Abstract
The spatiotemporal organization and dynamics of the plasma membrane and its constituents are central to cellular function. Fluorescence-based single-particle tracking has emerged as a powerful approach for studying the single molecule behavior of plasma-membrane-associated events because of its excellent background suppression, at the expense of imaging speed and observation time. Here, we show that interferometric scattering microscopy combined with 40 nm gold nanoparticle labeling can be used to follow the motion of membrane proteins in the plasma membrane of live cultured mammalian cell lines and hippocampal neurons with up to 3 nm precision and 25 μs temporal resolution. The achievable spatiotemporal precision enabled us to reveal signatures of compartmentalization in neurons likely caused by the actin cytoskeleton.
Introduction
Single-particle tracking (SPT) generally involves tagging an object of interest, such as a single molecule, with a probe and determining its position with subpixel localization precision in a series of images (1). These trajectories contain information on the diffusion coefficient of the molecule and, for deviations from purely Brownian motion, clues regarding the origin of additional factors controlling its motion (2, 3, 4, 5). Although SPT was first established using gold beads (6), current implementations largely make use of quantum dots (QDs) or single fluorescent dyes as probes, leading to the development of high-density methods (7, 8, 9).
The advantage of fluorescence emission as a contrast mechanism with respect to background suppression is somewhat offset, which is caused by photochemical and photophysical effects. Scattering labels, such as gold nanoparticles, are not subject to the limitations of fluorescent dyes and can thus in principle achieve much higher imaging speeds without loss of localization precision. SPT in living cells has been achieved with gold nanoparticle labels and dark-field microscopy with up to 25 μs temporal resolution and ∼17 nm spatial precision, revealing signatures of hop diffusion, albeit requiring comparatively large 40 nm labels (10). Although improvements to this approach have been reported in vitro (11), the seminal work by Kusumi and co-workers has remained state-of-the-art in terms of simultaneous imaging speed and localization precision for SPT on living cells.
More recently, interferometric scattering microscopy (iSCAT) (12) has demonstrated even better capabilities down to a precision of a few nanometers (13) and a temporal resolution of a few microseconds even with 20 nm diameter labels in vitro (14, 15). Given the importance of background suppression and the fact that iSCAT has to date only been demonstrated in environments optimized to provide minimal background scattering, it is unclear to what degree the technology is applicable to studies of live cells. To address this, we used gold nanoparticles (AuNPs) coupled to GFP-tagged or YFP-tagged model transmembrane proteins to demonstrate iSCAT-based SPT of membrane proteins with nanometer precision at multikilohertz speeds in live epithelial cells and cultured hippocampal neurons. We use this approach to investigate the effect of an increase in spatiotemporal precision on the appearance of anomalous diffusion at the nanoscale. Such deviation from Brownian motion could arise from interactions between the plasma membrane and the cortical cytoskeleton recently described in hippocampal neurons as a periodic actin-spectrin lattice (16). Here, we apply iSCAT to perform SPT with simultaneous 3 nm spatial precision and up to 25 μs temporal resolution on live cells, including human osteosarcoma cells and rat neurons. We find evidence of confined diffusion, including signatures reminiscent of the periodic actin-spectrin network in axons.
Materials and Methods
Cell culture
Primary hippocampal neurons were prepared from embryonic day 18 Sprague-Dawley rats (Charles River, Wilmington, MA). Neurons were maintained in neurobasal medium with B27 supplement and GlutaMAX (Thermo Fisher Scientific, Waltham, MA) on polylysine (Sigma-Aldrich, St. Louis, MO)-coated μGrid No. 1.5 glass bottom petri dishes (ZellKontakt, Nörten-Hardenberg, Germany) at 37°C in a CO2-controlled humidified incubator. After 3 days, the cytostatic cytosine β-d-arabinofuranoside (Sigma-Aldrich) was added at a final concentration of 5 μM. Neurons were transiently transfected with protein fusion constructs between day in vitro (DIV) 2 and DIV 8 using Lipofectamine 2000 (Thermo Fisher Scientific). The glycosylphosphatidylinositol (GPI)-GFP protein fusion construct was a gift from the Helenius laboratory (ETH Zurich, Zurich, Switzerland). L-YFP-GT46 (TM) was a gift from P. Keller (Max Planck Institute for Cell Biology and Genetics, Dresden, Germany). The dsRed-derived β-barrel fluorophore mHoneydew3 was subcloned into the plasmid to yield L-YFP-GTmHoneydew46 (TMX) with a larger cytosolic domain. Neurites were identified as either axons or dendrites based on morphology.
