Abstract
Translational recoding of mRNA through a –1 ribosomal slippage mechanism has been observed in RNA viruses and retrotransposons of both eukaryotes and prokaryotes. Whilst this provides a potentially powerful mechanism of gene regulation, the utilization of –1 translational frameshifting in regulating mammalian gene expression has remained obscure. Here we report a mammalian gene, Edr, which provides the first example of –1 translational recoding in a eukaryotic cellular gene. In addition to bearing functional frameshift elements that mediate expression of distinct polypeptides, Edr bears both CCHC zinc-finger and putative aspartyl protease catalytic site retroviral-like motifs, indicative of a relic retroviral-like origin for Edr. These features, coupled with conservation of Edr as a single copy gene in mouse and man and striking spatio-temporal regulation of expression during embryogenesis, suggest that Edr plays a functionally important role in mammalian development.
INTRODUCTION
In all species, triplet codon sequences on mRNA are accurately translated into polypeptides through recognition by aminoacyl tRNAs, such that the open reading frame (ORF) of the mRNA is maintained. In addition, non-standard decoding mechanisms have been identified in the form of programmed translational frameshifting, in which translating ribosomes are induced to slide 1 nt forward (+1 frameshifting) or backwards (–1 frameshifting) at a specific recoding site on the mRNA. As a consequence, multiple ORFs can be utilised from a single mRNA so diversifying the polypeptide coding sequence information of gene transcription units.
Programmed frameshifting has been found to occur in a variety of viral genomes (1–3), but is rare amongst prokaryotic and eukaryotic cellular genes. +1 programmed frameshifting has been observed in the prfB gene of Escherichia coli, which encodes peptide release factor 2 (RF2), and mammalian ornithine decarboxylase (ODC) antizymes (4–8). Interestingly, RF2 and antizyme genes use +1 frameshifting to modulate their own expression by negative feedback mechanisms. The concentration of RF2 products directly regulates the efficiency of such programmed frameshifting. ODC is the first and rate-limiting enzyme in the pathway of polyamine biosynthesis in mammalian cells. Antizymes not only inhibit the enzymatic activity of ODC but also mediate its degradation. The efficiency of frameshifting by antizyme genes that is required to synthesise the active form of antizyme is governed by the amount of polyamine present. These regulatory mechanisms of ODC are conserved from yeast to humans (9).
Programmed –1 frameshifting is much more prevalent among viruses than other species. For example, retroviruses, coronaviruses, toroviruses, arteriviruses and paromyxoviruses of mammals all utilise programmed frameshifts in decoding compact genomes (1,2,10). Programmed –1 ribosomal frameshifting in viruses conforms to a single model in which an N- and C-terminal fusion protein is encoded by two distinct, overlapping ORFs. Two basic sequence elements are required to promote efficient levels of –1 frameshifting. The first is a slippery heptamer sequence, X XXY YYZ (the 0 frame is indicated by the spaces) that allows the simultaneous slippage of ribosome-bound A- and P-site tRNAs by one base in the 5′ direction. The second is a RNA structural element downstream from the slippery sequence, consisting of either a pseudoknot structure or a simple stem–loop, which induces a ribosomal pause over the slippery site and increases the probability of 5′ ribosomal movement.
In E.coli, the dnaX gene utilises –1 translational frameshifting as a regulatory mechanism for coordinated expression of τ and γ subunits of DNA polymerase III holoenzyme (11–13). The dnaX frameshifting occurs on the slippery heptamer sequence, A AAA AAG, stimulated by two elements; an upstream hairpin and an upstream Shine–Dalgarno interaction site. Amongst eukaryotes, a recent bioinformatics based approach searching for consensus –1 ribosomal frameshift signals, including a heptamer sequence X XXY YYZ and a putative RNA pseudoknot structure, identified a number of cellular genes from several species including human, mouse, rat, chick and yeast with the potential to utilise –1 translational frameshifting (14). In a number of cases, identified frameshift signals were conserved in homologous genes from separate species. For example, the activity of –1 frameshifting in Saccharomyces cerevisiae was demonstrated in two selected motifs from yeast RAS1 and human CCR5 mRNA. However, these genes contained frameshift signals positioned to cause premature stoppage of the reading frames (10). Thus, the significance and origin of –1 frameshifting in these examples remains obscure.
Here we report a single copy mammalian gene, Edr (embryonal carcinoma differentiation regulated), which utilises a –1 ribosomal recoding mechanism to encode distinct polypeptides. Together with the presence of a CCHC zinc-binding motif and a putative aspartyl protease catalytic site, Edr is a new member of an emerging class of eukaryotic genes that bear relic motifs of retroviral genomes. The conservation of Edr in mouse and human, together with spatially and temporally regulated expression during embryogenesis suggest that Edr may play an important role in mammalian development.
MATERIALS AND METHODS
Isolation of Edr cDNA clones and northern blot analysis
The 2.4 kb partial Edr cDNA clone pGR165 was identified as one of several clones isolated by differential screening of a PCC3 cDNA library, as described previously (15). Total RNA was prepared from PCC3 cells, mouse embryos and adult tissues. The probe DNA was prepared from a 2.4 kb Edr cDNA clone (pGR165) isolated from PCC3 cDNA library, and labelled with [α-32P]dCTP. Mouse HPRT cDNA clone pHPT4 has been reported previously. Human β-actin cDNA was purchased from Clontech.
The cDNA insert of pGR165 was used to screen 4 × l05 clones from an E12.5 CD1 mouse embryo cDNA library (λZapII). Phage DNA was screened with the radiolabelled probe for 12 h at 42°C in 1× hybridization buffer (50 mM phosphate buffer pH 7.5, 5× SSC, 4× Denhardt’s reagent and 100 µg/ml herring sperm DNA, and 50% [v/v] formaldehyde), and washed twice for 30 min at 60°C in 0.2× SSC, 0.1% [v/v] SDS. Nested deletion derivatives of cDNA clones were obtained by unidirectional deletion using exonuclease III. Nucleotide sequence determination was performed by chain termination method using [α-35S]dATP (Amersham International, >400 Ci/mmol) and by Dye termination sequencing (Applied Biosystems) using an ABI 373A automated DNA sequencer. Confirmation of the nucleotide sequence of the palindrome-rich region (nucleotides 1875–2820), that was unstable on cloning in E.coli, was obtained by direct nucleotide sequence determination of RT–PCR fragments generated as follows. First strand cDNA was synthesised with primer P2 (Edr cDNA nucleotides 2820–2804) and amplified by PCR using P2 primer and P1 primer (Edr cDNA nucleotides 1875–1899). Sequence analysis was then performed using primers P1 and P2, as well as primers P3 (Edr cDNA nucleotides 2098–2124), P4 (Edr cDNA nucleotides 2228–2252) and P5 (Edr cDNA nucleotides 2720–2697) by Dye termination (Applied Biosystems) using an ABI 373A automated DNA sequencer. Nucleotide sequences were verified by analysis of independent first strand cDNA products.
