Abstract
Interest in how proline contributes to cancer biology is expanding because of the emerging role of a novel proline metabolic cycle in cancer cell survival, proliferation, and metastasis. Proline biosynthesis and degradation involve the shared intermediate Δ1-pyrroline-5-carboxylate (P5C), which forms L-glutamate-γ-semialdehyde (GSAL) in a reversible non-enzymatic reaction. Proline is synthesized from glutamate or ornithine through GSAL/P5C, which is reduced to proline by P5C reductase (PYCR) in a NAD(P)H-dependent reaction. The degradation of proline occurs in the mitochondrion and involves two oxidative steps catalyzed by proline dehydrogenase (PRODH) and GSAL dehydrogenase (GSALDH). PRODH is a flavin-dependent enzyme that couples proline oxidation with reduction of membrane-bound quinone, while GSALDH catalyzes the NAD+-dependent oxidation of GSAL to glutamate. PRODH and PYCR form a metabolic relationship known as the proline–P5C cycle, a novel pathway that impacts cellular growth and death pathways. The proline–P5C cycle has been implicated in supporting ATP production, protein and nucleotide synthesis, anaplerosis, and redox homeostasis in cancer cells. This Perspective details the structures and reaction mechanisms of PRODH and PYCR and the role of the proline–P5C cycle in cancer metabolism. A major challenge in the field is to discover inhibitors that specifically target PRODH and PYCR isoforms for use as tools for studying proline metabolism and the functions of the proline–P5C cycle in cancer. These molecular probes could also serve as lead compounds in cancer drug discovery targeting the proline–P5C cycle.
Graphical Abstract
The pyrrolidine ring makes proline a unique proteogenic amino acid with a distinctive role in protein folding and secondary structures. In addition to being required for protein biosynthesis, L-proline has critical roles in cellular bioenergetics,1–5 osmoregulation,5,6 stress protection,7–10 cellular signaling processes such as apoptosis,3,11,12 and cancer cell metabolism.3,13,14 Studies over the last two decades by Phang and co-workers have established a specialized role for proline in cancer metabolism.3,12,13,15–17 Further, recent discoveries of the broad effects of proline metabolism on cancer cell growth and survival have implicated proline metabolic enzymes as potential targets for therapeutic intervention.14,18–21 This Perspective examines the structures and mechanisms of two key proline metabolic enzymes, proline dehydrogenase (PRODH) and Δ1-pyrroline-5-carboxylate (P5C) reductase (P5CR, a.k.a. PYCR), and their role in cancer metabolism. The potential for designing inhibitors of PRODH and P5CR for cancer therapeutics is also discussed.
PROLINE METABOLISM
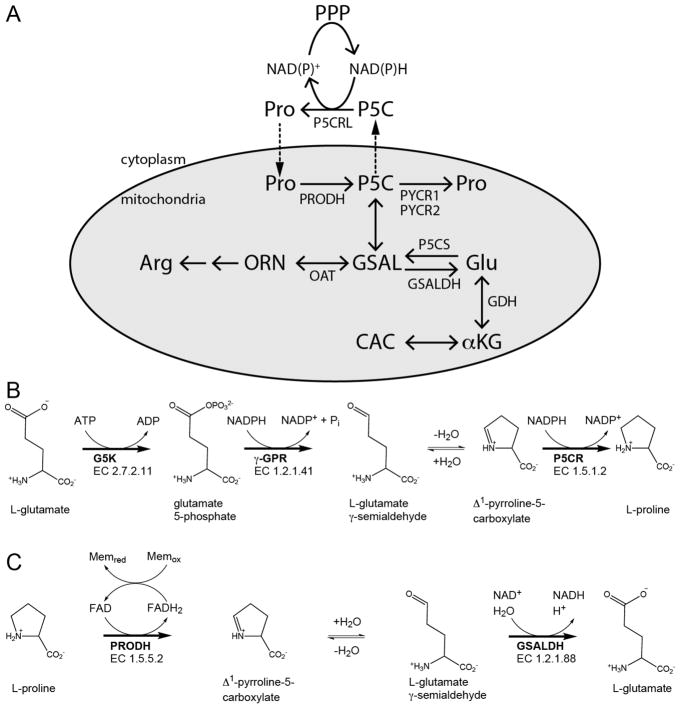
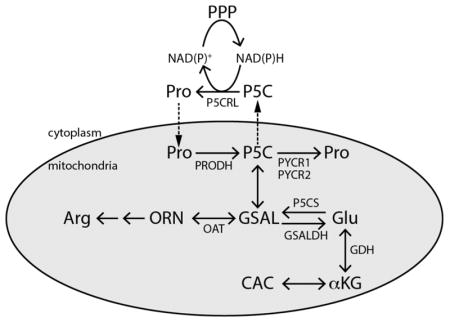
Proline is synthesized from ornithine or glutamate, with both precursors leading to L-glutamate-γ-semialdehyde (GSAL), an intermediate that spontaneously cyclizes to P5C with loss of water (Figure 1A).1,4,22 The formation of GSAL from ornithine is catalyzed by ornithine δ-amino acid transferase (EC 2.6.1.13). The route to GSAL from glutamate requires glutamate 5-kinase (G5K; EC 2.7.2.11) and γ-glutamyl phosphate reductase (γ-GPR; EC 1.2.1.41) (Figure 1B). G5K and γ-GPR are separate enzymes in bacteria and lower eukaryotes, whereas in higher organisms, such as plants and humans, G5K and γ-GPR are fused together in the bifunctional enzyme P5C synthase (P5CS).4,22 The final step of both proline biosynthetic routes is the reduction of P5C to proline catalyzed by NAD(P)H-dependent P5CR (EC 1.5.1.2). In humans, P5CR is known as PYCR, with isoforms PYCR1 and PYCR2 in the mitochondrion and isoform PYCRL in the cytosol.
Figure 1.
Reactions and enzymes of proline metabolism in higher organisms. (A) Summary of proline catabolism and biosynthesis. The gray oval represents the mitochondrion. (B) The reactions of proline biosynthesis from glutamate. (C) The reactions of proline catabolism. Abbreviations that are not listed in the text: ORN, ornithine; OAT, ornithine δ-aminotransferase; CAC, citric acid cycle; αKG, α-ketoglutarate; GDH, glutamate dehydrogenase; PPP, pentose phosphate pathway. P5CS is a bifunctional enzyme with G5K and γ-GPR fused together. Adapted with permission from ref 4. Copyright 2017 Mary Ann Liebert, Inc.
Breakdown of proline occurs strictly in the mitochondria, with PRODH and GSAL dehydrogenase (GSALDH) localized at the inner mitochondrial membrane and matrix, respectively (Figure 1A). PRODH (EC 1.5.5.2) performs the first step by generating P5C, which upon non-enzymatic hydrolysis forms GSAL (Figure 1C). PRODH is a flavin-containing enzyme that couples the oxidation of proline with reduction of membrane-bound ubiquinone or Coenzyme Q. Humans have two genes annotated as PRODH: PRODH1 (chromosome 22q11.21; NCBI Accession NM_016335) and PRODH2 (chromosome 19q13.12; NCBI Accession NM_021232). Deficiencies in PRODH1 manifest in type-I hyperprolinemia,23 whereas the loss of PRODH2 activity leads to hydroxyprolinemia,24 consistent with PRODH2 catalyzing the oxidation of trans-4-hydroxy-L-proline to Δ1-pyrroline-3-OH-5-carboxylate (3-OH-P5C).
GSALDH (EC 1.2.1.88) catalyzes the NAD+-dependent oxidation of GSAL to form glutamate (Figure 1C). Structural and kinetic analysis of human GSALDH shows that it follows a bi-bi-ordered mechanism with NAD+ binding and NADH product release being the first and last steps of the reaction, respectively.25 The products of the GSALDH reaction, glutamate and NADH, contribute to energy and nitrogen metabolism in the cell. NADH feeds electron equivalents to the respiratory chain via mitochondrial complex I, whereas glutamate is converted to α-ketoglutarate by glutamate dehydrogenase, thereby providing carbon to the tricarboxylic acid cycle (TCA) and ammonia for recycling of nitrogen.1,26,27
PRODH STRUCTURE AND MECHANISM
Kinetics of Proline Oxidation
Using rat mitochondria, Johnson and Strecker28 showed that proline is oxidized to P5C in a reaction that is linked to the respiratory chain. A Km of 2.3 mM proline was later determined with a solubilized enzyme preparation from rat liver mitochondria.29 The first characterization of a purified PRODH was the native proline utilization A (PutA) enzyme from Escherichia coli. PutA is a bacterial bifunctional enzyme in which PRODH and GSALDH activities are combined into a single polypeptide.30 In some bacteria, such as E. coli, PutAs also have an N-terminal DNA binding domain and regulate transcription of genes putP (proline transporter) and putA.30 PutA from E. coli (EcPutA) was found to be a dimer and to require FAD and electron acceptors for PRODH activity.31
Steady-state kinetic assays of EcPutA have shown that the PRODH reaction follows a two-site ping-pong mechanism,32 similar to that previously observed for the PRODH enzyme from Saccharomyces cerevisiae (Put1p).33 Reduction of the flavin cofactor by proline (reductive half-reaction) is followed by oxidation of reduced flavin by ubiquinone in the membrane (oxidative half-reaction).32,33 EcPutA PRODH exhibits a kcat of 5.2 s−1 for the overall reaction with Km values of 42 mM proline and 112 μM CoQ1.32 Microscopic rate constants of 27.5 s−1 for proline reduction of the FAD cofactor and 5.4 s−1 for oxidation of reduced FAD by CoQ1 were determined by stopped-flow kinetic measurements, indicating that the oxidative half-reaction is rate-limiting for the overall reaction.34 In a study by Serrano and Blanchard35 on monofunctional PRODH from Mycobacte-rium tuberculosis (MtbPRODH), primary kinetic isotope effects on V/Km(pro) and V gave evidence that hydride transfer from the proline C5 to the N5 of FAD is rate-limiting for the reductive half-reaction. Similar to EcPutA PRODH, the oxidative half-reaction was concluded to be rate-limiting for the overall MtbPRODH reaction.35
Serrano and Blanchard also showed that a highly a conserved Lys residue (Lys110 in MtbPRODH; Lys329 in EcPutA; Lys234 in human PRODH1) functions as a general base and is critical for catalysis.35 Figure 2A shows a proposed mechanism in which the conserved Lys residue first abstracts an amino proton from proline. The ensuing hydride transfer from the C5 of proline to the N5 of FAD is then shown as a stepwise transfer as proposed by Serrano and Blanchard.35
Figure 2.
