Abstract
The interaction between the (epi)genetic makeup of an individual and his/her environmental exposure record (exposome) is accepted as a determinant factor for a significant proportion of human malignancies. Recent evidence has highlighted the key role of epigenetic mechanisms in mediating gene–environment interactions and translating exposures into tumorigenesis. There is also growing evidence that epigenetic changes may be risk factor-specific (“fingerprints”) that should prove instrumental in the discovery of new biomarkers in cancer. Here, we review the state of the science of epigenetics associated with environmental stimuli and cancer risk, highlighting key developments in the field. Critical knowledge gaps and research needs are discussed and advances in epigenomics that may help in understanding the functional relevance of epigenetic alterations. Key elements required for causality inferences linking epigenetic changes to exposure and cancer are discussed and how these alterations can be incorporated in carcinogen evaluation and in understanding mechanisms underlying epigenome deregulation by the environment.
Keywords: epigenetics, environment, cancer, molecular mechanisms, research gaps, perspectives, biomarkers
Epidemiological studies have uncovered robust and consistent associations between environmental factors and cancer risk. However, these associations provide little information on the mechanism by which a given exposure leads to cancer. The interaction between the (epi)genetic makeup of an individual and his/her environmental exposure record (exposome)1 may determine a large fraction of human malignancies.
Epigenetic disruption is a near-universal feature of human malignancy and a key driver of many cancers.2 In recent years, accumulating evidence has highlighted the key role of epigenetics in mediating gene–environment interactions and their effect throughout the tumorigenesis process3 (Fig. 1). This progress has been catalyzed by advances in the epigenomic field, including the emergence of powerful technologies and state-of-the-art in vitro and in silico computational approaches. Well-established risk factors of cancer, such as age, inflammation, diet, and smoking have been studied in the context of epigenome deregulation, along with some less widely studied exposures and lifestyle factors such as air and water pollution, fungal toxins and endocrine disruptors (Fig. 1). Notably, numerous international cohorts have been established enabling an investigation of life course exposures on epigenetic profiles in the context of large-scale epidemiological studies.4
Figure 1.
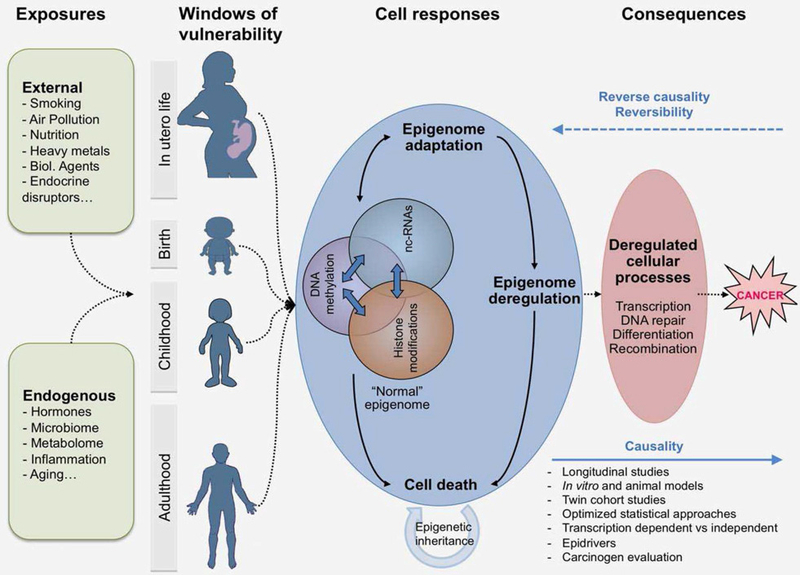
Exposures arising from external sources (environmental chemicals, air pollution, infectious agents, diet, tobacco, alcohol, endocrine disruptors) and internal processes (metabolism, hormones, inflammation, gut microflora, aging) may induce stable and potentially reversible changes in the epigenome. The patterns (“signatures”) and persistence of these alterations depend on multiple factors, including the type of epigenetic changes (some genomic regions remain methylated for longer periods than others), the dosage and duration of the exposure (longer and more intense exposures could minimize reversibility of DNA methylation), the tissue type and the developmental stage (in utero life or puberty may be particularly sensitive periods to some exposure). Thus, epigenetic mechanisms may represent “sensors” of exposure and “mediators” of the outcomes, including cancer development. Epigenome alterations should prove instrumental in discovery of new biomarkers for risk stratification and early detection and attractive targets for novel therapies and preventive strategies.