RNA in situ hybridisation
Fixation, embedding, sectioning, hybridisation, washes and autoradiography of mouse embryos for RNA in situ hybridisation were performed essentially as described previously (16). A 1.2 kb PstI DNA fragment corresponding to nucleotides 3333–4450 of Edr cDNA was subcloned into pBSM13+, and [α-35S]UTP-labelled single strand antisense and sense RNA probes were synthesised using T3 or T7 RNA polymerases.
Plasmid construction
Plasmids pRF1/RF2, pRF1 and pRF2 were constructed in the vector pSP64T (17) from Edr cDNA clones and encompass nucleotides 340–3501, 340–1382 and 1359–3501, respectively. An in-frame deletion of 135 nt (Edr cDNA nucleotides 2311–2445) within the unstable palindrome-rich region of RF2, is present in both pRF1/RF2 and pRF2 plasmids. PCR-mediated site-directed mutagenesis was performed as described previously (18) using a series of Edr-specific oligonucleotide primers. PCR-mediated site-directed mutagenesis of Edr was carried out by the same strategy as described previously using Vent DNA polymerase (New England Biolabs). The following oligonucleotides were used for the mutagenesis: mt1, 5′-GCCGGGGAACTCCCCGGCC-3′; mt1R, complementary to mt1; mt2, 5′-GCCGCCGGGAAATTCCCCG-3′; mt2R, complementary to mt2; mt3, 5′-GCCGGGGAACTCCGGCCCCGCTGTAG-3′; mt3R, complementary to mt3. PCR cloning oligonucleotides used were: P1R, 5′-CCCAGAGCCTTGGTGATGCC-3′; P2R, 5′-GCCAACGAATTCCTAGGAC-3′. PCR reactions were performed using the following conditions: 94°C for 1 min, 55°C for 1 min and 72°C for 2 min for 30 cycles. The reaction mixture was prepared as recommended by the manufacturer. pRF1/RF2-mt1, pRF1/RF2-mt2 and pRF1/RF2-mt3 were constructed as follows. pRF1/RF2-mt1: PCR performed using pRF1/RF2 as template with P1R-mt1R and P2R-mt1 oligo pair. Products were pooled and used in the second PCR with P1R and P2R oligo pair prior to digestion with EcoRI and replacement with corresponding EcoRI fragment of pRF1/RF2. pRF1/RF2-mt2: P1R-mt2R and P2R-mt2 oligo pair for the first PCR, prior to subsequent PCR and cloning as described above. pRF1/RF2-mt3: P1R-mt3R and P2R-mt3 oligo pair, prior to subsequent PCR and cloning as described above.
In vitro translation reactions
In vitro transcription of Edr RNAs was performed using SP6 polymerase (Promega) with the template plasmid DNA. The synthesised RNAs were then added to 16.5 µl of rabbit reticulocyte lysate (Promega), supplemented with 10 µCi of [35S]methionine (Amersham, 1000 Ci/mmol) in a final reaction volume of 25 µl, essentially as recommended by the manufacturer. Products were then subjected to SDS–PAGE, and gels incubated in Amplify (Amersham) prior to autoradiography.
RESULTS
Cloning of the Edr gene and expression in embryonic and adult tissues
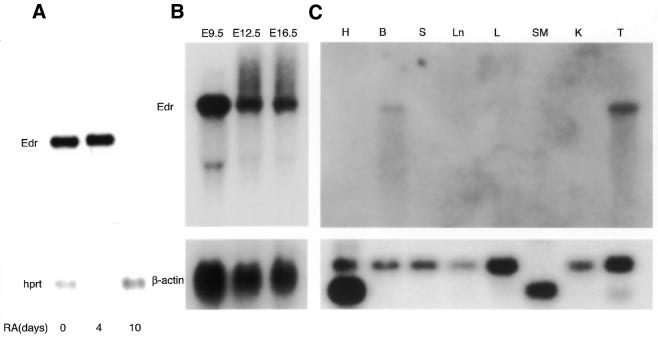
The Edr gene was identified initially by differential screening of an embryonal carcinoma (EC) cDNA library for genes expressed at a reduced level following retinoic acid (RA)-induced differentiation (15). In agreement with the screening procedure used to isolate this gene, the 2.4 kb cDNA insert of the Edr clone identified a 7 kb RNA species, abundant in undifferentiated PCC3 EC stem cells but barely detectable in 10 day RA-treated cultures by northern blot analysis (Fig. 1A). Subsequently, expression in the development of normal mice was examined by northern blot analyses using the cDNA probe. As seen in Figure 1B, the 7 kb Edr transcript was detected in whole embryo RNA at all stages between E9.5 and E16.5 days gestation. In contrast, of adult tissues screened, a low level of Edr mRNA was observed only in brain and testis (Fig. 1C). A second Edr hybridising RNA species of 2 kb was observed in E9.5 embryo RNA, but the nature of this RNA is presently unclear.
Figure 1.
Expression of Edr transcripts in PCC3 cells, adult mouse tissues and embryos. (A) Two micrograms of mRNA prepared from undifferentiated and differentiated PCC3 cells induced by RA for 4–10 days, were electrophoresed, blotted to nitrocellulose membrane and hybridised with [α-32P]dCTP-labelled probes for Edr (pGR165) and HPRT. (B) Ten micrograms of total cellular RNA prepared from mouse embryos at 9.5 (E9.5) to 16.5 (E16.5) days gestation were electrophoresed, blotted to nitrocellulose membrane and hybridised with the same probes as above. (C) Ten micrograms of total cellular RNA prepared from each of adult heart (H), brain (B), spleen (S), lung (Ln), liver (L), skeletal muscle (SM), kidney (K) and testis (T) were electrophoresed, blotted to nitrocellulose membrane, and hybridised with probes for Edr (pGR165) and human β-actin cDNA. In the heart, 2 and 1.6kb isoforms of β-actin were observed, whereas a 1.6 kb isoform was detected in skeletal muscle.
