Abstract
Homooligomerization of proline utilization A (PutA) bifunctional flavoenzymes is intimately tied to catalytic function and substrate channeling. PutA from Bradyrhizobium japonicum (BjPutA) is unique among PutAs in that it forms a tetramer in solution. Curiously, a dimeric BjPutA hot spot mutant was previously shown to display wild-type catalytic activity despite lacking the tetrameric structure. These observations raised the question of what is the active oligomeric state of BjPutA. Herein, we investigate the factors that contribute to tetramerization of BjPutA in vitro. Negative-stain electron microscopy indicates that BjPutA is primarily dimeric at nanomolar concentrations, suggesting concentration-dependent tetramerization. Further, sedimentation-velocity analysis of BjPutA at high (micromolar) concentration reveals that although the binding of active-site ligands does not alter oligomeric state, reduction of the flavin adenine dinucleotide cofactor results in dimeric protein. Size-exclusion chromatography coupled with multiangle light scattering and small-angle x-ray scattering analysis also reveals that reduced BjPutA is dimeric. Taken together, these results suggest that the BjPutA oligomeric state is dependent upon both enzyme concentration and the redox state of the flavin cofactor. This is the first report, to our knowledge, of redox-linked oligomerization in the PutA family.
Introduction
Catabolism of the amino acid proline is a crucial and tightly regulated process across all kingdoms of life. For example, in plants, regulation of proline catabolism is important in both abiotic (1, 2) and biotic (3) stresses as well as senescence (4). In humans, proline catabolism is involved in tumorigenesis and cancer metastasis (5, 6), susceptibility to schizophrenia (7, 8), and inherited hyperprolinemia disorders (9). Further, proline catabolism is vital for virulence of both pathogenic bacteria (10, 11, 12) and fungi (13).
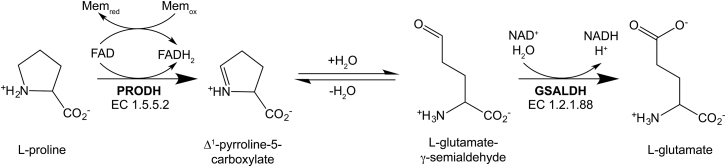
Proline catabolism, the four-electron oxidation of proline to glutamate, is carried out in two enzyme-catalyzed steps (14) (Fig. 1). In the first step, proline is oxidized to Δ1-pyrroline-5-carboxylate (P5C) by the flavoenzyme proline dehydrogenase (PRODH). The cyclic P5C intermediate undergoes nonenzymatic hydrolysis to L-glutamate-γ-semialdehyde (GSAL). GSAL is then oxidized to glutamate via the action of the NAD+-dependent GSAL dehydrogenase (GSALDH; also known as P5CDH and ALDH4A1).
Figure 1.
The reactions of proline catabolism.
In some microbes, PRODH and GSALDH are combined into the bifunctional flavoenzyme proline utilization A (PutA). There are three PutA domain architectures (PutA types A, B, and C) (15). The simplest of these architectures is type A PutA, which has an N-terminal PRODH module and a C-terminal GSALDH module but lacks additional structural and DNA-binding domains found in other types of PutAs. Structural and biophysical characterization of type A PutAs revealed a conserved homodimeric core structure that is thought to be essential for enzymatic activity and substrate channeling (14, 15, 16). The conserved type A PutA dimer is a domain-swapped assembly in which the oligomerization flap of one protomer contacts the catalytic modules of the other protomer. These quaternary structural interactions appear to serve two main purposes: stabilizing the aldehyde substrate site of the GSALDH module and sealing the substrate-channeling tunnel that links the two active sites within a protomer. Thus, oligomeric state is intimately tied to enzyme function in type A PutA.
Whereas all studied type A PutAs form this conserved dimeric quaternary structure, the type A PutA from Bradyrhizobium japonicum (BjPutA) is unique in that two dimers assemble into a ring-shaped tetramer (16, 17). Curiously, the dimer-dimer interface of the tetramer is remote from the active sites and substrate-channeling tunnel, which raised the question of whether the tetramer is functionally significant. Interestingly, the mutation of Arg51 in the dimer-dimer interface of BjPutA to Glu (Fig. S1) resulted in a cleanly dimeric BjPutA hot spot variant (R51E) that exhibited wild-type catalytic activity, suggesting that the tetramer is not essential for functionality (16). Because of these observations, the functional significance of BjPutA tetramer formation is unclear.
Herein, we further investigate BjPutA quaternary structure at various concentrations and in response to treatments that mimic steps in the catalytic mechanism. Negative-stain electron microscopy (EM) data suggest that BjPutA is dimeric at very low concentration, as expected for a reversible dimer-tetramer equilibrium. Further, analytical ultracentrifugation analysis at higher concentration suggests that reduction of the flavin adenine dinucleotide (FAD) cofactor abrogates tetramerization, producing primarily dimeric enzyme. BjPutA tetramer disruption in response to flavin reduction is also confirmed using analytical size-exclusion chromatography (SEC), multiangle light scattering (MALS), and small-angle x-ray scattering (SAXS). Finally, we provide data revealing that dimeric BjPutA can physically associate with lipid bilayers and transfer electrons to membrane-bound acceptors, further suggesting that dimeric BjPutA is functionally replete. Taken together, our data confirm that the type A PutA conserved dimer is the minimal relevant functional form of these flavoenzymes.
Materials and Methods
Expression and purification of BjPutA
BjPutA was expressed and purified as previously described (18). The final storage buffer for protein samples—i.e., the SEC mobile phase—contained 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-caboxyethyl)phosphine (TCEP), and 5% (v/v) glycerol. For analytical ultracentrifugation analysis, protein was used without freezing. For all other applications, protein was aliquoted in polymerase chain reaction strip tubes, flash-cooled in liquid nitrogen, and stored at −80°C.
Negative staining and transmission EM data acquisition and analysis
Preparation of negative-stained grids was carried out as previously described (19). Briefly, carbon-coated copper grids (Electron Microscopy Sciences, Hatfield, PA) were glow discharged, and a 5 μL drop of protein was applied to the grid and incubated for 2 min at room temperature. Excess protein was removed by blotting with Whatman P4 filter paper. This step was immediately followed by two washes in water and a final incubation for 2 min in 0.75% (w/v) uranyl formate (UF; Ted Pella, Reading, CA). Excess UF was removed from the grid with Whatman P4 filter paper, and the grid was air-dried.
Protein particles were observed using a JEOL JEM 1400 transmission electron microscope (Peabody, MA) at 25,000× nominal magnification at 120 kV. BjPutA was diluted in water to a concentration of 0.006 mg mL−1 (∼3 nM, dimer Mr) and stained with 0.75% w/v UF. Contrast transfer function correction was performed using CTFFind 4.1.8 (20) using a spherical aberration of 3.2 mm, amplitude contrast of 0.5, and magnified pixel size of 5.226 Å. The resolution range used for fitting was 10–30 Å. The defocus range was 5000–50,000 Å with a defocus step of 500 Å and an astigmatism of 100 Å. Manual picking was performed with a box size of 128 pixels. A total of 5066 particles was manually picked and processed using RELION 2.1 (21) software to determine three-dimensional (3D) reconstruction of the enzyme. Two-dimensional classification was performed with a 200 Å diameter mask and resulted in 30 classes that were all used for 3D classification. The initial model was determined by ab initio methods using RELION 2.1. One 3D class (5066 particles) was calculated, resulting in a final map with an estimated resolution of 28 Å. Chimera (22) was used to render and visualize the initial and refined 3D models. The threshold for 3D class visualization corresponded to the volume of the crystallographic dimer downsampled to 28 Å resolution using Chimera.
