Abstract
Background
OSA is a highly prevalent condition that is associated with a wide range of long-term morbidities including metabolic, cardiovascular, and cognitive alterations, possibly via activation of systemic inflammatory and oxidative stress pathways. Implementation of positive airway pressure (PAP) is the first-line treatment for OSA, as well as for obesity hypoventilation syndrome (OHS), its most severe phenotype. However, the molecular and cellular mechanisms underlying OHS-induced morbidities and their response to PAP treatment remain unclear, and could be mediated, in part, by OSA-induced epigenetic changes.
Methods
Blood was collected before starting PAP treatment (PRE group), as well as 6 weeks after PAP treatment (POST group) in 15 adult patients with OHS. DNA methylation profiles were studied by methylated DNA immunoprecipitation coupled to microarrays (MeDIP-chip) in six representative patients and further verified in a cohort of 15 patients by MeDIP-quantitative PCR.
Results
We identified 1,847 regions showing significant differential DNA methylation (P < .001; model-based analysis of tiling arrays score, > 4) between the groups. Analysis of biochemical pathways and gene networks demonstrated that differentially methylated regions were associated with immune responses, and particularly with mechanisms governing gene regulation by peroxisome proliferation-activated receptors (PPARs). Single-locus quantitative PCR analysis revealed that DNA methylation was increased at the PPAR-responsive elements (PPAREs) of eight genes in the post-treatment samples (PRE/POST fold changes: ABCA1, 3.11; ABCG1, 1.72; CD36, 5.04; FABP4, 2.49; HMOX, 2.74; NOS2, 7.78; PEPCK, 9.27; and ADIPOQ, 1.73), suggesting that PAP treatment leads to an increase in DNA methylation at PPAREs, possibly affecting the binding of the PPAR-γ complex and downstream gene expression.
Conclusions
Our work provides initial evidence of epigenetic regulation particularly involving metabolic pathways in patients with OHS who are responsive to PAP treatment.
Key Words: DNA methylation, monocytes, OSA, peroxisome proliferation-activated receptor γ, positive air pressure
Abbreviations: AHI, apnea-hypopnea index; DMR, differentially methylated region; HTN, hypertension; MeDIP, methylated DNA immunoprecipitation; OHS, obesity hypoventilation syndrome; PAP, positive airway pressure; PPAR, peroxisome proliferation-activated receptor; PPARE, peroxisome proliferation-activated receptor-responsive element; PPARG, peroxisome proliferation-activated receptor γ
We postulate that differentially methylated regions in blood monocytes may serve as potential biomarkers in clinical practice for monitoring the disease and the associated comorbidities.
The prevalence of obesity hypoventilation syndrome (OHS) is likely to increase with high rates of severe obesity.1 OHS is defined as daytime hypercapnia (daytime Paco2 ≥ 45 mm Hg) in an obese patient with sleep-disordered breathing in the absence of any other cause of hypoventilation.2 More than 90% of patients with OHS have concomitant OSA. Approximately 9% to 20% of patients with OSA have OHS.3
Both OHS and OSA have been associated with long-term morbidities including metabolic, cardiovascular, and cognitive alterations, ultimately increasing mortality and health-care costs.4 Previous studies indicated that chronic intermittent hypoxia, a hallmark of OSA, induces hyperlipidemia and insulin resistance as well as marked cellular inflammatory changes that account for a substantial proportion of OSA morbidities.5, 6, 7, 8, 9, 10, 11 Thus, improved understanding of the cellular and molecular mechanisms by which OSA affects physiological functions, and leads to the development of the associated morbidities, could open new venues for detecting, preventing, and treating them.
The current “gold standard” treatment for OSA and OHS consists in the application of positive airway pressure (PAP) during sleep.12, 13, 14, 15 Indeed, there is now ample evidence indicating the favorable impact of PAP treatment on the outcome of nearly all of the associated morbidities, such as neurocognitive changes,16, 17 cardiovascular diseases,18, 19 blood pressure,20, 21 serum lipid profiles,22 and insulin sensitivity and glucose metabolism.23, 24, 25, 26, 27 However, the cellular and molecular mechanisms that underlie the beneficial effects of PAP treatment remain to be determined.
Changes in epigenetic mechanisms may provide a plausible explanation of the beneficial effects of PAP treatment on OSA. For example, altered epigenetic regulation has been implicated in the emergence of metabolic dysfunction and atherosclerosis, two major morbidities of OSA.28, 29 Furthermore, considering the prominent role of monocytes and macrophages in OSA-induced morbidities,30, 31, 32, 33, 34, 35 it is noteworthy that monocyte/macrophage phenotype and function depend highly on the microenvironment, and that their activation, polarization, and inactivation involve transcriptional regulation and chromatin remodeling.36 In this context, epigenetic mechanisms have been implicated in the polarization of macrophages involved in atherogenesis and tissue insulin resistance.29, 37, 38 Work from our laboratory has shown that exposures to perturbations mimicking OSA in mice lead to insulin resistance and a proinflammatory macrophage phenotype, potentially involving epigenetic modifications.10, 36, 39, 40, 41
On the basis of the aforementioned considerations and since epigenetic signatures also hold promise as potential biomarkers that can be translated into clinical practice,42 we hypothesized that epigenetic changes are present in OHS, and therefore conducted epigenomic profiling of blood monocytes in patients with OHS before and after PAP treatment. We identified regions of differential DNA methylation that revealed specific genomic loci and candidate biochemical pathways that appear to be epigenetically dysregulated in hypercapnic OSA and targeted by PAP treatment.
Materials and Methods
A detailed description of the materials and methods for this study is provided as supplemental material (e-Appendix 1).
Patient Population and Sample Preparation
The patient population consisted of 15 adult patients who had received a diagnosis of OHS. Apnea-hypopnea index (AHI) testing was done before and 6 weeks after initiation of treatment, by overnight in-laboratory polysomnography. All the participants provided written informed consent and the research protocol was approved by the research ethics board at the University of Chicago (protocol 10-702-A-CR004). For each patient, blood was collected before starting PAP treatment (PRE group), as well as 6 weeks after PAP treatment (POST group). Peripheral blood mononuclear cells were isolated by gradient centrifugation, the CD14+ monocytes fraction was separated, and DNA was isolated.
Microarray-Based DNA Profiling
Fragmented DNA was immunoprecipitated with an antibody against 5-methylcytosine (AnaSpec, Inc.), according to the methylated DNA immunoprecipitation (MeDIP) protocol.43 Immunoprecipitated DNA was amplified using a adaptor-mediated PCR strategy, as described elsewhere,44 and target was prepared and hybridized on Affymetrix GeneChip human promoter array 1.0R (Affymetrix) and scanned. Microarray data were processed as previously described.45 Data preprocessing consisted of probe sequence adjustment, robust multichip averaging (RMA)46 background correction, quantile normalization, and log2 transformation. Regions of differential DNA methylation were identified using the model-based analysis of tiling arrays47 algorithm. Networks and pathways significantly enriched in the genes of interest were identified through Ingenuity Pathway Analysis (IPA) (Ingenuity Systems). The significance of the enrichment was calculated by Fisher’s exact test (right-tailed), with a cutoff P value = .05. The data set is available in the NCBI Gene Expression Omnibus (GEO) repository (accession number, GSE73053).