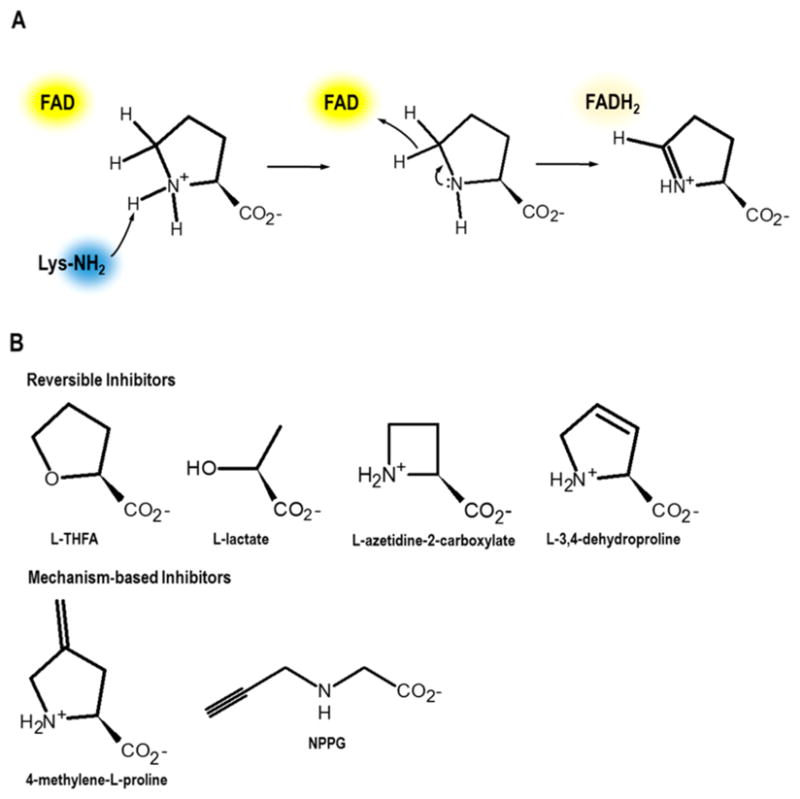
Proposed PRODH mechanism and inhibitors. (A) A conserved active-site Lys residue (Lys234 in human PRODH1) is proposed to deprotonate the proline amine. A stepwise hydride transfer from the C5 of proline to the N5 of FAD then generates P5C and FADH2. (B) Competitive reversible inhibitors and mechanism-based inhibitors of PRODH. Structures are known for PutAs non-covalently inhibited by L-THFA (PDB IDs 1TIW,40 4NMA,41 5KF6, and 5KF742) and L-lactate (PDB IDs 4O8A39 and 4NMB41) and covalently inactivated by NPPG (PDB IDs 3ITG43 and 4NME41).
Characterization of purified human recombinant PRODH136 and PRODH237 enzymes, which were significantly truncated to achieve solubility in E. coli, have confirmed they are flavin-dependent enzymes that catalyze the oxidation of proline and trans-4-hydroxy-L-proline, respectively. PRODH1 and PRODH2 are both mitochondrial enzymes and share 45% amino acid sequence identity.38 Using CoQ1, PRODH2 was–reported to have a 12-fold higher kcat/Km (0.93 M 1 s−1) with trans-4-hydroxy-L-proline relative to L-proline.37 The product of the PRODH2 reaction with 4-hydroxy-L-proline is 3-OH-P5C, which undergoes non-enzymatic hydrolysis to 4-hydroxyglutamate-γ-semialdehyde.38 GSALDH is then able to convert 4-hydroxyglutamate-γ-semialdehyde to 4-hydroxyglutamate.37 Because 4-hydroxyglutamate feeds glyoxylate production and ultimately oxalate, inhibition of PRODH2 has been suggested as a treatment for primary hyperoxaluria in patients with disorders in glyoxylate metabolism.37
The substrate selectivity of PRODH1 and PRODH2 was explored by site-directed mutagenesis of the PRODH domain in EcPutA, which, similar to PRODH1, utilizes 4-hydroxypro-line poorly as a substrate.31 X-ray crystal structures (2.0 Å resolution) of the PRODH domain from EcPutA in complex with the proline analogue L-tetrahydro-2-furoic acid (L-THFA) were examined to identify active-site residues critical for substrate recognition.39,40 In EcPutA, Tyr540 was found to be in close proximity to the C4 of proline, indicating that it could block the binding of 4-hydroxyproline.38 Interestingly, Tyr540 is conserved in PRODH1 (Tyr548), whereas the corresponding residue in PRODH2 is Ser485.38–40 Wild-type – EcPutA-PRODH has kcat/Km values of 492 M 1 s−1 for proline and 3 M−1 s−1 for 4-hydroxyproline.38 An EcPutA-PRODH – Tyr540Ser mutant exhibited kcat/Km values of 11 M 1 s−1 for 4-hydroxyproline and 79 M−1 s−1 for proline.38 Replacing Tyr540 with Ser increased the activity with 4-hydroxyproline, but the preference was still for proline, although the activity was >6-fold lower.38
PRODHs are also highly selective for the five-membered ring of proline. Studies of EcPutA-PRODH44 and human PRODH136 have found that pipecolate, a six-membered proline analogue, is neither a substrate nor an inhibitor. Thus, the upper limit of ring size for potential inhibitor molecules is considered to be five.
PRODH Structure
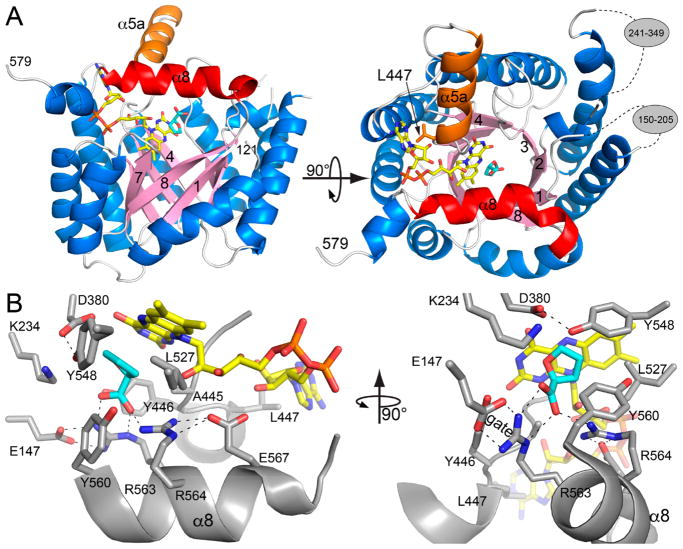
Currently no structures of PRODH enzymes from eukaryotes are available. This is largely due to the fact that eukaryotic PRODH is an inner mitochondrial protein, and its expression as a soluble protein in E. coli has been problematic. In contrast, the structures of bacterial PRODHs have been extensively characterized by high-resolution crystallography, and these structures provide a reliable template for modeling of human PRODH (Figure 3A).
Figure 3.
Homology model of residues 121–579 of human PRODH1 made with SWISS-MODEL using the default parameters.45 The template chosen by SWISS-MODEL was PDB ID 5KF6.42 (A) The PRODH (βα)8-barrel fold. FAD is colored yellow. The proline analogue L-THFA is colored cyan. The α8 and α5a helices are highlighted in red and orange, respectively. The gray ovals in the right-hand panel indicate residues omitted from the model because of high uncertainty. (B) Close-up views of the predicted active site of human PRODH1. FAD is colored yellow, and L-THFA is colored cyan.
