Abstract
The human gut microbiome has a significant role in host physiology; however its role in gluten catabolism is debatable. Present study explores the role of human gut microbes in gluten catabolism and a native human gut microbe Cellulomonas sp. HM71 was identified. SSU rDNA analysis has described human gut microbiome structure and also confirmed the permanent residentship of Cellulomonas sp. HM71. Catabolic potential of Cellulomonas sp. HM71 to cleave antigenic gluten peptides indicates presence of candidate gene encoding biocatalytic machinery. Genome analysis has identified the presence of gene encoding S9A serine protease family—prolyl endopeptidase, with Ser591, Asp664 and His685 signature residues. Cellulomonas sp. HM71 prolyl endopeptidase activity was found optimal at pH 7.0 and 37 °C with a KM of 35.53 μmol and specifically cleaves at proline residue. Current study describes the gluten catabolism potential of Cellulomonas sp. HM71 depicting possible role of human gut microbes in gluten catabolism to confer resistance mechanisms for the onset of celiac diseases in populations with gluten diet.
Electronic supplementary material
The online version of this article (10.1007/s12088-018-0732-2) contains supplementary material, which is available to authorized users.
Keywords: Human gut microbes, Culture dependent techniques, SSU rDNA, Gluten protein, Celiac disease
Introduction
More than a billion microbial cells are residing all over the human body, which are higher in number compared to human cells [1–3]. Human microbiome is extremely diverse with spatial abundance across the body locations [1–4]. Amidst other body locations, human gut microbiome is highly dense and comprises of trillions of bacterial cells, approximately ten times more than the human cells [1, 3, 5]. Bacteroidetes and Firmicutes are dominant microbial groups within gut [2, 4], followed by Proteobacteria, Verrucomicrobia, Actinobacteria, Fusobacteria, and Cyanobacteria in lesser proportions [1–3, 5]. A variety of molecular studies have decoded the impact of the gut microbiota in cardiovascular diseases, irritable bowel syndrome, diabetes, and cancer [5–7]. A number of association studies link human nutrition and health with the metabolic capacity of gut microbiota [6–10]. However, insights into role of the human gut microbiome in gluten catabolism await further understanding.
Wheat gluten is one of the major constituent of the total dietary protein consumption. Gluten has high content of proline and glutamine residues in their structure, which are largely resistant to cleavage by the major human gastrointestinal (GI) digestive enzymes [11]. Partial cleavage of gluten generates immunogenic gluten peptides [11, 12]. These peptides activate CD+T cells in the lamina propria of HLA-DQ2 mediated immune system, followed by immunological inflammation of mucosa leading to gluten sensitivity and subsequent onset of celiac disease [11, 13, 14]. Recently, various oral cavity microbes and gut microbes were reported for their bio-catalytic role in gluten catabolism [15–19]. These studies emphasize on the role of human gut microbes in gluten catabolism and their possible role in ameliorating celiac disease [15–18]. Recent culture dependent and culture independent studies has decoded the functional role of a fraction of the human gut microbes (~ 10–20%) and indicates that majority of human gut microbiome still awaits functional elucidation. Present study was aimed to explore the functional role of human gut microbes in gluten catabolism. A native human gut microbe Cellulomonas sp. HM71 was identified for its gluten catabolic potential, illustrating possible role of human gut microbes in dietary gluten catabolism to avoid onset of celiac disease.
Materials and Methods
Sample Collection
Fecal sample was collected in a sterile container from a healthy individual (Age 29, Male, Blood Sugar 100–120 mg/dl, BP 120/80), with no past history of prolonged illness or antibiotic treatment. Fresh sample was used to culture microbes and isolate metagenomic DNA. Human ethical guidelines were followed strictly before engaging individual for current study. The study has been conducted after ethical clearance from the human ethical committee at Maharshi Dayanand University, Rohtak, Haryana, India.