Single-Locus Analysis
Microarray data were verified by single-locus analysis using SYBR green-based real-time PCR analysis of the MeDIP DNA. Specific primers for the candidate loci are provided as supplemental material (e-Table 1).
Results
DNA Methylation Profiling
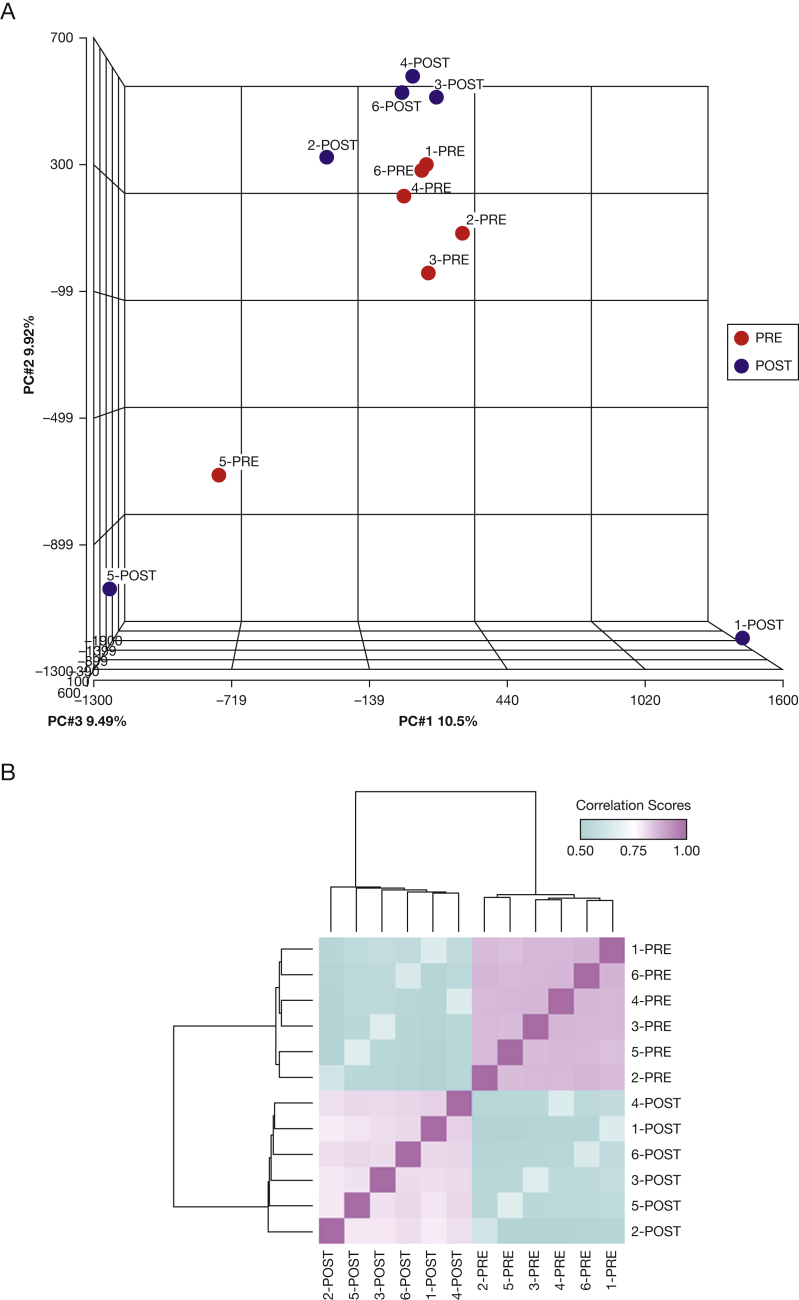
We selected a representative subgroup of six patients with OHS who showed the largest improvements in AHI before (PRE group) and after (POST group) PAP treatment, out of 15 patients severely affected with OHS and included in the study (Table 1 and e-Table 2). Mean treatment adherence values for all patients who provided studied samples (n = 15) and for those who provided samples selected for the microarray study (n = 6) were as follows: number of days used, 5.45 ± 1.37 and 5.90 ± 1.04 d/wk, respectively; percentage of days used, 88.92 ± 15.32% and 97.45 ± 4.09%, respectively; and percentage of days used for more than 4 h: 67.71 ± 25.04% and 81.05 ± 17.64%, respectively. We assessed DNA methylation profiles in blood monocytes by MeDIP and interrogated large-scale profiles using promoter arrays. All microarrays in the data set (n = 12) passed the technical quality controls and were included in the analyses (e-Fig 1). Principal component analysis (Fig 1A) showed two clearly independent clusters containing the PRE (red) and POST (blue) samples. One sample pair corresponding to the same patient (5-PRE and 5-POST) clustered separately from the rest of the samples. Post hoc review of demographic and clinical records (e-Table 2) showed that this patient was the youngest (41 years old) and the only subject among the six subjects who had received a diagnosis of congestive heart failure. Furthermore, one post-treatment sample (1-POST) was clearly separated from the rest of the samples. The patient providing this sample was the only current cigarette smoker and was also the only patient among those in the microarray study in whom type 2 diabetes mellitus was diagnosed. Despite the intragroup variation, DNA methylation profiles distinguished samples in the PRE and POST groups (Fig 1B).
Table 1.