The first high-resolution PRODH crystal structure was of the PRODH domain from EcPutA.39 The structure revealed that PRODH is a (βα)8-barrel with a non-covalently bound FAD cofactor. Conservation of the PRODH (βα)8-barrel was confirmed by several structures of full-length PutAs.41–43,47,48 The structures of stand-alone PRODH enzymes from Thermus thermophilus and Deinococcus radiodurans also feature a (βα)8-barrel.49,50
Homology modeling using the most recent update of the PDB predicts that the (βα)8-barrel fold is present within residues 121–579 of human PRODH1 (Figure 3A). In addition, the modeling predicts two large inserts of uncertain structure corresponding to residues 150–205 and 241–349. The (βα)8-barrel fold and inserts are consistently predicted by multiple homology modeling servers, such as SWISS-MODEL45 and Phyre2.46 Human PRODH2 is also predicted to contain the PRODH (βα)8-barrel fold; however, it appears to have only one inserted region of unknown structure (240–290).
An important structural difference between the PutA PRODH domain and stand-alone bacterial PRODHs is the insertion of an α-helix between β-strand 5 and α-helix 5 in PutA. Homology modeling predicts that this helix (α5a) is present in human PRODHs (Figure 3A). In PutAs, the α5a helix contains a nonpolar residue (Trp or Leu) that packs against the adenine of the FAD, which is presumed to be important for establishing the correct conformation of the cofactor. In human PRODH, this critical residue is predicted to be Leu447 (Figure 3A). The prediction of α5a in human PRODH suggests that PutAs may be preferred over stand-alone bacterial PRODHs as a model system for discovering PRODH inhibitors.
Crystal structures of PutAs in complex with L-lactate and the proline analogue L-THFA have revealed the role of active-site residues in substrate binding and conformational dynamics,4,30 and all of these residues are predicted to be in the active site of human PRODH (Figure 3B). The conserved sequence motif YXXRRXXE on α8 of the barrel is a key element in the PRODH active site. The two Arg residues of the motif (Arg563-Arg564 in human PRODH1) ion-pair with the proline carboxylate (Figure 3B) and are necessary for proline binding,4,30 while the Tyr residue of the motif (Tyr560) packs against the ring of the proline (Figure 3B). Leu527 is another highly conserved residue that shapes the active site and provides nonpolar interactions with the proline ring. The Glu residue of the motif stabilizes the second Arg residue via ion-pairing. The first Arg residue of the YXXRRXXE motif plays an important role in substrate binding and active-site dynamics by forming an ion-pair gate with a glutamate residue located on the β1–α1 loop—this interaction is also predicted to be present in human PRODHs (Figure 3B). Some of the other active-site residues that are conserved in bacterial and human PRODHs are shown in Figure 3B. Structural and mutational analysis of these residues in different PRODHs have shown they have important roles in catalytic steps,35,39,51–53 shaping the proline binding site,38,39 FAD conformation and redox potential,51,53 and conformational dynamics.51,52,54 The high structural and sequence conservation in the PRODH active site suggests that these residues have similar catalytic roles in human PRODH.
PRODH Inhibition
Consistent with a conserved PRODH domain structure across species, L-lactate, pyruvate, and the proline analogue L-THFA39,44 are reversible competitive inhibitors in several bacterial PutAs/PRODHs31,32 and human PRODHs (Figure 2B).36,37 L-THFA has also been used to selectively inhibit proline metabolism in bacterial10 and mammalian cells.8,14 L-THFA inhibits EcPutA PRODH with a Ki of 1.6 mM,32 and a Kd of 1.5 mM was estimated for binding of L-THFA to human PRODH1.36 L-Azetidine-2-carboxylate, a four-membered-ring proline analogue (Figure 2B), is a weak competitive inhibitor of EcPutA PRODH activity (Ki = 20 mM).31 Another potential inhibitor is L-3,4-dehydroproline (Figure 2B), which was reported to be a competitive inhibitor of PRODH activity in liver mitochondria (Ki = 0.16 mM)55 and has been used to inhibit proline catabolism in cell cultures.56,57 However, L-3,4-dehydroproline is also a substrate for EcPutA-PRODH,58 so the efficacy of L-3,4-dehydroproline as a specific inhibitor of PRODH activity is unclear and requires further investigation. Furthermore, L-3,4-dehydroproline should be used with caution especially since it is also a substrate for PYCR76 (in the reverse of the direction shown in Figure 1) and can be incorporated into newly synthesized proteins and impact collagen formation.60
Mechanism-based inhibitors for PRODH have also been discovered, such as N-propargylglycine (NPPG)61 and 4-methylene-L-proline.62 With both compounds, inactivation proceeds after the enzyme oxidizes the inhibitor, leaving the flavin reduced. High-resolution structures of a stand-alone bacterial PRODH61 and three PutAs41,48,54 have shown that inactivation by NPPG results in a three-carbon covalent link between an active-site lysine residue (Lys329 in EcPutA, equivalent to Lys234 in human PRODH1) and the FAD N5 atom. The NPPG-inactivated form of PutA has been a useful model for analyzing conformational changes that occur upon proline reduction of the FAD cofactor.41,48,54
In contrast to proline-analogue-based inhibitors, compounds that target the ubiquinone binding site have generally not been explored in PutAs/PRODHs, with the exception of atpenin A5. Atpenin A5, which is a ubiquinone analogue, was shown to be an EcPutA-PRODH competitive inhibitor (Kic = 97 μM) versus CoQ1 and an uncompetitive inhibitor (Kiu = 124 μM) with respect to proline.32 Considerably more work is needed to explore ubiquinone-based inhibition of PRODHs.
PYCR STRUCTURE AND MECHANISM
Kinetic Studies of PYCR
Early biochemical studies of PYCR were conducted on enzymes purified from a wide range of tissues and cell lines, such as liver from rat and calf, human erythrocytes, rat lens, bovine retina, human fibroblasts, and human lymphoblastoid cell lines.63–70 The kinetic measurements from these studies reveal general trends and hints about tissue-specific variations. The Km for P5C is fairly consistent at 0.1–1.0 μM, regardless of the cofactor used. The Km for NADPH tends to be lower than that for NADH, sometimes by as much as a factor of 10. The fibroblast and lymphoblastoid enzymes are exceptions, having nearly equal Km values for the two cofactors. The maximum velocity typically is higher with NADH than with NADPH; however, the ratio of V with NADH to V with NADPH varies substantially, from ~1 for retina to 10 for erythrocytes. Also, the sensitivity to product inhibition by proline and NADP+ is tissue-specific. For example, the erythrocyte and lens enzymes are inhibited by NADP+ but not by proline, whereas the fibroblast and lymphoblastoid enzymes are inhibited by proline but not by NADP+. Moreover, some enzymes are inhibited by ATP, but others are not.
The differences in cofactor preference and product inhibition caused Dougherty, Brandriss, and Valle63 to posit the existence of multiple forms of PYCR. Indeed, the use of modern molecular cloning and sequencing tools has revealed three PYCR genes, known as PYCR1, PYCR2, and PYCR3 (a.k.a. PYCRL), which produce a total of nine PYCR enzymes. (Christensen et al.71 show an amino acid sequence alignment of the nine PYCR isozymes.)
Because it is unclear which isoforms were being assayed in the early studies, it would be useful to have complete comparative kinetic analyses of all of the PYCRs using recombinant enzymes. However, only a few papers have reported the kinetic properties of recombinant human PYCR enzymes. De Ingeniis et al.72 measured the kinetic parameters of recombinant PYCR1, PYCR2, and PYCRL using P5C and NAD(P)H as the substrates. Christensen et al.71 reported values for PYCR1, also using the forward reaction. Meng et al.59 studied the reverse reaction of PYCR1 using thioproline and NAD+ as substrates.
The study by De Ingeniis et al.72 is the only one that compared PYCR isoforms. The Km values for P5C are in the low millimolar range, while those of NAD(P)H are a few hundred micromolar. The highest catalytic efficiency (kcat/Km) was observed with PYCR2 and NADH as the cofactor, while the lowest efficiency was seen with PYCR1 and NADPH. They also studied product inhibition by proline. PYCR2 is the most sensitive to inhibition by proline, with an apparent Ki of ~0.1 mM. Because this value is within the range of normal plasma proline levels in humans (0.05–0.3 mM73), the inhibition of PYCR2 by proline has physiological significance. In contrast, the weak inhibition of PYCR1 (Ki = 0.6 mM) and PYCRL (Ki = 8.5 mM) by proline may not be physiologically significant.
PYCR1 may also play a role in lysine catabolism. Struys et al.74 asked whether PYCR1 accepts Δ1-piperideine-6-carboxylate (P6C) as a substrate. P6C is the six-membered-ring counterpart of P5C and is an intermediate in lysine catabolism. P6C exists in equilibrium with the open-chain α-aminoadipate semialdehyde, which is the substrate for the lysine catabolic enzyme aldehyde dehydrogenase 7A1 (ALDH7A1). ALDH7A1 catalyzes the NAD+-dependent oxidation of α-aminoadipate semialdehyde to α-aminoadipate.75 Struys et al.74 showed that human PYCR1 reduces P6C to L-pipecolic acid with kinetic constants similar to those of the natural substrate. These studies suggest the intriguing idea that PYCR1 may have more than one metabolic function.
Much remains unknown about the biochemical properties of human PYCR. For example, studies are needed to establish the binding order of substrates. The inhibition of the recombinant enzymes by NAD(P)+ and ATP has not been studied. Only the three major isoforms have been studied; none of isozymes that result from alternative splicing have been expressed and purified. The impact of inherited disease-causing mutations on the structure and catalytic function of PYCR has yet to be determined. Finally, investigating the role of PYCR1 in lysine catabolism is an intriguing line of future research.
