Abstract
The ability to switch between yeast and hyphal morphologies is an important virulence factor for the opportunistic pathogen Candida albicans. Although the kinetics of appearance of the filamentous ring that forms at the incipient septum differ in yeast and cells forming hyphae (germ tubes) (Soll and Mitchell, 1983), the molecular mechanisms that regulate this difference are not known. Int1p, a C. albicans gene product with similarity in its C terminus to Saccharomyces cerevisiae Bud4p, has a role in hyphal morphogenesis. Here we report that in S. cerevisiae, Int1p expression results in the growth of highly polarized cells with delocalized chitin and defects in cytokinesis and bud-site selection patterns, phenotypes that are also seen in S. cerevisiae septin mutant strains. Expression of high levels of Int1p in S. cerevisiae generated elaborate spiral-like structures at the periphery of the polarized cells that contained septins and Int1p. In addition, Int1p coimmunoprecipitated with the Cdc11p and Cdc12p septins, and Cdc12p is required for the establishment and maintenance of these Int1p/septin spirals. Although Swe1p kinase contributes to INT1-induced filamentous growth in S. cerevisiae, it is not required for the formation of ectopic Int1p/septin structures. In C. albicans, Int1p was important for the axial budding pattern and colocalized with Cdc3p septin in a ring at the mother-bud neck of yeast and pseudohyphal cells. Under conditions that induce hyphae, both Cdc3p and Int1p localized to a ring distal to the junction of the mother cell and germ tube. Thus, placement of the Int1p/septin ring with respect to the mother–daughter cell junction distinguishes yeast/pseudohyphal growth from hyphal growth in C. albicans.
INTRODUCTION
Candida albicans is a multimorphic opportunistic fungal pathogen of humans. The ability to change morphology between ovoid yeast forms and several filamentous forms (germ tubes, pseudohyphae, and true hyphae) contributes to C. albicans virulence (Odds, 1988, 1994). Switching between growth forms is influenced by many factors including temperature, pH, carbon source, nitrogen source, and cell concentration (Odds, 1988). Analysis of the molecular mechanism(s) of morphogenesis has been difficult in C. albicans because it is asexual, and thus not amenable to genetic analysis, and because its codon usage is nonstandard, interfering with heterologous gene expression (De Backer et al., 2000).
INT1 encodes a protein (Int1p) that functions in morphogenesis. C. albicans int1/int1 strains have a reduced ability to form hyphae on Milk-Tween and Spider media but form apparently normal hyphae in the presence of serum (Gale et al., 1998). In addition, int1/int1 strains have attenuated virulence in a mouse model of systemic candidiasis (Gale et al., 1998). When INT1 is expressed in S. cerevisiae, it causes a morphological switch to highly polarized filamentous cells that resemble C. albicans germ tubes and hyphae (Gale et al., 1996). In S. cerevisiae, filamentous growth (termed “pseudohyphal growth” and most similar in morphology to C. albicans pseudohyphae) occurs in some diploid strains under nitrogen deprivation conditions (Gimeno and Fink, 1994) and in some haploid strains in rich medium (Roberts and Fink, 1994). Several C. albicans homologues of genes required for S. cerevisiae pseudohyphal growth (e.g., STE20 and STE12) are also required for filamentous growth in C. albicans (Liu et al., 1994; Kohler and Fink, 1996; Leberer et al., 1996; Lo et al., 1997); however, unlike pseudohyphal growth, INT1-induced filamentous growth (I-IFG) of S. cerevisiae is independent of STE20 and STE12 (Gale et al., 1996, 1998) and occurs in all haploid and diploid strains grown in rich or minimal medium. Because Int1p alters S. cerevisiae morphology, we hypothesized that it interacts with proteins conserved between the two fungi, so that studies of Int1p function in S. cerevisiae would provide insights into its function in C. albicans.
In S. cerevisiae, morphogenesis depends on the actin cytoskeleton, bud-site selection proteins, the cell cycle machinery, and cytokinetic structures at the bud neck. For example, cytokinesis requires septin proteins, components of a filament ring that localizes to the cytoplasmic face of the plasma membrane in the mother-bud neck (Longtine et al., 1996) and functions as a scaffold (Field and Kellogg, 1999) for proteins involved in bud-site selection (Chant et al., 1995; Sanders and Herskowitz, 1996), chitin deposition (DeMarini et al., 1997), cell cycle regulation (Barral et al., 1999; Longtine et al., 2000), and cytokinesis (Bi et al., 1998; Lippincott and Li, 1998a,b). Loss of any one of four septins (Cdc3, Cdc10, Cdc11, or Cdc12) results in delocalization of the septins and associated proteins from the mother-bud neck and in the formation of elongated buds that cannot complete cytokinesis (reviewed in Longtine et al., 1996).
Many of the S. cerevisiae proteins involved in morphogenesis are also found in C. albicans. For example, the C. albicans genome sequence includes sequences with similarity to CDC3, CDC10, CDC11, and CDC12, and the C. albicans homologues of CDC3 and CDC10 complement the morphogenesis defects of S. cerevisiae cdc3 and cdc10 strains (DiDomenico et al., 1994). In classic studies of C. albicans morphogenesis, the site of yeast cell septation was found to be characterized by a filament ring that appears at the mother-bud neck at the time of bud emergence (Soll and Mitchell, 1983). In contrast, a filament ring (detected by electron microscopy and Calcofluor staining) appears in hyphae ∼30 min after bud emergence and distant from the mother-bud junction. These studies suggested that the localization of septin proteins may be different in yeast and hyphal cells.
In a BLAST search, C. albicans Int1p is most similar to S. cerevisiae Bud4p and is the only predicted protein with significant similarity to Bud4p in the Candida genome sequence (http://sequence-www.stanford.edu/group/candida/search.html). Although the two proteins are only 24% identical over their entire sequence, they are 35% identical and 45% similar in the C-terminal ∼375 amino acids. In S. cerevisiae, Bud4p is found with the septin ring at the mother-bud neck and is required for the haploid-specific axial budding pattern, but it is not required for septin ring localization (Sanders and Herskowitz, 1996). Specific factors required for bud site selection in C. albicans, an asexual diploid, have not been studied. C. albicans cells exhibit a predominantly bipolar budding pattern at temperatures >30°C and an increasing predominance of the axial budding pattern at temperatures <30°C (Chaffin, 1983; Herrero et al., 1999).
In this study, we analyzed the effects of Int1p expression in S. cerevisiae to identify proteins that interact with Int1p to effect changes in bud morphology. We then used these insights to develop (and test) hypotheses regarding Int1p localization and interactions in C. albicans.
