Abstract
Spinal muscular atrophy (SMA) is one of the major genetic disorders associated with infant mortality. More than 90% cases of SMA result from deletions or mutations of Survival Motor Neuron 1 (SMN1) gene. SMN2, a nearly identical copy of SMN1, does not compensate for the loss of SMN1 due to predominant skipping of exon 7. However, correction of SMN2 exon 7 splicing has proven to confer therapeutic benefits in SMA patients. The only approved drug for SMA is an antisense oligonucleotide (Spinraza™/Nusinersen), which corrects SMN2 exon 7 splicing by blocking intronic splicing silencer N1 (ISS-N1) located immediately downstream of exon 7. ISS-N1 is a complex regulatory element encompassing overlapping negative motifs and sequestering a cryptic splice site. More than 40 protein factors have been implicated in the regulation of SMN exon 7 splicing. There is evidence to support that multiple exons of SMN are alternatively spliced during oxidative stress, which is associated with a growing number of pathological conditions. Here, we provide the most up to date account of the mechanism of splicing regulation of the SMN genes.
Keywords: SMN, SMA, Splicing, ISS-N1, ISS-N2, Cryptic splice site, U1 snRNA snRNA
1. Introduction
Pre-mRNA splicing is an essential process in eukaryotic cells during which noncoding (intronic) sequences are removed and coding (exonic) sequences are joined together to generate mRNA. The complex reaction of splicing is catalyzed by a spliceosome, a macromolecular machinery [1]. The most critical step of a splicing reaction is the accurate determination of the 5′ and 3′ splice sites (5′ss and 3′ss) that mark the beginning and end of an intron, respectively [2]. All intron-containing human genes have potential to be alternatively spliced, generating multiple mRNA isoforms from a single gene [3]. Decision to include or exclude an exon during pre-mRNA splicing is dictated by a combinatorial control of cis -elements and transacting factors. The same cis -element when presented in a different context may have different effects on splicing [4, 5]. Hence, the relative impact of a cis -element cannot be accurately predicted, it [the impact] requires experimental validation. Interpreting the consequences of a splicing-associated mutation remains a puzzle, since a single mutation can cause at least one of the following changes: loss of a positive element, gain of a negative element, change of a structural context, and nonsense-mediated decay (NMD) due to creation of an in-frame premature termination codon (PTC) [6, 7]. Rules of splicing are quite flexible and are heavily influenced by the relative abundance of various splicing factors in different tissues [8]. Further, splicing is coupled to other events including transcription, the 5′ capping, and the 3′ polyadenylation [6, 9]. Therefore, deciphering the mechanism by which a given exon is alternatively spliced remains a daunting task. A growing number of disorders are linked to aberrant splicing [10, 11]. Each case of aberrant splicing calls for an in-depth analysis of the context-specific rules so that strategies to manipulate splicing could be devised in a gene-specific manner.
Humans carry two near identical copies of the Survival Motor Neuron gene: SMN1 and SMN2 [12]. Both SMN genes code for SMN, a multifunction protein essential for the survival of all animal cells. The ability of SMN to interact with nucleic acids and proteins allows it to participate in various cellular processes, including but not limited to transcription, splicing, translation, macromolecular trafficking, and signal transduction [13]. The critical difference between SMN1 and SMN2 is the splicing of exon 7. Unlike SMN1 exon 7, SMN2 exon 7 is predominantly skipped in most tissues, except in testis [14]. The exon 7-skipped transcript generated by SMN2 codes for SMNΔ 7, a partially functional and unstable protein [15–17]. Loss of SMN1 creates SMN deficit, leading to spinal muscular atrophy (SMA), a major genetic disease of children and infants [18, 19]. Aberrant expression and/or localization of SMN have been associated with several other diseases, including amyotrophic lateral sclerosis (ALS), metabolic disorders, male infertility, and stress-associated disorders [14, 20–22]. Correction of SMN2 exon 7 splicing has proven to confer therapeutic benefits in mouse models of SMA [23, 24]. The first approved drug for SMA, Nusinersen (Spinraza™), is an antisense oligonucleotide (ASO) that promotes inclusion of SMN2 exon 7 by sequestering an inhibitory cis -element called Intronic Splicing Silencer N1 or ISS-N1 [25, 26]. In this review, we describe studies that culminated in the discovery of ISS-N1 and analyze how the characterization of ISS-N1 paved the way for a better understanding of pre-mRNA splicing in the context of a human disease. We summarize the role of various cis-elements and transacting factors that regulate SMN exon 7 splicing. We also discuss how lessons learnt from the SMN genes will help find effective therapies for genetic diseases associated with aberrant splicing.
2. Organization of Human SMN Genes
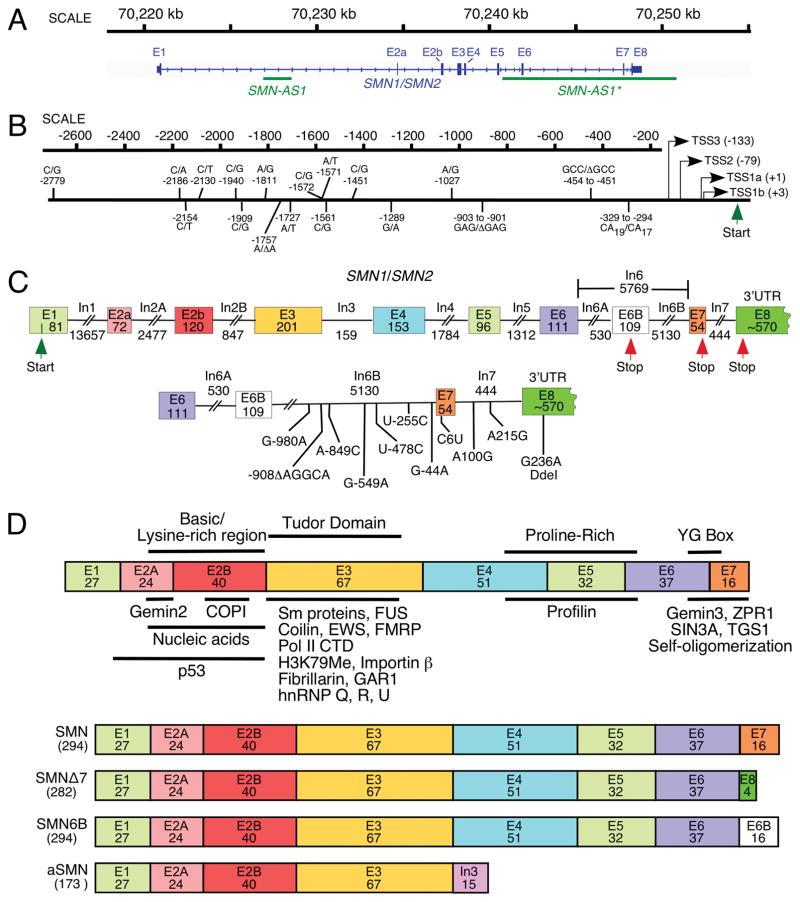
The presence of two SMN genes in humans is attributed to the intrachromosomal duplication of ~500 kb segment at the 5q13.3 locus on chromosome 5 (Fig. 1A; [12, 35, 36]). Despite conservation of the coding region of SMN between human and rodents, there are substantial differences in the promoter, intronic, and the untranslated regions (UTRs). The abundance of Alu elements in human SMN genes suggests a distinct regulation of transcription and splicing of SMN in primates. Both SMN genes are ~34 kb long including ~6 kb long promoter sequence. Several mutations within the promoter region distinguish SMN1 from SMN2, suggesting that transcription of these genes might be differentially regulated under certain conditions, such as stress (Fig. 1B). Each SMN gene is comprised of 10 exons, that is, 1, 2A, 2B, 3, 4, 5, 6, 6B, 7 and 8 (Fig. 1C). About 2/3rd of exon 1 serves as the 5′UTR, whereas the remaining 1/3rd serves as the coding sequence. Exon 8 is the longest exon that encodes the 3′UTR. SMN2 intronic sequences flanking exon 7 contain several substitutions and a 5-nt deletion (Fig. 1C). A C-to-T substitution at the sixth position (C6U) of exon 7, a G-to-A substitution at the -44th position (G-44A) of intron 6, and an A-to-G substitution at the 100th position (A100G) of intron 7 are associated with skipping of SMN2 exon 7 [37–40]. Recently discovered exon 6B is generated by exonization of an Alu element within intron 6 [33]. Another alternative transcript is generated by intron 3 retention. It codes for a short protein called axonal SMN or aSMN [34]. Considering intron 3 is conserved between human and mouse, expression of aSMN has been detected in mice as well. SMN contains several functional domains and interacts with various proteins. All isoforms of SMN possess identical N-terminus that is involved in interactions with both proteins and nucleic acids (Fig. 1D; [13]). Recent reports reveal that two antisense transcripts, which function as long noncoding RNAs (lncRNAs), are generated from SMN locus. One of these lncRNAs termed SMN-AS1 is ~1.6 kb long; it starts and finishes within intron 1 (Fig. 1A; [27]). Other one termed SMN-AS1* is ~10 kb long; it starts within intergenic region downstream of exon 8 and extends till intron 5 (Fig. 1A; [28]). These lncRNAs are specific to humans and their expressions appear to downregulate SMN levels through transcriptional control. The significance of fine-tuning of SMN levels within cells is underscored by a recent study that showed the pathogenesis of osteoarthritis caused by aberrantly high expression of SMN [41]. Factors that regulate SMN transcription and splicing modulate SMN levels in a cell-specific manner. Testis happens to be one of the tissues with a very high SMN demand. This demand is met by an entirely different set of rules that govern transcription and splicing of the SMN genes in testis. Here, we describe a critical role of the context-specific cis-elements in SMN splicing and outline the emerging rules that are likely to be applicable in most cell types.
Fig. 1.
Organization of SMN gene. (A) A view of human SMN1/SMN2 gene(s) located on chromosome 5. Exons and introns are shown as boxes and lines, respectively. Loci of antisense RNAs, SMN-AS1 [27], and SMN-AS1* [28] are marked with bars. (B) Diagrammatic representation of human SMN promoter region. Multiple transcription start sites (TSS) identified so far are indicated using arrows. Numbers in brackets correspond to their position relative to TSS1a (+1). TSS1a and TSS2 were identified in [29] as transcription start sites preferentially used in adult and fetal tissues, respectively. TSS1b was mapped in Echaniz-Laguna et al. [30], and TSS3 was identified in Monani et al., [31]. Nucleotide differences between the SMN1 and SMN2 promoters are indicated based on Monani et al., [31]; [29, 32]), where nucleotide positions were calculated from TSS1a. Translation initiation site is marked as Start. (C) Diagrammatic representation of the SMN1/SMN2 pre-mRNA. Exons and introns are shown as boxes and lines, respectively. Sizes of exons and introns are indicated in nucleotides (nts). The translation initiation and termination sites are marked as Start and Stop, respectively. Exon 8 is mostly used as the 3′ untranslated region (UTR). The bottom panel indicates nucleotides differences between SMN1 and SMN2 in the region located downstream of exon 6B. The last position of intron 6B is designated as −1. For exons 7 and 8, as well as intron 7, counting starts with the first position of the respective exon or intron. (D). Diagrammatic representation of SMN protein isoforms. Protein regions encoded by each exon are shown as colored boxes with the number of amino acids given. In the top panel, protein domains are indicted above, while SMN interacting partners are shown below the diagrammatic representation of the full-length SMN. For further details see Singh et al. [13]. The bottom panel shows the known SMN isoforms as compared to the full-length SMN protein. These isoforms are generated either due to exon 7 skipping or exonization of a region within intron 6 [33] or intron 3 retention [34]. The size of each isoform (in amino acids) is given in brackets. Abbreviations are given in Table 2
3. Regulation of SMN Exon 7 Splicing
Our understanding of SMN exon 7 skipping is continuing to evolve as more and more regulatory elements are being discovered within this relatively short exon and its flanking intronic sequences. Early studies established that the C6U substitution is the primary cause of SMN2 exon 7 skipping [38, 39]. It was also shown that the 3′ss of SMN2 exon 7 is weakened by the C6U substitution; but the usage of this 3′ss was enhanced when the downstream 3′ss of exon 8 was blocked [42]. Based on bioinformatics predictions and in vitro studies, it was proposed that C6U abrogates an enhancer associated with SRSF1 (ASF/SF2), a member of the highly conserved family of serine/arginine (SR)-rich proteins (Fig. 2A; [46]). However, this simple “SRSF1 abrogation” hypothesis did not hold true in a subsequent cell-based study, where the depletion of SRSF1 did not cause the expected enhancement of SMN1 exon 7 skipping (Fig. 2A; [47]). A more recent study suggests a surprising dual role of SRSF1 in regulation of SMN2 exon 7 splicing, as both overexpression and depletion of SRSF1 caused enhanced skipping of SMN2 exon 7 [48]. An alternative hypothesis that C6U creates a silencer associated with hnRNP A1/A2 was proposed to explain the skipping of SMN2 exon 7 [47]. Supporting this hypothesis, depletion of hnRNP A1/A2 promoted SMN2 exon 7 inclusion [47, 49, 50]. Subsequent studies implicated the role of multiple hnRNP A1/A2 sites in the regulation of SMN exon 7 splicing [37, 51–53]. These findings brought additional complexity to the interpretations of the hnRNP A1/A2 depletion experiments, since the observed effect could be attributed to abrogation of hnRNP A1/A2 binding to any/all of these sites within SMN2 pre-mRNA. Interestingly, hnRNP A1 knockout mice show muscle-specific developmental defects [54]. Hence, depletion of hnRNP A1 cannot be exploited for a potential therapy of SMA.
