Abstract
Introduction
Heat shock protein 90 (HSP90) regulates protein homeostasis in eukaryotes. As a ‘professional interactor’, HSP90 binds to and chaperones many proteins and has both housekeeping and disease-related functions but its regulation remains in part elusive. HSP90 complexes are a target for therapy, notably against cancer, and several inhibitors are currently in clinical trials. Proteomic studies have revealed the vast interaction network of HSP90 and, in doing so, the extent of cellular processes the chaperone takes part in, especially in yeast and human cells. Furthermore, small-molecule inhibitors were used to probe the global impact of its inhibition on the proteome.
Areas covered
We review here recent HSP90-related interactomics and total proteome studies and their relevance for research on cancer, neurodegenerative and pathogen diseases.
Expert commentary
Proteomics experiments are our best chance to identify the context-dependent global proteome of HSP90 and thus uncover and understand its disease-specific biology. However, understanding the complexity of HSP90 will require multiple complementary, quantitative approaches and novel bioinformatics to translate interactions into ordered functional networks and pathways. Developing therapies will necessitate more knowledge on HSP90 complexes and networks with disease relevance and on total proteome changes induced by their perturbation. Most work has been done in cancer, thus a lot remains to be done in the context of other diseases.
Keywords: HSP90 interactome, HSP90 networks, human disease, chaperones, epichaperome, cancer, proteomics, chaperome
1. Introduction
Heat shock protein 90 (HSP90) is a small family of 80–90 kDa chaperones, highly conserved across species and almost ubiquitously expressed. The general structure comprises an N-terminal ATPase domain with an ATP-binding pocket adopting the so-called Bergerat fold, followed by a flexible charged linker region connecting to a middle domain that is mainly dedicated to binding to other proteins, and a C-terminal domain which mediates dimerization. HSP90 proteins also possess at their C-terminus a MEEVD motif that enables binding of tetratricopeptide (TPR) domain containing proteins [1,2].
HSP90 has been extensively studied in yeast, where it exists as two isoforms: the constitutively expressed Hsc82 and the stress-inducible isoform Hsp82. Their homologs in mammalians are, respectively, HSP90β and HSP90α which can be found in the cytoplasm, nucleus as well as on the cell surface and extracellular space (for HSP90α [3]). TRAP1 and GRP94 (also called HSP90B1 or endoplasmin) are the mammalian mitochondrial and endoplasmic reticulum family members, respectively [4]. In this review, we will cover exclusively the cytosolic/nuclear HSP90 proteins, as they are much better characterized than the organellar paralogs. We will thus refer collectively to HSP90β and HSP90α as ‘HSP90’ unless specified.
Three decades of work have revealed that HSP90 is a master regulator of protein folding and homeostasis (‘proteostasis’) in the eukaryotic cell. HSP90 interacts with hundreds or possibly thousands of proteins and is involved in the maturation, stabilization, and regulation of specific classes of molecules. Its folding activity is connected with a complex ATP-driven cycle characterized by large conformational changes, which correlate with binding and dissociation of interactors [5]. HSP90 interactors are classified in three major categories: other chaperones, co-chaperones and cofactors, and clients [6]. Clients form the most diverse and numerous categories of interactors. Clients interact with HSP90 to achieve proper folding, to reach an activated state, to be transported to a cellular location, or to assemble in oligomeric structures, and this class includes proteins such as kinases, transcription factors (TFs), or steroid receptors, among others [7–9]. Chaperones and co-chaperones form various complexes with HSP90 to assist client function. Given the diversity of the functions of HSP90 clients, HSP90 emerged as a major player in many diverse cellular processes, such as signal transduction, proteostasis, DNA repair, stress response, and protein trafficking [10]. In addition to these roles, and under conditions of cellular stress, such as it occurs during disease initiation and progression, HSP90 may take on additional, context-dependent functions [11]. Evidence is building up that to accommodate such functions, the HSP90 chaperone becomes biochemically altered, and that certain small molecules, but not all, may be able to discriminate between the disease-associated roles of HSP90 and its essential housekeeping functions [12–17]. When one considers the potential context- and disease phenotype-dependent roles of HSP90, and the complex interactome of this protein, it appears just natural to use proteomics tools to probe the functions of this multitasking protein family.
This review aims first to summarize the recent proteomic work done on HSP90 ‘interactomics’ and second to discuss its therapeutic relevance and insights for cancer, neurodegenerative disorders, and a few bacterial and viral pathogens. Next, we will review the literature describing the impact of HSP90 inhibition on the proteome and post-translational modifications (PTMs) at proteome level, also in relation with the diseases cited above. We will finally discuss and conclude this review by highlighting the major conclusions from recent proteomic studies and how this field can contribute further to elucidate HSP90 and its functions in disease.
2. HSP90 interactomics
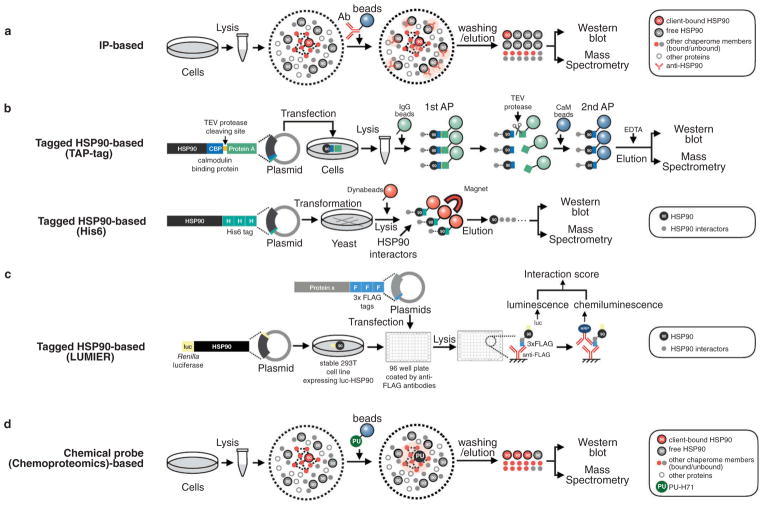
The need to define broad repertoires of proteins HSP90 interacts with has led to the development of several tools, each with their own advantages and drawbacks. A comprehensive review of earlier studies has been done in 2012 by Hartson and Matts [18]. The main tools available today comprise not only affinity pulldowns coupled to unbiased mass spectrometry-based proteomics (AP-MS), LUminescence-based Mammalian IntERactome (LUMIER) technology [19] but also two hybrid screens and chemical genetic interactions analysis. Various types of AP have been employed, from classical immunoprecipitation (IP), to affinity purification with bead-immobilized inhibitors, to tagged HSP90 capture, and have been used in combination with both MS and LUMIER (Figure 1).
Figure 1.
Strategies to interrogate the HSP90 interactome.
Technically, untargeted AP-MS approaches have the advantage to look at all proteins without any bias. The associated drawbacks are that low-abundance proteins will be poorly covered and that sample preparation conditions (e.g. lysis buffers) will impact the results, leading for example to loss of low-affinity interactors during washing steps. LUMIER overcomes some of these limitations as the method is more sensitive. Also, the nature of the reporter (photon) allows for a more accurate quantification than what can be achieved by MS. However, LUMIER needs a prey construct and a library of bait proteins and thus requires significant preliminary work and cannot be considered an unbiased technique. Finally, it should be remembered that neither technique can easily discriminate between direct (first order) and indirect (second and higher order) interactors that are detected as part of a larger complex.
Studying highly abundant, ubiquitous, and dynamic proteins such as HSP90α/β presents some specific challenges. First, as HSP90 is involved in stress responses, the growth and physiological state of the cells used can heavily impact the interactome, and such parameters are easily influenced by the experimental conditions. Second, subcellular fractions of the different isoforms are also a variable and indeed it has been shown that distinct interaction networks of HSP90 exist in particular cell lines [15]. Also, a range of conformers and complexes containing HSP90 exists, which may display preferential binding to some affinity tools. While such events can be by themselves biologically highly significant (discussed below), they can greatly complicate result interpretation. The possibility that, for example, the lysis method used privileges a certain complex, or that an antibody is specific for a given conformation, should be taken into account.
Over the last decade, the online resources of Didier Picard’s lab, notably the interactome database (HSP90Int.db), compiled the results of many types of interaction studies and have been of great value for anyone interested in HSP90 interactions (www.hsp90.org; www.picard.ch [20]). For example, based on the data in HSP90Int.db, Zuehlke et al. [21] compiled a comparison of the HSP90α versus HSP90β interactomes. Other extensive reviews on HSP90 including interactors are those of Refs. [10,22].
2.1. Recent HSP90 interactome studies
2.1.1. IP-based interactomics
IP is heavily dependent on the properties of the antibody used. One major issue in the case of HSP90 is if the antibody recognizes any HSP90 or binds preferentially to a specific conformation or a specific HSP90-containing complex, and where the epitope lies. Unfortunately, many antibodies for HSP90 IP are not well characterized from this point of view and this is an important limitation. For these and other reasons, investigators now probably prefer other capture methods such as tandem affinity purification (TAP) and LUMIER. While antibodies remain widely used for IPs followed by Western blot, we were unable to find significant interactomics studies using antibodies as a capture mean after 2012. Nevertheless, between 2005 and 2010, several studies based on antibodies as affinity tools and MS for identification were performed and helped to define the first set of core HSP90 interactors and co-chaperones [23–26] (Figure 1(a)). These were reviewed in 2012 [18].