We then isolated a series of overlapping Edr cDNA clones from a mouse E12.5 embryo cDNA library. The coding sequence of the Edr mRNA was determined by assimilating sequence data from these overlapping clones (DDBJ/EMBL/GenBank accession no. AJ006464; see Supplementary Material). A palindrome-rich region of ∼350 nt within the Edr cDNA was found to be highly polymorphic among individual cDNA clones. RT–PCR confirmed that this region was subject to deletion and rearrangement on cloning in E.coli, so the coding information in this region of the Edr transcription unit was determined by direct nucleotide sequencing of DNA fragments generated from independent RT–PCR experiments (see Materials and Methods).
Edr contains structural motifs characteristic of some retroviral genomes
Although no single ORF was detectable within the Edr cDNA sequences, two long and overlapping ORFs were identified. Searching EMBL, GenBank and SWISS-PROT databases with the full-length Edr nucleotide and putative polypeptide sequences revealed no overall homology with previously characterised genes or proteins, but several features of the putative ORFs were apparent.
The first reading frame (RF1) was predicted to encode a 320 amino acid polypeptide containing a putative zinc-binding domain of the CCHC subclass with the high content of basic residues commonly found in retroviral Gag genes (19,20; see Supplementary Material). Retroviral Zn2+-binding motif sequences are composed of a highly conserved amino acid sequence with invariably spaced cysteine (Cys) and histidine (His) residues of the form Cys-X2-Cys-X4-His-X4-Cys- (CCHC) flanked by many basic residues including arginine and lysine (21–23), often in contiguous doublets or triplets. The RF1 coding region of Edr (nucleotides 656–1357) also exhibited close identity with a 0.69 kb partial cDNA clone MyEF-3 cloned from mouse brain (24). Like Edr, MyEF-3 mRNA was detected in adult brain, but not adult heart, lung, liver, spleen or kidney, suggesting that MyEF-3 represents a partial cDNA clone of Edr RF1.
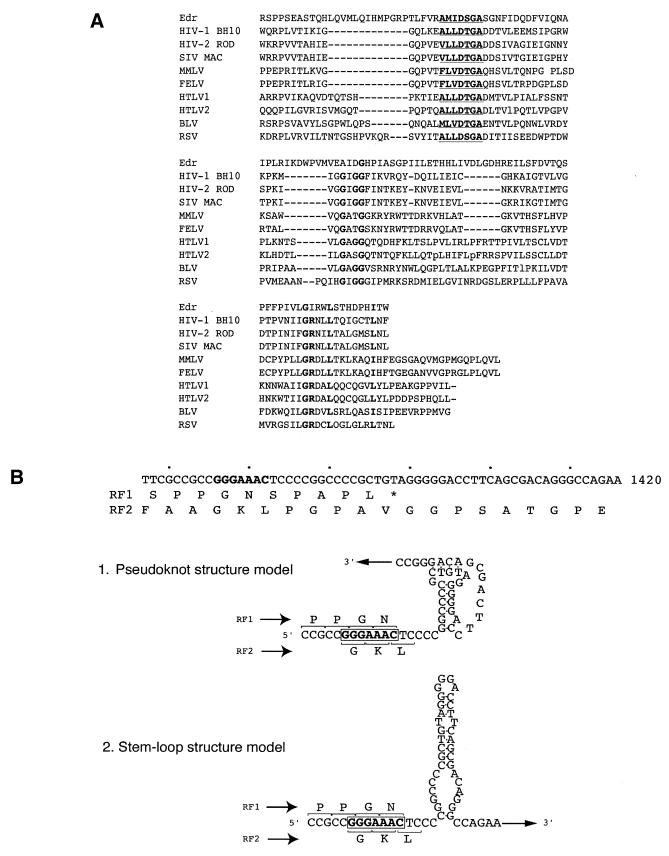
The second reading frame (RF2) had the potential to encode a 631 amino acid polypeptide containing a consensus motif for an aspartyl protease catalytic site, a characteristic feature of retroviral protease genes (25). Amino acid sequence alignments between Edr and retroviral proteases demonstrated that the structural motif of viral aspartyl proteases was highly conserved in Edr (Fig. 2A). Furthermore, as detailed in Figure 2B, the region of overlap between RF1 and RF2 contained the heptameric sequence GGGAAAC, and two pairs of short, or a pair of longer, complementary sequence motifs capable of self-annealing to form a stable secondary structure. There may exist other possible foldings downstream of the slippery sequences. However, either model has features characteristic of the stimulatory RNA secondary structures of –1 ribosomal frameshifting, as reported previously in RNA viral genomes (2). These findings raised the possibility that a –1 translational frameshift recoding mechanism could operate within the Edr mRNA to encode a full-length RF1/RF2 polypeptide.
Figure 2.
Identification of retroviral-like motifs in Edr. (A) Amino acid sequence alignment of Edr and retroviral protease motifs. Consensus sequence of aspartyl protease active sites [LIVMFGAC]-[LIVMTADN]-[LIVFSA]-D-[ST]-G-[STAV]-[STAPDENQ]-X-[LIVMFSTNC]-[LIVMFGTA] are underlined. Conserved residues are shown in bold. HIV-1, human immunodeficiency virus type 1 (BH10 isolate); HIV-2, human immunodeficiency virus type 2 (ROD isolate); SIV, simian immunodeficiency virus (MAC isolate); MMLV, Moloney murine leukaemia virus; FELV, feline leukaemia virus; HTLV1, human T-cell leukaemia virus type I; HTLV2, human T-cell leukaemia virus type II; BLV, bovine leukaemia virus. (B) Diagrammatic representation of the predicted –1 ribosomal slippage heptamer and two models of RNA secondary structure downstream from the slippery sequence— a putative pseudoknot and a simple stem–loop structure. The slippage sequence, the potential pseudoknot and a simple stem–loop structures are shown in relation to predicted amino acid sequences of RF1 and RF2. The heptameric –1 slippage sequence is denoted in bold.
