Abstract
Replication of the coronavirus genome requires continuous RNA synthesis, whereas transcription is a discontinuous process unique among RNA viruses. Transcription includes a template switch during the synthesis of subgenomic negative-strand RNAs to add a copy of the leader sequence. Coronavirus transcription is regulated by multiple factors, including the extent of base-pairing between transcription-regulating sequences of positive and negative polarity, viral and cell protein–RNA binding, and high-order RNA-RNA interactions. Coronavirus RNA synthesis is performed by a replication-transcription complex that includes viral and cell proteins that recognize cis-acting RNA elements mainly located in the highly structured 5′ and 3′ untranslated regions. In addition to many viral nonstructural proteins, the presence of cell nuclear proteins and the viral nucleocapsid protein increases virus amplification efficacy. Coronavirus RNA synthesis is connected with the formation of double-membrane vesicles and convoluted membranes. Coronaviruses encode proofreading machinery, unique in the RNA virus world, to ensure the maintenance of their large genome size.
Keywords: nidovirus, positive-strand RNA viruses, replication, transcription, virus-host interaction, RNA proofreading
CORONAVIRUS REPLICATION AND TRANSCRIPTION
Coronaviruses are enveloped, positive-strand RNA viruses with genomes approximately 30 kb in length that belong to the family Coronaviridae in the order Nidovirales (1). Coronaviruses infect a wide variety of mammalian and avian species, in most cases causing respiratory and intestinal tract disease. Human coronaviruses (HCoVs), such as HCoV-229E, HCoV-OC43, HCoV-NL63, and HKU1, have long been recognized as major causes of the common cold. Two recent HCoVs, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), emerged in 2002 and 2012, respectively, causing life-threatening disease in humans (2). In addition, novel animal coronaviruses, such as the porcine deltacoronavirus (PDCoV) (3) and the porcine epidemic diarrhea virus (PEDV) (4), have recently emerged, causing great economic loss in China and the United States.
The 5′-proximal two-thirds of the coronavirus genome encodes the replicase gene, which contains two open reading frames, ORF1a and ORF1b (Figure 1a). Translation of ORF1a yields polyprotein 1a (pp1a), and −1 ribosomal frameshifting allows translation of ORF1b to yield pp1ab (5, 6). Together, these polyproteins are co- and posttranslationally processed into 16 nonstructural proteins (nsps), most of them driving viral genome replication and subgenomic mRNA (sgmRNA) synthesis (Figure 1a). The 3′ third of the genome encodes the structural and accessory proteins, which vary in number among the different coronaviruses (Figure 1a) (1).
Figure 1.
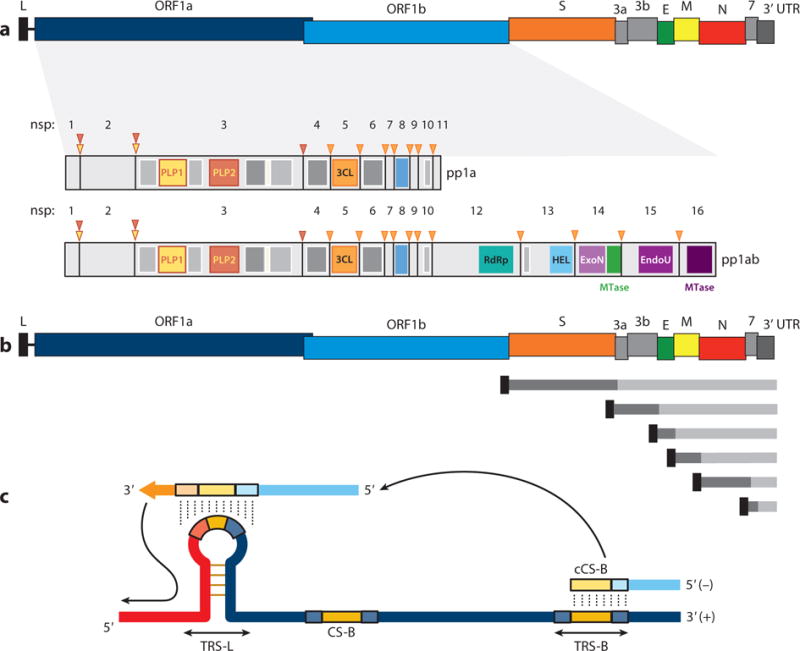
Coronavirus genome structure and gene expression. (a) Coronavirus genome structure. The upper scheme represents the TGEV genome. Labels indicate gene names; L corresponds to the leader sequence. Also represented are the nsps derived from processing of the pp1a and pp1ab polyproteins. PLP1, PLP2, and 3CL protease sites are depicted as inverted triangles with the corresponding color code of each protease. Dark gray rectangles represent transmembrane domains, and light gray rectangles indicate other functional domains. (b) Coronavirus genome strategy of sgmRNA expression. The upper scheme represents the TGEV genome. Short lines represent the nested set of sgmRNAs, each containing a common leader sequence (black) and a specific gene to be translated (dark gray). (c) Key elements in coronavirus transcription. A TRS precedes each gene (TRS-B) and includes the core sequence (CS-B) and variable 5′ and 3′ flanking sequences. The TRS of the leader (TRS-L), containing the core sequence (CS-L), is present at the 5′ end of the genome, in an exposed location (orange box in the TRS-L loop). Discontinuous transcription occurs during the synthesis of the negative-strand RNA (light blue), when the copy of the TRS-B hybridizes with the TRS-L. Dotted lines indicate the complementarity between positive-strand and negative-strand RNA sequences. Abbreviations: EndoU, endonuclease; ExoN, exonuclease; HEL, helicase; MTase, methyltransferase (green, N7-methyltransferase; dark purple, 2′-O-methyltransferase); nsp, nonstructural protein; PLP, papain-like protease; RdRp, RNA-dependent RNA polymerase; sgmRNA, subgenomic RNA; TGEV, transmissible gastroenteritis virus; TRS, transcription-regulating sequence; UTR, untranslated region.
Coronavirus RNA-dependent RNA synthesis includes two differentiated processes: genome replication, yielding multiple copies of genomic RNA (gRNA), and transcription of a collection of sgmRNAs that encode the viral structural and accessory proteins (7, 8).
Like that of other positive-strand RNA viruses, coronavirus genome replication is a process of continuous synthesis that utilizes a full-length complementary negative-strand RNA as the template for the production of progeny virus genomes. The initiation of negative-strand synthesis involves access of the RNA-dependent RNA polymerase (RdRp) to the 3′ terminus of the genome, promoted by 3′- end RNA sequences and structures (5). There is evidence that both 5′- and 3′- end RNA elements are required for the production of progeny positive-strand RNA from the intermediate negative-strand RNA, suggesting that interactions between the 5′ and 3′ ends of the genome contribute to replication (9).
In contrast to replication, coronavirus transcription includes a discontinuous step during the production of sgmRNAs (10, 11). This process, unique among known RNA viruses, is a hallmark of the order Nidovirales and ultimately generates a nested set of sgmRNAs that are 5′ and 3′ coterminal with the virus genome (Figure 1b). These sgmRNAs all include at their 5′ end a common leader sequence, whose length ranges from 65 to 98 nt in different coronaviruses (12). This common leader sequence is present only once at the very 5′ end of the genome, which implies that sgmRNAs are synthesized by the fusion of noncontiguous sequences, the leader and the 5′ end of each mRNA coding sequence, called the body (B). The transcription mechanism in coronaviruses is seemingly complicated as compared with the transcription mechanisms in other positive-strand RNA viruses, such as internal initiation and premature termination (13). In fact, in contrast to coronavirus and arterivirus sgmRNAs, subgenomic transcripts of other Nidovirales, such as toroviruses and roniviruses, do not have a common 5′ leader sequence (14). This observation raises the question of whether the presence of the leader sequence in coronavirus sgmRNAs provides any selective advantage to the virus. The presence of the 5′ leader sequence was shown to protect SARS-CoV mRNAs from nsp1-induced endonucleolytic cleavage of capped mRNAs, providing a strategy for the efficient accumulation of viral mRNAs and viral proteins during infection (15). Moreover, as noted below, the complement of the leader sequence supports initiation of positive-strand RNA synthesis, making the negative-strand subgenomic RNAs (sgRNAs) a template for further amplification of positive-strand sgmRNAs.
RNA Sequences Regulating Transcription
The transcription process is controlled by transcription-regulating sequences (TRSs) located at the 3′ end of the leader sequence (TRS-L) and preceding each viral gene (TRS-B) (Figure 1c). TRSs include a conserved core sequence (CS) 6–7 nt in length and variable 5′ and 3′ flanking sequences (the 5′ TRS and 3′ TRS, respectively) (16). Because the CS is identical for the genome leader (CS-L) and all mRNA coding sequences (CS-B), the CS-L could base-pair with the nascent negative strand complementary to each CS-B (cCS-B), allowing for leader-body joining (Figure 1c). By engineering the base-pairing between the CS-L and the cCS-B in infectious genomic cDNAs of coronaviruses (17) and arteriviruses (18, 19), it was formally demonstrated that (a) the discontinuous step of transcription occurs during the synthesis of the negative-strand RNA, and (b) base-pairing between the CS-L and the cCS-B is required to drive the template switch of the nascent negative-strand RNA to the leader. Additionally, the stability (free energy, G) of the extended duplex between the TRS-L and the complement of the TRS-B (cTRS-B), including 5′ and 3′ TRS flanking sequences, was confirmed as a critical regulatory factor for the synthesis of sgmRNAs (20, 21).