Microbial Community Structure of Human Gut Microbiome
A 100 mg of fresh feces was used for extraction of metagenomic DNA [4]. The qualitative and quantitative analysis of the metagenomic DNA was performed with NanoQuant (Tecangrp. Ltd, Switzerland) and Qubit® dsDNA HS Assay Kit (Life technologies, USA). Metagenomic DNA isolated from healthy human stool sample was amplified for SSU rRNA gene [20]. An amplified product was sequenced with next generation sequencing using Roche 454 GS FLX+ system. Quantitative Insights Into Microbial Ecology (QIIME) 1.9.1 pipeline was implemented for data analysis. SSU rRNA gene sequences were quality filtered [20, 21].
Glutenase Activity in Stool Microbial Pellet
To confirm the role of human gut microbes in gluten catabolism, microbial pellet was purified from fresh human feces [4]. Bacterial cells were ruptured by ultra-sonication at 4 °C for 10 min with an output of 650 W (on/off pulse of 9 s). The lysed cells were centrifuged at 13,000 rpm for 20 min at 4 °C to collect the supernatant, which was used for checking biocatalytic activity against the Z-Gly-Pro-pNA substrate [15].
Screening of Gluteanse Positive Microbe
Gluteanse active human gut microbes were isolated from fresh stool. A 50 mg of human feces were suspended in 1 ml phosphate buffer saline (PBS) pH 7.4. Suspension was centrifuged at 3000 rev min−1. Debris free fecal suspension was serially diluted and plated on to glutenase screening medium (Luria–Bertani broth (LB) supplemented with 1% gluten). Screening medium plates were incubated at 37 °C and observed for the presence of a hydrolytic halo surrounding the colony. Glutenase active microbial culture was purified with the dilution culturing method and glutenase activity of microbes was reassessed.
Phylogenetic Analysis of Glutenase Positive Microbe
Genomic DNA was isolated from overnight grown culture of glutenase positive microbe [22]. The SSU rRNA gene was amplified from the microbial genomic DNA to analyze its phylogeny [23]. Phylogenetic tree was constructed by using maximum likelihood method with help of MEGA-6 Package software version 6.06. The reliability of each branch was checked after performing bootstrap for 500 replicates [23].
Analysis for Gluten Catabolic Potential of Cellulomonas sp. HM71
Antigenic fraction of gluten was prepared from wheat gluten [24]. Gluten catabolic potential of Cellulomonas sp. HM71 was checked against synthetic substrate [15] and gluten antigenic fraction. Catabolism of antigenic gluten fraction was observed in a reaction mixture (2 ml) containing actively growing culture [A600nm(0.5)], antigenic peptide (5 mg/ml) in PBS (pH 7.4). Reaction mixture was incubated at 37 °C and fractions were collected at different time intervals (0, 2, 4, 6 and 24 h). Fractions were heat inactivated and catabolism of antigenic peptide was observed with reverse-phase HPLC using a C-18 column [RP18 column (3.9 × 150 mm)] (Waters, USA) [16].
In Silico Characterization of Cellulomonas PEP Gene
Prolyl endopeptidase (PEP) gene was retrieved through genome survey of Cellulomonas sp and PEP gene sequence was characterized by using different open source bioinformatic tools [25].
Enzyme Activity and Substrate Specificity of Cellulomonas sp HM71 Prolylendopeptidase
Kinetic properties of Cellulomonas sp HM71 prolylendopeptidase were analyzed with Z-Gly-Pro-4-nitroanilide substrate [15]. All assays were carried out in the triplicate to calculate standard deviation. Substrate specificity study of Cellulomonas sp HM 71 was checked with Suc-Ala-Pro-pNA, Z-QQP-pNA, Z-YPQ-pNA (Helix Biosciences, India) and Z-Gly-Pro-4-nitroanilide (Sigma Aldrich) [15].