Human osteosarcoma (U2OS) cells were maintained in Dulbecco’s modified eagle medium supplemented with fetal calf serum, glutamax, and pen/strep (All Life Technologies, Carlsbad, CA) at 37°C in a CO2-controlled humidified incubator.
Immunogold labeling
40 nm AuNPs were functionalized with streptavidin in-house, or commercially available 40 nm AuNPs (Sigma) were incubated with biotinylated anti-GFP nanobodies at molar ratios of 3–5:1. AuNPs were pelleted by centrifugation at 5000 rcf and resuspended by sonication in imaging buffer before addition to samples. At this stoichiometry, streptavidin-binding sites were not saturated.
SPT
SPT on live cells was performed in live cell imaging buffer (145 mM NaCl, 5 mM KCl, 10 mM glucose, 25 mM HEPES, 2 mM CaCl2, 1 mM MgCl2, (w/v) bovine serum albumin, 10 mM ascorbate). Functionalized AuNPs were then added before image acquisition. Multiple image series of frames were recorded at kHz. Imaging was performed at room temperature. Floating AuNPs were readily observed in the medium whenever they came close to the coverslip. Few AuNPs bound to the transfected cells, and typically a neurite was followed until a mobile AuNP was found.
iSCAT
For imaging at 40 kHz, a small area of the sample (fullwidth at half-maximum of 5 μm) was illuminated with high average power density by underfilling the back aperture of the objective with the collimated output of a fiber-coupled 635 nm diode laser. An output power of 6 mW was used at 40 kHz, corresponding to a power density at the sample of kW/cm2. At the same time, a large area of the sample was illuminated by scanning the collimated output of a 660 nm laser diode across the back focal plane of the objective with an orthogonal pair of acousto-optical deflectors. The two channels for illumination and imaging were combined and separated via dichroic mirrors (Fig. S1). The incident power densities lead to negligible local heating, which in turn could affect the observed mobilities. Previous studies (17) have observed surface heating on the order of 10 K for continuous wave illumination at 532 of 40 nm diameter AuNPs at MW/cm2 illumination densities. Given that our intensities are more than two orders of magnitude lower and preresonant (635 nm) rather than fully on-resonant (532 nm) with the plasmon resonance of the particle, we conclude that temperature-induced artifacts can be excluded in this study.
The position of 40 nm AuNPs was fitted using a two-dimensional (2D) Gaussian function to images, which were corrected using a temporal median filter. The localization error was estimated in each frame using the covariance-weighted mean-square error of the fit. Inaccurate localizations were removed (preserving the time coordinate) by filtering out data points with localization errors of nm. Noise levels of up to allow for the visualization of 40 nm AuNPs with an average localization error of 3 nm and a signal-to-noise ratio of 25 at 40 kHz frame rates. Trajectories were calculated by connecting localizations in subsequent frames. Background noise from cells is reduced substantially by temporal averaging; for example, data recorded on neurons at 40 kHz and averaged to 2 kHz have a background noise level of . This indicates that the background scattering noise fluctuates on fast timescales.
Data analysis
Trajectory and data analysis were performed along the axis of the neurite. Typically, the neurite was straight over the length of the trajectory because of the image acquisition parameters ( μm field of view). Where necessary, the trajectory was fragmented in time such that the region of the neurite traversed by the probe was approximately straight. One-dimensional diffusion coefficients were calculated as follows: .
