Abstract
Previously, we have shown that all class-1 polypeptide release factors (RFs) share a common glycine-glycine-glutamine (GGQ) motif, which is critical for RF activity. Here, we subjected to site-directed mutagenesis two invariant amino acids, Gln185 and Arg189, situated in the GGQ minidomain of human eRF1, followed by determination of RF activity and the ribosome binding capacity for mutant eRF1. We show that replacement of Gln185 with polar amino acid residues causes partial inactivation of RF activity; Gln185Ile, Arg189Ala and Arg189Gln mutants are completely inactive; all mutants that retain partial RF activity respond similarly to three stop codons. We suggest that loss of RF activity for Gln185 and Arg189 mutants is caused by distortion of the conformation of the GGQ minidomain but not by damage of the stop codon recognition site of eRF1. Our data are inconsistent with the model postulating direct involvement of Gln185 side chain in orientation of water molecule toward peptidyl-tRNA ester bond at the ribosomal peptidyl transferase centre. Most of the Gln185 mutants exhibit reduced ability to bind to the ribosome, probably, to rRNA and/or (peptidyl)-tRNA(s). The data suggest that the GGQ motif is implicated both in promoting peptidyl-tRNA hydrolysis and binding to the ribosome.
INTRODUCTION
Termination of protein synthesis requires the presence of one out of the three termination (stop, nonsense) codons at the ribosomal A site and of class-1 polypeptide release factor (RF) located at the same site (reviewed in 1). Under these conditions the ester bond in peptidyl-tRNA is hydrolysed at the ribosomal peptidyl transferase centre and free polypeptide is released. The hydrolytic reaction proceeds in vitro within the ribosome in the absence of energy source or any additional protein factors (2).
None of the hypotheses postulating the mechanism of decoding the termination codons has been proved directly. It is assumed that within the ribosome, stop codons are recognised by class-1 termination factors, RF1, RF2 and eRF1 (3). The main argument is the very tight contact between class-1 RFs and stop codons within the ribosome revealed by photocrosslinking both in prokaryotes (4,5) and eukaryotes (6). Another argument came from experiments showing that mutagenesis of class-1 RF sequences resulted in the modification of their stop codon recognition pattern (7,8). Alternatively, it was proposed that stop codons could be recognised by specific sequences in ribosomal RNAs (9).
Although amino acid sequences of prokaryotic and mitochondrial class-1 RFs differ considerably from that of eukaryotic/archaeal RFs (2,10), one common GGQ motif has been identified in all known class-1 RFs regardless of their origin (11). This GGQ tripeptide has been found to be functionally essential in eukaryotes both in vitro (11) and in vivo (12).
The eukaryotic class-1 RF (eRF1) is composed of three separate domains, termed N (or 1), M (or 2) and C (or 3) as follows from crystallographic data (12). Two domains (N and M, or 1 plus 2) form an enzymatically active ‘core’, whereas the C-terminal domain is not essential for termination reaction in vitro (13). The GGQ tripeptide occupies the tip of the M domain forming a minidomain with highly exposed amino acid residues (12).
Prokaryotic and eukaryotic class-1 RFs differ not only structurally but also functionally. RF1 and RF2 respond to UAA/UAG and UAA/UGA, respectively, whereas eRF1 responds to three stop codons. This difference implies that the GGQ motif being universal for all class-1 RFs could be implicated in peptidyl-tRNA hydrolysis rather than in stop codon recognition. It was assumed that this motif is involved in the recognition of the CCA-end of the terminal peptidyl-tRNA (11). This hypothesis was strongly supported by the peculiar shape of the human eRF1, in which the N domain mimics the anticodon arm of tRNA while the M domain with the GGQ tip mimics the acceptor stem with the CCA-end (12).
The aim of this work was to elucidate the potential role of two invariant amino acid residues, Gln185 and Arg189, located at the GGQ minidomain, in the structure and function of the human eRF1. To reach this goal, site-directed mutagenesis was applied coupled with determination of RF activity and the ribosome binding ability for all eRF1 mutants. We arrived at the conclusion that both amino acid residues are essential for maintenance of eRF1 function, possibly due to their involvement in stabilisation of the GGQ minidomain conformation. Moreover, the Gln185 and Arg189 residues seem to be implicated in ribosome binding but not in stop codon recognition.