Synthesis of the Edr RF1/RF2 polypeptide is mediated by –1 ribosomal frameshifting
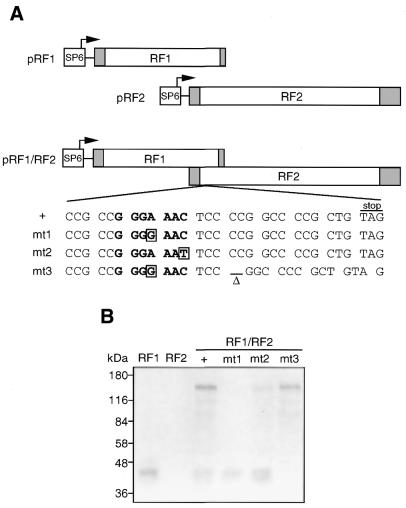
To obtain direct evidence that distinct polypeptides could be translated from the Edr mRNA using a –1 ribosomal frameshifting mechanism, in vitro translation assays were performed using a set of recombinant Edr cDNA expression vectors. Edr products encoded by the expression vector containing both RF1 and RF2 were compared with those generated from RF1 or RF2 alone. Three mutant RF1/RF2 expression vectors containing specific nucleotide changes within the predicted slippage and pseudoknot elements were also employed (Fig. 3A). An expression construct pRF1/RF2-mt1 contained a single A→G mutation within the putative –1 slippage site. This mutation was predicted to prevent maintenance of 2 of 3 bp at the anticodon of the tRNAasn at the P ribosomal site after shifting to the –1 frame of RF1. Thus, no RF1/RF2 protein would be generated, but synthesis of the RF1 polypeptide would remain unaffected. In contrast, the C→T point substitution in the pRF1/RF2-mt2 mutant was expected to still maintain the translation of RF1/RF2 expression by –1 ribosomal slippage, although the efficiency would be expected to be reduced by mismatch of 1 of 3 bp after shifting to the –1 frame of RF1. A third Edr expression vector mutant, pRF1/RF2-mt3 was derived from pRF1/RF2-mt1 by deletion of two cytosine residues within the predicted pseudoknot structure. In this way, a single ORF was created that combined RF1 plus RF2 with no requirement for –1 frameshifting.
Figure 3.
Edr utilises –1 ribosomal frameshifting to encode two polypeptides. (A) Plasmid templates for in vitro translation. pRF1, pRF2 and pRF1/RF2 expression plasmids were constructed by cloning Edr cDNA fragments into pSP65T as described in Materials and Methods. Plasmids pRF1/RF2-mt1 and pRF1/RF2-mt2 were derived from pRF1/RF2 by introduction of A→G and C→T nucleotide substitutions, respectively, within the putative slippage sequence. pRF1/RF2-mt3 was derived from pRF1/RF2-mt1 and contains an additional deletion of 2 nt predicted to contribute to the stable secondary structure of the pseudoknot structure. Shaded boxes denote the 5′- and 3′-untranslated regions within RF1 and RF2. The heptameric –1 slippage site is denoted in bold. (B) In vitro translation. Separate in vitro transcription and translation of plasmid templates from (A), was performed using rabbit reticulocyte lysate system in the presence of [35S]methionine. Products were subjected to 10% SDS–PAGE prior to autoradiography.
As shown in Figure 3B, plasmid pRF1 generated a close doublet of [35S]methionine-labelled polypeptides of ∼45 kDa. A doublet may result from translation initiation at each of the candidate in-frame methionine codons at the start of the putative RF1 ORF. The relative migration was somewhat slower than that predicted from the primary sequence alone (36.8 kDa). In contrast, no [35S]methionine-labelled proteins were synthesised with the plasmid pRF2, indicating that the putative initiating methionine of the second ORF in Edr was not efficiently utilised for translation. As predicted for a –1 ribosomal slippage mechanism, the plasmid pRF1/RF2 encoded both the 45 kDa RF1 polypeptide doublet and a larger protein species—a 140kDa Edr product. The observed migration of this protein species was also slower than the predicted molecular mass of an RF1/RF2 Edr product (112.6 kDa). The reasons for such differences in Edr RF1 and RF1/RF2 polypeptide sizes are presently unclear but may reflect distinctive conformations of Edr. We estimated the frameshift efficiency of Edr by comparison of the intensity of RF1 and RF1/RF2 products in relation to the number of methionine residues predicted in each polypeptide (Fig. 3B). In this way efficiency was estimated as at least 30%.
The Edr protein resulting from translation experiments in vitro using each of the three pRF1/RF2 mutant templates was consistent with utilisation of a –1 translational frameshift mechanism. The mt1 mutant template encoded the RF1-specific polypeptide doublet, but no 140 kDa Edr protein was detectable. In contrast, both RF1 and RF1/RF2 polypeptides were encoded by the mt2 mutant. Moreover, the intensity of RF1/RF2 polypeptides was reduced as expected. Finally, deletion of two residues in mt1 to create a single full-length ORF restored expression of the 140 kDa Edr product with the concommitant loss of the 45 kDa RF1 encoded polypeptides.
Spatial and temporal expression of Edr during mouse embryonic development
As northern blot analysis indicated that Edr expression was restricted mainly to fetal development, we sought further information on the spatial distribution of Edr transcripts during embryogenesis. For this purpose, a series of RNA in situ hybridisation experiments were performed on sections of mouse embryos between 7.5 and 17.5 days of gestation. This analysis revealed prominent expression of Edr in the mesenchymal components of a number of developing tissues.
