Abstract
Non-alcoholic fatty liver disease (NAFLD) is the accumulation of extra fat in liver cells not caused by alcohol. Elevated transaminase levels are common indicators of liver disease, including NAFLD. Previously, we demonstrated that PNPLA3 (rs738409), LYPLAL1 (rs12137855), PPP1R3B (rs4240624), and GCKR (rs780094) are associated with elevated transaminase levels in overweight/obese Mexican adults. We investigated the association between 288 SNPs identified in genome-wide association studies and risk of elevated transaminase levels in an admixed Mexican-Mestizo sample of 178 cases of NAFLD and 454 healthy controls. The rs2896019, rs12483959, and rs3810622 SNPs in PNPLA3 and rs1227756 in COL13A1 were associated with elevated alanine aminotransferase (ALT, ≥40 IU/L). A polygenic risk score (PRS) based on six SNPs in the ADIPOQ, COL13A1, PNPLA3, and SAMM50 genes was also associated with elevated ALT. Individuals carrying 9–12 risk alleles had 65.8% and 48.5% higher ALT and aspartate aminotransferase (AST) levels, respectively, than those with 1–4 risk alleles. The PRS showed the greatest risk of elevated ALT levels, with a higher level of significance than the individual variants. Our findings suggest a significant association between variants in COL13A1, ADIPOQ, SAMM50, and PNPLA3, and risk of NAFLD/elevated transaminase levels in Mexican adults with an admixed ancestry. This is the first study to examine high-density single nucleotide screening for genetic variations in a Mexican-Mestizo population. The extent of the effect of these variations on the development and progression of NAFLD in Latino populations requires further analysis.
Keywords: Alanine aminotransferase (ALT), Aspartate aminotransferase (AST), Mexican adults, Polymorphisms, polygenic risk score
Introduction
Non-alcoholic fatty liver disease (NAFLD) is defined by the accumulation of fat in the liver (≥5% of fat in hepatocytes), which can develop into a more serious condition known as non-alcoholic steatohepatitis (NASH). Progression from NAFLD to NASH increases risk of cirrhosis, liver failure, and hepatocellular carcinoma (Wiegand et al., 2007). According to the World Gastroenterology Organization, NAFLD has been increasing during the past 20 years and is now one of the most common types of liver disease in Western countries (Wiegand et al., 2007). The gold standard for the diagnosis of NAFLD/NASH is the liver biopsy, but it is an invasive procedure that requires a highly experienced hepatopathologist. Since performing a liver biopsy can be impractical, unnecessary, and cost prohibitive, especially in underdeveloped countries, other clinical approaches including laboratory tests and imaging studies can be used to detect NAFLD (Chalasani et al., 2012; Nalbantoglu and Brunt, 2014). The most common tests to evaluate the degree of liver injury or liver disease are alanine aminotransferase (ALT) and aspartate aminotransferase (AST) (Kang, 2013; Thapa and Walia, 2007). Growing evidence suggests that ALT/AST levels can be considered biomarkers of liver metabolic function, which may be indicative of a normal response to high fat intake or liver damage, including NAFLD (Jadaho et al., 2004; Kechagias et al., 2008; Sookoian et al., 2016; Sookoian and Pirola, 2012). The AST/ALT ratio represents the time course and disease aggressiveness that can be predicted from the relatively short half-life of AST (18 h), as compared to ALT (36 h) (Botros and Sikaris, 2013).
As a lipid metabolism disorder, NAFLD has been associated with both strong environmental risk factors and a genetic component. The first genome-wide association study (GWAS) of NAFLD identified the rs738409 G allele of the patatin-like phospholipase domain-containing-3 (PNPLA3) gene as significantly associated with increased hepatic fat, inflammation, hepatic enzyme levels, and susceptibility to NAFLD (Romeo et al., 2008). These associations have been replicated in multiple populations (Chambers et al., 2011; Hotta et al., 2010; Kollerits et al., 2010; Kotronen et al., 2009; Zain et al., 2012). A subsequent study, which used data from the Genetics of Obesity-related Liver Disease (GOLD) Consortium, identified four additional NAFLD risk genetic variants located in or near the neurocan (NCAN), glucokinase regulatory protein (GCKR), lysophospholipase-like 1 (LYPLAL1), and protein phosphatase 1 regulatory subunit 3b (PPP1R3B) genes (Speliotes et al., 2011). However, most studies to date that report an association between specific genetic variants and increased risk of NAFLD or elevated transaminase levels have been performed in predominantly European and Asian populations, and it remains unclear if other loci contribute to the excess of NAFLD and elevated transaminase levels observed in the Mexican population. In a previous study, we demonstrated that PNPLA3 (rs738409), LYPLAL1 (rs12137855), PPP1R3B (rs4240624), GCKR (rs780094), are associated with elevated transaminase levels in overweight/obese Mexican adults (Flores et al., 2016).
Since Mexicans and other Latino populations have been underrepresented in GWAS, it is critical to investigate the genetic variants and genes that are shared in these diverse populations. Additionally, it is important to determine if the presence of these variants is correlated with readily detectable biomarkers, such as liver enzymes. Our aim was to extend the finding of our previous study and investigate the association between NAFLD and SNPs that were previously identified by GWAS in other populations, in an admixed sample of Mexican adults.
Materials and Methods
Human subjects and phenotype data
Subjects
The Mexican Health Worker Cohort Study (MHWCS) is a long-term study of workers from the Mexican Institute of Social Security (IMSS) and the National Institute of Public Health (INSP) in Cuernavaca, Morelos (located in central Mexico) that focuses on the association between certain lifestyle factors and the development of chronic diseases (Denova-Gutiérrez et al., 2016).
A baseline assessment was conducted from 2004 to 2006 (Wave 1) and approximately 4,000 participants enrolled in the MHWCS. A second assessment from 2010 to 2013 served as a follow-up for the original participants and provided baseline information for new subjects who were enrolled in the MHWCS. For the follow-up phase, 2,500 MHWCS participants who were initially enrolled were invited to participate, and 1,855 (74%) took part in the second evaluation (Denova-Gutiérrez et al., 2016). Study participants completed several self-reported questionnaires that collected information about demographics, overall health status, and behavioral factors (eg. diet, physical activity, and alcohol consumption), at each follow-up period. They also underwent a complete physical examination and blood tests following an overnight fast, including transaminase levels (ALT and AST), cholesterol (total, HDL and LDL), triglycerides, glucose, proportion of body fat (DEXA), etc. at each follow-up phase. During Wave 2, the participants also provided a blood sample for genetic testing, after an overnight fast. The clinical procedures, data coding, entry, and participant follow-up practices have been standardized and validated (Denova-Gutiérrez et al., 2011; Morales et al., 2014).
A total of 632 MHWCS participants aged 18 to 85 years were selected for this case-control study. The controls included 454 participants who had at least two consecutive normal alanine aminotransferase (ALT <40 IU/L) results in both Wave 1 (2004–2006) and Wave 2 (2011–2013). The 178 cases of NAFLD (ALT ≥40 IU/L) were confirmed by ultrasound to identify the accumulation of fat in the liver. Participants who self-reported as heavy or binge drinkers, (Jiles et al., 2005) were infected with HBV or HCV, or had a prior liver disease diagnosis were excluded from this study. The Ethics Committee of IMSS and all participating institutions approved the study and the MHWCS subjects provided written informed consent prior to initiating any study activities. The study was performed according to the principles of the 1975 Declaration of Helsinki.
