Abstract
Meier-Gorlin syndrome (MGS) is an autosomal recessive disorder characterized by microtia, primordial dwarfism, small ears, and skeletal abnormalities. Patients with MGS often carry mutations in the genes encoding the components of the pre-replicative complex such as Origin Recognition Complex (ORC) subunits Orc1, Orc4, Orc6, and helicase loaders Cdt1 and Cdc6. Orc6 is an important component of ORC and has functions in both DNA replication and cytokinesis. Mutation in conserved C-terminal motif of Orc6 associated with MGS impedes the interaction of Orc6 with core ORC. In order to study the effects of MGS mutation in an animal model system we introduced MGS mutation in Orc6 and established Drosophila model of MGS. Mutant flies die at third instar larval stage with abnormal chromosomes and DNA replication defects. The lethality can be rescued by elevated expression of mutant Orc6 protein. Rescued MGS flies are unable to fly and display multiple planar cell polarity defects.
Keywords: ORC, Orc6, Drosophila, Meier-Gorlin syndrome
INTRODUCTION
Meier-Gorlin syndrome (MGS) is an autosomal recessive disorder comprising primordial dwarfism, small ears and aplastic or hypoplastic patella. Recently, mutations in genes encoding the members of the pre-replication complex (pre-RC) such as Orc1, Orc4, Orc6, Cdc6, and Cdt1 were identified in many MGS patients [Bicknell et al., 2011a,b; Guernsey et al., 2011]. Since observed mutations cluster in pre-RC components, it has been suggested that the clinical phenotype is caused by defects in DNA replication initiation. The pre-RC complex is essential for DNA replication and mutations in its components are expected to impair cell proliferation and to reduce growth.
The Origin Recognition Complex (ORC) plays a central role in the initiation of DNA replication but is also involved in non-replicative functions [Bell and Stillman, 1992; Bell, 2002; Chesnokov, 2007; Sasaki and Gilbert, 2007]. Orc6 protein, the smallest subunit of ORC, is important for DNA replication in all species [Lee and Bell, 1997; Chesnokov et al., 2001, 2003; Semple et al., 2006; Chen et al., 2007; Balasov et al., 2007, 2009]. Orc6 is also essential for cytokinesis in Drosophila and human cells [Prasanth et al., 2002; Chesnokov et al., 2003]. In Drosophila, Orc6 stimulates septin complex GTPase activity and polymerization during filament assembly through protein–protein interactions [Huijbregts et al., 2009; Akhmetova et al., 2015]. Metazoan Orc6 proteins consist of two functional domains: a larger N-terminal domain important for binding of DNA and smaller C-terminal domain important for protein-protein interactions [Chesnokov et al., 2003; Balasov et al., 2007; Duncker et al., 2009; Bleichert et al., 2013]. It has been shown that in metazoan species N-terminal domain of Orc6 carries a structural homology with TFIIB transcription factor [Chesnokov et al., 2003; Balasov et al., 2007; Liu et al., 2011]. The conserved motif in C-terminus of Orc6 is responsible for the interaction with Orc3 subunit of ORC [Bleichert et al., 2013] and tyrosine 232 to serine mutation in this region of the protein is linked to the MGS in humans [Bicknell et al., 2011a; de Munnik et al., 2012].
In order to study the effects of MGS mutation in animal model system we introduced MGS mutation in Drosophila Orc6 and established fruit fly model of MGS. Our findings support the hypothesis that impaired DNA replication underlies the developmental defects characteristic of this disorder. The Drosophila model of MGS will be useful to uncover molecular mechanisms of MGS and provide number of insights into disease origin and development.
MATERIALS AND METHODS
Site-Directed Mutagenesis
The amino acid substitution mutants Orc6-W228A/K229A, Orc6-D224A/Y225A, Orc6-Y225F, and Orc6-Y225S were generated following the Stratagene site-directed mutagenesis protocol (http://www.chem.agilent.com/Library/usermanuals/Public/200518.pdf). The deletion Orc6-200 mutant was created by PCR [Balasov et al., 2007].