MATERIALS AND METHODS
Cloning and mutagenesis of human eRF1
The full-length cDNA encoding eRF1 with C-terminal His-tag fusion was cloned into pET23b(+) vector (Novagen) under control of phage T7 RNA polymerase promoter. For this, the coding region of the cDNA from TB3-1 clone (14) was amplified using the forward primer (RFNde) 5′-GAGATATACATATGGCGGACGACCC-3′ (NdeI site underlined) and the reverse primer, 5′-GTGGTGCTCGAGGTAGTCATCAGGTC-3′ (XhoI site underlined). Then, the PCR product was subsequently treated with the restriction endonucleases NdeI and XhoI and inserted into pET23b(+) vector treated with the same endonucleases. The resulting construct was verified by DNA sequencing and used to perform mutagenesis.
The mutagenesis procedure was simplified by introducing into human eRF1 cDNA a unique Bst98I site affecting neither amino acid sequence nor the reading frame of human eRF1 using GeneEditor in vitro site-directed mutagenesis kit (Promega). For this purpose the RFBst primer 5′-CCATTCTTAAGCGGGCAAAACGCAAGG-3′ (Bst98I site underlined) was used. The resulting construct pERF4B containing the unique Bst98I site within the gene encoding human eRF1 at positions 576–581 (T576C substitution) from the start ATG codon was used for mutagenesis of human eRF1.
For mutagenesis of Gln185 and Arg189 the following primers were synthesised:
Q185R, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGAGCGACCTCC-3′; Q185E, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGATTCACCTCC-3′; Q185K, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGATTTACCTCC-3′; Q185I, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGAAATACCTCC-3′; Q185D, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGAGTCACCTCC-3′; Q185G, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGACCCACCTCC-3′; Q185N, 5′-CCATTCTTAAGCGGGCAAAACGCAAGGCTGAGTTACCTCC-3′; R189A, 5′-CCATTCTTAAGCGGGCAAAAGCCAAGGCTG-3′; R189K, 5′-CCATTCTTAAGCGGGCAAACTTCAAGGCTG-3′; R189Q, 5′-CCATTCTTAAGCGGGCAAACTGCAAGGCTG-3′.
The direct primer, RFNde, and one of the above mentioned primers containing a Bst98I site as the reverse primer were used in PCR. DNA amplifications were carried out in 50-µl reaction mixtures containing 100 ng of pERF4B, 0.4 µM each primer, 0.24 mM each deoxynucleoside triphosphate, 1× commercial Pfu DNA polymerase reaction buffer and 2.5 U of Pfu Turbo DNA polymerase (Stratagene). Amplifications were run for 1 cycle at 95°C for 3 min, 25 cycles at 95°C for 30 s, 43°C for 30 s, 72°C for 45 s in a thermocycler. The PCR product was purified in low-melting NuSieve GTG agarose (FMC Bioproducts), hydrolysed with NdeI and Bst98I and ligated with pERF4B vector treated with the same endonucleases. The ligated mixture was transformed into Escherichia coli, strain JM109. The resulting cloned DNAs were sequenced and appropriate clones used for expression of the mutant eRF1.
Expression and purification of human eRF1
Wild-type human eRF1 and its mutants containing His-tag at the C-terminus were expressed in E.coli, strain BL21(DE3), and purified using Ni–NTA resin, Superflow (Qiagen), as described (2,13).
Cloning and expression of human eRF3 in E.coli and purification of eRF3
Cloning of the full-length human eRF3 was performed as described earlier for the C-terminal part of human eRF3 (eRF3Cp) (15), using eRF3 cDNA inserted into the plasmid pUC19 (16) as a template for the PCR reaction. The forward primer was 5′-CCCGAATTCATATGGATCCGGGCGG-3′ (NdeI site underlined). Human eRF3 containing His-tag at the C-terminus was synthesised in E.coli, strain BL21(DE3), and purified as previously described (15).
Ribosomes
Rabbit reticulocyte 80S ribosomes washed with 0.5 M KCl were treated with puromycin and GTP for dissociation into subunits which were subsequently resolved by centrifugation in a 10–25% (w/v) sucrose gradient containing 0.3 M KCl, 3 mM MgCl2, 1 mM DTT and 20 mM Tris–HCl, pH 7.6. Before addition to the incubation mixtures, the subunits were combined in an equimolar ratio.