Characteristics of Patients Providing Samples for Full Data Set and Selected Samples for Microarray Analysis
| Characteristic | Full Sample Set (n = 15) | Samples in Microarray Study (n = 6) |
|---|---|---|
| Age, y | 50.9 ± 10.7 | 52.3 ± 9.1 |
| Sex | 6 male/9 female | 1 male/5 female |
| AHI, events/h TST | ||
| Pretreatment (PRE group) | 85.0 ± 30.9 | 91 ± 28.5 |
| Post-treatment (POST group) | 11.3 ± 11.4 | 3.8 ± 3.3 |
| Difference (PRE − POST) | 73.6 ± 31.2 | 87.3 ± 28.2 |
| 4% ODI, events/h TST | ||
| Pretreatment (PRE group) | 95.73 ± 35.13 | 108.22 ± 34.36 |
| Post-treatment (POST group) | 12.37 ± 13.99 | 4.43 ± 4.17 |
| Difference (PRE − POST) | 83.35 ± 36.39 | 103.78 ± 33.90 |
| T90, min | ||
| Pretreatment (PRE group) | 220.61 ± 95.40 | 221.60 ± 82.72 |
| Post-treatment (POST group) | 32.11 ± 37.70 | 7.17 ± 9.59 |
| Difference (PRE − POST) | 188.51 ± 104.28 | 214.43 ± 87.76 |
| Spo2 nadir, % | ||
| Pretreatment (PRE group) | 56.90 ± 8.12 | 57.78 ± 4.96 |
| Post-treatment (POST group) | 78.81 ± 10.91 | 83.45 ± 10.45 |
| Difference (PRE − POST) | 21.91 ± 10.82 | 25.67 ± 8.93 |
| Paco2, mm Hg | ||
| Pretreatment (PRE group) | 50.91 ± 7.11 | 48.35 ± 2.48 |
| Post-treatment (POST group) | 42.82 ± 6.58 | 41.03 ± 4.74 |
| Difference (PRE − POST) | 8.09 ± 4.63 | 7.32 ± 3.20 |
| BMI, kg/m2 | ||
| Pretreatment (PRE group) | 49.38 ± 10.23 | 50.70 ± 14.33 |
| Post-treatment (POST group) | 48.36 ± 8.31 | 48.97 ± 10.57 |
| Difference (PRE − POST) | 1.02 ± 2.85 | 1.73 ± 4.50 |
| Smoking status | 3 crt, 3 pst, 9 nvr | 1 crt, 1 pst, 4 nvr |
| Hypertension | 13/15 patients (86.7%) | 4/6 patients (66.7%) |
| Type 2 diabetes mellitus | 5/15 patients (33.3%) | 1/6 patients (16.7%) |
| Congestive heart failure | 6/15 patients (40.0%) | 1/6 patients (16.7%) |
| Dyslipidemia | 4/15 patients (26.7%) | 1/6 patients (16.7%) |
Data are shown as means ± SD. crt = current smoker; nvr = never smoker; ODI = oxygen desaturation index; POST group = patients from whom blood was collected 6 weeks after positive airway pressure treatment; PRE group = patients from whom blood was collected before starting positive airway pressure treatment; pst = past smoker; Spo2 = oxygen saturation as determined by pulse oximetry; T90 = time spent at less than 90% oxygen saturation.
Figure 1.
Differential DNA methylation profiles in PRE and POST groups. A, Principal component analysis was performed using the full microarray data from PRE samples (red points) and POST samples (blue points). Five samples from the same group clustered together, with the exception of one patient (Patient 5), whose PRE and POST samples clustered separately from the rest. Three principal components determine sample clustering: PC1 (10.5%, x axis), PC2 (9.92%, y axis), and PC3 (9.49%, z axis). B, Pairwise sample correlation was performed using significant DMRs (P < .001 and MAT score > 4). Bidimensional unsupervised clustering was performed in samples from the PRE and POST groups. Correlation coefficients are shown as a color gradient ranging from light blue (0.5) over white (0.75) to light pink (1.0). DMR = differentially methylated region; MAT = model-based analysis of tiling arrays; PRE group = blood samples collected from patients before starting positive airway pressure treatment; POST group = blood samples collected from patients 6 weeks after positive airway pressure treatment.
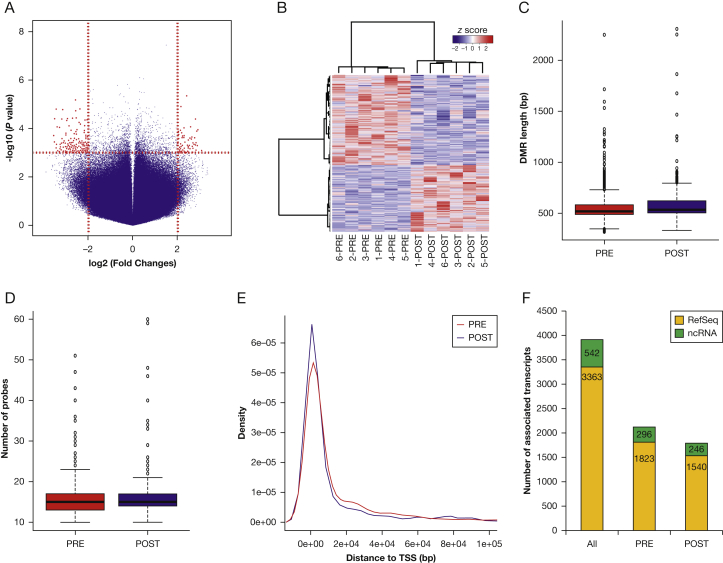
We identified 1,847 significant differentially methylated regions (DMRs) between the PRE and POST groups (P < .001 and absolute model-based analysis of tiling arrays score > 4; paired t test) (Fig 2A, e-Table 3). Unsupervised clustering analysis showed that PRE and POST group samples clustered together, based solely on the DMRs (Fig 2B). DMRs with higher methylation in the POST group were larger (PRE, 553.48 ± 5.39 bases; POST, 582.32 ± 4.78 bases; P = 6.5 × 10−5; t = –4.01; 95% CI , –42.96 to –14.70; paired t test) (Fig 2C) and contained more probes (POST, 16.09 ± 0.15 probes/region; PRE, 15.33 ± 0.13 probes/region; P = 7.4 × 10−5; t = –3.97; 95% CI , –1.14 to –0.38; paired t test) (Fig 2D) than those with higher methylation in the PRE group. We did not detect significant differences in the distance of the DMRs in each group to the transcription start site of the nearest gene (mean distance: POST, 31,575.72 ± 2,473.07 bp; PRE, 31,280 ± 2,729.61 bp; P = .949; t = 0.06; 95% CI, –8,885.27 to 9,476.06; paired t test) (Fig 2E).
Figure 2.
Characterization of differential DNA methylation between PRE and POST groups. A, Volcano plot of microarray data at probe level. The x axis represents the magnitude of the difference in signal intensity between the PRE and POST groups for each probe in the microarray, expressed as fold changes in log2 scale. Probes with increased microarray signals in POST and PRE groups had positive and negative values on the x axis, respectively. The y axis represents the significance of the difference in signal intensity between the PRE and POST groups for each probe in the microarray, expressed as the –log10-transformed P values. The vertical dashed red lines depict the cutoff values for the fold changes [log2(4) = 2]. Probes showing significant differences (P < .001 and fold change > 4) are shown in red. B, DNA methylation differences in the top 100 DMRs distinguished between the PRE and POST groups. Samples belonging to the same group clustered together by unsupervised clustering based solely on the DNA methylation differences of those 100 DMRs. Samples are accommodated in columns and DMRs in rows in the matrix. DNA methylation differences (expressed as z score) between the groups for each DMR in each sample are represented by a color gradient ranging from blue (negative z scores, meaning higher DNA methylation in the PRE group than in the POST group) through white (no differences) to red (positive z scores, meaning higher DNA methylation in the POST group than in the PRE group). C and D, DMRs with higher DNA methylation in the POST group were longer (C) and contained more probes (D) than those with higher DNA methylation in the PRE group (P < .0001, paired t test). E, Distance to TSS did not differ significantly between DMRs more highly methylated in the POST and PRE groups (P = .949; paired t test). The distance from the beginning of each region to the closest TSS is shown on the x axis. Red and blue lines represent the PRE and POST groups, respectively. F, Association with RefSeq (yellow column portions) and ncRNAs (green column portions) did not significantly differ between DMRs with higher DNA methylation in the POST or PRE groups (P = .889; Fisher’s exact test). ncRNAs = noncoding RNAs; TSS = transcription start site. See Figure 1 legend for expansion of other abbreviations.