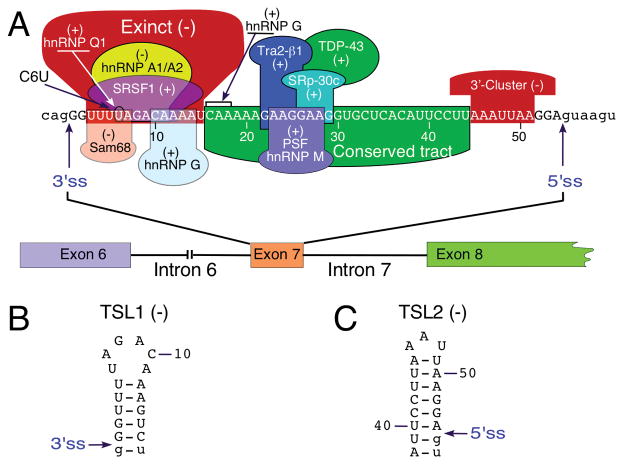
Fig. 2.
Exon 7 splicing regulation. (A) Diagrammatic representation of cis-elements and transacting factors that modulate SMN exon 7 splicing. Positive and negative elements are indicated by (+) and (−), respectively (For further details see [43]). Numbering of nucleotides starts with the first position of exon 7. Exonic and intronic sequences are shown in upper- and lower-case letters, respectively. The 3′ and 5′ss are indicated by arrows. (B) Terminal stem-loop structure, TSL1, formed at the beginning of SMN2 exon 7 as determined by enzymatic structure probing [44, 45]. Both TSL1 and TSL2 are marked by (−) because they contribute toward exon 7 skipping. Numbering of nucleotides starts with the first position of the exon. Exonic and intronic sequences are shown in upper- and lower-case letters, respectively. The 3′ ss is indicated by an arrow. (C) Terminal stem-loop structure, TSL2, formed at the end of SMN2 exon 7 as determined by enzymatic structure probing [44, 45]. (−) indicates that TSL1 contributes to exon 7 skipping. Numbering of nucleotides starts with the first position of the exon. Exonic and intronic sequences are shown in upper- and lower-case letters, respectively. The 5′ss is indicated by an arrow. Abbreviations are given in Table 2
The hnRNP A1/A2 model has been subsequently modified to include Sam68 as an additional factor associated with the inhibitory effect of C6U (Fig. 2A; [55]). Consistent with the role of hnRNP A1 and Sam68 in SMN2 exon 7 splicing, low extracellular pH that increased the nuclear concentrations of hnRNP A1 and Sam68 was found to enhance SMN2 exon 7 skipping [56]. Another mechanism by which C6U might affect SMN2 exon 7 splicing is through creation of an extended inhibitory context (Exinct) that consists of overlapping negative motifs [57]. Interestingly, C6U also strengthens a predicted terminal stem-loop structure, TSL1 (Fig. 2B). Supporting the distinct inhibitory role of TSL1, mutations that disrupted TSL1 without abrogating C6U-associated hnRNP A1/A2 motif promoted SMN2 exon 7 inclusion [57]. It should be noted that the proposed hypotheses associated with the inhibitory effect of C6U are not mutually exclusive. Recent years have witnessed a shift in the debate as critical roles of several other negative elements located away from the C6U site have been discovered. As per the exon definition model, positive factors bridge cross-exon interactions before splicing takes place [58]. An early study implicated SFRS10 (Tra2-beta1) as one of the factors that interacts directly with a GA-rich sequence located in the middle of exon 7 (Fig. 2A; [59]). Several other proteins, including TDP43, SRSF9 (SRp30c), PSF and hnRNP M, were subsequently shown to stimulate exon 7 inclusion through a direct or indirect interaction with exon 7 (Fig. 2A; [60–64]). Surprisingly, a follow-up study in a mouse model of SMA established that SFRS10 is dispensable for SMN exon 7 splicing [65]. This finding underscored the complexity of splicing regulation when the loss of a positive factor could be tolerated due to the presence of other factors with redundant/overlapping functions. Thus far, studies suggest that skipping of SMN2 exon 7 is driven largely by the occurrence of negative interactions. The list of factors that regulate SMN2 exon 7 is large and continues to grow (Table 1). However, interaction sites for most of the identified transacting factors remain unknown. There have been very limited attempts to correlate the effect of the naturally occurring mutation within a given factor and splicing of SMN exon 7.
Table 1.
Factors tested for an effect on SMN2 exon 7 splicing
| Effect of overexpression | Effect of depletion | ||||||
|---|---|---|---|---|---|---|---|
| Factor (Gene) | Effect on exon 7 splicing | Binding Location | SMN2m | SMN2g | SMN2m | SMN2g | References |
| ASF/SF2 (SRSF1) | Positive Neutral |
Exon 7 | No | Yes | No | Yes No |
[46, 47, 66,37, 53, 67, 68, 48] |
| SC35 (SRSF2) | Negative Neutral |
– | No | Yes | ND | Yes | [64, 47, 48] |
| SRp20 (SRSF3) | Negative Neutral |
– | No | Yes | ND | Yes | [64, 48] |
| SRp75 (SRSF4) | Negative | – | ND | No | ND | Yes | [48] |
| SRp40 (SRSF5) | Negative Neutral |
– | No | Yes | ND | Yes | [64, 48] |
| SRp55 (SRSF6) | Negative Neutral |
– | No | No | ND | Yes | [64, 48] |
| 9G8 (SRSF7) | Negative Neutral |
– | No | Yes | Yes | Yes | [47, 64, 69, 48] |
| SRp30c (SRSF9) | Positive Neutral |
– | Yes | No | ND | No | [64, 67, 48] |
| SRp38 (SRSF10) | Neutral | – | ND | No | ND | No | [48] |
| SRSF11 (SRSF11) | Negative | – | ND | Yes | ND | Yes | [48] |
| Tra2-β1 (TRA2B) | Positive | Exon 7 | Yes | Yes | No | ND | [59, 64, 60, 70] |
| ZIS/ZNF265 (ZRANB2) | Negative | – | Yes | ND | ND | ND | [71] |
| hnRNP A1 (HNRNPA1) | Negative | Exon 7, Intron 7, 3′ss | ND | Yes | Yes | Yes | [47, 66, 37, 53, 60, 67, 52, 51, 69, 50] |
| hnRNPA2/B1 (HNRNPA2/B1) | Negative | – | ND | ND | Yes | Yes | [47, 37, 67, 52, 69, 50, 48] |
| hnRNPC (HNRNPC) | Positive Negative Neutral |
I6–E7 junction | ND | ND | No; Yes | Yes | [68, 69, 48] |
| hnRNP D (HNRNPD) | Neutral | – | ND | ND | ND | No | [48] |
| hnRNP F (HNRNPF) | Neutral | – | ND | ND | No | No | [69, 48] |
| hnRNP G (RBMX) | Positive | Exon 7 | Yes | ND | ND | ND | [63, 72, 70, 60, 73] |
| hnRNP H (HNRNPH1) | Neutral | – | ND | ND | Yes; No | No | [51, 69, 48] |
| hnRNP K (HNRNPK) | Neutral | – | ND | ND | No | ND | [69] |
| hnRNP L (HNRNPL) | Neutral | – | ND | ND | No | ND | [69] |
| hnRNP M (HNRNPM) | Positive | – | Yes | ND | Yes | Yes | [67, 69, 62] |
| RALY (RALY) | Neutral | – | ND | ND | No | ND | [69] |
| hnRNP Q (SYNCRIP) | Positive | Exon 7 | Yes | Yes | Yes | ND | [67] |
| hnRNP U (HNRNPU) | Negative | – | ND | Yes | ND | Yes | [69, 48] |
| CHERP (CHERP) | Negative | – | ND | ND | Yes | ND | [69] |
| HuR (ELAVL1) | Negative | 3′-UTR | ND | ND | ND | Yes | [48] |
| PSF (SFPQ) | Positive | Exon 7 | Yes | Yes | ND | Yes | [67, 62] |
| PUF60 (PUF60) | Negative | 3′ss | ND | ND | Yes | Yes | [69, 74] |
| TDP-43 (TARDBP) | Positive | – | Yes | ND | No | ND | [60] |
| TIA1 (TIA1) | Positive | Intron 7 | Yes | Yes | Yes | Yes | [75] |
| RBM10 (RBM10) | Negative | – | ND | ND | Yes | Yes | [69, 76] |
| Sam68 (KHDRBS1) | Negative | Exon 7 | Yes | ND | Yes | ND | [55] |
| SF1 (SF1) | Negative | Branch Point | ND | ND | Yes | ND | [69] |
| SmD3 (SNRPD3) | Positive | – | ND | ND | ND | Yes | [77] |
| SON (SON) | Negative | – | ND | ND | Yes | ND | [69] |
| U1–70K (SNRNP70) | Positive | – | ND | ND | ND | Yes | [77] |
| U2AF35 (U2AF1) | Negative | – | ND | ND | Yes | Yes | [69] |
| U2AF65 (U2AF2) | Negative | 3′ss | ND | ND | Yes | Yes | [67, 69, 74] |
| U2B″ (U2B″) | Positive | – | ND | ND | ND | Yes | [77] |
Abbreviations: positive, positive effect on exon 7 splicing; negative, negative effect on exon 7 splicing; neutral, neutral effect on exon 7 splicing; 3′ss, 3′ splice site; 3′-UTR, 3′-untranslated region; ND, not performed or assayed; Yes, observed; No, not observed; SMN2m, SMN2 minigene; SMN2g, Endogenous SMN2 gene
3.1. In Vivo Selection of Exon 7
In vivo selection is a powerful method to determine the position-specific role of every exonic residue on splicing of a given exon. The feasibility of in vivo selection for an entire exon was first demonstrated in the context of SMN1 exon 7 [78]. The method employed a partially randomized exon 7 and repeated rounds of selection for sequences that promoted exon 7 inclusion [78]. The approach was modeled on in vitro selection of a large sequence used for the simultaneous identification of cis-elements and structural motifs critical for RNA-protein interaction [45, 79]. The results of in vivo selection confirmed the presence of “Exinct” in the beginning of exon 7 (Fig. 2A; [78]). The findings of in vivo selection also uncovered the role of a “conserved tract,” a long stretch of nucleotides in the middle of exon 7 that constituted a number of overlapping positive cis-elements (Fig. 2A; [78]). In addition, the results of in vivo selection revealed the existence of a negative cis-element, the “3′-cluster,” located toward the end of exon 7 (Fig. 2A; [78]). Of note, the “3′-cluster” overlaps with the exonic region that is not conserved between human and rodents, suggesting that human SMN exon 7 acquired this negative regulator of splicing after the divergence from the common rodent ancestor ~80 million years ago. Major findings of in vivo selection were independently confirmed by an antisense microwalk as well as by a machine-learning-based simulation study [80, 81]. The most surprising finding of in vivo selection was the overwhelming selection of a non-wild type G residue (A54G) at the last position of exon 7 [78]. Validating experiments confirmed the strong stimulatory effect of A54G substitution on SMN2 exon 7 splicing. For instance, substitutions abrogating various positive cis-elements of exon 7 were fully tolerated in the presence of 54G. Numerous mechanisms by which 54G imparts such a strong stimulatory effect on SMN2 exon 7 splicing could be envisioned. For example, 54G is predicted to disrupt an inhibitory structure (terminal stem loop 2 or TSL2) that sequesters the 5′ss of exon 7 (Fig. 2C). In addition, 54G increases the base pairing between U1 snRNP and the 5′ss of exon 7. Indeed, both of these predictions turned out to be true [82]. Hence, findings of in vivo selection had a transformative effect on our understanding of SMN exon 7 splicing. In particular, they revealed that the 5′ ss of exon 7 is weak in both SMN1 and SMN2. Subsequent studies focused on the mechanism that defines the 5′ss of exon 7 [43, 44, 83, 84]. These studies culminated in discoveries that led to the first therapy for SMA.