In vitro RF assay
The eRF1 activity was measured as described (2,17) at saturating level (50 µM) of one out of the three stop-codon-containing tetraplets. The incubation mixture (25 µl) contained 20 mM Tris–HCl, pH 7.5, 15 mM MgCl2, 8 mM NH4Cl, 1.5 pmol f[35S]Met–tRNAfMet–AUG ribosome complex and 4 pmol eRF1. The background was measured without tetraplet and subtracted from all values. The amount of f[35S]Met released without stop codon was 500–800 c.p.m. AUG and ribotetraplets were synthesised by A. Veniaminova and M. Ryabkova (Institute of Bioorganic Chemistry, Novosibirsk).
Assays for GTPase activity
GTPase activity was followed by accumulation of [32P]Pi using a modified charcoal precipitation assay as described (18). Incubation mixture (12.5 µl) contained 1 µM [γ-32P]GTP (10 000 c.p.m./pmol), 20 mM Tris–HCl, pH 7.5, 30 mM NH4Cl, 15 mM MgCl2, 0.36 µM ribosomes, 0.24 µM human eRF1 and 0.24 µM human eRF3. The reaction was run at 30°C for 20 min, stopped by adding 0.5 ml of a 5% charcoal suspension in 50 mM NaH2PO4 on ice. The mixture was vortexed and centrifuged at 10 000 r.p.m. for 10 min at 4°C, and the [32P]Pi released into 375 µl of supernatant was quantified by liquid scintillation counting. Release of [32P]Pi in the absence of human eRF1 was measured simultaneously and this value (background 2000–3000 c.p.m.) was subtracted from all samples.
RESULTS
As shown in Figure 1, part of the M (middle) domain of eRF1 encompassing the universal GGQ tripeptide is highly conserved within eukaryotes and archaebacteria (10) and forms a minidomain separated from the rest of the eRF1 molecule (12). As it is already known that Gly183 and Gly184 are critically essential for the RF activity in vitro (11) and in vivo (12) we have focused here on mutagenesis of two invariant residues, Gln185 and Arg189.
Figure 1.
Highly conserved region in eukaryotic and archaeal eRF1s encompassing the GGQ tripeptide common to all class-1 translation termination factors. Positions are numbered for the human eRF1. Accession numbers are given after the names of the species. Numbers in brackets correspond to NCBI-Entrez-Protein database accession numbers. Amino acids in alignment were shaded according to their identity percentage (white letters, black shading, 100%; white letters, dark-grey shading, 80%; black letters, light-grey shading, 70%).
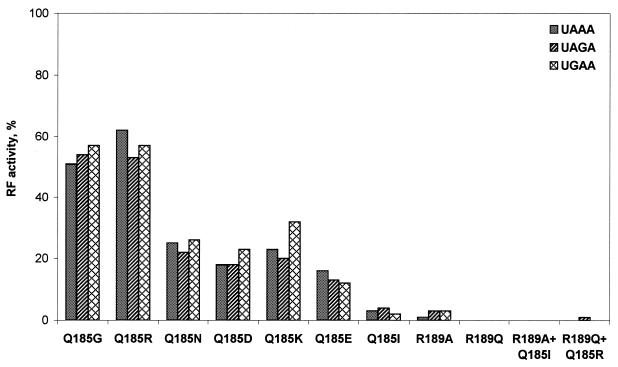
As is evident from Figure 2, all mutations of Gln185 and Arg189 caused a decrease in RF activity in vitro, but to different levels. Three groups of mutations were found. In the first group, complete inactivation was observed (Gln185Ile, Arg189Ala, Arg189Gln, Arg189Ala + Gln185Ile and Arg189Gln + Gln185Arg). The second group of mutations caused profound inactivation and the mutants retained 10–25% of the residual RF activity (Gln185Asp, Gln185Asn, Gln185Lys and Gln185Glu). The third group of mutants (Gln185Gly and Gln185Arg) retained no less than half of the initial RF activity (Fig. 2).
Figure 2.