Coronavirus transcription resembles high-frequency, similarity-assisted copy-choice RNA recombination, requiring sequence identity between donor and acceptor RNAs and hairpin structures present in the acceptor RNA (22), in which the TRS-L would act as an acceptor for the cTRS-B donor sequence (Figure 1c). Secondary structure analysis of the TRS-L region from transmissible gastroenteritis virus (TGEV) (23) and bovine coronavirus (BCoV) (24) showed that the CS-L is exposed in the loop of a structured hairpin that is relevant for replication and transcription (23). These observations provided experimental evidence for the selection of the TRS-L during the template switch, excluding other genome TRS-Bs that contain the CS. Only the CS-L, located in a sequence context with optimal secondary structure and stability for template switching, may act as a landing site for the newly synthesized negative-strand RNA.
The coronavirus discontinuous transcription process implies a premature termination during the synthesis of the negative-strand RNAs and a template switch of the nascent negative-strand RNA to the leader (Figure 1c). This switch requires long-distance RNA-RNA interactions, probably assisted by RNA-protein complexes that would bring into close proximity the 5′- end TRS-L and the TRS-B preceding each gene. These complexes, presumably formed prior to the template switch, might contribute to the stoppage of negative-strand RNA synthesis at the TRS-B (7). In TGEV, two intragenomic, long-distance RNA-RNA interactions have been described to regulate the transcription of sgmRNA N [coding for the nucleocapsid protein (N protein)], which is the most abundant sgmRNA during viral infection despite its low G value for TRS-L–TRS-B duplex formation (25). The first interaction is established between two complementary 9-nt cis-acting elements preceding the CS of the N gene, the proximal element (pE) and the distal element (dE) (Figure 2), which are located 7 and 449 nt upstream of the CS-N, respectively (25). The amount of sgmRNA N produced is directly proportional to the extent of the complementarity between pE and dE and inversely proportional to the distance between them (26). This interaction is probably necessary to relocate the active domain, another cis-acting RNA motif, consisting of a 173-nt region at the 5′ flank of dE, immediately preceding the CS-N (Figure 2) (26). The second long-distance RNA-RNA interaction is held between a 10-nt sequence within the active domain and a complementary RNA motif located at the 5′ end of the viral genome (nucleotides 477 to 486), more than 25,000 nt apart (27), and represents the longest-distance RNA-RNA interaction reported so far in the RNA virus world (Figure 2). This interaction could bring into physical proximity the leader sequence, at the genome 5′ end, and the TRS-N, which would promote the template switch during synthesis of the negative-strand sgRNAs (Figure 2). This long-distance RNA-RNA interaction provided for the first time experimental support of the physical proximity between the TRS-L and a TRS-B during discontinuous transcription in order to promote efficient RdRp transfer. The secondary structure of the active domain and the high-order structure formed by the RNA-RNA interactions could also promote the slowdown and stoppage of the transcription complex at the CS-N, as described for tombusvirus transcription (28). The sequences and secondary structures of the RNA motifs involved in these long-distance interactions are conserved among members of the species Alphacoronavirus I, suggesting a functional similarity (27).
Figure 2.
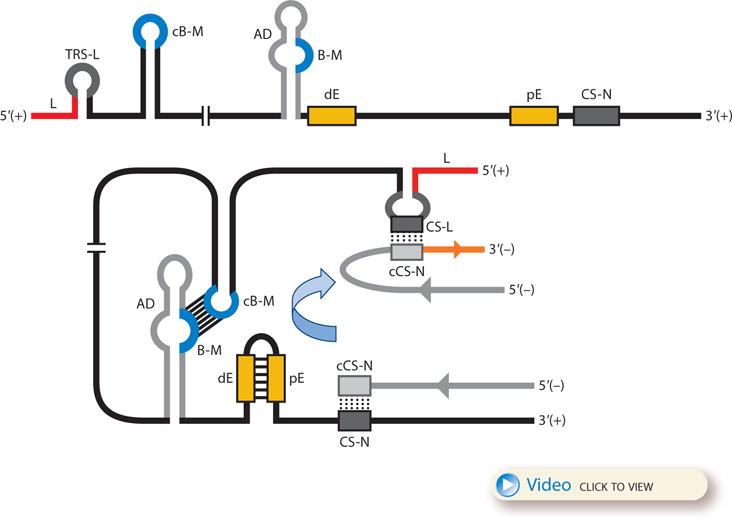
Model for the formation of genome high-order structures regulating N gene transcription. The upper linear scheme represents the coronavirus genome. The red line indicates the leader sequence in the 5′ end of the genome. The hairpin indicates the TRS-L. The gray line with arrowheads represents the nascent negative-sense RNA. The curved blue arrow indicates the template switch to the leader sequence during discontinuous transcription. The orange line represents the copy of the leader added to the nascent RNA after the template switch. The RNA-RNA interactions between the pE (nucleotides 26894 to 26903) and dE (nucleotides 26454 to 26463) and between the B-M in the active domain (nucleotides 26412 to 26421) and the cB-M in the 5′ end of the genome (nucleotides 477 to 486) are represented by solid lines. Dotted lines indicate the complementarity between positive-strand and negative-strand RNA sequences. Abbreviations: AD, active domain secondary structure prediction; B-M, B motif; cB-M, complementary copy of the B-M; cCS-N, complementary copy of the CS-N; CS-L, conserved core sequence of the leader; CS-N, conserved core sequence of the N gene; dE, distal element; pE, proximal element; TRS-L, transcription-regulating sequence of the leader. For an animated version of the model, see Video 1 or download a PowerPoint slideshow.
Coronavirus Transcription Model
Experimental data on transcription in coronaviruses (7, 17, 21, 25, 27) and the related arteriviruses (14) can be integrated into a transcription model that includes three steps (Figure 3): (a) First, gRNA forms transcription initiation precomplexes, bringing into physical proximity the distal TRS-L and TRS-B. RNA-RNA, RNA-protein, and protein-protein interactions might maintain these precomplexes in a dynamic equilibrium. (b) These precomplexes act as slowdown and detachment signals for the transcription complex during the synthesis of negative-strand RNA. (c) Once the TRS-B has been copied, if the ΔG of duplex formation between the cTRS-B (in the nascent negative-strand RNA) and the TRS-L exceeds a minimum threshold, a template switch to the leader takes place, adding a copy of the TRS-L to complete the negative-strand sgRNA. These negative-strand sgRNAs subsequently serve as templates for the synthesis of multiple copies of sgmRNAs.
Figure 3.
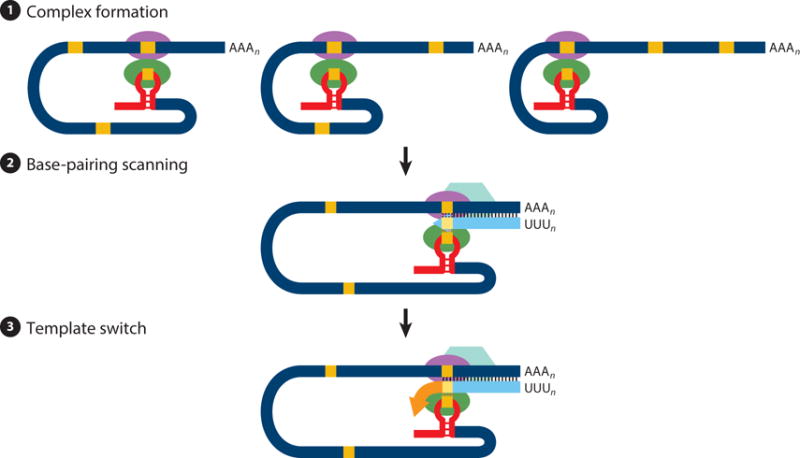
Three-step model of coronavirus transcription. (❶) Complex formation. Proteins binding transcription-regulating sequences are represented by ellipsoids, the leader sequence is indicated with a red bar, and core sequences are indicated with orange boxes. (❷) Base-pairing scanning. Negative-strand RNA is shown in light blue; the transcription complex is represented by a hexagon. Vertical lines indicate complementarity between the genomic RNA and the nascent negative strand. (❸) Template switch. Due to the complementarity between the newly synthesized negative-strand RNA and the transcription-regulating sequence of the leader, template switch to the leader is made by the transcription complex to complete the copy of the leader sequence.