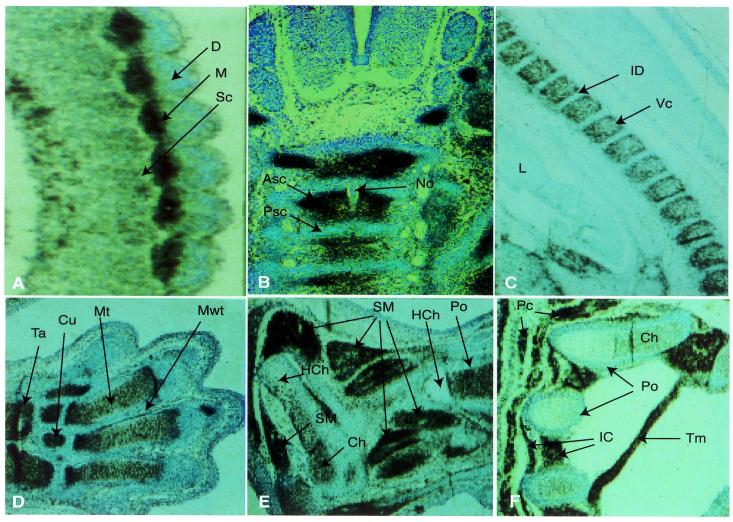
A striking differential pattern of Edr expression was observed in developing somites. High-level expression was restricted to the myotome, with lower levels in the migrating sclerotome, but no detectable transcripts in the dermatome (Fig. 4A). Subsequently, Edr transcripts were seen to be abundant in cells of the axial and appendicular chondrogenic skeletons (Fig. 4B–F) and during skeletal myogenesis (Fig. 4E and F). In the axial skeleton, segmental restriction of Edr transcripts to the loosely packed anterior half of developing vertebral sclerotome was distinct at day E12.5 (Fig. 4B), and expression persisted at day E14.5 within the derivative vertebral cartilage, although no transcripts were detected in the intervertebral discs (Fig. 4C). Expression profiles were similar in other fetal skeletal structures, including ribs, humerus, radius, ulna, smaller bones of fore and hind limbs and neural crest-derived craniofacial skeletal tissues (Fig. 4D–F; data not shown). No expression was apparent in perichondrial cells surrounding the developing bones. Moreover, loss of Edr expression correlated with chondrocyte hypertrophy and ossification in each bone. Edr expression was maintained in myotome derivative fetal muscles, for example the cutaneous muscles of the thorax and trunk regions (panniculus carnosus), transverse thoracis muscle and intercostal muscles at 17.5 days of gestation (Fig. 4E and F). Smooth muscle of the fetal gut and walls of major blood vessels also expressed Edr, whilst no Edr transcripts were detected in cardiac muscle at any time from E8.5 onward (data not shown).
Figure 4.
Edr expression during musculo-skeletal development. Sagittal (A,C,D–F), or transverse (B) sections of E10.5 (A), E12.5 (B), E14.5 (C), E17.5 (D–F) mouse embryos were hybridised with antisense 35S-radiolabelled RNA probes derived from the Edr cDNA clone pGR165, subjected to autoradiography for 48 h and counter-stained with toluidine blue. (A–C) 100× magnification. (D–F) 40× magnification. Some folding of tissue is apparent in (D). ASc, anterior sclerotome; Ca, calcaneus; Ch, chondrocytes; Cu, cuneiforme; D, dermatome; HCh, hypertrophied chondrocytes; IC, intercostal muscle; ID, intervertebral disc; M, myotome; Mt, cartilage primordium of metatarsale; Mwt, mesodermal web tissue; No, notochord; Pc, cutaneous muscle of thorax and trunk (panniculus carnosus); Po, periosteum; PSc, posterior sclerotome; Ra, radius; Sc, sclerotome; SM, fetal limb skeletal muscle; Ta, talus; Tm, Transversus thoracis muscle; VC, vertebral cartilage.
Edr transcripts were also observed in extra-embryonic tissues at all stages between E7.5–E17.5. High levels of Edr expression were detected in trophectoderm-derived giant cells, diploid trophoblast of the ectoplacental cone and trophoblastic labyrinth layer as well as chorion, amnion, visceral endoderm, parietal endoderm, the mesoderm-derived allantois, and visceral yolk sac at 14.5 days of gestation. In contrast, maternal spongiotrophoblast and uterine muscle layer did not express Edr at this stage of development (data not shown). Other mesenchymal cells of developing organs also contained Edr transcripts during fetal development, including those of lung, kidney, gonad and gut; these transcripts were restricted to the mesenchymal components of the tissue. Similarly, some neural and endocrine tissues including the endocrine (Rathke’s pouch) component of the developing pituitary expressed Edr (data not shown).
Edr maps to the proximal region of mouse chromosome 6
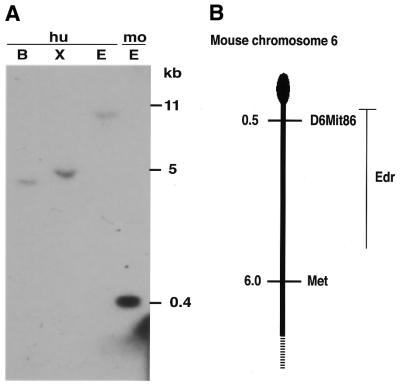
Genomic Southern blotting revealed that Edr is a single copy locus in the mouse genome and suggested conservation in the human genome (Fig. 5A). A BamHI restriction fragment length polymorphism (RFLP), identified with the Edr cDNA clone in C57BL6 and DBA2J inbred strains, was used to map the Edr locus in the BxD series of recombinant inbred (RI) mice (26). Comparison of the strain distribution pattern (SDP) of this RFLP with other loci mapped in BxD RI mice revealed identity with the SDP of D6Mit86 and 4/26 recombinants with Met proto-oncogene. This comparison places Edr at the proximal end of mouse chromosome 6 (Fig. 5B).
Figure 5.
Edr is a conserved mammalian gene and maps to the proximal region of mouse chromosome 6. (A) Genomic Southern blot analysis: 10 µg genomic DNA prepared from human term placenta (hu) or M.musculus (mo) was digested with BamHI (B), XbaI (X) or EcoRI (E) restriction endonucleases, prior to electrophoresis and transfer to nitrocellulose. The filter was hybridised with 32P-labelled Edr partial cDNA (395 bp EcoRI fragment encompassing nucleotides 1358–1753 in Edr cDNA) and washed to 0.1× SSC 0.1% SDS 65°C prior to autoradiography. (B) Comparison of the SDP of a BamHI RFLP at the Edr locus with others mapped in the BxD series of RI mice revealed linkage with D6Mit86 (0/26 recombinants and Met 4/26 recombinants). These linkage data map Edr to the proximal end of mouse chromosome 6.
