Abstract
Background
Reliable prognostic markers for predicting severity of allergic reactions during oral food challenges (OFC) have not been established.
Objective
We sought to develop a predictive algorithm of a food challenge severity score (CSS) to identify those at higher risk for severe reactions to a standardized peanut OFC.
Methods
Medical history and allergy tests were obtained for 120 peanut-allergic participants who underwent double-blind, placebo-controlled food challenges (DBPCFCs). Reactions were assigned a CSS between 1 to 6 based on cumulative tolerated dose and a “severity clinical indicator.” Demographic characteristics, clinical features, peanut component IgE values, and a basophil activation marker were considered in a multi-step analysis to derive a flexible decision rule to understand risk during peanut of OFC.
Results
18.3% participants had a severe reaction (CSS >4). The decision rule identified the following three variables (in order of importance) as predictors of reaction severity: ratio of %CD63hi stimulation with peanut to %CD63hi anti-IgE (CD63 ratio), history of exercise-induced asthma, and forced expiratory volume in 1 sec/forced vital capacity (FEV1/FVC) ratio. The CD63 ratio alone was a strong predictor of CSS (p<0.001).
Conclusion
The CSS is a novel tool that combines dose thresholds and allergic reactions to understand risks associated with peanut OFCs. Lab-values (CD63 ratio), along with clinical variables (exercise-induced asthma and FEV1/FVC ratio) contribute to the predictive ability of the severity of reaction to peanut OFC. Further testing of this decision rule is needed in a larger external data source before it can be considered outside of research settings.
Keywords: asthma, basophil activation testing, biomarkers, CD63, classification and regression tree, component resolved diagnostics, decision trees, food challenge, peanut allergy, random forest, severity
INTRODUCTION
Epidemiologic studies indicate that the prevalence of food allergies, especially to peanuts, has increased worldwide in the last few decades.1–3 Food and peanut allergies are now estimated to affect about 6–10% and 1–3% of the overall population, respectively. Among children with food allergies, about 30% of food-allergic individuals have allergies to multiple foods.4–7 Emergency department visits and hospitalizations due to food-induced anaphylaxis have significantly increased in recent years8 and, in the US, these now number approximately 200,000 per year with further increases expected.9 The risk of a severe reaction is higher for those with peanut and tree-nut allergies and it is estimated that more than 50% of these patients experience at least one severe allergic reaction.4
The diagnostic approach to food allergies can be complex.10, 11 Skin prick tests (SPTs) and measurements of food allergen-specific immunoglobulin E (IgE) are commonly performed; however, these tests are not definitive because they lack specificity and result in a high number of false-positives.10, 11 The current gold standard for confirming or excluding a food allergy is the double-blind, placebo-controlled food challenge (DBPCFC).10, 11 However, DBPCFCs carry the risk of systemic and potentially life-threatening reactions, including severe anaphylaxis, requiring hospitalization or intensive care.12
The development of clinically useful tools for stratifying patients according to potential risk of severe reactions during peanut OFC would therefore be of great clinical value. Predictive models using multivariate logistic regression analysis already have been developed for egg, wheat, and milk allergy and these are promising in efforts to stratify risk for severity of allergic reaction during peanut OFC with these foods.13–15 For peanut allergy, certain biomarkers have been associated with reaction severity during peanut OFCs, such as isolated sensitization to Ara h 2, epitope diversity (i.e., extent of IgE binding to the combination of Ara h 1, 2, and 3), and results of changes in levels of basophil CD63 or CD203c staining in basophil activation tests (BATs) upon ex vivo activation of basophils with peanut allergen.16–19 In this study, we sought to develop a predictive algorithm to identify individuals with peanut allergy who are at higher risk for severe reactions during standardized peanut OFC. To do this, we tested data obtained from participants eligible for a large phase 2, randomized clinical study using peanut oral immunotherapy, including well-characterized clinical data and the participant’s responses to validated and standardized peanut OFCs, as well as results of complete blood counts, SPTs, BATs, measurements of peanut-specific IgE (sIgE) and IgG4 (sIgG4), total IgE (tIgE) and peanut component resolved diagnostics (CRDs) (sIgE to Ara h 1, 2, 3, 8, 9). This permitted us to define biomarkers, which we incorporated into a unique food challenge severity score (CSS) and a predictive algorithm to identify those individuals at higher risk for severe reactions to a standardized peanut OFC.
METHODS
Study population and preliminary visit
All aspects of the study were authorized by the Stanford University School of Medicine Institutional Review Board (Stanford, CA) and were overseen in collaboration with NIAID and the DAIT Data Safety Monitoring Board. One hundred and twenty patients between the ages of 7 to 55 years with a convincing history of peanut allergy were recruited to undergo standardized DBPCFCs at the Sean N. Parker Center for Allergy and Asthma Research at Stanford University as part of screening for clinical trial enrollment (POISED: Peanut Oral Immunotherapy: Safety, Efficacy and Discovery; Clinical Trials Identifier: NCT02103270). A detailed medical history, physical examination, spirometry, SPT, sIgE and tIgE testing, BATs, and CRDs were completed at screening. Patients with a prior history of a peanut-allergic reaction requiring intubation or eliciting hypotension were excluded, while patients with previous reactions to peanut requiring epinephrine for other severe symptoms were eligible.
DBPCFCs
Standardized DBPCFCs to peanut and oat (placebo) were performed according to validated guidelines.11, 20, 21 A normal physical examination and Forced Expiratory Volume in 1 second (FEV1) ≥ 80% were confirmed prior to the start of each OFC. The protocol consisted of 7 escalating doses (5, 20, 50, 100, 100, 100, 125 mg) of the food protein in flour form, concealed in either applesauce or pudding, ingested by the participant every 15 minutes, as tolerated. Vital signs and pertinent physical examinations were repeated every 15 minutes, or more frequently at the discretion of the clinician. If a positive allergic reaction was observed during OFC20, 22, the OFC was immediately discontinued and appropriate treatments were administered as per guidelines and published procedures.20, 23, 24 Reaction severity was scored according to a combination of Bock and Practall grading scale (range 0–3) 22,24 and assigned a severity clinical indicator (SCI) of negative or positive. A positive SCI categorized those participants who had any of the following reactions: severe urticaria (involving more than 2/3 of body surface area), severe/disabling abdominal pain, vomiting and diarrhea in severe abdominal symptoms, any wheezing, any voice changes (airway obstruction), or hypotension. A negative SCI score was assigned for milder reactions. None of the patients in this cohort experienced voice changes or hypotension. To evaluate for sensitivity to peanut protein quantitatively, the cumulative tolerated dose (CTD; the amount of peanut ingested prior to treating the participant for objective symptoms) was also quantified.