2.1.2. Tagged HSP90-based interactomics
Various affinity tags have been used on HSP90 for interaction studies: TAP-tag, His-tag, and FLAG-tag (Figure 1(b)). Studies using these techniques are still growing in number, and recent ones benefit from improvements in the protein identification techniques, namely higher speed and sensitivity of MS instrumentation.
The group of Walid Houry did some early work [27,28] in yeast using TAP along with MS identification together with genetic screens, to map the physical and genomic interactome. Gano et al. [29] also used TAP with MS identification to study the effect of ligand binding on HSP90α interactions. This work is unique among AP-MS studies in that it included N- and C-terminal tagging and affinity purification of HSP90 bound to ATP, ADP, and inhibitors. The data obtained indicated specificity of binding of several co-chaperones for either the ATP or the ADP form, corroborating other studies [30–32] and providing new mechanistic insights.
Truman et al. [16] used AP-MS to study in yeast the Hsp82 interactome during DNA damage. They identified 147 interacting proteins enriched for cytoplasmic translation, nuclear transport, and response to starvation. DNA damage by methyl methanesulfonate treatment resulted in 70% of interactions unchanged, 2% increased, while the rest decreased. Among the few increased interactors, Rnr4 was found to require Hsp82 for function, and inhibition of Hsp82 resulted in decreased Rnr4 levels, suggesting it is a client. The human homolog of Rnr4, RRM2, is a known drug target in cancer therapy, and its status as an HSP90 client was then confirmed in the MCF-7 human breast cancer cells. HSP90 inhibition sensitized the cells to RRM2 inhibition, suggesting HSP90–RRM2 combinatorial inhibition as a new strategy for cancer treatment.
More recently, groundbreaking work was carried out by Taipale and colleagues [33,34]. They introduced a modified, quantitative version of the LUMIER assay (Figure 1(c)) to screen the interaction between 314 kinases, 843 TFs, 372 E3 ligase clones, and HSP90β in human 293T cells. They found that HSP90 interacted with >60% of kinases, <7% of TFs, and 31% of E3 ligases. Moreover, by using LUMIER, authors were able to experimentally validate that CDC37 acts as a co-chaperone adapter for most protein kinases. Precise determinants of kinase recognition by CDC37 could not be identified but several lines of evidence pointed toward intrinsic kinase domain stability as the major factor. Taipale et al. also confirmed that, upon HSP90 inhibition, HSP90–kinases interactions are globally reduced, and among the kinases that dissociate from HSP90 by treatment, some are degraded while others aggregate. Regarding the E3 ligases, those with Kelch or WD40 domains were found to bind to HSP90β in high proportions. Hence, this work provides not only identity of HSP90 interactors, but also mechanistic insights by which HSP90 recognizes its clients and knowledge on how HSP90 inhibition affects these interactions.
Taipale and coworkers further extended the study by integrating 54 additional proteins, mostly not only HSP90 co-chaperones but also HSP70 and some of its cofactors [34]. Here, they assayed a large set of interactions by combining both LUMIER and AP-MS techniques, to obtain an extensive protein–protein interaction (PPI) network. The latter appeared to consist of two major subnetworks, centered respectively around HSP90 and HSP70, and a bridge group of proteins connecting the subnetworks. Results revealed that some chaperones were more specific for their clients than others, that interactions of HSP90β correlated with interactions of HSP90α, and that treatment with the HSP90 inhibitor ganetespib resulted in decreased interaction of HSP90 with 46% of the identified proteins.
2.1.3. HSP90 interactomics by capture with immobilized inhibitors
Small molecules, i.e. chemical probes, have been used to probe HSP90 function in a number of studies (reviewed by Shrestha et al. [35]). Most often, they are used not only to inhibit HSP90 activity but they can also serve as capturing agents. Similarly to HSP90 antibodies, the use of these agents for interactomics investigations is determined by the inhibitor’s preference or lack of for a client protein-bound HSP90 conformation. For example, Tsaytler et al. [25] used biotinylated geldanamycin (GA) and streptavidin beads to capture HSP90 and its binders, but the pull-down efficacy was low and in turn, the identified interactome, was limited to abundant proteins. An improvement in the identified interactomes came upon the use of immobilized inhibitors that enrich in the active, client-protein bound, HSP90 (Figure 1(d)). One such inhibitor is PU-H71, and chemical probes using immobilized PU-H71 have been used in a variety of cancer interactome investigations, both in non-biased, MS-based inquiries [14,15,36–38], and in biased, interactome validation studies by Western blot [35,39–42], as we detail below.
Use of similar chemical tools also led to important discoveries and some debates in the cancer-related HSP90 field, the starting point being work by Kamal et al. in 2003 [13] supporting the existence of a unique high-affinity conformation of HSP90 in cancer cells.
Moulick et al., in 2011 [14], used repeated pulldowns with PU-H71- and GA-conjugated beads to show that these beads could not deplete HSP90 in K562 chronic myeloid leukemia cells and thus bind only a specific fraction of the protein. This was confirmed by Beebe and colleagues later [43]. The authors also showed that the fraction of HSP90 binding to PU-H71 beads was enriched in several co-chaperones. Crucially, MS analysis of PU-H71 beads pulldowns revealed a fraction highly enriched in proteins involved in key oncogenic signaling pathways such as the PI3K–AKT–mTOR, JAK–STAT, and Raf–MAPK pathways. Further forays into the interactome revealed the mechanism by which HSP90 may enhance the activity of such pathways. For example, HSP90 enhanced STAT5 signaling by binding to and influencing the conformation of STAT5 toward that facilitating phosphorylation and by maintaining STAT5 in an active conformation directly within STAT5-containing transcriptional complexes. The results suggested that, more than just folding individual clients, a fraction of HSP90 may act to stabilize and maintain the activity of entire oligomeric ‘signalosomes’ and transcriptional complexes.
More recently, Rodina and coworkers [15] used PU-H71 immobilized on beads and MS to study the HSP90 interactome in a large set of tumor cell lines. In addition to identifying distinct interactomes across the several cell types, they showed that tumors could be classified into two subtypes. These were differentiated by the connectivity (type 1) or the lack of (type 2) between the major chaperone machineries, HSP90 and HSP70, and also by their native isoelectric focusing signature, where type 1, but not type 2, contained HSP90 species with an isoelectric point (pI) >4.9. Bioinformatics analysis, validated by numerous lines of biochemical analyses, revealed that type 1 cells were enriched for networks with multiple connections between the HSP90 and the HSP70 systems compared to type 2 cells (the authors termed these intricate networks the epichaperome). These findings, discussed more in detail below in relationship with cancer therapy, may help to explain the conflicting results obtained on the affinity and activity of HSP90 inhibitors in cancer versus normal cells. They also provide a first characterization of a fraction of HSP90 with distinct, biologically relevant properties.
2.2. Therapeutically relevant HSP90 interactomics studies
2.2.1. Cancer
The implication of HSP90 (and chaperones in general) in tumorigenesis is based on a number of facts: (1) HSP90 is frequently (though not always) upregulated in tumors, (2) many (wild-type or mutated) oncogenes and growth-promoting proteins are HSP90 clients, and (3) the reasoning that cancer cells are often subjected to various types of stress and therefore need stress response proteins to survive and grow.
Studies of HSP90’s role and function progressively identified the chaperone as a potential drug target in cancer cells. The literature on HSP90 as a target in cancer is abundant [9,11,44–51]. Several inhibitors have been or are tested in clinical trials, with mixed results so far. Clearly, there is an immediate need to understand better the function(s) and biochemical nature of HSP90 in neoplastic cells and determine to what extent this protein is different, biochemically and functionally, from normal cells.
The relevance of proteomic interrogation of HSP90 in cancer originated not only from these considerations but also from the early work of Kamal et al. [13] who identified a distinct HSP90 fraction in cancer cells, which binds inhibitors with higher affinity and co-precipitates more co-chaperones than in normal cells, suggesting a higher degree of interactions for this HSP90 fraction in cancer. Though somewhat controversial, this critical discovery calls for the need to characterize the specific interactome of HSP90 in cancer and to find the determinants for low- versus high-affinity HSP90-inhibitor binding. The last major contributions on this point have been the work of Moulick et al. in 2011 [14] and Rodina et al. in 2016 [15] that established PU-H71 inhibitor AP as a mean to study a defined, cancer-enriched HSP90 complexes. Notably, the classification [15] of tumor cells into either type 1 cells that show a dramatic decrease in cell viability upon HSP90 inhibition versus type 2 cells, which stop proliferating but do not undergo apoptosis, could provide a first framework to try and explain data from clinical trials. Furthermore, the same study pointed to MYC transcriptional activity as a potential signature of a type 1 phenotype, and indeed MYC knockdown in type 1 cells transformed them in type 2 ones, and thus desensitized them to PU-H71 inhibition, while MYC expression in type 2 transformed them in type 1; the authors thus conclude that MYC could be one main trigger responsible for the formation of the epichaperome. Further on, they investigated the epichaperome in human breast cancer clinical samples and also found that sensitivity to inhibition correlated with abundance of type1-like HSP90 species.