REGULATION OF CORONAVIRUS PROTEIN STOICHIOMETRIC RATIOS
Viruses have developed diverse strategies to ensure the optimal expression ratio of each viral protein. In the case of coronaviruses, replicase proteins are expressed from a full-length gRNA by translation of two polyproteins that are proteolytically cleaved. In contrast, structural and accessory proteins are expressed from a nested set of sgmRNAs. Therefore, the abundance of each sgmRNA must be tightly regulated during the discontinuous transcription process to ensure appropriate viral protein ratios.
Multiple factors regulate the transcription process by modulating the template switch frequency during discontinuous transcription (9, 29). The most important one is the complementarity between the TRS-L and the cTRS-B (17, 21). In a study of several coronaviruses, most sgmRNAs synthesized could be predicted in silico by local base-pairing calculations (17). Additional factors may regulate sgmRNA levels, such as TRS secondary structure, proximity to the 3′ end, and RNA-RNA or protein-RNA interactions (7, 9). In this sense, coronavirus N protein is required for efficient sgmRNA transcription (30, 31). Coronavirus N protein has RNA chaperone activity that drives template switching in vitro and may also facilitate template switching during coronavirus transcription (31). Although nonessential for RNA synthesis, coronavirus nsp1 is associated with viral components of the replication-transcription complex (RTC) (32). Therefore, it may modulate coronavirus RNA synthesis similarly to arterivirus nsp1 protein, which modulates the relative abundance of sgmRNAs and gRNA (33).
As components of the coronavirus RTC, cell proteins can also modulate sgmRNA ratios. Infectious bronchitis virus (IBV) N protein was recently shown to recruit cellular helicase DDX1 to viral RTCs, facilitating TRS read-through and synthesis of long sgmRNAs (34). Interestingly, DDX1 recruitment requires N protein phosphorylation by cellular GSK3 kinase (34). Thus, the cell factor DDX1, attracted by phosphorylated N protein, provides a unique strategy for the transition from discontinuous to continuous transcription in coronaviruses to ensure balanced sgmRNA and full-length gRNA synthesis.
Coronavirus protein ratios are also posttranscriptionally regulated. Most sgmRNAs are structurally polycistronic but functionally monocistronic, with only the 5′-most ORF being translated into a viral protein. The clearest example of coronavirus translational regulation is the expression of the polyprotein pp1ab, which is generated by a programmed −1 ribosomal frame-shifting mechanism (35). This process leads to minor levels of most of the RNA-modifying enzymes, encoded by ORF1b, in comparison with those of other replicase enzymes, such as proteases, encoded by ORF1a. Alteration of coronavirus frame-shifting efficiency modified the ratio of replicase proteins, affecting viral RNA synthesis and virus production (36). In this sense, regulation of the ratio between the two viral polymerases nsp8 and nsp12, encoded by ORF1a and ORF1b, respectively, may be involved in controlling the levels of the different sgmRNAs during viral RNA synthesis (37).
ROLE OF DOUBLE-MEMBRANE VESICLES
Like that of other positive-strand RNA viruses, coronavirus RNA synthesis is associated with extensively rearranged intracellular membranes (38). High-resolution three-dimensional images obtained by electron tomography in SARS-CoV-infected cells showed a unique reticulovesicular network of modified endoplasmic reticulum that integrated convoluted membranes (CMs), interconnected double-membrane vesicles (DMVs), and vesicle packets apparently arising from DMV merger. Viral replicase subunits (nsp3, nsp5, and nsp8) localized to CMs, whereas dsRNA, presumably the replicative intermediate, mainly localized to the DMV interior, supporting the concept that the membrane network would contribute to protecting replicating RNA from antiviral defense mechanisms (38). In mouse hepatitis virus (MHV)-infected cells, newly synthesized RNA was detected in close proximity to DMVs and CMs (39), and viral RNA levels correlated with the number of DMVs (40–42). However, other data do not necessarily support the active contribution of DMVs to viral RNA synthesis. Nascent MHV RNAs colocalize with dsRNA only at early times postinfection; at later times, the dsRNA distributed throughout the cell is apparently transcriptionally inactive (43). Furthermore, RdRp or nascent viral RNA has not been detected inside DMVs, and ultrastructural analysis could not confirm any connection between the DMV interior and the cytoplasm (38), raising questions about the import and export of ribonucleotide precursors and produced RNAs exported from RNA synthesis areas (44). The coexpression of the SARS-CoV transmembrane nonstructural proteins nsp3, nsp4, and nsp6 resulted in the formation of CMs and DMVs (45), suggesting a function in the biogenesis of the membranous replicative structures, and also in the anchoring of the RTC (46–48).
In addition to DMVs, the gammacoronavirus IBV also induces different membrane structures such as spherules tethered to zippered endoplasmic reticulum (49). Unlike any previously identified coronavirus-induced structure, IBV spherules contain a pore connecting their interior to the cell cytoplasm (50).
The function and dynamics of DMVs and CMs and the precise localization of the sites of active viral RNA synthesis are still unresolved questions, and further studies are required. A possible model proposes that DMVs may be the initial sites of active RNA synthesis early in infection, whereas at later times, after membrane connections are lost, RNA synthesis shifts to the CMs, and DMVs become end-stage products that sequester nonfunctional dsRNAs to prevent the stimulation of the innate immune response (51, 52).
STRESS GRANULES AND PROCESSING BODIES IN REPLICATION-TRANSCRIPTION COMPLEX ACTIVITY
Stress granules and processing (P) bodies are cytoplasmic RNA granules that contain translationally silenced messenger ribonucleoproteins, contributing to translation regulation in cells. Whereas P bodies are constitutively expressed and include components involved in mRNA decay, stress granules are thought to be sites of mRNA storage and triage formed in response to stress conditions. Stress granules represent an intermediate stage in the dynamic equilibrium between active translation on free polysomes and mRNA decay in P bodies (53, 54).
During infection, RNA viruses dynamically interact with stress granules and P bodies (55), leading to varying stress granule phenotypes. Many viruses have evolved mechanisms to antagonize the formation of stress granules, suggesting that stress granules are involved in restricting virus replication through RNA silencing (56, 57). In contrast, other RNA viruses, such as respiratory syncytial virus, induce stress granule formation and take advantage of stress granule responses as part of the infectious cycle (58). For coronaviruses, MHV replication was found to be enhanced in cells deficient in stress granule formation, implying that stress granules contribute to viral inhibition (59). TGEV induced stress granules that persisted from 7 to 16 hpi, which was correlated with a decrease in viral replication and transcription (60). These granules contained the stress granule markers T cell intracellular antigen 1 (TIA-1), TIA-1-related protein (TIAR), and polypyrimidine tract–binding protein (PTB) in association with viral gRNA and sgmRNAs. TGEV-induced stress granules might contribute to the spatiotemporal regulation of viral RNA synthesis. Several stress granule proteins (including caprin and G3BP) have been associated with IBV N protein, pointing to the relevance of these RNA-protein complexes in the regulation of coronavirus gene expression (61).
A new hypothesis postulates that stress granules are involved in an integrated stress–innate immunity activation response (57, 62). In this pathway, viral RNA and proteins, along with host pathogen-sensing factors, such as the dsRNA-binding protein kinase R (PKR) and the RNA helicases retinoic acid–induced gene 1 (RIG-I) and melanoma differentiation–associated gene 5 (MDA5), can be sequestered in stress granules (63). Additional insight into the relevance of stress granules and P bodies for the regulation of coronavirus RNA synthesis is still required.
RELEVANCE OF THE CELL NUCLEUS IN CORONAVIRUS RNA SYNTHESIS
All positive-strand RNA viruses that infect animals replicate in the cytoplasm of the infected host cell. However, there is ample evidence that implicates the nucleus and nuclear proteins in the replication and pathogenesis of positive-strand RNA viruses, including coronaviruses (64). The replication of these RNA viruses in enucleated cells is variable, ranging from 10% to 100% of that in nucleated controls (65, 66). The relocation of nuclear proteins to the cytoplasm and of viral proteins to the nucleus during virus replication (7, 64, 67) (Table 1) highlights the relevance of this organelle during the coronavirus infectious cycle and raises important questions: What is the role of nuclear factors in the replication of these viruses, and do viral proteins traveling to the nucleus participate in RTC activity?
Table 1.
Nidovirus proteins that localize to the nucleus
| Family | Virus | Protein | Reference(s) |
|---|---|---|---|
| Coronaviridae | IBV | N | 69 |
| TGEV | N | 69 | |
| MHV | N | 69 | |
| IBV | 3b | 74 | |
| SARS-CoV | 3b | 76 | |
| MERS-CoV | 4b | 75, 78 | |
| SARS-CoV | 6 | 79, 159 | |
| SARS-CoV | 9b | 81, 160 | |
| TGEV | nsp1 | 82 | |
| Arteriviridae | EAV | N | 85 |
| PRRSV | N | 88, 92, 93, 161 | |
| EAV | nsp1 | 85 | |
| PRRSV | nsp1 | 89, 91 |
Virus structural proteins: nucleocapsid (N), 3b, 4b, 6, and 9b. Virus nonstructural protein: nonstructural protein 1 (nsp1). Virus name abbreviations: EAV, equine arteritis virus; IBV, infectious bronchitis virus; MERS-CoV, Middle East respiratory syndrome coronavirus; MHV, mouse hepatitis virus; PRRSV, porcine reproductive and respiratory syndrome virus; SARS-CoV, severe acute respiratory syndrome coronavirus; TGEV, transmissible gastroenteritis virus.