Structures of PYCR1
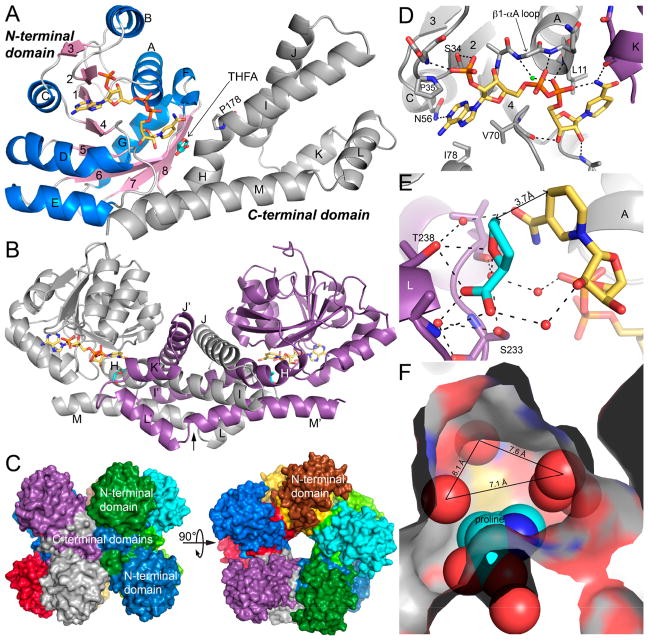
High-resolution crystal structures of PYCR1 were determined recently.71 These include structures of the NADPH substrate complex, proline product complex, and a ternary complex with NADPH and the P5C/proline analogue L-THFA. Low-resolution structures were reported over a decade ago; however, readers are cautioned that the assignment of the active site in the low-resolution structures was incorrect.59
The fold of PYCR1 has two domains (Figure 4A). The N-terminal domain has the Rossmann fold and binds NAD(P)H. The C-terminal domain consists of several α-helices and functions in oligomerization and substrate binding. Two protomers combine to form an interlocked dimer mediated by the C-terminal domains (Figure 4B). The dimers assemble further around a fivefold axis to form a pentamer-of-dimers decamer (Figure 4C). The fold of PYCR1, the interlocking dimer, and the cylindrical decamer are also observed in P5CRs from microorganisms.76
Figure 4.
Structure of PYCR1. (A) The fold of PYCR1 as seen in the ternary complex with NADPH and the proline/P5C analogue L-THFA (PDB ID 5UAV). The N-terminal NAD(P)H binding domain is colored according to secondary structure, with β-strands in pink and α-helices in blue. The C-terminal oligomerization domain is colored gray. NADPH appears in gold sticks. L-THFA is shown as cyan sticks. β-strands are labeled 1–8; α-helices are labeled A–M. Helix-disrupting Pro178 is shown. (B) The structure of the dimer. The α-helices of the C-terminal domain are labeled H–M for the gray protomer and H′–M′ for the purple protomer. NADPH and L-THFA are colored gold and cyan, respectively. The arrow represents the twofold axis of the dimer. (C) The PYCR1 pentamer-of-dimers decamer, with each chain colored differently. (D) The NADPH binding site (PDB ID 5UAT). Selected α-helices and β-strands are labeled as in (A). Helix K (in purple) is from the opposite protomer of the dimer. The conserved water molecule of the dinucleotide-binding Rossmann fold is colored green.77 (E) The active site of the ternary complex. NADPH and L-THFA are colored gold and cyan, respectively. The two protomers of the dimer are colored purple and gray. The A and L α-helices are labeled. (F) Depiction of the open space in the P5C/proline pocket (PDB ID 5UAU). The product proline is shown in cyan spheres. Water molecules are represented by red nonbonded spheres. Center-to-center distances between water molecules are indicated. (A–E) adapted with permission from ref 71. Copyright 2017 American Society for Biochemistry and Molecular Biology.
The substrate binding sites have been characterized at high resolution (1.9 Å). NADPH binds in an extended conformation at the C-terminal edge of the β-sheet of the Rossmann fold domain (Figure 4D). This is the canonical pose for NAD(P)-(H) bound to the Rossmann fold. Proline (as well as P5C) binds in the dimer interface and interacts primarily with the C-terminal domains of the dimer (Figure 4B). The hydrophilic parts of proline bind to the αK–αL loop and conserved Thr238 (shown for a proline analogue in Figure 4E). We note that Thr238 is present in all PYCR isoforms. The nonpolar ring of proline contacts the kink between helices H and I. The kink is caused by the presence of Pro178 and disrupts what would be a very long α-helix (Figure 4A). We note that Pro178 is replaced by Val in PYCRL.
The structure of PYCR1 complexed with NADPH and L-THFA provides a high-resolution (1.85 Å) model of the ternary E·S complex (Figure 4E). The nicotinamide of NADPH meets P5C in the junction between the dimer interface and the Rossmann domain. The rings of the two substrates stack in parallel, positioning the hydride donor atom of NADPH (C4) 3.7 Å from the acceptor atom of P5C. This arrangement is consistent with a direct hydride transfer mechanism. The structure implies the stereochemistry of hydride transfer. Because the B-side of the nicotinamide contacts L-THFA, PYCR1 is predicted to catalyze the transfer of the pro-4S hydrogen to P5C.
The crystal structures should be useful for inhibitor design. One could target either the P5C site or the NADPH site; however, the former may be preferred because NADPH binds to a conserved fold that is present in many enzymes. The structure of PYCR1 complexed with proline shows a pocket with available space that could be exploited (Figure 4F). This pocket binds the nicotinamide riboside of NADPH in the ternary complex. However, in the proline complex, the pocket contains five water molecules. A possible strategy could be to design proline analogues with functional groups that extend into the open space, displacing the water molecules and forming interactions with the amino acid residues that line the pocket.
THE PROLINE–P5C CYCLE
A unique aspect of proline metabolism is the cycling of proline and P5C to maintain redox homeostasis between the cytosol and mitochondria.2,3 As originally conceived, the proline cycle consists of catabolic and synthetic half-cycles that occur in different subcellular locations. The catabolic half-cycle is the oxidation of proline to P5C catalyzed by PRODH in mitochondria (Figure 1A). The synthetic half-cycle is the reduction of P5C back to proline catalyzed by PYCR (Figure 1A). If the reductive step occurs outside mitochondria, the net effect of the cycle is to transfer reducing equivalents from cytosolic NADPH into the mitochondrial respiratory chain. Evidence for the proline–P5C cycle was first reported by Hagedorn and Phang,78 who showed that NADPH could drive respiratory activity in mammals without stoichiometric consumption of proline. Evidence for a proline–P5C cycle has also been shown for plants.79
Whether the proline–P5C cycle is truly directional is complicated by uncertainties about the subcellular locations of the PYCR isoforms. If it is directional, then the reductive half-cycle should be catalyzed by PYCRL, which is known to be cytosolic.72 However, Fendt and co-workers concluded that PYCR1, the isoform thought to be localized in mitochondria,72 had the most significant role in proline–P5C cycling during spheroidal growth of breast cancer cells and was responsible for sustaining PRODH activity upon proline withdrawal.14 Phang et al.80 have suggested that PYCR1 may not be truly mitochondrial but could localize with mitochondrial markers as a result of association with mitochondrial outer membranes. Clarification of the subcellular localization of PYCR1 would be helpful in understanding the role of the proline–P5C cycle in normal redox homeostasis and cancer.
Regardless of which PYCR is involved, the proline–P5C cycle has been shown to enhance oxidative phosphorylation, maintain cytosolic pyridine nucleotide levels, and generate reactive oxygen species (ROS) leading to activation of various cell signaling pathways.3,14,15,78,79 The maintenance of NADP+ in the cytosol is proposed to link proline metabolism to the pentose phosphate pathway and nucleotide biosynthesis.3,15,78,81
PROLINE IN CANCER METABOLISM
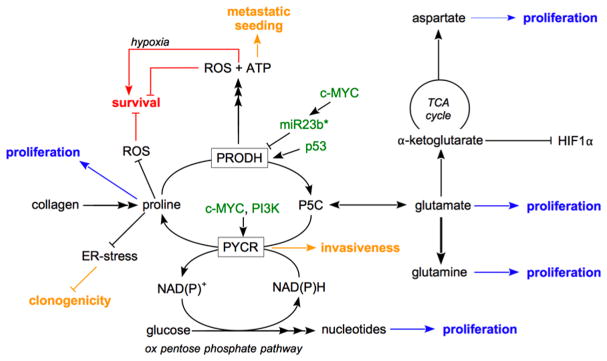
Cancer cells alter their metabolism to increase survival during cellular stress, to undergo uncontrolled proliferation, and to progress toward metastasis formation.82,83 Proline biosynthesis, catabolism, and cycling have been implicated as metabolic pathways selectively altered in cancer cells providing ATP, macromolecules, and redox cofactors.3 In the following, we will discuss the emerging role of proline metabolism in light of cancer cell proliferation, survival, and metastasis formation as illustrated in Figure 5.
Figure 5.