MATERIALS AND METHODS
Strains, Growth Conditions, and Plasmids
S. cerevisiae and C. albicans strains used in this study are described in Table 1. Yeast media (rich medium [YPAD], synthetic complete medium, and synthetic minimal medium lacking specific nutrients) have been described previously (Sherman, 1991). S. cerevisiae was grown at 30°C except where noted. C. albicans strains were grown in YPAD at 30°C to promote yeast form growth and at 37°C in YPAD containing 20% serum to promote hyphal growth. In Figure 1B, 10 μg/ml α-factor mating pheromone (Sigma, St. Louis, MO) was added to YJB5763 cells growing exponentially in YPAD + 2% glucose and incubation was continued until >90% of cells were forming mating projections (∼3 h).
Table 1.
Yeast strains constructed for this study
| Name | Genotype |
|---|---|
| S. cerevisiae | |
| M-652a | a/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 GIN4-GFP:kan/GIN4-GFP:kan |
| M-912a | a/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 HSL1-GFP:kan/HSL1-GFP:kan |
| M-768b | a his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-5 SEP7-GFP:kan |
| M-780b | a his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-5 MYO1-GFP:kan |
| YJB1843c | a/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1+ [pGAL1-INT1-URA3] |
| YJB1844c | a/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1+ [pBM272] |
| YJB1875d | a ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1+ [pGAL1-INT1-URA3] |
| YJB1876d | a ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1+ [pBM272] |
| YJB2681 | as M-768 except + [pGAL1-INT1-URA3] |
| YJB3362e | a leu2-Δ1 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 gal2+ [pGAL1-INT1-GFP-URA3-TRP1] |
| YJB3363e | a leu2-Δ1 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 gal2+ [pGAL1-INT1-HA-HIS3-URA3] |
| YJB3527f | a leu2-Δ1 ura3-52 can1 ade2 his3-Δ200 trp1-Δ63 ssd1 ho cla4∷URA3 Δclb1 clb3∷TRP1 clb4∷HIS3 bar1+ [p1227] |
| YJB3542g | a ade2-10 cry1 his4 leu2-Δ1 trp1-Δ63 ura3-52 SUP4-3ts cdc12-6ts+ [pMM19] + [pGAL1-INT1-URA3] |
| YJB3958e | a leu2-Δ1 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 gal2+ [pGAL1-INT1-3HA-HIS3-URA3] + [p1227] |
| YJB4840g | a ade2-10 cry1 his4 leu2-Δ1 trp1-Δ63 ura3-52 SUP4-3 cdc12-6+ [pGAL1-INT1-GFP-URA3-TRP1] |
| YJB5001e | a leu2-Δ1 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 gal2+ [p1227] + [pGAL1-INT1-URA3] |
| YJB5170h | a his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-5 MYO1-GFP:kan+ [pGAL1-INT1-URA3] |
| YJB5172 | as M-652 except + [pGAL1-INT1-URA3] |
| YJB5174 | as M-912 except + [pGAL1-INT1-URA3] |
| YJB5505j | a/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 CDC12/CDC12-YFP:TRP1+ [pGAL1-INT1-CFP-URA3-TRP1] |
| YJB5763k | a his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-5 2+ [pGAL1-INT1-URA3] |
| YJB5765e | a leu2-Δ1 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 gal2+ [pGAL1-INT1-URA3] |
| YJB5766e | a leu2-Δ1 trp1-Δ63 ura3-52 prb1-1122 pep4-3 prc1-407 gal2+ [pMM19] + [pGAL1-INT1-URA3] |
| YJB6003i | a bar1 leu2-Δ1 ura3-52 trp1-Δ63 his2 ade1 swe1∷LEU2+ [pCDC3-GFP-URA3] + [pGAL1-INT1-TRP1] |
| C. albicans | |
| CAF2l | URA3/ura3∷imm434 |
| CAG3m | ura3∷imm434/ura3∷imm434 int1∷hisG-URA3-hisG/int1∷hisG |
| CAG5m | ura3∷imm434/ura3∷imm434 int1∷hisG/int1∷INT1:URA3 |
| YJB5850n | ura3∷imm434/ura3∷imm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG CDC3/CDC3-CFP:HIS1 INT1/INT1-YFP:URA3 |
Provided by John Pringle (University of North Carolina, Chapel Hill, NC).
Derived from YPH499 (Sikorski and Hieter, 1989).
Derived from YPH501 (Sikorski and Hieter, 1989).
Derived from YJB365 (Berman strain collection).
Derived from HT9 (Tjandra et al., 1998).
Derived from JKY81-5-1 (provided by James Konopka (SUNY, Stonybrook, NY).
Derived from M-780 (Bi et al., 1998).
Derived from DLY1028 (McMillan et al., 1998).
Derived from YEF473 (Bi and Pringle, 1996).
Derived from YEF473A (Bi and Pringle, 1996).
Derived from BWP17 (Wilson et al., 1999).
Figure 1.
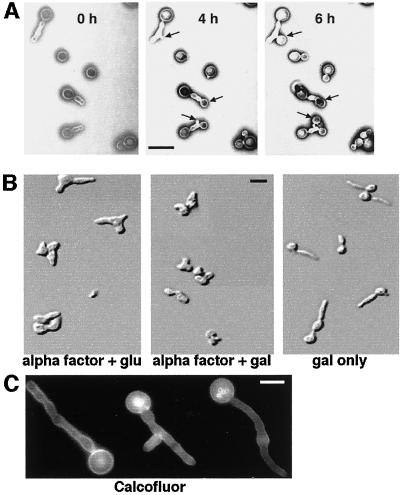
Characterization of INT1-filaments in S. cerevisiae. (A) The filaments induced by INT1 expression are not in a terminal physiological state. YJB5763 was grown in galactose to induce INT1-filaments and then immobilized onto agar containing glucose to repress GAL1-INT1. Images were then obtained at the times indicated. Sites of new bud growth are indicated by arrows. Bar, 10 μm. (B) INT1-filaments do not form in α-factor–arrested cells. DIC micrographs of YJB5763 were arrested with α-factor as described in MATERIALS AND METHODS, washed once with sterile water, and resuspended in medium containing α-factor with glucose (glu) or galactose (gal), or galactose without α-factor, as indicated. Bar, 10 μm. (C) Chitin localization is diffuse in INT1-induced filaments. Shown is fluorescence micrograph of strain YJB5763 expressing INT1 and stained with Calcofluor white. Bar, 5 μm.
Plasmid pGAL1-INT1-URA3 (pCG01 [Gale et al., 1996]) drives expression of INT1 from the GAL1 promoter from vector pBM272 (Johnston and Davis, 1984). To achieve high-level expression of proteins from the GAL1 promoter, strains pregrown in glucose-containing medium were subcultured into medium containing raffinose and galactose. p1227 (CDC12-GFP-LEU2) was provided by J. Konopka (State University of New York, Stony Brook, NY). pML726 (CDC3-GFP-URA3) was provided by J. Pringle (University of North Carolina, Chapel Hill, NC). pMM19 (CDC3-GFP-LEU2) was constructed from pML726, and pGAL1-INT1-TRP1 was constructed from pCG01 (Gale et al., 1996) by marker swapping URA3 to LEU2 (for pMM19) or URA3 to TRP1 (for pCG01) with the use of pUL7 and pUT11, respectively (Cross, 1997).