Clinical and anthropometric measurements
Trained nurses used standardized procedures to obtain the anthropometric measures of all MHWCS participants. Weight (kg) and height squared (m2) were used to calculate body mass index (BMI, kg/m2). The World Health Organization (WHO) guidelines were used to classify study subjects as): normal weight (18.5–24.9 kg/m2), overweight (25.0–29.9 kg/m2), or obese (≥30.0 kg/m2), based on their BMI (WHO, 2000). Biochemical parameters were measured after 8–10 hours of overnight fasting. The following clinical measures were determined using commercial tests: serum glucose, triglycerides, cholesterol (total, HDL and LDL), ALT and AST.
Genotyping of SNPs in candidate genes
A commercial isolation kit (QIAGEN systems Inc., Valencia, CA) was used to extract the genomic DNA from the peripheral blood of the study participants. Based on prior GWAS and candidate gene studies, we compiled a list of 288 SNPs in 60 genes that are associated with the development NAFLD, elevated transaminase levels, liver disease severity and metabolic comorbidities, which were included in a GoldenGate BeadArray (Illumina). Because the Mexican-Mestizo population is admixed, ancestry informative markers (AIMs) also were incorporated in the study design to assess whether any association could be confounded by population stratification. A panel of 96 AIMs distributed across the genome was selected from previous reports to mainly distinguish between three continental populations (American, European and African) (Flores et al., 2016; Kosoy et al., 2009).
Quality control
After genotype calling, the following quality control criteria were applied using the PLINK software (Purcell et al., 2007): (1) subject and SNP genotyping success rate ≥95 %; (2) minor allele frequency ≥0.05 %; and (3) departure from Hardy–Weinberg equilibrium (HWE) at P value ≥0.001. A total of 314 SNPs including the 96 AIMs, met the quality control criteria and were further analyzed.
Construction of the polygenic risk score
To evaluate the combined effect of the SNPs that were significantly associated with higher ALT and AST levels, we constructed a polygenic risk score (PRS) (Dudbridge, 2013) for each individual, which included six SNPs: ADIPOQ rs17366743, COL13A1 (rs7101190 and rs1227756), PNPLA3 (rs3810622 and rs738409) and SAMM50 rs2143571. The PRS was constructed by summing the number of risk alleles from these six SNPs for each individual. PNPLA3 rs12483959 and rs2896019 were excluded in the construction of the PRS because rs738409 was selected as a tag SNP in the haplotype block 1. The weighted PRS was calculated by multiplying the number of alleles for each SNP by the estimated effect (beta) obtained from the association analysis for ALT or AST levels. Genotypes for each SNP were scored using an additive model (0 for homozygous for the non-risk allele, 1 for heterozygous, and 2 for homozygous for the risk allele).
Statistical analyses
Ancestry from principal component analysis (PCA) was estimated using the smartpca program in the Eigensoft 3.0 package (Price, 2006), and ancestry estimates were included as confounding factors to correct for population stratification. The association between each SNP and elevated ALT or AST levels (≥40 IU/L) was tested using logistic regression analysis, adjusting for age, sex, BMI, principal components of ancestry, and number of tests performed. All associations were tested for additive, dominant, and recessive inheritance models, with the most significant model being reported. Multiple linear regression analysis was performed to test the independent effect of each risk allele or PRS on biochemical parameters. Because most of the SNPs we analyzed are well-validated variants, a P-value threshold of 0.05 was used to determine a significant association. Haploview software was used to construct haplotype blocks with strong linkage disequilibrium (LD) for which the one-sided upper 95% confidence bound on D′ was >0.98. All statistical analyses were performed using SPSS (version 16.0; Chicago, IL).
Results
Characteristics of the study population
The demographic and clinical characteristics of the study population are described in Table 1. A total of 632 study participants were included, of which 454 controls (71.8%) had persistently normal ALT levels (<40 IU/L) and 178 cases (28.2%) had at least two consecutive elevated ALT levels (≥40 IU/L). The mean age of the study subjects was 46.8 ± 11.2 and 49.5 ± 13.3 years, 42.7% of the cases and 19.4% of the controls were males, and BMI was 29.4 ± 4.5 and 26.2 ± 4.5 kg/m2 among the cases and controls, respectively. No significant differences were found between cases and controls in terms of hypertension, total cholesterol and LDL cholesterol (P>0.05). Significant differences were observed in age, sex, education level and certain clinical measures between the cases and controls. As expected, cases had a significantly higher mean BMI, glucose, triglycerides, ALT and AST levels, as compared to controls (P<0.01). Mean HDL-C levels were significantly lower among the cases (P<0.001).
Table 1.
Demographic and clinical characteristics of the study population
| All | Cases ALT ≥40 IU/L |
Controls ALT <40 IU/L |
P-value | |
|---|---|---|---|---|
| N (%) | 632 | 178 (28.2) | 454 (71.8) | |
| Age (years) | 48.7 ± 12.8 | 46.8 ± 11.2 | 49.5 ± 13.3 | 0.009 |
| Sex (male %) | 25.95 | 42.70 | 19.38 | 1.8×10−9 |
| Education Level | ||||
| ≤ 6 years | 17.09 | 11.80 | 19.16 | 0.041 |
| ≤ 12 years | 39.08 | 42.70 | 37.67 | |
| > 12 years | 42.41 | 42.70 | 42.29 | |
| Hypertension | ||||
| No | 74.37 | 71.35 | 75.55 | 0.276 |
| Yes | 25.63 | 28.65 | 24.45 | |
| BMI (kg/m2) | 27.1 ± 4.7 | 29.4 ± 4.5 | 26.2 ± 4.5 | 1.5×10−14 |
| Glucose (mg/dl) | 99.1 ± 33.5 | 105.2 ± 33.5 | 96.8 ± 33.2 | 0.004 |
| Total cholesterol (mg/dl) | 200.3 ± 39.7 | 199.0 ± 36.2 | 200.8 ± 41.0 | 0.599 |
| Triglycerides (mg/ml) | 169.2 ± 119.1 | 200.0 ± 102.3 | 157.1 ± 123.1 | 8.5×10−10 |
| HDL-C (mg/ml) | 38.7 ± 11.9 | 36.4 ± 9.6 | 39.5 ± 12.6 | 8.8×10−4 |
| LDL-C (mg/dl) | 124.6 ± 36.9 | 123.6 ± 35.1 | 125.0 ± 37.5 | 0.666 |
| ALT (IU/L) | 31.9 ± 26.5 | 64.8 ± 29.5 | 19.0 ± 7.0 | 2.2×10−49 |
| AST (IU/L) | 26.0 ± 16.1 | 44.6 ± 19.4 | 18.6 ± 5.1 | 9.9×10−42 |
Results are presented as means ± standard deviations or n (%).
P values were obtained by comparing cases to controls.
Differences between proportions were performed using chi-square tests of homogeneity and differences between means were performed using t-test. Triglycerides were log-transformed before the analysis.
Abbreviations: BMI, body mass index; HDL-C, High Density Lipoprotein Cholesterol; HDL-C, Low Density Lipoprotein Cholesterol; ALT, alanine transaminase; AST, aspartate transaminase.