Inactivation of BjPutA by N-propargylglycine
N-propargylglycine (NPPG) (Best of Chemical Sciences, Shirley, NY) was added to purified BjPutA at a concentration of 133 mM. The inactivation reaction was allowed to proceed overnight at 4°C. On the following day, protein was passed over a Superdex 10–300 200 pg SEC column (General Electric Healthcare Life Sciences, Marlborough, MA) using a buffer containing 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol as the mobile phase.
Sedimentation-velocity analytical ultracentrifugation
Sedimentation-velocity experiments were performed at 20°C in a Beckman XL-I analytical ultracentrifuge using an An50Ti rotor. Reference buffer and protein samples were loaded into a sedimentation-velocity cell bearing a two-sector charcoal-Epon centerpiece. Before centrifugation, the sample was allowed to equilibrate for 2 h. The sample was then centrifuged at 35,000 rpm for a total of 300 radial scans spaced at 2 min intervals and acquired using Rayleigh interference optics. Scans 10–300 were used in the analysis. The distributions of apparent sedimentation coefficients, c(s), were generated using SEDFIT (23).
The apparent c(s) distributions were determined for oxidized BjPutA, NPPG-inactivated BjPutA, BjPutA reduced with sodium dithionite, BjPutA reduced with proline, and oxidized BjPutA complexed with noncovalent, nonreducing active site ligands. The preparation of NPPG-inactivated BjPutA is described in the preceding section. For analysis of the c(s) distribution of BjPutA in the presence of active site ligands, protein samples were supplemented with 10 mM L-tetrahydrofuroic acid (THFA) and 1 mM NAD+ and dialyzed for 4 h with two buffer exchanges in a slide-a-lyzer minidialysis device (Thermo Fisher, Waltham, MA) against 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, 10 mM THFA, and 1 mM NAD+. For analysis of the c(s) distribution of BjPutA reduced with sodium dithionite or proline, purified BjPutA was first treated with the indicated concentrations of dithionite or proline. After treatment, the protein was immediately run over an SEC column with a mobile phase containing 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol supplemented with the indicated concentrations of sodium dithionite or proline. For the untreated and NPPG-inactivated experiments, the reference buffer used was 50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol. For the experiment performed in the presence of active site ligands, the dialysate served as the reference buffer. For the dithionite and proline treatment experiments, the reference buffer used was the aforementioned SEC mobile phase.
SEC-MALS-SAXS data acquisition and analysis
SAXS experiments were performed at Sector 18-ID (Advanced Photon Source, Chicago, IL) using a 3.5 m camera that gives access to a q range of ∼0.005–0.38 Å−1. To ensure optimal sample quality, an in-line SEC setup was employed. The sample at ∼8 mg mL−1 was loaded onto a Wyatt SEC column with an exclusion limit of 1.25 MDa run on an Infinity II high-performance liquid chromatography unit (Agilent, Santa Clara, CA). A MALS instrument along with a Wyatt DAWN Helios II QELS detector and a Wyatt Optilab T-rEX dRI detector (Wyatt, Goleta, CA) were included in the trajectory between the UV monitor on the high-performance liquid chromatography unit and the SAXS flow cell. SAXS data acquisition occurred simultaneously with sample elution. 0.5 s exposures were collected every 3 s. MALS and DLS data were processed using the ASTRA software package (Wyatt), and the SAXS data were reduced to I(q) vs. q curves (q = 4πsinθ/λ, where 2θ = scattering angle, and λ = 1.03 Å) and processed using the ATSAS program suite (24). Exposures flanking the elution peak were averaged to generate the I(q) vs. q curve for the buffer. The buffer I(q) vs. q curve was then subtracted from the elution peak curves to obtain the sample SAXS curve.
Scattering curves were averaged using PRIMUS (25). Guinier analysis was also performed in PRIMUS. GNOM (26) was used to calculate the distance distribution functions from the averaged scattering curves. Theoretical scattering curves for the dimeric and tetrameric crystal structure models of BjPutA (Protein Data Bank (PDB): 3HAZ) and the goodness-of-fit parameter (χ) were calculated using FoXS (27). MultiFoXS (27) was used to calculate theoretical SAXS curves from dimer-tetramer ensemble models.
DAMMIF (28) was used to generate shape reconstructions. For each reconstruction, 20 calculations were performed. Twofold symmetry was enforced both during the reconstruction calculations and during model averaging and filtering using DAMAVER (29). The averaged and filtered dummy atom models were superimposed onto either the dimeric or tetrameric crystal structure of BjPutA (PDB: 3HAZ) using SUPCOMB (30). The pdb2vol utility of situs (31) was used to convert dummy atom models into volumetric maps. The averaged SAXS curves have been deposited in the SASBDB (32) under the accession codes SASDDQ3 (oxidized BjPutA) and SASDDP3 (NPPG-inactivated BjPutA).
Lipid partitioning and functional membrane association assays
Lipid partitioning assays were performed as previously described for Escherichia coli PutA (33). Briefly, wild-type BjPutA, BjPutA R51E, and NPPG-inactivated BjPutA (0.3 mg mL−1) were incubated with 1 mg mL−1 liposomes prepared from E. coli polar lipids in 10 mM HEPES and 150 mM NaCl (pH 7.5) buffer with and without 20 mM proline for 1 h at room temperature. Samples were then centrifuged, and the soluble and lipid fractions analyzed by 4–20% gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels.
The ability of wild-type BjPutA and BjPutA R51E to functionally associate with membranes was measured by measuring the production of P5C using inverted E. coli JT31 putA− membrane vesicles as a terminal electron acceptor, as described previously (33). P5C was detected as an adduct formed by reaction with o-aminobenzaldehyde (o-AB) at 443 nm (ε = 2590 M−1cm−1). The assays were performed with 4 mM o-AB, 60 mM proline, and 0.02 mg mL−1 membrane vesicles in 50 mM 3-(N-morpholino)-propanesulfonic acid buffer (10 mM MgCl2, 10% glycerol (pH 7.4)).
Results
Analysis of BjPutA oligomeric state by EM
Previous oligomeric state studies of BjPutA revealed stable in vitro tetramer formation under all assayed conditions (16, 17). Methods typically employed to explore protein oligomeric state examine proteins at micromolar concentrations. Thus, analysis of a self-association regime with submicromolar affinity requires visualization by other methods. We sought to understand whether the BjPutA oligomeric state could be concentration-dependent at much lower concentrations (i.e., nanomolar). One method that provides information about protein oligomeric state at nanomolar protein concentrations is negative-stain EM.