Here, critical knowledge gaps and research needs are discussed and advances in epigenomics that may help an understanding of the functional relevance of epigenetic alterations induced by environmental exposures. All co-authors of this work have met during the first Environmental and Epigenetics Origin of Cancer meeting, held at IARC, Lyon, in June 2016 and have extensively interacted during and after the meeting to concretize in this article the valuable conclusions, arguments and highlights in the field. Accordingly, this manuscript is not intended to be a meeting report as it does not merely summarize different scientific opinions nor does it represent a review of the literature. Instead, it is intended to bring forward critical questions that need to be answered, approaches and study designs that could help answering them, methodology developments that could be implemented, important findings attained so far as examples, future utilities of the field and the direction(s) toward which all these developments could steer the field.
Risk Factors Associated With Epigenome Deregulation and Cancer
The mechanisms by which environmental factors can have long lasting effects on cancer outcome remain poorly understood (Fig. 1). For example, tobacco smoke has well-established effects on blood DNA methylation of newborns, children and adults,5,6 though it remains unclear how these effects contribute to tumorigenesis. In addition, nutrition was shown to affect metastable epialleles (MEs), exhibiting systemic (not cell type-specific) interindividual variation in DNA methylation7; however, whether these epigenetic polymorphisms may be useful as a predictor of cancer risk remains to be tested. Associations between folate status, methylation and human colon cancer have been established in the prevention of malignancy,8 but a protective role for folate against carcinogenesis has recently been questioned, with increasing evidence that excessive intake of synthetic folic acid may actually increase the risk of certain human malignancies.9
Environmental contaminants (such as inorganic arsenic) were shown to be associated with methylation changes in infant cord blood,10 suggesting the “transcription factor occupancy theory” as an underlying mechanism.11 Air pollution represents another epigenome disruptor; a recent meta-analysis showed that nitrous dioxide exposure during pregnancy is associated with cord blood differential DNA methylation in mitochondrialrelated genes.12 Endocrine-disrupting chemicals (EDCs) represent another example of pollutants that may deregulate the epigenome13 and contribute to the development of specific malignancies, especially hormone-deregulated cancers, although mechanism remains largely undetermined.14
Infection agents and chronic inflammation are also known to affect epigenetic states. For example, the maternal microbiome and the postnatal gut microbiome seem to play a role in modulating intestinal mucosal epigenetic patterning and consequent susceptibility to inflammatory bowel disease (IBD) and young-onset colorectal cancer.15,16 Another example is the epigenetic field cancerization observed in gastric cancer, where chronic inflammation induced by Helicobacter pylori is responsible for aberrant DNA methylation.17 In addition, oncogenic viruses such as Hepatitis B virus and Epstein–Barr virus are known to hijack the host epigenetic machinery to promote its replication and to cloak itself from the host surveillance system, but potentially leaving a recognizable epigenetic signature.18 The fact that infection-related cancers are often characterized by DNA methylation changes extending to noncancer adjacent tissues suggests that these alterations may be the result of a complex process involving chronic inflammation, immune response and changes in cell distribution in addition to possible direct effects of infectious agents or their mediators (e.g., viral proteins)
Exposure Timing and Epigenome Deregulation
In addition to the type of environmental exposure, timing also plays an important role in influencing disease risk. Embryonic life and fetal life comprise sensitive periods in the human life cycle due to the capacity for changes in cell fate during embryonic development, with potentially lifelong health outcomes. Epigenetic mechanisms represent likely “mediators” of these outcomes because they are implicated in (i) pathways driving embryogenesis, including tissue differentiation, (ii) mitotically heritable mechanisms with long lasting effects, and (iii) environmentally sensitive and potentially reversible molecular drivers of disease. There is increasing evidence showing how in utero exposure leaves epigenetic marks in the fetus, and these include food contaminants such as arsenic and heavy metals,19 aflatoxin B120 and tobacco smoke.5 The influence of many of these environmental contaminants on childhood cancer has yet to be evaluated. These findings do suggest, however, that critical time points for intervention and prevention strategies may occur early in life.
In addition to the embryonic period, environmental and epigenetic influences may alter other developmental stages, such as childhood and puberty, especially in females. In males, spermatogenesis starts at puberty and continues throughout life; whereas, in females, oogenesis begins before birth and is arrested in the prophase of meiosis until puberty. Hence, in girls, oocytes remain until puberty in a haploid demethylated state, which is more susceptible to environmental stressors than the diploid methylated state of the male germline. Later during adulthood, women may exhibit other susceptible windows of exposure during the menstrual cycle, pregnancy or menopause. These timing windows of exposures must be considered when analyzing the interaction between the environment, epigenetics and cancer.