Conservation and expression of the human Edr gene
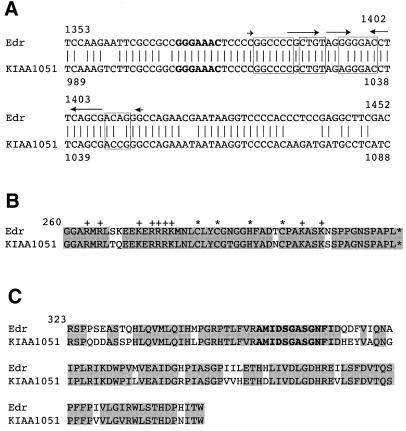
We then searched DNA databases to analyse whether the Edr gene is evolutionarily conserved in the genomes of humans, Drosophila, nematode and yeast. This analysis revealed a 6188 bp human cDNA clone (KIAA1051) that has >80% identity with Edr. This gene was mapped to human chromosome 7 in a region syntenic with mouse chromosome 6. Moreover, consistent with the expression data reported here, KIAA1051 expression was detected in embryos, placenta, adult brain and testes (27). Nucleotide and amino acid sequence comparison between Edr and KIAA1051 revealed that the –1 slippery sequence, the CCHC motif and the aspartyl protease motif are highly conserved in KIAA1051 (Fig. 6), although KIAA1051 is not a full-length cDNA clone. The nucleotide sequence differences in KIAA1051 in the region of both stems would be predicted to destabilise the possible pseudoknot structure. On the other hand, only a single nucleotide difference would be predicted to occur near the top of the stem in the putative simple stem–loop structure model. We further confirmed the conservation of frameshift signals in the human genome by PCR cloning and subsequent sequence analysis (data not shown). Together, with the results reported here, these data strongly suggest that KIAA1051 is the human orthologue of Edr. Within currently available databases, no genes with significant homology to Edr were identified in genomes of other species.
Figure 6.
Conservation of the –1 ribosomal frameshift signals and the aspartyl protease motif in human KIAA1051 gene. (A) Alignment of the nucleic acid sequences of Edr and KIAA1051 cDNAs. The heptameric –1 slippage sequence is denoted in bold. Conserved primary sequences for the potential pseudoknot and the simple stem–loop structures are shown in closed boxes, overlined (arrowed), respectively (KIAA1051 Genbank accession no. AB028974). (B) Comparison of the CCHC motif from Edr and KIAA1051. Conserved CCHC motifs are shown by asterisks. Basic residues outside CCHC motifs are marked with +. Identical residues are shaded. (C) Comparison of the putative aspartyl protease active sites from Edr and KIAA1051. Conserved residues for the aspartyl protease active sites are shown in bold. Identical residues are shaded.
DISCUSSION
We report here the identification and developmental expression of Edr, a mammalian gene containing two large overlapping ORFs and a functional signal sequence for ribosomal frameshifting in the –1(5′) direction. Edr represents the first example of a eukaryotic cellular gene that uses this mechanism to translate a fused protein from overlapping reading frames as observed previously for some retroviral and bacterial genes.
Edr cDNA was isolated initially from an EC cDNA library using a differential hybridisation method (15). Analysis of full-length cDNA clones failed to reveal any overall homology between Edr and genes of known function. Intriguingly for a single copy gene conserved between mouse and man, Edr was found to contain three motifs characteristic of gag-pro genes of some retroviral genomes. Most striking of these was the presence of two large ORFs, with the region of overlap containing a heptameric slippage sequence consisting of G GGA AAC and a sequence that can form two possible defined secondary structures, such as an RNA pseudoknot or simple stem–loop (28). Such features are necessary components of a –1 ribosomal slippage mechanism of translation (1,2,10,29) that has been reported previously in some retroviruses and the E.coli dnaX gene. In all cases, constraints required for –1 frameshifting include (i) a heptamer sequence X XXY YYZ, which allows translocation of both ribosome-bound tRNAs by 1 base in the 5′ direction by leaving their non-wobble bases correctly paired with the mRNA in the new reading frame, and (ii) a downstream stimulatory structure that can be either a stem–loop or pseudoknot within 9 nt 3′ of the slippery sequence (11–13). Furthermore, we have demonstrated that Edr can utilise these slippage signal sequences to promote the translation in vitro of two disinct polypeptides of 45 kDa (RF1) and a larger 140 kDa RF1/RF2 fusion protein. Using a series of site-directed mutations in and around the slippage sequence, we demonstrated –1 ribosomal frameshifting at the slippery site to encode the 140 kDa Edr protein. Thus, as with retroviral genomes, the cis-acting translational regulatory elements in Edr are necessary for efficient –1 ribosomal frameshifting. The estimation of the frameshift efficiency by in vitro translation analysis is at least 30%, but the effiicency of such a process in vivo remains to be determined.
In addition to –1 translational frameshifting, Edr coding regions contain two additional motifs that are highly conserved in retroviruses (25); a putative CCHC Zn2-binding motif with a high content of basic residues in RF1, and a putative aspartyl protease catalytic site in the RF1/RF2 translational slippage reading frame. Furthermore, this organisation of motifs is similar to that of retroviral gag-pro genes, in which a –1 translational frameshift mechanism is also utilised (1,2,10,29). Other regions of Edr contain no homology to retroviral genomes, there being no capacity to encode pol-like or env-like proteins in any reading frame and no long terminal repeat sequences.
As such, Edr is a member of a small emerging family of mammalian genes in which the relics of retroviral-like motifs have become subsumed within an otherwise conventional cellular gene. Other examples include Fv1 and Fv4 retroviral restriction loci, described in mice, and syncytin, reported in man and monkey. Fv1 is a single copy gene that encodes a novel Gag-like polypeptide (30). Similarly, the Fv4 locus uses a cellular promoter to express a truncated endogenous retrovirus encoding an Env gene and deleted Pol coding region that interferes with retroviral infection in some Mus musculus subspecies (31). However, neither Fv1 nor Fv4 is conserved across Mus species, or in humans, suggesting a relatively late acquisition and utilisation in Mus with respect to susceptibility to retroviral infection. Syncytin was recently identified as the Env gene of a human endogenous retrovirus and functions in placental morphogenesis (32). This gene is conserved in monkey but not other species, suggestive of the late acquisition of a viral-derived gene. It is important to note that Edr appears to differ from these other examples in two important respects. First, Edr is conserved between mouse and man. Secondly, to our knowledge, Edr is the first reported example of a mammalian gene that utilises –1 translational frameshifting.
Northern blot analyses presented here indicate that Edr is expressed at high levels during mouse embryogenesis, but amongst adult tissues low-level expression was detected only in brain and testes. Within the embryo, RNA in situ hybridisation revealed distinct temporal and spatial patterns of Edr mRNA distribution in the developing musculo-skeletal system. Transcripts were readily detectable during skeletal myogenesis and also in precartilage primordia and derivative chondrogenic cells of the developing skeleton. However, expression was not observed at stages of chondrocyte hypertrophy and ossification of bone tissues. Additional sites of expression were detected in mesenchymal tissues of developing lung, kidney, gonad, gut and placenta.