Results and Discussion
Microbial Community Structure of an Individual Gut Microbiome
Various attempts have been made to decipher human gut microbial community structure and found significant presence of Firmicutes, Bacteriodetes, Proteobacteria, Actinobacteria and Acidiobacteria [1–6, 26–28]. These microbes were characterized for their physiological functions, however a large portion of them still remains untapped. In the current study, an effort has been made to understand the functional role of resident gut microbes in gluten catabolism by decoding its native gut microbiome structure and identifying microbes with gluten catabolic potential. We obtained 46317 SSU rRNA gene sequences from gut metagenomic DNA. These sequences were curetted for V1–V4 regions of the SSU rRNA gene and ambiguous, low quality (< Q30) and chimeric sequences were removed, resulting in a total of 8602 high quality reads for downstream analysis. All sequences were processed using QIIME 1.9.1 with de Novo clustering option, resulting in 696 OTUs. The average Shannon diversity index was 6.34. The even rate of identification of new OTUs, in reference to the number of SSU rRNA gene sequences was 12.35. Reads were assigned into operational taxonomic units (OTUs) using a closed reference OTU picking protocol in QIIME 1.9.1, uclust to search sequences against a subset of the Greengenes database, version (13_8) with a filtration parameter of 97% sequence identity. This resulted in 11 microbial phyla, inclusive of Bacteriodetes (88.7), Firmicutes (7.54%) and Proteobacteria (2.64%), followed by Lentisphaerae (0.21%), Actinobacteria (0.21%), Gemmatimonadetes (0.19%), Acidobacteria (0.07%), Chloroflexi (0.03%), Verrucomicrobia (0.01%), Planctomycetes (0.01%) and Nitrospirae (0.01%). Only 0.29% of the reads matched with unclassified/unknown microbial groups. Abundance of Bacteriodetes, Firmicutes and Proteobacterial microbial groups within human gut microbiome is in agreement with other human gut microbiome studies [29]. This corroborates that most of the individuals share a common pool of microorganisms with inter-individual variability/individuality [30, 31]. This variability could be accounted by diet, physiology, geography and ethnicity [28].
Human gut microbes have efficient enzymatic machinery for catabolism of various dietary components like carbohydrates [32], polyphenols [33] and lipids [34]. However a limited information is available about the functional role of human gut microbes in gluten catabolism [16–19]. Accordingly, the human gut microbes were purified and checked for their gluten catabolic potential. Enzymatic assay with the microbial pellet lysate, hydrolyzed Z-Gly-Pro-pNA (synthetic enzymatic substrate for prolyl endopeptidase) and released 182 µM free pNA. Similar enzymatic activity was observed with agar plate assay, where gluten (1%) was supplemented in agar medium. The results highlighted the possible role of human gut microbes in gluten metabolism.
Glutenase activity screening identified a highly active gluten catabolic microbial strain, HM71. The glutenase positive microbe HM71 was checked for its phylogeny using SSU rRNA gene analysis. SSU rRNA gene from glutenase positive HM71 was sequenced and assembled to generate a sequence of 1384 bp that shared a good homology (98–99.3%) with various Cellulomonas species like Cellulomonas firmi and Cellulomonas biazotea. Even similar phylogenetic affiliation was observed within Phylogenetic tree (Fig. 1a). Accordingly, human gut microbe HM71 was identified as Cellulomonas species strain HM71. Previous studies have also shown the presence of various Cellulomonas sp. from human feces [29] and characterized their celluloytic machinery which was also found associated with Cellulomonas species HM71. A significant number of BLAST Hits of Cellulomonas species HM71 SSU rRNA gene sequences were observed within in-house SSU rRNA gene dataset, as well as in SSU rRNA gene dataset of Human Microbiome Project (HMP) (hmpdacc.org/) during abundance analysis (Fig. 1b). These results clearly demonstrate its nativity as a human gut microbe.
Fig. 1.