The coronavirus protein most frequently associated with the host cell nucleus is the N protein, and its transport to the nucleus is regulated by phosphorylation (68). N protein nuclear localization is associated with induction of cell cycle arrest and inhibition of cytokinesis (68–72) and is involved in recruitment to the cytoplasm of cell nuclear proteins, such as heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and the helicase DDX1 (34, 73). As noted above, N protein–recruited DDX1 functions in the RTC in facilitating TRS read-through and synthesis of long sgmRNAs (34). The 3b proteins of IBV (74) and SARS-CoV (75, 76), though different in nature, have also been located in part in the nuclei of transfected or infected cells. Following nuclear localization, SARS-CoV 3b protein traffics to the outer membrane of mitochondria, where it inhibits the induction of type 1 interferon (IFN) elicited by RIG-I and the mitochondrial antiviral signaling protein (77). Similarly, the 4b proteins of MERS-CoV, bat coronavirus (BtCoV)-HKU4, and BtCoV-HKU5 also localize to the nucleus and inhibit type 1 IFN induction and, less efficiently, NF-κB signaling pathways (78).
SARS-CoV proteins 6 and 9b affect nucleocytoplasmic transport. Protein 6 impedes nuclear import of factors such as STAT1 (79) and antagonizes IFN signaling pathways (80). Protein 9b shuttles from the nucleus by its interaction with cellular exportin 1 (Crm1), which is essential for proper protein 9b degradation, and blocking nuclear export of protein 9b induces cell apoptosis (81).
In TGEV-infected cells, nsp1 is distributed in both the nucleus and the cytoplasm (82), which is not surprising as it can freely diffuse into the nucleus because of its small molecular weight (~9 kDa) (83). In contrast to TGEV nsp1, both MHV nsp1 and SARS-CoV nsp1 are localized exclusively in the cytoplasm of virus-infected cells (83). Due to its binding to the 40S ribosomal subunit, nsp1 inhibits cellular mRNA translation in some cases (HCoV-229E and HCoV-NL63) but not in others (TGEV) (82, 83). In addition, nsp1 inhibits IFN induction and signaling (83).
Arterivirus nsp1 and N proteins also localize in the cytoplasm and the nucleus of infected cells (84, 85). Porcine reproductive and respiratory syndrome virus (PRRSV) N protein accumulates in the nucleoli of infected cells, where it interacts with the host cell proteins fibrillarin, nucleolin, and poly(A)-binding protein (PABP), the latter of which is transported to the nucleus during infection (86, 87). PRRSV N protein also activates the NF-κB pathway and enhances its nuclear localization. The presence of N protein in the nucleus seems important for PRRSV, as removal of its nuclear localization signal significantly attenuates the virus (88). The nsp1 protein interferes with IRF3-mediated IFN activity in the nucleus and with the NF-κB-mediated pathway in the cytoplasm (89–91). The nsp1β subunit of nsp1 suppresses the JAK-STAT pathway and also interacts with protein inhibitor of activated STAT1 (PIAS1) (92, 93). Because PIAS1 is a nuclear protein with multiple functions, its interaction with nsp1 may lead to the modulation of several host cell pathways.
Coronavirus and arterivirus proteins, like those from other cytoplasmic RNA viruses (65), interact with host cell proteins, modifying their nuclear-cytoplasmic localization and thereby affecting viral replication levels and modulating innate immune responses. Thus, nuclear proteins such as hnRNP A1 and PTB accumulate in the cytoplasm of cells infected by MHV and TGEV, respectively (60, 94). These proteins bind to TRSs and to the 5′ end of the viral genome (95, 96); PTB additionally reduces coronavirus RNA accumulation (60).
Other nuclear proteins, including the p100 transcriptional coactivator, PABP, and certain members of the hnRNPs such as hnRNP Q, showed preferential binding to the 3′ end of the coronavirus genome and a positive effect on coronavirus RNA synthesis (95, 97). The contribution of these proteins to host cell interactions in TGEV infection is supported by the formation of a complex including glyceraldehyde 3-phosphate dehydrogenase (GAPDH), glutamyl-prolyl-tRNA synthetase (EPRS), hnRNP Q, and the ribosomal protein L13a, which regulates the expression of inflammatory genes (98, 99). Similarly, in infections by other RNA viruses, several nuclear proteins (La, Sam68, PTB, proteasome activator PA28γ, and nucleolin) also relocalized to the cytoplasm and were involved in virus replication (65).
RNA GENOME 5 AND 3 CIS-ACTING ELEMENTS INVOLVED IN CORONAVIRUS RNA SYNTHESIS
Similar to that of other positive-strand RNA viruses, coronavirus RNA synthesis requires the specific recognition of cis-acting RNA elements, which are mainly located in the highly structured 5′ and 3′ untranslated regions (UTRs), although they may also extend into the adjacent coding sequences (9, 100, 101). Such cis-acting RNA elements in the 5′ end of the coronavirus genome were first studied in BCoV using defective interfering RNAs. More recently, these studies have been extended to other betacoronaviruses and, to a lesser extent, to alpha- and gammacoronaviruses. These RNA elements mainly consist of stem-loop (SL) structures that present a varying degree of conservation among different coronaviruses (9, 101–103). In order to consolidate the information from various publications, we adopt a uniform nomenclature for the 5′ cis-acting RNA elements (Figure 4) based on that used by the Leibowitz, Giedroc, and Olsthoorn laboratories (102, 103). SL1 and SL2 are conserved in all coronaviruses (101, 102). SL1 adopts a bipartite structure (104), and SL2 presents a highly conserved loop sequence that adopts a YNMG- or CUYG-type tetraloop conformation (105, 106). Both SLs are required for genome replication, and they may have a specific role in sgmRNA synthesis (104–106). SL3, which contains the leader CS, is involved in discontinuous transcription as a receptor for nascent negative-strand RNA. Downstream of the leader CS is SL4, a long hairpin that is structurally conserved in all coronavirus genera (101, 102). SL4 is essential for replication of BCoV defective interfering RNA (107), and it was proposed to function as a spacer element that controls the orientation of upstream SLs driving sgmRNA synthesis (108). Finally, a higher-order RNA structure (SL5) that extends into ORF1a seems to be conserved within specific coronavirus genera (102). In alphacoronaviruses, SL5 may be further subdivided into three hairpins (SL5a, SL5b, and SL5c), which are partially conserved in betacoronaviruses (101). In gammacoronaviruses, a possible SL5 is predicted to adopt a rod-like structure (102).
Figure 4.
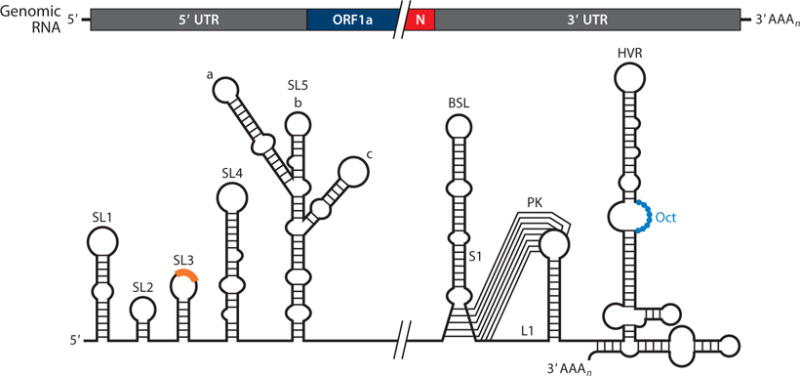
Coronavirus cis-acting RNA elements. The higher-order RNA structures indicated in the diagram are mainly based on studies done in betacoronaviruses. The core sequence within the leader transcription-regulating sequence is shown as an orange box on the top of SL3. Abbreviations: BSL, bulged stem loop; HVR, hypervariable region; L1, loop 1 of the pseudoknot; N, nucleocapsid; Oct, conserved octanucleotide; PK, pseudoknot; S1, stem 1 of the pseudoknot; SL, stem loop; UTR, untranslated region.