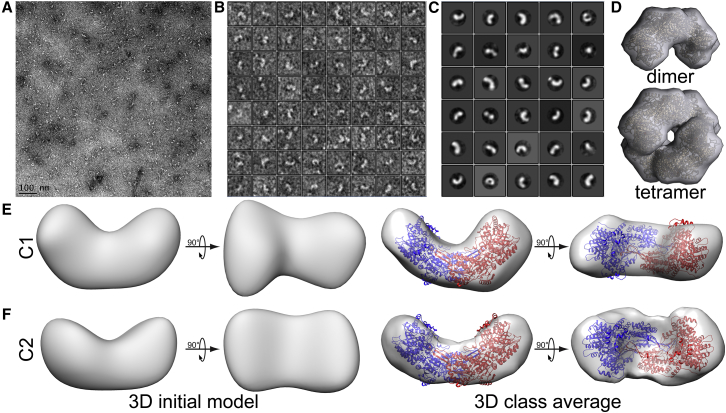
BjPutA was visualized by negative-stain EM at ∼3 nM (based on the dimer molecular mass (Mr) of 215 kDa). The micrographs (Fig. 2 A) revealed a preponderance of semicircular-shaped particles (Fig. 2, A and B). This shape is expected for the BjPutA domain-swapped dimer, which is shown for reference in Fig. 2 D. In contrast, the tetramer is expected to appear as a ring in side projection (Fig. 2 D). Ring-shaped particles were also evident in the micrographs, albeit at low frequency compared to the apparent dimer (Fig. 2, A and B).
Figure 2.
Negative-stain EM of BjPutA. (A) A representative electron micrograph is shown. A larger version of this micrograph is shown in Fig. S2. (B) A selection of picked particles from a representative micrograph is shown. (C) Two-dimensional class averages are shown. (D) A theoretical electron density envelope (25 Å resolution) is calculated from the crystallographic dimer and tetramer of BjPutA (PDB: 3HAZ). A cartoon representation of each crystal structure is shown inside the maps. (E) A three-dimensional initial model and class average of BjPutA is generated using all 30 two-dimensional class averages with no symmetry operators enforced (C1). (F) A three-dimensional initial model and class average of BjPutA is generated using all 30 two-dimensional class averages with C2 symmetry enforced. Chimera (22) was used to render and visualize the volumetric maps. To see this figure in color, go online.
Analysis of the grids resulted in identification of 5066 particles. The ring-shaped particles were included during particle picking to reduce bias toward the dimer. The generation of reference-free two-dimensional class averages in RELION resulted in 30 classes (Fig. 2 C). Of these classes, 28 strongly resemble the semicircular shape consistent with the expected shape of the BjPutA dimer. These 28 classes account for 4917 (97%) of the selected particles. The remaining two classes somewhat resemble the ring-shaped tetramer. These two classes account for 149 (3%) of the selected particles. This result suggests that wild-type BjPutA exists primarily as a dimer when assayed at nanomolar protein concentration.
3D class averages were calculated using all 30 two-dimensional class averages. All two-dimensional class averages were included to account for the low concentration of apparent tetramer visible in the micrographs. The two-dimensional classes were used to generate an ab initio reference model for refinement in RELION 2.1 (Fig. 2, E and F). Reference models were generated in the absence (Fig. 2 E) and presence (Fig. 2 F) of a twofold symmetry constraint. The two reference models were each used to calculate a single 3D class average. Interestingly, in both cases, the 3D class average exhibited a semicircular shape that resembles the BjPutA dimer. Further, superimposition of the BjPutA dimer crystal structure (PDB: 3HAZ) shows an excellent fit to the generated 3D envelope. This result suggests that the BjPutA dimer predominates in solution at low nanomolar concentrations, as expected for a reversible dimer-tetramer equilibrium.
Influence of active-site ligands and reduction of the FAD on oligomeric state
Negative-stain EM suggests that wild-type BjPutA is primarily dimeric at nanomolar enzyme concentration, and previously reported analysis of a dimeric BjPutA hot spot mutant (R51E) revealed similar catalytic activity as wild-type (16). These results suggest that the dimer is an active oligomeric state of BjPutA and that tetramerization is not necessary for in vitro catalytic function. To further explore the question of which oligomeric states of BjPutA are present in kinetics assays, we tested the effects of active site ligands and FAD redox state on oligomeric structure using sedimentation-velocity analytical ultracentrifugation.
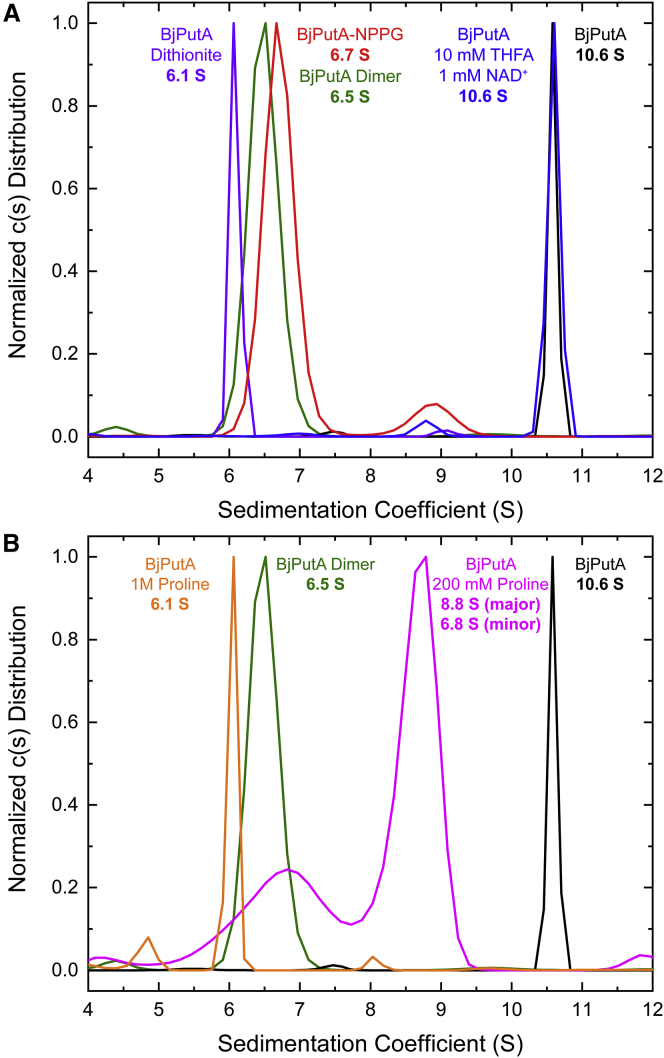
Sedimentation-velocity experiments performed on wild-type BjPutA (23 μM, dimer Mr) revealed a single major peak at apparent sedimentation coefficient 10.6 S (Fig. 3 A). This result is in excellent agreement with the previously reported value of 10.8 S, which corresponds to Mr of 429 kDa, a value that is essentially identical to the theoretical Mr of the BjPutA tetramer (430 kDa) (16).
Figure 3.