At the present time, the cellular functions of Edr remain to be determined. However, a number of possible roles can be envisaged. As proposed for Fv1 and Fv4 genes, Edr proteins encoding gag-like proteins may contribute to host defence mechanisms against retrovirus infection. Zn2+-binding motifs in gag precursor and nucleocapsid (NC) proteins are known to specifically bind retroviral nucleic acids at both assembly and infectivity stages of the viral replication cycle (21,33) and the NC domain may also promote annealing of primer tRNA to binding sites facilitating reverse transcription (34). There is some evidence to suggest that NC proteins can have a relaxed specificity for RNAs and recognise different viral genomes among a certain subfamily of retroviruses. For example, NC proteins of the mouse mammary tumor virus (MMTV) and human immunodeficiency virus type I (HIV-1) are known to recognise and package the other’s genome, even though the amino acid sequences differ (20). Additionally, the NC protein of M-MuLV can encapsidate HIV-1 RNA. Thus, Edr gag-like proteins could inhibit retroviral life cycles.
On the basis of the presence of a putative nucleic acid-binding motif, a second possible role of Edr proteins could lie in the regulation of expression of endogenous mammalian genes. Zinc-binding motifs of the CCHC subclass have been identified in a variety of eukaryotic proteins that regulate transcription, RNA splicing and translation. These include the developmentally regulated transcription factors Xpo (35) and X-cat2 (36), glh (37) and the Caenorhabditis elegans heterochronic gene, lin-28 (38). The mammalian CCHC-containing protein CNBP is a zinc-dependent, single-stranded DNA-binding protein implicated in transcriptional regulation (39–41). The CCHC motif of the yeast RNA splicing factor, SLU7, is required for the efficiency of 3′ splice site selection (42), and this nucleic acid-binding motif is also present in the human SR splicing factor 9G8 (43). In Drosophila, the translational suppressor nanos contains RNA-binding CCHC motifs, mutation of which can inhibit translational regulatory activity in vivo (44).
Whilst the RNA distribution of Edr is strikingly regulated during embryonic development it is also interesting to note that, as utilised by retroviruses, –1 translational frameshifting provides a potentially powerful mechanism to regulate the stoichiometry of related polypeptides in the cell. RNA viral frameshift signals determine the stoichiometric ratio of viral proteins such as Gag and Gag–Pol fusion proteins, and the efficiency of these products must be maintained for viral particle morphogenesis. Moreover, trans-acting factors that modulate –1 ribosomal frameshifting have been identified in yeast and man (45). Yeast strains harbouring the mof2-1 allele undergo a significant increase of –1 ribosomal frameshifting with associated inhibition of propagation of the translational frameshift dependent yeast M1 killer virus. Significantly, recombinant expression of the human homologue SU1 in a mof2-1 strain lowered programmed –1 ribosomal frameshifting efficiency to levels observed in wild-type cells and restored the ability of cells to propagate the M1 virus. However, it remains to be determined whether Edr provides an endogenous target for SU1 function in mammals.
Finally, it should be noted that pharmacological modulation of the –1 translational frameshifting mechanism has been proposed as a strategy towards development of antiviral agents. Whilst peptidyl transferase inhibitors clearly possess antiviral activity through targeting –1 translational frameshifting (46), the identification of Edr as a conserved mammalian cellular gene indicates that –1 frameshifting is not unique to viruses. This raises the possibility that antiviral agents that target this mechanism may have previously unanticipated consequences on cellular metabolism in non-infected cells.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to David Melton and Bernhard Herrmann for reagents and Ben Taylor for analysis of Rl SDPs. During the initial stages of this work A.D.R. and C.J.W. were supported by research studentships from the MRC and SERC, respectively. This work was paid for by the CRC, MRC and Ludwig Institute for Cancer Research. K.S. was supported partly by a Daiwa Anglo-Japan postdoctoral fellowship and Ludwig Institute for Cancer Research.
References
- 1.Atkins J.F., Weiss,R.B. and Gesteland,R.F. (1990) Ribosome gymnastics—degree of difficulty 9.5, style 10.0. Cell, 62, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farabaugh P.J. (1996) Programmed translational frameshifting. Annu. Rev. Genet., 30, 507–528. [DOI] [PubMed] [Google Scholar]
- 3.Gesteland R.F. and Atkins,J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- 4.Belcourt M.F. and Farabaugh,P.J. (1990) Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell, 62, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craigen W.J. and Caskey,C.T. (1986) Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature, 322, 273–275. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov I.P., Gesteland,R.F. and Atkins,J.F. (1998) A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics, 52, 119–129. [DOI] [PubMed] [Google Scholar]
- 7.Ivanov I.P., Matsufuji,S., Murakami,Y., Gesteland,R.F. and Atkins,J.F. (2000) Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J., 19, 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsufuji S., Matsufuji,T., Miyazaki,Y., Murakami,Y., Atkins,J.F., Gesteland,R.F. and Hayashi,S. (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell, 80, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov I.P., Rohrwasser,A., Terreros,D.A., Gesteland,R.F. and Atkins,J.F. (2000) Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc. Natl Acad. Sci. USA, 97, 4808–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson G.M. and Brewer,G. (1999) Slip-sliding the frame: programmed –1 frameshifting on eukaryotic transcripts. Genome Res., 9, 393–394. [PubMed] [Google Scholar]
- 11.Blinkowa A.L. and Walker,J.R. (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res., 18, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flower A.M. and McHenry,C.S. (1990) The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl Acad. Sci. USA, 87, 3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchihashi Z. and Kornberg,A. (1990) Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Natl Acad. Sci. USA, 87, 2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammell A.B., Taylor,R.C., Peltz,S.W. and Dinman,J.D. (1999) Identification of putative programmed –1 ribosomal frameshift signals in large DNA databases. Genome Res., 9, 417–427. [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman C.M., Lane,D.P., Watson,C.J. and Rigby,P.W. (1985) The regulation of gene expression in murine teratocarcinoma cells. Cold Spring Harb. Symp. Quant. Biol., 50, 701–706. [DOI] [PubMed]