3.2. Effect of Terminal Stem Loop 2
In order to demonstrate the role of an RNA structure in pre-mRNA splicing, one must first perform structure probing to definitively confirm the existence of a specific RNA structure. In addition, using site-specific mutagenesis one must then show a correlation between disruption of the structure and altered splicing. Validating experiments must also demonstrate that the splicing pattern is restored when the structure is reinstated. Thus far only a handful of studies have fulfilled the above-mentioned requirements to conclusively establish the role of an RNA structure in pre-mRNA splicing. Inspired by the results of in vivo selection, we performed a systematic study uncovering the role of the terminal stem-loop 2 (TSL2) predicted to partially sequester the 5′ss of exon 7 in splicing regulation of this exon (Fig. 2C). Enzymatic structure probing confirmed the existence of both TSL1 and TSL2 [82]. Supporting the inhibitory role of TSL2, U40G or A54C substitution that disrupted TSL2 was found to promote SMN2 exon 7 inclusion. As expected, when U40G and A54C substitutions were combined to reinstate the TSL2 structure, a strong inhibitory effect on SMN2 exon 7 splicing was restored [82]. These results unequivocally confirmed that TSL2 plays the inhibitory role in the regulation of SMN exon 7 splicing. One of the mechanisms by which TSL2 prevents SMN2 exon 7 inclusion is through poor recruitment of U1 snRNP at the 5′ss of exon 7. Consistent with this argument, a mutated U1 snRNA with extended complementarity to the 5′ss of exon 7 was found to restore SMN2 exon 7 inclusion [82]. Independently validating these findings, an ASO-mediated depletion of endogenous U1 snRNP was found to promote skipping of exon 7 from both SMN1 and SMN2 [49]. However, the effect of U1 snRNP depletion was less pronounced in case of SMN1 exon 7 than SMN2 exon 7. This could be due to C6U substitution strengthening TSL1 and as a consequence stabilizing TSL2. It is also possible that the stimulatory factor(s) interacting with SMN1 exon 7 disrupt TSL2.
3.3. Effect of Intronic Splicing Silencer N1
In an effort to identify additional cis-elements that might suppress the recognition of the 5′ss of SMN2 exon 7, we analyzed the intronic sequences immediately downstream of the 5′ss of exon 7. Using the SMN2 minigene we generated a set of mutants with overlapping deletions and tested their splicing pattern. Our results revealed that the sequence spanning from the 10th to 24th positions of intron 7 is highly inhibitory for exon 7 inclusion [85]. We termed this sequence as intronic splicing silencer N1 or ISS-N1 (Fig. 3; [85]). ISS-N1 deletion obviated the requirement for several positive cis -elements responsible for SMN exon 7 inclusion. We next employed type 1 SMA patient fibroblasts (GM03813) to validate the inhibitory effect of ISS-N1 in the context of the endogenous SMN2. Of note, GM03813 cells carry only SMN2 and offer an invaluable tool to examine the effect of compounds on spicing of SMN2 exon 7. As expected, an ASO that blocked ISS-N1 fully restored SMN2 exon 7 inclusion in GM03813 cells [85]. Importantly, ISS-N1-targeting ASO had a pronounced stimulatory effect on SMN2 exon 7 splicing even at a low concentration of 5 nM. This could be due to strong inhibitory nature of ISS-N1 combined with its high accessibility for an ASO that targets it. Among several hundred targets examined thus far, ISS-N1 remains the most effective target for an ASO-mediated stimulation of SMN2 exon 7 inclusion [89]. Numerous studies employing various mouse models have independently validated the in vivo efficacy of ISS-N1-targeting ASOs [23]. The recently approved ISS-N1-targeting drug for SMA, Nusinersen (synonyms: ISIS-SMNRx, IONIS-SMNRx and Spinraza™), is a modified oligonucleotide that carries phosphorothioate backbone and encompasses methoxyethyl modification at the 2′-hydroxyl position of the revealed that the sequence spanning from the 10th to 24th positions of intron 7 is highly inhibitory for exon 7 inclusion [85].
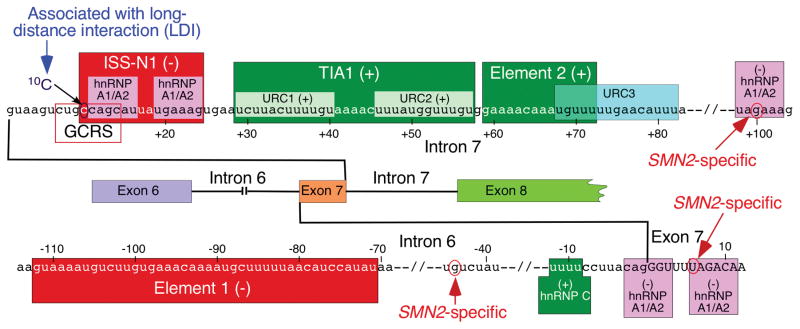
Fig. 3.
Diagrammatic representation of intronic cis-elements and transacting factors that modulate SMN exon 7 splicing. Positive and negative elements are indicated by (+) and (−), respectively. Positive and neutral numbers indicate nucleotide positions within intron 7 and exon 7, respectively, starting with the first intronic/exonic position. Negative numbers indicate nucleotide positions within intron 6, starting with the last intronic position. Exonic and intronic sequences are shown in upper- and lower-case letters, respectively. Exons and introns are also shown as colored boxes and lines. SMN2-spesific single nucleotide substitutions are indicated. Intron 7-located ISS-N1, the overlapping GC-rich sequence (GCRS) and 10C contribute to skipping of exon 7 [43]. ISS-N1 harbors two hnRNP A1/A2B1-binding sites that are highlighted in pink. An SMN2-specific C6U substitution in exon 7 and A100G substitution in intron 7 create additional binding sites for hnRNP A1 [37, 47]. Another hnRNP A1-binding site is located at the junction of intron 6 and exon 7 [51]. Element 2 and U-rich clusters (URC1 and URC2) are positive cis-elements [75, 86]. TIA1 interacts with URC1 and URC2 and promotes exon 7 inclusion [75]. Intron 6-located Element 1 is highlighted in red [87]. It serves as a binding site for PTB and FUSE-BP [88]. A binding site for the stimulatory hnRNP C1/C2 within intron 6 is highlighted in green [68].
Among several hundred targets examined thus far, ISS-N1 remains the most effective target for an ASO-mediated stimulation of SMN2 exon 7 inclusion [89]. Numerous studies employing various mouse models have independently validated the in vivo efficacy of ISS-N1-targeting ASOs [23]. The recently approved ISS-N1-targeting drug for SMA, Nusinersen (synonyms: ISIS-SMNRx, IONIS-SMNRx and Spinraza™), is a modified oligonucleotide that carries phosphorothioate backbone and encompasses methoxyethyl modification at the 2′-hydroxyl position of the sugar moiety [23]. The above-mentioned modifications are known to enhance the in vivo stability of oligonucleotides. Multiple reports published recently discuss different aspects of the drug development process that led to the FDA approval of Nusinersen [25, 26, 49, 90–93]. More than a dozen independent studies employing ASOs with different chemistries have validated the stimulatory effect of ISS-N1 sequestration on SMN2 exon 7 splicing [89, 94]. An in-depth analysis of these studies for an improved future ASO-based therapy is beyond the scope of this review.
Several studies have been performed to uncover the mechanism of ISS-N1 function. The inhibitory effect of ISS-N1 was only partially maintained in a heterologous background, suggesting that the context of SMN2 makes ISS-N1 a strong negative regulator of splicing [85]. An early report implicated two putative-binding sites of hnRNP A/A2 within ISS-N1 as the major cause of the inhibitory effect of this cis -element (Fig. 3; [52]). This model has been recently revised to suggest that two RNA-recognition motifs (RRMs) of a single hnRNP A1 molecule interact with two putative sites within ISS-N1 [95]. Noticeably, the cytosine residue at the first position (10C) of ISS-N1 does not fall within the putative hnRNP A1/A2-binding site. Yet, sequestration of 10C was found to be absolutely critical for an ASO-mediated splicing correction of SMN2 exon 7 (Fig. 3 ; [96]). It has been also confirmed that ASO-mediated sequestration of two putative hnRNP A1/A2-binding sites within ISS-N1 is not enough to produce a stimulatory effect on SMN2 exon 7 splicing [50, 96]. Overall, several studies suggest a more complex mode of ISS-N1 action. Furthermore, motifs upstream and downstream of ISS-N1 appear to be involved in it as well [49, 50, 96, 97].
In search for the shortest ASO that effectively restores SMN2 exon 7 inclusion, we performed an ultra-refined antisense microwalk within and around ISS-N1 sequence [97]. Of note, ASO sizes and their respective targets in our ultra-refined antisense microwalk differed by single nucleotides. Such approach unequivocally guarantees success for the identification of the shortest therapeutic ASO [98]. Our results showed that sequestration of a GC-rich sequence (GCRS) by an 8-mer ASO fully restored SMN2 exon 7 inclusion (Fig. 3; [97]). Interestingly, GCRS-targeting ASO was found to be more specific than an ISS-N1-targeting ASO, particularly at higher concentrations [97]. This is not entirely surprising, since long ASOs can tolerate mismatched base pairs, whereas as shorter ASOs require total complementarity. Subsequent studies confirmed the therapeutic efficacy of a GCRS-targeting ASO in both mild and severe mouse models of SMA [99]. Although GCRS partially overlaps with ISS-N1, it may represent a distinct negative element. Future studies will determine if a specific factor associates with GCRS.
3.4. Effect of U-Rich Clusters Within Intron 7
SMN intron 7 contains multiple U-rich clusters (URCs). URC1 and URC2 are located next to each other immediately downstream of ISS-N1 (Fig. 3). Element 2, the very first intronic cis-element shown to promote exon 7 inclusion, is located downstream of URC2 [86]. It partially overlaps with the third U-rich cluster, URC3 (Fig. 3). Overlapping deletions in the SMN2 minigene confirmed the strong stimulatory nature of the above URCs and Element 2. Subsequent experiments linked the stimulatory effect of URC1 and URC2 with TIA1, a glutamine-rich RNA-binding protein [75]. TIA1 and its related protein TIAR generally interact with URCs immediately downstream of a 5′ss and stimulate exon inclusion by promoting recruitment of U1 snRNP to suboptimal 5′ss [100]. However, the context of TIA1/TIAR interactions in SMN2 intron 7 is somewhat different due to the presence of ISS-N1 between the 5′ ss of exon 7 and URC1/URC2 sites to which TIA1 binds. Overexpression of TIA1 fully restored SMN2 exon 7 inclusion, suggesting that factors that interact with ISS-N1 interfere with recruitment of TIA1 to URC1/URC2 [75]. Supporting the role TIA1 in SMN exon 7 splicing in the context of a human disease, Welander distal myopathy (WDM) patients carrying a TIA1 mutation display an elevated level of SMN exon 7 skipping [101]. Recently, mutations in TIA1 have been also linked to frontotemporal dementia (FTD) and ALS [102]. However, it is not known if FTD/ALS patients carrying TIA1 mutations display SMN exon 7 skipping in any of their tissues. Notably, nervous tissue of Tia1 knockout mouse shows dysregulated expression of lipid storage and membrane dynamics factors [103]. However, effect of Tia1 deletion on SMN2 exon 7 splicing cannot be evaluated because mice lack SMN2. To obviate this problem, we generated a Tia1 knockout mouse in the context of a mild SMA model harboring SMN2 alleles [104]. Interestingly, loss of Tia1 in this mouse model did not show changes in SMN2 exon 7 splicing, although the severity of the SMA disease was affected in a gender-specific manner [104]. Several reasons may account for the discrepancy between the effects of Tia1 deletion (in mouse) and TIA1 mutation (in human). For instance, TIA1 is involved in various types of protein-protein and RNA-protein interactions during pre-mRNA splicing, stress granule formation, and mRNA trafficking [105, 106]. It is likely that a mutant TIA1 protein perturbs protein-protein and RNA-protein interactions in the above-mentioned processes. On the other hand, the complete loss of Tia1 in the mouse model is tolerated due to the presence of its related protein Tiar and/or other glutamine-rich RNA-binding protein.