Results and Discussion
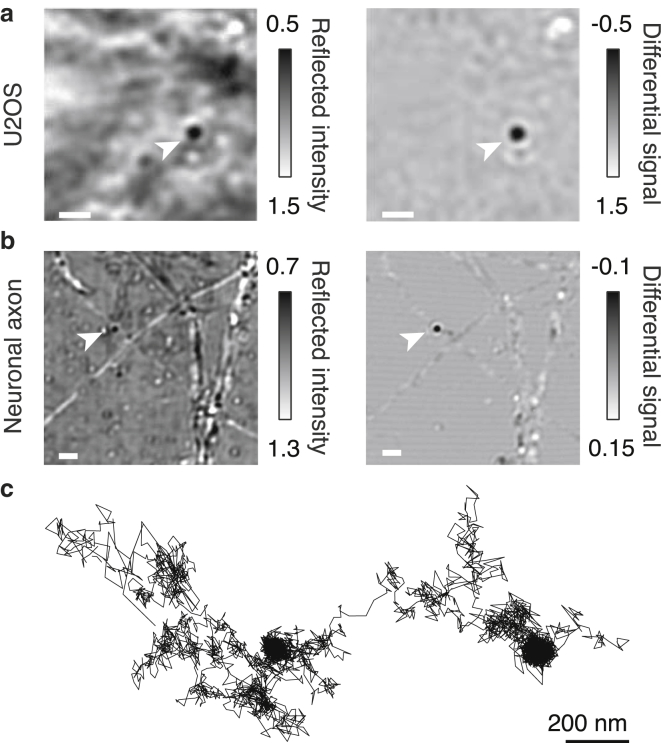
Our 40 nm AuNP labels could be observed directly on U2OS human osteosarcoma cells (Fig. 1 a) or on cultured hippocampal rat neurons (Fig. 1 b) in unprocessed frames collected at 0.5 ms exposure time. Removing the background caused by diffracting material in cells (left and right panels) significantly improved the image quality, resulting in an average localization error of 3 nm. By constructing a trajectory from the localizations of an AuNP-labeled GPI-GFP on a U2OS cell at 2 kHz, we observed regions where the protein halts for up to 50 ms (100 frames) in regions that are a few tens of nanometers in diameter (Fig. 1 c; Video S1).
Figure 1.
iSCAT images of a 40 nm AuNP-labeled membrane protein on U2OS cells (a) and on neuronal cells (b). The gold particle is marked by an arrow in both the raw (left) and background-subtracted (right) images. The reflected intensity was normalized in each case. Scale bars, 2 μm. (c) Representative trajectory of a 40 nm AuNP-labeled protein in the U2OS cell membrane recorded at 2 kHz (10,000 frames total) is shown.
We observed some scattering fluctuations from cells that varied in amplitude depending on the thickness and shape of the cell. On thin regions of U2OS cells and on neuronal axons or dendrites, we routinely observed background fluctuations of root mean-square (RMS) of the background light intensity. For comparison, the shot noise-induced background fluctuations under the current imaging conditions amounted to root mean-square (18). The variations in background scattering caused by fluctuating cell material were thus the limiting factor preventing the use of smaller scattering labels if nanometer localization precision was to be maintained.
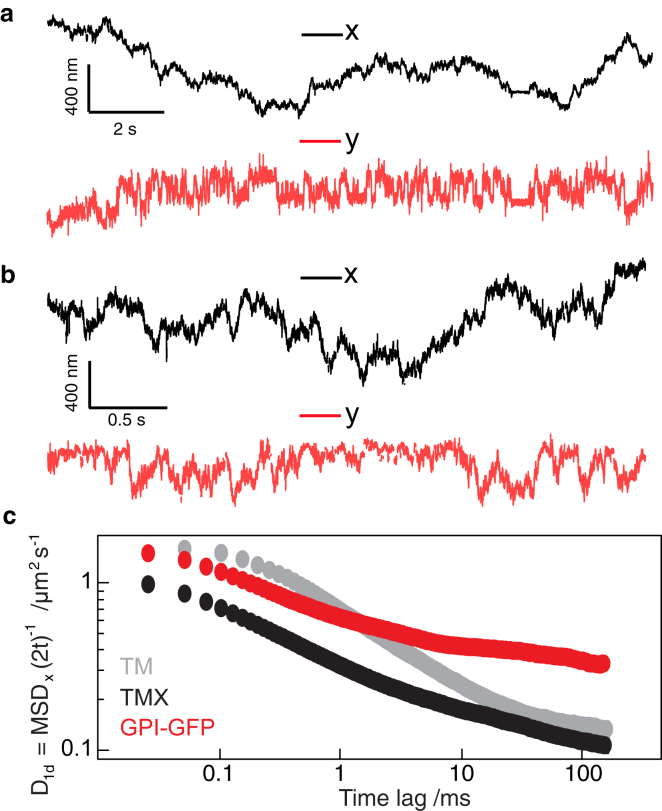
We then performed SPT experiments on neurites of cultured hippocampal neurons (Fig. 2). Neurons were imaged after DIV 6 because by that point, neuronal cells were well developed, and the axon extended far from the soma (Fig. S2). This greatly increased the probability of finding a suitable region of the cell with a weakly fluctuating background. Trajectories for 40 nm AuNPs attached to membrane proteins were oriented along the long axis of the neurite (Fig. 2, a and b; Fig. S3). The time-dependent one-dimensional diffusion coefficients (: longitudinal motion) for our trajectories revealed a significant reduction in for all membrane probes with increasing time lags (Fig. 2 c). This indicates that molecules exhibited subdiffusive motion, which was consistent with previous reports (19). We found subdiffusive behavior for all probes tested: the transmembrane proteins L-YFP-GT46—consisting of the single transmembrane domain of the low-density lipoprotein receptor and the intracellular domain of CD46 (TM)—and L-YFP-GTmHoneydew46, which additionally contained a bulky intracellular fluorescent protein (TMX), as well as a GFP attached to the outer membrane leaflet via a glycosyl-phosphatidylinositol anchor (GPI-GFP).