Rescue of the orc6 Mutant
The orc6 native promoter was cloned upstream of GAL4 inducible UAS promoter into a pUAST vector. Both promoters were able to drive expression of the GFP-Orc6 fused proteins (wild-type and mutants). All constructs were injected into Drosophila embryos and individual transgenic strains were set up. Expression of transgenic proteins was verified by Western blots. In the rescue experiments with native promoter, progeny from heterozygo us Δorc6/Cy;GFP-orc6 was analyzed for the presence of Δorc6/Δorc6;GFP-orc6 adult flies, where Δorc6 represents orc6 deletion [Balasov et al., 2009]. The UAS promoter was activated by the expression of GAL4 with a strong constitutive tubulin promoter. In this experiment, females of the genotype Δorc6/Cy;GFP-orc6 were crossed to males Δorc6/Cy;tubulin-GAL4/TM3,Sb, and resulting progeny was analyzed for the presence of Δorc6/Δorc6;GFP-orc6/tubulin-GAL4 adults. tubulin-GAL4 fly strain was obtained from Bloomington Stock Center.
Mitotic Chromosome Preparation
Preparation of mitoses was described previously [Lebedeva et al., 2000]. Briefly, third instar larval neural ganglia were incubated in 0.075M KCl for 5 min, fixed in methanol with acetic acid (3:1) for 20 min and then dispersed in a drop of 50% propionic acid on a slide. Slides were dried and stained with 5% Giemsa’s solution.
Immunoprecipitation (IP)
Ten fresh dissected ovaries or 20 salivary glands were crushed with glass homogenizer in 100 μl of high salt IP buffer (25 mM Hepes pH 7.6, 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 450 mM NaCl, 0.01% Triton X100) and extracted for 1 hr at 4°C with continuous rotation. Extracts were centrifuged at 15,000g for 15 min and supernatants were diluted threefold with low salt IP buffer (25 mM Hepes pH 7.6, 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 0.01% Triton X100). Protein A-Sepharose (Bio Vision Cat.6501-5) and rabbit polyclonal Orc2 antibodies were incubated with supernatant for 3 hr, washed 3 times with IP buffer and diluted in 10 ml of IP buffer. Samples were separated with SDS-PAGE and analyzed with Western blot. GFP-Orc6 fusion proteins were detected with anti-GFP monoclonal antibody (Clontech Laboratories).
Western Blot of Larval Neural Ganglia
Neural ganglia of third instar larvae were dissected and imaginal discs removed. Soluble fraction of MCM2–7 was extracted with 450 mM NaCl, 0.5% NP-40 in PBS for 1 hr. Insoluble proteins were pelleted, the pellet washed three times, and pellets from 8 to 10 ganglia per lane were loaded onto a SDS-PAGE gel. Mcm5 was detected by Western blotting with Mcm5 rabbit polyclonal antibodies (gift from Dr. Michael Botchan).
BrdU Labeling and Immunostaining of Polytene Chromosomes
Larval neural ganglia or testes were incubated with 1 μM BrdU in PBS for 30 min at 25°C. BrdU incorporation was detected by monoclonal antibodies (Becton Dickinson) following manufacturer recommendations. Polytene chromosomes of the salivary glands from third instar larvae were squashed in 45% acetic acid, frozen in liquid nitrogen and immunostained according to previously described protocol [Sullivan et al., 2000; Balasov et al., 2009].
RESULTS AND DISCUSSION
In our previous work we generated a deletion of the orc6 gene in Drosophila using the method of P element imprecise excision. Lethal alleles of orc6 are defective in DNA replication and also show abnormal chromosome condensation and segregation [Balasov et al., 2009]. Further sequence analyses of metazoan Orc6s revealed extremely conserved motifs within C-terminal domain of the protein including amino acids D224/Y225 and W228/K229. Mutations of these residues to alanines resulted in reduced DNA replication, chromosomal abnormalities, and lethality [Balasov et al., 2009; Bleichert et al., 2013]. Corresponding amino acids in humans are localized at position D231/Y232 and W235/K236. Sequence analysis of genes encoding for Orc6 from individuals with MGS identified a pathogenic missense mutation resulting in the substitution of tyrosine at position 232 to serine (Y232S) referred further as MGS mutation. To create Drosophila model of MGS we mutated tyrosine at position 225 to serine (Y225S) in C-terminus of Drosophila Orc6 protein. The resulting mutant transgene was introduced into orc6-deleted background and a fly strain expressing mutant protein was created. We compared the effect of the Orc6 carrying MGS mutation (Orc6-Y225S) with earlier reported orc6 transgenes carrying knockout mutations Orc6-D224A/Y225A, Orc6-W228A/K229A (described in detail in [Balasov et al., 2009; Bleichert et al., 2013]), Orc6-Y225F and Orc6-200 deletion missing whole C-terminal domain. Mutant orc6 transgenes used in this study are presented in Figure 1. All transgenic constructs contained GFP fused to N-terminus of Orc6 which did not affect normal Orc6 function [Balasov et al., 2009].