3.5. Effect of Long-Distance Interactions Within Intron 7
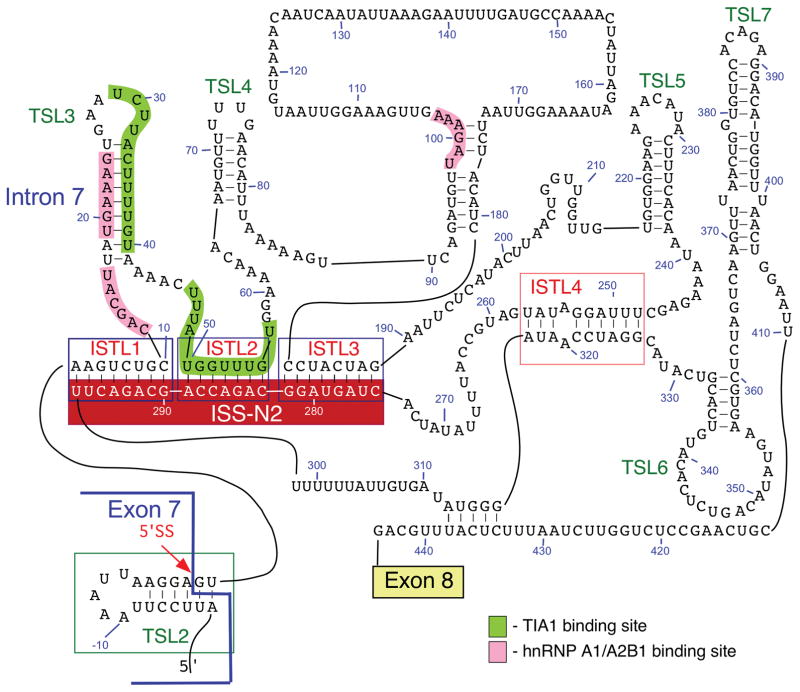
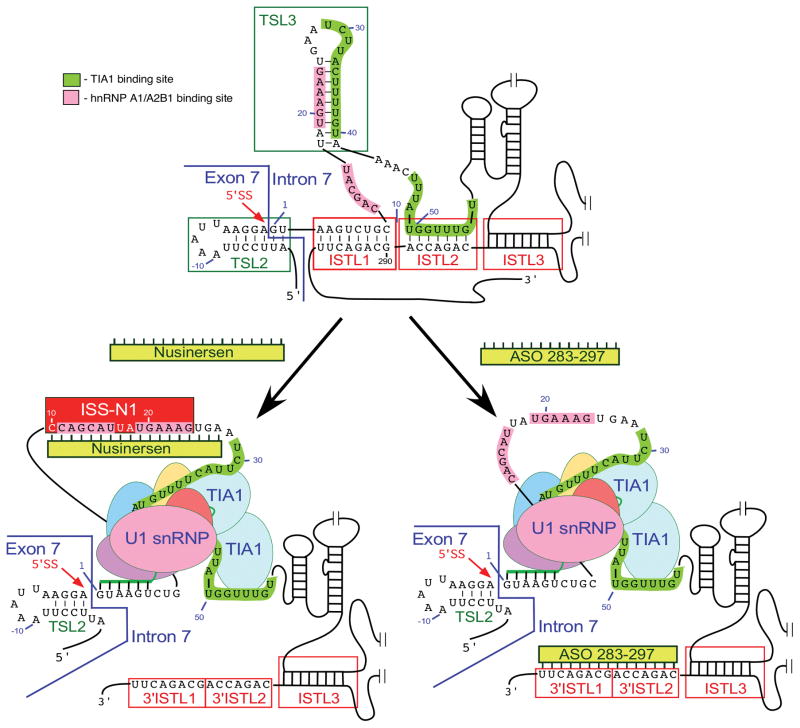
Splicing of SMN exon 7 is modulated by a unique RNA structure formed by long-distance interactions (LDI) within intron 7 [43, 50, 96]. This structure is termed as “Internal-Stem formed by LDI 1” or ISTL1 (Fig. 4; [50]). Chemical structure probing confirmed the formation of ISTL1 along with several other structures within intron 7 (Fig. 4). Two strands of ISTL1 are separated from each other by 279-nts, of which 189 residues are located within the independently folded modules. The 5′ strand of ISTL1 overlaps with the 5′ss of exon 7 as well as 10C, which occupies the first position of ISS-N1. It appears that the formation of ISTL1 strengthens TSL2. Consistently, F14, a 14-mer ASO that sequesters the first 14 residues of ISS-N1, including 10 C, destabilizes both ISTL1 and TSL2 [50, 96]. On the contrary, L14, a 14-mer ASO that sequesters the last 14 residues of ISS-N1, but not 10 C, strengthens both ISTL1 and TSL2. Consequently, F14 and L14 have opposite effects on SMN2 exon 7 splicing: F14 promotes SMN2 exon 7 inclusion, while L14 causes skipping of this exon [50, 96]. The opposite effects of F14 or L14 were found to be independent of the oligonucleotide chemistry, suggesting that ASO-induced structural rearrangement at the 5′ ss of exon 7 was the driving force behind the splicing outcomes [96]. This is a rare example in which two ASOs of identical size annealing to sequences differing only by a single nucleotide produce opposite effects on pre-mRNA splicing. The 3′ strand of ISTL1 overlaps with ISS-N2, a negative element located deep within intron 7 (Fig. 4; [50]). ISS-N2 also participates in the formation of ISTL2 and ISTL3, other intra-intronic structures formed by LDIs (Fig. 4). Formation of ISTL2 sequesters URC2, one of the binding sites of TIA1. Similar to ISS-N1, deletion or an ASO-mediated sequestration of ISS-N2 restores SMN2 exon 7 inclusion. Interestingly, ASO-mediated sequestration of ISS-N1 and ISS-N2 brings the similar structural changes at the 5′ ss of SMN2 exon 7, suggesting a common mechanism of action. It appears that both ISS-N1- and ISS-N2-targeting ASOs promote inclusion of SMN2 exon 7 through abrogation of ISTL1 and an improved recruitment of TIA1 (Fig. 5). In vivo study with an ISS-N2 targeting ASO was recently shown to confer gender-specific therapeutic benefits in a mild mouse model of SMA [107].
Fig. 4.
Secondary structure of SMN2 intron 7 derived from chemical probing. Numbering starts from the first position of intron 7. Negative numbers represent upstream sequences within exon 7. TSLs, ISTLs and binding sites for TIA1 and hnRNP A1/A2B1 are shown and highlighted. ISS-N2 is composed of the 3′ strands of ISTL1, ISTL2 and ISTL3 [43, 50]. The 5′ss of exon 7 is indicated by a red arrow. Abbreviations are given in Table 2.
Fig. 5.
ASO-based mechanism of SMN2 exon 7 splicing correction. Only the relevant sequences of exon 7/intron 7 are given. Nucleotide numbering starts from the first position of intron 7. ISS-N1 and the binding sites for TIA1 and hnRNP A1/A2B1 are marked by colored boxes. The 5′ ss of exon 7 is indicated by a red arrow. The annealing positions of U1 snRNA to this 5′ ss are shown. TSL2 and 3 are local RNA secondary structures, while ISTL1, 2 and 3 are the structures formed by long-distance interactions. These structures are boxed. Nusinersen and ASO 283–297 are shown as yellow bars [25, 107]. Their annealing positions within intron 7 are indicated. Targeting of the corresponding intronic sequences by Nusinersen and ASO 283–297 causes massive structural rearrangements, including disruption of TSL3 and ISTL1. As the results TIA1-binding sites become accessible, the recruitment of U1 snRNP to the 5′ ss of exon 7 is increased and, in case of Nusinersen, the binding of hnRNP A1/A2 to ISS-N1 is blocked. Abbreviations are given in Table 2.
3.6 Extension of Exon 7 by the Activation of a Cryptic 5′ss
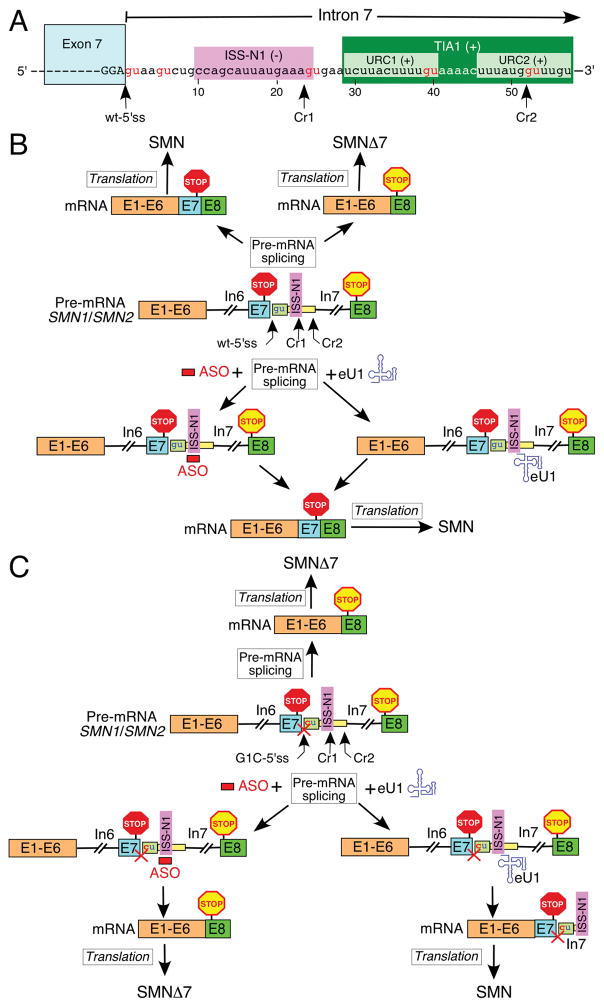
Various instances of SMA caused by enhanced exon 7 skipping triggered by mutations at the 3′ or the 5′ss of SMN1 exon 7 have been reported [12, 108, 109]. Such patients cannot benefit from Nusinersen or any other therapeutic approach requiring the fully functional splice sites of exon 7. However, these patients can take advantage of an engineered U1 snRNA (eU1)-based approach aimed at the activation of a cryptic 5′ss located downstream of the natural 5′ss of exon 7. The proof of principle has recently been established in the context of a pathogenic G-to-C mutation at the first position (G1C) of SMN1 intron 7 (Fig. 6; [49]). As expected, SMN1 exon 7 carrying G1C substitution undergoes complete skipping of exon 7 with or without an ISS-N1-targeting ASO. However, eU1s targeting ISS-N1 or sequences upstream or downstream of this cis-element activate a cryptic 5′ss (Cr1) leading to the inclusion of an “extended” exon 7. Of note, another cryptic 5′ss, Cr2, located within URC2 could also be activated by a different set eU1s, albeit with less efficiency [49]. Cr1 and Cr2 usage increases the length of exon 7 by 23 and 51 nts, respectively (Fig. 6). Since the stop codon of SMN is located within exon 7, activation of Cr1 or Cr2 will have no consequences for the protein. Indeed, the activation of Cr1 in SMN1 construct carrying pathogenic G1C mutation led to the production of SMN, confirming that transcripts generated by Cr1 activation are stable and translation competent (Fig. 6; [49]).
Fig. 6.