Research Gaps and Needs
Until very recently, there was a major gap in our understanding of the “normal” epigenome and its normal variability.21 As the capacity to map the epigenome continues to increase, the catalog of epigenetic variations associated with adverse environmental exposures will undoubtedly expand.22 The specific research questions highlighted below warrant particular attention in that they remain equivocal or have not been fully addressed.
Strengthening causal inference
To better infer causality of epigenetic associations linking environmental exposure and cancer, several critical scientific approaches are needed (Figs. 1 and 2)
Figure 2.
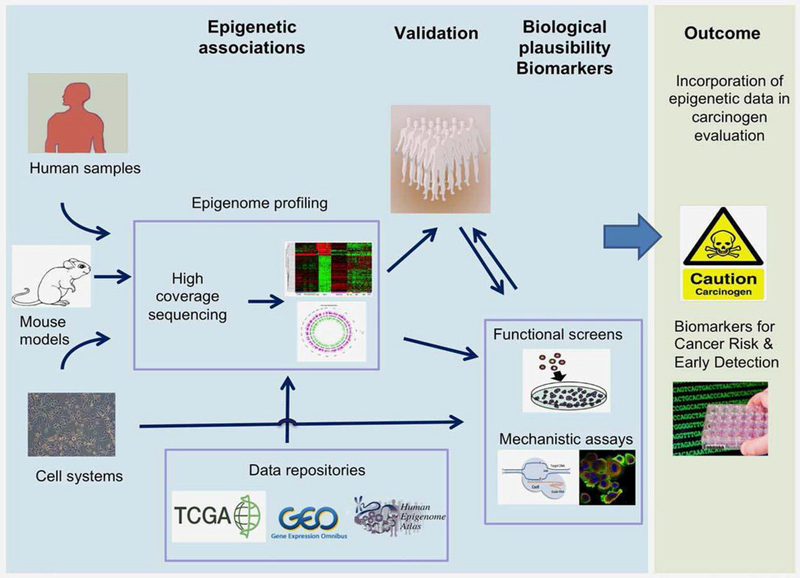
An integrated approach for the production and integration of epigenetic data in carcinogen identification and evaluation. This approach implies the use of cutting edge epigenomics, population-based cohorts, and innovative bioinformatics tools for the identification, quantification, mapping of changes in the epigenome induced by known and suspected carcinogens. Human tumor samples from case– control and population-based cohorts are used in combination with in vitro cell systems and mouse models to perform epigenomic profiling to identify signatures, genes and pathways that are deregulated by specific risk exposure. This is followed by validation in population-based cohorts and where appropriate the data are crossed with the epigenomic databases. Identification of genes and pathways is followed by functional studies to provide biological plausibility to associations that are observed. The outcome is providing evidence base for studies directly relevant to cancer causation and prevention and identification of markers for early detection and cancer risk stratification.
Establishing mechanistic causes through the use of cellular and animal models, which allow the systematic manipulation of variables (Fig. 2)
Based on mouse models, an important question of epigenetic cause versus consequence is being addressed across several windows of mouse development, showing that developmental reprogramming of H3K4me3 is acutely induced by EDCs, persists across the life-course, increases responsiveness to hormones without being dependent on abnormal transcription and promotes the development of hormone dependent tumors.13
(2) Coupling epigenetic mechanisms to other molecular players (including cross-omics)
For example, epigenetic marks can be functionally annotated to gene expression data and can be associated with causality through genetic variant randomization. Epigenetic variants that are causal to cancer would likely demonstrate functional consequences on gene activity or cellular function. Optimized statistical approaches are equally important and this is demonstrated through the example on the aryl-hydrocarbon receptor repressor (AHRR) methylation, which is to date the most consistent epigenetic signature of tobacco smoking. Although cigarette smoke is the strongest exposure factor causing lung cancer, the role of AHRR methylation in the causal pathway from smoking to lung cancer (as estimated by mediation analysis23) would require further evidence by Mendelian Randomization.24,25
(3) Integrating epigenetics within well-designed epidemiological studies, particularly prospective cohort designs (Fig. 1)
Cohort studies enable the identification of “driver” epigenetic alterations that occur prior to disease onset, and hence, avoid confounding by “passenger” events that are induced by the disease (reverse causality). Moreover, longitudinal cohorts that start in early life can contribute to our understanding of how the epigenome changes over critical periods throughout life, while cohort studies based on twin pairs can help disentangle the causal contribution of genetics relative to epigenetics in mediating the response to environmental cues and risk to cancer (Fig. 1). Evidence from the Peri/Postnatal Epigenetic Twins Study showed the role of both environment and genetic variation in determining neonatal epigenetic profile, with the heritability of DNA methylation profiles estimated at 15–20%.26 The environmental exposures per se also should be better estimated, especially given that long-term exposures cannot be measured with the same degree of accuracy as in short-term experimental studies. Another criterion in well designed studies is their ability to reproduce observed associations in multiple cohorts and large sample sizes. The Pregnancy and Childhood Epigenetics Consortium (PACE) and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortia provide interesting examples of the largest studies to date analyzing the effects of environmental exposure on epigenetic alterations in birth and adult cohorts, respectively.