Using similar tools, Caldas-Lopes and colleagues [52] used PU-H71 inhibitor for treatment and for capture to study the effect of HSP90 inhibition in triple-negative breast cancer (TNBC) cells. PU-H71 induced downregulation of proteins involved in the Ras–Raf–MAPK pathway, degradation of activated AKT and Bcl-xL, and inhibition of activated NF-κB, AKT, ERK2, PKC, and TYK2. Several oncoproteins known to drive TNBCs were captured by PU-H71 beads, suggesting these are HSP90 clients, e.g. Bcl-xL and AKT. Downregulation of these proteins alone is sufficient to induce apoptosis in TNBCs, and HSP90 inhibition led to their degradation. As HSP90 chaperones these two proteins and therefore participates in their regulation, this work hints at TNBC sensitization to Bcl-xL or AKT inhibitors through HSP90 inhibition.
In 2015, Zong et al. [40] used inhibitor capture and biochemical analysis techniques to study HSP90 interactors in acute myeloid leukemia (AML) and AML models in mice. The JAK–STAT and PI3K–Akt–mTOR signaling pathways are crucial in AMLs, and some of their components were found bound to HSP90. Interestingly, the degree of activation of these pathways and sensitivity to inhibition correlated well. STAT5 activation sensitized cells, and additional activation of AKT led to increased cell death upon drug treatment. Expression of FLT3 (an activator of both JAK–STAT and PI3K–Akt pathways) in low-sensitivity cells conferred a growth benefit but increased sensitivity to inhibition. Since many components of these pathways are HSP90 clients, and that the higher their activity the higher the need for HSP90, the authors suggest that the activated state of these pathways and the cell’s reliance on them is the cause of sensitivity to HSP90 inhibition. In other words, it is the activity of signaling networks, rather than of single signaling molecule alone, that accounts for cellular dependence on HSP90.
Again with similar tools, Goldstein et al. [36] showed that the inhibitor-binding HSP90 fraction in diffuse large B-cell lymphoma (DLBCLs) OCI-Ly1 and OCI-Ly7 cells is enriched for proteins implicated in the B-cell receptor (BCR) signaling pathway. HSP90 is additionally involved in the signaling by facilitating phosphorylation of BCR signalosome components such as SYK or BTK. Treatment with PU-H71 decreased BCR signaling, calcium flux, NF-κB signaling and led to growth arrest. Combining the treatment with ibrutinib, a BTK inhibitor, was more effective than either drug alone, and this was also observed in some DLBCLs clinical samples.
Using the same technique, Culjkovic-Kraljacic et al. [53] carried on this work in OCI-Ly1 and OCI-Ly18 DLBCLs. The authors show that HSP90 modulates the activity of EIF4E, which is pulled down along with HSP90 and, as EIF4E regulates mRNA export and translation of various genes, thus controls the posttranscriptional fate of crucial mRNA transcripts, among which the oncogenes BCL2, BCL6, and MYC. Interestingly, the mRNA encoding HSPA6, a member of the HSP70 family, is found to be a target of EIF4E as well. Inhibition of EIF4E with ribavirin impairs tumor growth decreases the cytosolic to nuclear ratio of BCL2, BCL6, MYC and even decreases mRNA levels of HSPA6. As HSP70 often drives resistance to HSP90 inhibitors, combined inhibition of EIF4E and HSP90 may improve the effectiveness of the therapy and, indeed, their results in mice support this strategy, since HSP70 export and translation was reduced under combined treatment.
2.2.2. Neurodegenerative disorders
Neurodegenerative disorders are essentially proteinopathies. As a chaperone, HSP90 plays an important role in proteostasis, and logically the role of HSP90 in such diseases has been the focus of several studies [54–57], though mostly not using proteomics techniques. For example, HSP90 interacts with both tau and the hyper-phosphorylated proteoform of tau, which plays a key role in Alzheimer’s disease (AD). Also, known tau kinases GSK3β, CDK5, and MARK2 are HSP90 clients. Treatment with an HSP90 inhibitor resulted in decreased levels of phosphorylated tau, and in induction of HSP70 which binds aberrant tau for ubiquitination by CHIP, providing a potential strategy for the treatment of AD [58].
In 2010, Riedel et al. [59] showed that treatment of the OLN-93 cell line expressing the A53T α-synuclein mutant with an HSP90 inhibitor reduced aggregation of the mutant protein. Combined inhibition of HSP90 and lysosomal degradation pathways restored aggregate formation. This, together with an observed increase in LC3-II, indicated that A53T α-synuclein is degraded by autophagy. Here, a proteomic untargeted approach would provide more insights on broader effects of 17-AAG in this context and, using a targeted approach, provide more information on the changes associated with delivery of A53T α-synuclein to the autophagosomes.
Gunawardana and coworkers [60] used tau-targeted affinity capture after mild in vivo cross-linking to probe the tau interactome in a human neuroblastoma SH-SY5Y cell-line model. Interestingly, several heat shock proteins were identified by tandem MS, including HSP90. Repeating the experiment with the P301L tau mutant resulted in altered amounts of interacting HSP90 and heat shock proteins, confirming a role of the C-terminal region of tau in interacting with these proteins that was previously suggested [61]. Since HSP90 chaperones kinases that phosphorylate tau, it would be very interesting in such a set-up to study effects of HSP90 inhibition on the tau interactome.
Recently, Shelton et al. [62] specifically studied the interaction of HSP90α and P301L tau with Aha1, p23, CDC37, FKBP4, and FKPB5 in the context of tau fibril formation. Their results show that only Aha1 enhanced the fibril formation in vitro, and that it required ATP. In mice, overexpression of Aha1 significantly increased insoluble sarkosyl-tau levels but not insoluble phosphorylated tau. The toxic T22-tau oligomer levels were also increased, and mice overexpressing Aha1 had altered memory. KU-177, a small molecule inhibiting Aha1-HSP90α interaction (but not HSP90α activity), reduced tau fibril formation while increasing soluble phosphorylated tau levels in vitro and in cultured HEK-P301L cells. This study highlights the specific function HSP90α can gain through its interactions with co-chaperones and supports a therapeutic approach aiming at inhibiting such specific functions by targeting particular HSP90 complexes or interactions.
Overall, interactome studies by proteomics techniques could bring a major contribution to the molecular characterization of HSP90’s role in neurodegenerative diseases, but much remains to be done.
2.2.3. Pathogens
Pathogens such as viruses, bacteria, fungi, and parasites hijack the biosynthetic machinery of the host to expand their population. Considering the structural as well as functional intricacies of microbial proteins, it is not surprising that they, like cellular proteins, rely on HSP90 for their folding and function. Although proteomic approaches are a small fraction of the work on this subject, they have generated valuable data. Geller et al. [63] reviewed in 2012 the literature on HSP90 in viral replication, its viral clients, and the possibility to use HSP90 inhibitors as broad antiviral drugs; therefore, in this section, we will only focus on more recent findings.
In 2013, Nayar and colleagues studied the HSP90 interactome in BC3 primary effusion lymphoma (PEL) cells infected with either Epstein–Barr virus or Kaposi sarcoma-associated herpes virus (KHSV) using inhibitor (PU-H71)-capture [37]. Pathway analysis of the interactome revealed enrichment of proteins involved in NF-κB activation by viruses, apoptosis, and autophagy. The viral oncoprotein vFLIP, too, but not the endogenous cFLIP, was found in HSP90 complexes captured by the inhibitor. vFLIP has been shown to be required for KHSV-infected PEL cells survival and antiapoptotic signaling induced by the NF-κB pathway. As the TF for vFLIP, SP1, was also an HSP90 client, treatment with PU-H71 led to reduced vFLIP mRNA levels, vFLIP degradation, and reduced NF-κB activity. This study also used the HSP90 interactome to derive potential combinatorial strategies for HSP90 inhibitors, and as such it found that dual inhibition of BCL2-antiapoptotic proteins, using obatoclax, and of HSP90, using PU-H71, to have synergistic effects on PEL cells.
In 2014, Nuss et al. [64] purified rift valley fever virus (RVFV) particles and subjected them to proteomic analysis. Chaperones HSP90α, HSP90β, HSPA5, HSPA8, CCT2, and CCT6A were found to be enriched. Interestingly, siRNA treatment against the HSP90β isoform reduced RVFV infection, while treatment against the HSP90α isoform had no perceptible effect. One however cannot exclude the participation of the HSP90α isoform, as often, phenotypes upon dowregulation of one paralog is masked or compensated by the upregulation of the other, as has been reported [15,65]. Inhibition with 17-AAG at various time points after Vero cells infection with RVFV unveiled early stage involvement of HSP90, as infection was reduced by 17-AAG 2–4 h postinfection but not after 14 h.