Gene-Associated DMRs
DMRs located within a 2-kb range surrounding the transcription start site are usually associated with differences in gene expression of the cognate gene.48, 49 Out of the full list of 1,847 DMRs, we selected 855 DMRs that were located within such segments (e-Table 4). These DMRs were associated with 3,905 annotated transcripts (Genome Reference Consortium human genome [build 37]/human genome version 19 [GRCh37/hg19] assembly) including 3,363 RefSeq (Reference Sequence) genes and 542 noncoding RNAs (Fig 2F). We did not find significant differences in the number of DMR-associated RefSeq genes and noncoding RNAs showing higher DNA methylation in the PRE and POST groups (P = .889; OR, 1.02; 95% CI, 0.84-1.22; Fisher’s exact test).
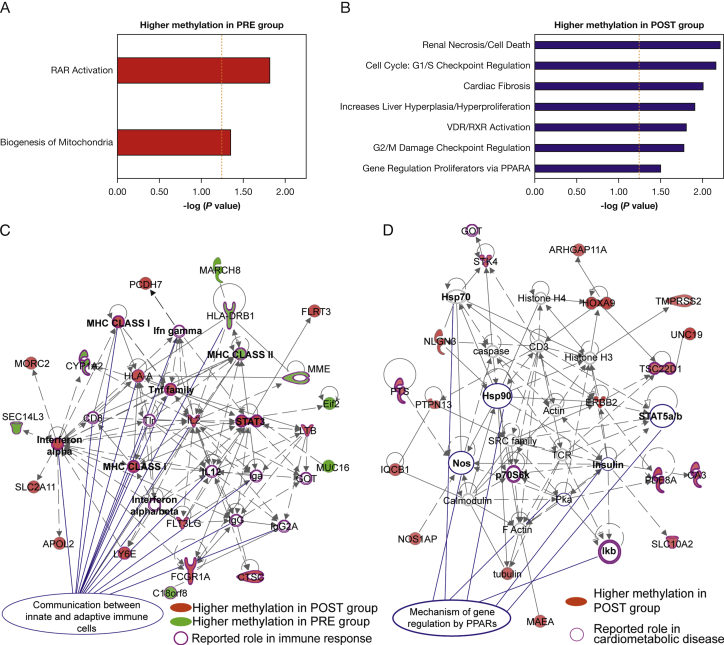
Pathways associated with retinoic acid receptor (RAR) activation (P = .015; Fisher’s exact test, right-tailed) and biogenesis of mitochondria (P = .043) were overrepresented among DMRs with higher DNA methylation in the PRE group (Fig 3A, e-Table 5). In contrast, pathways related to cell cycle and proliferation, as well as vitamin D receptor/retinoid X receptor (VDR/RXR) activation (P = .015) and regulation by peroxisome proliferation-activated receptor-α (P = .033), were overrepresented among DMRs with higher DNA methylation in the POST group (Fig 3B, e-Table 5). Importantly, unsupervised gene network analysis showed that DMRs were associated with molecules with reported roles in immune response, particularly for the communication between immune cells (interferon-α, -β, and -γ; MHC class I and II; tumor necrosis factor family, etc.) (Fig 3C). Furthermore, we found that several DMRs with higher methylation in the POST group corresponded to molecules involved in mechanisms of gene regulation by peroxisome proliferation-activated receptors (PPARs) and associated with cardiometabolic diseases (Fig 3D).
Figure 3.
Pathway and biological processes associated with DMRs. Biologically relevant gene interaction networks were identified by statistically significant overrepresentation in genes associated with the DMRs. Pathways and biological processes associated with DMRs show higher DNA methylation in the PRE group (red bars, panel A) and POST group (blue bars, panel B). Vertical orange dashed bars depicts the significance cutoff value for the overrepresentation test [–log10(P = .05) = 1.3; hypergeometric test). C, DMR-associated gene networks. Genes associated with DMRs with higher DNA methylation in POST and PRE samples are shown in red and green, respectively. Molecules with a reported function in immune response are circled in purple. Molecules with a role in communication between immune cells are indicated with blue lines. D, Gene network corresponding to mechanisms of gene regulation by peroxisome proliferation-activated receptors (PPARs) overrepresented in DMRs with high DNA methylation in the POST group (shown in red). Molecules with a reported role in cardiometabolic diseases are circled in purple. Molecules associated with PPAR pathways are indicated with blue lines. PPARA = peroxisome proliferation-activated receptor α; RAR = retinoic acid receptor; RefSeq = Reference Sequence; VDR/RXR = vitamin D receptor/retinoid X receptor. See Figure 1 legend for expansion of other abbreviations.
Single-Locus Analysis
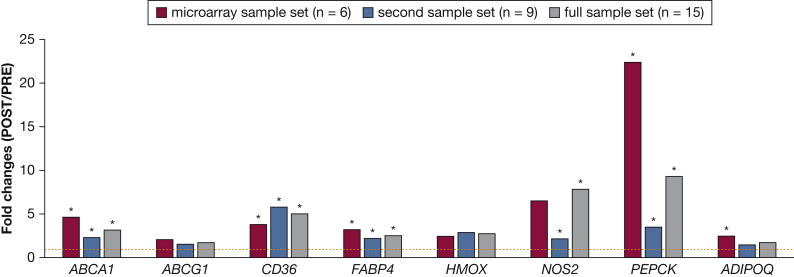
Next, we assessed DNA methylation changes that occur following PAP treatment in eight genes previously identified as PPAR targets and that have a reported role in inflammation and macrophage biology (e-Table 1). We defined MeDIP-quantitative PCR-based assays for these genes containing PPAR-responsive elements (PPAREs) and interrogated samples from patients studied by microarray analysis (n = 6) as well as in a second sample set (n = 9) (e-Table 2). DNA methylation was increased (fold changes [PRE/POST], > 1) in all genes in the POST group in both sample sets (Fig 4), suggesting that PAP treatment leads to an increase in DNA methylation at PPAREs, possibly affecting the binding of the peroxisome proliferation-activated receptor γ (PPARG) complex and downstream gene expression.
Figure 4.
Single-locus DNA methylation analysis in peroxisome proliferation-activated receptor-responsive elements (PPAREs) of candidate genes. Single-locus MeDIP-quantitative PCR results for the samples in the microarray study (red columns), the second sample set (blue columns), and the full sample set (gray columns). Quantitative PCR assays were defined in PPAREs of eight peroxisome proliferation-activated receptor γ target genes. Fold changes (POST/PRE) were calculated as the ratio of the percentage of INPUT recovery in each group and are shown on the y axis as means. Error bars correspond to the SEM. *P < .05, t test. MeDIP = methylated DNA immunoprecipitation. See Figure 1 legend for expansion of other abbreviations.