Morphological Observations
Chitin/bud scars were stained by adding 100 μg/ml Calcofluor white (Sigma) to the growth medium for 15–30 min. Bud scar patterns were scored for cells with 3–5 bud scars and were considered “axial” if all bud scars were in a single chain, “bipolar” if at least two bud scars were at opposite ends of a cell, and “random” if the pattern was neither bipolar nor axial. The scorer was blinded as to the identity of the strains being analyzed. The χ2 test of goodness to fit (Snedecor and Chochran, 1980) was performed by taking the distribution of wild-type into three classes as the null model and testing each strain against this model. Similarly, pairs of mutant strains were tested against one another. Samples were considered significantly different at the P < 0.005 level. Nuclear DNA was detected with DAPI (Sigma).
Differential interference contrast (DIC) and epifluorescence microscopy were performed with the use of a Nikon Eclipse E800 photomicroscope equipped with standard UV and FITC filter sets. Cyan fluorescent protein (CFP) (excitation filter 380–400 nm, barrier 435–485 nm) and yellow fluorescent protein (YFP) (excitation filter 490–510 nm, barrier 520–550 nm) filter sets were obtained from Chroma Technology Corporation (Brattleboro, VT). Digital images were collected with the use of a CoolCam liquid-cooled, three-chip color CCD camera (Cool Camera Company, Decatur, GA), captured to a Pentium II 300 MHz computer with the use of Image Pro Plus version 4.1 software (Media Cybernetics, Silver Spring, MD), and processed with the use of Adobe Photoshop. Images for YFP and CFP were converted to red and green, respectively. In merged images, overlapping YFP and CFP signals appear yellow or orange.
Indirect immunofluorescence was performed as described previously (Pringle et al., 1991), with the use of mouse monoclonal anti-hemagglutinin (HA) antibody (1:250) (Berkeley Antibody Co., Richmond, CA) and rabbit polyclonal anti-Cdc3p antibody (1:10) (Kim et al., 1991). Secondary antibodies (goat anti-mouse IgG tagged with CY3 and goat anti-rabbit IgG tagged with CY2; both from Jackson ImmunoResearch, West Grove, PA) were used at 1:300 dilution.
Construction of Fluorescent Protein and HA Fusions
Protein tags were introduced by PCR-mediated gene modifications (Wach et al., 1994, 1997; Longtine et al., 1998b; Gerami-Nejad et al., 2001) with the use of synthetic oligonucleotides (IDT, Coralville, IA). PCR products were used to transform yeast strains directly, and transformants were screened by PCR for correct insertion of the tag. Plasmids and genomic sequences were isolated (Hoffman and Winston, 1987) and sequenced (University of Minnesota Microchemical Facility) to verify that fusions were in-frame.
To tag the 3′-end of sequences in S. cerevisiae, we used pFA6a-GFP(S65T)-TRP1 (Longtine et al., 1998b), pDH3 (Yeast Resource Center, University of Washington, Seattle, WA), or pFA6a-3HA-TRP1 (Longtine et al., 1998b) as template with primers A Forward and A Reverse (Table 2) to generate INT1-GFP, INT1-CFP, and INT1-HA transformation cassettes for integration into the plasmid contained in strain YJB5765. pYFP-URA3 (Gerami-Nejad et al., 2001) was used as the template with primers C Forward and C reverse (Table 2) to generate a CDC12-YFP transformation cassette for YEF473.
Table 2.
Oligonucleotide primer sequences used in this study
| Name | Sequence (5′ → 3′) |
|---|---|
| A Forward | ACAACAACAACAACAACAACAACAACAAAGCTCCCAACAGCGGATCCCCGGGTTAATTAA |
| A Reverse | GCATATATTTGCCAATTAAAATTAAAAAAATCAAAAGTAGGAATTCGAGCTCGTTTAAAC |
| C Forward | AAAACTAGAAGAGCAGGTCAAAAGCTTGCAAGTAAAAAAATCCCATTTAAAAGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| C Reverse | GAAGAGACAAAGAGATAGGCGTTGAAATTCACGAGACAAAGAGGAAGACATTAACGACGGCCAGTGGAATTC |
| D Forward | ACAAAAATTATTACCACAAGACCCACCAGCACAACCAGCTCCACAAAAGAGTCGTAAAGGATTTTTACGTGGTGGTGGTTCTAAAGGTGAAGAATTATT |
| D Reverse | AATTAAACAAACAGATTAACAAACAAATAAACTAAATTAAGTTACATACTATTTAGCTATACCTCGGCCCTCTAGA-AGGACCACCTTTGATTG |
| E Forward | ACGTCAACCATGGGTAAATTTGATGCTTCAACAACAACAACAACAACAACAACAACAAAGCTCCCAACAGGGTG-GTGGTTCTAAAGGTGAAGAATTATT |
| E Reverse | AAAACGCTATTAGACTAAAAGATAATACAAAATGGGCATATATTTGCCAATTAAAATTAAAAAAATCAAAAGTAG-GAATTCCGGAATATTTATGAGAAAC |
To tag sequences in C. albicans, we used pCFP-HIS1 and pYFP-URA3 (Gerami-Nejad et al., 2001) as templates with primers D Forward and D Reverse and E Forward and E Reverse (Table 2) to generate CDC3-CFP and INT1-YFP transformation cassettes for use with strain BWP17.
Immunoprecipitation
Cells expressing INT1 for 8 h were lysed by the method of Frazier et al. (1998), except that cell debris was removed by three 10 min centrifugations at 12,000 × g followed by three 5 min centrifugations at 12,000 × g. Lysates were precleared by mixing for 1 h at 4°C with a combination of protein A- and protein G-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Each lysate was split into four aliquots to which were added rabbit anti-Rap1 antiserum (Enomoto et al., 1997), rabbit anti-Cdc11 IgG (Santa Cruz Biotechnology), mouse anti-HA IgG (12CA5) (Roche Molecular Biochemicals, Indianapolis, IN), or mouse anti-green fluorescent protein (GFP) IgG (Roche). After mixing for 1.5 h at 4°C, Protein A-Agarose beads or Protein G-Agarose beads were added and incubated for 1 h at 4°C. The beads were then washed by centrifugation four times with cold lysis buffer, and protein was eluted by boiling in reducing protein electrophoresis buffer (Laemmli, 1970) for 5 min. Samples were separated on a 7.5% acrylamide gel (Laemmli, 1970), blotted to polyvinylidene difluoride membrane (Millipore, Bedford, MA), and detected with peroxidase-conjugated anti-HA antibody (12CA5) (Roche) and “Supersignal” chemiluminescent substrate (Pierce Chemical Company, Rockford, IL).