Figure 2.
Analysis of 2D trajectories of 40 nm AuNP-labeled membrane protein probes. (a) Displacement in x (in direction of neurite propagation, black) and y (perpendicular to direction of neurite propagation, red) for a bead tracked at 2 kHz is shown. (b) The same scenario is shown for a bead tracked at 40 kHz. (c) Time-dependent one-dimensional diffusion coefficient (x dimension) plotted against the time-lag used for analysis. Shown are plots for membrane proteins differing in their cytosolic domains and membrane anchors. TM = L-YFP-GT46, TMX = L-YFP-GTmHoneydew46, GPI-GFP = glycosylphosphatidylinositol-anchored green fluorescent protein. To see this figure in color, go online.
A critical and recurring question relates to the validity of SPT results obtained with nanoparticle labels. As in our case, their much larger size compared to that of fluorescent dyes enables higher tracking speed, precision, and duration at the expense of introducing a much more significant perturbation to the system. In the past, attempts to address this issue have focused on controlling the number of potential interaction points (20). Although it is well established that nanoparticle labels slow down the diffusion of lipids, for example by multiple interactions depending on labeling stoichiometry, there is little direct evidence that the label can switch the mode of motion from Brownian to anomalous. Our recent observations of anomalous subdiffusion in the presence of transmembrane coupling and Brownian motion in its absence with identical nanoparticle labels (13) support this notion, as does the recently demonstrated comparability of SPT and fluorescence correlation spectroscopy performed with different methodologies and label sizes (21). Although the complexity of the cellular environments precludes the level of control achievable in an artificial system, we argue that the current evidence suggests that the observed nanoparticle dynamics is representative of the underlying receptor motion even if the absolute diffusion coefficients may be influenced by factors such as cross-linking.
As shown previously (22), the diffusion coefficient of GPI-GFP was about four times higher than for transmembrane proteins on timescales of ms, indicating that the gold particle label is not dominant in determining the mobility of the moiety to which it is attached. It is widely accepted that GPI-anchored proteins exhibit subdiffusive motion in the plasma membrane and at longer time lags show increased mobility compared to transmembrane proteins. The motion of GPI-anchored proteins is reportedly influenced by transmembrane interactions with the cytoskeleton that may or may not be cholesterol dependent (23, 24).