SSU rRNA gene sequence analysis. Phylogenetic analysis of glutenase positive human gut microbiome HM71 (a), abundance of Cellulomonas sp. HM71 SSU rRNA gene sequence homologs within NIH- HMP stool SSU rRNA gene sequence dataset and in house human fecal SSU rRNA gene dataset (b)
Gluten Catabolic Potential of Cellulomonas sp. HM71
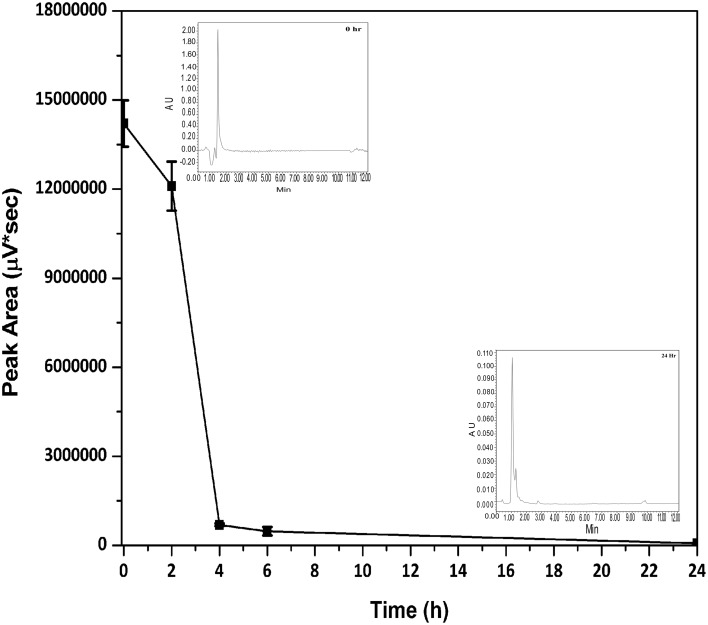
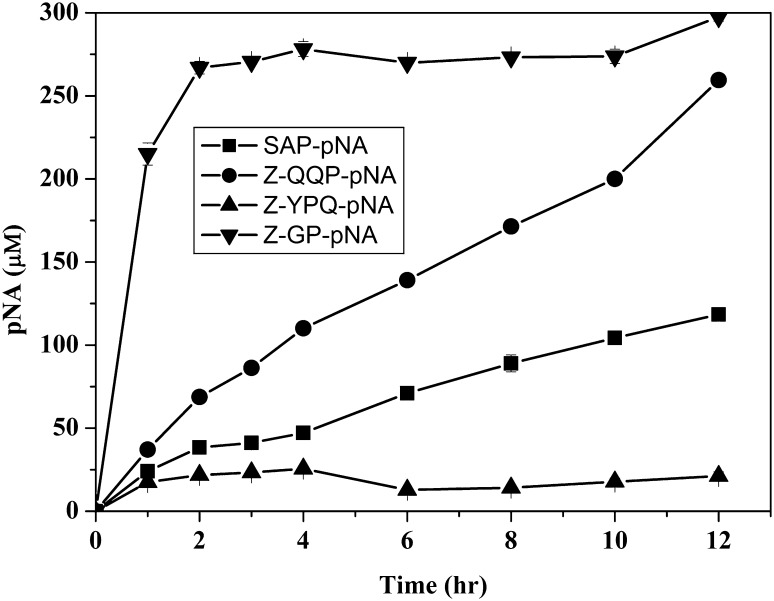
Time series analysis of antigenic gluten catabolism with Cellulomonas sp. HM71 has indicated a significant degradation of antigenic peptide (> 95% %) within 4 h of incubation (Fig. 2). Catabolism of antigenic gluten fraction confirms gluten catabolic potential of Cellulomonas sp. HM71. Cellulomonas sp. HM71 also showed a remarkable biocatalytic activity against Z-GP-pNA, SAP-pNA and Z-QQP-pNA synthetic substrates for glutenase [35], while a poor activity against Z-YPQ-pNA (Fig. 3). These results indicate its proline specific biocatalytic activity with potential to hydrolyze the immunogenic gliadins that could lead to the onset of celiac disease [12, 29, 30]. Cellulomonas sp. HM71 possibly has hosts bioactive S9A serine proteases. Presence of gluten catabolic potential in a human gut microbe indicates possible role of human gut microbes to confer resistance mechanisms for declining onset of celiac diseases in populations with gluten diet.
Fig. 2.
Cellulomonas sp. HM71 hydrolytic activity against gluten antigenic fraction. HPLC chromatogram and peak area were checked after incubating gluten antigenic fraction with Cellulomonas sp. HM71 for different time period (0, 2, 4, 6 and 24 h)
Fig. 3.