- 16.Wilkinson D.G., Bailes,J.A., Champion,J.E. and McMahon,A.P. (1987) A molecular analysis of mouse development from 8 to 10 days post coitum detects changes only in embryonic globin expression. Development, 99, 493–500. [DOI] [PubMed] [Google Scholar]
- 17.Krieg P.A. and Melton,D.A. (1984) Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res., 12, 7057–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reith A.D., Ellis,C., Lyman,S.D., Anderson,D.M., Williams,D.E., Bernstein,A. and Pawson,T. (1991) Signal transduction by normal isoforms and W mutant variants of the Kit receptor tyrosine kinase. EMBO J., 10, 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bess J.W.,Jr, Powell,P.J., Issaq,H.J., Schumack,L.J., Grimes,M.K., Henderson,L.E. and Arthur,L.O. (1992) Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I and other retroviruses. J. Virol., 66, 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Summers M.F., Henderson,L.E., Chance,M.R., Bess,J.W.,Jr, South,T.L., Blake,P.R., Sagi,I., Perez-Alvarado,G., Sowder,R.C.D., Hare,D.R. et al. (1992) Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci., 1, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berkowitz R.D., Ohagen,A., Hoglund,S. and Goff,S.P. (1995) Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol., 69, 6445–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowzard J.B., Bennett,R.P., Krishna,N.K., Ernst,S.M., Rein,A. and Wills,J.W. (1998) Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol., 72, 9034–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon D.T., Li,G. and Aldovini,A. (1998) Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J. Virol., 72, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steplewski A., Krynska,B., Tretiakova,A., Haas,S., Khalili,K. and Amini,S. (1998) MyEF-3, a developmentally controlled brain-derived nuclear protein which specifically interacts with myelin basic protein proximal regulatory sequences. Biochem. Biophys. Res. Commun., 243, 295–301. [DOI] [PubMed] [Google Scholar]
- 25.Rao J.K., Erickson,J.W. and Wlodawer,A. (1991) Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry, 30, 4663–4671. [DOI] [PubMed] [Google Scholar]
- 26.Taylor B.A. (1978) In Morse,H.C. (ed.), Origin of Inbred Mice. Academic Press, New York, NY.
- 27.Kikuno R., Nagase,T., Ishikawa,K., Hirosawa,M., Miyajima,N., Tanaka,A., Kotani,H., Nomura,N. and Ohara,O. (1999) Prediction of the coding sequences of unidentified human genes. XIV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res., 6, 197–205. [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Chamorro,M., Lee,S.I., Shen,L.X., Hines,J.V., Tinoco,I.,Jr and Varmus,H.E. (1995) Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J., 14, 842–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacks T. (1990) Translational suppression in gene expression in retroviruses and retrotransposons. Curr. Top. Microbiol. Immunol., 157, 93–124. [DOI] [PubMed] [Google Scholar]
- 30.Best S., Le Tissier,P., Towers,G. and Stoye,J.P. (1996) Positional cloning of the mouse retrovirus restriction gene Fv1. Nature, 382, 826–829. [DOI] [PubMed] [Google Scholar]
- 31.Gardner M.B., Kozak,C.A. and O’Brien,S.J. (1991) The Lake Casitas wild mouse: evolving genetic resistance to retroviral disease. Trends Genet., 7, 22–27. [DOI] [PubMed] [Google Scholar]
- 32.Mi S., Lee,X., Li,X., Veldman,G.M., Finnerty,H., Racie,L., LaVallie,E., Tang,X.Y., Edouard,P., Howes,S., Keith,J.C.,Jr and McCoy,J.M. (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature, 403, 785–789. [DOI] [PubMed] [Google Scholar]
- 33.Lapadat-Tapolsky M., De Rocquigny,H., Van Gent,D., Roques,B., Plasterk,R. and Darlix,J.L. (1993) Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle [published erratum appears in Nucleic Acids Res., (1993) 21, 2024]. Nucleic Acids Res., 21, 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Y.X., Campbell,S., Harvin,D., Ehresmann,B., Ehresmann,C. and Rein,A. (1999) The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol., 73, 4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato S.M. and Sargent,T.D. (1991) Localized and inducible expression of Xenopus-posterior (Xpo), a novel gene active in early frog embryos, encoding a protein with a ‘CCHC’ finger domain. Development, 112, 747–753. [DOI] [PubMed] [Google Scholar]
- 36.Mosquera L., Forristall,C., Zhou,Y. and King,M.L. (1993) A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development, 117, 377–386. [DOI] [PubMed] [Google Scholar]
- 37.Gruidl M.E., Smith,P.A., Kuznicki,K.A., McCrone,J.S., Kirchner,J., Roussell,D.L., Strome,S. and Bennett,K.L. (1996) Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 93, 13837–13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moss E.G., Lee,R.C. and Ambros,V. (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell, 88, 637–646. [DOI] [PubMed] [Google Scholar]
- 39.Flink I.L. and Morkin,E. (1995) Alternatively processed isoforms of cellular nucleic acid-binding protein interact with a suppressor region of the human beta-myosin heavy chain gene. J. Biol. Chem., 270, 6959–6965. [DOI] [PubMed] [Google Scholar]
- 40.Michelotti E.F., Tomonaga,T., Krutzsch,H. and Levens,D. (1995) Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem., 270, 9494–9499. [DOI] [PubMed] [Google Scholar]
- 41.Rajavashisth T.B., Taylor,A.K., Andalibi,A., Svenson,K.L. and Lusis,A.J. (1989) Identification of a zinc finger protein that binds to the sterol regulatory element. Science, 245, 640–643. [DOI] [PubMed] [Google Scholar]
- 42.Frank D. and Guthrie,C. (1992) An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev., 6, 2112–2124. [DOI] [PubMed] [Google Scholar]
- 43.Cavaloc Y., Popielarz,M., Fuchs,J.P., Gattoni,R. and Stevenin,J. (1994) Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J., 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis D., Treiber,D.K., Tao,F., Zamore,P.D., Williamson,J.R. and Lehmann,R. (1997) A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J., 16, 834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui Y., Dinman,J.D., Kinzy,T.G. and Peltz,S.W. (1998) The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol., 18, 1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinman J.D. and Kinzy,T.G. (1997) Translational misreading: mutations in translation elongation factor 1alpha differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA, 3, 870–881. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.