Food Challenge Severity Score
We developed a clinical severity scoring system, with scores between 1 and 6. This new scoring system combines two important factors, the SCI and the CTD, as detailed in Table 1; e.g., CSS 1 indicates a mild reaction on ingestion of a large amount of peanut and CSS 6 indicates a severe reaction following ingestion of a small amount of peanut.
Table 1.
Distribution of Challenge Severity Score
| Cumulative Tolerated Dose (mg) | Severity Challenge Indicator(SCI) | Challenge Severity Score (CSS) | Number of Subjects (%) |
|---|---|---|---|
| 175 – 375 | Negative | 1 | 15 (12.5%) |
| 25 – 75 | Negative | 2 | 44 (36.7%) |
| < 25 | Negative | 3 | 39 (32.5%) |
| 175 – 375 | Positive | 4 | 9 (7.5%) |
| 25 – 75 | Positive | 5 | 12 (10%) |
| < 25 | Positive | 6 | 1 (0.8%) |
Note: SCI is a measure of reaction severity.
Basophil Activation Tests
BATs were performed as previously described.25 Whole blood was collected in heparin tubes, kept at 4°C for 24 hours, and then stimulated for 30 minutes with peanut (prepared from Byrd Mill peanut flour) as previously described 25 (peanut concentrations of 1000, 100, 10, 1, and 0.1 ng/mL) at 37°C. Anti-IgE, IL-3, or Roswell Park Memorial Institute (RPMI) media alone were used as controls. Cells were stained with CD203c, CD63, CD123, and HLA-DR. Basophils were gated as CD123+HLA-DR−. Basophil expression of CD63 and CD203c was analyzed with FlowJo (Ashland, Oregon). In total, we assessed 15 BAT variables as presented in eTable 1. As previously reported, the following forms of CD203c and CD63 were evaluated: the ratio of CD203c stimulation with peanut to CD203c RPMI (negative control), %CD63hi stimulation with peanut, and the ratio of %CD63hi stimulation with peanut to %CD63hi anti-IgE.26 The ratio of the percentages of CD63hi basophils after stimulation with varying concentrations of peanut vs. anti-IgE (CD63 ratio) were also included to assess the magnitude of the response.25
Statistical analysis and variable selection process
Descriptive statistics such as means, medians, standard deviations and interquartile ranges were used to compare baseline characteristics by CSS. Due to only one subject with an assigned CSS of 6, scores 5 and 6 were collapsed when reporting demographics. Graphical tools were used to assess distributional aspects. Log-transformations were applied when normality assumptions were violated. Kruskal-Wallis and Fisher’s exact tests were used to draw comparisons on CSS by continuous and categorical characteristics, respectively.
Our variable selection decisions are outlined in eFigure 1. Among the 94 available clinical, laboratory, and demographic features, self-reported overall and detailed allergic rhinitis variables were excluded (n=7) due to non-concordance against clinical testing (eTable 2), as our interest was in arriving at an objective decision rule. Further, absolute counts were included for analysis from the CBC (excluded n=4 variables).
We then took a three-staged approached to arriving at our model. In the first stage, one BAT variable out of 15 possible BATs was pre-selected using an Area under the Curve (AUC), as most laboratories would have resources to measure one BAT (either CD203c or CD63) following stimulation with one concentration of peanut. Logistic regression models were fit to SCI as a function of each log-transformed BAT, and the BAT with the highest AUC was selected to be included as a potential predictor in the second stage of the process.
In the second stage, variables of importance for CSS were identified from the subset of 69 clinical, laboratory, and demographic features, including the pre-selected BAT from the first stage, using conditional random forest (RF) approaches.27 Conditional RF methods use a permutation-based significance testing procedure to select variables as opposed to the default approach that utilizes the Gini statistic, avoiding the bias of selecting variables that have many possible splits found in traditional RF methods.28 Variables were chosen as relevant if their importance value was above the absolute value of the lowest negative-scoring variable.29
In the third stage, a clinically interpretable decision rule using classification and regression trees (CART) was derived from the subset of variables deemed important via RF in the second stage. CART is a tree-based learning technique for classifying observations.30
To compare the results derived from variable selection via RF and prediction via CART to more widely used statistical methods, the least absolute shrinkage and selection operator (LASSO) was applied to the 69 pre-selection features. LASSO is a regression analysis method that performs both variable selection and regularization in order to gain higher predictive ability of the final fitted model.31 The Root Mean Squared Error (RMSE) from CART and the LASSO regression model were compared.
Two supplementary analyses were also performed. First, CSS was fit as a function of log-transformed CD63 ratio alone in a linear regression model, and the change in CSS and p-value were reported. Second, four logistic regression models were fit to an indicator for whether or not a subject reacted to the peanut OFCs at or before 175mg as a function of the CD63 ratio, Ara h 2, sIgE, and SPT, respectively. A cutoff of 175mg peanut protein was chosen to be comparable to other published literature, looking at a dose less than a full peanut (240mg).32, 33 Youden’s index was used to determine a threshold for each predictor which maximized the AUC.34 The threshold, and AUC (95% CI) were reported for each of the four predictors.
The entire three-stage process and LASSO were internally validated using repeated 10-fold cross validation as commonly performed 35. All analyses were conducted using R, version 3.2.2.36, 37 Additional details of statistical methods can be found in eMethods.
RESULTS
Total study population
The median participant age was 11 years. Baseline demographics are reported in Table 2. Eighty-two percent of the cohort was under 18 years of age; 33% of the cohort was female. Participants had comorbid atopic conditions including other food allergies (70%), asthma (67%), atopic dermatitis (73%), and allergic rhinitis (AR) (75%).
Table 2.