FIG. 1.

Mutations in a conserved C-terminal helical domain of Orc6 used in this study. Black box represents DNA binding N-terminal domain of Orc6. Conservatives amino acids of the C-terminus are shown in insert. Mutated amino acids are colored in red. Arrow indicates Meier-Gorlin syndrome associated mutation.
First, we tested the orc6 MGS transgene for ability to rescue third instar lethality associated with deletion of orc6 gene. We found that the expression of orc6-Y225S under orc6 native promoter was not enough to rescue flies to viability. In human Orc6, Y232 to S substitution corresponds to a mild clinical appearance of MGS [Bicknell et al., 2011a; de Munnik et al., 2012], however, in Drosophila, phenotype was similar to orc6 deleted mutant. Molecular and cell analysis of tissues derived from these animals revealed significantly reduced DNA replication judged by BrdU incorporation in either larval neural ganglia (Fig. 2A) or testes (Fig. 2B).
FIG. 2.
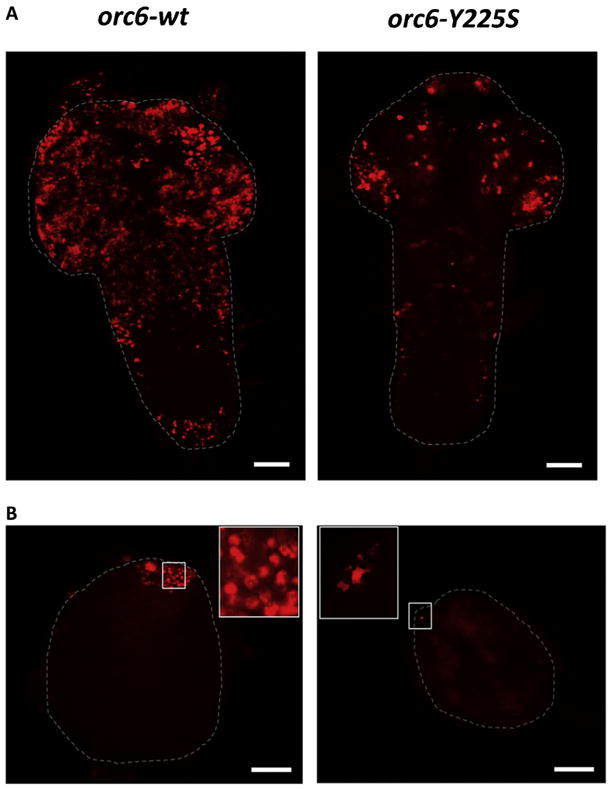
Replication defects in Drosophila MGS mutant. Both neural ganglia (A) and testes (B) of the Orc6-Y225S mutant show significantly reduced BrdU incorporation level. Testes are smaller and never reach normal size. Testes tips with BrdU incorporation spots are also shown in the enlarged inserts. Scale bar 50 μm.
As the next step, we analyzed mitotic chromosomes from neural ganglia of MGS mutant larvae. Normal Drosophila melanogaster karyotype consists of four pairs of chromosomes (Fig. 3A, wildtype). In cells carrying Orc6-Y225S mutation chromosomes lost parts of their arms, appeared aberrantly condensed, fragmented, misaligned and sometimes polyploid (Fig. 3A, lower row). In our earlier analysis of Orc6 functions in vivo, C-terminal Orc6 deletion (Orc6-200) as well as Orc6-D224A/Y225A and Orc6-W228A/ K229A knockout Orc6 mutations also resulted in third instar lethality associated with defects in DNA replication and chromosome abnormalities [Balasov et al., 2009]. Similar chromosome defects were observed for other ORC mutants in flies [Landis et al., 1997; Pinto et al., 1999; Loupart et al., 2000; Pflumm and Botchan, 2001; Park and Asano, 2008].
FIG. 3.