Substrate specificity of Cellulomonas sp. HM71 prolylendopeptidase
In Silico Characterization of Cellulomonas sp. HM71 PEP Gene
Many bacterial and fungal species are reported to harbor gene for prolyl endopeptidase [3, 14]. A genome survey of Cellulomonas species has identified PEP genes within Cellulomonas microbial clade. PEP gene encodes a protein of 728 amino acids and molecular weight of 78kD. The encoded protein belongs to serine protease subfamily S9A within the esterase super family (Supplementary Fig. 1). The PEP protein was checked for various physiochemical characteristics (Table 1). These analyses indicated its aqueous solubility and stability with cytosolic topology. Similar characteristics were observed for Myxococcus xanthus prolyl endopeptidase [19]. Multiple sequence alignment of Cellulomonas PEP with other characterized PEP protein has indicated the presence of three active residue Ser591, Asp664, His685, as described in other studies [35].
Table 1.
Physiochemical characterization of PEP sequence
| Characteristics | PEP |
|---|---|
| Protein super family | Esterase lipase |
| Protein family | Serine 9A |
| EC number | EC 3.4.21.26 |
| Amino acids | 728 |
| Molecular weight | 78,393.1 Da |
| Theoretical pI | 5.07 |
| Ext. coefficient (all pairs of Cys residues form cystines) | 138,895 |
| Ext. coefficient (all Cys residues are reduced) | 138,770 |
| Estimated half-life | 30 h (mammalian reticulocytes, in vitro) |
| > 20 h (yeast, in vivo) | |
| > 10 h (Escherichia coli, in vivo) | |
| Instability index | 35.11 |
| Subcellular localization | Cytosolic |
| Hydrophobicity | No |
| N-Glycosylation site | No sites predicted |
| Prolyl endopeptidase serine family | 509–727 |
| Active site residues | Ser591, Asp664, His685 (Position as per gene encoded sequence) |
| Conserved domains | Prolyl oligopeptidase |
| Signal peptide | No |
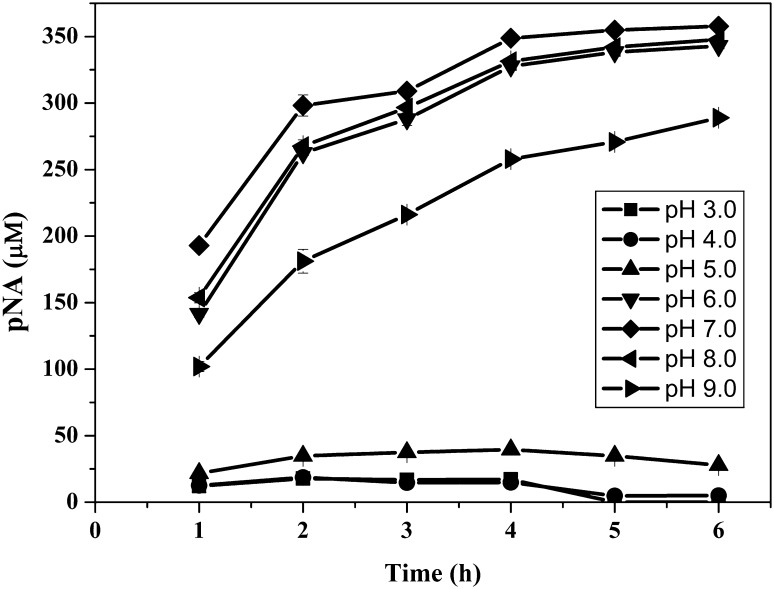
Kinetic Properties of Cellulomonas sp HM71 Prolylendopeptidase
Prolyl-endopeptidase of Cellulomonas sp. HM71 showed its biocatalytic activity in the pH range of 6.0–9.0 (Fig. 4), with an optimum pH of 7.0 at 37 °C in 50 mM phosphate buffer, with unit activity of 3.22 μM min−1 with KM and Vmax of 35.53 μmol and 0.362 μmol min−1 respectively. We further investigated whether the function of microbial prolyl endopeptidase was pH dependent. The enzyme pH activity profile was matched with gently acidic environment of the upper small intestine (pH 6–8) that provides the clue about the metabolic site of gluten in human intestine. The cleavage site specificity of the PEP enzyme is at SAP↓, GGP↓, TP↓G and GP↓. The cleavage activities are highly active against dipeptide GP↓ and tripeptide GGP↓ at proline residue site. On the basis of its active sites and cleavage properties, it was confirmed that our microbial enzyme has sufficient potential for degrading the gluten. Findings in present study have potential role of human gut microbes in management of celiac disease. Till date, a limited information is available about the existence of gluten-degrading microorganisms in the human gut [16–19] and absorption/detoxification of gluten in human gut is still a mystery. The current study has identified a human gut microbe with a potential to detoxify the antigenic gluten peptides. The present study highlights the role of human gut microbes in gluten catabolism and identifies faecal bacteria that secretes gluten degrading enzyme. These bacteria and its secreted enzyme could lead to novel and effective strategies to detoxify immunogenic gluten peptides prior to reaching the proximal small intestine.