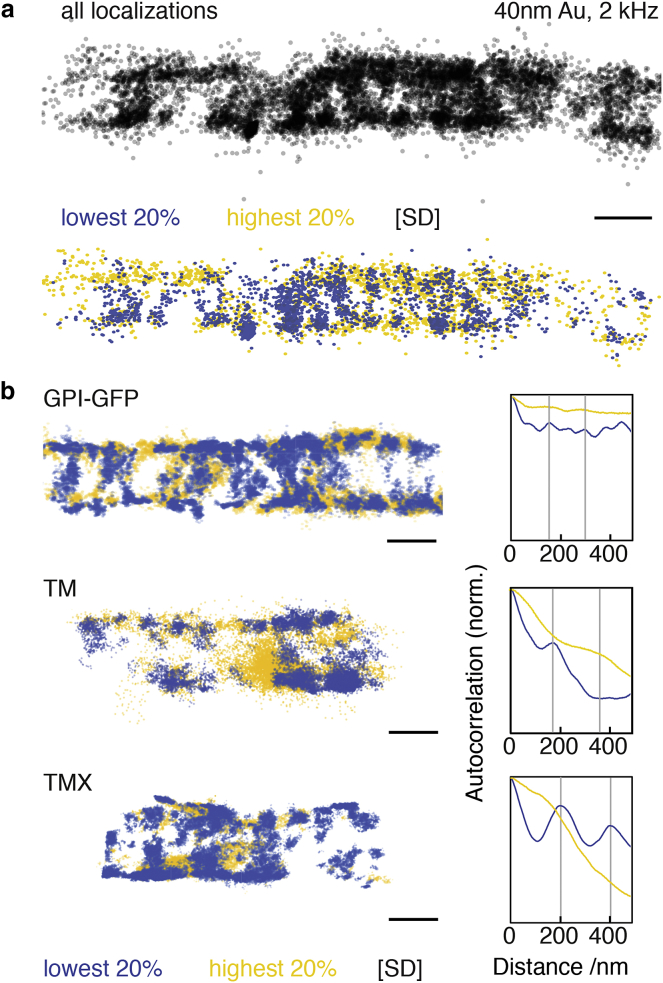
When we analyzed individual trajectories measured in neuronal cells, we did not observe transient periods of confinement in neurons as we did in U2OS cells (Fig. 3; Fig. S4), indicating that these events are unlikely to be caused by the label. By visual examination, the iSCAT trajectories did not generally contain signatures expected from compartmentalized motion. On some occasions, however, a seemingly periodic array of preferential residence areas of membrane molecules became apparent (Fig. 3, a and b), similar to our previous observation of membrane compartments in the axon initial segment with QD-labeled membrane proteins (25). Numerous reports highlight the existence of a periodic cortical actin-spectrin network in axons (16, 26, 27, 28) and possibly dendrites (29, 30), and we found the periodic arrangement of membrane compartments to be alternating with such actin rings in our fluorescence imaging studies (25).
Figure 3.
Analysis of membrane protein mobility on neurites. (a) localizations from a 2 kHz tracking experiment of a 40 nm AuNP-labeled GPI-GFP protein (black, top). The of data points exhibiting the lowest confinement are shown in yellow, and the with highest confinement are shown in blue (below). ((b), left) Confinement analysis of different membrane proteins in neurites. TM = L-YFP-GT46, TMX = L-YFP-GTmHoneydew46, GPI-GFP = glycosylphosphatidylinositol-anchored green fluorescent protein. ((b), right) Autocorrelations along the propagation direction of the neurite of the most confined (blue) and least confined (yellow) of datapoints from the data set are shown. Scale bars, 200 nm. To see this figure in color, go online.
We thus asked whether we could find evidence for the same nm periodicity in our iSCAT trajectory data. We applied a moving window of 5 ms over the frame-to-frame displacement of the particles and plotted the of positions with the lowest SD over this window in blue (Fig. 3 a). Such a statistical analysis preferentially detects weak confinement events, which are otherwise overshadowed by random motion (see also Fig. S5). For these datapoints, a repetitive pattern was clearly visible from the plotted positions in several trajectories. On the other hand, when we plotted the of positions that showed the lowest confinement in this analysis (yellow), we found that no such pattern was detectable. The repetitive pattern seen in the autocorrelation analysis of confined localizations followed a nm periodicity (Fig. 3 b), which was comparable to that of the actin-spectrin cytoskeleton in neurites. This observation agrees with our previous description of a periodic pattern in localizations from SPT with QDs on live neurons (25). We remark that we here only tracked single AuNPs over a few seconds, compared to multiple QDs being tracked over several minutes in our previous work. This demonstrates the high sensitivity of the iSCAT approach, enabled by the comparatively large number of datapoints retrievable from individual trajectories. Compared to U2OS cells, neurons presented additional challenges as the diameter of a neurite is typically nm with an approximately circular shape and therefore high curvature.