Effect of an ASO and eU1 on splicing of exon 7. (A) Diagrammatic representation of exon 7/intron 7 junction. Exonic and intronic sequences are shown in upper- and lower-case letters, respectively. Exon7 is also shown as a blue box. Nucleotide numbering starts from the first position of intron 7. ISS-N1 and URC1 and URC2 are marked by colored boxes. The wild type and the cryptic 5′ ss of exon 7 (Cr1 and 2) are indicated by arrows. GU dinucleotides are highlighted in red. (B) Model of how in the context of the intact 5′ ss of exon 7 an ASO and eU1 promote production of the full-length SMN protein (Adapted from [49]). The ASO block ISS-N1 and eU1 activates usage of the wild-type 5′ ss of exon 7. Exons and introns are indicated by the colored boxes and lines, respectively. The ASO is shown as a red bar, and eU1 as a blue structure. ISS-N1, stop codons in exon 7 and 8 and the 5′ ss of exon 7, wild type and cryptic, are indicated. (C) Model of how in the context of the mutated 5′ ss of exon 7 only eU1 promotes production of the full-length SMN protein (Adapted from [49]). The G to C mutation at the first position of intron 7 is shown in red. The inactivation of the 5′ ss is signified by a red cross. The ASO blocks ISS-N1 and eU1 activate usage of the cryptic 5′ ss of exon 7, Cr1. Exons and introns are indicated by the colored boxes and lines, respectively. The ASO is shown as a red bar, and eU1 as a blue structure. ISS-N1, stop codons in exon 7 and 8 and the 5′ ss of exon 7, wild type and cryptic, are indicated. Abbreviations are given in Table 2.
The discovery of Cr1 and Cr2 brings new perspective to our understanding of SMN exon 7 splicing regulation. Cr1 partially overlaps with ISS-N1, suggesting that the factors interacting with ISS-N1 are likely to suppress the activation of Cr1 as well. Interestingly, Cr1 is efficiently activated even by those eU1s that did not anneal to Cr1 directly [49]. Also, activation of Cr1 does not require assistance of the endogenous U1 snRNP, suggesting that usage of Cr1 can occur in the absence of the typical RNA:RNA duplex formed between the 5′ss and the U1 snRNA. This finding has broad implications as it suggests that the U1 snRNP can affect selection of a 5′ss from distance. It appears that positive cis-elements required for inclusion of SMN exon 7 are dispensable for Cr1 activation. For instance, point mutations that activated Cr1 in SMN2 tolerated the loss of the enhancer associated with Tra2-beta1. Further, eU1s targeting Cr1 prevented skipping of exon 7 associated with the pathogenic mutation at the 3′ss of SMN1 exon 7. Overall, these findings suggest that the activation of Cr1 might employ an entirely different set of rules.
3.7 Role of cis-Elements Within Intron 6
Various mutations at the 3′ss of SMN1 intron 6 have been found to be associated with SMA pathogenesis [12, 110, 111]. However, very limited studies have been done to uncover the role of cis-elements within SMN intron 6. Element 1, an extended inhibitory sequence situated immediately upstream of the 3′ss of exon 7, was the first cis-element to be reported within intron 6 (Fig. 3; [87]). Deletion or an ASO-mediated sequestration of Element 1 promoted SMN2 exon 7 inclusion [87, 112]. A recent report demonstrated an in vivo efficacy of an Element 1-targeting ASO in a severe mouse model of SMA [112]. Another negative cis -element at the junction of intron 6 and exon 7 has been suggested to constitute a binding site for hnRNP A1 (Fig. 3; [51]). The location of this site right next to the other hnRNP A1-binding site created by the C6U mutation within exon 7 strikingly resembles the arrangement of two putative hnRNP A1 sites within ISS-N1. As recently proposed, close proximity of the two hnRNP A1 sites is conducive for a tight interaction involving two RRMs of a single hnRNP A1 molecule [95]. The polypyrimidine tract (PPT) at the 3′ ss of exon 7 has been suggested to harbor a positive element associated with hnRNP C (Fig. 3; [68]). However, the role of hnRNP C in SMN exon 7 splicing could not be independently validated by depletion experiments [48, 113]. Interestingly, an A-to-G substitution at the -44th position (A-44G) of intron 6 has been found to promote SMN2 exon 7 inclusion (Fig. 3; [40]). The A-44G substitution is naturally present in human population and SMA patients carrying A-44G substitution show mild phenotype [40].
4. Exonization of an Intronic Alu-Element
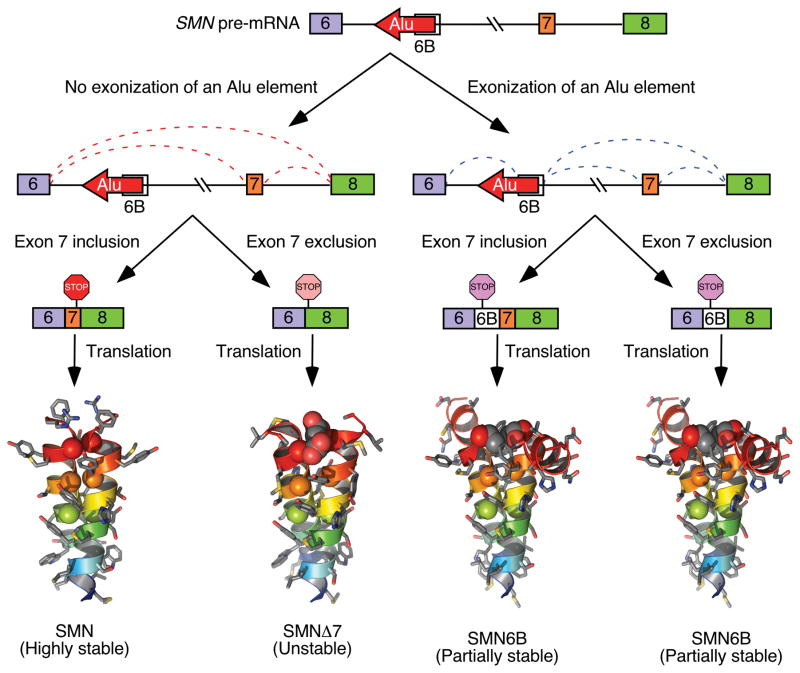
Alu elements are primate-specific transposable elements encompassing ~300 bp bipartite motifs derived from the 7SL RNA, an essential component of the protein signal recognition complex [114]. Insertion of Alu elements has played a significant role in primate evolution due to their drastic effect on chromatin remodeling, transcription and generation of novel exons [115, 116]. Multi-exon skipping detection assay (MESDA) is a powerful technique that simultaneously detects most SMN splice isoforms in a single reaction [117]. Employing MESDA, we have recently reported a novel exon, exon 6B, generated by the exonization of an Alu element located within intron 6 [33]. Expression of exon 6B-containing transcripts has been confirmed in various tissues of a mouse model of SMA as well as in human tissues examined [33]. Both SMN1 and SMN2 produce exon 6B-containing transcripts. Generally, the right arm of an antisense sequence of an Alu is used for exonization [118]. However, the 109-nt long exon 6B originated from the left antisense arm of an Alu element. The low expression of exon 6B-containing transcripts is attributed to various factors, including suppression by hnRNP C and degradation by Nonsense
Mediated Decay (NMD). An overwhelming 39% of SMN sequence is occupied by >40 Alu elements located within introns. Exon 6B is the first and only known example of SMN exon derived from the exonization of an intronic Alu element. Due to its location upstream of exons 7, it is likely that splicing of exon 6B is influenced by exon 7 and vice versa. However, the mechanism of exon 6B splicing regulation remains to be determined. Amino acids coded by exon 7 define the critical C-terminus of SMN and confer protein stability. The loss of amino acids coded by exon 7 is the primary reasons why SMNΔ7 is less stable than SMN (Fig. 7; [16, 119]). Irrespective of exon 7 inclusion or skipping, the exon 6B-containing transcripts code for SMN6B protein in which the last 16 amino acids are coded by exon 6B. The altered C-terminus makes SMN6B less stable than SMN. However, SMN6B was found to be more stable than SMNΔ7, suggesting that the altered C-terminus of SMN6B is not deleterious as observed in case of SMNΔ7 (Fig. 7; [33]). As expected, SMN6B retains the ability to interact with Gemin2, a key protein required for most SMN functions. Similar to SMN, SMN6B localizes to both, nuclear and cytosolic compartments. Hence, it is likely that SMN6B will be able to ameliorate SMA pathology if expressed at sufficient levels.
Fig. 7.
A model showing skipping and inclusion of SMN exon 6B. Exon 6B is derived from an Alu element located within SMN intron 6 [33]. Transcripts that include exon 7 but exclude exon 6B produce full-length SMN, a highly stable protein. Transcripts that lack both exons 6B and exon 7 produce SMNΔ7, an unstable and partially functional protein. Transcripts that include exon 6B produce SMN6B protein irrespective of inclusion or exclusion of exon 7. SMN6B protein is more stable than SMNΔ7 [33]
5. Alternative Splicing of Other SMN Exons
The diversity of SMN splice isoforms is best demonstrated by MESDA, which captures susceptibility of various SMN exons to skipping under normal and stress-associated conditions [117]. Low levels of exon 3 and exon 5-skipped transcripts are generated under normal conditions in most tissues from both SMN1 and SMN2 [117]. SMN2 exons 5 and 7 become highly susceptible to skipping under the conditions of oxidative stress, although skipping of SMN1 exon 5 is also enhanced by oxidative stress. A recent study examined the effect of paraquat, an oxidative-stress-causing agent, on splicing of various SMN2 exons in different tissues of a transgenic mouse model harboring SMN2 [120]. Findings of this study revealed tissue-specific effect of oxidative stress on splicing of various SMN2 exons. For instance, skipping of SMN2 exons 3, 5, and 7 was found to be substantially increased under oxidative stress in lung as compared to brain and spinal cord, which instead showed significant enhancement of SMN2 exons 5 and 7 skipping. The study also captured individual differences of the effect of oxidative stress on splicing of various SMN2 exons. For example, one of the four animals examined showed enhanced co-skipping of exons 3, 4, 5, 6 and 7 in liver at 8 h post paraquat treatment. Another animal showed enhanced co-skipping of exons 3, 5, 6 and 7 in liver at 12 h post paraquat treatment. While reasons for these individual differences remain unknown, findings underscore that the rules of stress-associated splicing regulation should be interpreted with caution.
Depletion of U1 snRNP creates a stress on the splicing machinery as well as on other co-transcriptional events dependent upon the availability of U1 snRNP [121]. A diverse set of SMN transcripts is generated upon depletion of U1 snRNP by an ASO that sequesters the 5′ end of endogenous U1 snRNA [49]. MESDA profile of SMN transcripts generated under U1 snRNP depletion condition is distinct from those observed under the conditions of oxidative stress. For example, splicing of all exons was affected under U1 snRNP depletion, whereas splicing of SMN2 exons 5 and 7 was the most affected under oxidative stress condition [49, 117]. Interestingly, skipping of exon 6 was the least among all other internal exons of SMN under both U1 snRNP depletion and oxidative stress conditions [49, 117]. This could be attributed to relatively high accessibility of the 5′ ss of exon 6 coupled with a strong duplex between U1 snRNP and the 5′ ss of exon 6. It is likely that the energy (ATP) deficit created by oxidative stress downregulates the biogenesis of snRNPs, particularly U1 snRNP, which is generally maintained at a higher level than other snRNPs. It has been recently shown that the depletion of DHX9, an RNA helicase that resolves the double-stranded RNA structures, enhances the Alu-induced RNA processing defects, including aberrant pre-mRNA splicing and circRNA production from transcripts harboring Alu repeats [122]. Similar to snRNP biogenesis, RNA helicases require ATP for their function. Therefore, it is likely that large RNA:RNA duplexes formed by Alu elements positioned in opposite orientations in SMN pre-mRNA are not appropriately resolved by RNA helicases under the conditions of oxidative stress. Preliminary analysis of the publicly available circRNA database suggests production of circRNAs by SMN [123]. However, it is not known what fraction of SMN transcripts make circRNAs and which of the circRNAs are predominantly expressed in most cell types. Future studies will determine how Alu elements might impact generation of SMN circRNAs under normal and stress-associated conditions in a cell-specific manner.