RF activity in an in vitro assay for human eRF1 mutants toward three stop codons. For details see Materials and Methods.
The mutant eRF1s respond similarly to three stop codons irrespective of the degree of inactivation and of the nature of the substituting amino acids (Fig. 2). The complete or partial loss of RF activity might be caused by damage of the eRF1 ribosome-binding site (RBS) (11) due to mutations. We tested this possibility by following the eRF3 GTPase activity, which entirely depends on the complex formation between eRF1 and eRF3 within the ribosome (13,15,18,19). As is evident from Table 1, eRF1 mutants are able to stimulate the eRF3 GTPase activity showing that even inactive eRF1 mutants partly retained their ability to bind to the ribosome and to eRF3. This result is in full agreement with the previous data where it was shown that Gly183 and Gly184 mutants maintain their binding potential toward the ribosome (11). Partial impairment of the ribosome-binding ability for the most mutant eRF1s implies that the GGQ minidomain is essential not only for RF activity as shown earlier (11,12), but also for interaction with the ribosome. On the other hand, the results indicate that reduced RF activity of some mutants (Fig. 2) is only partially associated with weaker binding to the ribosome and Gln185 contributes also to conservation of the RF activity of eRF1.
Table 1. GTPase activity of human eRF3 promoted by the human eRF1 containing mutations of Gln185 and Arg189.
| eRF1 mutants |
eRF3 GTPase activity, % |
| Gln185 (wild-type) | 100 |
| Gln185Arg | 100 |
| Arg189Gln + Gln185Arg | 100 |
| Gln185Asn | 75 |
| Gln185Gly | 60 |
| Gln185Ile | 60 |
| Arg189Gln | 56 |
| Gln185Lys | 48 |
| Arg189Ala | 45 |
| Arg189Ala + Gln185Ile | 40 |
| Gln185Glu | 35 |
| Gln185Asp | 30 |
The eRF1-dependent and ribosome-dependent GTPase activity of human eRF3 was equal to 36 000–44 000 c.p.m. The SD was ±10%. Average from at least three independent measurements is presented. For further details see Materials and Methods.
DISCUSSION
The GGQ motif is present in all class-1 RFs (11), including prokaryotic RF1 and RF2 that differ in codon specificity. In organisms with variant genetic codes, where one or two out of three stop codons are reassigned for sense codons, the GGQ motif is also present (20–22). In these organisms, eRF1 responds only to stop codons but not to the reassigned codon (23). All mutations of Gln185 (Fig. 2), Gly183 and Gly184 (11) cause reduction or abolishment of RF activity toward all three stop codons without manifestation of any stop codon preference. Taken together, sequence and mutagenesis data support the notion that the GGQ minidomain is not implicated in stop codon recognition.
Three main arguments were forwarded to support the idea that the GGQ motif is implicated in promotion of hydrolysis of the ester bond in the terminal peptidyl-tRNA. First, ubiquitous conservation of this motif throughout the living matter regardless of the stop codon recognition properties of class-1 RFs coupled with its critical functional significance (11). Secondly, correlation between the universal occurrence of the GGQ motif in class-1 RFs and the universality of the CCA-end of tRNA irrespective of the enormous variability of the peptidyl and tRNA moieties of peptidyl-tRNA (11). Thirdly, a very peculiar positioning of the GGQ minidomain in the crystal structure of human eRF1 (12) occupying one of the extremities of the Y-like shaped eRF1 molecule remote from the N domain which is involved in stop codon recognition (7). This postulated interaction of the GGQ motif with peptidyl transferase centre implies that the GGQ domain should display RNA-binding properties toward peptidyl-tRNA (mostly toward the CCA-end) and large rRNA which forms the backbone of the peptidyl transferase centre (24). Therefore, promotion of peptidyl-tRNA hydrolysis offered by class-1 RFs requires the interaction with RNAs at least at two ribosomal sites, with mRNA (stop codon recognition) and with rRNA/tRNA at the peptidyl transferase centre. In fact, as shown in Table 1 most of the mutants possess reduced ability to stimulate eRF3 GTPase activity, which reflects the ribosome-binding potential of the mutant eRF1. Remarkably, the introduction of acidic amino acid residues (aspartic and glutamic acids) causes the most dramatic effect while replacements with basic (arginine and lysine), or polar (asparagine) or aliphatic (isoleucine) residues is less significant. The acidic amino acid residues should interfere with the RNA binding while other residues seem to be compatible or even favourable as, for example, in the case of arginine known to be widely implicated in RNA binding (25). This result is anticipated in view of the postulated role of the GGQ minidomain in binding to RNAs within the ribosome.