Genetic association with elevated ALT levels
Of the 218 SNPs tested, 24 were significantly associated with elevated ALT levels (≥40 IU/L) (P<0.02, after adjusting for age, sex, BMI, and principal components of ancestry) (Supplementary Table 1). However, after correcting for number of tests performed, only COL13A1 rs1227756 remained significantly associated with a higher risk of elevated ALT levels, as did PNPLA3 (rs12483959 and rs2896019) with a lower risk of having elevated ALT levels (P=0.022, 0.004 and 0.018, respectively). The association of the PNPLA3 SNPs (rs3810622 and rs738409) with risk of elevated ALT levels was found to have a borderline statistical significance (P=0.052 and 0.070, respectively) (Table 2). After stratifying by sex, the associations of the four PNPLA3 variants (rs3810622, rs12483959, rs2896019 and rs738409) with elevated ALT levels were only significant among females (OR=3.01, 95% CI 1.81–5.00, P=0.005; OR=0.36, 95% CI 0.22–0.59, P=0.010; OR=0.36, 95% CI 0.22–0.58, P=0.009 and OR=2.57, 95% CI 1.59–4.16, P=0.028, respectively; adjusted for age, sex, BMI, principal components of ancestry, and number of tests).
Table 2.
Association between the COL13A1 and PNPLA3 SNPs with elevated ALT levels, by sex. n (%)
| COL13A1 | PNPLA3 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||
| SNP | rs1227756 | rs3810622 | rs12483959 | rs2896019 | rs738409 | |||||||||||||||
|
| ||||||||||||||||||||
| AA | AG | GG | Prec | CC | CT | TT | Prec | GG | GA | AA | Pdom | TT | TG | GG | Pdom | AA | AG | GG | Prec | |
| Total (N=632) | ||||||||||||||||||||
| Cases ALT ≥40 IU/L |
20 (11.4) | 66 (37.7) | 89 (50.9) | 0.022 | 8 (4.5) | 54 (30.7) | 114 (64.8) | 0.052 | 79 (44.6) | 71 (40.1) | 27 (15.3) | 0.004 | 75 (42.6) | 74 (42.0) | 27 (15.3) | 0.018 | 21 (11.9) | 73 (41.2) | 83 (46.9) | 0.070 |
| Controls ALT <40 IU/L |
65 (14.4) | 232 (51.4) | 154 (34.1) | 30 (6.6) | 194 (42.9) | 228 (50.4) | 125 (27.6) | 232 (51.2 | 96 (21.1) | 121 (26.7) | 236 (52.1) | 96 (21.2) | 79 (17.5) | 232 (51.4) | 140 (31.0) | |||||
|
| ||||||||||||||||||||
| Male (N=164) | ||||||||||||||||||||
| Cases ALT ≥40 IU/L |
11 (14.9) | 28 (37.8) | 35 (47.3) | NS | 6 (8.0) | 25 (33.3) | 44 (58.7) | NS | 32/42.7) | 28 (37.3) | 15 (20.0) | NS | 29 (38.7) | 31 (41.3) | 15 (20.0) | NS | 10 (13.2) | 33 (43.3) | 33 (43.4) | NS |
| Controls ALT <40 IU/L |
14 (16.1) | 44 (50.6) | 29 (33.3) | 4 (4.5) | 30 (34.1) | 54 (61.4) | 32 (36.4) | 42 (47.7) | 14 (15.9) | 32 (36.4) | 42 (47.7) | 14 (15.9) | 12 (13.6) | 39 (44.3) | 37 (42.0) | |||||
|
| ||||||||||||||||||||
| Female (N=468) | ||||||||||||||||||||
| Cases ALT ≥40 IU/L |
9 (8.9) | 38 (37.6) | 54 (53.5) | 0.245 | 2 (2.0) | 29 (28.7) | 70 (69.3) | 0.005 | 47 (46.1) | 43 (42.2) | 12 (11.8) | 0.010 | 46 (45.5) | 43 (42.6) | 12 (11.9) | 0.009 | 11 (10.9) | 40 (39.6) | 50 (49.5) | 0.028 |
| Controls ALT <40 IU/L |
51 (14.0) | 188 (51.6) | 125 (34.3) | 26 (7.1) | 164 (45.1) | 174 (47.8) | 93 (25.5) | 190 (52.1) | 82 (22.5) | 89 (24.4) | 194 (53.2) | 82 (22.5) | 67 (18.5) | 193 (53.2) | 103 (28.4) | |||||
P values were calculated by logistic regression analysis, and the statistically significant results are shown in bold.
All models were adjusted for age, BMI, ancestry and number of tests. The analyses with the total population were additionally adjusted for sex.
SNP, single nucleotide polymorphism; ALT, alanine transaminase; BMI, body mass index.
Genetic association with ALT and AST levels
A significant association was observed between 16 variants and higher ALT or AST levels, and three SNPs were found to be associated with higher AST levels (P<0.05 after adjusting for age, sex, BMI, and principal components of ancestry) (Supplementary Table 2). However, after additionally controlling for number of tests, only two variants in PNPLA3 (rs3810622 and rs738409) and COL13A rs1227756 remained significantly associated with higher ALT or AST levels, while SAMM50 rs2143571 was only associated with higher AST levels (P<0.05). PNPLA3 rs3810622 showed the strongest and most significant effect, an increase of 8.0 and 5.7 IU/L in ALT and AST levels, respectively (P≤0.001) (Table 3).
Table 3.
Association between the ADIPOQ, COL13A1, PNPLA3 and SAMM50 SNPs with select clinical measures
| Gene SNP |
ADIPOQ rs17366743 |
COL13A1 rs7101190 |
COL13A1 rs1227756 |
PNPLA3 rs3810622 |
PNPLA3 rs12483959 |
PNPLA3 rs2896019 |
PNPLA3 rs738409 |
SAMM50 rs2143571 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| Effect (SE) | Padd | Effect (SE) | Pdom | Effect (SE) | Prec | Effect (SE) | Prec | Effect (SE) | Pdom | Effect (SE) | Pdom | Effect (SE) | Prec | Effect (SE) | Padd | |
| BMI (kg/m2) | −1.2 (0.6) | NS | −0.5 (0.4) | NS | 0.3 (0.4) | NS | −0.8 (0.4) | NS | 0.6 (0.4) | NS | 0.4 (0.4) | NS | −0.2 (0.4) | NS | 0.1 (0.3) | NS |
| TG (mg/dl) | −4.8 (13.9) | NS | −22.8 (9.3) | NS | 22.9 (9.6) | NS | 2.2 (9.4) | NS | −3.5 (10.1) | NS | −3.7 (10.2) | NS | −2.2 (9.9) | NS | −5.6 (6.9) | NS |
| HDL-C (mg/dl) | −1.2 (1.4) | NS | −0.9 (1.0) | NS | −0.6 (1.0) | NS | 1.4 (1.0) | NS | −2.5 (1.0) | NS | −2.4 (1.0) | NS | 2.4 (1.0) | NS | 0.7 (0.7) | NS |
| Glucose (mg/dl) | −6.1 (3.9) | NS | −2.8 (2.6) | NS | 3.8 (2.9) | NS | −3.1 (2.6) | NS | −0.1 (2.8) | NS | 0.8 (2.9) | NS | 2.3 (2.8) | NS | −1.3 (1.9) | NS |
| ALT (U/L) | −10.9 (2.8) | 0.014 | −6.6 (1.9) | 0.045 | 7.3 (1.9) | 0.003 | 8.0 (1.9) | 0.001 | −8.6 (2.1) | 0.002 | −7.6 (2.1) | 0.008 | 6.8 (2.0) | 0.023 | 5.1 (1.4) | 0.077 |
| AST (U/L) | −6.1 (1.8) | 0.120 | −4.1 (1.2) | 0.074 | 4.8 (1.2) | 7.4×10−4 | 5.7 (1.2) | 8.2×10−5 | −6.2 (1.3) | 1.6×10−4 | −5.5 (1.3) | 0.001 | 4.7 (1.2) | 0.015 | 7.5 (1.8) | 0.032 |
P values were calculated by linear regression analysis, and the statistically significant results are shown in bold.