Fig. 4.
Enzymatic analysis of Cellulomonas sp. HM71 prolylendopeptidase at different pH
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1: Phylogenetic analysis of Cellulomonas PEP with different members of other groups of serine family
Acknowledgements
Funding for this work was obtained from the CSIR sponsored research Scheme 60(0099)/11/EMRII and UGC Major Research project 41-1256/2012 SR. Jitendra Kumar and Dr Manoj Kumar thank CSIR, New Delhi and Department of Science and Technology (SB/YS/LS-201/2013), New Delhi for the fellowships, respectively.
Compliance with Ethical Standards
Ethical Statement
The current study describes the functional role of human gut microbe isolated from human faeces of a human participant. Accordingly, human ethical guidelines were followed strictly and an ethical clearance was sought from human ethical committee of Maharishi Dayanand University, Rohtak, Haryana, India for conducting the present study. Along with this, this article does not contain any studies with animals performed by any of the authors.
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12088-018-0732-2) contains supplementary material, which is available to authorized users.
References
- 1.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purohit HJ. Gut-bioreactor and human health in future. Indian J Microbiol. 2018;58:3–7. doi: 10.1007/s12088-017-0697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yadav M, Verma MK, Chauhan NS. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. 2018;200:203–217. doi: 10.1007/s00203-017-1459-x. [DOI] [PubMed] [Google Scholar]
- 4.Kumar J, Kumar M, Gupta S, Ahmed V, Bhambi M, Pandey R, Chauhan NS. An improved methodology to overcome key Issues in human fecal metagenomic DNA extraction. Genom Proteom Bioinform. 2016;14:371–378. doi: 10.1016/j.gpb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 6.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verma MK, Ahmed V, Gupta S, Kumar J, Pandey R, Mandhan V, Chauhan NS. Functional metagenomics identifies novel genes ABCTPP, TMSRP1 and TLSRP1 among human gut enterotypes. Sci Rep. 2018;8:1397. doi: 10.1038/s41598-018-19862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Requena T, Martínez-Cuesta MC, Peláez C. Diet and microbiota linked in health and disease. Food Funct. 2018;9:688–704. doi: 10.1039/C7FO01820G. [DOI] [PubMed] [Google Scholar]
- 9.Dutton RJ, Turnbaugh PJ. Taking a metagenomic view of human nutrition. Curr Opin Clin Nutr Metab Care. 2012;15:448–454. doi: 10.1097/MCO.0b013e3283561133. [DOI] [PubMed] [Google Scholar]
- 10.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 11.Kumar J, Kumar M, Pandey R, Chauhan NS. Physiopathology and management of gluten-induced celiac disease. J Food Sci. 2017;82:270–277. doi: 10.1111/1750-3841.13612. [DOI] [PubMed] [Google Scholar]
- 12.Rey M, Yang M, Lee L, Zhang Y, Sheff JG, Sensen CW, Mrazek H, Halada P, Man P, McCarville JL, Verdu EF, Schriemer DC. Addressing proteolytic efficiency in enzymatic degradation therapy for celiac disease. Sci Rep. 2016;6:30980. doi: 10.1038/srep30980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sollid LM, Khosla C. Future therapeutic options for celiac disease. Nat Clin Pract Gastroenterol Hepatol. 2005;2:140–147. doi: 10.1038/ncpgasthep0111. [DOI] [PubMed] [Google Scholar]