Baseline demographics
| Challenge Severity Score (CSS) | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| 1 (n=15) | 2 (n=44) | 3 (n=39) | 4 (n=9) | 5,6† (n=13) | Total (N=120) | P-value* | |
| Age at enrollment (y), median (IRQ) | 9 (4) | 11 (7) | 11 (13) | 13 (18) | 9 (2) | 11 (7) | 0.10 |
| Child (less than 18 yo), no. (%) | 14 (93.3) | 38 (86.4) | 28 (71.8) | 6 (66.7) | 12 (92.3) | 98 (81.7) | 0.15 |
| Female, no. (%) | 6 (40.0) | 8 (18.2) | 17 (43.6) | 4 (44.4) | 4 (30.8) | 39 (32.5) | 0.10 |
| Race, no. (%) | |||||||
| Caucasian | 14 (93.3) | 34 (77.3) | 21 (53.9) | 7 (77.8) | 9 (69.2) | 85 (70.8) | 0.04 |
| Asian | 3 (20.0) | 11 (25.0) | 22 (56.4) | 2 (22.2) | 4 (30.8) | 42 (35.0) | 0.02 |
| Black or African American | 0 | 3 (6.8) | 0 | 0 | 0 | 3 (2.5) | 0.48 |
| Native Hawaiian or Pacific Islander | 0 | 0 | 1 (2.6) | 0 | 0 | 1 (0.8) | 0.63 |
| Not Hispanic or Latino, no. (%) | 15 (100.0) | 42 (95.5) | 39 (100.0) | 9 (100.0) | 12 (92.3) | 117 (97.5) | 0.43 |
| BMI (kg/m2), mean (SD) | 17.8 (3.2) | 18.6 (4.7) | 20.3 (6.0) | 20.5 (3.4) | 17.7 (2.5) | 19.1 (4.8) | 0.12 |
| Number of food allergies, mean (SD) | 3.1 (2.5) | 3.9 (2.5) | 4.0 (2.8) | 3.0 (2.3) | 3.6 (2.7) | 3.7 (2.6) | 0.71 |
| Subjects with food allergy to more than one food, n (%) | 11 (73.3) | 33 (75.0) | 27 (69.2) | 5 (55.6) | 8 (61.5) | 84 (70.0) | 0.72 |
| Asthma and allergy history, no. (%) | |||||||
| Asthma | 6 (40.0) | 26 (59.1) | 30 (76.9) | 7 (77.9) | 11 (84.6) | 80 (66.7) | 0.04 |
| Age of onset (y), mean (SD) | 2.9 (1.9) | 3.6 (2.7) | 3.7 (3.2) | 4.5 (3.3) | 3.2 (2.1) | 3.6 (2.8) | 0.91 |
| AD | 11 (73.3) | 33 (75.0) | 28 (71.8) | 4 (44.4) | 11 (84.6) | 87 (72.5) | 0.37 |
| Age of onset (y), mean (SD) | 0.8 (1.4) | 0.9 (1.2) | 2.6 (5.8) | 0.5 (0.4) | 0.6 (0.8) | 1.4 (3.5) | 0.83 |
| AR | 11 (73.3) | 34 (77.3) | 28 (71.8) | 8 (88.9) | 9 (69.2) | 90 (75.0) | 0.84 |
| Age of onset (y), mean (SD) | 5.5 (2.2) | 5.1 (4.0) | 5.7 (3.6) | 5.9 (3.3) | 3.3 (1.6) | 5.2 (3.5) | 0.19 |
AD, Atopic dermatitis; AR, allergic rhinitis; BMI, body mass indicator
Kruskal-Wallis and Fisher’s exact tests were used to calculate p-values for continuous and categorical variables by CSS, respectively.
Collapsed CSS 5 and 6 due to only 1 subject in the CSS 6 category.
Characteristics of study population and CSS outcomes
OFCs were discontinued after objective signs or symptoms of an allergic reaction were observed. Symptoms ranged from mild urticaria to more severe symptoms such as wheezing or diffuse urticaria. The mean CSS was 2.7 with a standard deviation of 1.2. The majority (82%) of subjects were assigned a CSS of 1–3, indicating less severe reactions in the DBPCFC (Table 1). All challenge reactions were treated with no sequelae. We observed no biphasic reactions. Eighteen children (7–17 years) and 4 adults (18–53 years) had more severe scores (CSS greater than 3). Of the 22 participants with a more severe CSS, 50% had wheezing, 45% had severe abdominal pain, and 5% had severe urticaria (eTable 3). Each of the 120 participants developed an objective allergic reaction to a CTD of ≤ 500 mg peanut protein during their DBPCFC. The median CTD was 75.0 mg.
A higher proportion of Caucasians were in the lowest CSS group (CSS 1, 93.3%, p = 0.04, Table 2). There were more patients with a history of asthma in the highest CSS group (CSS 5–6, 84.6%, p= 0.04), however, per our protocol, DBPCFCs were only conducted if FEV1% was ≥ 80%. Consequently, mean FEV1% was 95.5% and mean FEV1/FVC ratios 84% (Table 3). There was no evidence to suggest that peanut SPT average wheal size, sIgE, tIgE, CRD tests, or complete blood counts were significantly different across CSS levels (Table 3).
Table 3.
Clinical and lab characteristics
| Challenge Severity Score (CSS) | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| 1 (n=15) | 2 (n=44) | 3 (n=39) | 4 (n=9) | 5,6‡ (n=13) | Total (N=120) | P-value * | |
| FEV1 % at DBPCFC, mean (SD) | 94.5 (10.1) | 97.5 (12.9) | 94.7 (11.2) | 91.1 (7.6) | 94.7 (13.1) | 95.5 (11.7) | 0.72 |
| FEV1/FVC at DBPCFC, mean (SD) | 0.86 (0.07) | 0.86 (0.07) | 0.82 (0.07) | 0.80 (0.08) | 0.82 (0.06) | 0.84 (0.07) | 0.02 |
| Peanut wheal size (mm), mean (SD) | 12.2 (7.45) | 13.3 (6.4) | 13.4 (5.6) | 12.7 (5.8) | 13.5 (7.1) | 13.3 (6.1) | 0.96 |
| Complete Blood Count, mean (SD) | |||||||
| Absolute Basophil Count | 0.05 (0.03) | 0.04 (0.03) | 0.04 (0.02) | 0.04 (0.02) | 0.04 (0.01) | 0.04 (0.02) | 0.90 |
| Absolute Neutrophil Count | 3.3 (1.2) | 2.9 (1.3) | 3.2 (1.4) | 3.7 (1.1) | 3.2 (1.8) | 3.1 (1.4) | 0.17 |
| Absolute Lymphocyte Count | 2.4 (0.7) | 2.2 (0.6) | 2.2 (0.6) | 2.4 (0.8) | 2.2 (0.5) | 2.2 (0.6) | 0.78 |
| Absolute Monocyte Count | 0.5 (0.2) | 0.5 (0.2) | 0.5 (0.2) | 0.5 (0.1) | 0.5 (0.2) | 0.5 (0.2) | 0.92 |
| Absolute Eosinophil Count | 0.5 (0.2) | 0.4 (0.3) | 0.4 (0.2) | 0.4 (0.3) | 0.4 (0.2) | 0.4 (0.2) | 0.61 |
| CD203c Basophil Activation Test, mean (SD) † | |||||||