RNA-binding properties for the GGQ minidomain suggested here are also indirectly supported by recent findings with RNA–protein complexes. Sequence-specific RNA binding by a KH protein domain has been structurally characterised (26 and references therein). The KH domain exposes the invariant Gly-X-X-Gly tetrapeptide, in which X represents arginine or glycine in the loop connecting two helices. Remarkably, the GGQ minidomain contains a Gly-Arg-Gly-Gly tetrapeptide (Fig. 1, positions 181–184) unique in the eRF1 sequence.
The reduction of the ribosome-binding ability has never exceeded 3-fold (Table 1). Thus, the GGQ minidomain is not a single RBS in eRF1 and other site(s) contribute to the binding. This is consistent with the earlier finding where it was suggested that the RBSs are located both at the N and M domains of human eRF1 (13).
Our data on partial conservation of RF activity in mutant eRF1s (Fig. 2) are in agreement with recent observations in which Gln235 of E.coli RF1 equivalent to Glu185 in human eRF1 was mutated to glutamic acid (27). This amino acid change does not abolish the activity of the factor in vivo and the same mutation in RF2 does not impair RF activity in vitro although the kc was reduced 30-fold.
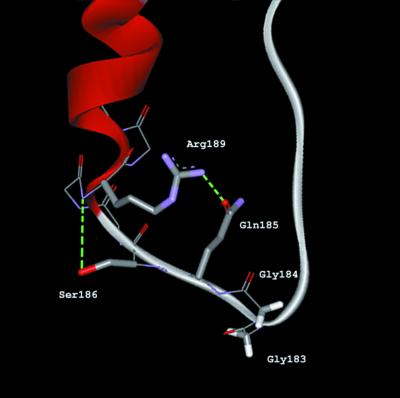
The conformation of the Gln185 residue is stabilised by a network of hydrogen bonds involving conserved residues of the GGQ minidomain (Fig. 3). The amide group of Gln185 accepts a hydrogen bond from the guanidinium group of Arg189. In turn, the main-chain amide group of Arg189 donates a hydrogen bond to the hydroxyl group of Ser186 (12). This non-covalent structure of minidomain explains why replacement of Arg189 with alanine or glutamine is harmful for RF activity (Fig. 2). These substitutions destroy the network of hydrogen bonds essential for stabilisation of the tertiary structure of the minidomain. We assume that partial inactivation of the eRF1 activity for polar Gln185 substitution is associated with the disruption or distortion of the hydrogen-bond network responsible for a local stability of eRF1 three-dimensional structure (12). A double mutant in which glutamine and arginine residues were mutually exchanged (Arg185–Gln189) was completely active in the ribosome binding (Table 1). This result argues in favour of the essential role played by non-covalent interactions between these residues in conservation of the three-dimensional structure of the GGQ minidomain, which is important for the ribosome binding.
Figure 3.
Ribbon diagram of the minidomain containing the GGQ tripeptide derived from crystallographic data (12) by WebLab ViewerLite program version 4.0 (Molecular Simulations Inc.). Side-chain residues of Gly183, Gly184, Gln185, Ser186 and Arg189 displayed in a stick mode and backbone in a line mode. Hydrogen bonds are coloured in green.
In E.coli RF2 Ser246, which is near the GGQ motif (positions 250–252), significantly affects RF activity and ribosome binding of the factor (27–30). These data are consistent with the notion (11,12) that the GGQ minidomain plays a pivotal role in class-1 RF functioning. Remarkably, at the same distance from the GGQ motif, a strongly conserved lysine–histidine dipeptide is present in eRF1/aRF1 protein family (Fig. 1), which may be an equivalent to essential Ser246 in E.coli RF2.