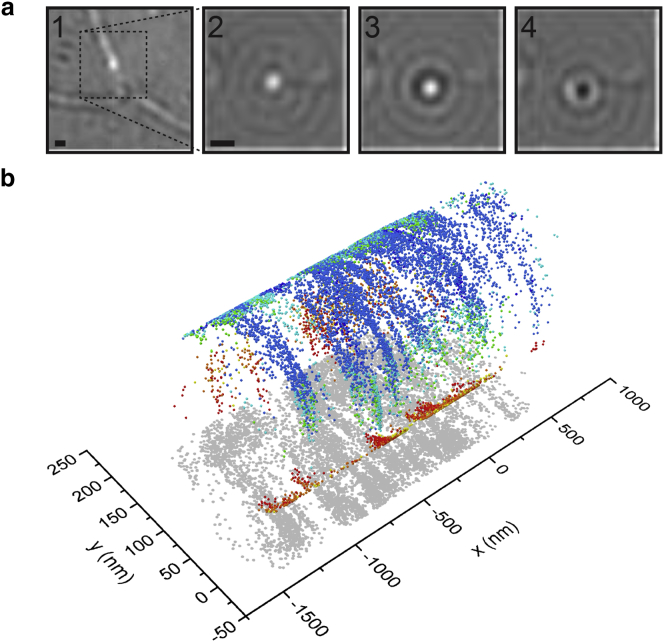
Because of the interferometric nature of iSCAT, the contrast of the AuNPs varied with their height above the coverglass (Fig. 4 a). For 40 nm AuNPs, the contrast ranged from to , yielding a maximal signal-to-noise ratio of at 635 nm. We took advantage of this observation and assigned a z-value to our 2D SPT data based on changes in contrast (Fig. S7) to render our localizations in three dimensions (3D) (Fig. 4 b). We note that the density in the y coordinate is less amenable to interpretation for several reasons: the projection of the cross section of the neurite onto the y axis produces a higher density of localizations at the circumference of the neurite. Furthermore, the localization density in y depends on the proximity to the zero-contrast plane, the gold particle being undetectable in this plane. We remark that the 3D information is only qualitative. An accurate and indeed even vaguely quantitative calibration would require the control and knowledge of two key parameters (31) that determine the amplitude and sign of the interference term measured by iSCAT. 1) A means to control the distance of an AuNP to the cover glass surface with nanometer precision is required. We remark that approaches based on translating the sample with a nanopositioner, as often used in fluorescence-based experiments, is not possible in iSCAT because the critical parameter is the particle-to-surface distance and not the position of the object in the focus. 2) The signal amplitude and sign is a consequence of several phase factors contributing to the interference term, including the Gouy phase (which is nontrivial to measure in a high numerical aperture system), the scattering phase (which varies from particle to particle), and the pathlength difference between the particle and the reference wave (which is also nontrivial to quantify on the nanometer scale even for a stationary particle). The resulting scatterplots nevertheless reproduce the pattern expected from the shape of the neurite (Fig. 4 b).
Figure 4.
3D iSCAT tracking on live neurons. (a) Individual frames from a video show a 40 nm AuNP diffusing on a neurite, exhibiting variations in contrast as a function of time. (b) A 3D trajectory shows a neurite reconstructed from the -coordinates using the circular geometry (Fig. S7). The sign of the z-coordinate was determined by applying the condition that contrast varies continuously with z. Contrast values are color-coded from red to blue. The recorded 2D data are displayed in gray and projected onto the plane. Scale bars, 500 nm. To see this figure in color, go online.
We have shown here that iSCAT SPT with a spatial precision of a few nanometers and a temporal resolution in the microsecond range is possible on live cells. The key difference that has allowed an improvement in spatiotemporal resolution over existing dark-field methods is likely a combination of the achievable power densities with lasers, as opposed to incoherent illumination (23), and a reduced requirement for high dynamic range detection. The latter stems from the weaker dependence of scattering intensity on object diameter for interferometric compared to dark-field detection, making it feasible to image a weak scatterer in the presence of significant background from large objects, such as axons, or the cell body. Using this approach, we found that the apparent diffusion coefficient of membrane molecules indeed depends on the timelag with which the mean-square displacement is measured and calculated, as proposed previously (10). This has been interpreted as indicative of membrane compartmentalization (19) because according to the Singer-Nicolson model, diffusion should be free in the plasma membrane (32). Kusumi et al. suggested a model in which membrane protein diffusion is free on the microsecond and nanometer scale but confined in domains of 10–100 s of nanometers in diameter in the plasma membrane, depending on the submembrane actin cytoskeleton (23). This notion is supported by recent observations in our laboratory revealing confinement of GPI-GFP motion between 200-nm-spaced actin rings in the axon initial segment (25). Using iSCAT, we did not observe clearly defined compartments, yet we still found signatures of subdiffusive motion on the timescale of tens of microseconds. Spatially resolved analysis of protein motion suggests a periodicity consistent with actin ring spacings, although we cannot here confirm axon initial segment localization. Measurements close to the soma where the axon initial segment is located were impeded by a highly fluctuating background and a low probability of encountering AuNPs.