(1) Epigenetic mechanisms in relevance to biochemical precursors of DNA methylation
Although folate, a methyl donor, has a strong impact on DNA methylation and cancer, the directionality of those effects remains questionable. For example, high folate levels may lead to both high and low DNA methylation and to both increased and decreased risk of cancer. These seemingly contradicting findings become more biologically plausible upon dissecting the effect of folate exposure by dosage, timing, target genes and cancer types. Mouse studies directly testing the effect of folic acid intake at various stages of the life course and on various tissues may be particularly important for fine tuning the intricate associations between folate exposure, epigenetics and cancer. Additionally, the influence of folate species on methylation and cancer risk remains to be established.
(2) Epigenetic mechanisms in relevance to transcriptional machinery
Although DNA hypermethylation in the promoters of many genes is generally associated with transcriptional silencing, the importance of the link between epigenetics and transcription remains an open question. CpG methylation that is not associated with RNA expression may have little functional relevance, but this is questioned by the evidence showing how developmental reprogramming involving the remodeling of chromatin marks may lead to increased responsiveness to hormones without necessarily altered transcription.13 It also remains to be established to what extent the link between methylation and expression is due to loss of transcription factor binding.27 Moreover, much remains to be learned about the functional regulation of ultralow methylation regions (ULMRs), which are methylated at 1–20% and rarely studied using traditional methodologies (J.P. Issa, unpublished data).
(3) Epigenetic mechanisms in relevance to chromatin landscape
While DNA methylation is known to be transmitted with high fidelity across cell divisions, the chromatin landscape is less characterized, and it is still unclear how a defined chromatin domain is reproduced following cell replication. Recently, the Polycomb Repressive Complex 2 has been implicated in the inheritance of histone modifications across cell divisions.28 However, none of the existing techniques for analysis of histone modifications is ready to use on the biospecimen types and sample size scales that are utilized in population-based studies. The advent of new technologies in chromatin biology holds promise for future studies aiming to investigate the role of chromatin in mediating effects of environmental exposures on cancer.
(4) Epigenetic mechanisms in relevance to epidrivers
The role of epidrivers (the genes involved in epigenetic regulation exhibiting recurrent disruption in cancer through mutational or nonmutational mechanisms) in carcinogenesis requires particular attention.29–31 The fact that >50% of human cancers harbor mutations in enzymes that are involved in chromatin organization32 argues that epidrivers may represent an early and central event in tumorigenesis. To confirm this, mechanistic studies of epidrivers altered by specific carcinogenic agents should be considered using in vitro human and mouse models and state-of-the-art approaches (epigenome-wide shRNA or CRISPR library screens, epigenome editing, and functional genomics). Characterization of epidriver events is expected to advance the knowledge of mechanisms of carcinogenesis and underpin studies of cancer etiology, therapy and prevention.
Analytical considerations
In population-based epigenome-wide studies, the generation, analysis and interpretation of data are not straightforward.33,34 Several studies demonstrated the robustness of wet lab and bioinformatics pipelines and capacity to perform epigenome analysis in high throughput and genome-wide settings; however, there is a lack of consensus on the pertinent optimal analytical approaches. While recent studies of epigenome-wide changes associated with some known risk factors used a GWAS-like strategy that treats individual CpG sites independently, there is wide recognition that more advanced approaches (including CpG regional clusters) may be more informative.34
Epigenetic reversibility, effect size and rate of change
Several studies highlight the reversibility or lifetime persistence of some specific epigenetic changes associated with environmental exposures (Fig. 1). This seems to depend on multiple factors, including the type of epigenetic signatures (some CpGs remain methylated for longer periods than others, given the same exposure), the level and duration of the exposure, the tissue type, and the developmental stage (in utero life or puberty are sensitive periods to exposure and can be prone to epigenetic alterations with long-term effects). More studies and cohorts with repeated time points are needed to enhance the resolution of the epigenetic snapshots taken at different developmental stages of life (Fig. 1). Such study designs can also enable the assessment of the “rate” of change (and not only the effect size) of DNA methylation in response to exposure over time. These studies may also need to consider if the reversal of the change leads to reversal of risk, as specific epigenetic events during a critical developmental period could initiate a program which later in life could be important, regardless of the continued presence or absence of that initiator (a “hit and run” effect).