Demographic and Clinical Variables
We studied whether demographic and clinical variation in the patients who provided our sample set influences the differential DNA methylation associated with PAP treatment at the eight PPAREs. We evaluated continuous variables: age, pretreatment AHI, differences in AHI at pre- and post-PAP treatment, PAP usage (mean of days used, percentage of days used, and percentage of days used for more than 4 h), and pre- and post-PAP treatment values for 4% oxygen desaturation index, time spent at less than 90% oxygen saturation, peripheral capillary oxygen saturation nadir, Paco2, and body mass index; as well as other discrete variables such as treatment modality, sex, smoking status, and diagnosis of hypertension, type 2 diabetes mellitus, congestive heart failure, or dyslipidemia.
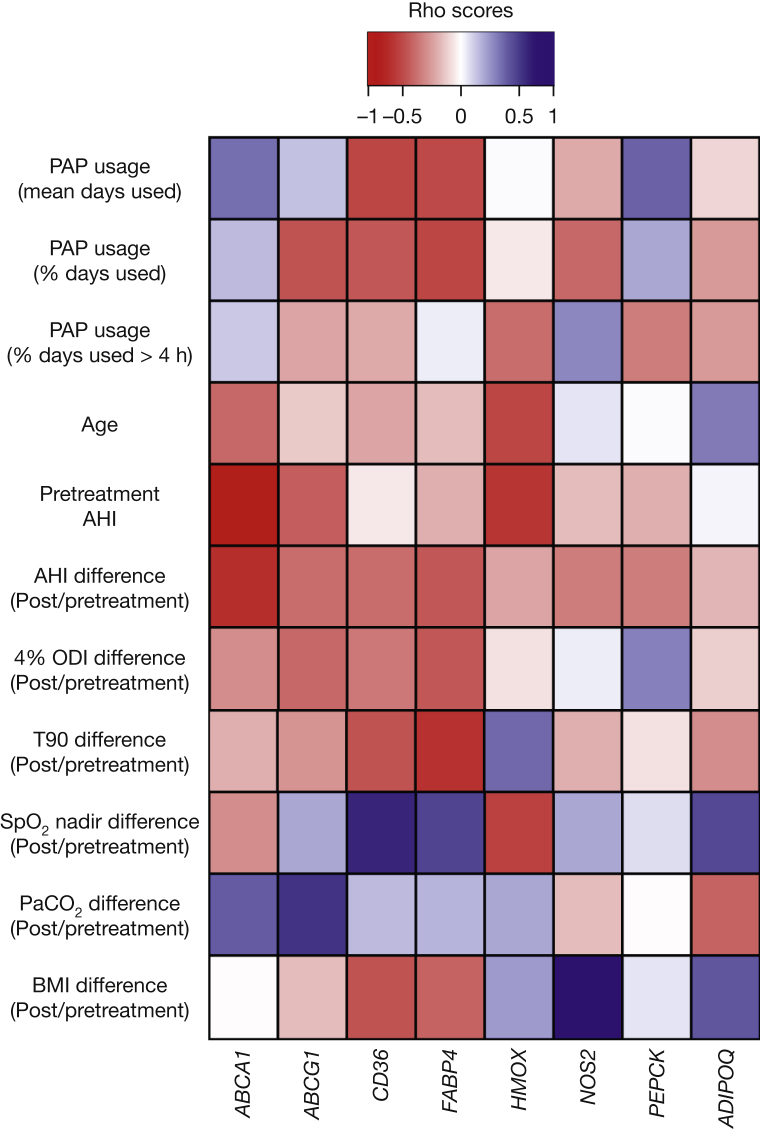
Figure 5 illustrates the results of the correlation analysis between the DNA methylation fold changes (PRE/POST) and the continuous variables. We detected a significant negative correlation (P < .05, Spearman’s rank correlation test) at the PPARE of the ABCA1 gene and the initial AHI value (ρ = −0.797, P = 1.8 × 10−3), as well as the differences in AHI at pre- and post-PAP treatment (ρ = –0.621, P = .027). In addition, we found a significant negative difference at the PPARE of the FABP4 gene and the pre- and post-PAP treatment differences in 4% oxygen desaturation index (ρ = –0.604, P = .032) and time spent at less than 90% oxygen saturation (ρ = –0.576, P = .042) values, as well as a positive correlation at the PPARE of the NOS2 gene with the pre- and post-PAP treatment differences in body mass index (ρ = 0.776, P = 1.7 × 10−3).
Figure 5.
Association with demographic and clinical variables. Matrix shows the correlation coefficients with continuous variables. Spearman’s correlation test was performed for each variable and the fold changes (POST/PRE) for DNA methylation in each target gene. ρ correlation coefficients are shown as a color gradient from red (negative correlation) through white (no correlation) to blue (positive correlation). AHI = apnea-hypopnea index; ODI = oxygen desaturation index; PAP = positive airway pressure; Spo2 = oxygen saturation as determined by pulse oximetry; T90 = time spent at less than 90% oxygen saturation.
Among discrete variables, we detected a significant association with hypertension (HTN) at the PPARE of the FABP4 gene. The medians of the HTN and no-HTN groups were 1.73 and 0.02, respectively (mean ranks: HTN = 8, no HTN = 4.5; U = 22, Z = –2.17, P = .025, r = 0.61) (e-Fig 2).
Discussion
This study shows that in a cohort of patients severely affected with OHS, extensive changes in DNA methylation occur following implementation of PAP therapy, and that among the PAP-responsive gene pathways, PPAR-related genes appear to be particularly responsive.
Before we discuss the potential implications of our findings, several methodological considerations deserve comment. First, we selected a cohort of patients with particularly severe hypercapnic OSA (OHS) as opposed to patients with eucapnic OSA, aiming to maximize the probability of positive findings in both epigenetic profiles and PAP-associated changes. Second, this study did not aim to address the variability of changes in a large population, or to identify and validate specific biomarkers corresponding to favorable response to treatment. Instead, the aim was to establish a proof of concept for the presence of epigenetic changes in OHS, whereby treatment of OSA by PAP and consequent improvements in the severity of OSA would elicit changes in epigenetic marks, and provide initial conceptual hypotheses that can be tested in future studies. Third, the lack of a control group (ie, nontreated patient sample) does not allow us to completely rule out the possibility of other effects acting in parallel with PAP treatment. Considering the large degree of phenotypic variance in both the clinical presentation of OSA and the risk for OSA-associated morbidities,50, 51, 52, 53 the current results would justify more detailed assessments of specific subphenotypes to gain increased insights into the potential pathways involved, and the role of epigenetics in this context.
Inflammation is one of the major cellular mechanisms affected by OSA,54 and monocyte/macrophage polarization is clearly affected by this disease.30, 31, 32, 33, 34, 35, 55 Analysis of blood monocytes provides a low-invasive approach to study cells that participate in the inflammatory response. Previous studies have shown that monocytes are activated in both OSA and other sleep disorders,33, 56, 57, 58 representing an appropriate cell population to study the epigenetic regulation of OSA-associated inflammatory responses. Further studies are, however, needed to determine specific epigenetic profiles in resident and bone marrow-derived tissue macrophages and circulating monocytes, and how each of these unique myeloid cell populations is altered by OSA and PAP treatment.