MGS mutations result in chromosome defects, disrupt the interaction of Orc6 with core ORC and impair MCM chromatin association. (A) Mitotic chromosomes of the neural ganglia from wild-type and mutant larvae. Upper row: karyotypes of orc6 deletion mutant, wild-type (Canton S) and orc6 deletion rescued with orc6 wild-type transgene are shown from left to right. Karyotypes of the MGS mutant orc6-Y225S mutant are shown in the lower row. Scale bar 10 mm. (B) Immunostaining of polytene chromosomes from salivary glands expressing GFP-Orc6-wt or MGS mutant GFP-Orc6-Y225S protein. Anti-Orc6 antibodies were used. (C) Western immunoblotting analysis of ORC complexes precipitated using anti-Orc2 antibody from extracts isolated from fly ovaries expressing different GFP-tagged Orc6 proteins under control of native orc6 promoter: GFP-Orc6-D224A/Y225A, GFP-Orc6-Y225S, GFP-Orc6-Y225F and GFP-Orc6-wt (wild-type). Canton S flies without any transgenes were used as a negative control. GFP-Orc6 fusion proteins were detected with anti-GFP monoclonal antibody, while Orc5 subunit of ORC was detected using anti-Orc5 antibodies. Ovary extract (In) and immunoprecipitated material (IP) are shown for each transgene. (D) Chromatin association of Mcm5, a member of the MCM2–7 complex, is reduced in orc6-null fly larvae expressing Orc6 mutants. Neural ganglia of homozygous, orc6-null Drosophila larvae expressing GFP-tagged Orc6 proteins under native orc6 promoter were isolated and subjected to salt extraction to solubilize cellular proteins. Insoluble and chromatin associated proteins were pelleted by centrifugation and analyzed by Western blotting using Mcm5 polyclonal antibodies. Pnut was used as a loading control.
The metazoan Orc6 consists of two domains, a larger N-terminal domain which carries homology with TFIIB transcription factor and is important for DNA binding [Chesnokov et al., 2003; Balasov et al., 2007; Liu et al., 2011] and a shorter C-terminal domain important for the function of Orc6 in cytokinesis [Prasanth et al., 2002; Chesnokov et al., 2003; Huijbregts et al., 2009; Bernal and Venkitaraman, 2011; Akhmetova et al., 2015]. The C-terminal domain of Orc6 is also essential and sufficient for the interaction of Orc6 with core ORC [Bleichert et al., 2013]. Specifically, the Y225S mutation in Drosophila Orc6 disrupted the interaction of the protein with Orc3 and with the rest of the ORC in vitro [Bleichert et al., 2013]. Mutating conserved D224/Y225 and W228/K229 amino acids within C-terminus into alanines had even more dramatic effect suggesting that these amino acids mediate Orc6 recruitment into ORC and moreover, are required for replicative helicase MCM2-7 loading onto chromatin [Balasov et al., 2009; Bleichert et al., 2013]. As expected, MGS mutation did not change DNA binding ability of Orc6 and its association with chromosomes in vivo when expressed in Drosophila salivary glands. Both GFP-Orc6 wild-type and GFP-Orc6-Y225S mutant proteins were found on chromosomes in a similar pattern (Fig. 3B) unlike Orc6 mutants carrying mutations in the N-terminal TFIIB-like domain which is important for DNA binding [Balasov et al., 2007; Liu et al., 2011].
However, the association with core ORC was disrupted for Orc6 carrying Y225S MGS mutation as is shown in Figure 3C. In a positive control experiment with flies carrying wild-type GFP-orc6, ORC was immunoprecipitated with the anti-Orc2 antibody from ovary extract and, as expected, “pulled down” material contained Orc5 and GFP–Orc6 wild-type subunits (Fig. 3C, lanes 7 and 8). In a negative control experiment with flies carrying GFP–orc6-D224A/ Y225A transgene, the amount of GFP–Orc6-D224A/Y225A mutant immunoprecipitated with the anti-Orc2 antibody was significantly diminished compared to wild-type GFP–Orc6, as shown by Western blotting (Fig. 3C, lanes 5 and 6; and [Bleichert et al., 2013]). Similarly, in flies carrying the GFP-orc6-Y225S MGS transgene, the amount of immunoprecipitated Orc6 mutant protein was greatly reduced (Fig. 3C, lanes 3 and 4), indicating that this mutation impedes Orc6 binding to core ORC.