Cell type heterogeneity
This remains a major concern in epigenetic studies, the deconvolution of which becomes more intricate in tumor tissues, which exhibit both clonal and genetic intratumor heterogeneity.35 For example, head and neck squamous cell carcinomas exhibit extensive heterogeneity in etiological, environmental, cellular and molecular features, hampering accurate prognosis, treatment planning and identification of causative genes that may serve as molecular drug targets.36–38 Recent advances in bioinformatics have helped correct for possible changes in cell subpopulations using DNA methylome-based prediction algorithms that rely on reference tissues (initially using peripheral blood39 and recently cord blood40) and reference-free methods (a recent but rapidly developing field.41,42 Emerging methods for single-cell epigenomics should also provide exciting tools for resolving the issues related to the variability of the epigenome among different cells and cell clones in complex tissues.43,44
Target tissue
Epigenetic changes are abundant and directly measureable in tumor biopsies, especially when compared with adjacent tissue. However, aberrant DNA methylation has also been observed in surrogate tissues such as peripheral blood of cancer patients. Epigenetic alterations can arise in early stages of embryonic development, when epigenetic patterns undergo large-scale reprogramming (Fig. 3), and, hence, may be propagated in most, if not all, tissues, thereby generating identical constitutional epimutations throughout the body7 or creating a mosaic pattern of the epigenome in a given organ.45 In this scenario, the timing of the epigenetic event and proliferation history of the affected cells will determine the proportion and distribution of the cells harboring epimutations across different tissues (Fig. 3). These considerations provide the basis for developing epigenetic biomarkers in blood, which can serve as a surrogate for diagnostics and risk stratification of cancer in other tissues. For example, methylation of SEPT9 has been shown to be a reliable and sensitive blood-based biomarkes for colorectal cancer detection.46
Figure 3.
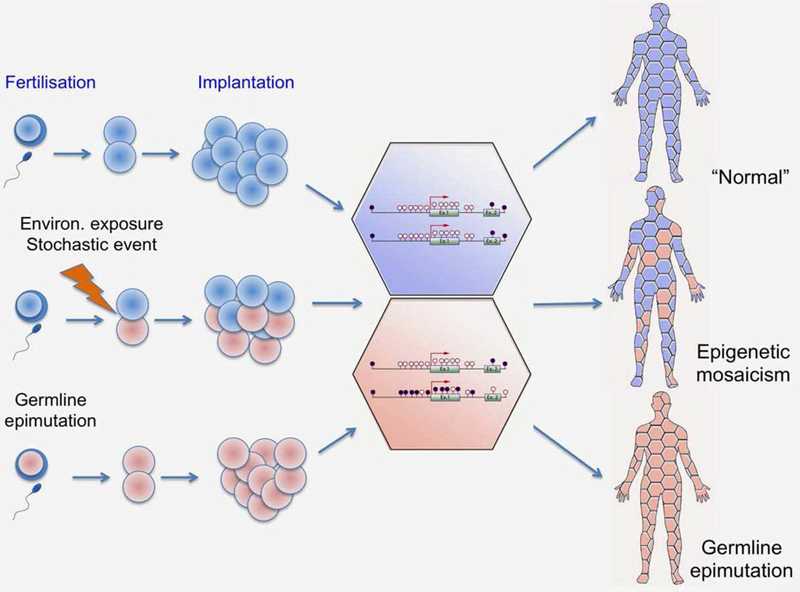
Constitutional epimutations and epigenetic mosaicism as a mechanism of cancer causality and targets for biomarker discovery. Although epigenetic patterns are tissue specific, interrogating the epigenome in tissues that are not the target tissue (surrogates) may be informative of exposure history and cancer risk. Environmental exposure, stochastic event, or even germline epimutation may be propagated over life course and result in epigenetic mosaicism or germline epimutations across tissues which may constitute an increased susceptibility to cancer.
Besides blood and urine samples, additional body fluids and different types of biospecimens collected through noninvasive or minimally invasive techniques, such as buccal swabs, breast lavage and cervical smears, may provide attractive targets for the discovery of biomarkers of exposure or early detection of cancer.