We found that pathways associated with biogenesis of mitochondria were overrepresented in regions with decreased DNA methylation in the POST group, suggesting potential activation on PAP treatment (Fig 3A). Mitochondrial dysfunction was reported in whole blood samples from patients with OSA, closely correlated with the degree of oxygen desaturation or severity of OSA and potentially reflecting excessive oxidative stress.59 Concordant with these findings, our results may suggest an epigenetically mediated mechanism by which PAP treatment attenuates the oxidative stress levels observed in patients with OHS.30 Moreover, PPAR pathways were overrepresented in regions with higher DNA methylation in the POST group (Figs 3B, 3D). PPARs are ligand-activated transcription factors that have a major role in lipid metabolism and inflammation.60, 61 In particular, the PPARG pathway represents one of the major molecular and biochemical pathways leading the transition from M2-like phenotypes to M1 in macrophages.36, 62, 63 Here, we observed increased DNA methylation after PAP treatment at PPAREs of eight PPARG-targeted genes (Fig 4). Thus, our microarray-based and single-locus results suggest that epigenetic (dys)regulation of PPAR pathways in macrophages may operate as a major component of OSA, leading to inflammation and macrophage activation. Moreover, we report here that PAP treatment leads to changes in the epigenetic profiles of the genes associated with the PPAR pathways, which may account for the reductions in inflammation occurring in patients with OHS who received this treatment.64 Further studies are warranted to unravel the molecular mechanisms involved in the epigenetic regulation of PPAR pathways in macrophages, and how they are affected by OSA and PAP treatment.
Variation of DNA methylation profiles among the samples within the same group was a common feature in both the microarray-based and single-locus analyses. Since the sample size in this study was limited, we could not formally stratify the patients according to clinical and demographic variables. Instead, we sought to identify associations that can influence the extent of variation in DNA methylation profiles on PAP treatment. Interestingly, we found that DNA methylation variation in only one gene (ABCA1) correlated significantly (P < .05, Spearman’s rank test) with initial AHI value and AHI pre/post treatment variation (Fig 5). Whereas initial AHI represents the severity of OSA before treatment, AHI pre-/post-treatment is a potential measure of PAP efficacy. Even though this is clearly beyond the scope of the present study, we hypothesize that assessing DNA methylation in blood monocytes may provide useful biomarker signatures that allow prediction of the efficiency of PAP treatment and its outcomes.53, 65 Longitudinal studies assessing DNA methylation of this and other genes in larger populations of patients with OSA will be required to formally discover and validate such potentially predictive markers.
Our findings suggest that PAP may act on metabolic function via epigenetic processes, specifically DNA methylation at PPAREs.66, 67, 68, 69, 70, 71
Acknowledgments
Author contributions: D. G. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis, including and especially any adverse effects. R. C. conceived the study with D. G., carried out the epigenetic studies, participated in data analysis and interpretation, and drafted the manuscript. C. Z., R. B., and J. A. were responsible for microarray data analysis. A. K. performed experiments and B. M. was involved in patient recruitment and sleep study interpretations. D. G. conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: B. M. has served as a consultant to Philips/Respironics and has received research support from Philips/Respironics. He has also received honorarium from Zephyr Medical Technologies and has served on the advisory board of Itamar Medical. None declared (R. C., C. Z., R. B., J. A., A. K., D. G.).
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: This work was supported by the Herbert T. Abelson Chair in Pediatrics and by National Institutes of Health [Grant R01HL119161 to B. M.].
Supplementary Data
References
- 1.Ogden C.L., Carroll M.D., Flegal K.M. Prevalence of obesity in the United States. JAMA. 2014;312(2):189–190. doi: 10.1001/jama.2014.6228. [DOI] [PubMed] [Google Scholar]
- 2.Mokhlesi B. Obesity hypoventilation syndrome: a state-of-the-art review. Respir Care. 2010;55(10):1347–1362. [PubMed] [Google Scholar]
- 3.Balachandran J.S., Masa J.F., Mokhlesi B. Obesity hypoventilation syndrome: epidemiology and diagnosis. Sleep Med Clin. 2014;9(3):341–347. doi: 10.1016/j.jsmc.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young T., Peppard P.E., Gottlieb D.J. Epidemiology of obstructive sleep apnea: a population health perspective. Am J Respir Crit Care Med. 2002;165(9):1217–1239. doi: 10.1164/rccm.2109080. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Savransky V., Nanayakkara A., Smith P.L., O’Donnell C.P., Polotsky V.Y. Hyperlipidemia and lipid peroxidation are dependent on the severity of chronic intermittent hypoxia. J Appl Physiol (1985) 2007;102(2):557–563. doi: 10.1152/japplphysiol.01081.2006. [DOI] [PubMed] [Google Scholar]
- 6.Gharib S.A., Khalyfa A., Abdelkarim A. Intermittent hypoxia activates temporally coordinated transcriptional programs in visceral adipose tissue. J Mol Med (Berl) 2012;90(4):435–445. doi: 10.1007/s00109-011-0830-7. [DOI] [PubMed] [Google Scholar]
- 7.Drager L.F., Li J., Reinke C., Bevans-Fonti S., Jun J.C., Polotsky V.Y. Intermittent hypoxia exacerbates metabolic effects of diet-induced obesity. Obesity (Silver Spring) 2011;19(11):2167–2174. doi: 10.1038/oby.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun J.C., Shin M.K., Yao Q. Acute hypoxia induces hypertriglyceridemia by decreasing plasma triglyceride clearance in mice. Am J Physiol Endocrinol Metab. 2012;303(3):E377–E388. doi: 10.1152/ajpendo.00641.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulain L., Thomas A., Rieusset J. Visceral white fat remodelling contributes to intermittent hypoxia-induced atherogenesis. Eur Respir J. 2014;43(2):513–522. doi: 10.1183/09031936.00019913. [DOI] [PubMed] [Google Scholar]
- 10.Carreras A., Zhang S.X., Almendros I. Resveratrol attenuates intermittent hypoxia-induced macrophage migration to visceral white adipose tissue and insulin resistance in male mice. Endocrinology. 2015;156(2):437–443. doi: 10.1210/en.2014-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carreras A., Kayali F., Zhang J., Hirotsu C., Wang Y., Gozal D. Metabolic effects of intermittent hypoxia in mice: steady versus high-frequency applied hypoxia daily during the rest period. Am J Physiol Regul Integr Comp Physiol. 2013;303(7):R700–R709. doi: 10.1152/ajpregu.00258.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan C.E., Berthon-Jones M., Issa F.G. Nocturnal nasal-airway pressure for sleep apnea. N Engl J Med. 1983;309(2):112. doi: 10.1056/NEJM198307143090215. [DOI] [PubMed] [Google Scholar]