Low amounts of hexameric ORC on DNA are expected to lead to impaired pre-RC formation. Therefore, next, we analyzed chromatin association of a member of the MCM2–7 complex, Mcm5, in Drosophila larval neural ganglia expressing either wild-type or mutant GFP–orc6 transgenes (Fig. 3D). Wild-type GFP-Orc6 and mutantGFP-Orc6-D224A/Y225Awereusedaspositiveandnegative controls. After salt extraction of larval neural ganglia extracts, the amount of Mcm5 in the pellet fraction was greatly decreased when GFP-orc6-Y225S (Fig. 3D, lane 3) or GFP-orc6-D224A/Y225A (Fig. 3D, lane 5) transgene was expressed as the sole source of Orc6 compared to larval neural ganglia expressing the wild-type GFP–orc6 (Fig. 3D, lane 1). Thus, a MGS mutation within Orc6 not only disrupts hexameric ORC formation but also prevents MCM loading onto chromatin, explaining the reduced DNA synthesis observed in mutant flies (Fig. 2 and [Balasov et al., 2009]).
Next, we tried to create less defective mutation for conserved tyrosine 225 within Orc6. Therefore, in addition to GFP-Orc6-Y225S with MGS mutation we also tested GFP-Orc6-Y225F construct where tyrosine (also known as 4-hydroxy-phenylalanine) at position 225 was replaced with phenylalanine which lacks the specific −OH group at carbon 4. Obtained transgene orc6-Y225F was introduced into orc6 deletion background and tested for viability. We found that the expression of orc6-Y225F restores viability of orc6 deletion and rescued flies have a phenotype indistinguishable from wild-type. Consistent with this result GFP-Orc6-Y225F protein was detected in IP with anti-Orc2 antibodies indicating that the interaction with core ORC was not affected (Fig. 3C, lanes 9 and 10). It is interesting to note that two organisms evolutionally distant from Drosophila such as D. discoideum and P.pallidum (slime molds) also contain phenylalanine instead of tyrosine at this position [Bleichert et al., 2013].
A single amino acid human MGS mutation in Orc6 resulted in a mild clinical appearance [Bicknell et al., 2011a; de Munnik et al., 2012], whereas the Drosophila Orc6-Y225S MGS mutation is lethal. We hypothesized that elevated level of expression of mutant Orc6 could improve survival rate of adult flies. We used the GAL4-UAS system to boost the expression of transgenes under a constitutive ubiquitous tubulin promoter [Brand and Perrimon, 1993; Duffy, 2002]. Percentage of rescued flies was calculated based on expected segregation of phenotypes (Table I). Under the native promoter GFP-orc6 wild-type rescued 92% of expected progeny to adult stage based on a total of 989 analyzed flies. GFP-orc6-Y225F rescued 50% (total progeny 214). Other mutant transgenes GFP-orc6-Y225S (total 515), GFP-orc6-D224A/Y225A (total 617), GFP-orc6-W228A/K229A (total 713), GFP-orc6-200 (total 820) did not rescue any flies to viability. However, when expressed under the strong tubulin promoter, GFP-orc6-Y225S rescued 16% of adults (total progeny 1281), GFP-orc6-D224A/Y225A rescued 20% (total 838) and GFP-orc6 wild-type rescued 91% of adults (total 704). GFP-orc6-W228A/K229A, and GFP-orc6-200 did not rescue any adult flies.
TABLE I.
Rescue of orc6 Deletion Mutant With Different orc6 Transgenes Under Native or tubulin Promoter
| GFP-orc6-wild-type | GFP-orc6-Y225F | GFP-orc6-Y225S | GFP-orc6-D224A/Y225A | GFP-orc6-W228A/K229A | GFP-orc6-200 | |
|---|---|---|---|---|---|---|
| Native | + | + | − | − | − | − |
| promoter | 92% (989) | 50% (214) | 0% (515) | 0% (617) | 0% (713) | 0% (820) |
| tubulin | + | + | + | + | − | − |
| promoter | 91% (704) | 55% (183) | 16% (1281) | 20% (838) | 0% (489) | 0% (504) |
In all rescue experiments of the orc6 deletion mutant we used transgenes expressing fused GFP-Orc6 proteins. Six different constructs were designed. GFP was attached to the N-terminus of either wild-type Orc6 or one of the mutants. All constructs contained two active promoters-native orc6 promoter and GAL4 inducible UAS promoter. (+), the expression of transgene restores viability of orc6-deleted flies; (−), no rescue. Percentage of rescued flies was calculated based on expected segregation of phenotypes. Total progeny analyzed is shown in parentheses.