6. Effect of Transcription on Splicing of Various SMN Exons
Transcription requires opening of chromatin structure followed by recruitment of transcription initiation factors [9]. Transcription in vivo is coupled to splicing through two likely mechanisms: “recruitment coupling” and “kinetic coupling” [124]. These two mechanisms are not mutually exclusive and it is often difficult to conclusively distinguish one mechanism from the other. In case of recruitment coupling, RNA polymerase II (pol II) recruits splicing factors at the promoter site and then transports it to the splice sites. In case of kinetic coupling, the rate of transcription elongation influences the outcome of splicing. The evidence that transcription affects splicing of SMN exon 7 comes from a promoter-swapping experiment performed in minigene systems. In particular, the replacement of the wild-type SMN promoter with CMV or TK promoter caused enhanced skipping of exon 7 in both SMN1 and SMN2 minigenes [117]. These results suggested that wild-type promoter harbors sequences that are stimulatory for exon 7 splicing. Additional evidence that transcription affects SMN splicing comes from small molecules that affect the activity of histone acetylases (HATs) and histone deacetylases (HDACs). The former and the latter enzymes activate and suppress transcription, respectively. Various HDAC inhibitors, including trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), and benzamide M344, have been shown to modulate splicing of SMN exon 7 [125]. Another mechanism by which transcription could modulate splicing of SMN exons is through the regulation of the formation of loops within pre-mRNA. PTB and hnRNP A1/A2 have been implicated in deciding splicing outcomes through looping out specific sequences [126, 127]. In particular, looping out of an exon promotes its skipping, whereas looping out of an intra-intronic sequence promotes exon inclusion. Furthermore, a slow elongating pol II might delay the formation of a specific loop. Considering that SMN pre-mRNA contains binding sites for the loop-forming hnRNP A1/A2 protein, it is highly likely that splicing of various SMN exons is regulated by transcription.
7. Conclusions
SMA is one of the leading genetic diseases associated with infant mortality. As soon as the association of SMA with SMN1 deletion/mutations was established in 1995, attempts began to find a potential cure/therapy for this disorder. Since SMN2 is almost universally present in SMA patients, it offers an obvious therapeutic target for exon 7 splicing correction. The major breakthrough came when the critical role of the context-specific cis-elements located away from the pathogenic mutations, such as C6U, was beginning to be established. In particular, the discovery of the intronic cis-element, ISS-N1, reported in 2006 produced an effective target, sequestration of which fully corrected SMN2 exon 7 splicing and restored SMN levels in SMA patient cells. General interest in ISS-N1 combined with subsequent independent validations of its therapeutic potential paved a way to the first FDA-approved drug for SMA. In addition, the detailed characterization of ISS-N1 led to the discovery of a unique RNA structure formed by long-distance intra-intronic interactions that contributes to exon 7 skipping. Interestingly, abrogation of a similar structure within intron 3 of the proteolipid protein 1 (PLP1) gene has been recently suggested to cause X-linked Pelizaeus–Merzbacher disease or PMD [128]. Growing evidence suggests that splicing of various exons is differentially regulated under the normal and stress-associated conditions. It is also becoming obvious that the intronic Alu elements are capable of increasing the diversity of SMN splice isoforms and may play an important role in the generation of circRNAs [123]. Furthermore, new findings that two antisense transcripts are produced from the SMN locus highlight the existence of an addition layer of SMN transcription and potentially splicing control. The development of novel tools and reliable assays that accurately capture transcription-coupled splicing events would tremendously advance our understanding of how expression of the SMN gene is regulated, including the pre-mRNA splicing step. This advancement would also uncover the likely mechanisms of the tissue-specific modulation of splicing of various SMN exons under the normal and stress-associated conditions. A better understanding of SMN splicing has implications for several diseases impacted by the low levels of the SMN protein. Lessons learnt from SMN would also provide unique insights into our understanding of a growing number of human diseases associated with aberrant splicing.
Table 2.
Abbreviations and terminology used in this study
| Abbreviation | Full Name | Relevant Figures |
|---|---|---|
| 3′ss | 3′ splice site | 2 |
| 3′-UTR | 3′ untranslated region | |
| 5′ss | 5′ splice site | 2 |
| 5′-UTR | 5′ untranslated region | |
| ASO | Antisense oligonucleotide | 5 |
| bp | Base pair | |
| C6U | A C-to-U substitution at the 6th position of SMN2 exon 7 | 2 |
| Element 1 | Negative cis-element located within SMN intron 6 | 3 |
| Element 2 | Positive cis-element located within SMN intron 7 | 3 |
| eU1 | Engineered U1 snRNA | 6 |
| hnRNP | Hetero-nuclear ribonucleoprotein | 2 |
| ISS-N1 | Intronic splicing silencer N1 (located within SMN intron 7) | 3,5 |
| ISS-N2 | Intronic splicing silencer N2 (located within SMN intron 7) | 4,5 |
| ISTL1 | Internal stem formed by LDI-1 (located within SMN intron 7) | 4,5 |
| ISTL2 | Internal stem formed by LDI-2 (located within SMN intron 7) | 4 |
| ISTL3 | Internal stem formed by LDI-3 (located within SMN intron 7) | 4 |
| ISTL4 | Internal stem formed by LDI-4 (located within SMN intron 7) | 4 |
| nt | Nucleotide | |
| LDI | Long-distance interaction (located within SMN intron 7) | 3,4,5 |
| lncRNA | Long non-coding RNA | 1 |
| Nusinersen | An ASO drug that targets ISS-N1 sequence (synonym of Spinraza™) | 5 |
| SMA | Spinal Muscular Atrophy | |
| SMN (Italics) | Survival motor neuron gene or transcript | |
| SMN-AS1 (Italics) | Antisense transcript (lncRNA) generated from SMN locus | 1 |
| SMN2m | SMN2 minigene | |
| SMN2g | Endogenous SMN2 gene | |
| SMN-AS1* (Italics) | Antisense transcript (lncRNA) generated from SMN locus | 1 |
| SMN | Survival motor neuron protein | |
| SMN6B | SMN6B protein | 7 |
| Spinraza™ | An ASO drug that targets ISS-N1 sequence (synonym of Nusinersen) | 5 |
| TSL1 | Terminal stem-loop 1 located within SMN exon 7 | 2 |
| TSL2 | Terminal stem-loop 2 located within SMN exon 7 | 2,4,5 |
| TSS | Transcription start site | 1 |
| U1 or U1 snRNA | U1 small nuclear RNA | 5,6 |
| U1 snRNP | U1 small nuclear ribonucleoprotein | 5,6 |
| URC1 | U-rich cluster 1 located within intron 7 | 3,5 |
| URC2 | U-rich cluster 2 located within intron 7 | 3,5 |
| URC3 | U-rich cluster 3 located within intron 7 | 3,5 |
| UTR | Untranslated region | |
| wt | Wild-type |
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 NS055925 and R21 NS101312), Iowa Center for Advanced Neurotoxicology (ICAN), and Salsbury Endowment (Iowa State University, Ames, IA, USA) to RNS. The authors acknowledge and regret not being able to include several references due to lack of space.
Footnotes
Disclosures and Competing Interests:
The ISS-N1 target (US Patent# US7838657) was discovered in the Singh laboratory at UMass Medical School (MA, USA). Inventors, including RN Singh, NN Singh and UMASS Medical School, are currently benefiting from licensing of the ISS-N1 target to Ionis Pharmaceuticals and Biogen. Iowa State University holds intellectual property rights on GC-rich and ISS-N2 targets. Therefore, inventors including RN Singh, NN Singh and Iowa State University could potentially benefit from any future commercial exploitation of GC-rich and ISS-N2 targets.
References
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. https://doi.org/10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283(3):1211–5. doi: 10.1074/jbc.R700035200. https://doi.org/10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 3.Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. https://doi.org/10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Erkelenz S, Mueller WF, Evans MS, Busch A, Schöneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19(1):96–102. doi: 10.1261/rna.037044.112. https://doi.org/10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, Ares M, Jr, Yeo GW. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1(2):167–78. doi: 10.1016/j.celrep.2012.02.001. https://doi.org/10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu RevBiochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. https://doi.org/10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14(8):1463–9. doi: 10.1261/rna.1069408. https://doi.org/10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu XD, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. https://doi.org/10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saldi T, Cortazar MA, Sheridan RM, Bentley DL. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J Mol Biol. 2016;428(12):2623–35. doi: 10.1016/j.jmb.2016.04.017. https://doi.org/10.1016/j.jmb.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136(4):777–93. doi: 10.1016/j.cell.2009.02.011. https://doi.org/10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deschênes M, Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell. 2017;16(5):918–33. doi: 10.1111/acel.12646. https://doi.org/10.1111/acel.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Paslier D, Frézal J, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–65. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 13.Singh RN, Howell MD, Ottesen EW, Singh NN. Diverse role of survival motor neuron protein. Biochim Biophys Acta. 2017;1860(3):299–315. doi: 10.1016/j.bbagrm.2016.12.008. https://doi.org/10.1016/j.bbagrm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ottesen EW, Howell MD, Singh NN, Seo J, Whitley EM, Singh RN. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Sci Rep. 2016;6:17. doi: 10.1038/srep20193. https://doi.org/10.1038/srep20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29(5):1107–15. doi: 10.1128/MCB.01262-08. https://doi.org/10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho SC, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24(5):438–42. doi: 10.1101/gad.1884910. https://doi.org/10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J. Refined characterization of the expression and stability of the SMN gene products. Am J Pathol. 2007;171(4):1269–80. doi: 10.2353/ajpath.2007.070399. https://doi.org/10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad S, Bhatia K, Kannan A, Gangwani L. Molecular Mechanisms of Neurodegeneration in Spinal Muscular Atrophy. J Exp Neuro. 2016;10:39–49. doi: 10.4137/JEN.S33122. https://doi.org/10.4137/jen.s33122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nash LA, Burns JK, Chardon JW, Kothary R, Parks RJ. Spinal muscular atrophy: more than a disease of motor neurons? Curr Mol Med. 2016;16(9):779–92. doi: 10.2174/1566524016666161128113338. https://doi.org/10.2174/1566524016666161128113338. [DOI] [PubMed] [Google Scholar]
- 20.Bowerman M, Michalski JP, Beauvais A, Murray LM, DeRepentigny Y, Kothary R. Defects in pancreatic development and glucose metabolism in SMN-depleted mice independent of canonical spinal muscular atrophy neuromuscular pathology. Hum Mol Genet. 2014;23(13):3432–44. doi: 10.1093/hmg/ddu052. https://doi.org/10.1093/hmg/ddu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez CE, Cunningham D, Chandler DS. SMN regulation in SMA and in response to stress: new paradigms and therapeutic possibilities. Hum Genet. 2017;136:1173. doi: 10.1007/s00439-017-1835-2. https://doi.org/10.1007/s00439-017-1835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Muela N, Litterman NK, Norabuena EM, Mull JL, Galazo MJ, Sun C, Ng SY, Makhortova NR, White A, Lynes MM, Chung WK, Davidow LS, Macklis JD, Rubin LL. Single-cell analysis of SMN reveals its broader role in neuromuscular disease. Cell Rep. 2017;18(6):1484–98. doi: 10.1016/j.celrep.2017.01.035. https://doi.org/10.1016/j.celrep.2017.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howell MD, Singh NN, Singh RN. Advances in therapeutic development for spinal muscular atrophy. Future Med Chem. 2014;6(9):1081–99. doi: 10.4155/fmc.14.63. https://doi.org/10.4155/fmc.14.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo J, Howell MD, Singh NN, Singh RN. Spinal muscular atrophy: an update on therapeutic progress. Biochim Biophys Acta. 2013;1832(12):2180–90. doi: 10.1016/j.bbadis.2013.08.005. https://doi.org/10.1016/j.bbadis.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ottesen EW. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl Neurosci. 2017;8:1–6. doi: 10.1515/tnsci-2017-0001. https://doi.org/10.1515/tnsci-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh NN, Howell MD, Androphy EJ, Singh RN. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017b;24:520–6. doi: 10.1038/gt.2017.34. https://doi.org/10.1038/gt.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.d’Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, Rigo F, Bennett CF, Sumner CJ. Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy. Neuron. 2017;93(1):66–79. doi: 10.1016/j.neuron.2016.11.033. https://doi.org/10.1016/j.neuron.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo CJ, Maier VK, Davey R, Brennan J, Li G, Brothers J, 2nd, Schwartz B, Gordo S, Kasper A, Okamoto TR, Johansson HE, Mandefro B, Sareen D, Bialek P, Chau BN, Bhat B, Bullough D, Barsoum J. Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc Natl Acad Sci U S A. 2017;114(8):E1509–18. doi: 10.1073/pnas.1616521114. https://doi.org/10.1073/pnas.1616521114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene. 2001;279(2):109–17. doi: 10.1016/s0378-1119(01)00758-2. https://doi.org/10.1016/S0378-1119(01)00758-2. [DOI] [PubMed] [Google Scholar]