Proline metabolism in cancer cell proliferation, survival, and metastasis formation. Proline metabolism supports cancer cell proliferation via biomass precursor production (indicated in blue). Proline metabolism supports or inhibits cancer cell survival via ROS (indicated in red). Proline metabolism supports metastasis formation by promoting invasiveness, clonogenicity, and metastatic seeding (indicated in yellow). Green indicates regulators of proline catabolism and biosynthesis. P5C refers to Δ1-pyrroline-5-carboxylate.
Cancer cell proliferation depends on biomass production, i.e., the generation of macromolecules such as DNA and protein from amino acids and other metabolites.83 Proline biosynthesis has been shown to fuel protein production, which is needed for cell proliferation. Accordingly, it has been found that c-MYC and PI3K signaling, which supports cell proliferation, increases the gene expression of P5CS, PYCR1, PYCR2, and PYCRL.13,15 In kidney cancer, proline is a limiting amino acid for protein synthesis, and the knockout of PYCR1 is sufficient to impair in vivo proliferation in these cancers.84 Accordingly, PYCR1 is overexpressed in tumors of human non-small cell lung carcinoma patients, and knockdown of the enzyme impairs proliferation in cell lines.19 Moreover, proline derived from collagen has been found to support pancreatic cancer cell proliferation.20 The importance of proline for the proliferation of pancreatic cancer cells was indicated not only by direct incorporation of proline into protein but also via proline catabolism resulting in glutamine, glutamate, and aspartate production, which in turn are precursors of DNA and/or protein. Finally, proline biosynthesis can also indirectly support biomass production by generating the redox cofactor NADP+, which increases the activity of the oxidative pentose phosphate pathway, which in turn generates precursors of nucleotide biosynthesis15 needed for DNA and RNA production. Moreover, it was found that in cancer cells expressing c-MYC, knockdown of proline biosynthesis resulted in decreased glycolysis and ATP production.15,85 Interestingly, it was recently shown that in isocitrate dehydrogenase 1 (IDH1) mutant cancer cells, proline biosynthesis is increased compared with wild-type IDH cancer cells, which results in a partial decoupling of the electron transport chain from the TCA cycle.86 Accordingly, ATP-coupled oxygen consumption increased in IDH1 mutant cancer cells upon proline biosynthesis inhibition. It is therefore tempting to speculate that both observations can impact cancer cell proliferation by perturbing the cellular redox balance. In summary, proline metabolism can support cancer proliferation and therefore constitutes an interesting intervention point to inhibit the growth of certain tumor types.
Cancer cells have an increased capacity to survive under harsh conditions and to evade apoptosis.87 One important mechanism to induce apoptosis and decrease cell survival is the generation of ROS. Strikingly, proline metabolism can contribute to ROS scavenging and generation by different means. First, proline itself has some antioxidant capacity. Thus, increasing proline levels by proline supplementation and overexpression of PYCR1 or decreasing them by over-expression of the proline catabolism enzyme PRODH increases or decreases cellular ROS scavenging, respectively.7,88 Second, the PRODH-catalyzed reaction results not only in ATP production but also ROS generation.12 Therefore, high proline catabolism can induce apoptosis and cell senescence, which has been shown to be counteracted by superoxide dismutase expression in colorectal cancer cells11 or antioxidants in osteosarcoma cells.89 Accordingly, PRODH expression is positively regulated by the tumor suppressor p53,12 while the oncogene c-MYC negatively regulates PRODH expression via miR23b*.13,16 Moreover, oral cancer overexpressed 1 (ORAOV1) protein has been suggested to bind to PYCR1, resulting in decreased ROS production, presumably through activation of proline biosynthesis.90 Additionally, proline catabolism can result in production of α-ketoglutarate, which is a metabolic substrate of HIF1α prolyl hydroxylases that targets HIF1α for degradation,17 and hypoxia can activate proline biosynthesis in liver cancer cells and might support HIF1α stabilization.91 These effects could contribute to the antitumor effect of PRODH expression in some cancers.17 Controversial to this potential tumor suppression function of proline catabolism is that in a hypoxic environment PRODH expression has been shown to be increased and contribute to the survival of cancer cells by inducing autophagy.92 Thus, this suggests that the environment needs to be considered when evaluating the role of PRODH for cancer cell survival. Taken together, these results indicate that proline biosynthesis inhibition targets cancer cell survival while proline catabolism inhibition can have context-dependent pro- or antisurvival effects in cancer cells.
One of the most lethal capacities of cancer cells is their ability to metastasize to distant organs. During this process, cancer cells change their metabolism to invade surrounding tissue, survive in the circulation, and colonize distant organs.83,93,94 Recently, it has been shown that proline metabolism supports this metastatic cascade leading to secondary tumors, i.e., metastases. It has been shown that human metastasis tissue exhibits upregulated expression of PRODH compared with primary breast tumor tissue.14 Accordingly, it has been found that PRODH inhibition impairs metastasis formation in different metastatic breast cancer mouse models without adverse effects on normal cells and tissues with high PRODH expression.14 This selectivity toward breast cancer cells can be explained on the one hand by the observation that (at least in vitro) normal mammary epithelial cells display low PRODH expression and on the other hand by the finding that PRODH inhibitor doses required to impair proline catabolism in metastasis tissue are lower than those needed to inhibit PRODH in liver, heart, and brain.14 Mechanistically, the proline cycle composed of PRODH and PYCR1 activity allows metastasizing breast cancer cells to produce ATP at the expense of NADPH, fostering metastatic seeding.14 Additionally, inhibition of proline biosynthesis has been found in some cancer cells and yeast to result in unresolved endoplasmic reticulum (ER) stress.9,85 In cancer cells, inhibition of PYCR1 hampers clonogenicity, which can support the ability of cancer cells to initiate new tumors.85 Consistent with this observation, it was found that in human breast cancers PYCR1 gene expression is correlated with invasiveness,18 which is an early event of the metastatic cascade. While some of these effects could be alleviated by exogenous proline, the dependence of metastasizing breast cancer cells on PRODH and PYCR1 could not be rescued by proline supplementation or recapitulated by depletion of exogenous proline. In conclusion, inhibition of proline metabolism by targeting the respective enzymes has emerged as an interesting target to interfere with the metastatic cascade and prevent metastasis formation.
CONCLUSIONS
Over the past decade, several major advances have been achieved in understanding the structures and mechanisms of proline metabolism and its specialized role in cancer metabolism. Structures of bacterial PRODHs with active-site ligands are available that enable modeling and structure-based inhibitor design studies of human PRODH1 and PRODH2. Structures of human PYCR1 in ternary complex with NADPH and a product analogue provide a template for the design of novel inhibitors. A major challenge that remains is solving high-resolution structures of human PRODH1, PRODH2, PYCR2, and PYCRL. Structures of the these enzymes along with biochemical characterization of the enzyme mechanisms will be important for designing inhibitors specific for PRODH1 over PRODH2 and for isoform-specific inhibitors of PYCRs. Covalent inactivation of PRODH by targeting the FAD cofactor could be a novel approach by combining specificity of binding and chemical reactivity. Historically pharma has been skeptical of covalent inactivators, but opinions are changing.95 Having small molecules with different modes of inhibition would be powerful tools for investigating proline metabolism in cancer.
Although structure-based inhibitor discovery is now possible, some practical hurdles need to be overcome. One issue is whether the enzymes can be generated in sufficient quantity for high-throughput screening of compound libraries. Recombinant PYCR1 can be expressed and purified from E. coli and is amenable to inhibitor screening. Also, fusion-tagged PYCRs with high activity have been reported and may be suitable as well. Screening for inhibitors of human PRODHs will be more challenging because of the lack of a convenient method for generating recombinant enzyme. A workaround strategy could be to use PutA PRODHs as surrogates for human PRODH in screening of compound libraries, followed by testing of hit compounds for inhibition of PRODH activity in mitochondrial extracts. Additionally, robust assays need to be developed for drug screening. PRODH activity is typically monitored by measuring reduction of an artificial electron acceptor such as dichlorophenolindolphenol or the formation of a chromogenic adduct between o-aminobenzaldehyde and P5C. PYCR activity can be measured in the forward direction by following the decrease in NAD(P)H absorbance, but some laboratories also measure PYCR activity in the reverse direction using 3,4-dehydroproline as a substrate and monitoring NADH formation.59
Understanding the mechanisms of proline metabolic enzymes can be leveraged to design and discover molecular probes that could be used to study the functions of the proline–P5C cycle in cancer metabolism and as lead compounds for cancer therapy. Because the proline–P5C cycle has been shown to significantly influence cellular growth and death pathways, the potential of this novel cycle in tumorigenesis and cancer needs to be further explored.3,14,81 More biochemical details of the proline–P5C cycle are needed to fully understand its role in cancer metabolism. For example, does the proline–P5C cycle depend on a mitochondrion–cytosol shuttle or is it contained within the mitochondrion? If mitochondrial uptake is required, how is P5C imported into the mitochondrion? Also, are PYCR1 and PYCR2 strictly localized in the inner mitochondrial matrix? Does each PYCR have a specialized role in the proline–P5C cycle depending on the cellular context and growth conditions? Dissecting the individual roles of PYCRs and PRODH in the proline cycle as it relates to cancer will be an important area for future research.