- 14.Rizzello CG, De Angelis M, Di Cagno R, Camarca A, Silano M, Losito I, De Vincenzi M, De Bari MD, Palmisano F, Maurano F, Gianfrani C, Gobbetti M. Highly efficient gluten degradation by Lactobacilli and fungal proteases during food processing: new perspectives for celiac disease. Appl Environ Microbiol. 2007;73:4499–4507. doi: 10.1128/AEM.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG, Helmerhorst EJ. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS ONE. 2011;6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutiérrez S, Pérez-Andrés J, Martínez-Blanco H, Ferrero MA, Vaquero L, Vivas S, Casqueiro J, Rodríguez-Aparicio LB. The human digestive tract has proteases capable of gluten hydrolysis. Mol Metab. 2017;6:693–702. doi: 10.1016/j.molmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caminero A, Herrán AR, Nistal E, Pérez-Andrés J, Vaquero L, Vivas S, Ruiz de Morales JM, Albillos SM, Casqueiro J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. 2014;88:309–319. doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- 18.Herrán AR, Pérez-Andrés J, Caminero A, Nistal E, Vivas S, Ruiz de Morales JM, Casqueiro J. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res Microbiol. 2017;168:673–684. doi: 10.1016/j.resmic.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Kocazorbaz EK, Zihnioglu F. Purification, characterization and the use of recombinant prolyl oligopeptidase from Myxococcus xanthus for gluten hydrolysis. Protein Expr Purif. 2017;129:101–107. doi: 10.1016/j.pep.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Gupta S, Kumar M, Kumar J, Ahmad V, Pandey R, Chauhan NS. Systemic analysis of soil microbiome deciphers anthropogenic influence on soil ecology and ecosystem functioning. Int J Environ Sci Technol. 2017;14:2229–2238. doi: 10.1007/s13762-017-1301-7. [DOI] [Google Scholar]
- 21.Morowitz MJ, Denef J, Costello EK, Thomas BC, Poroyko V, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 23.Kumar Mondal A, Kumar J, Pandey R, Gupta S, Kumar M, Bansal G, Mukerji M, Dash D, Chauhan NS. Comparative genomics of host-symbiont and free-living Oceanobacillus species. Genome Biol Evol. 2017;9:1175–1182. doi: 10.1093/gbe/evx076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazer AC, Fletcher RF, Ross CA, Shaw B, Sammons HG, Schneidner R. Gluten-induced enteropathy: the effect of partially digested gluten. Lancet. 1959;2:252–255. doi: 10.1016/S0140-6736(59)92051-3. [DOI] [PubMed] [Google Scholar]
- 25.Yandamuri RC, Gautam R, Darkoh C, Dareddy V, Bouhssni M, Clack B. Cloning, expression, sequence analysis and homology modeling of the prolyl endoprotease from Eurygaster integriceps puton. Insects. 2014;5:762–782. doi: 10.3390/insects5040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microbes Environ. 2017;32:300–313. doi: 10.1264/jsme2.ME17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature. 2012;12:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierer N, Ferrenberg S, Flores GE, Gonzalez A, Kueneman J, et al. From animalcules to an ecosystem: application of ecological concepts to the human microbiome. Annu Rev Ecol Evol Sys. 2012;43:137–155. doi: 10.1146/annurev-ecolsys-110411-160307. [DOI] [Google Scholar]
- 32.Tasse L, Bercovici J, Pizzut-Serin S, Robe P, Tap J, Klopp C, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuervo A, Hevia A, Lopez P, Suarez A, Sanchez B, Margolles A, Gonzalez S. Association of polyphenols from oranges and apples with specific intestinal microorganisms in systemic lupus erythematosus patients. Nutrients. 2015;7:1301–1317. doi: 10.3390/nu7021301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohira H, Tsutsui W, Fujioka Y. Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis? J Atheroscler Thromb. 2017;24:660–672. doi: 10.5551/jat.RV17006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helmerhorst EJ, Zamakhchari M, Schuppan D, Oppenheim FG. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PLoS ONE. 2010;5:e13264. doi: 10.1371/journal.pone.0013264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1: Phylogenetic analysis of Cellulomonas PEP with different members of other groups of serine family