Rescue to viability also correlated with a restoration of normal karyotype as larval mitotic chromosomes from animals with boosted expression of GFP-orc6-Y225S and GFP-orc6-D224A/ Y225A were normal as compared to the wild-type chromosomes (Fig. 4A). Mitotic chromosomes from cells with elevated expression of GFP-orc6-W228A/K229A or GFP-orc6-200 remained fragmented and misaligned indicating that severe defects caused by these particular mutations cannot be relieved by over-expression. The recently published structure of Drosophila ORC revealed fine details about protein-protein interactions between Orc6 C-terminus and core ORC, showing the importance of Orc6 amino acids Y225, W228, M232, and A236 for hydrophobic binding site in Orc3 [Bleichert et al., 2015]. Orc6 W228 residue is localized in the core of Orc6-Orc3 binding site. This might explain the more severe phenotype associated with W228A/K229A mutations observed in our study.
FIG. 4.

The elevated expression of Orc6 MGS transgenes rescues normal karyotype and restores Orc6 association with core ORC. (A) The expression of orc6-Y225S and orc6-D224A/Y225A transgenes under strong tubulin promoter restores normal karyotype. orc6-W228A/K229A and orc6-200 mutants are not able to rescue chromosome defects under either promoter. Scale bar 10 μm. (B) Western immunoblotting analysis of ORC complexes precipitated using anti-Orc2 antibody from extracts isolated from fly ovaries over-expressing different GFP-tagged Orc6 proteins under control of strong tubulin promoter. (+), tubulin boost; (−), no tubulin boost. In, input; IP, immunoprecipitated material.
The elevated expression of GFP-orc6-Y225S and GFP-orc6-D224A/Y225A allowed detection of mutant Orc6 proteins when immunoprecipitated with anti-Orc2 antibodies (Fig. 4B) indicating that increasing the level of expression of Orc6 mutants was enough to force the equilibrium toward the formation of six-subunit ORC. C-terminus of both Drosophila and human Orc6s contains conservative motifs responsible for association with ORC complex. Both flies and humans with C-terminal truncations do not survive to adult stage [Balasov et al., 2009; Shalev et al., 2015]. However, point mutation in human (Y232S) demonstrates mild postnatal phenotype [Bicknell et al., 2011a; de Munnik et al., 2012], while the Drosophila Y225S MGS mutation shows no difference from lethal orc6 deletion. Importantly, human Orc6 is loosely associated with the core ORC [Vashee et al., 2001, 2003; Ranjan and Gossen, 2006], whereas Drosophila Orc6 associates with other ORC subunits more tightly [Gossen et al., 1995; Chesnokov et al., 1999, 2001]. This might explain why the Y225S MGS mutation has more dramatic effect for survival of Drosophila than in humans. In this study we found that elevated expression of mutant protein rescues third instar lethality, restores normal karyotype and allows detection of Orc6 on DNA with the rest of ORC complex. It would be interesting to test if patients with a Y225S MGS mutation have an increased level of mutant Orc6 expression, resulting in reduced growth retardation and milder clinical manifestation of symptoms.