Early-life exposures
As described earlier, “windows of vulnerability” exist during in utero development, within which maternal exposure factors may alter the fetal epigenome, increasing susceptibility to later-onset diseases, including cancer.47 A recent example illustrates the complex association between in utero exposure to tobacco smoke and childhood cancer. A study of neonatal blood spots showed that DNA methylation at birth was altered in association with early pregnancy maternal folate status.48 DNA methylation marks of smoking demonstrated a difference between cases and controls (J. Wiemels, unpublished data), consistent with the interaction between maternal smoking in cancer risk in the offspring.49 International collaboration on such a rare disease (to assimilate large samples and replicate findings in multiple cohorts) may help decipher this complex exposure-to-phenotype pattern.
Epigenetic clock and cancer risk
One of the best-characterized DNA methylation signatures in population-based studies is chronological age. Age-associated epigenetic changes have been identified and provide the basis for an “epigenetic clock.”50 Age is the strongest demographic risk factor for cancer, indicating that molecular changes upon aging trigger malignant transformation.51 DNA methylation clock may be affected by different external and endogenous factors. Those exposures may contribute to methylation drift52 and “accelerated” aging, emphasizing that the often ignored rate of change in methylation can be important even though the magnitude of methylation differences might be minimal. As DNA methylation landscape is altered as a function of age (independently of exposures), there is a need to explore synergistic epigenetic effects between age and environmental exposures. For instance, DNA methylation profiling in a large prospective cohort revealed an association between the epigenetic age acceleration and breast cancer risk,53 although further studies are needed to establish the synergy between exposure and age. Importantly, age-associated epigenetic silencing of HAND2 seems to be an early event in endometrial carcinogenesis, leading to gradual inactivation of the progesterone tumor suppressor pathway and sensitizing endometrial epithelial cells to oncogenic estrogen.54 Therefore, this may serve as a paradigm for aging-associated epigenetic changes sensitizing (priming) the cells for subsequent exposure to oncogenic stimuli. Further studies are needed to test the presence of synergistic age-exposure mediating effects on the DNA methylome and cancer risk.
Toward incorporating epigenetic data into carcinogen identification and evaluation
Recent advances in epigenetics represent an exciting opportunity toward the incorporation of epigenetic mechanisms into carcinogen evaluation and safety assessment (Fig. 2). In spite of recent data on epigenetic mechanisms as biological mediators of certain exposures (such as EDCs discussed above), evidence for a causal role of epigenetic changes in carcinogenesis is limited. Although the incorporation of epigenetic mechanisms into carcinogen evaluation is at an early stage,21,55 important data have been generated, and valuable scientific resources could be applied in the main international programs of carcinogen evaluation (such as the IARC Mono graphs Programs and National Toxicology Programs in the US). There will be value in designing integrated approaches aiming to interrogate all layers of the epigenome in response to carcinogen exposure in populations followed by validation in population-based studies and functional characterization in in vitro model systems (Fig. 2). There is an urgent need to develop epigenetic assays that incorporate scientifically sound experimental designs with particular consideration for dose and route of exposure. Identifying a set of priority carcinogens to be studied in detail will be an important start. We propose that particular attention should be paid to potential “epigenetic carcinogens” (such are those classified by IARC as probably carcinogenic or possibly carcinogenic to humans [Groups 2 A and 2B] that seem to act through nonmutational mechanisms), as opposed to established mutagens.
Conclusions
Remarkable progress in the field of epigenetics provides a better understanding of the etiology of human cancers and suggests a potential causal role for epigenetic disruptions linking environmental exposure to tumorigenesis. The emergence of powerful sequencing technologies has enabled the analysis of the epigenome with high resolution in both genome-wide and high-throughput settings, thus dramatically accelerating investigations in cancer biology and molecular epidemiology. Major international efforts have brought about critical advances, with the establishment of reference epigenomes for many normal cell types and cancer-specific epigenomes for several tumor types. Recent studies contributed to the identification of epigenetic events deregulated by specific environmental and lifestyle stressors, supporting the hypothesis that the epigenome may function as an interface between environmental factors and the genome. Importantly, many studies provided evidence that environmental exposures can induce specific changes in the epigenome. Such epigenetic “fingerprints” will prove instrumental in carcinogen evaluation and identification and in the discovery of new biomarkers for risk stratification and novel interventions for prevention.