| PF 0.1 | 1.28 (0.40) | 1.14 (0.15) | 1.20 (0.18) | 1.42 (0.24) | 1.17 (0.17) | 1.20 (0.22) | 0.01 |
| PF 1 | 1.28 (0.27) | 1.28 (0.22) | 1.34 (0.25) | 1.49 (0.27) | 1.38 (0.24) | 1.32 (0.24) | 0.09 |
| PF 10 | 1.52 (0.52) | 1.66 (0.36) | 1.66 (0.40) | 1.88 (0.42) | 1.72 (0.40) | 1.67 (0.40) | 0.11 |
| PF 100 | 1.74 (0.66) | 1.85 (0.49) | 1.86 (0.51) | 2.02 (0.44) | 1.91 (0.36) | 1.86 (0.50) | 0.44 |
| PF 1000 | 1.73 (0.63) | 1.82 (0.48) | 1.75 (0.47) | 1.97 (0.44) | 1.86 (0.28) | 1.80 (0.47) | 0.39 |
| %CD63high Basophil Activation Test, median (IQR)† | |||||||
| PF 0.1 | 0.48 (1.02) | 0.60 (1.24) | 0.54 (0.81) | 0.45 (0.66) | 0.90 (1.18) | 0.55 (1.25) | 0.75 |
| PF 1 | 0.31 (0.62) | 1.66 (4.35) | 2.21 (3.24) | 2.06 (3.35) | 4.83 (5.83) | 1.92 (4.20) | 0.01 |
| PF 10 | 3.33 (6.63) | 10.70 (25.97) | 14.20 (18.37) | 12.60 (15.80) | 24.40 (19.80) | 11.90 (20.83) | 0.004 |
| PF 100 | 10.50 (25.61) | 24.00 (31.68) | 20.50 (30.99) | 9.59 (18.70) | 36.80 (22.50) | 22.40 (29.48) | 0.02 |
| PF 1000 | 6.82 (22.54) | 14.85 (25.04) | 13.50 (13.63) | 11.30 (17.68) | 14.90 (16.79) | 13.80 (18.85) | 0.24 |
| PF10/%CD63high anti-IgE | 0.46 (1.59) | 0.70 (1.42) | 1.51 (1.84) | 2.42 (7.57) | 1.93 (5.07) | 1.21 (1.82) | 0.01 |
| Antibodies (ng/L), median (IQR) | |||||||
| Peanut IgG4 (ug/L) | 430.0 (1870.0) | 1110.0 (2345.0) | 640.0 (930.0) | 450.0 (620.0) | 1600.0 (1220.0) | 830 (1,600) | 0.05 |
| Peanut IgE | 77.1 (277.7) | 178.9 (412.5) | 193.0 (478.0) | 21.9 (217.3) | 383.1 (614.6) | 192.9 (458.0) | 0.13 |
| Total IgE | 1342.0 (2364.4) | 1321.3 (2526.6) | 1149.2 (1766.6) | 592.9 (1461.6) | 1351.8 (2249.7) | 1284.7 (2379) | 0.49 |
| Peanut components (ng/L), median (IQR) | |||||||
| Ara h 1 | 27.3 (166.3) | 21.4 (115.1) | 57.1 (191.2) | 3.6 (15.5) | 179.6 (212.5) | 34.9 (177.8) | 0.18 |
| Ara h 2 | 39.3 (107.8) | 70.6 (209.5) | 87.1 (197.9) | 12.8 (140.9) | 196.7 (216.9) | 85.0 (207.1) | 0.09 |
| Ara h 3 | 0.9 (37.2) | 3.2 (22.2) | 9.7 (32.2) | 1.1 (5.3) | 25.9 (39.3) | 5.7 (38.6) | 0.31 |
| Ara h 8 | 0.0 (2.5) | 0.0 (2.5) | 0.0 (1.6) | 0.0 (0.6) | 0.0 (0.4) | 0.0 (1.8) | 0.95 |
| Ara h 9 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.16 |
BAT, Basophil activation test; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; DBPCFC, double-blind placebo-controlled food challenge
Kruskal-Wallis and Fisher’s exact tests were used to calculate p-values for continuous and categorical variables by CSS, respectively.
PF 100 indicates values for basophils stimulated with peanut flour at the 100 ng/mL concentration, etc.
Collapsed CSS 5 and 6 due to only 1 subject in the CSS 6 category.
Preselection of Basophil Activation Test
The %CD63hi response to 10 ng/mL peanut divided by %CD63hi anti-IgE (CD63 ratio) was chosen among all BATs to be further considered in RF based on having the highest AUC (0.67) (eTable 1).
Predictive algorithm for the CSS
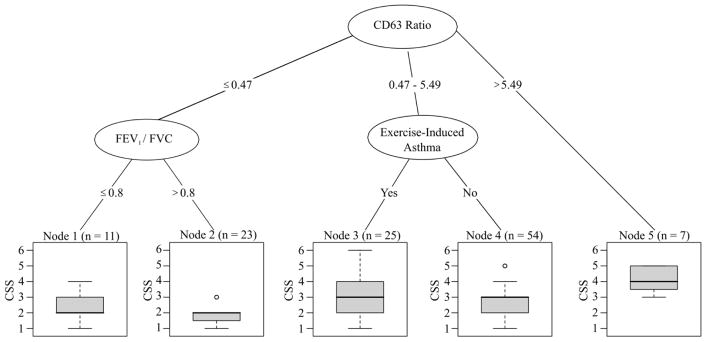
Of the set of 69 features, 4 features were identified as important predictors of the CSS using RF selection and the threshold criteria previously described (Figure 1). The CD63 ratio, history of exercise-induced asthma (EIA), the ratio of FEV1 to FVC at DBPCFC, and absolute blood basophil count were then entered into a CART model, with three appearing in the final decision rule (Figure 2). Our analysis demonstrated that the CD63 ratio, a history of EIA, and the FEV1/FVC ratio at DBPCFC are important predictors of reaction severity. Participants with a CD63 ratio greater than 5.49 were at highest risk of experiencing a severe reaction with a median CSS of 4. For CD63 ratios between 0.47 and 5.49, a history of EIA may increase the risk of a severe reaction. For CD63 ratios ≤ 0.47, an FEV1/FVC ratio of ≤ 0.8 may result in slightly higher risk of a severe reaction compared in participants with higher FEV1/FVC ratios. To assess the validity of this model, a repeated 10-fold cross-validation was used and yielded an RMSE of 1.16 for the three-staged modeling process.
FIG. 1.
Random forest plot highlights variables which were deemed important: the CD63 ratio, a history of exercise-induced asthma (EIA), FEV1/FVC at DBPCFC, and the absolute blood basophil count.
Included variables are listed on the y-axis (n=69), with corresponding variable importance on the x-axis. Variables were deemed important predictors of the severity score if their importance value was larger than the absolute value of the lowest negative-scoring variable (to the right of the dashed red line). Therefore, the ratio of %CD63hi peanut protein concentration 10 to anti-IgE, a history of exercise-induced asthma (EIA), forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) at double-blind placebo-controlled food challenge (DBPCFC), and absolute blood basophil count were considered important variables. sIgE: peanut-specific IgE; sIgG4: peanut-specific IgG4; tIgE: total IgE; SPT: skin prick test; BMI = body mass index.