All models were adjusted for, age, sex, BMI, ancestry and number of tests.
TG, ALT and AST were log transformed for the analysis.
SNP, single nucleotide polymorphism; SE, standard error; BMI, body mass index; TG, triglycerides; HDL-C, high-density lipoprotein cholesterol; ALT, alanine transaminase; AST, aspartate transaminase.
After adjusting for age, sex, BMI, and principal components of ancestry; twelve SNPs were significantly associated with lower ALT and AST levels (P<0.05) (Supplementary Table 2). As shown in Table 3, two PNPLA3 SNPs (rs12483959 and rs2896019) were associated with lower ALT and AST levels (P<0.01), with PNPLA3 rs12483959 having the most significant effect on decreased ALT and AST levels by 8.6 and 6.2 IU/L, respectively (P≤0.002). In addition, COL13A1 rs7101190 and ADIPOQ rs17366743 also remained significantly associated with lower ALT levels (P<0.05). Interestingly, ADIPOQ rs17366743 has the strongest effect, decreasing ALT levels by 10.9 IU/L (P=0.014) (Table 3).
Genetic association with other metabolic parameters
Since the variants we analyzed have also been linked to other metabolic traits, we decide to evaluate their association with clinical variables that have been associated with NAFLD. We found a significant association between SLC2A1 (rs841848 and rs841858) and higher serum glucose concentrations (P<0.001, after adjusting for age, sex, BMI and principal components of ancestry) (Supplementary Table 3), but only SLC2A1 rs841848 remained significantly associated after correcting for number of tests (P=0.048). Of the all-biological candidate variants, six were significantly associated with BMI, four with triglycerides, and four with HDL-C levels (P<0.05, after adjusting for age, sex, BMI, and principal components of ancestry) (Supplementary Table 3). After adjusting for number of tests, only the association of TGM5 rs748404 with triglyceride levels remained significant, for each copy of the T risk allele, triglyceride levels increased 26.7 mg/ml (P=0.026).
Association between PNPLA3 haplotype and risk of elevated ALT levels
The LD pattern of the PNPLA3 polymorphisms was evaluated to determine haplotype blocks. Two independents blocks were detected, block 1 contained rs12483959, rs2896019 and rs738409, while block 2 contained rs3810622 (Supplementary Figure 1). The haplotype GGG in block 1 was significantly associated with risk of elevated ALT (≥40 IU/L) (P=0.035). The SNP rs738409 was selected as a haplotype-tag SNP.
Association of the PRS with elevated ALT levels
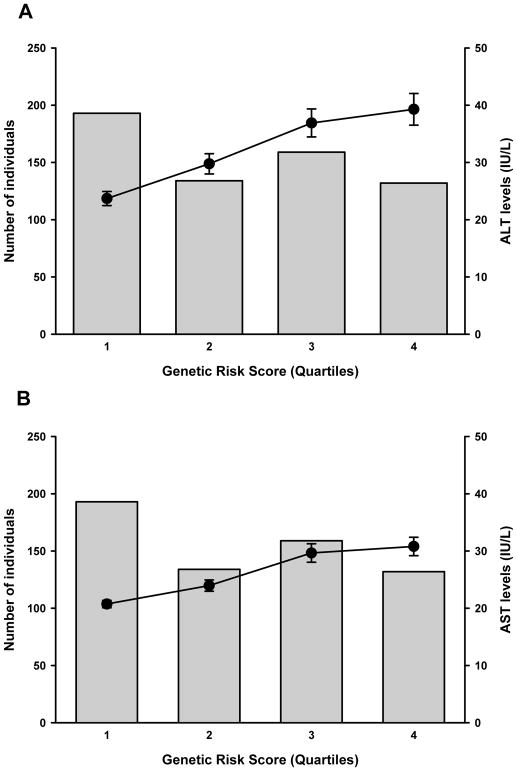
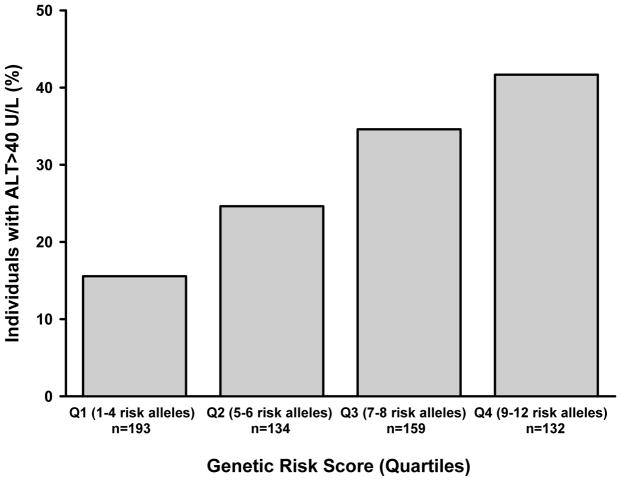
As the PRS increases, ALT and AST levels also rise as a function of the number of risk alleles (P<0.0001, respectively, adjusted for age, sex, BMI, and principal components of ancestry) Figure 1. The analysis using the weighted PRS showed similar results (P<0.0001, respectively). The PRS was also strongly associated with elevated ALT levels (OR=1.70, 95% CI 1.41–2.05, P<0.0001) (Figure 2).
Fig. 1.
Distribution of the weighted genetic risk score and cumulative effects of the risk alleles from six SNPs on ALT and AST serum levels. Mean ALT (A) and AST (B) levels significantly increase as a function of the number of risk alleles (quartiles) (P=1.0×10−9 and 7.7×10−11), respectively, adjusted for age, sex, BMI, and admixture.
Fig. 2.
Association between the weighted genetic risk score from six SNPs and proportion of individuals with elevated ALT levels. The percentage of subjects with ALT ≥40 IU/L increased significantly as a function of the number of risk alleles (quartiles) (P<0.001 adjusted for age, sex, BMI, and admixture).
Discussion
In this case-control study, we analyzed 218 SNPs that were associated with the development of NAFLD, elevated transaminase levels, liver disease severity, and metabolic comorbidities in previous studies (Macaluso et al., 2015; Speliotes et al., 2011). After adjusting for number of tests performed, COL13A1 rs1227756 was significantly associated with a higher risk of elevated ALT levels, while individuals with the PNPLA3 variants rs12483959 and rs2896019 had a lower risk of elevated ALT levels. Two PNPLA3 SNPs (rs3810622 and rs738409) were also associated with a greater risk of elevated ALT levels, but only in females. Our results support the association between PNPLA3 variant rs738409 (I148M) and elevated levels of ALT that has been observed in other studies with Mexican-Mestizo populations (Flores et al., 2016; Larrieta-Carrasco et al., 2014, 2013). The lack of significance among males could be due to the lower number of male participants in the MHWCS, as compared to females (70% vs. 30%, respectively) (Denova-Gutiérrez et al., 2016). Our results also support previous findings regarding sexual dimorphism in the genetic association between PNPLA3 rs738409 and liver transaminases (Larrieta-Carrasco et al., 2014; Li et al., 2012; Sookoian and Pirola, 2011).