Initial studies using defective interfering RNAs from alpha-, beta-, and gammacoronaviruses delimitated the 3′ cis-acting RNA elements required for coronavirus RNA synthesis to the 3′ UTR plus the poly(A) tail (100). Further investigations allowed the identification and functional characterization of these 3′ cis-acting RNA elements (reviewed in 9, 101) (Figure 4). Immediately downstream of the N gene stop codon, there are two overlapping, essential RNA structures consisting of a bulged stem loop (BSL) and a hairpin-type RNA pseudoknot (PK), which are structurally and functionally conserved in all betacoronaviruses (9, 100). An intriguing property of the BSL and the PK is that, because they overlap by 5 nt, they cannot simultaneously fold up completely, which has led to the speculation that each element may adopt alternate configurations, acting as a molecular switch that operates at some stage of RNA synthesis (109). In addition, evidence for a direct interaction of the PK loop 1 (L1) with the 3′ end of the genome and with nsp8 and nsp9 has been reported (110). Based on these studies, a model for the initiation of coronavirus negative-strand RNA synthesis was proposed (Figure 5). In this model, the binding of a protein complex (including nsp8 and nsp9) to the stem formed by base-pairing of the PK L1 and the 3′ end of the genome may cause a conformational shift that leaves free the 3′ end of the genome and disrupts the lower stem of the BSL, leading to the formation of the PK. In this new conformation, the 3′ end of the genome is recognized by the RdRp and associated factors, promoting the initiation of negative-strand RNA synthesis (110). Further studies are required to confirm this model and to analyze whether or not alpha- and gammacoronaviruses, which lack either the BSL or the PK, respectively (101, 111, 112), employ a similar molecular switch mechanism.
Figure 5.
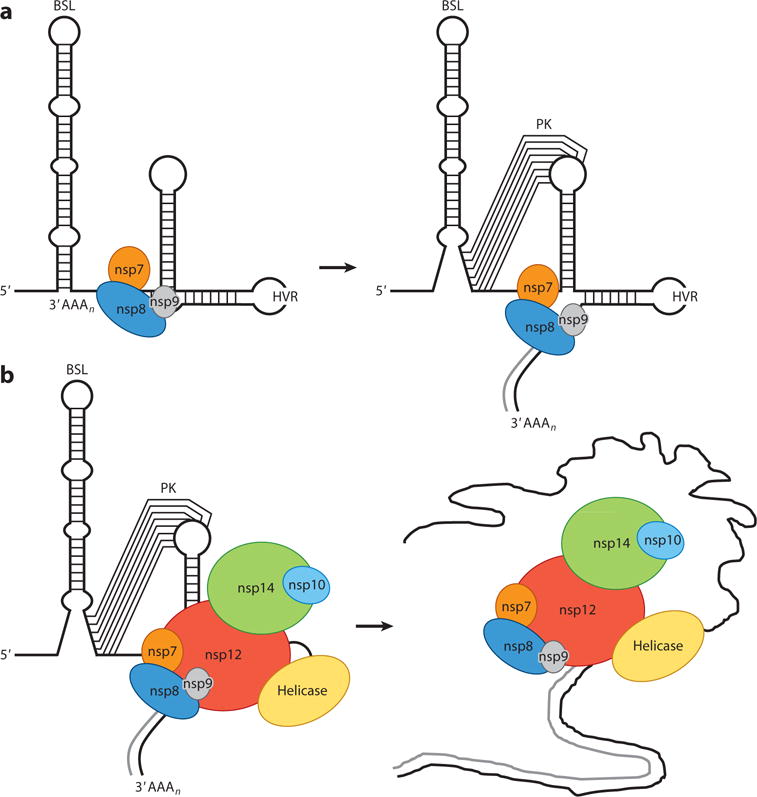
Coronavirus replication-transcription complex. (a) After binding of the nsp8, nsp7, and nsp9 complex to the genomic RNA 3′ end, the nsp8 primase activity initiates RNA synthesis de novo. This leads to a conformational change in the 3′-end RNA structure, allowing transition from a BSL to a PK folding. (b) PK formation allows binding of the RNA-dependent RNA polymerase (nsp12) complex, including helicase and nsp14-nsp10. This core replication-transcription complex includes polymerase activity (nsp12 and nsp8), processivity factors (nsp7 and nsp8), and proofreading activity (nsp14 and nsp10). Abbreviations: BSL, bulged stem loop; HVR, hypervariable region; nsp, nonstructural protein; PK, pseudoknot.
Downstream of the PK there is a hypervariable region (HVR) that is highly divergent in sequence and structure among coronaviruses but contains a universally conserved octanucleotide sequence in a single-stranded region (Figure 4) (101, 112–114). In MHV, the HVR was predicted to fold in a complex multiple stem-loop structure, which is nonessential for RNA synthesis in cell culture but affects pathogenicity in vivo (110, 115). Although the strict conservation of the octanucleotide sequence suggests an important functional role, the activity of this sequence remains to be determined. Finally, MHV and BCoV defective interfering RNA studies have provided evidence that the poly(A) tail is another cis-replication signal that requires a minimum of 5 to 10 adenylate residues to be functional, probably via interaction with PABP (116).
CELLULAR AND VIRAL PROTEINS OF THE CORONAVIRUS REPLICATION-TRANSCRIPTION COMPLEX
Most of the nsps encoded in the replicase gene, together with the N protein and an unknown number of cellular proteins, assemble into a membrane-associated viral RTC that mediates both genome replication and the synthesis of the nested set of sgmRNAs (5, 7, 9, 30, 31, 117–119). From these, the key enzymes involved in coronavirus RNA synthesis are the RdRp (nsp12), the helicase (nsp13), and the nsp7-nsp8 complex, which is a processivity factor for the RdRp (Figure 5).
The RdRp domain, located in the C-terminal region of nsp12, contains all conserved motifs of canonical RdRps, including the palm, fingers, and thumb domains (120, 121). In addition to the RdRp domain, nsp12 also contains an N-terminal domain that is essential for RdRp activity (122) and probably interacts with nsp5, nsp8, and nsp9 (123). In vitro, full-length nsp12 drives RNA synthesis in a primer-dependent manner on both homo- and heteropolymeric RNA templates (124, 125). However, the in vitro nsp12 RdRp activity is weak and nonprocessive, in contrast with the efficient replication of the RNA genome in vivo.
Coronavirus nsp8 bears a second, noncanonical RdRp activity that synthesizes short oligonucleotides (<6 nt), acting as an RNA primase that produces the primers required for nsp12-mediated RNA synthesis (37, 126). Structural studies have shown that nsp8 interacts with nsp7, forming a hexadecameric protein complex (eight molecules of each nsp) that contains a channel capable of encircling RNA due to its internal dimensions and electrostatic properties (127, 128). This complex, which is active in both de novo initiation and primer extension (126), confers processivity to the RdRp in an in vitro assay using purified proteins (129). The nsp7-nsp8-nsp12 complex formed in vitro is able to catalyze de novo synthesis of relatively long RNAs (up to 340 nt) in a processive manner. Also interacting with nsp8 is nsp9, a small protein that binds ssRNA without sequence specificity (130–132). Dimerization of nsp9 is critical for virus replication (133), and its structural and functional features strongly suggest that it could be a component of the RTC catalytic core, stabilizing viral RNAs during RNA synthesis and processing.
Coronavirus nsp13 contains a superfamily 1 helicase domain linked to an N-terminal zinc-binding domain (134) that is essential for helicase activity in vitro (135). The protein is able to unwind dsRNA and dsDNA in a 5′-to-3′ direction with the energy obtained by the hydrolysis of all NTPs and dNTPs (136–140). It was proposed that the resulting ssRNAs probably serve as templates for RNA synthesis by the RdRp. Besides NTPase and dNTPase activities, coronavirus helicases also possess RNA 5′-triphosphatase (RTPase) activity, which may be involved in viral mRNA capping (136, 138). Obviously, the nsp13 5′-to-3′ helicase activity does not fit the expected 3′-to-5′ polarity required to separate secondary structures in the RNA template during negative-strand synthesis (118). Thus, cellular helicases may bind the RTC to assist coronavirus proteins in 3′-to-5′ unwinding. SARS-CoV nsp13 has been shown to interact specifically with the cellular RNA helicase DDX5, which is involved in coronavirus RNA synthesis (141). In addition, the cellular helicase DDX1 is recruited to RTCs, and the effects of DDX1 expression knockdown indicate that it might be an essential cofactor for coronavirus RNA replication and transcription (34, 142). Interestingly, the helicase activity of nsp13 is enhanced 2-fold by nsp12 through direct protein-protein interaction (143), suggesting that interaction of these proteins in a functional RTC improves the efficiency of viral RNA synthesis.
Coronavirus Proofreading System
The replication and maintenance of the coronavirus genome, the largest known viral RNA, is a hallmark of nidoviruses. These viruses encode a unique set of RNA-modifying activities that are not present in other viral RNA genomes. One of them is the exonuclease activity of nsp14 (ExoN), related to the DEDD superfamily of exonucleases (144). In addition to the N-terminal ExoN domain, nsp14 also contains a C-terminal N7-methyltransferase (N7-MTase) domain, which is involved in viral mRNA capping (145).
Coronavirus nsp14 ExoN activity was proposed to be part of an RNA proofreading machinery during coronavirus replication (146), and accumulating data support this role (147). Phylogenetically, only long-size nidovirus genomes encode an ExoN activity, and acquisition of this activity seems crucial to allow nidovirus genome expansion. The discovery of insect nidoviruses with a 20-kb RNA genome, encoding ExoN function, strongly reinforced this idea (148). In addition, point mutations in the catalytic ExoN residues led to coronaviruses with altered replication fidelity and a mutator phenotype, as they have 15- to 20-fold higher mutation accumulation rates (149, 150). As a proofreading component, ExoN activity should contribute to the removal of misincorporated nucleotides. In fact, nsp14 ExoN activity efficiently removes a mismatched 3′-end nucleotide, mimicking an RdRp misincorporation product (151). At the same time, coronaviruses are relatively resistant to mutagens such as ribavirin and 5-fluorouracil, whereas coronaviruses with reduced ExoN activity are highly susceptible to these agents (152). These findings were the first experimental evidence supporting nsp14 ExoN activity in a coronavirus proofreading system.