FIG. 2.
Using classification and regression trees (CART), 3 features were used to classify participants and assess risk of severity of reaction (CSS): the CD63 ratio, a history of exercise-induced asthma (EIA), and FEV1/FVC at DBPCFC.
ratio of heparin %CD63hi peanut protein concentration 10 to anti-IgE (CD63 Ratio), forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) at double-blind placebo-controlled food challenge (DBPCFC), and a history of exercise-induced asthma.
Features stratified by the terminal nodes of the CART are displayed in Table 4. Notably, node 5, which has a median CSS of 4 is characterized by a population of participants with the highest IgE values (total, peanut, Ara h 1, 2, 3, 8, and 9) and highest CD63 ratios.
Table 4.
Baseline characteristic differences by CART terminal nodes
| CV RMSE = 1.16 | |||||
|---|---|---|---|---|---|
|
| |||||
| Characteristic | Node 1 (n=11) | Node 2 (n=23) | Node 3 (n=25) | Node 4 (n=54) | Node 5 (n=7) |
| Continuous variables, Mean (SD) | |||||
| Peanut SPT | 4.27 (1.31) | 5.78 (2.81) | 4.88 (1.84) | 5.08 (2.12) | 5.50 (1.89) |
| FEV1 at DBPCFC | 92.18 (9.53) | 99.4 (13.87) | 91.24 (8.85) | 96.5 (11.59) | 94.57 (14.30) |
| FEV1/FVC ratio at DBPCFC | 0.75 (0.05) | 0.88 (0.05) | 0.81 (0.07) | 0.85 (0.06) | 0.80 (0.10) |
| Number of food allergies | 3.00 (3.22) | 4.00 (2.59) | 4.36 (2.66) | 3.65 (2.43) | 2.57 (2.15) |
| BMI | 18.63 (3.91) | 20.12 (6.79) | 19.44 (4.48) | 18.32 (3.89) | 21.09 (6.24) |
| CD63 ratio | 0.20 (0.16) | 0.20 (0.17) | 1.73 (1.45) | 2.01 (1.12) | 10.72 (4.41) |
| Continuous Variables, Median (1st quartile, 3rd quartile) | |||||
| Age at enrollment | 11 (9, 16) | 9 (8, 14.5) | 12 (9, 16) | 10.5 (8, 14) | 13 (10.5, 2.5) |
| Peanut diagnosis age | 2 (1, 3) | 2 (1, 3) | 1 (0.8, 2) | 1.5 (1, 2) | 1.5 (1, 3.5) |
| Peanut allergy duration | 9.2 (6.8, 13.3) | 8 (6, 13) | 9.3 (7.5, 13.3) | 8.7 (7, 12) | 11.5 (9.8, 29.5) |
| Ara h 1 (ng/L) | 1.2 (0, 3.6) | 0.6 (0, 26.0) | 52.5 (3.0, 135) | 82.6 (10.2, 224) | 134.7 (60.0, 232) |
| Ara h 2 (ng/L) | 6.3 (1.8, 14.0) | 8.9 (1.4, 64.8) | 75.6 (25.1, 169) | 152.9 (52.6, 409) | 193.5 (104.1, 439) |
| Ara h 3 (ng/L) | 0 (0, 1.3) | 0.4 (0, 3.0) | 4.1 (0.5, 40.3) | 18.2 (3.0, 46.4) | 25.9 (14.0, 43.4) |
| Ara h 8 (ng/L) | 0 (0, 1.3) | 0.3 (0, 12.2) | 0 (0, 0.7) | 0 (0, 1.5) | 0.4 (0.1, 2.2) |
| Ara h 9 (ng/L) | 0 (0, 12.1) | 0 (0, 0.2) | 0 (0, 0) | 0 (0, 0) | 0.5 (0, 2.6) |
| Peanut IgG4 (μg/L) | 480 (255, 715) | 360 (210, 1520) | 940 (350, 1600) | 910 (402, 2388) | 2460 (1055, 4535) |
| Peanut IgE (ng/L) | 14.4 (2.9, 65.8) | 47.3 (8.5, 219) | 192.8 (36.1, 312) | 316.0 (147, 767) | 446.5 (218, 1252) |
| Total IgE (ng/L) | 1342.0 (708, 3370) | 1193.2 (603, 2302) | 1281 (683, 2403) | 1339.6 (699, 3226) | 3991.8 (473, 8808) |
| Categorical Variables, Percent | |||||
| Male | 81.8 | 82.6 | 68 | 61.1 | 42.9 |
| Caucasian | 90.9 | 73.9 | 64 | 68.5 | 71.4 |
| Asian | 18.2 | 43.5 | 32 | 37 | 28.6 |
| Black | 0 | 4.3 | 4 | 1.9 | 0 |
| Non-Hispanic | 100 | 95.7 | 100 | 96.3 | 100 |
| Peanut Dx only | 54.5 | 17.4 | 24 | 29.6 | 57.1 |
| Tree nut Dx | 36 | 70 | 68 | 59 | 43 |
| History of Asthma | 63.6 | 52.2 | 100 | 53.7 | 100 |
| History of AR | 72.7 | 87 | 84 | 66.7 | 71.4 |
| History of AD | 63.6 | 73.9 | 84 | 72.2 | 42.9 |
AD, Allergic dermatitis; AR, allergic rhinitis; BMI, body mass indicator; CART, classification and regression trees; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; DBPCFC, double blind placebo controlled food challenge; SPT, skin prick test.
To determine the predictive ability of the CD63 ratio using standard regression techniques without the presence of other features, we also fit a linear regression model for CSS as a function of log-transformed CD63 ratio. We found that the CD63 ratio was a significant predictor of CSS alone, with a cross-validated root-mean-square-error (RSME) of 1.08 (eTable 4).
LASSO regression
After including all 69 pre-selected features into a LASSO model, history of EIA and the CD63 ratio were selected as statistically significant predictors of CSS (eTable 5). The repeated 10-fold cross-validation RMSE for this model was 2.47.