To the best of our knowledge, this is the first study to identify the association of the PNPLA3 rs3810622 and COL13A1 rs1227756 variants with a higher risk of elevated ALT levels, and PNPLA3 rs2896019 and rs12483959 with a lower risk of elevated ALT levels in an admixed sample of Mexican adults. Previously, rs12483959 has been associated with obesity and insulin resistance in children (Johansson et al., 2009), while rs2896019 and rs3810622 have been linked to steatosis grade (Kitamoto et al., 2013), decreased serum triglycerides, hepatocyte ballooning and NAFLD activity score (NAS) (Kitamoto et al., 2013). The presence of COL13A1 rs1227756 has been associated with lobular inflammation in NAFLD patients (Chalasani et al., 2010). Furthermore, we also found that ALT and AST levels were significantly higher among the carriers of PNPLA3 variants rs3810622 and rs738409, and rs1227756 in COL13A1, while rs2143571 in SAMM50 was only associated with increased levels of AST. The link between PNPLA3 rs738409 and higher ALT levels has been consistently replicated in different populations including Mexican adults, children, and Indigenous groups (Flores et al., 2016; Larrieta-Carrasco et al., 2014, 2013; León-Mimila et al., 2015). The effect of PNPLA3 rs3810622 on transaminase levels observed in our study is similar to the findings of GWAS studies with Japanese (Kitamoto et al., 2013) and Chinese populations (Song et al., 2016), although our results indicate a stronger and more significant association.
This is the first study to report an association between rs1227756 COL13A1 and risk of increased ALT and AST levels. Prior to our study, this variant had only been associated with lobular inflammation in NAFLD patients (Chalasani et al., 2010). The collagen type XIII, α 1 (COL13A1) gene encodes the chain of a non fibrillar collagen and it has been linked with modifying the inflammatory response genes in the mouse intestine (Tuomisto et al., 2008). Since COL13A1 has been associated with the intestinal inflammatory response, it may also play a role in liver inflammation that has yet to be evaluated. Recently, a study of monozygotic twins with NASH-related cirrhosis found rs1227756 to be a contributing factor in disease progression (Grove et al., 2016). Additionally, we observed an association between rs1227756 COL13A and higher triglyceride levels, which supports the findings of a study conducted with NAFLD patients in India (Ravi Kanth et al., 2014). Our results differ from those of a recent study that did not observe an association between the rs1227756 variant and the presence of NAFLD in Chinese children (Shang et al., 2015). This leads us to consider that the effect of this SNP among the Mexican-Mestizo population may be more significant than in Europeans or other populations. Future studies need to examine the extent of the association between these variants and NAFLD in other populations.
The SAMM50 gene is located in the same genetic region as PNPLA3, and polymorphisms of this gene, including rs2143571, have been associated with NAFLD among Japanese, Chinese, and Indian populations (Chen et al., 2015; Kitamoto et al., 2013; Ravi Kanth et al., 2014). We found that the variant rs2143571 SAMM50 was associated with higher levels of AST in our sample of Mexican adults. A study with a Chinese population found that the variant rs2143571 was associated with elevated levels of both ALT and AST, as well as higher triglyceride levels (Chen et al., 2015). Moreover, the rs2143571 “A” allele was associated with an increased risk of developing NAFLD, compared with the non-carriers (Chen et al., 2015). Kitamoto et al. (Kitamoto et al., 2013) found that four variants of SAMM50, including variant rs2143571, were associated with histological severity and proposed that SAMM50 could be involved in necroinflamation and fibrosis. Mitochondrial abnormalities have been observed in the liver biopsies of NASH patients (Caldwell et al., 1999; Sanyal et al., 2001). In addition, several studies have recognized that mitochondrial dysfunction plays an important role in insulin resistance (Lowell and Shulman, 2005; Ma et al., 2012). These reports and our results suggest that the SAMM50 gene could be a key factor in mitochondrial dysfunction, due in part to a defective removal of reactive oxygen species, as a pathophysiological contributor to the development and progression of NAFLD (Rector et al., 2011).
There are several risk factors associated with the progression from NAFLD to NASH, which include obesity, insulin resistance, hypertriglyceridemia, and low high-density lipoproteins (HDL) cholesterol (Fazel et al., 2016; Than and Newsome, 2015). To further investigate these pathways, we evaluated the relationship between certain genetic variants and specific metabolic measures. We found an association between SLC2A1 (rs841848 and rs841858) and higher levels of glucose. SLC2A1 is a facilitative glucose transporter responsible for constitutive or basal glucose uptake (Rhoads, 1994). In the context of fatty liver, a correct balance in glucose uptake and subsequent metabolism prevents the development of insulin resistance, thereby avoiding the accumulation of fat. Two SNPs in SLC2A1 (rs4658 and rs841856) were found to be associated with NAFLD in a Spanish population. Interestingly, in the liver biopsies of patients with NAFLD, SLC2A1 expression was down-regulated and homozygous carriers of the rs4658 G-allele had lower expression of SLC2A1 messenger. In addition, silencing SLCA2A1 in THLE2 cells leads to an increased presence of lipid droplets when oleic acid is added to culture medium (Vazquez-Chantada et al., 2013). SLC2A1 variants could be important in the balance of glucose uptake and in fat accumulation.
We also found an association between TGM5 (rs748404) and high triglyceride levels. The TGM5 gene is a member of the transglutaminase family. Transglutaminases (TGs) are Ca(2+)-dependent enzymes that catalyze the formation of covalent bonds between glutamine and lysine residues, and contribute to fibrotic diseases via crosslinking-mediated stabilization of extracellular matrix (Eckert et al., 2014; Iismaa et al., 2009; Lorand and Graham, 2003). TGM1, TGM3 and TGM5 have been associated with mouse liver fibrosis and altered healing processes (De Koning et al., 2012; Tatsukawa et al., 2017), which is interesting since NAFLD can progress to cirrhosis. According to the tissue-specific pattern of mRNA expression profiles from 79 human tissues and cell types (http://biogps.org), TGM5 is highly expressed in the liver, although the precise role of TGM5 in NAFLD is not known.
Although the rs17366743 variant of ADIPOQ was not associated with elevated levels of ALT or AST in our study, this gene may also play a key role in the development of NAFLD. ADIPOQ, also known as adiponectin, is an important adipokine involved in the control of fat metabolism and insulin sensitivity, and its variant rs17366743 has been associated with diabetes (Hivert et al., 2008). Other variants of ADIPOQ located in regulatory regions have been associated with low levels of adiponectin (Heid et al., 2006; Hivert et al., 2008; Menzaghi et al., 2007), which is particularly relevant in the context of metabolic syndrome and diabetes since they are both important risk factors in the development of NAFLD.
Since NAFLD is a complex disease involving multiple factors, it is important to understand that a single SNP cannot be responsible for the development of the disease, and much less for its progression to NASH, cirrhosis, or hepatocarcinoma. GWAS studies have attempted to evaluate different combinations of SNPs that may help explain the various genetic risk factors for developing NAFLD, as well as its progression to more serious liver disease. In our study, we found that a PRS based on six SNPs in the ADIPOQ, COL13A1, PNPLA3, and SAMM50 genes was associated with the greatest risk of elevated ALT levels. Individuals carrying 9–12 risk alleles had 65.8% and 48.5% higher ALT and AST levels, respectively, than those with 1–4 risk alleles. The PRS explained the variance of ALT levels with a higher level of significance than the individual variants, though less strongly when compared with PNPLA3 rs3810622.