Sedimentation-velocity analysis of BjPutA after treatment with active site ligands or NPPG. (A) The distribution of sedimentation coefficients for untreated BjPutA (black), BjPutA supplemented with active site ligands (10 mM THFA and 1 mM NAD+; blue), NPPG-inactivated BjPutA (red), dithionite-reduced BjPutA (purple), and the previously reported BjPutA hot spot mutant dimer (green) (16) are shown. (B) The distribution of sedimentation coefficients for untreated BjPutA (black), BjPutA supplemented with 200 mM proline (magenta), BjPutA supplemented with 1 M proline (orange), and the previously reported BjPutA hot spot mutant dimer (green) (16) are shown. All experiments were run at a protein concentration of 5 mg mL−1 (23 μM based on dimer Mr). To see this figure in color, go online.
To test whether the binding of active site ligands influences the oligomeric state of BjPutA, a sedimentation-velocity experiment was performed on BjPutA (23 μM, dimer Mr) in the presence of 1 mM NAD+ and 10 mM THFA, a proline analog and competitive inhibitor of PRODHs and PutAs (33, 34, 35, 36, 37, 38, 39, 40). The distribution of sedimentation coefficients revealed a single major peak at an apparent sedimentation coefficient of 10.6 S, which was consistent with the presence of a tetramer (Fig. 3 A). This result suggests that the binding of these active site ligands does not disrupt the BjPutA tetramer.
Because the first reaction catalyzed by PutA results in the two-electron reduction of the FAD cofactor, we wanted to study the influence of flavin reduction on the oligomeric state. To ensure flavin reduction on the timescale of the sedimentation-velocity assay (>10 h), BjPutA was inactivated using NPPG. Inactivation by NPPG results in the irreversible two-electron reduction of the FAD and the covalent linkage between the FAD N5 and an active site lysine. Previous studies have shown that NPPG-inactivated PutA is a good surrogate for the proline-reduced enzyme both in terms of the structure of the reduced FAD and the biochemical properties of the inactivated enzyme (16, 34, 41, 42).
Sedimentation-velocity experiments were performed on NPPG-inactivated BjPutA to determine any effects on oligomeric state. Interestingly, the distribution of sedimentation coefficients revealed a single major peak at apparent sedimentation coefficient 6.7 S (Fig. 3 A). By comparison, the reported apparent sedimentation coefficient of the dimeric BjPutA hot spot mutant is 6.5 S, which corresponds to Mr of 216 kDa, a value that is essentially identical to the theoretical Mr of the BjPutA dimer (215 kDa) (16), which is in good agreement with the current result. To determine whether the observed effect of NPPG-inactivation was due to reduction of the FAD or the covalent linkage resulting from NPPG-inactivation, oxidized BjPutA was treated with sodium dithionite, which has previously been shown to chemically reduce the FAD in PutA (34, 43). The distribution of sedimentation coefficients observed after dithionite treatment revealed a single peak at the apparent sedimentation coefficient of 6.1 S, which was consistent with the dimer (Fig. 3 A). Taken together, these data suggest that reduction of the flavin results in dissociation of the BjPutA tetramer into dimers. To our knowledge, this is the first example of PutA oligomeric state being sensitive to the FAD redox state.
In the PutA PRODH reaction, the substrate proline is oxidized to P5C with concomitant reduction of the FAD (Fig. 1). Therefore, to understand whether reduction of the enzyme by the natural substrate likewise induces a shift in oligomeric state, sedimentation-velocity experiments were performed on BjPutA after incubation with proline. First, BjPutA was incubated with 200 mM proline, which is approximately five times the previously reported Km for proline (44). The distribution of sedimentation coefficients revealed major and minor peaks at apparent sedimentation coefficients of 8.8 and 6.8 S, respectively (Fig. 3 B). The minor peak at 6.8 S may be interpreted as a pool of fully reduced dimeric protein. The major peak is approximately halfway between the apparent sedimentation coefficients of the dimer and tetramer, which could suggest the presence of a dimer-tetramer equilibrium. This equilibrium may be due to incomplete reduction, i.e., not all four active sites of the tetramer are reduced. Thus, one interpretation of this result is that the solution contains a pool of fully reduced dimeric protein and an equilibrium of differentially reduced dimers and tetramers in intermediate exchange.
To determine whether higher concentrations of proline can cleanly produce dimeric enzyme, BjPutA was treated with 1 M proline. Interestingly, the distribution of apparent sedimentation coefficients reveals a single peak at 6.1 S, which is consistent with other observed dimeric BjPutA species (Fig. 3 B). These results reveal that the observed shift in BjPutA oligomeric state upon flavin reduction is observed with its native reductant and is not limited only to nonnatural reducing agents.
Analysis of BjPutA and NPPG-inactivated BjPutA by light scattering and SAXS