DISCUSSION
The DBPCFC is currently the gold standard for diagnosing food allergies and is most often performed in a research setting where the patients are likely to be allergic. In these allergic patients, while a positive outcome confirming food allergy is likely on DBPCFC, severe symptoms can be life threatening.10, 11,12 Identification of possible prognostic indicators that are associated with severe outcomes during a peanut OFC may aid in appropriate risk stratification and management when a OFC is being considered by a trained professional. Previous studies for peanut allergy have focused on clinical features and certain tests, such as SPTs, 38 sIgE,11, or CRDs that can assist in distinguishing between the absence or presence of a food allergy.39 Many investigators have already shown that sIgE to Ara h 2, as opposed to sIgE to whole peanut, has greater specificity for predicting reactivity during peanut OFC (ranging from 85%–95%, depending on the cohort).40–46
Few studies have examined biomarkers to predict the severity of a reaction. Our approach was to examine demographics, clinical and laboratory data in relation to a potential new predictor tool using a CART statistical approach. BATs can be a useful tool in predicting the severity of reaction during peanut OFC in peanut-sensitized individuals,17, 19, 47 despite earlier studies to the contrary.48 Other authors have published evidence on severity scores; however, these did not incorporate history of exercise-induced asthma, CD63 ratio, or FEV1/FVC ratio at DBPCFC.17, 19 Some groups have also tried to use clinical characteristics to predict increased adverse events during oral immunotherapy (OIT). One study found baseline allergic rhinitis and peanut SPT wheal size were significant predictors of higher overall AE rates. SPT wheal size predicted increased gastrointestinal AEs, and AR predicted increased systemic reactions during OIT.49
Our study is a prospective study of a well-characterized population of peanut-allergic children and adults who underwent DBPCFCs. Patients with a prior history of severe reactions to peanut were included, except for those patients with a history of a peanut-allergic reaction requiring intubation or eliciting hypotension. We aimed to develop a possible method to predict severe allergic reactions, as management of these reactions is labor-intensive, requires increased observation, and can be life-threatening.
There is no universal consensus to characterize what constitutes a severe reaction during peanut OFC. While other grading systems to assign ordinal values to severity of individual symptoms have been developed, not all symptoms should be assessed based on similar grading. For instance, severe pruritus grade 3 has a different clinical significance compared to wheezing grade 3.50 Because there is no universal agreement on the criteria for administrating epinephrine during peanut OFC, the SCI was created to characterize symptoms that most practitioners would treat with epinephrine. The CSS is novel in that it not only groups similarly severe symptoms in a more clinically meaningful way using SCI, but it incorporates the amount of food allergen ingested.20, 51, Further, CSS provides multi-level phenotyping of clinical symptom severity on an ordinal scale that captured the allergic features of the reaction. For example, a subject who is able to consume the equivalent of one peanut (240mg) and develops limited hives would be assigned a CSS of 1, whereas a subject who can only consume a low dose of peanut (5mg) and develops wheezing would receive a CSS of 6. Clinically these are important distinctions that have heretofore not been successfully characterized.
To our knowledge, this is the first study to use machine learning procedures, i.e. random forest and CART, which incorporate a combination of clinical and laboratory based parameters to develop an internally validated decision rule that could potentially be used by clinicians to better understand a patient’s risk of having a severe allergic reaction during peanut OFC. Unlike CART, previous models using standard regression procedures are not well-equipped to discover, in an agnostic fashion, complex interactions among features that are not pre-specified using dichotomous branching logic and optimal cut-off values among the features.
The basophil activation ratio of CD63 was prominently featured in our predictive model, similar to work done by Santos et al.17 The ratio of CD63 positive basophils after stimulation with peanut vs. anti-IgE was selected in the CART model in three different ranges to be an important initial discriminator of the CSS. A CD63 ratio of greater than 5.49 identified a subset of patients who had the highest predicted CSS. The absolute basophil count was chosen as one of four predictors in the RF model, but then excluded in the CART model, likely as no significant cut points could be identified to risk stratify participants.
In the predictive model (Figure 2), not only was a history of EIA important, but the CART analysis also selected the FEV1/FVC ratio on the day of peanut OFC. Asthmatics are known to have higher risk for more severe allergic reactions.52, 53 Thus, it is not recommended to conduct a peanut OFC in a patient with respiratory symptoms or an active respiratory infection. Current research protocols specify an FEV1/FVC ratio ≥ 80% cutoff prior to the start of peanut OFCs; this should also be considered for peanut OFCs in the allergist’s office as well.
Interestingly, antibody tests, including sIgE, sIgG4, ratios of sIgE/sIgG4, and peanut CRDs were similar in peanut allergic participants who had mild or severe reactions, and thus were not identified as important variables relative to the ones ultimately selected, particularly, the CD63 ratio. However, our data shows that the participants with the highest median CSS of 4 (node 5 in Fig. 2, Table 4) had on average higher IgEs (total, peanut, and CRDs) compared to subjects in other nodes. Even though these biomarkers had higher expression in node 5 compared to other nodes, they were not as predictive as the CD63 ratio (Figure 1). Additionally, we fit several logistic regression models to determine if any known predictors of severity (ie, SPT, Ara h 2, and sIgE) were better than the CD63 ratio, and found that the CD63 ratio was the superior biomarker (eTable 6).
With more clinical trials studying different therapies in peanut-allergic individuals, our tool could be used to aid the researcher in phenotyping those patients at risk for a more severe reaction at DBPCFC. We propose to currently apply this predictive decision rule in clinical research only. It is important to note that if a patient has a concurrent illness 49, these co-existing conditions can increase the likelihood of positive outcomes during peanut OFC. Our DBPCFCs were done in controlled settings in which we excluded illnesses or other events known to put participants at risk; these are the same precautions one would take in an outpatient clinic or clinical research study.
To provide a comparison between CART and a more widely used method, we fit a LASSO regression model using the 69 features chosen in the pre-selection process. We found that a history of EIA and a higher CD63 ratio were significantly associated with higher CSS. Further, the RMSE of the LASSO model was higher compared to that of the CART (2.44 vs 1.16), suggesting that the CART method was a superior predictive model. Importantly, while the cross-validated RMSE of the linear regression model that only includes CD63 ratio was lower than that obtained using CART (1.08 vs 1.16), interpreting results from the CART model is arguably more intuitive than interpretation resulting from a standard regression model. From the regression model, a 10% increase in the CD63 ratio was associated with a 0.02 unit increase in CSS (eTable 4). Conversely, using CART, we see how CSS is predicted to increase when values of CD63 ratio exceed a threshold.
In conclusion, we present early findings from a well characterized, relatively large, peanut allergic cohort that underwent standardized OFCs as part of a large, phase 2 prospective study. The decision rule developed using the CSS may be considered as a predictive aid in clinical research settings to determined risk of severe reaction during peanut OFC However, further testing of this decision rule is needed in a larger validation cohort. One of the limitation of this study is that this study did not include children (who may have no clear history of EIA), and this model cannot, at this time, be applied in younger children. The severity predictors we discovered in our CART and RF analyses highlight the importance of the CD63 ratio which may aid the researcher in deciding to conduct peanut OFCs.