Figure 5 shows phenotype of the MGS model flies overexpressing GFP-orc6-Y225S as a sole source of Orc6. The surviving flies carrying this MGS mutation were shorter as compared to the flies carrying a copy of the wild-type orc6 transgene driven by tubulin-GAL4 (Fig. 5A). Measurements of head, thorax and abdomen lengths reveal that MGS flies have slightly longer heads (0.47 ± 0.05 mm vs. 0.40 ± 0.03 mm, P < 0.05) and shorter abdomens (1.38 ± 0.12 mm versus 1.50 ± 0.10 mm, P < 0.05). Mutant flies are not able to fly and all have upheld wing posture (Fig. 5A and compare Video MGS and Video WT). A similar flightless phenotype is a characteristic of muscle-defective mutations such as Mhc-myosin heavy chain [Homyk and Emerson, 1988] suggesting possible defects in muscle development. Other phenotypes include the loss of posterior scutellar bristles (Fig. 5B), the appearance of rough eyes spots (Fig. 5C) as well as irregular disoriented hairs on an abdomen (Fig. 5D). All these observed phenotypes are consistent with planar cell polarity defects [Devenport, 2014; Galic and Matis, 2015]. Genetic studies in Drosophila have identified two signaling systems, the Frizzled and Fat/Dachsous system, which are both required for PCP establishment in many different tissues and are conserved in metazoan species [Bosveld et al., 2012; Singh and Mlodzik, 2012]. The Fat/Dachsous cadherin system is known to control tissue size and shape in animals [Lawrence et al., 2008; Goodrich and Strutt, 2011]. How animal cells cooperate to build tissues of particular forms remains a fundamental unsolved problem in biology. It will be interesting to test how MGS mutation in Orc6 resulting in MGS phenotype might affect organ growth and ultimately tissue and organism size. We think that availability of the Drosophila model of MGS will allow thorough studies revealing crucial and conserved mechanisms of organism development and disease pathogenesis.
FIG. 5.
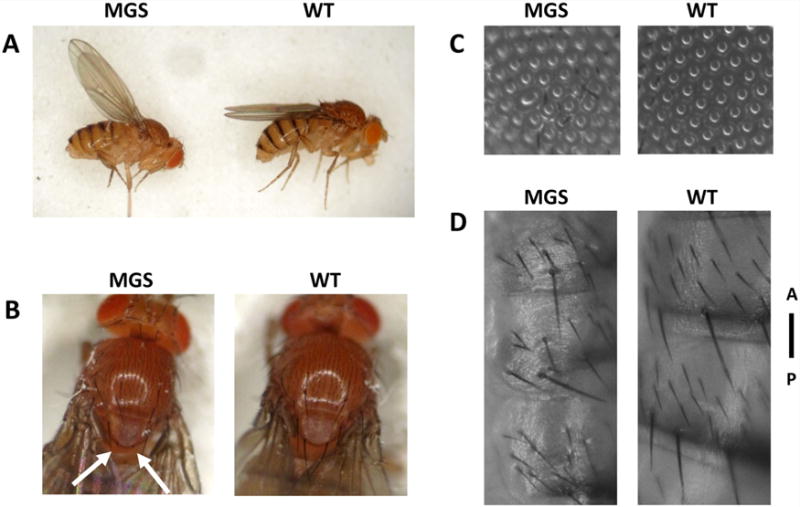
Mutant phenotype of orc6 deletion flies rescued by overexpression of orc6-Y225S under strong ubiquitous tubulin promoter. Mutant flies are not able to fly (compare Video MGS and Video WT) and have upheld wing posture (A). Other shown phenotypes include missing posterior scutellar bristles (B), rough spots on the eye (C) and irregular hair pattern on the abdomen (D). A-anterior, P-posterior.
Acknowledgments
We thank Dr. Michael Botchan for providing anti-Mcm5 antibodies. This work was supported by a grant from NIH to IC (GM097052).
Footnotes
Authors declare no conflict of interests.