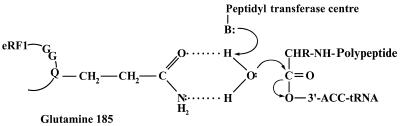
It has been proposed (12) that Gln185 of the GGQ tripeptide coordinates a catalytic water molecule near a hydrolysable ester bond in peptidyl-tRNA at the peptidyl transferase centre of the ribosome as shown below.
As is evident from the scheme, two structural features of the glutamine side chain, the carbonyl oxygen and the amide nitrogen, are critical for forming two hydrogen bonds with two hydrogen atoms of the water molecule. However, glycine residue replacing Gln185 in mutant eRF1 possesses none of these two groups and no side chain but, nevertheless, the remaining RF activity of the Gln185Gly mutant is substantial (Fig. 2). This 2-fold reduction is correlated with the reduced ribosome-binding capacity of the same mutant (Table 1). Therefore, presumably, the reduced RF activity for the Gln185Gly mutant is mostly due to weaker binding rather than loss of function.
This result strongly contradicts the above hypothesis as the glycine residue totally lacks the ability to coordinate a water molecule as shown on the above scheme. The Gln185Arg mutant also retains substantial RF activity (Fig. 2), although the ability of arginine residue to coordinate a water molecule seems questionable. In E.coli, glutamine in the GGQ tripeptide of RF2 is methylated and this post-translational modification enhances the RF activity of RF2 (29). It is unknown whether N5-methylglutamine residue is able to function as prescribed by the above scheme.
In yeast eRF1, structurally similar to human eRF1 (2), Gln182 in the GGQ tripeptide was replaced by leucine, arginine or proline residues and the activity of these mutants was tested in vivo using a genetic approach (12). It appeared that all these substitutions were lethal for yeast cells (12). The complete loss of the RF activity for human eRF1 Gln185Ile mutant (Fig. 2) is entirely consistent with the data for yeast eRF1 Gln182Leu mutant. The Gln182Pro mutation in yeast eRF1 could induce a serious distortion of the GGQ minidomain conformation, which in turn may cause a reduction of eRF1-binding ability toward the ribosome as we have shown here for other glutamine mutants (Table 1). The 2-fold reduction in RF activity observed for Gln185Arg mutant (Fig. 2) without change in ribosome-binding ability (Table 1) is inconsistent with a lethal effect observed in vivo with yeast eRF1 bearing the same mutation (12). It remains unknown whether the mutant eRF1s are present in yeast cells as the mutant proteins have not been detected in vivo and it was not ruled out that mutant yeast eRF1 proteins became highly susceptible to proteolysis. Furthermore, for yeast eRF1 mutants it remains unknown whether their ribosome-binding activity was affected or remained intact.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank N. Ivanova and A. Poltaraus for sequencing mutants of the eRF1 gene. This work was supported by Human Frontier Science Programme (grant 96-032), by Russian Foundation for Basic Research (grant to L.K.), by Programme for Support of Russian Scientific Schools (grant to L.K.) and by Danish Cancer Society (grant to J.J.).
References
- 1.Kisselev L. and Buckingham,R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- 2.Frolova L., Le Goff,X., Rasmussen,H.H., Cheperegin,S., Drugeon,G., Kress,M., Arman,I., Haenni,A.-L., Celis,J.E., Philippe,M., Justesen,J. and Kisselev,L. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura Y., Ito,K. and Ehrenberg,M. (2000) Mimicry grasps reality in translation termination. Cell, 101, 349–352. [DOI] [PubMed] [Google Scholar]
- 4.Brown C.M. and Tate,W.P. (1994) Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem., 269, 33164–33170. [PubMed] [Google Scholar]
- 5.Poole E.S., Brimacombe,R. and Tate,W.P. (1997) Decoding the translational termination signal: the polypeptide chain release factor in Escherichia coli crosslinks to the base following the stop codon. RNA, 3, 974–982. [PMC free article] [PubMed] [Google Scholar]
- 6.Chavatte L., Frolova,L., Kisselev,L. and Favre,A. (2001) The polypeptide chain release factor eRF1 specifically contacts the sUGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem., 268, 2896–2904. [DOI] [PubMed] [Google Scholar]