Acknowledgements
This article was instigated by the “Epigenetics and Environmental Origins of Cancer” (EEOC) meeting which was held with 140 international participants at the International Agency for Research on Cancer (IARC, Lyon, France) on the 11th and 12th of June, 2016. Presentations from leading scientists in the field were grouped thematically around the following topics: Environmental Agents and Lifestyles, Nutrition and Metabolism, Endocrine Disruptors, Carcinogen Evaluation, Early Life Exposures, and Infections and the Microbiome. The meeting highlighted important research advances, needs and gaps in the field, including critical assessments and interdisciplinary approaches. The EEOC was organized by the Epigenetics Group, IARC, Lyon, and supported by grants from the Institut National du Cancer (INCa, France), the European Commission (EC) seventh Framework Programme (FP7) Translational Cancer Research (TRANSCAN) Framework, the Fondation ARC pour la Recherche sur le Cancer (France), and Plan Cancer-Eva-Inserm research grant to Z.H. S.A. was supported by the postdoctoral fellowship from the International Agency for Research on Cancer, partially supported by the EC FP7 Marie Curie Actions – People – Co-funding of regional, national and international programmes (COFUND). A.S. is supported by the PhD fellowship from the Fonds National de la Recherche, Luxembourg (AFR Code: 10100060). Special thanks to Ms Elizabeth Page for her help in organizing the conference, to EpiGenie, represented by Dr Angelika Merkel, for covering the meeting and to the members of the Section of Mechanisms of Carcinogenesis at IARC: Dr Szilvia Ecsedi, Dr Nora Fernandez Jimenez, Ms Hana Huskova, Ms Diana Maria Narvaez Noguera, Dr Vibha Patil, Dr Fazlur Rahman Talukdar, Dr Hae Don Woo and Ms Maria Zhivagui for their help during the conference.
Grant sponsor: Institut National du Cancer (INCa, France); Grant sponsor: European Commission (EC) seventh Framework Programme (FP7) Translational Cancer Research (TRANSCAN) Framework; Grant sponsor: Fondation ARC pour la Recherche sur le Cancer (France); Grant sponsor: Plan Cancer-Eva-Inserm; Grant sponsor: International Agency for Research on Cancer; Grant sponsor: Marie Curie Actions – People – Co-funding of regional, national and international programmes (COFUND); Grant number: EC FP7; Grant sponsor: Fonds National de la Recherche, Luxembourg; Grant number: 10100060
Footnotes
Conflict of Interest
The authors declare that they have no competing financial interests.
References
- 1.Wild CP The exposome: from concept to utility. Int J Epidemiol 2012;41:24–32. [DOI] [PubMed] [Google Scholar]
- 2.Jones AP Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- 3.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 2011;13:97–109. [DOI] [PubMed] [Google Scholar]
- 4.Ng JW, Barrett LM, Wong A, et al. The role of longitudinal cohort studies in epigenetic epidemiology: challenges and opportunities. Genome Biol 2012;13:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 2016;98:680–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambatipudi S, Cuenin C, Hernandez-Vargas H, et al. Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics 2016. [DOI] [PubMed] [Google Scholar]
- 7.Silver JM, Kessler JN, Hennig JB, et al. Independent genomewide screens identify the tumor suppressor VTRNA2–1 as a human epiallele responsive to periconceptional environment. Genome Biol 2015;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duthie JS Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J Inherit Metab Dis 2011;34:101–9. [DOI] [PubMed] [Google Scholar]
- 9.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA 2007;297:2351–9. [DOI] [PubMed] [Google Scholar]
- 10.Laine EJ, Bailey AK, Rubio-Andrade M, et al. Maternal arsenic exposure, arsenic methylation efficiency, and birth outcomes in the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Mexico. Environ Health Perspect 2015; 123:186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin EM, Fry RC. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environ Epigenet 2016;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruzieva O, Xu CJ, Breton CV, et al. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Trevino SL, Lee Yean Wong R, et al. Reprogramming of the epigenome by MLL1 links early-life environmental exposures to prostate cancer risk. Mol Endocrinol (Baltimore, Md) 2016; me20151310-me. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleisch FA, Wright OR, Baccarelli AA. Environmental epigenetics: a role in endocrine disease? J Mol Endocrinol 2012;49:R61–R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah R, Cope JL, Nagy-Szakal D, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes 2016; 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahara T, Yamamoto E, Madireddi P, et al. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology 2014;146: 530–8e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattori N, Ushijima T. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med 2016;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res 2011; 727:55–61. [DOI] [PubMed] [Google Scholar]