To further assess the in-solution effects of NPPG inactivation on the BjPutA oligomeric state, SEC MALS tandem SAXS (SEC-MALS-SAXS) experiments were performed. In this set of experiments, purified protein is separated by SEC, and the eluent is routed directly to MALS and then SAXS for characterization of the molecular mass and in-solution structural properties, respectively.
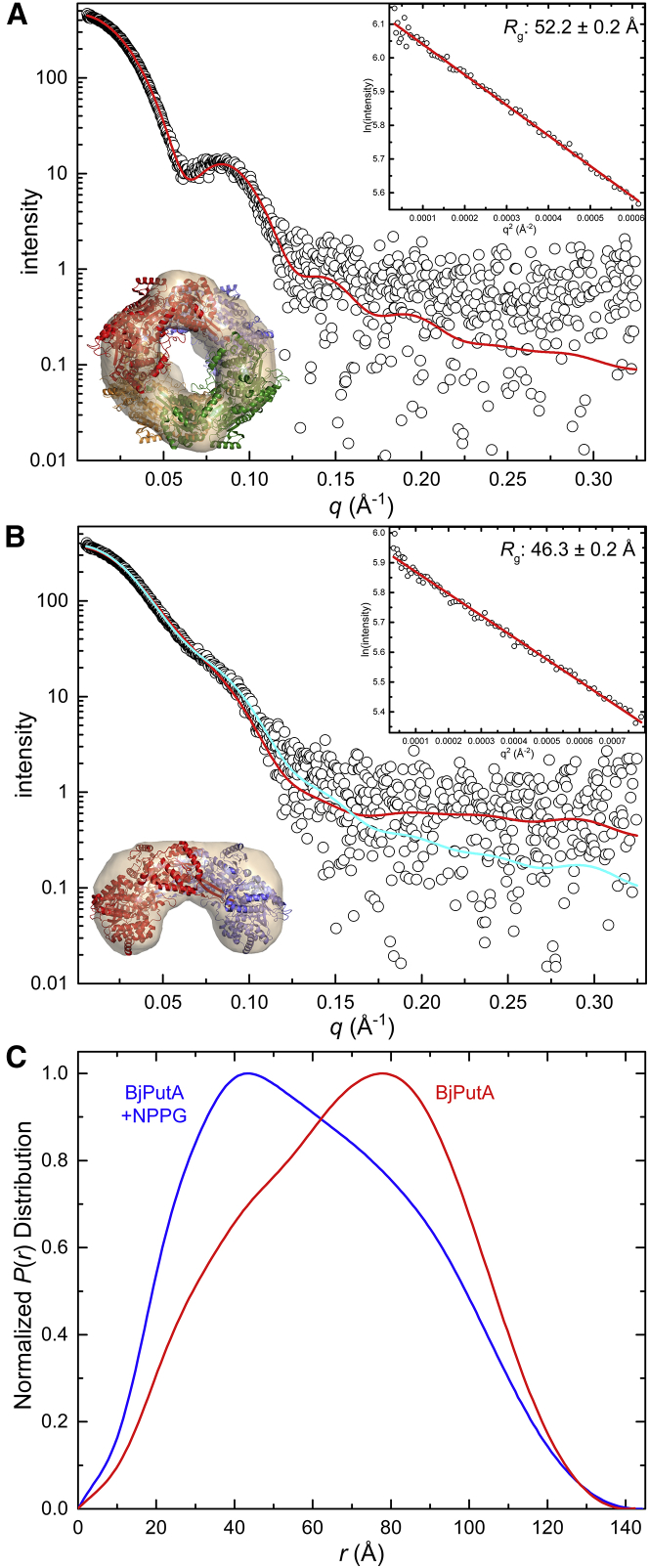
Wild-type, untreated BjPutA was subjected to SEC-MALS-SAXS analysis. Results from the MALS experiment revealed an average in-solution Mr of 398 kDa, which is 7% lower than the theoretical tetramer mass, and a hydrodynamic radius (Rh) of 68 Å (Table 1). Further, SAXS analysis revealed an experimental scattering curve with a Guinier radius of gyration (Rg) of 52.2 Å (Fig. 4 A). The theoretical scattering curve generated from the crystallographic tetramer shows good agreement with the experimental scattering profile (Fig. 4 A; χ: 1.1), and the shape reconstruction from DAMMIF exhibits the ring shape diagnostic of the tetramer (Fig. 4 A). Also, the Mr calculated from the SAXS curve using the SAXS MoW2 server (45) was 434 kDa, which is 1% higher than the theoretical tetramer Mr (Table 1). Previous SAXS analysis of wild-type BjPutA also revealed that the P(r) distribution has a most-probable real-space vector of ∼80 Å (16), and the current SAXS data are consistent with this observation (Fig. 4 C). Taken together, these data are consistent with oxidized wild-type BjPutA being tetrameric under the investigated solution conditions.
Table 1.
Structural Parameters from MALS and SAXS
| BjPutA | BjPutA + NPPG | |
|---|---|---|
| MALS Analysis | ||
| Rh (Å) | 68 ± 2 | 60 ± 2 |
| Mr (kDa) | 398 ± 2 | 237 ± 2 |
| SAXS Mr (kDa)a | 433.8 | 249.7 |
| Guinier Analysis | ||
| Rg (Å) | 52.2 ± 0.2 | 46.3 ± 0.2 |
| qmin (Å−1) | 0.0057 | 0.0057 |
| qRg range | 0.3–1.3 | 0.26–1.3 |
| Coefficient of correlation, R2 | 0.992 | 0.991 |
| P(r) Analysis | ||
| Rg (Å) | 52.0 ± 0.1 | 47.0 ± 0.1 |
| dmax (Å) | 142.3 | 144.15 |
| q range (Å−1) | 0.0057–0.3337 | 0.0057–0.3826 |
| GNOM total estimate | 0.7896 | 0.8840 |
| Porod volume (Å3) | 582,000 | 330,000 |
| SASBDB Code | SASDDQ3 | SASDDP3 |
The theoretical Mr values of the dimer and tetramer are 215 and 430 kDa, respectively.
Calculated using the SAXS MoW2 server.
Figure 4.
SAXS analysis of oxidized and NPPG-inactivated (reduced) BjPutA. (A) The composite experimental SAXS curve of untreated BjPutA obtained from SEC-MALS-SAXS is shown in open circles. The red curve was calculated from the crystallographic BjPutA tetramer (PDB: 3HAZ) using FoXS (χ: 1.1). The top right inset shows the Guinier plot and the calculated Rg. Also shown is a DAMMIF shape reconstruction superimposed with the crystallographic tetramer. (B) The composite experimental SAXS curve of NPPG-inactivated BjPutA obtained from SEC-MALS-SAXS is shown in open circles. The red curve was calculated from the crystallographic BjPutA dimer (PDB: 3HAZ) using FoXS (χ: 1.7). The cyan curve shows the two-body fit composed of 93%:7% BjPutA dimer:tetramer generated by MultiFoXS (χ: 1.3). The top right inset shows the Guinier plot and the calculated Rg. Also shown is a DAMMIF shape reconstruction superimposed with the crystallographic dimer. (C) Experimental distance distribution functions for untreated BjPutA (red) and NPPG-inactivated BjPutA (blue) are shown. To see this figure in color, go online.
NPPG-inactivated BjPutA was also subjected to SEC-MALS-SAXS analysis. Results from the MALS experiment revealed an average in-solution Mr of 237 kDa, which is 10% higher than the theoretical dimer mass, and an Rh of 60 Å (Table 1), which is consistent with reduction in the overall size of the particle compared to oxidized BjPutA. Further, SAXS analysis of the experimental scattering curve revealed a Guinier Rg of 46.3 Å (Fig. 4 B). For comparison, the experimental Guinier Rg of the dimeric BjPutA hot spot mutant (R51E) is 45 Å (16). The theoretical scattering curve generated from the crystallographic dimer shows good agreement with the experimental scattering profile (Fig. 4 B; χ: 1.7), and the shape reconstruction resembles the dimer (Fig. 4 B). We note that a two-body fit with 93%:7% dimer:tetramer slightly statistically improves the fit to the experimental scattering profile (Fig. 4 B; χ: 1.3). Improvement of the fit by introducing a small amount of tetramer may reflect either incomplete NPPG inactivation or a change in the in-solution structure that crystallographic models do not capture. The calculated average in-solution Mr using the SAXS MoW2 server (45) was 250 kDa, which is 16% higher than the theoretical dimer Mr (Table 1). Further, the P(r) distribution is qualitatively different from the tetrameric enzyme, showing a most-probable real-space vector of only ∼40 Å, compared to ∼80 Å for the tetramer (Fig. 4 C). The P(r) of the NPPG-inactivated enzyme is very similar to that of the dimeric BjPutA hot spot mutant (R51E) (16). Overall, these data are consistent with the sedimentation-velocity data. Taken together, these results suggest that flavin reduction via NPPG inactivation disrupts the BjPutA tetramer into dimers.