- 7.Bertram G., Bell,H.A., Ritchie,D.W., Fullerton,G. and Stansfield,I. (2000) Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA, 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K., Uno,M. and Nakamura,Y. (2000) A tripeptide anticodon deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- 9.Arkov A.L. and Murgola,E.J. (1999) Ribosomal RNAs in translation termination: facts and hypotheses. Biochemistry (Moscow), 64, 1354–1359. [PubMed] [Google Scholar]
- 10.Kisselev L.L., Oparina,N.Y. and Frolova,L.Y. (2000) Class 1 translation termination factors are structurally and functionally similar to suppressor tRNA and are related to various structural-functional families (prokaryotes and mitochondria–eukaryotes and archaebacteria). Mol. Biol. (Moscow), 34, 427–442. [PubMed] [Google Scholar]
- 11.Frolova L.Y., Tsivkovskii,R.Y., Sivolobova,G.F., Oparina,N.Y., Serpinsky,O.I., Blinov,V.M., Tatkov,S.I. and Kisselev,L.L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song H., Mugnier,P., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factors eRF1 – mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- 13.Frolova L., Merkulova,T.I. and Kisselev,L. (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA, 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grenett H.E., Bounelis,P. and Fuller,G.M. (1992) Identification of a human cDNA with high homology to yeast omnipotent suppressor 45. Gene, 110, 239–243. [DOI] [PubMed] [Google Scholar]
- 15.Frolova L.Y., Simonsen,J.L., Merkulova,T.I., Litvinov,D.Y., Martensen,P.M., Rechinsky,V.O., Camonis,J.H., Kisselev,L.L. and Justesen,J. (1998) Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem., 15, 36–44. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino S., Miyazawa,H., Enomoto,T., Hanaoka,F., Kikuchi,Y., Kikuchi,A. and Ui,M. (1989) A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J., 8, 3807–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caskey C.T., Beaudet,A.L. and Tate,W.P. (1974) Mammalian release factor: in vitro assay and purification. Methods Enzymol., 30, 293–303. [DOI] [PubMed] [Google Scholar]
- 18.Frolova L., Le Goff,X., Zhouravleva,G., Davydova,E., Philippe,M. and Kisselev,L. (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA, 2, 334–341. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhouravleva G., Frolova,L., Le Goff,X., Le Guellec,R., Inge-Vechtomov,S., Kisselev,L. and Philippe,M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang A., Brunen-Nieweler,C., Muramatsu,T., Kuchino,Y., Beier, H. and Heckmann,K. (2001) The ciliate Euplotes octocarinatus expresses two polypeptide release factors of the type eRF1. Gene, 262, 161–168. [DOI] [PubMed] [Google Scholar]
- 21.Inagaki Y. and Doolittle,W.F. (2001) Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res., 29, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C.A., Knight,R.D. and Landweber,L.F. (2001) The molecular basis of nuclear genetic code change in ciliates. Curr. Biol., 11, 65–74. [DOI] [PubMed] [Google Scholar]
- 23.Kervestin S., Frolova,L., Kisselev,L. and Jean-Jean,O. (2001) Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO Rep., 2, 680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–929. [DOI] [PubMed] [Google Scholar]
- 25.Jones S., Daley,D.T.A., Luscombe,N.M., Berman,H.M. and Thornton,J.M. (2001) Protein-RNA interactions: a structural analysis. Nucleic Acids Res., 29, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Canadillas J.-M. and Varani,G. (2001) Recent advances in RNA–protein recognition. Curr. Opin. Struct. Biol., 11, 53–58. [DOI] [PubMed] [Google Scholar]
- 27.Dincbas-Renqvist V., Engstrom,A., Mora,L., Heurgue-Hamard,V., Buckingham,R. and Ehrenberg,M. (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J., 19, 6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uno M., Ito,K. and Nakamura,Y. (1996) Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie, 78, 935–943. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura Y., Kawazi,Y., Uno,M. and Ito,K. (2000) Genetic probes to bacterial release factors: tRNA mimicry hypothesis and beyond. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. ASM Press, Washington, DC, pp. 519–526.
- 30.Wilson D.N., Guevremont,D.and Tate,W.P. (2000) The ribosomal binding and peptidyl-tRNA hydrolysis functions of Escherichia coli release factor 2 are linked through residue 246. RNA, 6, 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]