary for the chromatin immunoprecipitations. We also thank members of his laboratory for helpful discussions. This work was supported in part by the Bar Ilan University Health Sciences Research Centre.
References
- 1.Van Holde K.E. (1989) Chromatin. Springer-Verlag, New York, NY.
- 2.Kornberg R.D. (1974) Chromatin structure: a repeating unit of histones and DNA. Science, 184, 868–871. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg R.D. and Thomas,J.O. (1974) Chromatin structure; oligomers of the histones. Science, 184, 865–868. [DOI] [PubMed] [Google Scholar]
- 4.Graziano V., Gerchman,S.E., Schneider,D.K. and Ramakrishnan,V. (1994) Histone H1 is located in the interior of the chromatin 30-nm filament. Nature, 368, 351–354. [DOI] [PubMed] [Google Scholar]
- 5.Doenecke D., Tonjes,R. and Kress,H. (1988) The H1 and core histone subtypes: differential gene expression and varied primary structures. Adv. Enzyme Regul., 27, 107–120. [DOI] [PubMed] [Google Scholar]
- 6.Thomas J.O. and Furber,V. (1976) Yeast chromatin structure. FEBS Lett., 66, 274–280. [DOI] [PubMed] [Google Scholar]
- 7.Lohr D., Corden,J., Tatchell,K., Kovacic,R.T. and Van Holde,K.E. (1977) Comparative subunit structure of HeLa, yeast and chicken erythrocyte chromatin. Proc. Natl Acad. Sci. USA, 74, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horz W. and Zachau,H.G. (1980) Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J. Mol. Biol., 144, 305–327. [DOI] [PubMed] [Google Scholar]
- 9.Shen X., Yu,L., Weir,J.W. and Gorovsky,M.A. (1995) Linker histones are not essential and affect chromatin condensation in vivo. Cell, 82, 47–56. [DOI] [PubMed] [Google Scholar]
- 10.Sirotkin A.M., Edelmann,W., Cheng,G., Klein-Szanto,A., Kucherlapati,R. and Skoultchi,A.I. (1995) Mice develop normally without the H1(0) linker histone. Proc. Natl Acad. Sci. USA, 92, 6434–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prymakowska-Bosak M., Przewloka,M.R., Iwkiewicz,J., Egierszdorff,S., Kuras,M., Chaubet,N., Gigot,C., Spiker,S. and Jerzmanowski,A. (1996) Histone H1 overexpressed to high level in tobacco affects certain developmental programs but has limited effect on basal cellular functions. Proc. Natl Acad. Sci. USA, 93, 10250–10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stargell L.A., Karrer,K.M. and Gorovsky,M.A. (1990) Transcriptional regulation of gene expression in Tetrahymena thermophila. Nucleic Acids Res., 18, 6637–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen X.T. and Gorovsky,M.A. (1996) Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell, 86, 475–483. [DOI] [PubMed] [Google Scholar]
- 14.Dou Y., Mizzen,C.A., Abrams,M., Allis,C.D. and Gorovsky,M.A. (1999) Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell, 4, 641–647. [DOI] [PubMed] [Google Scholar]
- 15.Dou Y. and Gorovsky,M.A. (2000) Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell, 6, 225–231. [DOI] [PubMed] [Google Scholar]
- 16.Steinbach O.C., Wolffe,A.P. and Rupp,R.A. (1997) Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature, 389, 395–399. [DOI] [PubMed] [Google Scholar]
- 17.Bouvet P., Dimitrov,S. and Wolffe,A.P. (1994) Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev., 8, 1147–1159. [DOI] [PubMed] [Google Scholar]
- 18.Doenecke D., Albig,W., Bouterfa,H. and Drabent,B. (1994) Organization and expression of H1 histone and H1 replacement histone genes. J. Cell. Biochem., 54, 423–431. [DOI] [PubMed] [Google Scholar]
- 19.Moorman A.F., de Boer,P.A., Charles,R. and Lamers,W.H. (1987) The histone H1 degrees/H5 variant and terminal differentiation of cells during development of Xenopus laevis. Differentiation, 35, 100–107. [DOI] [PubMed] [Google Scholar]
- 20.Grunwald D., Lawrence,J.J. and Khochbin,S. (1995) Accumulation of histone H1(0) during early Xenopus laevis development. Exp. Cell Res., 218, 586–595. [DOI] [PubMed] [Google Scholar]
- 21.Takami Y. and Nakayama,T. (1997) A single copy of linker H1 genes is enough for proliferation of the DT40 chicken B cell line and linker H1 variants participate in regulation of gene expression. Genes Cell, 2, 711–723. [DOI] [PubMed] [Google Scholar]
- 22.Gunjan A. and Brown,D.T. (1999) Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res., 27, 3355–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barra J.L., Rhounim,L., Rossignol,J.L. and Faugeron,G. (2000) Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol. Cell. Biol., 20, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 546–547. [DOI] [PubMed] [Google Scholar]
- 25.Landsman D. (1996) Histone H1 in Saccharomyces cerevisiae—a double mystery solved. Trends Biochem. Sci., 21, 287–288. [PubMed] [Google Scholar]
- 26.Ushinsky S.C., Bussey,H., Ahmed,A.A., Wang,Y., Friesen,J., Williams,B.A. and Storms,R.K. (1997) Histone H1 in Saccharomyces cerevisiae. Yeast, 13, 151–161. [DOI] [PubMed] [Google Scholar]
- 27.Patterton H.G., Landel,C.C., Landsman,D., Peterson,C.L. and Simpson,R.T. (1998) The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem., 273, 7268–7276. [DOI] [PubMed] [Google Scholar]
- 28.Puig S., Matallana,E. and Perez-Ortin,J.E. (1999) Stochastic nucleosome positioning in a yeast chromatin region is not dependent on histone H1. Curr. Microbiol., 39, 168–172. [DOI] [PubMed] [Google Scholar]
- 29.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston J.R. (1994) Molecular Genetics of Yeast: A Practical Approach. IRL Press, Oxford, UK.