Twin studies suggest that transaminase levels have a genetic heritability irrespective of anthropometric features and environmental factors (Bathum et al., 2001; Rahmioglu et al., 2009), which means that certain genes may regulate liver enzyme levels. Studies of genetic regions like PNPLA3, SAMM50 and PARVB support this idea, since variants in these genes have been associated with NAFLD and elevated ALT and AST levels (Flores et al., 2016; Kitamoto et al., 2013; Ravi Kanth et al., 2014). Krawczyk et al. found that the combined effect of several SNPs was linked to the presence of elevated ALT levels, as well as NAFLD severity (Krawczyk et al., 2017). The SNPs included in the PRS we created are likely associated with higher levels of ALT/AST and fatty liver through different mechanisms, which account for the complexity of NAFLD. PNPLA3 variants may explain the hepatic fat accumulation that is essential for the development of NAFLD (Romeo et al., 2008; Speliotes et al., 2011). Additionally, SAMM50 and COL13A1 variants may promote inflammation (Ravi Kanth et al., 2014; Tuomisto et al., 2008), which also contributes to the development of NAFLD. Elevated fat intake increases ALT and AST levels as a normal metabolic response (Kechagias et al., 2008). A genetic condition could mimic or aggravate fat intake and might help to explain the elevated transaminase levels observed among NAFLD patients. Our results suggest that the combined presence of these six SNPs could have a greater impact on the development of NAFLD and subsequent liver damage than a single polymorphism.
This study has some limitations that should be considered. First, the diagnosis of NAFLD was based on the presence of persistently elevated transaminase levels and ultrasonography results. Although performing a liver biopsy is the gold standard for the diagnosis of NAFLD, it is expensive and in some cases may result in morbidity or very rarely death. According to the American Association for the Study of Liver Disease (AASLD) practice guidelines, a liver biopsy should only be obtained from patients who would receive a clear and significant benefit from a definitive diagnosis, treatment, and improved prognosis (Chalasani et al., 2012). Second, this study was based on a cross-sectional design and the selected SNPs were chosen from previous investigations that were conducted in mostly European populations. Third, we used data from a sample of Mexican adults, so our findings may not be representative of other Latino populations because of the heterogeneity observed among Latino groups. Fourth, the MHWCS participants are mostly female (70%) and health workers, who are likely more educated and healthier than the general population of Mexico. While the MHWCS is not a population-based sample, the participants are predominantly middle-class, urban adults from central Mexico, who are employed in the formal sector of the economy, and are representative of approximately 34% of the population (Secretaría de Economía (2014) Programa nacional de protección a los derechos del consumidor 2013–2018. Diario Oficial de la Federación, México., n.d.). Additionally, after adjusting our analyses for multiple testing, the only variants that remained significantly associated with elevated ALT levels were rs12483959 and rs2896019 in PNPLA3 and rs1227756 in COL13A1, probably due to our limited sample size and the elevated number of SNPs that were tested.
In conclusion, this study suggests that variants in the PNPLA3, SAMM50, COL13A1 and ADIPOQ genes are associated with the presence of elevated ALT levels, in a sample of admixed Mexican adults. This study represents the first high-density single nucleotide screening for variations in NAFLD carried out in a Mexican-Mestizo population. The combined effect of these SNPs is likely to have an impact on the development of NAFLD, but further studies are required to determine the magnitude of this association in other Latino populations. Our results independently support those of other studies that have identified loci associated with an increased risk of NAFLD, but more studies are needed to confirm these findings in diverse populations.
Supplementary Material
Acknowledgments
This research was supported by funding from the Instituto Nacional de Medicina Genomica (INMEGEN) grant #119-11/2012/I, the Consejo Nacional de Ciencia y Tecnología (CONACyT) grants #26267M and SALUD-2011-01-161930, and the Instituto Mexicano del Seguro Social (IMSS). Dr. Flores was supported by NIH/NCI 5K07CA197179. The authors express their gratitude to the Mexican Health Worker Cohort Study participants, their families, and the IMSS clinicians and staff. We also acknowledge the services provided by the Microarray Core Facility at the INMEGEN (MSc. Raul Mojica Espinosa).
Abbreviations
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- SNP
Single Nucleotide Polymorphism
- PRS
polygenic risk score
- NAFLD
Non-alcoholic fatty liver disease
Footnotes
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Bathum L, Petersen HC, Rosholm JU, Petersen PH, Vaupel J, Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: Results from a population-based Danish twin study. Clin Chem. 2001;47:81–87. [PubMed] [Google Scholar]
- Botros M, Sikaris KA. The de ritis ratio: the test of time. Clin Biochem Rev. 2013;34:117–30. [PMC free article] [PubMed] [Google Scholar]
- Caldwell SH, Swerdlow RH, Khan EM, Iezzoni JC, Hespenheide EE, Parks JK, Parker WD. Mitochondrial abnormalities in non-alcoholic steatohepatitis. J Hepatol. 1999;31:430–434. doi: 10.1016/S0168-8278(99)80033-6. [DOI] [PubMed] [Google Scholar]
- Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Cui J, Taylor KD, Wilson L, Cummings OW, Chen YDI, Rotter JI. Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology. 2010:139. doi: 10.1053/j.gastro.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Gieger C, Heard-Costa NL, Hottenga J-J, Kühnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Mateo Leach I, O’Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJC, Dixon AL, Döring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen A-L, Heath S, Hirschfield GM, Hofman A, Homuth G, Hyppönen E, Janssen HLA, Johnson T, Kangas AJ, Kema IP, Kühn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin J-P, Loos RJF, Martin NG, Moffatt MF, Montgomery GW, Munroe PB, Musunuru K, Nakamura Y, O’Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JNL, Seedorf U, Sen-Chowdhry S, Siminovitch KA, Smit JH, Spector TD, Tan W, Teslovich TM, Tukiainen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann H-E, Willemsen G, Würtz P, Xu C, Yerges-Armstrong LM, Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietiläinen KH, Schumann G, Snieder H, Sternberg MJE, Stolk RP, Thomas HC, Thorsteinsdottir U, Uda M, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JCM, Wolffenbuttel BHR, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Völzke H, Schadt EE, Scott J, Järvelin M-R, Elliott P, Kooner JS. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–8. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lin Z, Jiang M, Lu L, Zhang H, Xin Y, Jiang X, Xuan S. Genetic Variants in the SAMM50 Gene Create Susceptibility to Nonalcoholic Fatty Liver Disease in a Chinese Han Population. Hepat Mon. 2015;15 doi: 10.5812/hepatmon.31076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning HD, Van Den Bogaard EH, Bergboer JGM, Kamsteeg M, Van Vlijmen-Willems IMJJ, Hitomi K, Henry J, Simon M, Takashita N, Ishida-Yamamoto A, Schalkwijk J, Zeeuwen PLJM. Expression profile of cornified envelope structural proteins and keratinocyte differentiation-regulating proteins during skin barrier repair. Br J Dermatol. 2012;166:1245–1254. doi: 10.1111/j.1365-2133.2012.10885.x. [DOI] [PubMed] [Google Scholar]