Using a polyclonal serum anti-chikungunya virus nsP3 and nsP4 and anti-HSP90 antibodies, Rathore et al. [66] showed that nsP3 interacts with HSP90β and nsP4 with both isoforms. HEK293T cells infected with chikungunya virus had reduced viral RNA and protein levels in a dose-dependent manner when treated with GA. siRNA targeted at HSP90α only had similar effects, suggesting specific roles in viral replication for the two isoforms.
L-protein is part of the RNA-dependent RNA polymerase complex in the human respiratory syncytial virus, and tagging this subunit with EGFP for anti-GFP IP allowed Munday and coworkers [67] to identify L-protein interactors, among which were STIP1 (also known as HOP), HSP90α, DNAJA1 (HSP40 family), and HSPA1B (HSP70 family). HSP90 inhibition with 17-AAG resulted in decreased L-protein function and stability, without affecting the P-protein subunit of the polymerase complex. In this case and for the two precedent studies presented here, differences are observed in interactions between the HSP90 isoforms, and this calls for a clarification of the different functions of HSP90α versus HSP90β related to viral replication.
Following a riboproteomics experiment in the murine norovirus (MNV), which showed that HSP90 interacts with the 5′ and 3′ extremities of the MNV-1 genome [68], a focus on HSP90 interactors in MNV and human norovirus (HuNoV) was provided by Vashist and colleagues in 2015 [69]. Using recombinant His-tagged HSP90 and LUMIER, the authors proved that the capsid protein VP1 is an HSP90 client in both MNV and HuNoV and demonstrate that HSP90 inhibition results in reduced VP1 levels and MNV-1 infection in cell culture and in mice.
Considering the number of viruses and pathogens whose survival relies on HSP90 and the associated therapeutic potential, compared to the tremendous work carried out in oncology, it may be that this field is unjustifiably left behind.
3. Global impact of HSP90 inhibition on the proteome
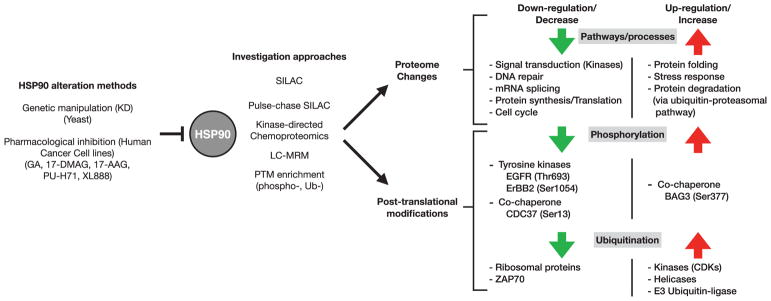
Analyzing the consequences on the proteome of reducing/removing HSP90 activity is another obvious way of assessing its physiological role. We will cover here only proteomics studies characterizing the global impact of HSP90 reduction or inhibition on the proteome and on its PTM status (Figure 2).
Figure 2.
Large scale methods to assess the global impact of HSP90 inhibition on the proteome and its post-translations modification status.
3.1. Large-scale proteome alterations following HSP90 inhibition
The presence of two functionally similar isoforms and the lethality of total KO’s has made it difficult to perform experiments based on genetic manipulations, with the exception of yeast systems. In 2014, Gopinath and coworkers [70] assayed the effects on the yeast proteome of an inducible knockdown of Hsc82 (in an Hsp82−/− background) using SILAC and MS along with transcriptomic analysis. A total of 3561 proteins were identified, of which 904 had their levels change more than 1.5-fold upon doxycycline treatment. The mRNA levels of 66% of these 904 misregulated proteins remained unaltered, indicating that the degradation machinery plays an active role under stress. Downregulated proteins were enriched for kinases and proteins involved in DNA repair, while upregulated ones were enriched for proteins involved in stress response, protein folding, and stabilization. Interestingly, downregulated proteins were also enriched for proteins with human homologs, proteins encoded by essential genes, proteins with high connectivity in the yeast PPI network, which highlights HSP90 as a major regulatory hub.
Due to the difficulty of performing genetic manipulations of HSP90 and also due to their intrinsic relevance for therapy, most experiments in mammalian cells used chemical inhibitors to block HSP90 and were carried out in cancer cell lines. So far, all studies were done using inhibitors that act by inserting into the ATP-binding pocket of HSP90 located in its N-terminal domain.
Sharma et al. [71] quantified more than 6000 proteins in HeLa cells and showed that HSP90 inhibition by 17-DMAG led to upregulation of chaperones (among which HSP40s, HSP70s, and both cytosolic HSP90 isoforms), components of the proteasome pathway, and more generally proteins involved in protein folding and unfolded protein binding. Not surprisingly, a substantial fraction of the observed upregulated proteins is downstream of HSF1, the master transcription factor activated by heat shock and many stress signals. HSP90 inhibition caused also downregulation of clients, including kinases, some of which play a role in RAS or AKT signaling pathways, and proteins involved in DNA repair and in sphingolipid metabolism.
Wu and coworkers [72] studied the effect of GA and PU-H71 treatment in K562, MDA-MB-231, Colo205, and Cal27 cancer cells. Of all proteins quantified (more than 6000), about 1600 had modified levels upon HSP90 inhibition and effects of GA and PU-H71 correlated well. Downregulation of 52–65% of the proteins that had reduced levels was dependent on the cell line, indicating differential effects of HSP90 inhibition. Kinases showed little discrepancy with 89–92% being downregulated, thus constituting the main class of downregulated proteins. Changes in half-lives were also measured and revealed overall shorter half-lives for kinases, predicting that kinase-mediated signaling pathways will be the first affected by HSP90 inhibition. ERK and MAPK signaling pathways were downregulated across all cell lines, while the mTOR pathway was more affected in Colo205 than in MDA-MB-213 cells. As expected, upregulated proteins mainly included chaperones.
Haupt et al. [73] compared the effect of HSP90 inhibition by GA in Hs68 fibroblast cells (non-transformed) and in SW480, U2OS, and A549 tumor cells specifically on the kinome, using kinases-targeted affinity beads. The extent of kinases degradation upon drug treatment varied from one cell type to another, both in time and space: about 70% were downregulated in Hs68 cells whereas it ranged from 80% to 90% for cancer cells, and downregulation was faster in Hs68 and SW480 cells than in U2OS and A549 cells. Kinases involved in the MAPK or TGF-β signaling pathways were less affected by the drug in Hs68 cells than in the cancer cells, but no such difference was observed for kinases involved in cell-cycle regulation. This work is also the first to report downregulation of BMP receptors, some of which fulfill proliferation promoting function in breast cancer or proliferation suppressive function in prostate cancer. Interestingly, simultaneous treatment with GA and the proteasomal inhibitor MG132 restored the levels of 86% of all kinases that are affected by GA alone in cancer cells, but only three kinases in Hs68 cells, indicating that downregulation in the latter is not dependent on this degradation pathway.
In 2013, Fierro-Monti and colleagues developed a method which allows to quantitate protein decay rate and synthesis rate in addition to protein levels, termed pulse-chase SILAC and described in Ref. [74], in order to study how HSP90 inhibition with GA affects the proteome in T cells [75]. Seventeen percent of all proteins were impacted by GA, with upregulation of proteins involved in protein folding and stress response, including HSP90 and co-chaperones AHSA1, CDC37, STIP1, and FKBP4, but intriguingly not AARSD or FKBP5, suggesting that GA may remodel the HSP90 chaperone complexes. Downregulated proteins comprised kinases and proteins involved in protein synthesis. More complex effects were observed for components of the ubiquitination machinery, with initial upregulation followed by downregulation. More generally, a strong reduction in the global synthesis rate and a strong increase in decay rate were observed, with the median protein half-life dropping from 55.9 to 32 h. Overall, changes in synthesis rates appeared to determine final changes in protein levels except for kinases and other HSP90 clients, for which decay rates played a major role. The changes observed for other groups of proteins appeared complex and multifactorial, e.g. chaperones had strongly not only increased synthesis (and mRNA levels) but also increased decay rates. Interestingly, some oncoproteins (e.g. Rab proteins) were upregulated and some tumor suppressors (e.g. retinoblastoma protein) were downregulated, highlighting some complex effects of HSP90 inhibition.
From a different angle, aiming at developing a method for routine evaluation of personalized treatments against melanoma, Rebecca et al. [76] studied the effect of HSP90 inhibition by targeted proteomics measuring specifically about 80 signaling proteins and a few heat shock proteins. HSP90 inhibition resulted in increase of HSP71 (an inducible cytosolic HSP70 isoform) and both HSP90 isoforms levels in a NRAS mutant cell line. Once again, kinases were downregulated, with tyrosine kinases being more affected. In xenografts of 1205LuR melanoma cells (vemurafenib, B-RAF inhibitor, resistant), HSP90 inhibition decreased the levels of proteins involved in the PI3K/AKT/mTOR signaling pathway such as mTOR, AKT1, and AKT2. While not a global study, this work provides a proof of concept that targeted proteomics can be used to assess global impacts of HSP90 inhibition on signa.ling pathways previously identified as essential for cancer survival.