RESULTS
INT1-induced Filaments Are Highly Polarized Buds with Defects in Cytokinesis
INT1 expression in S. cerevisiae causes the formation of a high proportion (>50–90%, depending on the strain background) of filamentous cells (INT1-filaments) (Gale et al., 1996). If INT1 expression is subsequently repressed, round buds emerge from the INT1-filaments (Figure 1A), indicating that the elongated cells were not in a terminal physiological state. To determine whether I-IFG is due to a cell cycle block that inhibits nuclear division, we analyzed the number of nuclei present in INT1-filaments. We observed an average of three nuclei per filament (range one to eight nuclei per filament) after ∼18 h of growth in galactose. Cells not expressing INT1 and growing in galactose-containing medium would have gone through approximately eight cell divisions. To determine whether I-IFG is restricted to a particular stage of the cell cycle, INT1 was expressed in MATa yeast cells arrested in G1 by treatment with α-factor. These cells did not produce filaments (Figure 1B, center panel). In contrast, cells expressing INT1 formed filaments in the absence of α-factor (Figure 1B, right panel). Thus, INT1-filaments, like normal buds, do not emerge until the cell cycle has traversed START. In addition, I-IFG is dependent on the ability to polarize the cytoskeleton: a cdc24–4ts strain did not form INT1-induced filaments at the restrictive temperature. Thus, INT1-filaments appear to be elongated buds that can progress through the nuclear cell cycle, albeit with an apparent delay.
To determine whether INT1-filaments contain septa, chitin was stained with Calcofluor. No distinct septa or chitin rings were observed even in very elongated INT1-filaments (Figure 1C). This diffuse localization of chitin appeared similar to that in S. cerevisiae strains with mutations in the septins or proteins involved in chitin deposition that localize to the mother-bud neck (DeMarini et al., 1997). The absence of septa and the presence of multiple nuclei suggest that INT1-expressing cells are defective in cytokinesis. To distinguish between a defect in cytokinesis and a defect in cell separation, cells expressing INT1 were fixed and treated after fixation with lyticase, which digests yeast cell walls and separates cells that have undergone cytokinesis (Hartwell, 1971; Pringle and Mor, 1975). We compared INT1-filaments with the aggregates of cells formed in a myo1 strain that is defective in cell separation but not in cytokinesis (Bi et al., 1998). In contrast to myo1 strains, which separated into individual cells, >95% of the INT1-filaments were not affected by lyticase treatment.
Unusual Septin Structures in Cells Expressing INT1
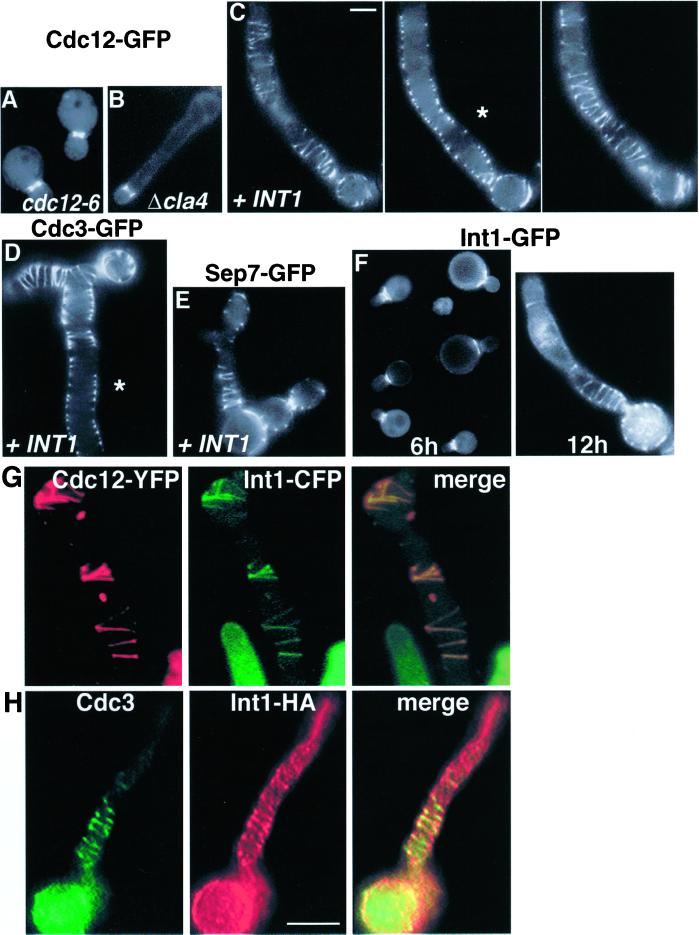
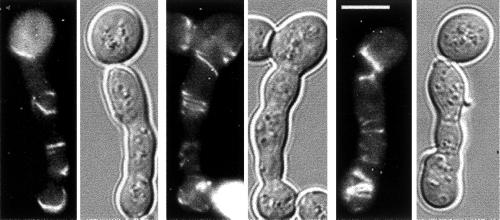
Septin localization to the mother-bud neck (Figure 2A) is disrupted in septin mutants. Because cells expressing INT1 resemble septin mutants, we analyzed septin localization in INT1-filaments with the use of functional GFP-tagged septins. All proteins that were examined localized as multiple rings and elaborate spiral-like structures that often appeared to be connected (Figure 2, C–E) and, like the normal septin ring, were localized near the cell periphery (Figure 2, C and D, asterisks). Similar structures were observed by indirect immunofluorescence with the use of septin-specific antibodies (Figure 2H). Thus, with the use of either fusion proteins or native proteins, we detect elaborate septin structures at the periphery of the polarized buds induced by INT1 expression. These structures were sharply distinct from the septin structures observed in other types of elongated S. cerevisiae cells such as cla4Δ cells and cells expressing high levels of SWE1 (Sia et al., 1998; Tjandra et al., 1998; Longtine et al., 2000) (Figure 2B). In addition, overexpression of Bud4p (the closest S. cerevisiae homologue of Int1p; see INTRODUCTION) only rarely (<1% of cells) induced elongated buds, and these had normal-looking septin rings at the mother-bud neck (our unpublished results).
Figure 2.
Septins and Int1p localize as spirals in INT1-filaments. Examples of images taken in the equatorial plane of a cell are noted by an asterisk (C and D). (A) YJB5001 grown in glucose and expressing Cdc12-GFP. (B) YJB3527 (cla4Δ) expressing Cdc12-GFP. (C) YJB5001 grown on galactose and thus expressing Cdc12-GFP and Int1p. Serial optical sections of one cell are shown. Bar (shown in C), 5 μm, is representative for A–F. (D) YJB5766 expressing Cdc3-GFP and Int1p. (E) YJB2681 expressing Sep7-GFP and Int1p. (F) YJB3362 expressing Int1-GFP for 6 or 12 h of growth on galactose, as indicated. (G) YJB5505 expressing both Cdc12-YFP and Int1-CFP after 12 h of INT1 induction. (H) Indirect immunofluorescence micrographs of YJB3363, after 12 h of INT1-HA induction, with the use of anti-HA and anti-Cdc3p antibodies as indicated. Bar (shown in H), 5 μm, is representative of G–H.