Supplementary Material
Acknowledgments
FUNDING SOURCE
Supported by NIH grant U19 AI104209 (RSC, KM, KCN, and SJG), T32 AR050942 (to JSR), Glen and Wendy Miller Family Foundation (RG and BMS), and the Sean N. Parker Center for Allergy and Asthma Research at Stanford University.
We thank all the participants and their families who are part of this phase 2 study. We also thank Dr. Vanitha Sampath for her critical read, review, and edits, Dr. Robert Tibshirani for his review and input on the statistical methods, and Dr. Marshall Plaut for his critical read, review, and input to the manuscript.
ABBREVIATIONS
- AUC
Area under the Curve
- BAT
Basophil Activation Test
- CART
Classification and Regression Tree
- CRDs
Component Resolved Diagnostics
- CSS
Challenge Severity Score
- DBPCFC
Double-Blind, Placebo-Controlled, Food Challenge
- EIA
Exercise-induced Asthma
- FEV1
Forced Expiratory Volume in 1 second
- FVC
Forced Vital Capacity
- LASSO
Least Absolute Shrinkage and Selection Operator
- RF
Random Forest
- RMSE
Root Mean Squared Error
- SCI
Severity Clinical Indicator
- sIgE
Peanut-specific Immunoglobulin E
- sIgG4
Peanut-specific Immunoglobulin IgG4
- SPT
Skin Prick Test
- tIgE
Total IgE
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
CLINICAL TRIALS REGISTRATION
https://clinicaltrials.gov; National Clinical Trial Identifier: NCT02103270
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rinaldi M, Harnack L, Oberg C, Schreiner P, St Sauver J, Travis LL. Peanut allergy diagnoses among children residing in Olmsted County, Minnesota. J Allergy Clin Immunol. 2012;130:945–950. doi: 10.1016/j.jaci.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sicherer SH, Munoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J Allergy Clin Immunol. 2010;125:1322–1326. doi: 10.1016/j.jaci.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 3.Jackson KD, Howie LD, Akinbami LJ. Trends in allergic conditions among children: United States, 1997–2011. NCHS Data Brief. 2013:1–8. [PubMed] [Google Scholar]
- 4.Gupta RS, Springston EE, Warrier MR, et al. The prevalence, severity, and distribution of childhood food allergy in the United States. Pediatrics. 2011;128:e9–17. doi: 10.1542/peds.2011-0204. [DOI] [PubMed] [Google Scholar]
- 5.Soller L, Ben-Shoshan M, Harrington DW, et al. Overall prevalence of self-reported food allergy in Canada. J Allergy Clin Immunol. 2012;130:986–988. doi: 10.1016/j.jaci.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 6.Nwaru BI, Hickstein L, Panesar SS, Roberts G, Muraro A, Sheikh A. Prevalence of common food allergies in Europe: a systematic review and meta-analysis. Allergy. 2014;69:992–1007. doi: 10.1111/all.12423. [DOI] [PubMed] [Google Scholar]
- 7.Osborne NJ, Koplin JJ, Martin PE, et al. Prevalence of challenge-proven IgE-mediated food allergy using population-based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol. 2011;127:668–676. e661–662. doi: 10.1016/j.jaci.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 8.Dyer AA, Lau CH, Smith TL, Smith BM, Gupta RS. Pediatric emergency department visits and hospitalizations due to food-induced anaphylaxis in Illinois. Ann Allergy Asthma Immunol. 2015;115:56–62. doi: 10.1016/j.anai.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Clark S, Espinola J, Rudders SA, Banerji A, Camargo CA., Jr Frequency of US emergency department visits for food-related acute allergic reactions. J Allergy Clin Immunol. 2011;127:682–683. doi: 10.1016/j.jaci.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 10.American College of Allergy, Asthma, & Immunology. Food allergy: a practice parameter. Ann Allergy Asthma Immunol. 2006;96:S1–68. [PubMed] [Google Scholar]
- 11.Sampson HA, Aceves S, Bock SA, et al. Food allergy: a practice parameter update-2014. J Allergy Clin Immunol. 2014;134:1016–1025. e1043. doi: 10.1016/j.jaci.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Cianferoni A, Khullar K, Saltzman R, et al. Oral food challenge to wheat: a near-fatal anaphylaxis and review of 93 food challenges in children. World Allergy Organ J. 2013;6:14. doi: 10.1186/1939-4551-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugiura S, Matsui T, Furuta T, Sasaki K, Kando N, Ito K. Development of a prediction model for severe wheat allergy. Pediatr Allergy Immunol. 2017 doi: 10.1111/pai.12806. [DOI] [PubMed] [Google Scholar]
- 14.Sugiura S, Matsui T, Nakagawa T, et al. Development of a prediction model of severe reaction in boiled egg challenges. Allergol Int. 2016;65:293–299. doi: 10.1016/j.alit.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Sugiura S, Sasaki K, Matsui T, Nakagawa T, Kando N, Ito K. Development of a prediction model for a severe reaction in cow’s milk challenges. Allergol Int. 2017;66:493–494. doi: 10.1016/j.alit.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Flinterman AE, Knol EF, Lencer DA, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121:737–743. e710. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 17.Santos AF, Du Toit G, Douiri A, et al. Distinct parameters of the basophil activation test reflect the severity and threshold of allergic reactions to peanut. J Allergy Clin Immunol. 2015;135:179–186. doi: 10.1016/j.jaci.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113:776–782. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 19.Song Y, Wang J, Leung N, et al. Correlations between basophil activation, allergen-specific IgE with outcome and severity of oral food challenges. Ann Allergy Asthma Immunol. 2015;114:319–326. doi: 10.1016/j.anai.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Bock SA, Sampson HA, Atkins FM, et al. Double-blind, placebo-controlled food challenge (DBPCFC) as an office procedure: a manual. J Allergy Clin Immunol. 1988;82:986–997. doi: 10.1016/0091-6749(88)90135-2. [DOI] [PubMed] [Google Scholar]