References
- Akhmetova K, Balasov M, Huijbregts RP, Chesnokov I. Functional insight into the role of Orc6 in septin complex filament formation in Drosophila. Mol Biol Cell. 2015;26(1):15–28. doi: 10.1091/mbc.E14-02-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27(8):3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M, Huijbregts RP, Chesnokov I. Functional analysis of an Orc6 mutant in Drosophila. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0902670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex [see comments] Nature. 1992;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16(6):659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Bernal JA, Venkitaraman AR. A vertebrate N-end rule degron reveals that Orc6 is required in mitosis for daughter cell abscission. J Cell Biol. 2011;192(6):969–978. doi: 10.1083/jcb.201008125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, van Bokhoven H, Dean J, Edrees AY, Feingold M, Fryer A, Hoefsloot LH, Kau N, Knoers NV, Mackenzie J, Opitz JM, Sarda P, Ross A, Temple IK, Toutain A, Wise CA, Wright M, Jackson AP. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet. 2011a;43(4):356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, Johnson D, Wise C, Jackson AP, O’Driscoll M, Jeggo PA. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011;43(4):350–355. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- Bleichert F, Balasov M, Chesnokov I, Nogales E, Botchan MR, Berger JM. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519(7543):321–326. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosveld F, Bonnet I, Guirao B, Tlili S, Wang Z, Petitalot A, Marchand R, Bardet PL, Marcq P, Graner F, Bellaiche Y. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science. 2012;336(6082):724–727. doi: 10.1126/science.1221071. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007;21(22):2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 1999;13(10):1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci USA. 2001;98(21):11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100(16):9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- Devenport D. The cell biology of planar cell polarity. J Cell Biol. 2014;207(2):171–179. doi: 10.1083/jcb.201408039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34(1–2):1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10(3):214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB, Clayton-Smith J, Edrees AY, Feingold M, Fryer A, van Hagen JM, Hennekam RC, Jansweijer MC, Johnson D, Kant SG, Opitz JM, Ramadevi AR, Reardon W, Ross A, Sarda P, Schrander-Stumpel CT, Schoots J, Temple IK, Terhal PA, Toutain A, Wise CA, Wright M, Skidmore DL, Samuels ME, Hoefsloot LH, Knoers NV, Brunner HG, Jackson AP, Bongers EM. Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Euv J Hum Genet. 2012;20(6):598–606. doi: 10.1038/ejhg.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic M, Matis M. Polarized trafficking provides spatial cues for planar cell polarization within a tissue. BioEssays. 2015 doi: 10.1002/bies.201400196. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138(10):1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex [see comments] Science. 1995;270(5242):1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, Perry S, Ferguson M, LeBlanc M, Paquette J, Patry L, Rideout AL, Thomas A, Orr A, McMaster CR, Michaud JL, Deal C, Langlois S, Superneau DW, Parkash S, Ludman M, Skidmore DL, Samuels ME. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43(4):360–364. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- Homyk T, Jr, Emerson CP., Jr Functional interactions between unlinked muscle genes within haploinsufficient regions of the Drosophila genome. Genetics. 1988;119(1):105–121. doi: 10.1093/genetics/119.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, Chesnokov I. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Molecular Biol Cell. 2009;20(1):270–281. doi: 10.1091/mbc.E08-07-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G, Kelley R, Spradling AC, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci USA. 1997;94(8):3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Struhl G, Casal J. Do the protocadherins Fat and Dachsous link up to determine both planar cell polarity and the dimensions of organs? Nat Cell Biol. 2008;10(12):1379–1382. doi: 10.1038/ncb1208-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva LI, Trunova SA, Omel’ianchuk LV. [Genetic control of mitosis. Adaptive modifications of v158 mutation expression] Genetika. 2000;36(10):1348–1354. [PubMed] [Google Scholar]
- Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17(12):7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci USA. 2011;108(18):7373–7378. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupart ML, Krause SA, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- Park SY, Asano M. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc Natl Acad Sci USA. 2008;105(34):12343–12348. doi: 10.1073/pnas.0805189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–16707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Pinto S, Quintana DG, Smith P, Mihalek RM, Hou ZH, Boynton S, Jones CJ, Hendricks M, Velinzon K, Wohlschlegel JA, Austin RJ, Lane WS, Tully T, Dutta A. Latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant [see comments] Neuron. 1999;23(1):45–54. doi: 10.1016/s0896-6273(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297(5583):1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA. 2006;103(13):4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19(3):337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Semple JW, Da-Silva LF, Jervis EJ, Ah-Kee J, Al-Attar H, Kummer L, Heikkila JJ, Pasero P, Duncker BP. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J. 2006;25(21):5150–5158. doi: 10.1038/sj.emboj.7601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev SA, Khayat M, Etty DS, Elpeleg O. Further insight into the phenotype associated with a mutation in the ORC6 gene, causing Meier-Gorlin syndrome 3. Am J Med Genet Part A. 2015;167A(3):607–611. doi: 10.1002/ajmg.a.36906. [DOI] [PubMed] [Google Scholar]
- Singh J, Mlodzik M. Planar cell polarity signaling: coordination of cellular orientation across tissues. 4. Vol. 1. Wiley Interdiscip Rev Dev Biol; 2012. pp. 479–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, Hawley RS. Drosophila Protocols. Cold Spring Harbor, New York: CSHL Press; 2000. p. 697. [Google Scholar]
- Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276(28):26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17(15):1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]