- 13.Stasche N. Selective indication for positive airway pressure (PAP) in sleep-related breathing disorders with obstruction. GMS Curr Top Otorhinolaryngol Head Neck Surg. 2006;5:Doc06. [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein L.J., Kristo D., Strollo P.J. Clinical guideline for the evaluation, management and long-term care of obstructive sleep apnea in adults. J Clin Sleep Med. 2009;5(3):263–276. [PMC free article] [PubMed] [Google Scholar]
- 15.Spicuzza L., Caruso D., Di Maria G. Obstructive sleep apnoea syndrome and its management. Ther Adv Chronic Dis. 2015;6(5):273–285. doi: 10.1177/2040622315590318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aloia M.S., Ilniczky N., Di Dio P., Perlis M.L., Greenblatt D.W., Giles D.E. Neuropsychological changes and treatment compliance in older adults with sleep apnea. J Psychosom Res. 2003;54(1):71–76. doi: 10.1016/s0022-3999(02)00548-2. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman M.E., Arnedt J.T., Stanchina M., Millman R.P., Aloia M.S. Normalization of memory performance and positive airway pressure adherence in memory-impaired patients with obstructive sleep apnea. Chest. 2006;130(6):1772–1778. doi: 10.1378/chest.130.6.1772. [DOI] [PubMed] [Google Scholar]
- 18.Marin J.M., Carrizo S.J., Vicente E., Agusti A.G. Long-term cardiovascular outcomes in men with obstructive sleep apnoea-hypopnoea with or without treatment with continuous positive airway pressure: an observational study. Lancet. 2005;365(9464):1046–1053. doi: 10.1016/S0140-6736(05)71141-7. [DOI] [PubMed] [Google Scholar]
- 19.Bradley T.D., Floras J.S. Obstructive sleep apnoea and its cardiovascular consequences. Lancet. 2009;373(9657):82–93. doi: 10.1016/S0140-6736(08)61622-0. [DOI] [PubMed] [Google Scholar]
- 20.Montesi S.B., Edwards B.A., Malhotra A., Bakker J.P. The effect of continuous positive airway pressure treatment on blood pressure: a systematic review and meta-analysis of randomized controlled trials. J Clin Sleep Med. 2012;8(5):587–596. doi: 10.5664/jcsm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iftikhar I.H., Valentine C.W., Bittencourt L.R. Effects of continuous positive airway pressure on blood pressure in patients with resistant hypertension and obstructive sleep apnea: a meta-analysis. J Hypertens. 2014;32(12):2341–2350. doi: 10.1097/HJH.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadeem R., Singh M., Nida M. Effect of CPAP treatment for obstructive sleep apnea hypopnea syndrome on lipid profile: a meta-regression analysis. J Clin Sleep Med. 2014;10(12):1295–1302. doi: 10.5664/jcsm.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorkova Z., Petrasova D., Molcanyiova A., Popovnakova M., Tkacova R. Effects of continuous positive airway pressure on cardiovascular risk profile in patients with severe obstructive sleep apnea and metabolic syndrome. Chest. 2008;134(4):686–692. doi: 10.1378/chest.08-0556. [DOI] [PubMed] [Google Scholar]
- 24.Chen L., Pei J.H., Chen H.M. Effects of continuous positive airway pressure treatment on glycaemic control and insulin sensitivity in patients with obstructive sleep apnoea and type 2 diabetes: a meta-analysis. Arch Med Sci. 2014;10(4):637–642. doi: 10.5114/aoms.2014.44854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jullian-Desayes I., Joyeux-Faure M., Tamisier R. Impact of obstructive sleep apnea treatment by continuous positive airway pressure on cardiometabolic biomarkers: a systematic review from sham CPAP randomized controlled trials. Sleep Med Rev. 2015;21:23–38. doi: 10.1016/j.smrv.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Pamidi S., Wroblewski K., Stepien M. Eight hours of nightly continuous positive airway pressure treatment of obstructive sleep apnea improves glucose metabolism in patients with prediabetes: a randomized controlled trial. Am J Respir Crit Care Med. 2015;192(1):96–105. doi: 10.1164/rccm.201408-1564OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salord N., Fortuna A.M., Monasterio C. A randomized controlled trial of continuous positive airway pressure on glucose tolerance in obese patients with obstructive sleep apnea. Sleep. 2016;39(1):35–41. doi: 10.5665/sleep.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinberg A.P., Irizarry R.A. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neele A.E., Van den Bossche J., Hoeksema M.A., de Winther M.P. Epigenetic pathways in macrophages emerge as novel targets in atherosclerosis. Eur J Pharmacol. 2015;763(Pt A):79–89. doi: 10.1016/j.ejphar.2015.03.101. [DOI] [PubMed] [Google Scholar]
- 30.Gozal D., Kheirandish-Gozal L. Cardiovascular morbidity in obstructive sleep apnea: oxidative stress, inflammation, and much more. Am J Respir Crit Care Med. 2008;177(4):369–375. doi: 10.1164/rccm.200608-1190PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kheirandish-Gozal L., Wang Y., Duggan R.C. Nitric oxide production by monocytes in children with OSA and endothelial dysfunction. Clin Sci (Lond) 2014;127(5):323–330. doi: 10.1042/CS20130679. [DOI] [PubMed] [Google Scholar]
- 32.Chuang L.P., Chen N.H., Lin Y., Ko W.S., Pang J.S. Increased MCP-1 gene expression in monocytes of severe OSA patients and under intermittent hypoxia. Sleep Breath. 2016;20(1):425–433. doi: 10.1007/s11325-015-1252-5. [DOI] [PubMed] [Google Scholar]
- 33.Chuang L.P., Chen N.H., Lin S.W. Increased C-C chemokine receptor 2 gene expression in monocytes of severe obstructive sleep apnea patients and under intermittent hypoxia. PLoS One. 2014;9(11):e113304. doi: 10.1371/journal.pone.0113304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briançon-Marjollet A., Henri M., Pépin J.L., Lemarié E., Lévy P., Tamisier R. Altered in vitro endothelial repair and monocyte migration in obstructive sleep apnea: implication of VEGF and CRP. Sleep. 2014;37(11):1825–1832. doi: 10.5665/sleep.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lattimore J.D., Wilcox I., Nakhla S., Langenfeld M., Jessup W., Celermajer D.S. Repetitive hypoxia increases lipid loading in human macrophages—a potentially atherogenic effect. Atherosclerosis. 2005;179(2):255–259. doi: 10.1016/j.atherosclerosis.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11(11):750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 37.Olefsky J.M., Glass C.K. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 38.Van den Bossche J., Neele A.E., Hoeksema M.A. Inhibiting epigenetic enzymes to improve atherogenic macrophage functions. Biochem Biophys Res Commun. 2014;455(3-4):396–402. doi: 10.1016/j.bbrc.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 39.Bonsignore M.R., Borel A.L., Machan E., Grunstein R. Sleep apnoea and metabolic dysfunction. Eur Respir Rev. 2013;22(129):353–364. doi: 10.1183/09059180.00003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S.X., Khalyfa A., Wang Y. Sleep fragmentation promotes NADPH oxidase 2-mediated adipose tissue inflammation leading to insulin resistance in mice. Int J Obes (Lond) 2014;38(4):619–624. doi: 10.1038/ijo.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gharib S.A., Khalyfa A., Abdelkarim A., Bhushan B., Gozal D. Integrative miRNA-mRNA profiling of adipose tissue unravels transcriptional circuits induced by sleep fragmentation. PLoS One. 2012;7(5):e37669. doi: 10.1371/journal.pone.0037669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levenson V.V. DNA methylation biomarkers of cancer: moving toward clinical application. Pharmacogenomics. 2004;5(6):699–707. doi: 10.1517/14622416.5.6.699. [DOI] [PubMed] [Google Scholar]