- Denova-Gutiérrez E, Castañón S, Talavera JO, Flores M, Macías N, Rodríguez-Ramírez S, Flores YN, Salmerón J. Dietary patterns are associated with different indexes of adiposity and obesity in an urban Mexican population. J Nutr. 2011;141:921–927. doi: 10.3945/jn.110.132332. [DOI] [PubMed] [Google Scholar]
- Denova-Gutiérrez E, Flores YN, Gallegos-Carrillo K, Ramírez-Palacios P, Rivera-Paredez B, Muñoz-Aguirre P, Velázquez-Cruz R, Torres-Ibarra L, Meneses-León J, Méndez-Hernández P, Hernández-López R, Salazar-Martínez E, Talavera JO, Tamayo J, Castañón S, Osuna-Ramírez I, León-Maldonado L, Flores M, Macías N, Antúnez D, Huitrón-Bravo G, Salmerón J. Health workers cohort study: methods and study design. Salud Publica Mex. 2016;58:708–716. doi: 10.21149/spm.v58i6.8299. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Power and Predictive Accuracy of Polygenic Risk Scores. PLoS Genet. 2013:9. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Kaartinen MT, Nurminskaya M, Belkin AM, Colak G, Johnson GVW, Mehta K. Transglutaminase regulation of cell function. Physiol Rev. 2014;94:383–417. doi: 10.1152/physrev.00019.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazel Y, Koenig AB, Sayiner M, Goodman ZD, Younossi ZM. Epidemiology and natural history of non-alcoholic fatty liver disease. Metabolism. 2016;65:1017–1025. doi: 10.1016/j.metabol.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Flores YN, Velázquez-Cruz R, Ramírez P, Bañuelos M, Zhang ZF, Yee HF, Chang SC, Canizales-Quinteros S, Quiterio M, Cabrera-Alvarez G, Patiño N, Salmerón J. Association between PNPLA3 (rs738409), LYPLAL1 (rs12137855), PPP1R3B (rs4240624), GCKR (rs780094), and elevated transaminase levels in overweight/obese Mexican adults. Mol Biol Rep. 2016;43:1359–1369. doi: 10.1007/s11033-016-4058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove JI, Austin M, Tibble J, Aithal GP, Verma S. Monozygotic twins with NASH cirrhosis: cumulative effect of multiple single nucleotide polymorphisms? Ann. Hepatol. 2016;15:277–82. doi: 10.5604/16652681.1193726. [DOI] [PubMed] [Google Scholar]
- Heid IM, Wagner SA, Gohlke H, Iglseder B, Mueller JC, Cip P, Ladurner G, Reiter R, Stadlmayr A, Mackevics V, Illig T, Kronenberg F, Paulweber B. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55:375–384. doi: 10.2337/diabetes.55.02.06.db05-0747. [DOI] [PubMed] [Google Scholar]
- Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J, Fox CS, O’Donnell CJ, Cupples LA, Meigs JB. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes. 2008;57:3353–9. doi: 10.2337/db08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Yoneda M, Hyogo H, Ochi H, Mizusawa S, Ueno T, Chayama K, Nakajima A, Nakao K, Sekine A. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iismaa SE, Mearns BM, Lorand L, Graham RM. Transglutaminases and disease: lessons from genetically engineered mouse models and inherited disorders. Physiol Rev. 2009;89:991–1023. doi: 10.1152/physrev.00044.2008. [DOI] [PubMed] [Google Scholar]
- Jadaho SB, Yang RZ, Lin Q, Hu H, Anania Fa, Shuldiner AR, Gong DW. Murine alanine aminotransferase: cDNA cloning, functional expression, and differential gene regulation in mouse fatty liver. Hepatology. 2004;39:1297–1302. doi: 10.1002/hep.20182. [DOI] [PubMed] [Google Scholar]
- Jiles R, Hughes E, Murphy W, et al. Surveillance for certain health behaviors among states and selected local areas--Behavioral risk factor surveillance system, United States, 2003. MMWR Surveil Summ. 2005;54:1–116. [PubMed] [Google Scholar]
- Johansson LE, Johansson LM, Danielsson P, Norgren S, Johansson S, Marcus C, Ridderstråle M. Genetic variance in the adiponutrin gene family and childhood obesity. PLoS One. 2009;4:1–6. doi: 10.1371/journal.pone.0005327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K-S. Abnormality on Liver Function Test. Pediatr Gastroenterol Hepatol Nutr. 2013;16:225–232. doi: 10.5223/pghn.2013.16.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechagias S, Ernersson A, Dahlqvist O, Lundberg P, Lindström T, Nystrom FH. Fast-food-based hyper-alimentation can induce rapid and profound elevation of serum alanine aminotransferase in healthy subjects. Gut. 2008;57:649–54. doi: 10.1136/gut.2007.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, Teranishi H, Mizusawa S, Ueno T, Chayama K, Nakajima A, Nakao K, Sekine A, Hotta K. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013;132:783–792. doi: 10.1007/s00439-013-1294-3. [DOI] [PubMed] [Google Scholar]
- Kollerits B, Coassin S, Kiechl S, Hunt SC, Paulweber B, Willeit J, Brandsttter A, Adams TD, Kronenberg F. A common variant in the adiponutrin gene influences liver enzyme levels. J Med Genet. 2010;47:116–119. doi: 10.1136/jmg.2009.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotronen A, Johansson LE, Johansson LM, Roos C, Westerbacka J, Hamsten A, Bergholm R, Arkkila P, Arola J, Kiviluoto T, Fisher RM, Ehrenborg E, Orho-Melander M, Ridderstråle M, Groop L, Yki-Järvinen H. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- Krawczyk M, Rau M, Schattenberg JM, Bantel H, Pathil A. Combined effects of the TM6SF2 rs58542926, PNPLA3 rs738409 and MBOAT7 rs641738 variants on NAFLD severity: multicentre biopsy-based study. J Lipid Res. 2017;58:247–255. doi: 10.1194/jlr.P067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieta-Carrasco E, Acuña-Alonzo V, Velázquez-Cruz R, Barquera-Lozano R, León-Mimila P, Villamil-Ramírez H, Menjivar M, Romero-Hidalgo S, Méndez-Sánchez N, Cárdenas V, Bañuelos-Moreno M, Flores YN, Quiterio M, Salmerón J, Sánchez-Muñoz F, Villarreal-Molina T, Aguilar-Salinas CA, Canizales-Quinteros S. PNPLA3 I148M polymorphism is associated with elevated alanine transaminase levels in Mexican Indigenous and Mestizo populations. Mol Biol Rep. 2014;41:4705–4711. doi: 10.1007/s11033-014-3341-0. [DOI] [PubMed] [Google Scholar]
- Larrieta-Carrasco E, León-Mimila P, Villarreal-Molina T, Villamil-Ramírez H, Romero-Hidalgo S, Jacobo-Albavera L, Gutiérrez-Vidal R, López-Contreras BE, Guillén-Pineda LE, Sánchez-Muñoz F, Bojalil R, Mejía-Domínguez AM, Méndez-Sánchez N, Domínguez-López A, Aguilar-Salinas CA, Canizales-Quinteros S. Association of the I148M/PNPLA3 variant with elevated alanine transaminase levels in normal-weight and overweight/obese Mexican children. Gene. 2013;520:185–188. doi: 10.1016/j.gene.2013.03.038. [DOI] [PubMed] [Google Scholar]