- 31.Lamb J.R., Michaud,W.A., Sikorski,R.S. and Hieter,P.A. (1994) Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J., 13, 4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ausubel F.M. (1987) Current Protocols in Molecular Biology. Greene Publishing & John Wiley, New York, NY.
- 33.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Lashkari D.A., DeRisi,J.L., McCusker,J.H., Namath,A.F., Gentile,C., Hwang,S.Y., Brown,P.O. and Davis,R.W. (1997) Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl Acad. Sci. USA, 94, 13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shalon D., Smith,S.J. and Brown,P.O. (1996) A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res., 6, 639–645. [DOI] [PubMed] [Google Scholar]
- 36.Lowary P.T. and Widom,J. (1989) Higher-order structure of Saccharomyces cerevisiae chromatin. Proc. Natl Acad. Sci. USA, 86, 8266–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Godde J.S. and Widom,J. (1992) Chromatin structure of Schizosaccharomyces pombe. A nucleosome repeat length that is shorter than the chromatosomal DNA length. J. Mol. Biol., 226, 1009–1025. [DOI] [PubMed] [Google Scholar]
- 38.Hecht A. and Grunstein,M. (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol., 304, 399–414. [DOI] [PubMed] [Google Scholar]
- 39.Gotta M., Strahl-Bolsinger,S., Renauld,H., Laroche,T., Kennedy,B.K., Grunstein,M. and Gasser,S.M. (1997) Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J., 16, 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philippsen P., Stotz,A. and Scherf,C. (1991) DNA of Saccharomyces cerevisiae. Methods Enzymol., 194, 169–182. [DOI] [PubMed] [Google Scholar]
- 41.Foley K.P., McArthur,G.A., Queva,C., Hurlin,P.J., Soriano,P. and Eisenman,R.N. (1998) Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J., 17, 774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 43.Allan J., Staynov,D.Z. and Gould,H. (1980) Reversible dissociation of linker histone from chromatin with preservation of internucleosomal repeat. Proc. Natl Acad. Sci. USA, 77, 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunjan A., Alexander,B.T., Sittman,D.B. and Brown,D.T. (1999) Effects of H1 histone variant overexpression on chromatin structure. J. Biol. Chem., 274, 37950–37956. [DOI] [PubMed] [Google Scholar]
- 45.Sandaltzopoulos R., Blank,T. and Becker,P.B. (1994) Transcriptional repression by nucleosomes but not H1 in reconstituted preblastoderm Drosophila chromatin. EMBO J., 13, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orlando V., Strutt,H. and Paro,R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- 47.Smith J.S., Caputo,E. and Boeke,J.D. (1999) A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol., 19, 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strathern J.N., Jones,E.W. and Broach,J.R. (1982) The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Krylov D., Leuba,S., van Holde,K. and Zlatanova,J. (1993) Histones H1 and H5 interact preferentially with crossovers of double-helical DNA. Proc. Natl Acad. Sci. USA, 90, 5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivanchenko M., Zlatanova,J. and van Holde,K. (1997) Histone H1 preferentially binds to superhelical DNA molecules of higher compaction. Biophys. J., 72, 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizzen C.A., Dou,Y., Liu,Y., Cook,R.G., Gorovsky,M.A. and Allis,C.D. (1999) Identification and mutation of phosphorylation sites in a linker histone. Phosphorylation of macronuclear H1 is not essential for viability in Tetrahymena. J. Biol. Chem., 274, 14533–14536. [DOI] [PubMed] [Google Scholar]