- 43.Mohn F., Weber M., Schubeler D., Roloff T.C. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 44.Cortese R., Kwan A., Lalonde E. Epigenetic markers of prostate cancer in plasma circulating DNA. Hum Mol Genet. 2012;21(16):3619–3631. doi: 10.1093/hmg/dds192. [DOI] [PubMed] [Google Scholar]
- 45.Cortese R., Khalyfa A., Bao R., Andrade J., Gozal D. Epigenomic profiling in visceral white adipose tissue of offspring of mice exposed to late gestational sleep fragmentation. Int J Obes (Lond) 2015;39(7):1135–1142. doi: 10.1038/ijo.2015.38. [published correction appears in Int J Obes (Lond). 2015;39(9):1432] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irizarry R.A., Hobbs B., Collin F. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 47.Johnson W.E., Li W., Meyer C.A. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103(33):12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eckhardt F., Lewin J., Cortese R. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortese R., Lewin J., Backdahl L. Genome-wide screen for differential DNA methylation associated with neural cell differentiation in mouse. PLoS One. 2011;6(10):e26002. doi: 10.1371/journal.pone.0026002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan C.M., Kendzerska T., Wilton K., Lyons O.D. The different clinical faces of obstructive sleep apnea (OSA), OSA in older adults as a distinctly different physiological phenotype, and impact of OSA on cardiovascular events after coronary artery bypass surgery. Am J Respir Crit Care Med. 2015;192(9):1127–1129. doi: 10.1164/rccm.201507-1472RR. [DOI] [PubMed] [Google Scholar]
- 51.Perri R.A., Kairaitis K., Wheatley J.R., Amis T.C. Anthropometric and craniofacial sexual dimorphism in obstructive sleep apnea patients: is there male-female phenotypical convergence? J Sleep Res. 2015;24(1):82–91. doi: 10.1111/jsr.12205. [DOI] [PubMed] [Google Scholar]
- 52.Edwards B.A., Wellman A., Sands S.A. Obstructive sleep apnea in older adults is a distinctly different physiological phenotype. Sleep. 2014;37(7):1227–1236. doi: 10.5665/sleep.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez-de-la-Torre M., Khalyfa A., Sánchez-de-la-Torre A. Precision medicine in patients with resistant hypertension and obstructive sleep apnea: blood pressure response to continuous positive airway pressure treatment. J Am Coll Cardiol. 2015;66(9):1023–1032. doi: 10.1016/j.jacc.2015.06.1315. [DOI] [PubMed] [Google Scholar]
- 54.Dewan N.A., Nieto F.J., Somers V.K. Intermittent hypoxemia and OSA: implications for comorbidities. Chest. 2015;147(1):266–274. doi: 10.1378/chest.14-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gileles-Hillel A., Almendros I., Khalyfa A., Zhang S.X., Wang Y., Gozal D. Early intermittent hypoxia induces proatherogenic changes in aortic wall macrophages in a murine model of obstructive sleep apnea. Am J Respir Crit Care Med. 2014;190(8):958–961. doi: 10.1164/rccm.201406-1149LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanke-Labesque F., Pépin J.L., de Jouvencel T. Leukotriene B4 pathway activation and atherosclerosis in obstructive sleep apnea. J Lipid Res. 2012;53(9):1944–1951. doi: 10.1194/jlr.P022814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akinnusi M., Jaoude P., Kufel T., El-Solh A.A. Toll-like receptor activity in patients with obstructive sleep apnea. Sleep Breath. 2013;17(3):1009–1016. doi: 10.1007/s11325-012-0791-2. [DOI] [PubMed] [Google Scholar]
- 58.Yamauchi M., Tamaki S., Tomoda K. Evidence for activation of nuclear factor κB in obstructive sleep apnea. Sleep Breath. 2006;10(4):189–193. doi: 10.1007/s11325-006-0074-x. [DOI] [PubMed] [Google Scholar]
- 59.Kim Y.S., Kwak J.W., Lee K.E. Can mitochondrial dysfunction be a predictive factor for oxidative stress in patients with obstructive sleep apnea? Antioxid Redox Signal. 2014;21(9):1285–1288. doi: 10.1089/ars.2014.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee C.H., Olson P., Evans R.M. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144(6):2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 61.Marx N., Duez H., Fruchart J.C., Staels B. Peroxisome proliferator-activated receptors and atherogenesis: regulators of gene expression in vascular cells. Circ Res. 2004;94(9):1168–1178. doi: 10.1161/01.RES.0000127122.22685.0A. [DOI] [PubMed] [Google Scholar]
- 62.Bouhlel M.A., Derudas B., Rigamonti E. PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature. 2007;447(7148):1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie X., Pan L., Ren D., Du C., Guo Y. Effects of continuous positive airway pressure therapy on systemic inflammation in obstructive sleep apnea: a meta-analysis. Sleep Med. 2013;14(11):1139–1150. doi: 10.1016/j.sleep.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 65.McEvoy R.D., Michael M.Z. Measuring blood microRNAs to provide personalized advice to sleep apnea patients with resistant hypertension: dreaming the future. J Am Coll Cardiol. 2015;66(9):1033–1035. doi: 10.1016/j.jacc.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 66.Hotamisligil G.S., Shargill N.S., Spiegelman B.M. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 67.Maedler K., Sergeev P., Ris F. Glucose-induced beta cell production of IL-1β contributes to glucotoxicity in human pancreatic islets. J Clin Invest. 2002;110(6):851–860. doi: 10.1172/JCI15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berk B.C., Weintraub W.S., Alexander R.W. Elevation of C-reactive protein in “active” coronary artery disease. Am J Cardiol. 1990;65(3):168–172. doi: 10.1016/0002-9149(90)90079-g. [DOI] [PubMed] [Google Scholar]
- 69.Ridker P.M., Cushman M., Stampfer M.J., Tracy R.P., Hennekens C.H. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N Engl J Med. 1997;336(14):973–979. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- 70.Navarro-González J.F., Mora-Fernández C. The role of inflammatory cytokines in diabetic nephropathy. J Am Soc Nephrol. 2008;19(3):433–442. doi: 10.1681/ASN.2007091048. [DOI] [PubMed] [Google Scholar]
- 71.Donath M.Y., Shoelson S.E. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11(2):98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.