Colocalization and Interaction of Int1p with Septins in S. cerevisiae
Induction of an Int1-GFP or an Int1p-HA fusion protein also produced many (∼85% of the cells) INT1-filaments. Int1-GFP was first visible ∼6 h after induction, when it localized primarily to the mother-bud neck (Figure 2F, left panel). After 6–8 h, elongated buds became apparent, and the majority of the Int1-GFP localized to the junction between the mother cell and the elongating bud. At later times, Int1-GFP appeared as rings and spiral-like structures similar to the septin structures in INT1-filaments (Figure 2F, right panel). Indeed, double-label experiments with the use of Int1-CFP and Cdc12-YFP (Figure 2G) or immunofluorescence on Int1p-HA–expressing cells (Figure 2H) revealed a high degree of colocalization of Int1p and the septins in the rings and spirals.
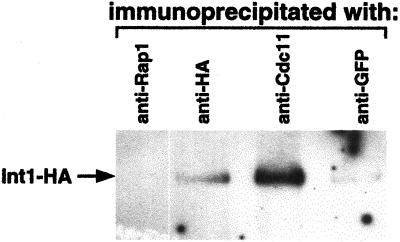
This colocalization suggested that Int1p may physically interact with one or more septin proteins. To test this, we immunoprecipitated septins and associated proteins from INT1-filaments with the use of antibodies directed against native Cdc11p or against GFP (for Cdc12-GFP). Int1p coprecipitated in both cases (although more effectively with Cdc11p), but not in a control experiment with the use of antibodies to Rap1p, an abundant nuclear protein (Figure 3). Although Int1p may have a higher affinity for Cdc11p than for Cdc12p, this result may also reflect different affinities of the anti-Cdc11p and anti-GFP antibodies used. Thus, Int1p and the septins appear to interact specifically in the “Int1p/septin spirals.”
Figure 3.
Int1p immunoprecipitates with Cdc11p and Cdc12p septins. Immunoblot of cell lysates from YJB3958 expressing both Int1-HA and Cdc12-GFP. Antibodies to Rap1p, HA, Cdc11p, or GFP were used to precipitate protein complexes as indicated. The precipitates were separated by SDS-PAGE, and Int1-HA was detected with anti-HA antibody.
To determine whether septin function is required for the establishment and/or maintenance of the Int1p/septin spirals, we used a cdc12–6 strain that forms normal septin rings at 23°C but delocalizes all septins after a brief incubation at 37°C (Kim et al., 1991; Longtine et al., 1996; Barral et al., 2000). In one experiment, cells were pregrown at 37°C for 1 h before inducing INT1 expression. Under these conditions, no bud neck or spiral localization of Int1-GFP or Cdc3-GFP was observed. Rather, both proteins appeared diffuse or formed ectopic aggregates (Figure 4A). In contrast, wild-type cells formed and maintained spirals at 37°C. In a second experiment, INT1 was induced for 12 h at 23°C so that elongated buds and spirals were evident, and then the cells were shifted to 37°C. The Cdc3-GFP structures disappeared, and the Int1-GFP structures appeared to become fragmented (Figure 4B). Thus, both the establishment and maintenance of intact spirals required functional Cdc12p. The fragmented appearance of Int1-GFP in cdc12–6 cells suggests that existing spirals remained partially intact in the absence of Cdc12p, perhaps because some of the Int1p within the spirals is stabilized by Int1p–Int1p interactions rather than by interactions only with the septins.
Figure 4.
Cdc12p septin is required for the establishment and maintenance of spirals. Bar, 10 μm. Fluorescence micrographs of cdc12–6 strains YJB4840 and YJB3542 expressing Int1-GFP (left) or Cdc3-GFP and Int1p (right). (A) Strains were pregrown at 37°C for 1 h to inactivate Cdc12–6p, subcultured to galactose-containing medium at 37°C to induce INT1 expression, and photographed after 12 h of growth. (B) Strains were pregrown in galactose at 23°C to induce INT1 and generate spirals, shifted to 37°C to inactivate Cdc12–6p, and photographed after 12 h at 37°C. Bar, 5 μm.
Even low levels of INT1 expression (that generated <5% filamentous cells) could apparently produce subtle disorganization of the septins. We observed that such expression disrupted both axial and bipolar bud-site selection patterns (Table 3) as previously observed for other alterations of septin organization (Flescher et al., 1993; Chant et al., 1995).
Table 3.
Effect of INT1 expression on the bud site selection pattern of S. cerevisiae strains
| Strain | Ploidy | Plasmid | % Axial | % Bipolar | % Random |
|---|---|---|---|---|---|
| YJB1876 | Haploid | pBM272 | 84 | 14 | 2 |
| YJB1875 | Haploid | pGAL1-INT1 | 40 | 8 | 52 |
| YJB1844 | Diploid | pBM272 | 3 | 84 | 13 |
| YJB1843 | Diploid | pGAL1-INT1 | <1 | 5 | 94 |
The indicated strains were grown overnight to saturation in minimal medium without uracil, lacking glucose, but containing raffinose (2%). Growth in raffinose is sufficient to induce a small amount of INT1 expression from pGAL1-INT1. Cells were subcultured 1:10 into fresh medium containing galactose to fully induce INT1 expression, grown for 5 h, and stained with Calcofluor white to visualize bud scars. A total of 200 cells was scored for each strain.