Finally, issues of how proline metabolic enzyme inhibitors may affect metabolism in healthy cells and tissues needs to be considered. Presently it appears that inhibiting proline catabolism would have less adverse effects than inhibiting proline biosynthesis. Inborn deficiencies of the proline catabolic enzymes PRODH1 and GSALDH lead to hyperprolinemia metabolic disorders,73 with loss of PRODH1 activity linked to increased susceptibility to schizophrenia.96,97 How harmful it would be to disrupt proline catabolism in healthy tissues during therapeutic treatment is not clear, but treatment of mice with the PRODH inhibitor L-TFHA blocked lung metastases with no negative effects on healthy tissue.14 These results show promise for tolerance of PRODH inhibition and proline buildup in healthy human cells. The situation for proline biosynthesis is more complicated, as not necessarily the products per se but the reactions of the synthetic pathway are sometimes most critical. This is illustrated by the different phenotypes of PYCR1 and PYCR2 inborn deficiencies. Loss of PYCR1 gene function is associated with autosomal recessive cutis laxa with clinical features of wrinkled skin, aged appearance, and connective tissue weakness.98 Mutations in the PYCR2 gene are associated with autosomal recessive neurologic disorder and syndrome of postnatal microcephaly.99,100 Decreased mitochondrial function and oxidative stress tolerance have been reported for PYCR1- and PYCR2-depleted human cells.98,99 Knockdown of PYCR1 or PYCR2 has been shown to have little impact on growth rates, whereas knockdown of PYCRL markedly inhibits proliferation.15 Interestingly, the growth of cells with PYCRL knockdown could not be rescued by exogenous proline.15 Considering these aforementioned effects and the potential impact of PYCRs on NAD(P)H redox homeostasis and energy metabolism (see Figure 5), the likelihood of significant side effects from drugs targeting proline biosynthesis seems greater than that from drugs targeting PRODH or GSALDH. A concern for both pathways, however, is the potential for cells to accumulate P5C, which was recently listed among the top 30 damage-prone endogenous metabolites.101 Thus, adverse metabolic effects in healthy tissues and organs will need to be examined with any drugs targeting proline metabolic enzymes. Hopefully, though, compounds can be designed to have high selectivity toward cancer cells.
Acknowledgments
Funding
This research was supported by NIGMS, National Institutes of Health, Grants R01GM061068 and R01GM065546, and by the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. S.-M.F. acknowledges funding from FWO – Odysseus II, FWO – Research Grants/Projects, and KU Leuven – Methusalem Co-Funding.
ABBREVIATIONS
- ALDH
aldehyde dehydrogenase
- GSAL
glutamate-γ-semi-aldehyde
- GSALDH
glutamate-γ-semialdehyde dehydrogenase
- G5K
glutamate 5-kinase
- γ-GPR
γ-glutamyl phosphate reductase
- NAD+
nicotinamide adenine dinucleotide
- NPPG
N-propargylglycine
- PRODH
proline dehydrogenase
- P6C
Δ1-piperideine-6-carboxylate
- P5C
Δ1-pyrroline-5-carboxylate
- P5CR or PYCR
Δ1-pyrroline-5-carboxylate reductase
- P5CS
Δ1-pyrroline-5-carboxylate synthase
- 3-OH-P5C
Δ1-pyrroline-3-OH-5-carboxylate
- PutA
proline utilization A
- ROS
reactive oxygen species
- L-THFA
L-tetrahydro-2-furoic acid
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- 2.Phang JM. The regulatory functions of proline and pyrroline-5-carboxylic acid. Curr Top Cell Regul. 1985;25:91–132. doi: 10.1016/b978-0-12-152825-6.50008-4. [DOI] [PubMed] [Google Scholar]
- 3.Phang JM. Proline metabolism in cell regulation and cancer biology: Recent advances and hypotheses. Antioxid Redox Signaling. 2012 doi: 10.1089/ars.2017.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanner JJ. Structural biology of proline catabolic enzymes. Antioxid Redox Signaling. 2012 doi: 10.1089/ars.2017.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christgen SL, Becker DF. Role of proline in pathogen and host interactions. Antioxid Redox Signaling. 2012 doi: 10.1089/ars.2017.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood JM. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol. 2011;65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan N, Dickman MB, Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radical Biol Med. 2008;44:671–681. doi: 10.1016/j.freeradbiomed.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natarajan SK, Zhu W, Liang X, Zhang L, Demers AJ, Zimmerman MC, Simpson MA, Becker DF. Proline dehydrogenase is essential for proline protection against hydrogen peroxide-induced cell death. Free Radical Biol Med. 2012;53:1181–1191. doi: 10.1016/j.freeradbiomed.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang X, Dickman MB, Becker DF. Proline biosynthesis is required for endoplasmic reticulum stress tolerance in Saccharomyces cerevisiae. J Biol Chem. 2014;289:27794–27806. doi: 10.1074/jbc.M114.562827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Alfano JR, Becker DF. Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. J Bacteriol. 2015;197:431–440. doi: 10.1128/JB.02282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Borchert GL, Donald SP, Surazynski A, Hu CA, Weydert CJ, Oberley LW, Phang JM. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis. 2005;26:1335–1342. doi: 10.1093/carcin/bgi083. [DOI] [PubMed] [Google Scholar]
- 12.Donald SP, Sun XY, Hu CAA, Yu J, Mei JM, Valle D, Phang JM. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61:1810. [PubMed] [Google Scholar]
- 13.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TWM, Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A. 2012;109:8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elia I, Broekaert D, Christen S, Boon R, Radaelli E, Orth MF, Verfaillie C, Grünewald TGP, Fendt SM. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat Commun. 2017;8:15267. doi: 10.1038/ncomms15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W, Hancock CN, Fischer JW, Harman M, Phang JM. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci Rep. 2015;5:17206. doi: 10.1038/srep17206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Zabirnyk O, Wang H, Shiao YH, Nickerson ML, Khalil S, Anderson LM, Perantoni AO, Phang JM. MicroRNA-23b* targets proline oxidase, a mitochondrial tumor suppressor protein in renal cancer. Oncogene. 2010;29:4914–4924. doi: 10.1038/onc.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Borchert GL, Donald SP, Diwan BA, Anver M, Phang JM. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res. 2009;69:6414. doi: 10.1158/0008-5472.CAN-09-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding J, Kuo ML, Su L, Xue L, Luh F, Zhang H, Wang J, Lin TG, Zhang K, Chu P, Zheng S, Liu X, Yen Y. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis. 2017;38:519–531. doi: 10.1093/carcin/bgx022. [DOI] [PubMed] [Google Scholar]
- 19.Cai F, Miao Y, Liu C, Wu T, Shen S, Su X, Shi Y. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol Lett. 2018;15:731–740. doi: 10.3892/ol.2017.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivares O, Mayers JR, Gouirand V, Torrence ME, Gicquel T, Borge L, Lac S, Roques J, Lavaut MN, Berthèzene P, Rubis M, Secq V, Garcia S, Moutardier V, Lombardo D, Iovanna JL, Tomasini R, Guillaumond F, Vander Heiden MG, Vasseur S. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat Commun. 2017;8:16031. doi: 10.1038/ncomms16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craze ML, Cheung H, Jewa N, Coimbra NDM, Soria D, El-Ansari R, Aleskandarany MA, Wai Cheng K, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. MYC regulation of glutamine-proline regulatory axis is key in luminal B breast cancer. Br J Cancer. 2018;118:258–265. doi: 10.1038/bjc.2017.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fichman Y, Gerdes SY, Kovacs H, Szabados L, Zilberstein A, Csonka LN. Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev Camb Philos Soc. 2015;90:1065–1099. doi: 10.1111/brv.12146. [DOI] [PubMed] [Google Scholar]
- 23.Efron ML. Familial hyperprolinemia. report of a second case, associated with congenital renal malformations, hereditary hematuria and mild mental retardation, with demonstration of an enzyme defect. N Engl J Med. 1965;272:1243–1254. doi: 10.1056/NEJM196506172722401. [DOI] [PubMed] [Google Scholar]
- 24.Efron ML, Bixby EM, Pryles CV. Hydroxyprolinemia. Ii A rare metabolic disease due to a deficiency of the enzyme “hydroxyproline oxidase”. N Engl J Med. 1965;272:1299–1309. doi: 10.1056/NEJM196506242722501. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava D, Singh RK, Moxley MA, Henzl MT, Becker DF, Tanner JJ. The three-dimensional structural basis of type II hyperprolinemia. J Mol Biol. 2012;420:176–189. doi: 10.1016/j.jmb.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao JR, Knight K, Engel AL, Jankowski C, Wang Y, Manson MA, Gu H, Djukovic D, Raftery D, Hurley JB, Du J. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J Biol Chem. 2017;292:12895–12905. doi: 10.1074/jbc.M117.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, Haigis MC. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. 2017;358:941–946. doi: 10.1126/science.aam9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson AB, Strecker HJ. The interconversion of glutamic acid and proline. IV The oxidation of proline by rat liver mitochondria. J Biol Chem. 1962;237:1876–1882. [PubMed] [Google Scholar]