All these studies share their main conclusions, i.e. HSP90 inhibition profoundly affects the proteome, causing depletion of clients – especially protein kinases – and activation of the integrated cellular stress response. The data also show that the changes induced are very complex and include primary (client degradation) as well as secondary, systemic effects due, among others, to cell-cycle arrest. Furthermore, the effects of HSP90 inhibitors seem to vary significantly depending on the cellular type and the drug concentrations used. Thus, while global proteomics contributed significantly to the understanding of the impact of HSP90 N-terminal domain inhibition, it also highlighted some crucial questions for cancer therapy that we will address in the discussion section.
Finally, the interest in HSP90 inhibitors for the treatment of infectious and neurodegenerative diseases calls for proteomics studies as those discussed in this section in the context of these pathologies.
While total proteome studies can be very informative, one should be aware of the challenges they present and their limitations. Indeed, it is often difficult to disentangle direct, primary effects due to HSP90 inhibition and secondary, systemic changes due e.g. to stress responses and cell-cycle arrest. For a better understanding, such studies should be complemented with determination of cell physiological parameters e.g. proliferation rate, apoptosis, and cell-cycle analysis. Also, as most often in proteomics, very low abundance proteins and membrane proteins are poorly sampled.
3.2. Proteome-wide analysis of PTMs upon HSP90 inhibition
MS is the most flexible and general tool available today for the analysis of PTMs. Multiple modifications have been identified on HSP90 itself by specific studies and as a by-product of numerous global PTM studies. Extensive data on HSP90 are compiled on public databases such as phosphosite.org which listed, as of 4th of May 2017 no less than 168 PTMs sites for HSP90β and 179 for HSP90α. The main PTMs known on HSP90 have been reviewed in detail before, together with their functional significance, when known [6,21,44,77–82]. Rather, here, we will cover the few studies assessing the impact of HSP90 inhibition on the global proteome PTMs (Figure 2).
Sharma et al. [71] analyzed, simultaneously to the total proteome, the phosphoproteome in response to HSP90 inhibition by 17-DMAG and observed a global decrease in phosphorylation levels. Thirty-four percent of all phosphopeptides were downregulated by more than twofold, whereas 6% were upregulated by the same amount. Half of the downregulated ones contained proline-directed motifs, and the authors suggest that this predominance may be due to the downregulation of cyclin-dependent kinases and reduced MAPK signaling. Since it is also known that phosphorylation levels peak at mitosis [83], and that HSP90 inhibition results in cell-cycle arrest, it cannot be excluded that the drop in phosphorylation is partially a secondary effect of the suppression of cell-cycle progression.
In a similar manner, Jin et al. [84] quantified 1150 phosphosites in DLD-1 cells treated with 17-AAG (but only for a short time, 3 h) and found only 1 upregulated phosphopeptide which belonged to BAG3, a HSP70 co-chaperone. About 10% of phosphopeptides had reduced levels, among which phosphosites CDC37 Ser13 (required for kinase binding to HSP90), ERBB2 Ser1054, and EGFR Thr693. The main hypothesis proposed for the global decrease in phosphoproteome is the downregulation of kinases or the downregulation of phosphorylated protein levels themselves. Overall, phosphoproteomics should be very informative for the functional evaluation of the effects of HSP90 inhibition, given that phosphosites of key signaling molecules can be taken as diagnostic of precise phenotypes (e.g. reduced signaling or reduced proliferation).
Other than phosphorylation, only ubiquitination has been analyzed on a proteome scale following HSP90 inhibition. Quadroni et al. [85] investigated the effects of HSP90 inhibition by GA on the ubiquitinome in Jurkat T cells and how it relates to the degradation and synthesis rates of proteins. While changes observed in proteins levels were maximal after 20 h of drug treatment, changes in lysine-linked ‘GlyGly’ modifications (diagnostic of ubiquitination post-trypsin digestion) were faster and appeared already at 2 h. The largest increase in GlyGly modification was seen for HSP90 clients, e.g. CDK1, CDK6, LCK – however, opposite unexpected effects were also observed, e.g. client ZAP70 which had reduced ubiquitination. Consistent with changes seen in total protein levels, proteins with increased GlyGly motifs were enriched for kinases and helicases. Globally, HSP90 inhibition also led to a reorganization of the ubiquitin pool with a quick depletion of K11- and K63-linked ubiquitin units along an increase in K48-linked chains, before a return to initial levels after 20 h of treatment. However, more complex and protein family-specific effects emerged from the data: ribosomal proteins had decreased GlyGly motifs along with a mild decrease in protein level, and several heat shock proteins had not only increased ubiquitination but also increased protein levels. Integrating the changes measured previously in the same system at the level of protein synthesis/decay rates, the authors suggest that globally, protein synthesis rate impacts ubiquitination levels, as a certain percentage of all newly synthesized proteins is immediately degraded [86,87]. Thus, correct understanding of the different ubiquitination patterns requires the knowledge of many parameters, as well as the state of the ubiquitin–proteasome system.
In conclusion, global PTM studies can open a window on a normally hidden cross-section of the proteome. For proteins impacted by HSP90 inhibition, this could be the best opportunity to understand the mechanism behind their changes. As any other approach, global PTM studies have some specific limitations and bottlenecks. First, it is often difficult to unambiguously assign the enzyme responsible for adding or removing a PTM mark on a given substrate site. Second, the functional impact of most PTMs on the protein carrying them is simply unknown; elucidating it is challenging and may require significant follow-up work. While providing a large amount of data, these analyses often produce only a few immediately exploitable answers, while yielding many new questions.
4. Expert commentary
In the past decade, a substantial amount of knowledge was gained through proteomic studies centered on HSP90. The number and the nature of interactors (in particular clients) discovered emphasized the numerous tasks HSP90 participates in besides protein folding. The subsequent development and use of specific inhibitors enabled researchers to reveal, through global proteomics measurements, the effects of blocking HSP90, namely client depletion, the induction of a broad stress response, a global drop in phosphorylation events together with context-specific sensitization effects. The mass of data obtained suggested numerous possible therapeutic usages of HSP90 inhibition, notably in combination with inhibitors of other pathways [16,36,52,88].
5. Five-year view
Nonetheless, a tangle of crucial, interconnected questions on the fundamental biology of HSP90 and the action of inhibitors remain at least partially unanswered and will need to be tackled in the next few years. First of all, discrepancies in the reported apparent affinities of HSP90 for N-terminal pocket inhibitors in cells, tissues (cancer vs. normal) and in vitro still remain largely unexplained, as is the issue of how the affinity varies in the presence or absence of co-chaperones [14,89,90]. This question is tightly linked to the key question of whether, where, and to what extent HSP90 is present as a distinct (more complexed or more networked?) form in cancer cells, which leads to a higher affinity of HSP90 for certain inhibitors [13]. Again, reports appear regularly, indicating that cancer and healthy cells can have HSP90 with similar affinity for some inhibitors [91]. Altogether, these seemingly discrepant reports suggest a complex situation – HSP90 is not a ‘single’ protein but rather a chameleonic chaperone whose biochemistry and function changes as dictated by its environment. In this intricate scenario, small molecule inhibitors appear as the most powerful tools to dissect functional differences. These may act as sensors of the HSP90 subspecies and their distinct thermodynamics, and over the time they spend inside the cell, may preferentially sample such HSP90s. The more the affinity of such molecules is tilted toward the ‘normal’ cell HSP90, the more likely the ‘housekeeping’ functions will also be affected at inhibitor concentrations needed to affect the ‘cancer functions’ of HSP90 and vice versa.
Indeed, pulldowns with some immobilized inhibitors were shown to capture only a fraction of cellular HSP90 in cancer cells [14]. The subsequent characterization of the ‘epichaperome’ as a high-affinity inhibitor-binding complexes of HSP90/70-linked proteins more abundant in some (but not all) cancer cells [15] offers thus a paradigm to try and explain a broad range of seemingly diverging data. Notably, proteomics analyses played a major role in characterizing the epichaperome. Still, the concept needs further, broader validation and by itself raises new questions, such as what the molecular determinants of the high- versus low-affinity binding of HSP90 to inhibitors are. One can hypothesize that either a precise cocktail of interactors/cofactors or some unique PTMs or again the targeting to a precise cellular location may trigger the formation of epichaperome complexes. The identification of such molecular determinants will be essential for both validation and further functional studies and here we anticipate that proteomics approaches will play a major role. The realization that HSP90 incorporated into epichaperomes and not HSP90 alone is needed for tumor survival may bring into question the clinical target for HSP90 agents. Is it HSP90 incorporated into the epichaperome, an entity biochemically and functionally distinct from the housekeeping, dynamic HSP90 complexes? And if yes, what are the implications for the development of such agents as therapeutics? Epichaperomes are unlikely to be unique to cancer, thus raising the question of what other diseases may use epichaperome networks to regulate their proteome under the respective pathologic conditions.