The nsp14 protein is part of the RTC core complex, formed by nsp12 (RdRp) and the nsp7-nsp8 processivity factor, providing proofreading and capping activities (129). Interestingly, nsp10 is able to enhance ExoN activity up to 35-fold in vitro (151), binding nsp14 either alone or in the nsp7-nsp8-nsp12-nsp14 complex (129). The involvement of nsp10 in coronavirus RNA synthesis was first reported from the analysis of MHV mutants (153). More recently, it has been shown that nsp10 acts as a cofactor of both nsp14 ExoN and nsp16 methyltransferase (MTase) activities (154). Moreover, as nsp14 and nsp16 bind to overlapping nsp10 sites, nsp10 might act as a molecular switch, mediating interactions between RNA and proteins from both proofreading and mRNA capping machineries.
Coronavirus RNA Capping Pathway
Capping of viral RNAs by conventional or unconventional pathways (reviewed in 155) leads to 5′-end cap structures that allow efficient viral protein synthesis and, in many cases, escape from the innate immune system. Coronaviruses follow the canonical capping pathway, which consists of four sequential enzymatic reactions: (a) RTPase, encoded by the nsp13 helicase, hydrolyzing the γ-phosphate of the mRNA; (b) an as-yet-unidentified guanylyltransferase (GTase) adding GMP to the 5′-diphosphate RNA; (c) nsp14 N7-MTase methylating the guanosine, leading to a cap-0 structure that is essential for efficient translation initiation; and (d) nsp16 2′-O-methyltransferase (2′-O-MTase) carrying out further methylations, leading to cap-1 and cap-2 structures, which are required to efficiently escape the nonself RNA recognition system of the host cell (156). Interestingly, whereas nsp10 binding has no effect on nsp14 N7-MTase activity, nsp10 is required for nsp16 2-O-MTase activity. These data, in conjunction with those in the preceding section, highlight the importance of nsp10 as modulator of two different activities in the coronavirus proofreading and capping machinery.
Encapsidation of the Coronavirus Replication-Transcription Complex
It is currently accepted that, unlike that in negative-strand RNA viruses, the RTC in positive-strand RNA viruses generally is not incorporated into viral particles. However, recent studies based on proteomic, biochemical, and immunoelectron microscopy assays reported the presence of RdRp, nsp2, nsp3, and nsp8 in TGEV particles (157) and nsp2, nsp3, and nsp5 in SARS-CoV particles (158). These data suggest that the RTC might be encapsidated in coronaviruses. It is speculated that the encapsidated RTC could act as a starting replication machinery, with a round of genome amplification before translation leading to improved efficiency of virus infection. Further studies are required to investigate whether other viral and cellular components of the RTC are also encapsidated and what biological role they play in the coronavirus life cycle.
SUMMARY POINTS.
Coronaviruses express their 3′-proximal ORFs through a collection of overlapping, nested sgmRNAs generated by a mechanism of discontinuous transcription unique among RNA viruses. This process includes a template switch during the synthesis of negative-strand sgRNAs to add a copy of the leader sequence, located at the genome 5′ end.
Coronavirus transcription is regulated by multiple factors, including the extent of base-pairing between the complement of the TRS-B in the nascent negative strand and the TRS-L as well as protein-RNA and RNA-RNA interactions. Moreover, coronavirus N protein RNA chaperone activity is essential for efficient transcription.
Coronavirus RNA synthesis is associated with extensive modification of intracellular membranes, including DMVs and CMs.
The requirement of the nucleus for coronavirus replication is variable, but optimum levels of progeny are obtained only in its presence. Several coronavirus proteins involved in RNA synthesis travel to the nucleus. Conversely, many nuclear proteins are transported to the cytoplasm to facilitate coronavirus RNA synthesis.
Coronavirus cis-acting RNA elements involved in RNA synthesis are mainly located in the highly structured 5′ and 3′UTRs.
The replicase proteins nsp7, nsp8, nsp12, and nsp14 may constitute an RTC core complex.
Coronaviruses encode a proofreading machinery, unique among the RNA viruses, to ensure the maintenance of their large genome size. The ExoN activity of nsp14 is a key element of the proofreading system.
FUTURE ISSUES.
Further research is required on cis-acting elements involved in replication and transcription, and on the viral and cellular proteins that bind them.
The function and dynamics of DMVs and CMs and the precise localization of the sites of active viral RNA synthesis remain unresolved questions.
The contribution of cytoplasmic RNA-protein complexes containing viral RNAs, such as stress granules, to the regulation of coronavirus RNA expression requires further research.
Limited information is available on the temporal regulation of viral translation, replication, and transcription over the course of infection and on how switching between these processes occurs.
In vitro reconstitution of the RTC core complex will allow the study of the coronavirus proofreading mechanism, the temporal or spatial regulation of proofreading and capping activities, which share several viral components, and the role of N protein RNA chaperone activity.
Understanding the interaction of cell and viral proteins within the nucleus, and of nuclear proteins traveling to the cytoplasm to interact with viral factors, may provide novel avenues to clarify coronavirus replication.
It is still unknown whether replication and transcription are simultaneous or sequential processes.
Acknowledgments
This work was supported by grants from the Ministry of Science and Innovation of Spain (MINECO project BIO2013-42869-R), the European Community’s Seventh Framework Programme (FP7/2007–2013; project EMPERIE EC grant agreement number 223498), and the US National Institutes of Health (NIH) (projects 2P01AI060699-06A1 and CRIP-HHSN266200700010C). S.Z. received a contract from NIH project 2P01AI060699-06A1. We thank Marga González for her technical assistance.
Glossary
- SARS-CoV
severe acute respiratory syndrome coronavirus
- MERS-CoV
Middle East respiratory syndrome coronavirus
- nsp
nonstructural protein
- sgmRNA
subgenomic mRNA
- RdRp
RNA-dependent RNA polymerase
- TRS
transcription-regulating sequence
- CS
conserved core sequence
- TGEV
transmissible gastroenteritis virus
- BCoV
bovine coronavirus
- N protein
nucleocapsid protein
- RTC
replication-transcription complex
- IBV
infectious bronchitis virus
- CMs
convoluted membranes
- DMVs
double-membrane vesicles
- MHV
mouse hepatitis virus
- ExoN
exonuclease
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, et al. Coronaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic; 2012. pp. 774–96. [Google Scholar]
- 2.Coleman CM, Frieman MB. Coronaviruses: important emerging human pathogens. J Virol. 2014;88:5209–12. doi: 10.1128/JVI.03488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20:1347–50. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4:e00737–13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93:1385–409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enjuanes L, Almazan F, Sola I, Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol. 2006;60:211–30. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 8.Lai MMC, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sola I, Mateos-Gomez PA, Almazan F, Zuñiga S, Enjuanes L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 2011;8:237–48. doi: 10.4161/rna.8.2.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enjuanes L, Gorbalenya AE, de Groot RJ, Cowley JA, Ziebuhr J, Snijder EJ. The Nidovirales. In: Mahy BWJ, Van Regenmortel M, Walker P, Majumder-Russell D, editors. Encyclopedia of Virology. 3rd Oxford, UK: Elsevier; 2008. pp. 419–30. [Google Scholar]
- 11.Sawicki SG, Sawicki DL. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–19. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 12.van der Most RG, Spaan WJM. Coronavirus replication, transcription, and RNA recombination. In: Siddell SG, editor. The Coronaviridae. New York: Plenum; 1995. pp. 11–31. [Google Scholar]
- 13.Miller WA, Koev G. Synthesis of subgenomic RNAs by positive-strand RNA virus. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- 14.Pasternak AO, Spaan WJ, Snijder EJ. Nidovirus transcription: how to make sense? J Gen Virol. 2006;87:1403–21. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 15.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLOS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso S, Izeta A, Sola I, Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J Virol. 2002;76:1293–308. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuñiga S, Sola I, Alonso S, Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J Virol. 2004;78:980–94. doi: 10.1128/JVI.78.2.980-994.2004. Proposes a working model of coronavirus transcription based on the role of transcription-regulating sequence complementarity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasternak AO, van den Born E, Spaan WJ, Snijder EJ. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–28. doi: 10.1093/emboj/20.24.7220. Describes the role of transcription-regulating sequence complementarity in arterivirus transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Marle G, Dobbe JC, Gultyaev AP, Luytjes W, Spaan WJM, Snijder EJ. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. PNAS. 1999;96:12056–61. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasternak AO, van den Born E, Spaan WJM, Snijder EJ. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J Virol. 2003;77:1175–83. doi: 10.1128/JVI.77.2.1175-1183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sola I, Moreno JL, Zuñiga S, Alonso S, Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J Virol. 2005;79:2506–16. doi: 10.1128/JVI.79.4.2506-2516.2005. Reports the relevance of sequence complementarity and transcription-regulating sequence composition in coronavirus transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy PD, Simon AE. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 23.Dufour D, Mateos-Gomez PA, Enjuanes L, Gallego J, Sola I. Structure and functional relevance of a transcription-regulating sequence involved in coronavirus discontinuous RNA synthesis. J Virol. 2011;85:4963–73. doi: 10.1128/JVI.02317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang RY, Krishnan R, Brian DA. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol. 1996;70:2720–29. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno JL, Zuñiga S, Enjuanes L, Sola I. Identification of a coronavirus transcription enhancer. J Virol. 2008;82:3882–93. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mateos-Gomez PA, Zuñiga S, Palacio L, Enjuanes L, Sola I. Gene N proximal and distal RNA motifs regulate coronavirus nucleocapsid mRNA transcription. J Virol. 2011;85:8968–80. doi: 10.1128/JVI.00869-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateos-Gomez PA, Morales L, Zuñiga S, Enjuanes L, Sola I. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J Virol. 