NPPG inactivation does not disrupt membrane association
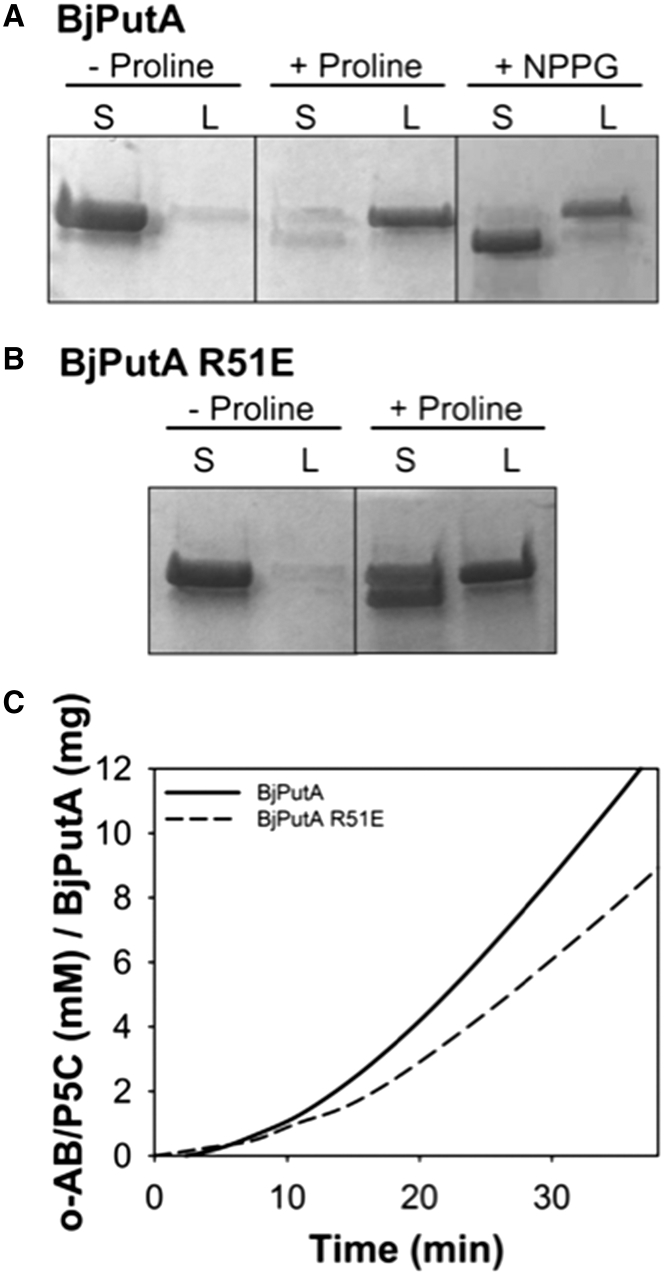
To determine the impact of NPPG inactivation on membrane association, lipid-partitioning assays were performed. Here, wild-type BjPutA alone, wild-type BjPutA reduced with 20 mM proline, and wild-type BjPutA inactivated with NPPG were incubated with polar E. coli lipids. Samples were then separated into soluble and lipid fractions, and the localization of the PutA protein was visualized with SDS-PAGE. Under oxidized conditions (-proline), the majority of the PutA protein is found in the soluble fraction, whereas in the presence of proline, the BjPutA distribution shifts dramatically to the lipid fraction (Fig. 5 A). This result indicates that reduction of the FAD enhances membrane association, which is in agreement with studies of BjPutA and other PutAs (46, 47, 48, 49, 50, 51). Inactivation of wild-type BjPutA with NPPG also increases the partitioning of protein into the lipid fraction, although not as extensively as observed with proline (Fig. 5 A). This result is consistent with NPPG locking the FAD in BjPutA into a reduced state.
Figure 5.
Membrane association of various forms of BjPutA. (A) Wild-type oxidized BjPutA, wild-type BjPutA reduced with 20 mM proline, and NPPG-inactivated BjPutA were incubated with 0.8 mg mL−1E. coli polar liposomes for 1 h in 10 mM HEPES buffer (150 mM NaCl (pH 7.5)). Soluble and lipid-associated fractions were then separated by air-fuge ultracentrifugation and analyzed using SDS-PAGE, as previously described (33). (B) Lipid partitioning assays were performed with BjPutA mutant R51E in the presence and absence of 20 mM proline, as described in (A). (C) The ability of the BjPutA R51E mutant to functionally associate with inverted membrane vesicles was measured by incubating BjPutA wild-type or mutant R51E mutant with 50 mM proline, 4 mM o-AB, and 0.1 mg mL−1 vesicles prepared from JT31 putA−E. coli. Reactions were performed in 20 mM 3-(N-morpholino)-propanesulfonic acid (pH 7.5) and o-AB/P5C adduct formation monitored at 443 nm.
Dimeric BjPutA is capable of in vitro physical and functional membrane association
The data presented above are consistent with the dimer being the active form of BjPutA. We next sought to test this idea more directly by studying the physical and functional membrane association capabilities of the dimeric BjPutA through use of the hot spot mutant BjPutA R51E. As described previously, BjPutA R51E is dimeric even at high protein concentration (10–30 μM using dimer Mr).
Lipid partitioning assays show that proline induces a shift in protein distribution, leading to increased amounts of BjPutA R51E in the lipid fraction (Fig. 5 B), similar to the pattern observed with wild-type BjPutA (Fig. 5 A), indicating a physical association of reduced dimeric BjPutA R51E with E. coli polar lipids.
Next, the functional association of BjPutA R51E with lipids was investigated. In this assay, catalytic turnover is measured using inverted membrane vesicles in place of an exogenously added terminal electron acceptor. Sustained catalysis requires the protein to form a productive electron transfer complex with the membrane-bound acceptors. Steady-state production of P5C is observed in this assay with the dimeric R51E variant, similar to wild-type BjPutA (Fig. 5 C). Taken together, the lipid partitioning and functional association results indicate that disruption of the tetrameric state to form dimeric BjPutA does not impact the ability of the protein to physically associate with membranes and functionally couple to membrane-bound electron acceptors.
Discussion
Approximately 50% of proteins oligomerize, implying that self-association is important for protein function (52, 53). The PutA family exemplifies this idea. PutAs exhibit a variety of oligomeric structures (15). Type A PutAs have the minimal domain architecture and exist primarily as domain-swapped dimers or, in the singular case of BjPutA, a ring-shaped dimer-of-dimers tetramer (16). Type B PutAs have an extra Rossmann fold domain in the C-terminal half of the polypeptide chain that mimics the dimerization domain of type A PutA (35, 54). The presence of this extra domain radically alters the quaternary structure; type B PutAs can form an ellipsoidal dimer that is unlike the semicircular type A dimer (Fig. 2 D). Type C PutAs form yet another distinct dimer that is mediated by an N-terminal ribbon-helix-helix DNA-binding domain (55).
How oligomerization underlies PutA function has been a subject of investigation. The functional relevance of the domain-swapped type A dimer seems obvious from structures of PutA (16, 17, 34) and monofunctional aldehyde dehydrogenases (56, 57). In these enzymes, the oligomerization domain of one protomer, which is a bipartite β-flap structure, forms a β-sheet interaction with the catalytic domain of the opposite protomer, and side chains of the oligomerization domain reach across the dimer interface to form stabilizing interactions with the aldehyde substrate binding site of the opposite protomer. Mutation of residues in the oligomerization domains of type A PutA and aldehyde dehydrogenases have confirmed the importance of these quaternary structural interactions for catalytic function (56, 58). Additionally, in type A PutAs, the oligomerization domain seals the substrate-channeling tunnel of the opposite protomer, ensuring that the intermediate is properly channeled between the active sites. Similarly, the functional importance of the type C PutA dimer is obvious. The ribbon-helix-helix DNA binding domains of type C PutAs must dimerize to bind DNA and repress transcription of the proline utilization regulon (55). In contrast, the functional significance of the type B dimer is less obvious. The active sites of type B PutAs are outside of the dimer interface; however, we recently showed that the binding of a proline analog enhances the dimerization of a type B PutA, implying that dimerization may occur during catalysis (54).