Other questions need to also be addressed to advance HSP90 as a therapeutic target. For example, what HSP90 inhibition induced downregulation of which protein or pathway components is critical for the killing of cancer cells or pathogens? To what extent does the induction of the stress response through HSF1 activation rescue cancer cells from HSP90 inhibition [92]? Proteomics analysis also showed the extent of the generated stress response, suggesting that HSP90 inhibition in cancer could be a double-edged sword if used in the wrong cellular context. As a consequence, several groups are trying to develop inhibitors that either target the C-terminal domain [93–97] or the middle domain [98] or aim at blocking interactions of HSP90 with co-chaperones, e.g. inhibiting interactions with the kinase adapter CDC37 [99–102] or inhibiting the ATPase stimulation by Aha1 [103]. The goal of these efforts is either to have inhibitors that block Hsp90 action without inducing the deleterious heat shock response and/or have more selective inhibitors with more restricted and better predictable effects. So far, the action of these C-terminal inhibitors has been mostly studied in a targeted manner, typically by assessing induction of HSP70 as marker of heat shock response and some client proteins to measure HSP90’s (loss of) folding activity. Clearly, proteome-wide measurements could provide a much more complete view of the real biological impact of these novel molecules and are thus urgently needed to compare them with the better known effects of N-terminal domain inhibitors. More in general, more studies on the impact of HSP90 inhibitors in normal cells/tissues will also be needed to assess realistically the specificity (or not) of the effects observed in cancer cells.
Overall, proteomics is probably the most suitable approach to thoroughly characterize the inhibitor binding and nonbinding fractions of HSP90 and, more generally, interactome-related questions. A number of additional studies are also needed to address other basic questions. For example, most proteomics studies so far focused on the cytosolic isoforms of HSP90; however, HSP90 inhibitors also bind the ER-located paralog, GRP94 (HSP90B1), and to a lesser extent the mitochondrial one, TRAP1 [104]. More paralog-specific inhibitors are needed to disentangle individual effects and some initial work has been done in this direction [65,105–110]. Specific interactome studies on all cellular paralogs are also needed, including some addressing explicitly the differences between HSP90α and HSP90β in the same system. Also, integrating spatial information (e.g. specify the cytosolic vs. the nuclear interactome of HSP90β, the cytosolic vs. extracellular interactome of HSP90α, etc.) will provide information essential to build a complete picture of HSP90 interaction networks and functions in cells.
To fulfill all these tasks and given the complexity and of HSP90 roles and functions, we anticipate that proteomics experiments will have to incorporate a quantitative dimension. Besides accurate measurements of relative proteins levels, experiments may also aim at measuring the stoichiometries of proteins in complexes, the occupancy of PTM sites on HSP90, and the general state of PTMs in HSP90-bound complexes. In this way, it will be possible to better evaluate the correlation of individual molecular species with the phenotypes observed in different cells and tissues.
Finally, proteomics also offer great promise for exploring the role of HSP90 in neurodegenerative diseases and pathogen-derived diseases. Here, not much has been done, so plenty of opportunities remain for exciting discoveries.
Key issues.
Considering the complexity of the HSP90 machinery, both at the biochemical and functional level, several complementary methods will be needed to fully understand its function in diseases.
One thing to keep in mind when designing interactomic experiments is to understand that the interactome of HSP90 is modulated by its environment. Disease-relevant proteomics studies on HSP90 need to be addressed in models that most closely recapture the cellular (and extra-cellular, though this becomes challenging) complement of the specific disease.
Bioinformatics methods need to be developed to understand not only individual HSP90 interactors, but rather how these interactors integrate into the complex functional networks and pathways that are cell and context-dependent.
For translational applications, a one-protein approach has failed in most part, highlighting the need for a network and interactome-approach, where the global implications of the HSP90 function over the cellular proteome are taken into account for a most judicious application of HSP90 into treatment. In practice, data on both interactome and total proteome changes upon HSP90 inhibition will be needed to understand response vs. nonresponse to drug treatment and develop suitable biomarkers.
Understanding the molecular basis for the distinct biochemical and functional nature of HSP90 in disease promises a path for the development of HSP90 agents with improved higher therapeutic index, and thus higher therapeutic potential.
HSP90 has important and validated roles in maintaining a disease phenotype in a variety of pathologies but interac-tomics studies in diseases other than cancer are scarce. In particular in pathogen-caused diseases, the role played by HSP90 could be simpler to dissect than e.g. in cancer and could thus offer an easier path to treatment.
Acknowledgments
Funding
This manuscript was supported by grants from the U.S. National Institutes for Health: R01 CA155226, R01 CA172546, P01 CA186866, P30 CA08748, P50 CA86438; Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Experimental Therapeutics Center of MSKCC, the Lymphoma Research Foundation, and the Swiss National Science Foundation: 31003A_166562.
Footnotes
Declaration of interest
The Memorial Sloan Kettering Cancer Centre holds the intellectual property rights to PU-H71 and its derivatives, and uses of such inhibitors. Samus Therapeutics Inc, of which G Chiosis has partial ownership, has licensed this portfolio. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta Mol Cell Res. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Eustace BK, Sakurai T, Stewart JK, et al. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 4.Sreedhar AS, Kalmár É, Csermely P, et al. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 5.Prodromou C. The “active life” of Hsp90 complexes. Biochim Biophys Acta Mol Cell Res. 2012;1823:614–623. doi: 10.1016/j.bbamcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 7.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 8.Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochim Biophys Acta Mol Cell Res. 2012;1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Trepel J, Mollapour M, Giaccone G, et al. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 11.Taldone T, Ochiana SO, Patel PD, et al. Selective targeting of the stress chaperome as a therapeutic strategy. Trends Pharmacol Sci. 2014;35:592–603. doi: 10.1016/j.tips.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickey CA, Kamal A, Lundgren K, et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Kamal A, Thao L, Sensintaffar J, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. Provides a first evidence for the possible biochemical difference between HSP90 in normal versus cancer cells. [DOI] [PubMed] [Google Scholar]
- 14•.Moulick K, Ahn JH, Zong H, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7:818–826. doi: 10.1038/nchembio.670. Characterizes in detail the biochemical nature of the HSP90 species isolated by PU-H71 in CML cell lines and primary specimens and demonstrates how its ability to select for active, tumor-specific HSP90 complexes may be used to investigate the complement of oncogenic proteins regulated by HSP90 in CML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Rodina A, Wang T, Yan P, et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 2016;538:397–401. doi: 10.1038/nature19807. Uses proteomics analyses to define cofactors that result in chaperome hyperconnectivity into the formation of epichaperome networks. Also, uses proteomics to discover molecular factors behind the rewiring of the HSP90 chaperome into epichaperome networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truman AW, Kristjansdottir K, Wolfgeher D, et al. Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase. J Proteomics. 2015;112:285–300. doi: 10.1016/j.jprot.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodford MR, Dunn D, Miller JB, et al. Impact of posttranslational modifications on the anticancer activity of Hsp90 Inhibitors. Adv Cancer Res. 2016;129:31–50. doi: 10.1016/bs.acr.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 18•.Hartson SD, Matts RL. Approaches for defining the Hsp90-dependent proteome. Biochim Biophys Acta Mol Cell Res. 2012;1823:656–667. doi: 10.1016/j.bbamcr.2011.08.013. Good earlier review article on proteomics approaches used to understand the biology of HSP90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrios-Rodiles M, Brown KR, Ozdamar B, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 20.Echeverría PC, Bernthaler A, Dupuis P, et al. An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One. 2011;6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuehlke AD, Beebe K, Neckers L, et al. Regulation and function of the human HSP90AA1 gene. Gene. 2015;570:8–16. doi: 10.1016/j.gene.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Silva VCH, Ramos CHI. The network interaction of the human cytosolic 90kDa heat shock protein Hsp90: a target for cancer therapeutics. J Proteomics. 2012;75:2790–2802. doi: 10.1016/j.jprot.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 23.Falsone SF, Gesslbauer B, Tirk F, et al. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579:6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Te J, Jia L, Rogers J, et al. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res. 2007;6:1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- 25.Tsaytler PA, Krijgsveld J, Goerdayal SS, et al. Novel Hsp90 partners discovered using complementary proteomic approaches. Cell Stress Chaperones. 2009;14:629–638. doi: 10.1007/s12192-009-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Heuvelman DM, Carroll JA, et al. Geldanamycin-induced PCNA degradation in isolated Hsp90 complex from cancer cells. Cancer Invest. 2010;28:635–641. doi: 10.3109/07357901003630983. [DOI] [PubMed] [Google Scholar]