- 21.Boyce JA, Assa’ad A, Burks AW, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol. 2010;126:S1. doi: 10.1016/j.jaci.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson HA, Gerth van Wijk R, Bindslev-Jensen C, et al. Standardizing double-blind, placebo-controlled oral food challenges: American Academy of Allergy, Asthma & Immunology-European Academy of Allergy and Clinical Immunology PRACTALL consensus report. J Allergy Clin Immunol. 2012;130:1260–1274. doi: 10.1016/j.jaci.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Lieberman P, Nicklas RA, Oppenheimer J, et al. The diagnosis and management of anaphylaxis practice parameter: 2010 update. J Allergy Clin Immunol. 2010;126:477–480. e471–442. doi: 10.1016/j.jaci.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 24.Nowak-Wegrzyn A, Assa’ad AH, Bahna SL, Bock SA, Sicherer SH, Teuber SS. Work Group report: oral food challenge testing. J Allergy Clin Immunol. 2009;123:S365–383. doi: 10.1016/j.jaci.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 25.Mukai K, Gaudenzio N, Gupta S, et al. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 hours before analysis. J Allergy Clin Immunol. 2017;139:889–899. e811. doi: 10.1016/j.jaci.2016.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos AF, Lack G. Basophil activation test: food challenge in a test tube or specialist research tool? Clin Transl Allergy. 2016;6:10. doi: 10.1186/s13601-016-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: Rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychological Methods. 2009;14:323–348. doi: 10.1037/a0016973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strobl C, Boulesteix AL, Kneib T, Augustin T, Zeileis A. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: Rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychological Methods. 2009;14:323–348. doi: 10.1037/a0016973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breiman L, Friedman J, Olshen R, CJS . Classification and Regression Trees. Belmont, CA: Wadsworth; 1984. [Google Scholar]
- 31.Tibshirani R. Regression Shrinkage and Selection via the Lasso. JR Statist Soc B. 1996;58:267–288. [Google Scholar]
- 32.Varshney P, Jones SM, Scurlock AM, et al. A randomized controlled study of peanut oral immunotherapy: clinical desensitization and modulation of the allergic response. J Allergy Clin Immunol. 2011;127:654–660. doi: 10.1016/j.jaci.2010.12.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bird JA, Spergel JM, Jones SM, et al. Efficacy and Safety of AR101 in Oral Immunotherapy for Peanut Allergy: Results of ARC001, a Randomized, Double-Blind, Placebo-Controlled Phase 2 Clinical Trial. J Allergy Clin Immunol Pract. 2017 doi: 10.1016/j.jaip.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Raton M, Rodríguez-Álvarez MX, Suárez CC, Sampedro FG. OptimalCutpoints: An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. J Stat Softw. 2014;61:1–36. [Google Scholar]
- 35.Kohavi R. [Accessed Aug 12, 2017];A study of cross-validation and bootstrap for accuracy estimation and model selection. 1995 https://pdfs.semanticscholar.org/0be0/d781305750b37acb35fa187febd8db67bfcc.pdf.
- 36.Hothorn T, Hornik K, Zeileis A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J Comp Graph Stat. 2006;15:651–674. [Google Scholar]
- 37.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2015. [Accessed Dec 3, 2017]. https://www.r-project.org/ [Google Scholar]
- 38.Sporik R, Hill D, Hosking C. Specificity of allergen skin testing in predicting positive open food challenges to milk, egg and peanut in children. Clin Exp Allergy. 2000;30:1541–1546. doi: 10.1046/j.1365-2222.2000.00928.x. [DOI] [PubMed] [Google Scholar]
- 39.Lack G. Clinical practice. Food allergy. N Engl J Med. 2008;359:1252–1260. doi: 10.1056/NEJMcp0800871. [DOI] [PubMed] [Google Scholar]
- 40.Beyer K, Grabenhenrich L, Hartl M, et al. Predictive values of component-specific IgE for the outcome of peanut and hazelnut food challenges in children. Allergy. 2015;70:90–98. doi: 10.1111/all.12530. [DOI] [PubMed] [Google Scholar]
- 41.Codreanu F, Collignon O, Roitel O, et al. A novel immunoassay using recombinant allergens simplifies peanut allergy diagnosis. International archives of allergy and immunology. 2010;154:216–226. doi: 10.1159/000321108. [DOI] [PubMed] [Google Scholar]
- 42.Dang TD, Tang M, Choo S, et al. Increasing the accuracy of peanut allergy diagnosis by using Ara h 2. Journal of Allergy and Clinical Immunology. 2012;129:1056–1063. doi: 10.1016/j.jaci.2012.01.056. [DOI] [PubMed] [Google Scholar]
- 43.Lieberman JA, Glaumann S, Batelson S, Borres MP, Sampson HA, Nilsson C. The utility of peanut components in the diagnosis of IgE-mediated peanut allergy among distinct populations. J Allergy Clin Immunol Pract. 2013;1:75–82. doi: 10.1016/j.jaip.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Nicolaou N, Poorafshar M, Murray C, et al. Allergy or tolerance in children sensitized to peanut: prevalence and differentiation using component-resolved diagnostics. J Allergy Clin Immunol. 2010;125:191–197. e191–113. doi: 10.1016/j.jaci.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Klemans RJ, Broekman HC, Knol EF, et al. Ara h 2 is the best predictor for peanut allergy in adults. J Allergy Clin Immunol Pract. 2013;1:632–638. e631. doi: 10.1016/j.jaip.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Klemans RJ, Otte D, Knol M, et al. The diagnostic value of specific IgE to Ara h 2 to predict peanut allergy in children is comparable to a validated and updated diagnostic prediction model. Journal of Allergy and Clinical Immunology. 2013;131:157–163. doi: 10.1016/j.jaci.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Ocmant A, Mulier S, Hanssens L, et al. Basophil activation tests for the diagnosis of food allergy in children. Clin Exp Allergy. 2009;39:1234–1245. doi: 10.1111/j.1365-2222.2009.03292.x. [DOI] [PubMed] [Google Scholar]
- 48.Blumchen K, Beder A, Beschorner J, et al. Modified oral food challenge used with sensitization biomarkers provides more real-life clinical thresholds for peanut allergy. J Allergy Clin Immunol. 2014;134:390–398. e394. doi: 10.1016/j.jaci.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Virkud YV, Burks AW, Steele PH, et al. Novel baseline predictors of adverse events during oral immunotherapy in children with peanut allergy. J Allergy Clin Immunol. 2017;139:882–888. e885. doi: 10.1016/j.jaci.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.US Department of Health and Human Services. [Accessed Dec 6, 2017];Common Terminology Criteria for Adverse Events (CTCAE), v4.03. 2010 https://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03_2010-06-14_QuickReference_8.5x11.pdf.
- 51.Yanagida N, Okada Y, Yukiko H, et al. Food allergy and anaphylaxis – 2054. Easy-to-use severity grading system for treatment of symptoms induced by oral food challenge. The World Allergy Organization Journal. 2013;6:P137–P137. [Google Scholar]
- 52.Bock SA, Muñoz-Furlong A, Sampson HA. Fatalities due to anaphylactic reactions to foods. J Allergy Clin Immunol. 2001;107:191–193. doi: 10.1067/mai.2001.112031. [DOI] [PubMed] [Google Scholar]
- 53.Bock SA, Muñoz-Furlong A, Sampson HA. Further fatalities caused by anaphylactic reactions to food, 2001–2006. J Allergy Clin Immunol. 2007;119:1016–1018. doi: 10.1016/j.jaci.2006.12.622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.