This study provides evidence that the tetrameric form of BjPutA is not essential for function despite being consistently observed by x-ray crystallography, SAXS, and analytical ultracentrifugation (16, 17, 44, 59). We previously showed that the dimeric R51E variant of BjPutA retains catalytic activity when assayed with exogenous model electron acceptors (16). Here, we extended this result by showing R51E associates physically and functionally with lipids. Furthermore, we showed that wild-type BjPutA is dimeric at the concentration used in activity assays, and reduction of the FAD, which occurs during catalysis, promotes disruption of the tetramer into dimers. Together, these results suggest that the BjPutA tetramer is not essential for catalytic function. Further, our results suggest that tetramerization could inhibit catalytic function. This conclusion is in agreement with previous results showing that all other type A PutAs studied to date are dimeric (16). It is possible that the BjPutA tetramer is an anomaly in the PutA family and possibly an artifact—albeit an interesting one—of the in vitro purified enzyme. An alternative interpretation of the data presented here is that the BjPutA tetramer plays a physiological role by sequestering the inactive form of the enzyme. Because of the crowded nature of the cell and plasma membrane, it could be advantageous to limit the heterointeractions of the inactive enzyme by making it tightly homooligomerize.
To our knowledge, ours is the first report of the flavin redox state altering oligomeric structure of PutA. It is known that reduction of the FAD in PutA—by proline, dithionite, or NPPG—dramatically changes the structure of the FAD by inducing large butterfly bending of the isoalloxazine ring system and crankshaft rotation of the ribityl chain (34, 41, 43). These conformational changes in the FAD are thought to initiate a more global protein conformational change that enhances membrane association of PutA (33, 49), known collectively as “functional switching” (60). Thus, the observation that reduction of BjPutA via NPPG induces membrane association is consistent with studies of other PutAs. We note there is precedent for the flavin redox state controlling the oligomerization of flavoenzymes. For example, the binding of NADH to monomeric apoptosis-inducing factor results in a dimeric FADH2-NAD+ charge-transfer complex (61), and the conformational changes that lead to dimerization have been studied with SAXS (62). Also, the transition from ring-shaped tetramer to dimer observed for BjPutA is reminiscent of the reverse of the redox-linked “dimers-to-donuts” transition of peroxiredoxins (not a flavoenzyme), in which oxidation favors dimers and reduction favors a ring-shaped pentamer-of-dimers decamer (63).
This study highlights the technical challenges of connecting enzyme function and oligomeric structure. Steady-state kinetics assays typically use the enzyme at relatively low concentration, such as nanomolar. Low enzyme concentration is required in practice for several reasons, including satisfying the assumptions of the Michaelis-Menten equation, avoiding saturation of the assay readout, and ensuring the linearity of the assay readout with time. In contrast, biophysical measurements of oligomeric structure typically are done at much higher protein concentrations, such as micromolar or higher. Crystallography is the extreme case, in which the initial protein concentration in the crystallization drop is on the order of 10 mg mL−1, which corresponds to 0.2 mM for a 50 kDa protein. Similarly, although SAXS is an excellent tool for determining protein oligomeric structure (64), the minimal protein concentration for SAXS is ∼1 mg mL−1. And the gold standard of protein molecular mass determination—analytical ultracentrifugation—is typically performed at micromolar protein concentration. Thus, it is challenging to identify the predominant oligomeric state of an enzyme under the precise conditions that are used in functional assays. An added complexity is that the binding of substrates or a change in redox state of the enzyme could influence the enzyme self-association equilibrium.
Here, we used EM to overcome the concentration mismatch of kinetics and biophysical assays. An advantage of EM is that individual particles can be visualized in grids prepared at the concentrations used in enzyme assays, and the solution conditions in EM can be similar to those used in the functional assays. Further, the determination of oligomeric structure is a relatively low-resolution application of EM, so negative-stain EM or moderate resolution cryo-EM is sufficient. Given the current enthusiasm about the “resolution revolution” in cryo-EM (65) and the attendant investment in high-end EM by universities and national funding agencies, we anticipate that EM will increasingly become an important method for investigating how oligomeric structure underlies enzyme function.
Author Contributions
D.A.K., A.C.C., S.L.C., and S.C. performed the experiments. All authors analyzed the data and designed the experiments. D.A.K. and J.J.T. wrote the manuscript.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award numbers R01GM065546 and R01GM061068 as well as an administrative supplement to R01GM065546 (Collaborative Supplements for Cryo-Electron Microscopy Technology Transfer). This research used resources from the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under contract number DE-AC02-06CH11357. This project was supported by grant 9 P41 GM103622 from the NIGMS of the National Institutes of Health. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS. The content is solely the responsibility of the authors and does not necessarily reflect the official views of NIGMS or the National Institutes of Health.
Editor: James Cole.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30568-X.
Supporting Material
References
- 1.Szabados L., Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Per T.S., Khan N.A., Anjum N.A. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: phytohormones, mineral nutrients and transgenics. Plant Physiol. Biochem. 2017;115:126–140. doi: 10.1016/j.plaphy.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Qamar A., Mysore K.S., Senthil-Kumar M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 2015;6:503. doi: 10.3389/fpls.2015.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Becker D.F. Connecting proline metabolism and signaling pathways in plant senescence. Front. Plant Sci. 2015;6:552. doi: 10.3389/fpls.2015.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phang J.M., Liu W., Christian K.J. The proline regulatory axis and cancer. Front. Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elia I., Broekaert D., Fendt S.M. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 2017;8:15267. doi: 10.1038/ncomms15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willis A., Bender H.U., Valle D. PRODH variants and risk for schizophrenia. Amino Acids. 2008;35:673–679. doi: 10.1007/s00726-008-0111-0. [DOI] [PubMed] [Google Scholar]
- 8.Straub R.E., Jiang Y., Kendler K.S. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquet H., Berthelot J., Frebourg T. The severe form of type I hyperprolinaemia results from homozygous inactivation of the PRODH gene. J. Med. Genet. 2003;40:e7. doi: 10.1136/jmg.40.1.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan N., Doster A.R., Becker D.F. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect. Immun. 2008;76:3037–3044. doi: 10.1128/IAI.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajima K., Inatsu S., Nagata K. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed. Res. 2008;29:9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- 12.Berney M., Weimar M.R., Cook G.M. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol. Microbiol. 2012;84:664–681. doi: 10.1111/j.1365-2958.2012.08053.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee I.R., Lui E.Y., Fraser J.A. Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway. Genetics. 2013;194:421–433. doi: 10.1534/genetics.113.150326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanner J.J. Structural biology of proline catabolic enzymes. Antioxid. Redox Signal. 2017 doi: 10.1089/ars.2017.7374. Published online November 13, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L.K., Becker D.F., Tanner J.J. Structure, function, and mechanism of proline utilization A (PutA) Arch. Biochem. Biophys. 2017;632:142–157. doi: 10.1016/j.abb.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korasick D.A., Singh H., Tanner J.J. Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure. FEBS J. 2017;284:3029–3049. doi: 10.1111/febs.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava D., Schuermann J.P., Tanner J.J. Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA. 2010;107:2878–2883. doi: 10.1073/pnas.0906101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuermann J.P., White T.A., Tanner J.J. Three crystal forms of the bifunctional enzyme proline utilization A (PutA) from Bradyrhizobium japonicum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008;64:949–953. doi: 10.1107/S174430910802842X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rames M., Yu Y., Ren G. Optimized negative staining: a high-throughput protocol for examining small and asymmetric protein structure by electron microscopy. J. Vis. Exp. 2014;90:e51087. doi: 10.3791/51087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 21.Kimanius D., Forsberg B.O., Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franke D., Petoukhov M.V., Svergun D.I. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konarev P.V., Volkov V.V., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 26.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 27.Schneidman-Duhovny D., Hammel M., Sali A. FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozin M.B., Svergun D.I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 31.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valentini E., Kikhney A.G., Svergun D.I. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2015;43:D357–D363. doi: 10.1093/nar/gku1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu W., Haile A.M., Becker D.F. Involvement of the β3-α3 loop of the proline dehydrogenase domain in allosteric regulation of membrane association of proline utilization A. Biochemistry. 2013;52:4482–4491. doi: 10.1021/bi400396g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh H., Arentson B.W., Tanner J.J. Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc. Natl. Acad. Sci. USA. 2014;111:3389–3394. doi: 10.1073/pnas.1321621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo M., Gamage T.T., Tanner J.J. Structures of proline utilization A (PutA) reveal the fold and functions of the aldehyde dehydrogenase superfamily domain of unknown function. J. Biol. Chem. 2016;291:24065–24075. doi: 10.1074/jbc.M116.756965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu W., Gincherman Y., Becker D.F. Effects of proline analog binding on the spectroscopic and redox properties of PutA. Arch. Biochem. Biophys. 2002;408:131–136. doi: 10.1016/s0003-9861(02)00535-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M., White T.A., Tanner J.J. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43:12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostrander E.L., Larson J.D., Tanner J.J. A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate. Biochemistry. 2009;48:951–959. doi: 10.1021/bi802094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White T.A., Krishnan N., Tanner J.J. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J. Biol. Chem. 2007;282:14316–14327. doi: 10.1074/jbc.M700912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo M., Arentson B.W., Tanner J.J. Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release. Biochemistry. 2012;51:10099–10108. doi: 10.1021/bi301312f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava D., Zhu W., Tanner J.J. The structure of the proline utilization a proline dehydrogenase domain inactivated by N-propargylglycine provides insight into conformational changes induced by substrate binding and flavin reduction. Biochemistry. 2010;49:560–569. doi: 10.1021/bi901717s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White T.A., Johnson W.H., Jr., Tanner J.J. Structural basis for the inactivation of Thermus thermophilus proline dehydrogenase by N-propargylglycine. Biochemistry. 2008;47:5573–5580. doi: 10.1021/bi800055w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W., Zhang M., Becker D.F. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA--membrane binding. Biochemistry. 2007;46:483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arentson B.W., Luo M., Becker D.F. Kinetic and structural characterization of tunnel-perturbing mutants in Bradyrhizobium japonicum proline utilization A. Biochemistry. 2014;53:5150–5161. doi: 10.1021/bi5007404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer H., de Oliveira Neto M., Craievich A.F. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Cryst. 2010;43:101–109. [Google Scholar]
- 46.Wood J.M. Membrane association of proline dehydrogenase in Escherichia coli is redox dependent. Proc. Natl. Acad. Sci. USA. 1987;84:373–377. doi: 10.1073/pnas.84.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown E.D., Wood J.M. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J. Biol. Chem. 1993;268:8972–8979. [PubMed] [Google Scholar]
- 48.Surber M.W., Maloy S. Regulation of flavin dehydrogenase compartmentalization: requirements for PutA-membrane association in Salmonella typhimurium. Biochim. Biophys. Acta. 1999;1421:5–18. doi: 10.1016/s0005-2736(99)00104-2. [DOI] [PubMed] [Google Scholar]
- 49.Zhu W., Becker D.F. Flavin redox state triggers conformational changes in the PutA protein from Escherichia coli. Biochemistry. 2003;42:5469–5477. doi: 10.1021/bi0272196. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W., Krishnan N., Becker D.F. Kinetic and thermodynamic analysis of Bradyrhizobium japonicum PutA-membrane associations. Arch. Biochem. Biophys. 2006;445:174–183. doi: 10.1016/j.abb.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 51.Arentson B.W., Hayes E.L., Becker D.F. Engineering a trifunctional proline utilization A chimaera by fusing a DNA-binding domain to a bifunctional PutA. Biosci. Rep. 2016;36:e00413. doi: 10.1042/BSR20160435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodsell D.S., Olson A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 53.Levy E.D., Teichmann S. Structural, evolutionary, and assembly principles of protein oligomerization. Prog. Mol. Biol. Transl. Sci. 2013;117:25–51. doi: 10.1016/B978-0-12-386931-9.00002-7. [DOI] [PubMed] [Google Scholar]
- 54.Korasick D.A., Gamage T.T., Tanner J.J. Structure and characterization of a class 3B proline utilization A: ligand-induced dimerization and importance of the C-terminal domain for catalysis. J. Biol. Chem. 2017;292:9652–9665. doi: 10.1074/jbc.M117.786855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh R.K., Larson J.D., Tanner J.J. Small-angle X-ray scattering studies of the oligomeric state and quaternary structure of the trifunctional proline utilization A (PutA) flavoprotein from Escherichia coli. J. Biol. Chem. 2011;286:43144–43153. doi: 10.1074/jbc.M111.292474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinmetz C.G., Xie P., Hurley T.D. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z.J., Sun Y.J., Wang B.C. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 58.Luo M., Christgen S., Tanner J.J. Evidence that the C-terminal domain of a type B PutA protein contributes to aldehyde dehydrogenase activity and substrate channeling. Biochemistry. 2014;53:5661–5673. doi: 10.1021/bi500693a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korasick D.A., Pemberton T.A., Tanner J.J. Structural basis for the substrate inhibition of proline utilization A by proline. Molecules. 2017;23:32. doi: 10.3390/molecules23010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Becker D.F., Zhu W., Moxley M.A. Flavin redox switching of protein functions. Antioxid. Redox Signal. 2011;14:1079–1091. doi: 10.1089/ars.2010.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Churbanova I.Y., Sevrioukova I.F. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 2008;283:5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 62.Brosey C.A., Ho C., Tainer J.A. Defining NADH-driven allostery regulating apoptosis-inducing factor. Structure. 2016;24:2067–2079. doi: 10.1016/j.str.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood Z.A., Poole L.B., Karplus P.A. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 64.Korasick D.A., Tanner J.J. Determination of protein oligomeric structure from small-angle X-ray scattering. Protein Sci. 2018;27:814–824. doi: 10.1002/pro.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callaway E. The revolution will not be crystallized: a new method sweeps through structural biology. Nature. 2015;525:172–174. doi: 10.1038/525172a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.