- 30.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet. 1999;64(5):1365–70. doi: 10.1086/302372. https://doi.org/10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monani UR, McPherson JD, Burghes AH. Promoter analysis of the human centromericand telomeric survival motor neuron genes (SMNC and SMNT) Biochim Biophys Acta. 1999b;1445(3):330–6. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 32.Boda B, Mas C, Giudicelli C, Nepote V, Guimiot F, Levacher B, Zvara A, Santha M, LeGall I, Simonneau M. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur J Hum Genet. 2004;12(9):729–37. doi: 10.1038/sj.ejhg.5201217. https://doi.org/10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- 33.Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN. A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein. Sci Rep. 2016a;6:14. doi: 10.1038/srep30778. https://doi.org/10.1038/srep30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G. Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc Nat Acad Sci U S A. 2007;104(6):1959–64. doi: 10.1073/pnas.0610660104. https://doi.org/10.1073/pnas.0610660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rochette CF, Gilbert N, Simard LR. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet. 2001;108(3):255–66. doi: 10.1007/s004390100473. https://doi.org/10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 36.Schmutz J, Martin J, Terry A, Couronne O, Grimwood J, Lowry S, Gordon LA, Scott D, Xie G, Huang W, Hellsten U, Tran-Gyamfi M, She X, Prabhakar S, Aerts A, et al. The DNA sequence and comparative analysis of human chromosome 5. Nature. 2004;431(7006):268. doi: 10.1038/nature02919. https://doi.org/10.1038/nature02919. [DOI] [PubMed] [Google Scholar]
- 37.Kashima T, Rao N, Manley JL. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci U S A. 2007b;104(9):3426–31. doi: 10.1073/pnas.0700343104. https://doi.org/10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–11. doi: 10.1073/pnas.96.11.6307. https://doi.org/10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999a;8(7):1177–83. doi: 10.1093/hmg/8.7.1177. https://doi.org/10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 40.Wu X, Wang SH, Sun J, Krainer AR, Hua Y, Prior TW. A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum Mol Genet. 2017;26(14):2768–80. doi: 10.1093/hmg/ddx166. https://doi.org/10.1093/hmg/ddx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cucchiarini M, Madry H, Terwilliger EF. Enhanced expression of the central survival of motor neuron (SMN) protein during the pathogenesis of osteoarthritis. J Cell Mol Med. 2014;18(1):115–24. doi: 10.1111/jcmm.12170. https://doi.org/10.1111/jcmm.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim SR, Hertel KJ. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J Biol Chem. 2001;276(48):45476–83. doi: 10.1074/jbc.M107632200. https://doi.org/10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 43.Singh NN, Lee BM, Singh RN. Splicing regulation in spinal muscular atrophy by a RNA structure formed by long distance interactions. Ann N Y Acad Sci. 2015b;1341:176–87. doi: 10.1111/nyas.12727. https://doi.org/10.1111/nyas.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh RN. Evolving concepts on human SMN Pre-mRNA splicing. RNA Biol. 2007a;4(1):7–10. doi: 10.4161/rna.4.1.4535. https://doi.org/10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- 45.Singh RN. Unfolding the mystery of alternative splicing through a unique method of in vivoselection. Front Biosci. 2007b;12:3263–72. doi: 10.2741/2310. https://doi.org/10.2741/2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–84. doi: 10.1038/ng854. https://doi.org/10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 47.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–3. doi: 10.1038/ng1207. https://doi.org/10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 48.Wee CD, Havens MA, Jodelka FM, Hastings ML. Targeting SR proteins improves SMN expression in spinal muscular atrophy cells. PLoS One. 2014;9(12):e115205. doi: 10.1371/journal.pone.0115205. https://doi.org/10.1371/journal.pone.0115205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh NN, Del Rio-Malewski JB, Luo D, Ottesen EW, Howell MD, Singh RN. Activation of a cryptic 5′ splice site reverses the impact of pathogenic splice site mutations in the spinal muscular atrophy gene. Nucleic Acids Res. 2017a;45:12214. doi: 10.1093/nar/gkx824. https://doi.org/10.1093/nar/gkx824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013;41(17):8144–65. doi: 10.1093/nar/gkt609. https://doi.org/10.1093/nar/gkt609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doktor TKd, Schroeder LD, Vested A, Palmfeldt J, Andersen HS, Gregersen N, Andresen BS. SMN2 exon 7 splicing is inhibited by binding of hnRNP A1 to a common ESS motif that spans the 3′ splice site. Hum Mutat. 2011;32(2):220–30. doi: 10.1002/humu.21419. https://doi.org/10.1002/humu.21419. [DOI] [PubMed] [Google Scholar]
- 52.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNPA1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82(4):834–48. doi: 10.1016/j.ajhg.2008.01.014. https://doi.org/10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007a;16(24):3149–59. doi: 10.1093/hmg/ddm276. https://doi.org/10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 54.Liu TY, Chen YC, Jong YJ, Tsai HJ, Lee CC, Chang YS, Chang JG, Chang YF. Muscle developmental defects in heterogeneous nuclear Ribonucleoprotein A1 knockout mice. Open Biol. 2017;7(1) doi: 10.1098/rsob.160303. pii: 160303. https://doi.org/10.1098/rsob.160303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010;29(7):1235–47. doi: 10.1038/emboj.2010.19. https://doi.org/10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YC, Yuo CY, Yang WK, Jong YJ, Lin HH, Chang YS, Chang JG. Extracellular pH change modulates the exon 7 splicing in SMN2 mRNA. Mol Cell Neurosci. 2008b;39(2):268–72. doi: 10.1016/j.mcn.2008.07.002. https://doi.org/10.1016/j.mcn.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Singh NN, Androphy EJ, Singh RN. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun. 2004a;315(2):381–8. doi: 10.1016/j.bbrc.2004.01.067. https://doi.org/10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 58.De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4(1):49–60. doi: 10.1002/wrna.1140. https://doi.org/10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 59.Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci U S A. 2000;97(17):9618–23. doi: 10.1073/pnas.160181697. https://doi.org/10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bose JK, Wang I-F, Hung L, Tarn W-Y, Shen C-KJ. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem. 2008;283(43):28852–9. doi: 10.1074/jbc.M805376200. https://doi.org/10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho S, Moon H, Loh TJ, Oh HK, Cho S, Choy HE, Song WK, Chun J-S, Zheng X, Shen H. hnRNP M facilitates exon 7 inclusion of SMN2 pre-mRNA in spinal muscular atrophy by targeting an enhancer on exon 7. Biochim Biophys Acta. 2014a;1839(4):306–15. doi: 10.1016/j.bbagrm.2014.02.006. https://doi.org/10.1016/j.bbagrm.2014.02.006.88. [DOI] [PubMed] [Google Scholar]
- 62.Cho S, Moon H, Loh TJ, Oh HK, Williams DR, Liao DJ, Zhou J, Green MR, Zheng X, Shen H. PSF contacts exon 7 of SMN2 pre-mRNA to promote exon 7 inclusion. Biochim Biophys Acta. 2014b;1839(6):517–25. doi: 10.1016/j.bbagrm.2014.03.003. https://doi.org/10.1016/j.bbagrm.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11(17):2037–49. doi: 10.1093/hmg/11.17.2037. https://doi.org/10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 64.Young PJ, DiDonato CJ, Hu D, Kothary R, Androphy EJ, Lorson CL. SRp30c-dependentstimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1. Hum Mol Genet. 2002;11(5):577–87. doi: 10.1093/hmg/11.5.577. https://doi.org/10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 65.Mende Y, Jakubik M, Riessland M, Schoenen F, Rossbach K, Kleinridders A, Köhler C, Buch T, Wirth B. Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing. Hum Mol Genet. 2010;19(11):2154–67. doi: 10.1093/hmg/ddq094. https://doi.org/10.1093/hmg/ddq094. [DOI] [PubMed] [Google Scholar]
- 66.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet. 2006;78(1):63–77. doi: 10.1086/498853. https://doi.org/10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen H-H, Chang J-G, Lu R-M, Peng T-Y, Tarn W-Y. The RNA Binding Protein hnRNP Q Modulates the Utilization of Exon 7 in the Survival Motor Neuron 2 (SMN2) Gene. Mol Cell Biol. 2008a;28(22):6929–38. doi: 10.1128/MCB.01332-08. https://doi.org/10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irimura S, Kitamura K, Kato N, Saiki K, Takeuchi A, Gunadi, Matsuo M, Nishio H, Lee MJ. HnRNP C1/C2 may regulate exon 7 splicing in the spinal muscular atrophy gene SMN1. Kobe J Med Sci. 2009;54(5):E227–36. [PubMed] [Google Scholar]
- 69.Xiao R, Tang P, Yang B, Huang J, Zhou Y, Shao C, Li H, Sun H, Zhang Y, Fu X-D. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol Cell. 2012;45(5):656–68. doi: 10.1016/j.molcel.2012.01.009. https://doi.org/10.1016/j.molcel.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cléry A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH-T. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-β1. Nat Struct Mol Biol. 2011;18(4):443–50. doi: 10.1038/nsmb.2001. https://doi.org/10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 71.Li J, Chen X, Xiao P, Li L, Lin W, Huang J, Xu P. Expression pattern and splicing function of mouse ZNF265. Neurochem Res. 2008;33(3):483–9. doi: 10.1007/s11064-007-9461-3. https://doi.org/10.1007/s11064-007-9461-3. [DOI] [PubMed] [Google Scholar]
- 72.Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, Sperling J, Sperling R, Stamm S. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem. 2009;284(21):14303–15. doi: 10.1074/jbc.M901026200. https://doi.org/10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moursy A, Allain FH-T, Cléry A. Characterization of the RNA recognition mode of hnRNPG extends its role in SMN2 splicing regulation. Nucleic Acids Res. 2014;42(10):6659–72. doi: 10.1093/nar/gku244. https://doi.org/10.1093/nar/gku244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF(65) PLoS One. 2007;2(6):e538. doi: 10.1371/journal.pone.0000538. https://doi.org/10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh NN, Seo JB, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31(5):935–54. doi: 10.1128/MCB.00945-10. https://doi.org/10.1128/mcb.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sutherland LC, Thibault P, Durand M, Lapointe E, Knee JM, Beauvais A, Kalatskaya I, Hunt SC, Loiselle JJ, Roy JG, Tessier SJ, Ybazeta G, Stein L, Kothary R, Klinck R, Chabot B. Splicing arrays reveal novel RBM10 targets, including SMN2 pre-mRNA. BMC Mol Biol. 2017;18(1):19. doi: 10.1186/s12867-017-0096-x. https://doi.org/10.1186/s12867-017-0096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jodelka FM, Ebert AD, Duelli DM, Hastings ML. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet. 2010;19(24):4906–17. doi: 10.1093/hmg/ddq425. https://doi.org/10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004b;10(8):1291–305. doi: 10.1261/rna.7580704. https://doi.org/10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh RN, Saldanha RJ, D’Souza LM, Lambowitz AM. Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol. 2002;318(2):287–303. doi: 10.1016/S0022-2836(02)00054-2. https://doi.org/10.1016/S0022-2836(02)00054-2. [DOI] [PubMed] [Google Scholar]