Int1p/Septin Spirals Do Not Contain Other Bud-neck Associated Proteins
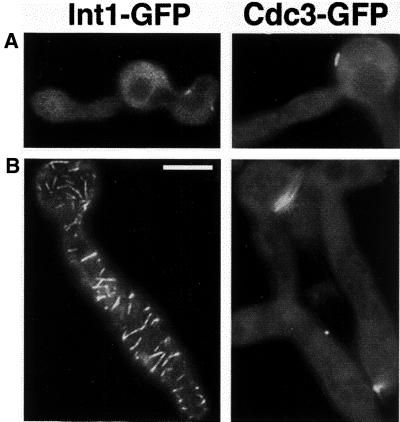
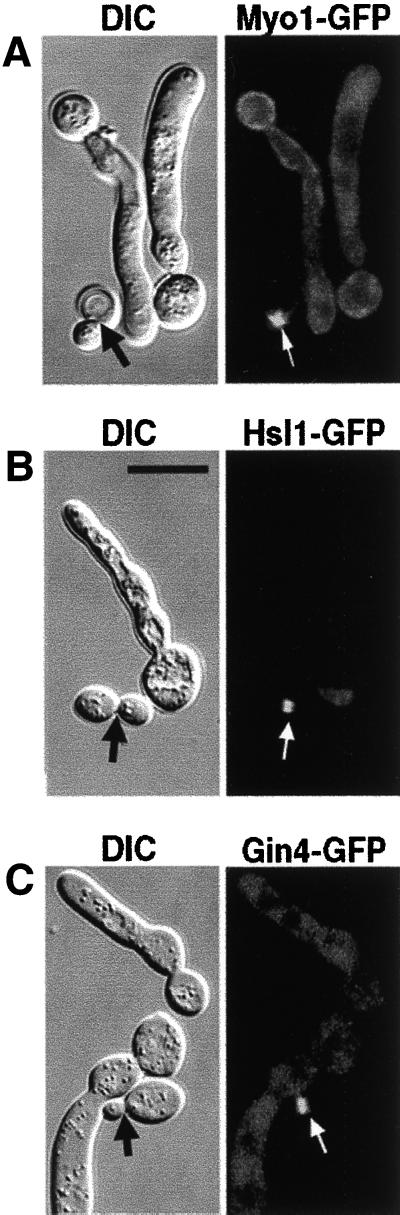
To determine whether expression of INT1 affects the localization of other bud-neck–associated proteins, we analyzed the localization of Myo1-GFP, Gin4-GFP, and Hsl1-GFP (Bi et al., 1998; Lippincott and Li, 1998b; Barral et al., 1999; Longtine et al., 2000). In INT1-filaments, no detectable signal was observed for Myo1-GFP, Gin4-GFP, or Hsl1-GFP either at the mother-bud neck or in spirals; instead, these proteins appeared diffuse and cytoplasmic (Figure 5). In the subset of INT1-expressing cells that had normal buds (presumably because they did not respond to Int1p), these proteins were at the neck (Figure 5, arrows). Thus, perturbation of septin organization by INT1 expression also disrupted the localization of Myo1p, Gin4p, and Hsl1p, consistent with previous reports that the localization of all three proteins requires normal septin structure (Bi et al., 1998; Longtine et al., 2000). Moreover, these results show that the Int1p/septin spirals do not contain all of the proteins normally associated with the septin ring.
Figure 5.
Expression of INT1 alters the localization of other bud-neck–associated proteins. DIC and fluorescence micrographs of strains expressing INT1 and Myo1-GFP (YJB5170) (A), Hsl1-GFP (YJB5174) (B), or Gin4-GFP (YJB5172) (C). Cells not forming polarized buds in response to INT1 expression (black arrows) have normal localization of the GFP-fusion proteins at the mother-bud neck (white arrows). Bar, 10 μm.
The Role of Swe1p in I-IFG and Int1p/Septin Spiral Formation
Because alterations of the septin ring can activate the Swe1p kinase and thus produce a cell-cycle delay that results in the formation of elongated buds (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000), we asked whether Swe1p is necessary for the effects seen during INT1 overexpression by inducing INT1 in swe1 and SWE1 strains expressing CDC12-GFP or INT1-GFP. In the swe1 strains, the proportion of cells forming polarized buds was significantly reduced but was not eliminated (<60% of the level in the SWE1 strain). Furthermore, in some of the swe1 cells with polarized buds, ectopic structures containing Int1-GFP and Cdc3-GFP were observed (Figure 6). In the swe1 strain, just as in wild-type strains, the spiral-like Int1p/septin structures are observed only in cells that form elongated buds. Thus, expression of INT1 generates polarized buds partly, but not entirely, through a Swe1-mediated delay of the cell cycle that presumably results from the alteration of septin structures; however, SWE1 does not appear to be required for the formation of Int1/septin spirals.
Figure 6.
The formation of Int1p spirals does not require SWE1. DIC and fluorescence micrographs of S. cerevisiae strain YJB6003 (swe1) expressing Cdc3-GFP and Int1p are shown. Bar, 5 μm.
Localization of Int1p in C. albicans
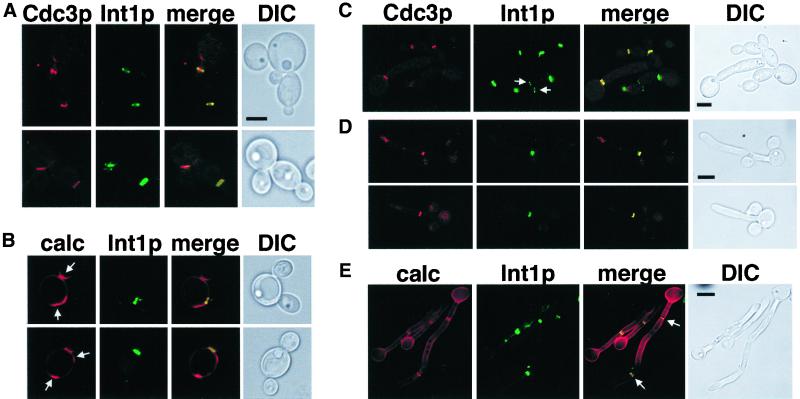
In C. albicans, INT1 only affects hyphal growth under a subset of hyphal induction conditions. To determine whether Int1p acts by interacting with the C. albicans septins, we first localized Int1p and septins in yeast, pseudohyphal, and hyphal cells by tagging one copy of INT1 with YFP and one copy of CDC3 with CFP. These cells formed yeast and hyphal cells at rates similar to those of the parental strains, indicating that the fluorescent protein tags did not interfere with septin or Int1p function. Both Int1-YFP and Cdc3-CFP localized to the mother-bud necks of small and large budded yeast cells (Figure 7A) and pseudohyphal cells (Figure 7C). Occasionally, we observed an Int1p signal without a corresponding Cdc3p signal on unbudded cells (Figure 7C, arrows). This may be caused by differing focal planes of localization or differing intensities of signals, or it may indicate that Int1p is present before Cdc3p at incipient bud sites. Additionally, Int1p colocalized with only one chitin-containing structure (Figure 7B; arrows indicate chitin signals without corresponding Int1p signals), suggesting that Int1p is present only at the necks of newly formed (or forming) buds.
Figure 7.
Colocalization of Int1p, Cdc3p septin, and septa in C. albicans. Bars (A–C), 5 μm; (D–E), 10 μm. YJB5850 grown at 30°C in minimal medium (A), 30°C in minimal medium with Calcofluor white (B), 37°C in YPAD to induce pseudohyphae (C), 37°C in YPAD, without shaking, to induce hyphae (D), or 37°C in YPAD, without shaking, and stained with Calcofluor white (E). Arrows, see description in RESULTS. The CFP and Calcofluor images were changed to red to facilitate merging.