2013;87:177–86. doi: 10.1128/JVI.01782-12. Describes the longest RNA-RNA interaction known so far, leading to a high-order structure controlling coronavirus transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson BL, White KA. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol. 2014;12:493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almazan F, Galán C, Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J Virol. 2004;78:12683–88. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuñiga S, Cruz JL, Sola I, Mateos-Gomez PA, Palacio L, Enjuanes L. Coronavirus nucle-ocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84:2169–75. doi: 10.1128/JVI.02011-09. Reports the relevance of the N protein in coronavirus transcription and template switching. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brockway SM, Lu XT, Peters TR, Dermody TS, Denison MR. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J Virol. 2004;78:11551–62. doi: 10.1128/JVI.78.21.11551-11562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nedialkova DD, Gorbalenya AE, Snijder EJ. Arterivirus nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLOS Pathog. 2010;6:e1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu CH, Chen PJ, Yeh SH. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–72. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plant EP, Dinman JD. The role of programmed −1 ribosomal frameshifting in coronavirus propagation. Front Biosci. 2008;13:4873–81. doi: 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plant EP, Sims AC, Baric RS, Dinman JD, Taylor DR. Altering SARS coronavirus frameshift efficiency affects genomic and subgenomic RNA production. Viruses. 2013;5:279–94. doi: 10.3390/v5010279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–42. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLOS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulasli M, Verheije MH, de Haan CA, Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12:844–61. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verheije MH, Raaben M, Mari M, te Lintelo EG, Reggiori F, et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLOS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stokes HL, Baliji S, Hui CG, Sawicki SG, Baker SC, Siddell SG. A new cistron in the murine hepatitis virus replicase gene. J Virol. 2010;84:10148–58. doi: 10.1128/JVI.00901-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagemeijer MC, Vonk AM, Monastyrska I, Rottier PJ, de Haan CA. Visualizing coronavirus RNA synthesis in time by using click chemistry. J Virol. 2012;86:5808–16. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–56. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 45.Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:e00524–13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oostra M, Hagemeijer MC, van Gent M, Bekker CP, te Lintelo EG, et al. Topology and membrane anchoring of the coronavirus replication complex: Not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol. 2008;82:12392–405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gadlage MJ, Sparks JS, Beachboard DC, Cox RG, Doyle JD, et al. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J Virol. 2010;84:280–90. doi: 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagemeijer MC, Monastyrska I, Griffith J, van der Sluijs P, Voortman J, et al. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–59:125–35. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maier HJ, Hawes PC, Cottam EM, Mantell J, Verkade P, et al. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4:e00801–13. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maier HJ, Hawes PC, Keep SM, Britton P. Spherules and IBV. Bioengineered. 2014;5:288–92. doi: 10.4161/bioe.29323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagemeijer MC, Rottier PJ, de Haan CA. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4:3245–69. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neuman BW, Angelini MM, Buchmeier MJ. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22:642–47. doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–83. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reineke LC, Lloyd RE. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–67. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE., Jr Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol. 2010;84:12274–84. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 2007;9:2218–29. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sola I, Galán C, Mateos-Gomez PA, Palacio L, Zuñiga S, et al. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J Virol. 2011;85:5136–49. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emmott E, Munday D, Bickerton E, Britton P, Rodgers MA, et al. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J Virol. 2013;87:9486–500. doi: 10.1128/JVI.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Courtney SC, Scherbik SV, Stockman BM, Brinton MA. West Nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J Virol. 2012;86:3647–57. doi: 10.1128/JVI.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Onomoto K, Yoneyama M, Fung G, Kato H, Fujita T. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014;35:420–28. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emmott E, Rodgers MA, Macdonald A, McCrory S, Ajuh P, Hiscox JA. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol Cell Proteomics. 2010;9:1920–36. doi: 10.1074/mcp.M900345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gustin KE, Sarnow P. Positive-strand RNA viruses and the nucleus. In: Hiscox JA, editor. Viruses and the Nucleus. Chichester, UK: Wiley; 2006. pp. 161–84. [Google Scholar]
- 66.Wilhelmsen KC, Leibowitz JL, Bond CW, Robb JC. The replication of murine coronaviruses in enucleated cells. Virology. 1981;110:225–30. doi: 10.1016/0042-6822(81)90027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emmett SR, Dove B, Mahoney L, Wurm T, Hiscox JA. The cell cycle and virus infection. Methods Mol Biol. 2005;296:197–218. doi: 10.1385/1-59259-857-9:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Surjit M, Liu B, Chow VT, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–81. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J Virol. 2001;75:9345–56. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cawood R, Harrison SM, Dove BK, Reed ML, Hiscox JA. Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle. 2007;6:863–67. doi: 10.4161/cc.6.7.4032. [DOI] [PubMed] [Google Scholar]
- 71.Ding L, Huang Y, Du Q, Dong F, Zhao X, et al. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem Biophys Res Commun. 2014;445:497–503. doi: 10.1016/j.bbrc.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu X, Zhang H, Zhang Q, Huang Y, Dong J, et al. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet Microbiol. 2013;164:212–21. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo H, Chen Q, Chen J, Chen K, Shen X, Jiang H. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 2005;579:2623–28. doi: 10.1016/j.febslet.2005.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen S, Wen ZL, Liu DX. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311:16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khan S, Fielding BC, Tan TH, Chou CF, Shen S, et al. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 2006;122:20–27. doi: 10.1016/j.virusres.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan X, Yao Z, Shan Y, Chen B, Yang Z, et al. Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res. 2005;114:70–79. doi: 10.1016/j.virusres.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Freundt EC, Yu L, Park E, Lenardo MJ, Xu XN. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83:6631–40. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol. 2014;95:874–82. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hussain S, Gallagher T. SARS-coronavirus protein 6 conformations required to impede protein import into the nucleus. Virus Res. 2010;153:299–304. doi: 10.1016/j.virusres.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol. 2007;81:9812–24. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma K, Akerstrom S, Sharma AK, Chow VT, Teow S, et al. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLOS ONE. 2011;6:e19436. doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol. 2011;85:638–43. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Zheng Z, Zhou P, Zhang B, Shi Z, et al. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J Gen Virol. 2010;91:2947–58. doi: 10.1099/vir.0.025205-0. [DOI] [PubMed] [Google Scholar]
- 85.Tijms MA, van der Meer Y, Snijder EJ. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J Gen Virol. 2002;83:795–800. doi: 10.1099/0022-1317-83-4-795. [DOI] [PubMed] [Google Scholar]
- 86.Burgess HM, Gray NK. An integrated model for the nucleo-cytoplasmic transport of cytoplasmic poly(A)-binding proteins. Commun Integr Biol. 2012;5:243–47. doi: 10.4161/cib.19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salaun C, MacDonald AI, Larralde O, Howard L, Lochtie K, et al. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J Virol. 2010;84:8539–48. doi: 10.1128/JVI.00668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee C, Hodgins D, Calvert JG, Welch SK, Jolie R, Yoo D. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology. 2006;346:238–50. doi: 10.1016/j.virol.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song C, Krell P, Yoo D. Nonstructural protein 1α subunit-based inhibition of NF-κB activation and suppression of interferon-β production by porcine reproductive and respiratory syndrome virus. Virology. 2010;407:268–80. doi: 10.1016/j.virol.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 90.Han M, Du Y, Song C, Yoo D. Degradation of CREB-binding protein and modulation of type I interferon induction by the zinc finger motif of the porcine reproductive and respiratory syndrome virus nsp1α subunit. Virus Res. 2013;172:54–65. doi: 10.1016/j.virusres.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 91.Kim O, Sun Y, Lai FW, Song C, Yoo D. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402:315–26. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patel D, Nan Y, Shen M, Ritthipichai K, Zhu X, Zhang YJ. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol. 2010;84:11045–55. doi: 10.1128/JVI.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen Z, Lawson S, Sun Z, Zhou X, Guan X, et al. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li HP, Huang P, Park S, Lai MMC. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J Virol. 1999;73:772–77. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Galán C, Sola I, Nogales A, Thomas B, Akoulitchev A, et al. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–14. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi KS, Huang P, Lai MM. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology. 2002;303:58–68. doi: 10.1006/viro.2002.1675. [DOI] [PubMed] [Google Scholar]