- 29.Kowaloff EM, Phang JM, Granger AS, Downing SJ. Regulation of proline oxidase activity by lactate. Proc Natl Acad Sci U S A. 1977;74:5368–5371. doi: 10.1073/pnas.74.12.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu LK, Becker DF, Tanner JJ. Structure, function, and mechanism of proline utilization A (PutA) Arch Biochem Biophys. 2017;632:142–157. doi: 10.1016/j.abb.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scarpulla RC, Soffer RL. Membrane-bound proline dehydrogenase from Escherichia coli. Solubilization, purification, and characterization. J Biol Chem. 1978;253:5997–6001. [PubMed] [Google Scholar]
- 32.Moxley MA, Tanner JJ, Becker DF. Steady-state kinetic mechanism of the proline:ubiquinone oxidoreductase activity of proline utilization A (PutA) from Escherichia coli. Arch Biochem Biophys. 2011;516:113–120. doi: 10.1016/j.abb.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wanduragala S, Sanyal N, Liang X, Becker DF. Purification and characterization of Put1p from Saccharomyces cerevisiae. Arch Biochem Biophys. 2010;498:136–142. doi: 10.1016/j.abb.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moxley MA, Becker DF. Rapid reaction kinetics of proline dehydrogenase in the multifunctional proline utilization A protein. Biochemistry. 2012;51:511–520. doi: 10.1021/bi201603f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrano H, Blanchard JS. Kinetic and isotopic characterization of L-proline dehydrogenase from Mycobacterium tuberculosis. Biochemistry. 2013;52:5009–5015. doi: 10.1021/bi400338f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tallarita E, Pollegioni L, Servi S, Molla G. Expression in Escherichia coli of the catalytic domain of human proline oxidase. Protein Expression Purif. 2012;82:345–351. doi: 10.1016/j.pep.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Summitt CB, Johnson LC, Jonsson TJ, Parsonage D, Holmes RP, Lowther WT. Proline dehydrogenase 2 (PRODH2) is a hydroxyproline dehydrogenase (HYPDH) and molecular target for treating primary hyperoxaluria. Biochem J. 2015;466:273–281. doi: 10.1042/BJ20141159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostrander EL, Larson JD, Schuermann JP, Tanner JJ. A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate. Biochemistry. 2009;48:951–959. doi: 10.1021/bi802094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10:109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, White TA, Schuermann JP, Baban BA, Becker DF, Tanner JJ. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43:12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh H, Arentson BW, Becker DF, Tanner JJ. Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc Natl Acad Sci U S A. 2014;111:3389–3394. doi: 10.1073/pnas.1321621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, Tanner JJ. Structures of proline utilization A (PutA) Reveal the fold and functions of the aldehyde dehydrogenase superfamily domain of unknown function. J Biol Chem. 2016;291:24065–24075. doi: 10.1074/jbc.M116.756965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srivastava D, Schuermann JP, White TA, Krishnan N, Sanyal N, Hura GL, Tan A, Henzl MT, Becker DF, Tanner JJ. Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 2010;107:2878–2883. doi: 10.1073/pnas.0906101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu W, Gincherman Y, Docherty P, Spilling CD, Becker DF. Effects of proline analog binding on the spectroscopic and redox properties of PutA. Arch Biochem Biophys. 2002;408:131–136. doi: 10.1016/s0003-9861(02)00535-0. [DOI] [PubMed] [Google Scholar]
- 45.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 46.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korasick DA, Gamage TT, Christgen S, Stiers KM, Beamer LJ, Henzl MT, Becker DF, Tanner JJ. Structure and characterization of a class 3B proline utilization A: Ligand-induced dimerization and importance of the C-terminal domain for catalysis. J Biol Chem. 2017;292:9652–9665. doi: 10.1074/jbc.M117.786855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ. Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure. FEBS J. 2017;284:3029–3049. doi: 10.1111/febs.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White TA, Krishnan N, Becker DF, Tanner JJ. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J Biol Chem. 2007;282:14316–14327. doi: 10.1074/jbc.M700912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo M, Arentson BW, Srivastava D, Becker DF, Tanner JJ. Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release. Biochemistry. 2012;51:10099–10108. doi: 10.1021/bi301312f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Zhang M, Zhu W, Zhou Y, Wanduragala S, Rewinkel D, Tanner JJ, Becker DF. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA–membrane binding. Biochemistry. 2007;46:483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu W, Haile AM, Singh RK, Larson JD, Smithen D, Chan JY, Tanner JJ, Becker DF. Involvement of the beta3-alpha3 loop of the proline dehydrogenase domain in allosteric regulation of membrane association of proline utilization A. Biochemistry. 2013;52:4482–4491. doi: 10.1021/bi400396g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moxley MA, Zhang L, Christgen S, Tanner JJ, Becker DF. Identification of a conserved histidine as being critical for the catalytic mechanism and functional switching of the multifunctional proline utilization A protein. Biochemistry. 2017;56:3078–3088. doi: 10.1021/acs.biochem.7b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srivastava D, Zhu W, Johnson WH, Jr, Whitman CP, Becker DF, Tanner JJ. The structure of the proline utilization a proline dehydrogenase domain inactivated by N-propargylglycine provides insight into conformational changes induced by substrate binding and flavin reduction. Biochemistry. 2010;49:560–569. doi: 10.1021/bi901717s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dashman T, Lewinson TM, Felix AM, Schwartz MA. L-3,4-Dehydroproline: hepatic mitochondrial metabolism and inhibition of proline oxidase. Res Commun Chem Pathol Pharmacol. 1979;24:143–157. [PubMed] [Google Scholar]
- 56.Pandhare J, Dash S, Jones B, Villalta F, Dash C. A novel role of proline oxidase in HIV-1 envelope glycoprotein-induced neuronal autophagy. J Biol Chem. 2015;290:25439–25451. doi: 10.1074/jbc.M115.652776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hancock CN, Liu W, Alvord WG, Phang JM. Co-regulation of mitochondrial respiration by proline dehydrogenase/oxidase and succinate. Amino Acids. 2016;48:859–872. doi: 10.1007/s00726-015-2134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood JM. Genetics of L-proline utilization in Escherichia coli. J Bacteriol. 1981;146:895–901. doi: 10.1128/jb.146.3.895-901.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng Z, Lou Z, Liu Z, Li M, Zhao X, Bartlam M, Rao Z. Crystal structure of human pyrroline-5-carboxylate reductase. J Mol Biol. 2006;359:1364–1377. doi: 10.1016/j.jmb.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 60.Kerwar SS, Felix AM. Effect of L-3,4-dehydroproline on collagen synthesis and prolyl hydroxylase activity in mammalian cell cultures. J Biol Chem. 1976;251:503–509. [PubMed] [Google Scholar]
- 61.White TA, Johnson WH, Jr, Whitman CP, Tanner JJ. Structural basis for the inactivation of Thermus thermophilus proline dehydrogenase by N-propargylglycine. Biochemistry. 2008;47:5573–5580. doi: 10.1021/bi800055w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tritsch D, Mawlawi H, Biellmann JF. Mechanism-based inhibition of proline dehydrogenase by proline analogues. Biochim Biophys Acta, Protein Struct Mol Enzymol. 1993;1202:77–81. doi: 10.1016/0167-4838(93)90065-y. [DOI] [PubMed] [Google Scholar]
- 63.Dougherty KM, Brandriss MC, Valle D. Cloning human pyrroline-5-carboxylate reductase cDNA by complementation in Saccharomyces cerevisiae. J Biol Chem. 1992;267:871–875. [PubMed] [Google Scholar]
- 64.Merrill MJ, Yeh GC, Phang JM. Purified human erythrocyte pyrroline-5-carboxylate reductase. Preferential oxidation of NADPH. J Biol Chem. 1989;264:9352–9358. [PubMed] [Google Scholar]
- 65.Matsuzawa T. Purification and characterization of pyrroline-5-carboxylate reductase from bovine retina. Biochim Biophys Acta, Gen Subj. 1982;717:215–219. doi: 10.1016/0304-4165(82)90172-6. [DOI] [PubMed] [Google Scholar]
- 66.Shiono T, Kador PF, Kinoshita JJ. Purification and characterization of rat lens pyrroline-5-carboxylate reductase. Biochim Biophys Acta, Gen Subj. 1986;881:72–78. doi: 10.1016/0304-4165(86)90098-x. [DOI] [PubMed] [Google Scholar]