In hyphal cells, Int1p localized to single or double rings that colocalized with Cdc3p and chitin (Figure 7, D and E) at a position distant from the junction of the mother cell and the germ tube. In general, the position of the first septin ring was observed 10–20 μm distant from the mother–daughter neck. In contrast to the solitary signal present in yeast cells, we observed Int1p and Cdc3p remaining at multiple septa within the growing hypha (Figure 7E, arrows, and unpublished observations). Thus, in C. albicans, septins and Int1p colocalize in a ring that becomes the site of septation and that probably corresponds to the filamentous ring observed by Soll and coworkers (Soll and Mitchell, 1983). Furthermore, the number and position of Int1p/septin ring(s) distinguish yeast/pseudohyphae from true hyphae.
Role of Int1p in C. albicans Bud Site Selection
Because Int1p localizes to septin rings and has similarity to S. cerevisiae Bud4p, we asked whether Int1p has a role in C. albicans bud site selection by comparing the bud site selection patterns of an int1/int1 strain (CAG3), an int1/int1::INT1 reintegrant strain (CAG5), and the parental strain (CAF2) at 28°C. CAF2 and CAG5 displayed approximately equal numbers of cells with axial and bipolar budding patterns. In contrast, the int1/int1 strain displayed a significant reduction in axial budding and an increase in bipolar budding (Table 4) when compared with both CAF2 and CAG5. Thus, like Bud4p, INT1 contributes to the axial bud-site selection pattern in budding C. albicans cells.
Table 4.
Effect of INT1 expression on the bud site selection pattern of C. albicans strains
| Strain (n) | Relevant genotype | % Axial | % Bipolar | % Random |
|---|---|---|---|---|
| CAF2 (490) | INT1/INT1 | 58 | 42 | 0 |
| CAG3 (531) | int1/int1 | 26 | 73 | 1 |
| CAG5 (317) | int1/int1∷INT1 | 47 | 51 | 2 |
Strains were grown overnight to saturation in rich (YPAD) medium at 28°C and stained with Calcofluor white to detect bud scars. χ2 analysis showed that CAF2, CAG3, and CAG5 were significantly different from each other at the p < 0.005 level.
DISCUSSION
Int1p and Septins in S. cerevisiae
C. albicans is an asexual diploid organism that forms budding yeast and pseudohyphal cells as well as elongated hyphal cells that generally remain attached to one another. Because the molecular mechanisms of morphogenesis are better understood in S. cerevisiae, and because Int1p expression in S. cerevisiae generated a highly filamentous phenotype, we used the analysis of Int1p in S. cerevisiae to guide our study of Int1p function in C. albicans. The effects of Int1p on yeast cell shape are almost certainly distinct from the pathways regulating pseudohyphal growth in S. cerevisiae or hyphal growth in C. albicans. Indeed, in S. cerevisiae, I-IFG occurs in the absence of genes required for pseudohyphal growth (Gale et al., 1996, 1998). Despite this, study of Int1p in S. cerevisiae provided us with insights into the potential role and function of Int1p in C. albicans, specifically, that Int1p interacts with the septin ring at the mother-bud neck. Although Int1p interacts with at least a subset of the septins, bud-neck proteins such as Myo1p, Gin4p, and Hsl1p do not associate with the Int1p/septin spirals. This implies that the spirals are ectopic septin structures rather than elaborate versions of essentially normal septin rings. Further study of the unusual spiral-like nature of the Int1p/septin structures in S. cerevisiae may provide additional information for models of septin organization.
Despite the similarity between Int1p and Bud4p, high levels of Bud4p did not change cell morphology or septin structure in the dramatic way that Int1p expression did. Lord et al. (2000) reported that very high levels of Bud3p induced a proportion (∼40%) of filamentous cells, some of which contained ectopic septin structures. Similarly, overexpression of the polo-like kinase Cdc5p, which may have a role at the bud neck in the initiation of cytokinesis (Lee et al., 1998, 1999; Song et al., 2000), results in hyperpolarized buds containing ectopic septin ring structures, although these do not appear as elaborate as the Int1p/septin spirals that we have observed. In addition, overexpression of Afr1p, a bud-neck–localized protein required for the formation of mating projections, also causes hyperpolarized bud growth (Konopka et al., 1995). Thus, high-level expression of some, but not all, bud-neck components can cause hyperpolarized bud growth and formation of ectopic septin structures.
Alteration of the septin ring can activate Swe1p and thus delay the transition from polarized to isotropic growth that normally occurs during activation of Clbp/Cdc28p kinase complexes (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). Because Swe1p contributes to INT1-induced filamentous growth (Asleson et al., 2001), disruption of the septin ring by Int1p probably prolongs the polarized growth phase of the cell cycle via Swe1p activation; however, Sla2p, a component of actin cortical patches, contributes to INT1-induced filamentous growth even in the absence of Swe1p (Asleson et al., 2001), indicating that the activation of Swe1p does not account for all of the polarized growth observed in cells expressing Int1p. In addition, we observed ectopic septin structures in polarized swe1 cells expressing INT1, indicating that SWE1 is not required for the disruption of the septin ring by Int1p.
Int1p and Septins in C. albicans
Int1p contributes to morphogenesis in C. albicans (Gale et al., 1998) and colocalizes with septins at the mother-bud neck of yeast and pseudohyphal cells as well as at the septa of hyphal cells. Classic studies of C. albicans morphogenesis suggested that the commitment to yeast versus hyphal growth is determined by the timing and position of cytokinetic structures (Mitchell and Soll, 1979a,b; Soll and Mitchell, 1983) and that the ultrastructure of septa differ in hyphal and yeast cells (Gow et al., 1980). During hyphal induction, we consistently observe septa, septins, and Int1p within the growing germ tube and at a distance of 10–20 μm from the mother–daughter cell junction. This is consistent with the recent observations of others (Sudbery, 2001; Warenda and Konopka, personal communication) with the use of anti-Cdc11p antibody and septin fluorescent protein fusions, respectively, and supports the model that yeast and pseudohyphae are fundamentally different from true hyphae. Our results are also consistent with the results of Mitchell and Soll (Mitchell and Soll, 1979a,b; Soll and Mitchell, 1983) except that in our study the septin ring appears at a greater distance from the mother–daughter cell junction. This disparity may be due to the difference in hyphal-inducing medium used or the growth temperature used, or both. Interestingly, Sudbery (2001) noted a transient Cdc11p signal at the mother–daughter junction that disappeared as germ tube growth progressed. In our experiments, we did not observe such a transient signal for Cdc3p. The discrepancy between the observations may be due to a difference in visualization between the fluorescent-tagged antibody used by Sudbery (2001) and our fluorescent protein fusions or may indicate a difference in the actual localization of Cdc11p versus Cdc3p in C. albicans hyphae.