- 97.Choi KS, Mizutani A, Lai MM. SYNCRIP, a member of the heterogeneous nuclear ribonucleo-protein family, is involved in mouse hepatitis virus RNA synthesis. J Virol. 2004;78:13153–62. doi: 10.1128/JVI.78.23.13153-13162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29:679–90. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem Sci. 2009;34:324–31. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Masters PS. Genomic cis-acting elements in coronavirus RNA replication. In: Thiel V, editor. Coronaviruses: Molecular and Cellular Biology. Norfolk, UK: Caister Academic; 2007. pp. 65–80. [Google Scholar]
- 101.Madhugiri R, Fricke M, Marz M, Ziebuhr J. RNA structure analysis of alphacoronavirus terminal genome regions. Virus Res. 2014;194:76–89. doi: 10.1016/j.virusres.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen SC, Olsthoorn RC. Group-specific structural features of the 5′ -proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang H, Feng M, Schroeder ME, Giedroc DP, Leibowitz JL. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J Virol. 2006;80:10600–14. doi: 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li L, Kang H, Liu P, Makkinje N, Williamson ST, et al. Structural lability in stem-loop 1 drives a 5′UTR–3′UTR interaction in coronavirus replication. J Mol Biol. 2008;377:790–803. doi: 10.1016/j.jmb.2008.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu P, Li L, Keane SC, Yang D, Leibowitz JL, Giedroc DP. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J Virol. 2009;83:12084–93. doi: 10.1128/JVI.00915-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee CW, Li L, Giedroc DP. The solution structure of coronaviral stem-loop 2 (SL2) reveals a canonical CUYG tetraloop fold. FEBS Lett. 2011;585:1049–53. doi: 10.1016/j.febslet.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raman S, Bouma P, Williams GD, Brian DA. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J Virol. 2003;77:6720–30. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang D, Liu P, Giedroc DP, Leibowitz J. Mouse hepatitis virus stem-loop 4 functions as a spacer element required to drive subgenomic RNA synthesis. J Virol. 2011;85:9199–209. doi: 10.1128/JVI.05092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goebel SJ, Hsue B, Dombrowski TF, Masters PS. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J Virol. 2004;78:669–82. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zust R, Miller TB, Goebel SJ, Thiel V, Masters PS. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J Virol. 2008;82:1214–28. doi: 10.1128/JVI.01690-07. Proposes a model for RNA synthesis initiation involving RNA structures and replicase components. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Williams GD, Chang RY, Brian DA. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J Virol. 1999;73:8349–55. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dalton K, Casais R, Shaw K, Stirrups K, Evans S, et al. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J Virol. 2001;75:125–33. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Q, Johnson RF, Leibowitz JL. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J Virol. 2001;75:12105–13. doi: 10.1128/JVI.75.24.12105-12113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Williams AK, Wang L, Sneed LW, Collisson EW. Analysis of a hypervariable region in the 3′ non-coding end of the infectious bronchitis virus genome. Virus Res. 1993;28:19–27. doi: 10.1016/0168-1702(93)90086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goebel SJ, Miller TB, Bennett CJ, Bernard KA, Masters PS. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J Virol. 2007;81:1274–87. doi: 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Spagnolo JF, Hogue BG. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol. 2000;74:5053–65. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J Virol. 2005;79:6620–30. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ziebuhr J. The coronavirus replicase. In: Enjuanes L, editor. Coronavirus Replication and Reverse Genetics. Berlin: Springer-Verlag; 2005. pp. 57–94. [Google Scholar]
- 119.Ulferts R, Imbert I, Canard B, Ziebuhr J. Expression and functions of SARS coronavirus replicative proteins. In: Lal SK, editor. Molecular Biology of the SARS-Coronavirus. Berlin: Springer-Verlag; 2010. pp. 75–98. [Google Scholar]
- 120.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu X, Liu Y, Weiss S, Arnold E, Sarafianos SG, Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 2003;31:7117–30. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng A, Zhang W, Xie Y, Jiang W, Arnold E, et al. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335:165–76. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brockway SM, Clay CT, Lu XT, Denison MR. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J Virol. 2003;77:10515–27. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–14. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ahn DG, Choi JK, Taylor DR, Oh JW. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch Virol. 2012;157:2095–104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–47. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhai Y, Sun F, Li X, Pang H, Xu X, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–86. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Imbert I, Snijder EJ, Dimitrova M, Guillemot JC, Lecine P, Canard B. The SARS-coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–48. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. PNAS. 2014;111:E3900–9. doi: 10.1073/pnas.1323705111. Describes the coronavirus core RNA synthesis complex integrating polymerase, proofreading, and capping activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. Comprehensive bioinformatic predictions on the presence of unique enzymatic RNA-modifying activities in coronavirus genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. PNAS. 2004;101:3792–96. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sutton G, Fry E, Carter L, Sainsbury S, Walter T, et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–53. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol. 2009;83:3007–18. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–61. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Seybert A, Posthuma CC, van Dinten LC, Snijder EJ, Gorbalenya AE, Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78:5619–32. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Seybert A, Hegyi A, Siddell SG, Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–68. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tanner JA, Watt RM, Chai YB, Lu LY, Lin MC, et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J Biol Chem. 2003;278:39578–82. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ivanov KA, Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J Virol. 2004;78:7833–38. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee NR, Kwon HM, Park K, Oh S, Jeong YJ, Kim DE. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsp13. Nucleic Acids Res. 2010;38:7626–36. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen JY, Chen WN, Poon KM, Zheng BJ, Lin X, et al. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch Virol. 2009;154:507–12. doi: 10.1007/s00705-009-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol. 2010;84:8571–83. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Adedeji AO, Marchand B, te Velthuis AJ, Snijder EJ, Weiss S, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLOS ONE. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. PNAS. 2006;103:5108–13. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen Y, Cai H, Pan J, Xiang N, Tien P, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. PNAS. 2009;106:3484–89. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–79. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Smith EC, Sexton NR, Denison MR. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol. 2014;1:111–32. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- 148.Smith EC, Denison MR. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr Opin Virol. 2012;2:519–24. doi: 10.1016/j.coviro.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, et al. Infidelity of SARS-CoV nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLOS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. Describes the relevance of coronavirus ExoN activity in replication fidelity and proofreading mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–44. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. PNAS. 2012;109:9372–77. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Smith EC, Blanc H, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLOS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Donaldson EF, Sims AC, Graham RL, Denison MR, Baric RS. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J Virol. 2007;81:6356–68. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, et al. Coronavirus nsp10, a critical co-factor for activation of multiple replicative enzymes. J Biol Chem. 2014;289:25783–96. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLOS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nogáles A, Marquez-Jurado S, Galán C, Enjuanes L, Almazan F. Transmissible gastroenteritis coronavirus RNA-dependent RNA polymerase and nonstructural proteins 2, 3, and 8 are incorporated into viral particles. J Virol. 2012;86:1261–66. doi: 10.1128/JVI.06428-11. Reports the incorporation of RTC components into coronavirus particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol. 2008;82:5279–94. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sims AC, Tilton SC, Menachery VD, Gralinski LE, Schafer A, et al. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol. 2013;87:3885–902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Moshynskyy I, Viswanathan S, Vasilenko N, Lobanov V, Petric M, et al. Intracellular localization of the SARS coronavirus protein 9b: evidence of active export from the nucleus. Virus Res. 2007;127:116–21. doi: 10.1016/j.virusres.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rowland RR, Kervin R, Kuckleburg C, Sperlich A, Benfield DA. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
RELATED RESOURCES
- CDC (Cent. Dis. Control Prev.) Coronavirus. Atlanta, GA: CDC; 2015. http://www.cdc.gov/coronavirus/ [Google Scholar]
- Enjuanes L, Sola I, Zuñiga S, Moreno JL. Coronavirus RNA synthesis: transcription. In: Thiel V, editor. Coronaviruses: Molecular and Cellular Biology. Norfolk, UK: Caister Academic; 2007. pp. 81–107. [Google Scholar]
- Graham R, Donalson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–48. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski LE, Baric RS. Molecular pathology of emerging coronavirus infections. J Pathol. 2015;235:185–95. doi: 10.1002/path.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C, Goeman JJ, del Carmen Parquet M, Thi Nga P, Snijder EJ, et al. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLOS Pathog. 2013;9:e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Baric RS. Bugs in the system. Immunol Rev. 2013;255:256–74. doi: 10.1111/imr.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–50. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organ.) global Alert and Response: Coronavirus Infections. Geneva: WHO; 2015. http://www.who.int/csr/disease/coronavirus_infections/en/ [Google Scholar]