- 19.Green BB, Karagas MR, Punshon T, et al. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire Birth Cohort Study (USA). Environ Health Perspect 2016;124:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez-Vargas H, Castelino J, Silver JM, et al. Exposure to aflatoxin B1 in utero is associated with DNA methylation in white blood cells of infants in The Gambia . Int J Epidemiol 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herceg Z, Lambert M- P, van Veldhoven K, et al. Towards incorporating epigenetic mechanisms into carcinogen identification and evaluation. Carcinogenesis 2013;34:1955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busche S, Shao X, Caron M, et al. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol 2015;16:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fasanelli F, Baglietto L, Ponzi E, et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Comms 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richmond RC, Hemani G, Tilling K, et al. Challenges and novel approaches for investigating molecular mediation. Hum Mol Genet 2016;25: R149–R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latvala A, Ollikainen M. Mendelian randomization in (epi)genetic epidemiology: an effective tool to be handled with care. Genome Biol 2016; 17:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon L, Joo JE, Powell JE, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res 2012;22: 1395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziller MJ, Gu H, Muller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013;500:477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumesic PA, Homer CM, Moresco JJ, et al. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 2015; 160:204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plass C, Pfister SM, Lindroth AM, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 2013;14:765–80. [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013; 339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Perez A, Jene-Sanz A, Lopez-Bigas N. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol 2013;14:r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones PA, Issa JP, Baylin S Targeting the cancer epigenome for therapy. Nat Rev Genet 2016;17: 630–41. [DOI] [PubMed] [Google Scholar]
- 33.Breton CV, Marsit CJ, Faustman E, et al. Small-magnitude effect sizes in epigenetic end points are important in Children’s Environmental Health Studies: the Children’s Environmental Health and Disease Prevention Researches Center’s epigenetics working group. Environ Health Perspect 2017;125:511–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mill J, Heijmans BT From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet 2013;14:585–94. [DOI] [PubMed] [Google Scholar]
- 35.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenberg MS, Ellisen WL The moleculars pathogenesis of head and neck squamous cell carcinoma. J Clin Invest 2012;122:1951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lima SC, Hernandez-Vargas H, Simao T, et al. Identification of a DNA methylome signature of esophageal squamous cell carcinoma and potential epigenetic biomarkers. Epigenetics 2011;6: 1217–27. [DOI] [PubMed] [Google Scholar]
- 38.Leemans RC, Braakhuis MBJ, Brakenhoff HR The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9–22. [DOI] [PubMed] [Google Scholar]
- 39.Houseman AE, Accomando PW, Koestler CD, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 2016;11:354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houseman EA, Molitor J, Marsit CJ Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 2014;30: 1431–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houseman EA, Kile ML, Christiani DC, et al. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 2016;17:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark SJ, Lee HJ, Smallwood SA, et al. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol 2016;17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herceg Z, Hernandez-Vargas H New concepts of old epigenetic phenomena and their implications for selecting specific cell populations for epigenomic research. Epigenomics 2011;12:383–6. [DOI] [PubMed] [Google Scholar]
- 45.Hitchins MP. Constitutional epimutation as a mechanism for cancer causality and heritability? Nat Rev Cancer 2015;15:625–34. [DOI] [PubMed] [Google Scholar]
- 46.Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011;9:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghantous A, Hernandez-Vargas H, Byrnes G, et al. Characterising the epigenome as a key component of the fetal exposome in evaluating in utero exposures and childhood cancer risk. Mutagenesis 2015;30:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonseth S, Roy R, Houseman EA, et al. Periconceptional folate consumption is associated with neonatal DNA methylation modifications in neural crest regulatory and cancer development genes. Epigenetics 2015;10:1166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stjernfeldt M, Berglund K, Lindsten J, et al. Maternal smoking during pregnancy and risk of childhood cancer. Lancet 1986;1:1350–2. [DOI] [PubMed] [Google Scholar]
- 50.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013;14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Q, Wagner W Epigenetic aging signatures are coherently modified in cancer. PLoS Genet 2015;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Issa JP Aging and epigenetic drift: a vicious cycle. J Clin Invest 2014;124:24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ambatipudi S, Horvath S, Perrier F, et al. DNA methylome analysis identifies accelerated epigenetic aging associated with postmenopausal breast cancer susceptibility. Eur J Cancer 2017;75: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones A, Teschendorff AE, Li Q, et al. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. PLoS Med 2013;10:e1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith TM, Guyton ZK, Gibbons FC, et al. Key characteristics of carcinogens as a basis for organizing data on mechanisms of carcinogenesis. Environ Health Perspect 2016;124:713–21. [DOI] [PMC free article] [PubMed] [Google Scholar]