In C. albicans, int1/int1 cells are defective in hyphal growth under some, but not all, environmental conditions (Gale et al., 1998). Thus, Int1p may play an important role in a subset of the environmental sensing and signal transduction pathways that trigger hyphal growth in C. albicans. Because several S. cerevisiae proteins (e.g., Swe1p, Hsl1p, Hsl7p, Gin4p, Bud3p, Bud4p, Myo1p, and Cdc5p) that influence morphogenesis also localize to the septin scaffold (Sanders and Herskowitz, 1996; Bi et al., 1998; Longtine et al., 1998a, 2000; Barral et al., 1999; Lord et al., 2000; Song et al., 2000), the colocalization of Int1p and septins in C. albicans suggests that Int1p influences the response to some hyphal induction conditions through its interactions with proteins in the septin ring.
Int1p affects axial bud site selection in C. albicans, as does Bud4p in S. cerevisiae, but INT1 does not complement a bud4Δ strain for bud-site selection (Gale et al., 1998), and high levels of Bud4p do not produce the aberrant morphology seen with Int1p overexpression. Thus, Int1p and Bud4p are not complete functional homologues. These proteins may have evolved different related functions that both occur at the cell membrane or the mother-bud neck, or both. It is notable that the only known role for Bud4p is in haploid bud-site selection, yet BUD4 is expressed in both haploid and diploid S. cerevisiae cells (Sanders and Herskowitz, 1996). This suggests that Bud4p may have additional, uncharacterized functions independent of its known role in bud-site selection. Just as studies of Int1p in S. cerevisiae have provided insights into Int1p function in C. albicans, possible roles for Bud4p in diploid S. cerevisiae cells may be revealed by studying Int1p functions in C. albicans.
ACKNOWLEDGMENTS
We thank E. Bi, B. Cormack, D. Kellogg, J. Konopka, J. Pringle, and the Yeast Research Center (University of Washington) for providing plasmids, strains, and antibodies; P. Sudbery, J. Konopka, and Sylvia Sanders for sharing results before publication; J. Asleson for technical assistance; M. Sanders and D. Gartner for assistance with microscopy and image processing; and J. Beckerman, C. Bendel, E. Bensen, S. Enomoto, and L. Glowczewski for helpful discussions and critical review of this manuscript. This work was approved for publication by the Director of the Oklahoma Agricultural Experiment Station. This work was supported by Child Health Research Center award P30 HD33692, March of Dimes Basil O'Connor Award 5-FY99-791, and National Institutes of Health Grant AI-01712-02 (C.G.), by project H-2410 (M.L.), by Burroughs-Wellcome Senior Scholar Award 0677 (J.B.), and by National Institutes of Health grant AI-25827 (contract to J.B.).
Abbreviations used:
- CFP
cyan fluorescent protein
- DIC
differential interference contrast
- GFP
green fluorescent protein
- HA
hemagglutinin epitope
- I-IFG
INT1-induced filamentous growth
- YFP
yellow fluorescent protein
REFERENCES
- Asleson CM, Bensen ES, Gale CA, Melms AS, Kurischko C, Berman J. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol Cell Biol. 2001;21:1272–1284. doi: 10.1128/MCB.21.4.1272-1284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin WL. Site selection for bud and germ tube emergence in Candida albicans. J Gen Microbiol. 1983;130:431–440. [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. “Marker swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- De Backer MD, Magee PT, Pla J. Recent developments in molecular genetics of Candida albicans. Annu Rev Microbiol. 2000;54:463–498. doi: 10.1146/annurev.micro.54.1.463. [DOI] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AE, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDomenico BJ, Brown NH, Lupisella J, Greene JR, Yanko M, Koltin Y. Homologs of the yeast neck filament associated genes: isolation and sequence analysis of Candida albicans CDC3 and. CDC10. Mol Gen Genet. 1994;242:689–698. doi: 10.1007/BF00283424. [DOI] [PubMed] [Google Scholar]
- Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- Field CM, Kellogg D. Septins: cytoskeletal polymers or signaling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier JA, Wong ML, Longtine MS, Pringle JR, Mann M, Mitchison TJ, Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale C, Finkel D, Tao N, Meinke M, McClellan M, Olson J, Kendrick K, Hostetter M. Cloning and expression of a gene encoding an integrin-like protein in Candida albicans. Proc Natl Acad Sci USA. 1996;93:357–361. doi: 10.1073/pnas.93.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale CA, Bendel CM, McClellan M, Hauser M, Becker JM, Berman J, Hostetter MK. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad M, Berman J, Gale CA. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR, Gooday GW, Newsam RJ, Gull K. Ultrastructure of the septum in Candida albicans. Curr Micro. 1980;4:357–359. [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Herrero AB, Lopez MC, Fernandez-Lago L, Dominguez A. Candida albicans and Yarrowia lipolytica as alternative models for analyzing budding patterns and germ tube formation in dimorphic fungi. Microbiology. 1999;145:2727–2737. doi: 10.1099/00221287-145-10-2727. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka JB, DeMattei C, Davis C. AFR1 promotes polarized apical morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:723–730. doi: 10.1128/mcb.15.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, Schmidt A, Gow NA, Brown AJ, Thomas DY. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Song S, Erikson RL. The polo-box-dependent induction of ectopic septal structures by a mammalian polo kinase, Plk, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:14360–14365. doi: 10.1073/pnas.96.25.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998a;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998b;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998a;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998b;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Yang MC, Mischke M, Chant J. Cell cycle programs of gene expression control morphogenetic protein localization. J Cell Biol. 2000;151:1501–1512. doi: 10.1083/jcb.151.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Sia RAL, Lew DJ. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LH, Soll DR. Commitment to germ tube or bud formation during release from stationary phase in Candida albicans. Exp Cell Res. 1979a;120:167–179. doi: 10.1016/0014-4827(79)90547-0. [DOI] [PubMed] [Google Scholar]
- Mitchell LH, Soll DR. Temporal and spatial differences in septation during synchronous mycelium and bud formation by Candida albicans. Exp Mycol. 1979b;3:298–309. [Google Scholar]
- Odds FC. Morphogenesis in Candida, with special reference to C. albicans. In: Candida and Candidosis. London: Tindall; 1988. pp. 42–59. [Google Scholar]
- Odds FC. Pathogenesis of Candida infections. J Am Acad Dermatol. 1994;31:S2–5. doi: 10.1016/s0190-9622(08)81257-1. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Mor J-R. Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem. Methods Cell Biol. 1975;11:131–168. doi: 10.1016/s0091-679x(08)60320-9. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Bardes ES, Lew DJ. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor GW, Chochran WG. Statistical Methods. Ames, IA: The Iowa State University Press; 1980. [Google Scholar]
- Soll DR, Mitchell LH. Filament ring formation in the dimorphic yeast Candida albicans. J Cell Biol. 1983;96:486–493. doi: 10.1083/jcb.96.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Molec Microbiol. 2001;41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- Tjandra H, Compton J, Kellogg D. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]