- 67.Yeh GC, Harris SC, Phang JM. Pyrroline-5-carboxylate reductase in human erythrocytes. J Clin Invest. 1981;67:1042–1046. doi: 10.1172/JCI110115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lorans G, Phang JM. Proline synthesis and redox regulation: differential functions of pyrroline-5-carboxylate reductase in human lymphoblastoid cell lines. Biochem Biophys Res Commun. 1981;101:1018–1025. doi: 10.1016/0006-291x(81)91850-7. [DOI] [PubMed] [Google Scholar]
- 69.Smith RJ, Downing SJ, Phang JM, Lodato RF, Aoki TT. Pyrroline-5-carboxylate synthase activity in mammalian cells. Proc Natl Acad Sci U S A. 1980;77:5221–5225. doi: 10.1073/pnas.77.9.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peisach J, Strecker HJ. The interconversion of glutamic acid and proline. V The reduction of Δ1-pyrroline-5-carboxylic acid to proline. J Biol Chem. 1962;237:2255–2260. [PubMed] [Google Scholar]
- 71.Christensen EM, Patel SM, Korasick DA, Campbell AC, Krause KL, Becker DF, Tanner JJ. Resolving the cofactor-binding site in the proline biosynthetic enzyme human pyrroline-5-carboxylate reductase 1. J Biol Chem. 2017;292:7233–7243. doi: 10.1074/jbc.M117.780288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Ingeniis J, Ratnikov B, Richardson AD, Scott DA, Aza-Blanc P, De SK, Kazanov M, Pellecchia M, Ronai Z, Osterman AL, Smith JW. Functional specialization in proline biosynthesis of melanoma. PLoS One. 2012;7:e45190. doi: 10.1371/journal.pone.0045190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Metabolic and Molecular Basis of Inherited Disease. McGraw-Hill; New York: 2001. pp. 1821–1838. [Google Scholar]
- 74.Struys EA, Jansen EE, Salomons GS. Human pyrroline-5-carboxylate reductase (PYCR1) acts on Δ1-piperideine-6-carboxylate generating L-pipecolic acid. J Inherited Metab Dis. 2014;37:327–332. doi: 10.1007/s10545-013-9673-4. [DOI] [PubMed] [Google Scholar]
- 75.Luo M, Tanner JJ. Structural basis of substrate recognition by aldehyde dehydrogenase 7A1. Biochemistry. 2015;54:5513–5522. doi: 10.1021/acs.biochem.5b00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nocek B, Chang C, Li H, Lezondra L, Holzle D, Collart F, Joachimiak A. Crystal structures of delta1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J Mol Biol. 2005;354:91–106. doi: 10.1016/j.jmb.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bottoms CA, Smith PE, Tanner JJ. A structurally conserved water molecule in Rossmann dinucleotide-binding domains. Protein Sci. 2002;11:2125–2137. doi: 10.1110/ps.0213502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hagedorn CH, Phang JM. Catalytic transfer of hydride ions from NADPH to oxygen by the interconversions of proline and delta 1-pyrroline-5-carboxylate. Arch Biochem Biophys. 1986;248:166–174. doi: 10.1016/0003-9861(86)90413-3. [DOI] [PubMed] [Google Scholar]
- 79.Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A. Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem. 2009;284:26482–26492. doi: 10.1074/jbc.M109.009340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phang JM, Liu W, Zabirnyk O. Proline metabolism and microenvironmental stress. Annu Rev Nutr. 2010;30:441–463. doi: 10.1146/annurev.nutr.012809.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elia I, Schmieder R, Christen S, Fendt SM. Organ-specific cancer metabolism and its potential for therapy. Handb Exp Pharmacol. 2015;233:321–353. doi: 10.1007/164_2015_10. [DOI] [PubMed] [Google Scholar]
- 83.Lunt SY, Fendt SM. Metabolism – A cornerstone of cancer initiation, progression, immune evasion and treatment response. Curr Opin Syst Biol. 2018;8:67–72. [Google Scholar]
- 84.Loayza-Puch F, Rooijers K, Buil LCM, Zijlstra J, Oude Vrielink FJ, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, van Tellingen O, Bex A, Agami R. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- 85.Sahu N, Dela Cruz D, Gao M, Sandoval W, Haverty PM, Liu J, Stephan JP, Haley B, Classon M, Hatzivassiliou G, Settleman J. Proline starvation induces unresolved ER stress and hinders mTORC1-dependent tumorigenesis. Cell Metab. 2016;24:753–761. doi: 10.1016/j.cmet.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 86.Hollinshead KER, Munford H, Eales KL, Bardella C, Li C, Escribano-Gonzalez C, Thakker A, Nonnenmacher Y, Kluckova K, Jeeves M, Murren R, Cuozzo F, Ye D, Laurenti G, Zhu W, Hiller K, Hodson DJ, Hua W, Tomlinson IP, Ludwig C, Mao Y, Tennant DA. Oncogenic IDH1 mutations promote enhanced proline synthesis through pycr1 to support the maintenance of mitochondrial redox homeostasis. Cell Rep. 2018;22:3107–3114. doi: 10.1016/j.celrep.2018.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 88.Guo JY, Teng X, Laddha SV, Ma S, Van Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD, White E. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. doi: 10.1101/gad.283416.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nagano T, Nakashima A, Onishi K, Kawai K, Awai Y, Kinugasa M, Iwasaki T, Kikkawa U, Kamada S. Proline dehydrogenase promotes senescence through the generation of reactive oxygen species. J Cell Sci. 2017;130:1413. doi: 10.1242/jcs.196469. [DOI] [PubMed] [Google Scholar]
- 90.Togashi Y, Arao T, Kato H, Matsumoto K, Terashima M, Hayashi H, de Velasco MA, Fujita Y, Kimura H, Yasuda T, Shiozaki H, Nishio K. Frequent amplification of ORAOV1 gene in esophageal squamous cell cancer promotes an aggressive phenotype via proline metabolism and ROS production. Oncotarget. 2014;5:2962–2973. doi: 10.18632/oncotarget.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang L, Zeng J, Geng P, Fang C, Wang Y, Sun M, Wang C, Wang J, Yin P, Hu C, Guo L, Yu J, Gao P, Li E, Zhuang Z, Xu G, Liu Y. Global metabolic profiling identifies a pivotal role of proline and hydroxyproline metabolism in supporting hypoxic response in hepatocellular carcinoma. Clin Cancer Res. 2018;24:474–485. doi: 10.1158/1078-0432.CCR-17-1707. [DOI] [PubMed] [Google Scholar]
- 92.Liu W, Glunde K, Bhujwalla ZM, Raman V, Sharma A, Phang JM. Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer Res. 2012;72:3677–3686. doi: 10.1158/0008-5472.CAN-12-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rinaldi G, Rossi M, Fendt SM. Metabolic interactions in cancer: Cellular metabolism at the interface between the microenvironment, the cancer cell phenotype and the epigenetic landscape. Wiley Interdiscip Rev: Syst Biol Med. 2018;10:e1397. doi: 10.1002/wsbm.1397. [DOI] [PubMed] [Google Scholar]
- 94.Fendt SM. Is there a therapeutic window for metabolism-based cancer therapies? Front Endocrinol. 2017;8:150. doi: 10.3389/fendo.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discovery. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 96.Clelland CL, Read LL, Baraldi AN, Bart CP, Pappas CA, Panek LJ, Nadrich RH, Clelland JD. Evidence for association of hyperprolinemia with schizophrenia and a measure of clinical outcome. Schizophr Res. 2011;131:139–145. doi: 10.1016/j.schres.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zarchi O, Carmel M, Avni C, Attias J, Frisch A, Michaelovsky E, Patya M, Green T, Weinberger R, Weizman A, Gothelf D. Schizophrenia-like neurophysiological abnormalities in 22q11.2 deletion syndrome and their association to COMT and PRODH genotypes. J Psychiatr Res. 2013;47:1623–1629. doi: 10.1016/j.jpsychires.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 98.Reversade B, Escande-Beillard N, Dimopoulou A, Fischer B, Chng SC, Li Y, Shboul M, Tham PY, Kayserili H, Al-Gazali L, Shahwan M, Brancati F, Lee H, O’Connor BD, Schmidt-von Kegler M, Merriman B, Nelson SF, Masri A, Alkazaleh F, Guerra D, Ferrari P, Nanda A, Rajab A, Markie D, Gray M, Nelson J, Grix A, Sommer A, Savarirayan R, Janecke AR, Steichen E, Sillence D, Hausser I, Budde B, Nurnberg G, Nurnberg P, Seemann P, Kunkel D, Zambruno G, Dallapiccola B, Schuelke M, Robertson S, Hamamy H, Wollnik B, Van Maldergem L, Mundlos S, Kornak U. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat Genet. 2009;41:1016–1021. doi: 10.1038/ng.413. [DOI] [PubMed] [Google Scholar]
- 99.Nakayama T, Al-Maawali A, El-Quessny M, Rajab A, Khalil S, Stoler JM, Tan WH, Nasir R, Schmitz-Abe K, Hill RS, Partlow JN, Al-Saffar M, Servattalab S, LaCoursiere CM, Tambunan DE, Coulter ME, Elhosary PC, Gorski G, Barkovich AJ, Markianos K, Poduri A, Mochida GH. Mutations in PYCR2, encoding pyrroline-5-carboxylate reductase 2, cause microcephaly and hypomyelination. Am J Hum Genet. 2015;96:709–719. doi: 10.1016/j.ajhg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zaki MS, Bhat G, Sultan T, Issa M, Jung HJ, Dikoglu E, Selim L, Mahmoud IG, Abdel-Hamid MS, Abdel-Salam G, Marin-Valencia I, Gleeson JG. PYCR2 mutations cause a lethal syndrome of microcephaly and failure to thrive. Ann Neurol. 2016;80:59–70. doi: 10.1002/ana.24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lerma-Ortiz C, Jeffryes JG, Cooper AJ, Niehaus TD, Thamm AM, Frelin O, Aunins T, Fiehn O, de Crecy-Lagard V, Henry CS, Hanson AD. ‘Nothing of chemistry disappears in biology’: the Top 30 damage-prone endogenous metabolites. Biochem Soc Trans. 2016;44:961–971. doi: 10.1042/BST20160073. [DOI] [PubMed] [Google Scholar]