- León-Mimila P, Vega-Badillo J, Gutiérrez-Vidal R, Villamil-Ramírez H, Villareal-Molina T, Larrieta-Carrasco E, López-Contreras BE, Kauffer LRM, Maldonado-Pintado DG, Méndez-Sánchez N, Tovar AR, Hernández-Pando R, Velázquez-Cruz R, Campos-Pérez F, Aguilar-Salinas CA, Canizales-Quinteros S. A genetic risk score is associated with hepatic triglyceride content and non-alcoholic steatohepatitis in Mexicans with morbid obesity. Exp Mol Pathol. 2015;98:178–183. doi: 10.1016/j.yexmp.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Li L, Qu HQ, Rentfro AR, Grove ML, Mirza S, Lu Y, Hanis CL, Fallon MB, Boerwinkle E, Fisher-Hoch SP, McCormick JB. PNPLA3 polymorphisms and liver aminotransferase levels in a Mexican American population. Clin Investig Med. 2012:35. doi: 10.1016/j.biotechadv.2011.08.021.Secreted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–7. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- Ma ZA, Zhao Z, Turk J. Mitochondrial dysfunction and β-cell failure in type 2 diabetes mellitus. Exp Diabetes Res. 2012 doi: 10.1155/2012/703538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso FS, Maida M, Petta S. Genetic background in nonalcoholic fatty liver disease: A comprehensive review. World J Gastroenterol. 2015 doi: 10.3748/wjg.v21.i39.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:1198–1209. doi: 10.2337/db06-0506. [DOI] [PubMed] [Google Scholar]
- Morales LS, Flores YN, Leng M, Sportiche N, Gallegos-Carrillo K, Salmeron J. Risk factors for cardiovascular disease among Mexican-American adults in the United States and Mexico: a comparative study. Salud Publica Mex. 2014;56:197–205. doi: 10.21149/spm.v56i2.7335. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu ILK, Brunt EM. Role of liver biopsy in nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20:9026–37. doi: 10.3748/wjg.v20.i27.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmioglu N, Andrew T, Cherkas L, Surdulescu G, Swaminathan R, Spector T, Ahmadi KR. Epidemiology and genetic epidemiology of the liver function test proteins. PLoS One. 2009:4. doi: 10.1371/journal.pone.0004435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi Kanth VV, Sasikala M, Rao PN, Avanthi US, Rao KR, Nageshwar Reddy D. Pooled genetic analysis in ultrasound measured nonalcoholic fatty liver disease in Indian subjects: A pilot study. World J Hepatol. 2014;6:435–442. doi: 10.4254/wjh.v6.i6.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector RS, Thyfault JP, Uptergrove GM, Morris EM, Naples P, Borengasser SJ, Mikus CR, Laye MJ, Harold M, Booth FW, Ibdah JA, Naples SP, Borengasser SJ, Mikus CR, Laye MJ, Laughlin MH, Booth FW, Ibdah JA. Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non- alcoholic fatty liver disease in an obese rodent model. J Hepatol. 2011;52:727–736. doi: 10.1016/j.jhep.2009.11.030.Mitochondrial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. James M. Decker. Hepatology. 1994;19:540–542. doi: 10.1002/hep.1840190237. [DOI] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML, Clore JN. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–92. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- Secretaría de Economía. Programa nacional de protección a los derechos del consumidor 2013–2018. Diario Oficial de la Federación; México: 2014. n.d. [Google Scholar]
- Shang X-R, Song J-Y, Liu F-H, Ma J, Wang H-J. GWAS-Identified Common Variants With Nonalcoholic Fatty Liver Disease in Chinese Children. J Pediatr Gastroenterol Nutr. 2015;60:669–674. doi: 10.1097/MPG.0000000000000662. [DOI] [PubMed] [Google Scholar]
- Song G, Xiao C, Wang K, Wang Y, Chen J, Yu Y, Wang Z, Deng G, Sun X, Zhong L, Zhou C, Qi X, Wang S, Peng Z, Wang X. Association of patatin-like phospholipase domain- containing protein 3 gene polymorphisms with susceptibility of nonalcoholic fatty liver disease in a Han Chinese population. 2016;33:0–5. doi: 10.1097/MD.0000000000004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Castaño GO, Scian R, Gianotti TF, Dopazo H, Rohr C, Gaj G, Martino JS, Sevic I, Flichman D, Pirola CJ. Serum aminotransferases in nonalcoholic fatty liver disease are a signature of liver metabolic perturbations at the amino acid and Krebs cycle level. Am J Clin Nutr. 2016;103:422–434. doi: 10.3945/ajcn.115.118695. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Pirola CJ. Alanine and aspartate aminotransferase and glutamine-cycling pathway: Their roles in pathogenesis of metabolic syndrome. World J Gastroenterol. 2012;18:3775–3781. doi: 10.3748/wjg.v18.i29.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, Eiriksdottir G, Garcia ME, Launer LJ, Nalls MA, Clark JM, Mitchell BD, Shuldiner AR, Butler JL, Tomas M, Hoffmann U, Hwang SJ, Massaro JM, O’Donnell CJ, Sahani DV, Salomaa V, Schadt EE, Schwartz SM, Siscovick DS, Voight BF, Carr JJ, Feitosa MF, Harris TB, Fox CS, Smith AV, Kao WHL, Hirschhorn JN, Borecki IB. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011:7. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsukawa H, Tani Y, Otsu R, Nakagawa H, Hitomi K. Global identification and analysis of isozyme-specific possible substrates crosslinked by transglutaminases using substrate peptides in mouse liver fibrosis. Sci Rep. 2017;7:45049. doi: 10.1038/srep45049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than NN, Newsome PN. A concise review of non-alcoholic fatty liver disease. Atherosclerosis. 2015 doi: 10.1016/j.atherosclerosis.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Thapa BR, Walia A. Liver function tests and their interpretation. Indian Journal of Pediatrics. 2007:663–671. doi: 10.1007/s12098-007-0118-7. [DOI] [PubMed] [Google Scholar]
- Tuomisto A, Sund M, Tahkola J, Latvanlehto A, Savolainen ER, Autio-Harmainen H, Liakka A, Sormunen R, Vuoristo J, West A, Lahesmaa R, Morse HC, Pihlajaniemi T. A mutant collagen XIII alters intestinal expression of immune response genes and predisposes transgenic mice to develop B-cell lymphomas. Cancer Res. 2008;68:10324–10332. doi: 10.1158/0008-5472.CAN-08-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Chantada M, Gonzalez-Lahera A, Martinez-Arranz I, Garcia-Monzon C, Regueiro MM, Garcia-Rodriguez JL, Schlangen KA, Mendibil I, Rodriguez-Ezpeleta N, Lozano JJ, Banasik K, Justesen JM, Joergensen T, Witte DR, Lauritzen T, Hansen T, Pedersen O, Veyrie N, Clement K, Tordjman J, Tran A, Le Marchand-Brustel Y, Buque X, Aspichueta P, Echevarria-Uraga JJ, Martin-Duce A, Caballeria J, Gual P, Castro A, Mato JM, Martinez-Chantar ML, Aransay AM. Solute carrier family 2 member 1 is involved in the development of nonalcoholic fatty liver disease. Hepatology. 2013;57:505–514. doi: 10.1002/hep.26052. [DOI] [PubMed] [Google Scholar]
- WHO. Obesity: preventing and managing the global epidemic. Report of a WHO consultation, World Health Organization technical report series. 2000 doi: 10.1016/S0140-6736(57)91352-1. [DOI] [PubMed] [Google Scholar]
- Wiegand J, Mössner J, Tillmann HL. Nichtalkoholische Fettleber und Steatohepatitis. Internist (Berl) 2007;48:154–163. doi: 10.1007/s00108-006-1796-3. [DOI] [PubMed] [Google Scholar]
- Zain SM, Mohamed R, Mahadeva S, Cheah PL, Rampal S, Basu RC, Mohamed Z. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum Genet. 2012 doi: 10.1007/s00439-012-1141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.