- 80.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. https://doi.org/10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RKC, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, Morris Q, Barash Y, Krainer AR, Jojic N, Scherer SW, Blencowe BJ, Frey BJ. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218):1254806. doi: 10.1126/science.1254806. https://doi.org/10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35(2):371–89. doi: 10.1093/nar/gkl1050. https://doi.org/10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh NN, Singh RN. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011;8(4):600–6. doi: 10.4161/rna.8.4.16224. https://doi.org/10.4161/rna.8.4.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Singh NN, Androphy EJ, Singh RN. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit Rev Eukaryot Gene Expr. 2004c;14(4):271–85. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30. https://doi.org/10.1615/CritRevEukaryotGeneExpr.v14.i4.30. [DOI] [PubMed] [Google Scholar]
- 85.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol CellBiol. 2006;26(4):1333–46. doi: 10.1128/MCB.26.4.1333-1346.2006. https://doi.org/10.1128/mcb.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miyaso H, Okumura M, Kondo S, Higashide S, Miyajima H, Imaizumi K. An intronicsplicing enhancer element in survival motor neuron (SMN) pre-mRNA. J Biol Chem. 2003;278(18):15825–31. doi: 10.1074/jbc.M209271200. https://doi.org/10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- 87.Miyajima H, Miyaso H, Okumura M, Kurisu J, Imaizumi K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J Biol Chem. 2002;277(26):23271–7. doi: 10.1074/jbc.M200851200. https://doi.org/10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- 88.Baughan TD, Dickson A, Osman EY, Lorson CL. Delivery of bifunctional RNAs that targetan intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet. 2009;18(9):1600–11. doi: 10.1093/hmg/ddp076. https://doi.org/10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh NN, Lee BM, DiDonato CJ, Singh RN. Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy. Future Med Chem. 2015a;7:1793–808. doi: 10.4155/fmc.15.101. https://doi.org/10.4155/fmc.15.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aartsma-Rus A. FDA approval of nusinersen for spinal muscular atrophy makes 2016 the year of splice modulating oligonucleotides. Nucleic Acid Ther. 2017;27(2):67–9. doi: 10.1089/nat.2017.0665. https://doi.org/10.1089/nat.2017.0665. [DOI] [PubMed] [Google Scholar]
- 91.Glascock J, Lenz M, Hobby K, Jarecki J. Cure SMA and our patient community celebrate the first approved drug for SMA. Gene Ther. 2017;24(9):498–500. doi: 10.1038/gt.2017.39. https://doi.org/10.1038/gt.2017.39. [DOI] [PubMed] [Google Scholar]
- 92.Wan L, Dreyfuss G. Splicing-correcting therapy for SMA. Cell. 2017;170(1):5. doi: 10.1016/j.cell.2017.06.028. https://doi.org/10.1016/j.cell.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 93.Wood MJA, Talbot K, Bowerman M. Spinal muscular atrophy: antisense oligonucleotidetherapy opens the door to an integrated therapeutic landscape. Hum Mol Genet. 2017;26(R2):R151–9. doi: 10.1093/hmg/ddx215. https://doi.org/10.1093/hmg/ddx215. [DOI] [PubMed] [Google Scholar]
- 94.Sivanesan S, Howell MD, DiDonato CJ, Singh RN. Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Transl Neurosci. 2013;4:1–7. doi: 10.2478/s13380-013-0109-2. https://doi.org/10.2478/s13380-013-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beusch I, Barraud P, Moursy A, Cléry A, Allain FH. Tandem hnRNP A1 RNA recognitionmotifs act in concert to repress the splicing of survival motor neuron exon 7. Elife. 2017:6. doi: 10.7554/eLife.25736. pii:e25736. https://doi.org/10.7554/eLife.25736. [DOI] [PMC free article] [PubMed]
- 96.Singh NN, Hollinger K, Bhattacharya D, Singh RN. An antisense microwalk reveals criticalrole of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010;16:1167–81. doi: 10.1261/rna.2154310. https://doi.org/10.1261/rna.2154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–50. doi: 10.4161/rna.6.3.8723. https://doi.org/10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seo J, Ottesen EW, Singh RN. Antisense methods to modulate pre-mRNA splicing. Methods Mol Biol. 2014;1126:271–83. doi: 10.1007/978-1-62703-980-2_20. https://doi.org/10.1007/978-1-62703-980-2_20. [DOI] [PubMed] [Google Scholar]
- 99.Kiel JM, Seo J, Howell MD, Hsu WH, Singh RN, DiDonato CJ. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol Ther Nucleic Acids. 2014;3:e174. doi: 10.1038/mtna.2014.23. https://doi.org/10.1038/mtna.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Förch P, Puig O, Martínez C, Séraphin B, Valcárcel J. The splicing regulator TIA-1 interactswith U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21(24):6882–92. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klar J, Sobol M, Melberg A, Mäbert K, Ameur A, Johansson ACV, Feuk L, Entesarian M, Orlén H, Casar-Borota O, Dahl N. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat. 2013;34(4):572–7. doi: 10.1002/humu.22282. https://doi.org/10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- 102.Hirsch-Reinshagen V, Pottier C, Nicholson AM, Baker M, Hsiung GR, Krieger C, Sengdy P, Boylan KB, Dickson DW, Mesulam M, Weintraub S, Bigio E, Zinman L, Keith J, Rogaeva E, Zivkovic SA, Lacomis D, Taylor JP, Rademakers R, Mackenzie IRA. Clinical and neuropathological features of ALS/FTD with TIA1 mutations. Acta Neuropathol Commun. 2017;5(1):96. doi: 10.1186/s40478-017-0493-x. https://doi.org/10.1186/s40478-017-0493-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heck MV, Azizov M, Stehning T, Walter M, Kedersha N, Auburger G. Dysregulated expression of lipid storage and membrane dynamics factors in Tia1 knockout mouse nervous tissue. Neurogenetics. 2014;15(2):135–44. doi: 10.1007/s10048-014-0397-x. https://doi.org/10.1007/s10048-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Howell MD, Ottesen EW, Singh NN, Anderson RL, Seo J, Sivanesan S, Whitley EM, Singh RN. TIA1 is a gender-specific disease modifier of a mild mouse model of spinal muscular atrophy. Sci Rep. 2017a;7:18. doi: 10.1038/s41598-017-07468-2. https://doi.org/10.1038/s41598-017-07468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Díaz-Muñoz MD, Kiselev VY, Novère NL, Curk T, Ule J, Turner M. Tia1 dependent regulation of mRNA subcellular location and translation controls p53 expression in B cells. Nat Commun. 2017;8(1):530. doi: 10.1038/s41467-017-00454-2. https://doi.org/10.1038/s41467-017-00454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vanderweyde T, Apicco DJ, Youmans-Kidder K, Ash PEA, Cook C, Lummertz da Rocha E, Jansen-West K, Frame AA, Citro A, Leszyk JD, Ivanov P, Abisambra JF, Steffen M, Li H, Petrucelli L, Wolozin B. Interaction of tau with the RNA-binding Protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 2016;15(7):1455–66. doi: 10.1016/j.celrep.2016.04.045. https://doi.org/10.1016/j.celrep.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Howell MD, Ottesen EW, Singh NN, Anderson RL, Singh RN. Gender-specific amelioration of SMA phenotype upon disruption of a deep intronic structure by an oligonucleotide. Mol Ther. 2017b;25(6):1328–41. doi: 10.1016/j.ymthe.2017.03.036. https://doi.org/10.1016/j.ymthe.2017.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ronchi D, Previtali SC, Sora MGN, Barera G, Del Menico B, Corti S, Bresolin N, Comi GP. Novel splice-site mutation in SMN1 associated with a very severe SMA-I phenotype. J Mol Neurosci. 2015;56:212–5. doi: 10.1007/s12031-014-0483-4. https://doi.org/10.1007/s12031-014-0483-4. [DOI] [PubMed] [Google Scholar]
- 109.Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schöneborn S, Wienker T, Zerres K. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999;64(5):1340–56. doi: 10.1086/302369. https://doi.org/10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sheng-Yuan Z, Xiong F, Chen YJ, Yan TZ, Zeng J, Li L, Zhang YN, Chen WQ, Bao XH, Zhang C, Xu XM. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. Eur J Hum Genet. 2010;18(9):978–84. doi: 10.1038/ejhg.2010.54. https://doi.org/10.1038/ejhg.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vezain M, Gérard B, Drunat S, Funalot B, Fehrenbach S, N’Guyen-Viet V, Vallat JM, Frébourg T, Tosi M, Martins A, Saugier-Veber P. A leaky splicing mutation affecting SMN1 exon 7 inclusion explains an unexpected mild case of spinal muscular atrophy. Hum Mutat. 2011;32(9):989–94. doi: 10.1002/humu.21528. https://doi.org/10.1002/humu.21528. [DOI] [PubMed] [Google Scholar]
- 112.Osman EY, Washington CW, 3rd, Kaifer KA, Mazzasette C, Patitucci TN, Florea KM, Simon ME, Ko CP, Ebert AD, Lorson CL. Optimization of morpholino antisense oligonucleotides targeting the intronic repressor element1 in spinal muscular atrophy. Mol Ther. 2016;24(9):1592–601. doi: 10.1038/mt.2016.145. https://doi.org/10.1038/mt.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct Competition between hnRNP C and U2AF65 Protects the Transcriptome from the Exonization of Alu Elements. Cell. 2013;152(3):453–66. doi: 10.1016/j.cell.2012.12.023. https://doi.org/10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12(12):12. doi: 10.1186/gb-2011-12-12-236. https://doi.org/10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bouttier M, Laperriere D, Memari B, Mangiapane J, Fiore A, Mitchell E, Verway M, Behr MA, Sladek R, Barreiro LB, Mader S, White JH. Alu repeats as transcriptional regulatoryplatforms in macrophage responses to M-tuberculosis infection. Nucleic Acids Res. 2016;44(22):10571–87. doi: 10.1093/nar/gkw782. https://doi.org/10.1093/nar/gkw782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daniel C, Silberberg G, Behm M, Ohman M. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol. 2014;15(2):17. doi: 10.1186/gb-2014-15-2-r28. https://doi.org/10.1186/gb-2014-15-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh NN, Seo J, Rahn SJ, Singh RN. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. Plos One. 2012;7(11):17. doi: 10.1371/journal.pone.0049595. https://doi.org/10.1371/journal.pone.0049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12(7):1060–7. doi: 10.1101/gr.229302. https://doi.org/10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19(1):63–6. doi: 10.1038/ng0598-63. https://doi.org/10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 120.Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN. Oxidative stresstriggers body-wide skipping of multiple exons of the spinal muscular atrophy gene. PLoS One. 2016b;11(4):31. doi: 10.1371/journal.pone.0154390. https://doi.org/10.1371/journal.pone.0154390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oh JM, Di C, Venters CC, Guo J, Arai C, So BR, Pinto AM, Zhang Z, Wan L, Younis I, Dreyfuss G. U1 snRNP telescripting regulates a size-function-stratified human genome. Nat Struct Mol Biol. 2017;24:993. doi: 10.1038/nsmb.3473. https://doi.org/10.1038/nsmb.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aktaş T, Ilik IA, Maticzka D, Bhardwaj V, Rodrigues CP, Mittler G, Manke T, Backofen R, Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–9. doi: 10.1038/nature21715. https://doi.org/10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 123.Ottesen EW, Seo J, Singh NN, Singh RN. A multilayered control of the human Survival Motor Neuron gene expression by Alu elements. Front Microbiol. 2017;8:2252. doi: 10.3389/fmicb.2017.02252. https://doi.org/10.3389/fmicb.2017.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Acuña LIG, Kornblihtt AR. Long range chromatin organization: a new layer in splicing regulation? Transcription. 2014;5(3):e28726. doi: 10.4161/trns.28726. https://doi.org/10.4161/trns.28726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh NN, Howell MD, Singh RN. Transcriptional and splicing regulation of spinal muscular atrophy genes. In: Charlotte SJ, Paushkin S, Ko C-P, editors. Spinal muscular atrophy: disease mechanisms and therapy. Amsterdam: Elsevier Inc; 2016. [Google Scholar]
- 126.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4(2):e21. doi: 10.1371/journal.pbio.0040021. https://doi.org/10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31(2):73–6. doi: 10.1016/j.tibs.2005.12.003. https://doi.org/10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 128.Taube JR, Sperle K, Banser L, Seeman P, Cavan BC, Garbern JY, Hobson GM. PMD patient mutations reveal a long-distance intronic interaction that regulates PLP1/DM20 alternative splicing. Hum Mol Genet. 2014;23(20):5464–78. doi: 10.1093/hmg/ddu271. https://doi.org/10.1093/hmg/ddu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liang DM, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–47. doi: 10.1101/gad.251926.114. https://doi.org/10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]