- 27•.Zhao R, Davey M, Hsu YC, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. An extensive study that integrates genomic and proteomic approaches to identify the global set of HSP90 interactomes suggested the plausible involvement of HSP90 in transcriptional regulation, cell cycle, DNA processing, and cellular transport. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y, Kakihara Y, Krogan N, et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gano JJ, Simon JA. A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol Cell Proteomics. 2010;9:255–270. doi: 10.1074/mcp.M900261-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siligardi G, Hu B, Panaretou B, et al. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J Biol Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin SH, Sobott F, Yao ZP, et al. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Richter K, Buchner J. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat Struct Mol Biol. 2011;18:61–66. doi: 10.1038/nsmb.1965. [DOI] [PubMed] [Google Scholar]
- 33••.Taipale M, Krykbaeva I, Koeva M, et al. Quantitative analysis of Hsp90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. Uses LUMIER-based proteomics to investigate the basis for HSP90 regulation of kinases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Taipale M, Tucker G, Peng J, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. Uses LUMIER-based proteomics to understand how co-chaperones link HSP90 to specific cellular processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Shrestha L, Patel HJ, Chiosis G. Chemical tools to investigate mechanisms associated with HSP90 and HSP70 in Disease. Cell Chem Biol. 2016;23:158–172. doi: 10.1016/j.chembiol.2015.12.006. Good review article on chemical probes useful to investigate the functional and biochemical nature of HSP90 in diseases includes those with use in proteomics investigations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein RL, Yang SN, Taldone T, et al. Pharmacoproteomics identifies combinatorial therapy targets for diffuse large B cell lymphoma. J Clin Invest. 2015;125:4559–4571. doi: 10.1172/JCI80714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nayar U, Lu P, Goldstein RL, et al. Targeting the Hsp90-associated viral oncoproteome in gammaherpesvirus-associated malignancies. Blood. 2013;122:2837–2847. doi: 10.1182/blood-2013-01-479972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giulino-Roth L, Van Besien HJ, Dalton T, et al. Inhibition of Hsp90 suppresses PI3K/AKT/mTOR signaling and has antitumor activity in Burkitt lymphoma. Mol Cancer Ther. 2017;16:1779–1790. doi: 10.1158/1535-7163.MCT-16-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo A, Lu P, Lee J, et al. HSP90 stabilizes B-cell receptor kinases in a multi-client interactome: PU-H71 induces CLL apoptosis in a cytoprotective microenvironment. Oncogene. 2017;36:3441–3449. doi: 10.1038/onc.2016.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Zong H, Gozman A, Caldas-Lopes E, et al. A hyperactive signalosome in acute myeloid leukemia drives addiction to a tumor-specific Hsp90 species. Cell Rep. 2015;13:2159–2173. doi: 10.1016/j.celrep.2015.10.073. Indicates hyperactivity of certain signaling networks, rather than single kinase activity, to account for sensitivity to inhibition of HSP90 in AML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucine N, Marubayashi S, Bhagwat N, et al. Tumor-specific HSP90 inhibition as a therapeutic approach in JAK-mutant acute lymphoblastic leukemias. Blood. 2015;126:2479–2483. doi: 10.1182/blood-2015-03-635821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerchietti LC, Lopes EC, Yang SN, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med. 2009;15:1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beebe K, Mollapour M, Scroggins B, et al. Post-translational modification and conformational state of Heat Shock Protein 90 differentially affect binding of chemically diverse small molecule inhibitors. Oncotarget. 2013;4:1065–1074. doi: 10.18632/oncotarget.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alarcon SV, Mollapour M, Lee M-J, et al. Tumor-intrinsic and tumor-extrinsic factors impacting hsp90- targeted therapy. Curr Mol Med. 2012;12:1125–1141. doi: 10.2174/156652412803306729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrott JJ, Haystead TAJ. Hsp90, an unlikely ally in the war on cancer. Febs J. 2013;280:1381–1396. doi: 10.1111/febs.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong DS, Banerji U, Tavana B, et al. Targeting the molecular chaperone heat shock protein 90 (HSP90): lessons learned and future directions. Cancer Treat Rev. 2013;39:375–387. doi: 10.1016/j.ctrv.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Jego G, Hazoumé A, Seigneuric R, et al. Targeting heat shock proteins in cancer. Cancer Lett. 2013;332:275–285. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Jhaveri K, Ochiana SO, Dunphy MP, et al. Heat shock protein 90 inhibitors in the treatment of cancer: current status and future directions. Expert Opin Investig Drugs. 2014;23:611–628. doi: 10.1517/13543784.2014.902442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyata Y, Nakamoto H, Neckers L. The therapeutic target Hsp90 and cancer hallmarks. Curr Pharm Des. 2013;19:347–365. doi: 10.2174/138161213804143725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J, Liu T, Rios Z, et al. Heat shock proteins and cancer. Trends Pharmacol Sci. 2016;38:226–256. doi: 10.1016/j.tips.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Wong DS, Jay DG. Emerging roles of extracellular Hsp90 in cancer. Adv Cancer Res. 2016;129:141–163. doi: 10.1016/bs.acr.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Caldas-Lopes E, Cerchietti L, Ahn JH, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci U S A. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Culjkovic-Kraljacic B, Fernando TM, Marullo R, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood. 2016;127:858–868. doi: 10.1182/blood-2015-05-645069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo W, Sun W, Taldone T, et al. Heat shock protein 90 in neurode-generative diseases. Mol Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salminen A, Ojala J, Kaarniranta K, et al. Hsp90 regulates tau pathology through co-chaperone complexes in Alzheimer’s disease. Prog Neurobiol. 2011;93:99–110. doi: 10.1016/j.pneurobio.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Inda C, Bolaender A, Wang T, et al. Stressing out Hsp90 in neurotoxic proteinopathies. Curr Top Med Chem. 2016;16:2829–2838. doi: 10.2174/1568026616666160413141350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reis SD, Pinho BR, Oliveira JMA. Modulation of molecular chaperones in huntington’s Disease and other polyglutamine disorders. Mol Neurobiol. 2017;54:5829–5854. doi: 10.1007/s12035-016-0120-z. [DOI] [PubMed] [Google Scholar]
- 58.Carman A, Kishinevsky S, Koren J, III, et al. Chaperone-dependent neurodegeneration: a molecular perspective on therapeutic intervention. J Alzheimer’s Dis Park. 2013;2013 doi: 10.4172/2161-0460.S10-007. pii007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riedel M, Goldbaum O, Schwarz L, et al. 17-AAG induces cytoplasmic α-synuclein aggregate clearance by induction of autophagy. PLoS One. 2010;5:e8753. doi: 10.1371/journal.pone.0008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gunawardana CG, Mehrabian M, Wang X, et al. The human tau interactome: binding to the ribonucleoproteome, and impaired binding of the P301L mutant to chaperones and the proteasome. Mol Cell Proteomics. 2015;14:3000–3014. doi: 10.1074/mcp.M115.050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karagöz GE, Duarte AMS, Akoury E, et al. Hsp90-tau complex reveals molecular basis for specificity in chaperone action. Cell. 2014;156:963–974. doi: 10.1016/j.cell.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shelton LB, Baker JD, Zheng D, et al. Hsp90 activator Aha1 drives production of pathological tau aggregates. Proc Natl Acad Sci U S A. 2017;114:9707–9712. doi: 10.1073/pnas.1707039114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geller R, Taguwa S, Frydman J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta Mol Cell Res. 2012;1823:698–706. doi: 10.1016/j.bbamcr.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nuss JE, Kehn-Hall K, Benedict A, et al. Multi-faceted proteomic characterization of host protein complement of rift valley fever virus virions and identification of specific Heat Shock Proteins, including HSP90, as important viral host factors. PLoS One. 2014;9:e93483. doi: 10.1371/journal.pone.0093483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel PD, Yan P, Seidler PM, et al. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat Chem Biol. 2013;9:677–684. doi: 10.1038/nchembio.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rathore APS, Haystead T, Das PK, et al. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antiviral Res. 2014;103:7–16. doi: 10.1016/j.antiviral.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 67.Munday DC, Wu W, Smith N, et al. Interactome analysis of the human respiratory syncytial virus RNA polymerase complex identifies protein chaperones as important cofactors that promote L-protein stability and RNA synthesis. J Virol. 2015;89:917–930. doi: 10.1128/JVI.01783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vashist S, Urena L, Chaudhry Y, et al. Identification of RNA-Protein Interaction Networks Involved in the Norovirus Life Cycle. J Virol. 2012;86:11977–11990. doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vashist S, Urena L, Gonzalez-Hernandez MB, et al. The molecular chaperone Hsp90 is a therapeutic target for noroviruses. J Virol. 2015;89:6352–6363. doi: 10.1128/JVI.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gopinath RK, You S-T, Chien K-Y, et al. The Hsp90-dependent proteome is conserved and enriched for hub proteins with high levels of protein-protein connectivity. Genome Biol Evol. 2014;6:2851–2865. doi: 10.1093/gbe/evu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71••.Sharma K, Vabulas RM, Macek B, et al. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol Cell Proteomics. 2012;11:M111.014654. doi: 10.1074/mcp.M111.014654. The first total SILAC-based deep proteome study of the impact of HSP90 inhibition with an ansamycin compound (17-DMAG) in HeLa cells, complemented with an extensive mapping of the effects on the phosphoproteome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Wu Z, Moghaddas Gholami A, Kuster B. Systematic identification of the HSP90 regulated proteome. Mol Cell Proteomics. 2012;11:M111.016675. doi: 10.1074/mcp.M111.016675. Analysis of full proteome and kinome (by chemical affinity) changes following HSP90 inhibition by geldanamycin in four different cancer cell lines. Also included pulse-chase measurements to determine changes in protein turnover. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Haupt A, Joberty G, Bantscheff M, et al. Hsp90 inhibition differentially destabilises MAP kinase and TGF-beta signalling components in cancer cells revealed by kinase-targeted chemoproteomics. BMC Cancer. 2012;12:38. doi: 10.1186/1471-2407-12-38. Measured the changes in total proteins and protein kinases (also using kinase affinity beads) in three cancer and one nontransformed cell line. The authors used proteasome inhibition to define a list of protein kinases that are bona fide HSP90 clients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fierro-Monti I, Racle J, Hernandez C, et al. A novel pulse-chase SILAC strategy measures changes in protein decay and synthesis rates induced by perturbation of proteostasis with an Hsp90 inhibitor. PLoS One. 2013;8:e80423. doi: 10.1371/journal.pone.0080423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Fierro-Monti I, Echeverria P, Racle J, et al. Dynamic impacts of the inhibition of the molecular chaperone Hsp90 on the T-cell proteome have implications for anti-cancer therapy. PLoS One. 2013;8:e80425. doi: 10.1371/journal.pone.0080425. Mild HSP90 inhibition in a T-cell line was assessed at two time points. The results showed not only depletion of clients and oncoproteins but also a pro-survival response and depletion of some tumor suppressors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rebecca VW, Wood E, Fedorenko IV, et al. Evaluating melanoma drug response and therapeutic escape with quantitative proteomics. Mol Cell Proteomics. 2014;13:1844–1854. doi: 10.1074/mcp.M113.037424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta Mol Cell Res. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayer MP, Le Breton L. Hsp90: breaking the symmetry. Mol Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 79.Prodromou C. Regulatory mechanisms of Hsp90. Biochem Mol Biol J. 2017;3:1–13. doi: 10.21767/2471-8084.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walton-Diaz A, Khan S, Bourboulia D, et al. Contributions of co-chaperones and post-translational modifications towards Hsp90 drug sensitivity. Future Med Chem. 2013;5:1059–1071. doi: 10.4155/fmc.13.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soroka J, Wandinger SK, Mäusbacher N, et al. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 82.Woodford MR, Truman AW, Dunn DM, et al. Mps1 mediated phosphorylation of Hsp90 confers renal cell carcinoma sensitivity and selectivity to Hsp90 inhibitors. Cell Rep. 2016;14:872–884. doi: 10.1016/j.celrep.2015.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 84.Jin J, Tian R, Pasculescu A, et al. Mutational analysis of glycogen synthase kinase 3β protein kinase together with kinome-wide binding and stability studies suggests context-dependent recognition of kinases by the chaperone heat shock protein 90. Mol Cell Biol. 2016;36:1007–1018. doi: 10.1128/MCB.01045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85••.Quadroni M, Potts A, Waridel P. Hsp90 inhibition induces both protein-specific and global changes in the ubiquitinome. J Proteomics. 2015;120:215–229. doi: 10.1016/j.jprot.2015.02.020. Enrichment of modified peptides was used to measure the impact of HSP90 inhibition on the ubiquitinome in Jurkat T cells. Correlation of the data with previously acquired total proteome changes shows not only that HSP90 clients are, as expected, mostly rapidly degraded by ubiquitination but also that levels of ubiquitin modifications are impacted by many factors, notably basal synthesis rates. [DOI] [PubMed] [Google Scholar]
- 86.Kim W, Bennett EJ, Huttlin EL, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Huang W, Wu QD, Zhang M, et al. Novel Hsp90 inhibitor FW-04-806 displays potent antitumor effects in HER2-positive breast cancer cells as a single agent or in combination with lapatinib. Cancer Lett. 2015;356:862–871. doi: 10.1016/j.canlet.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 89.Gooljarsingh LT, Fernandes C, Yan K, et al. A biochemical rationale for the anticancer effects of Hsp90 inhibitors: slow, tight binding inhibition by geldanamycin and its analogues. Proc Natl Acad Sci U S A. 2006;103:7625–7630. doi: 10.1073/pnas.0602650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woodford MR, Dunn DM, Blanden AR, et al. The FNIP co-chaperones decelerate the Hsp90 chaperone cycle and enhance drug binding. Nat Commun. 2016;7:12037. doi: 10.1038/ncomms12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, Koay YC, McAlpine SR. How selective are heat shock protein 90 (Hsp90) inhibitors for cancer cells over normal cells? Chem Med Chem. 2017;12:353–357. doi: 10.1002/cmdc.201600595. [DOI] [PubMed] [Google Scholar]
- 92.Mendillo ML, Santagata S, Koeva M, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garg G, Zhao H, Blagg BSJ. Design, synthesis and biological evaluation of alkylamino biphenylamides as Hsp90 C-terminal inhibitors. Bioorg Med Chem. 2017;25:451–457. doi: 10.1016/j.bmc.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Byrd KM, Subramanian C, Sanchez J, et al. Synthesis and biological evaluation of novobiocin core analogues as Hsp90 Inhibitors. Chemistry. 2016;22:6921–6931. doi: 10.1002/chem.201504955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bopp B, Ciglia E, Ouald-Chaib A, et al. Design and biological testing of peptidic dimerization inhibitors of human Hsp90 that target the C-terminal domain. Biochim Biophys Acta. 2016;1860:1043–1055. doi: 10.1016/j.bbagen.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 96.Koay YC, Wahyudi H, McAlpine SR. Reinventing Hsp90 Inhibitors: blocking C-Terminal binding events to Hsp90 by using dimerized inhibitors. Chemistry. 2016;22:18572–18582. doi: 10.1002/chem.201603464. [DOI] [PubMed] [Google Scholar]
- 97.Dlugosz A, Janecka A. Novobiocin analogs as potential anticancer agents. Mini Rev Med Chem. 2017;17:728–733. doi: 10.2174/1389557516666161223155525. [DOI] [PubMed] [Google Scholar]
- 98.Ramsey DM, McConnell JR, Alexander LD, et al. An Hsp90 modulator that exhibits a unique mechanistic profile. Bioorg Med Chem Lett. 2012;22:3287–3290. doi: 10.1016/j.bmcl.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang W, Ye M, Zhang L-R, et al. FW-04-806 inhibits proliferation and induces apoptosis in human breast cancer cells by binding to N-terminus of Hsp90 and disrupting Hsp90-Cdc37 complex formation. Mol Cancer. 2014;13:150. doi: 10.1186/1476-4598-13-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Li L, Fu WT, et al. Optimization and bioevaluation of Cdc37-derived peptides: an insight into Hsp90-Cdc37 protein-protein interaction modulators. Bioorg Med Chem. 2017;25:233–240. doi: 10.1016/j.bmc.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 101.Wang L, Li L, Zhou Z-H, et al. Structure-based virtual screening and optimization of modulators targeting Hsp90-Cdc37 interaction. Eur J Med Chem. 2017;136:63–73. doi: 10.1016/j.ejmech.2017.04.074. [DOI] [PubMed] [Google Scholar]
- 102.Wang L, Li L, Gu K, et al. Targeting Hsp90-Cdc37: a promising therapeutic strategy by inhibiting Hsp90 chaperone function. Curr Drug Targets. 2017;18:1572–1585. doi: 10.2174/1389450117666160527125522. [DOI] [PubMed] [Google Scholar]
- 103.Stiegler SC, Rübbelke M, Korotkov VS, et al. A chemical compound inhibiting the Aha1-Hsp90 chaperone complex. J Biol Chem. 2017 doi: 10.1074/jbc.M117.797829. jbc.M117.797829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taldone T, Patel PD, Patel M, et al. Experimental and structural testing module to analyze paralogue-specificity and affinity in the Hsp90 inhibitors series. J Med Chem. 2013;56:6803–6818. doi: 10.1021/jm400619b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim H, Yang J, Kim MJ, et al. Tumor necrosis factor receptor-associated protein 1 (TRAP1) mutation and TRAP1 inhibitor gami-trinibtriphenylphosphonium (G-TPP) induce a forkhead box O (FOXO)-dependent cell protective signal from mitochondria. J Biol Chem. 2016;291:1841–1853. doi: 10.1074/jbc.M115.656934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Condelli V, Maddalena F, Sisinni L, et al. Targeting TRAP1 as a downstream effector of BRAF cytoprotective pathway: a novel strategy for human BRAF-driven colorectal carcinoma. Oncotarget. 2015;6:22298–22309. doi: 10.18632/oncotarget.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee C, Park H-K, Jeong H, et al. Development of a mitochondria-targeted Hsp90 inhibitor based on the crystal structures of human TRAP1. J Am Chem Soc. 2015;137:4358–4367. doi: 10.1021/ja511893n. [DOI] [PubMed] [Google Scholar]
- 108.Crowley VM, Khandelwal A, Mishra S, et al. Development of glucose regulated protein 94-selective inhibitors based on the BNIM and radamide scaffold. J Med Chem. 2016;59:3471–3488. doi: 10.1021/acs.jmedchem.6b00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu BX, Hong F, Zhang Y, et al. GRP94/gp96 in cancer: biology, structure, immunology, and drug development. Adv Cancer Res. 2016;129:165–190. doi: 10.1016/bs.acr.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 110.Patel HJ, Patel PD, Ochiana SO, et al. Structure-activity relationship in a purine-scaffold compound series with selectivity for the endoplasmic reticulum Hsp90 paralog Grp94. J Med Chem. 2015;58:3922–3943. doi: 10.1021/acs.jmedchem.5b00197. [DOI] [PMC free article] [PubMed] [Google Scholar]