It will be exciting to investigate if membrane compartmentalization can be correlated to specific locations in neurons. Furthermore, the basis for subdiffusive motion on longer timescales could be transmembrane interactions with other submembrane agents. In previous experiments in supported lipid bilayers, we showed that the motion of cross-linked lipids in the top bilayer leaflet can be influenced by the immobilization of lipids in the lower bilayer leaflet (13). Future work aimed at identifying the specific factors that control transmembrane confinement of membrane molecules in cells will be an important contribution to the understanding of the dynamics and organization of the plasma membrane. The high spatiotemporal precision of iSCAT together with the potential for 3D tracking is a powerful approach to SPT in live cells.
It is important here to emphasize the complementarity of different technologies. SPT based on iSCAT on cells will always suffer from scattering background from the cell body and any associated temporal fluctuations, which is a factor that has also made it impossible to use labels smaller than 40 nm AuNPs in this study. High-speed mobility observations are now possible using single-molecule fluorescence, for example through the use of stimulated emission depletion (33) or nanophotonic approaches (34), although the combination of spatiotemporal resolution and observation time demonstrated here is unlikely to be matched by single-molecule fluorescent labels in the absence of dramatic improvement of the associated photochemistry and photophysics. We therefore believe that the true additional benefit of iSCAT for SPT on living cells is in combination with existing and future single-molecule fluorescence methods in a complementary rather than a competitive fashion.
Author Contributions
H.E. and P.K. conceptualized the study. G.d.W., D.A., H.E., and P.K. devised the methodology. G.d.W. and D.A. utilized the software. G.d.W., D.A., H.E., and P.K. conducted validation. G.d.W. and D.A. performed the formal analysis. G.d.W. and D.A. conducted the investigation. D.A. and H.E. offered resources. G.d.W., D.A., H.E., and P.K. wrote the original draft of the manuscript. G.d.W., D.A., H.E., and P.K. reviewed and edited the article. G.d.W., D.A., H.E., and P.K. created the visualizations. H.E. and P.K. supervised the project.
Acknowledgments
H.E. was supported by Deutsche Forschungsgemeinschaft grant SFB 958 INST 130/827-2. P.K. was supported by a European Research Council Starting Investigator grant (nanoscope, 337757).
Editor: Katharina Gaus.
Footnotes
Eight figures and one video are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30582-4.
Supporting Material
References
- 1.Chenouard N., Smal I., Meijering E. Objective comparison of particle tracking methods. Nat. Methods. 2014;11:281–289. doi: 10.1038/nmeth.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewers H., Smith A.E., Helenius A. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl. Acad. Sci. USA. 2005;102:15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmuth J.A., Burckhardt C.J., Sbalzarini I.F. A novel supervised trajectory segmentation algorithm identifies distinct types of human adenovirus motion in host cells. J. Struct. Biol. 2007;159:347–358. doi: 10.1016/j.jsb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Persson F., Lindén M., Elf J. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods. 2013;10:265–269. doi: 10.1038/nmeth.2367. [DOI] [PubMed] [Google Scholar]
- 5.Holcman D., Hoze N., Schuss Z. Analysis and interpretation of superresolution single-particle trajectories. Biophys. J. 2015;109:1761–1771. doi: 10.1016/j.bpj.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geerts H., De Brabander M., Hollenbeck P. Nanovid tracking: a new automatic method for the study of mobility in living cells based on colloidal gold and video microscopy. Biophys. J. 1987;52:775–782. doi: 10.1016/S0006-3495(87)83271-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cognet L., Leduc C., Lounis B. Advances in live-cell single-particle tracking and dynamic super-resolution imaging. Curr. Opin. Chem. Biol. 2014;20:78–85. doi: 10.1016/j.cbpa.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Triller A., Choquet D. New concepts in synaptic biology derived from single-molecule imaging. Neuron. 2008;59:359–374. doi: 10.1016/j.neuron.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Giannone G., Hosy E., Cognet L. High-content super-resolution imaging of live cell by uPAINT. Methods Mol. Biol. 2013;950:95–110. doi: 10.1007/978-1-62703-137-0_7. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara T., Ritchie K., Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno H., Nishikawa S., Noji H. Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys. J. 2010;98:2014–2023. doi: 10.1016/j.bpj.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kukura P., Ewers H., Sandoghdar V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Meth. 2009;6:923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- 13.Spillane K.M., Ortega-Arroyo J., Kukura P. High-speed single-particle tracking of GM1 in model membranes reveals anomalous diffusion due to interleaflet coupling and molecular pinning. Nano Lett. 2014;14:5390–5397. doi: 10.1021/nl502536u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh C.L., Spindler S., Sandoghdar V. Tracking single particles on supported lipid membranes: multimobility diffusion and nanoscopic confinement. J. Phys. Chem. B. 2014;118:1545–1554. doi: 10.1021/jp412203t. [DOI] [PubMed] [Google Scholar]
- 15.Wu H.M., Lin Y.H., Hsieh C.L. Nanoscopic substructures of raft-mimetic liquid-ordered membrane domains revealed by high-speed single-particle tracking. Sci. Rep. 2016;6:20542. doi: 10.1038/srep20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu K., Zhong G., Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda M., Saito Y., Kawata S. Nanoscale heating of laser irradiated single gold nanoparticles in liquid. Opt. Express. 2011;19:12375–12383. doi: 10.1364/OE.19.012375. [DOI] [PubMed] [Google Scholar]
- 18.Ortega-Arroyo J., Kukura P. Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Phys. Chem. Chem. Phys. 2012;14:15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- 19.Murase K., Fujiwara T., Kusumi A. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 2004;86:4075–4093. doi: 10.1529/biophysj.103.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G.M., Ishihara A., Jacobson K.A. Direct observation of brownian motion of lipids in a membrane. Proc. Natl. Acad. Sci. USA. 1991;88:6274–6278. doi: 10.1073/pnas.88.14.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagerholm B.C., Andrade D.M., Eggeling C. Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED-FCS. J. Phys. D Appl. Phys. 2017;50:063001. doi: 10.1088/1361-6463/aa519e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albrecht D., Winterflood C.M., Ewers H. Dual color single particle tracking via nanobodies. Methods Appl. Fluoresc. 2015;3:024001. doi: 10.1088/2050-6120/3/2/024001. [DOI] [PubMed] [Google Scholar]
- 23.Kusumi A., Nakada C., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 24.Saha S., Anilkumar A.A., Mayor S. GPI-anchored protein organization and dynamics at the cell surface. J. Lipid Res. 2016;57:159–175. doi: 10.1194/jlr.R062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albrecht D., Winterflood C.M., Ewers H. Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. J. Cell Biol. 2016;215:37–46. doi: 10.1083/jcb.201603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong G., He J., Zhuang X. Developmental mechanism of the periodic membrane skeleton in axons. eLife. 2014;3:e04581. doi: 10.7554/eLife.04581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J., Zhou R., Zhuang X. Prevalent presence of periodic actin-spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc. Natl. Acad. Sci. USA. 2016;113:6029–6034. doi: 10.1073/pnas.1605707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leterrier C., Potier J., Dargent B. Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 2015;13:2781–2793. doi: 10.1016/j.celrep.2015.11.051. [DOI] [PubMed] [Google Scholar]
- 29.D’Este E., Kamin D., Hell S.W. STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 2015;10:1246–1251. doi: 10.1016/j.celrep.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Bär J., Kobler O., Mikhaylova M. Periodic F-actin structures shape the neck of dendritic spines. Sci. Rep. 2016;6:37136. doi: 10.1038/srep37136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan M., Mojarad N., Sandoghdar V. Geometry-induced electrostatic trapping of nanometric objects in a fluid. Nature. 2010;467:692–695. doi: 10.1038/nature09404. [DOI] [PubMed] [Google Scholar]
- 32.Singer S.J., Nicolson G.L. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 33.Eggeling C., Ringemann C., Hell S.W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 34.Punj D., Ghenuche P., Wenger J. Plasmonic antennas and zero-mode waveguides to enhance single molecule fluorescence detection and fluorescence correlation spectroscopy toward physiological concentrations. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2014;6:268–282. doi: 10.1002/wnan.1261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.