Abstract
The complex cell envelopes of Corynebacterineae contribute to the virulence of pathogenic species (such as Mycobacterium tuberculosis and Corynebacterium diphtheriae) and capacity of nonpathogenic species (such as Corynebacterium glutamicum) to grow in diverse niches. The Corynebacterineae cell envelope comprises an asymmetric outer membrane that overlays the arabinogalactan-peptidoglycan complex and the inner cell membrane. Dissection of the lipid composition of the inner and outer membrane fractions is important for understanding the biogenesis of this multilaminate wall structure. Here, we have undertaken the first high-resolution analysis of C. glutamicum inner and outer membrane lipids. We identified 28 lipid (sub)classes (>233 molecular species), including new subclasses of acylated/acetylated trehalose mono/dicorynomycolic acids, using high-resolution LC/MS/MS coupled with mass spectral library searches in MS-DIAL. All lipid subclasses exhibited polarized distributions across the inner and outer membrane fractions generated by differential solvent extraction. Strikingly, deletion of the TmaT protein, which is required for transport of trehalose corynomycolates across the inner membrane, led to the accumulation of triacylglycerols in the inner membrane and to suppressed synthesis of phosphatidylglycerol and alanylated lipids. These analyses indicate unanticipated connectivity in the synthesis and/or transport of different lipid classes in C. glutamicum.
Keywords: bacterial membranes, mycobacterium, trehalose lipids, lipidomics
Mycobacteria and Corynebacteria are Gram-positive bacteria of the suborder Corynebacterineae. This group includes medically important pathogens, such as Mycobacterium tuberculosis, Mycobacterium leprae (1), and Corynebacterium diphtheriae (2), the causative agents of tuberculosis, leprosy, and diphtheria, respectively. M. tuberculosis alone is estimated to chronically infect one-third of the world’s population and is the leading cause of mortality and morbidity from a bacterial pathogen, with around 1.5 million deaths per year (3, 4). All these bacteria synthesize a complex multilaminate cell envelope that contributes to the intrinsic resistance of these bacteria to many antibiotics and their success as pathogens. The mycobacterial/corynebacterial cell envelopes comprise the inner (cell) membrane (IM), an intermediate layer of covalently linked peptidoglycan, arabinogalactan (AG), and long-chain mycolic acids (the mAGP complex), and an outer membrane (OM) comprising free (glyco)lipids that interdigitate with the underlying AG-linked mycolic acids. Genetic or chemical inhibition of many of these cell envelope lipids leads to loss of viability and/or virulence in pathogenic mycobacterial species, suggesting that pathways involved in cell envelope synthesis are potential drug targets (5, 6). While considerable progress has been made in defining the structure and many of the enzymes involved in mycobacterial and corynebacterial cell envelope synthesis, less is known about how the synthesis of different classes of cell envelope lipids is inter-connected and how cell envelope assembly is coordinated with bacterial growth.
The nonpathogenic species Corynebacterium glutamicum is an indispensable workhorse of the biotechnological industry (7, 8) and a genetically tractable model system for investigating mycobacterial/corynebacterial cell envelope synthesis. Significantly, this species is permissive to loss of key cell envelope components, facilitating functional studies on the role of individual cell envelope components and interconnection (9–11). Studies undertaken by many groups have led to the identification of ∼50 distinct mycobacterial/corynebacterial lipid (sub)classes that exhibit polarized distributions in different cell envelope layers (12–14). Specifically, the C. glutamicum IM is thought to comprise phosphatidylglycerol (PG), phosphatidylinositol (PI), and cardiolipin (CL), as well as the phosphatidylinositol mannosides (PIMs) and glucuronic acid diacylglycerols (Gl-A and Gl-X to -Z) that function as membrane anchors for the lipomannan (LM) and lipoarabinomannan (LAM). In contrast, the OM consists of the glycosylated corynomycolic acids, glucosylmonocorynomycolate (GMM), trehalose monocorynomycolate (TMCM), and trehalose dicorynomycolate (TDCM) as well as of free mycolic acids (MAs), and phospholipids (9, 15–18). LM and LAM are also exposed on the bacteria surface, suggesting a subpool may also be embedded in the OM (14, 15, 19).
A number of studies have exploited mass spectrometry-based lipidomic approaches to define global changes in the lipid composition of mycobacterial and corynebacterial cell envelopes under different growth or drug treatments (14, 15, 20–22). These studies have been greatly facilitated by the development of lipidomics databases, such as Mtb LipidDB (13), MycoMass, and MycoMaps (12) which allow rapid matching of species and facilitate downstream data analysis. While these databases provide information on the accurate masses of many corynebacterial lipid (sub)classes, they generally do not include tandem mass spectrometry (MS/MS) fragmentation data, which is required to distinguish between isobaric lipid species. Other libraries, such as LipidBlast (23), contain in silico predicted MS/MS fragmentation patterns for 119,200 lipid species across 26 lipid (sub)classes, but do not cover many lipid classes unique to C. glutamicum and other Corynebacterineae. Similarly, Lipid Maps, the first internationally accepted lipid classification, nomenclature, and structural representation system (24), also does not cover all mycobacterial/corynebacterial lipid classes.
In this study, we describe a high-resolution lipidomics analysis of C. glutamicum IM and OM fractions using LC-quadrupole TOF (QTOF)-MS and customized in silico MS/MS libraries using MS-DIAL and the LipidBlast template. MS-DIAL is an open-source software for data-independent acquisition-based identification and quantification of small molecules by mass spectral deconvolution (25). Our C. glutamicum libraries contain over 90,000 molecular species and can be used to rapidly process LC-QTOF-MS/MS derived data from C. glutamicum IM and OM fractions and other Corynebacterineae. These analyses allowed for the identification of three new subclasses of trehalose corynomycolates: acyl-trehalose monohydroxylcorynomycolate (Acyl-hTMCM), acyl-trehalose monoacetylcorynomycolate (Acyl-AcTMCM), and trehalose acetyl-hydroxylcorynomycolate (Ac1-hTDCM), in addition to previously characterized C. glutamicum lipids.
We have previously shown that the C. glutamicum NCgl2759 encodes a TMCM mycolyl acetyltransferase (TmaT) that modifies the single MA of TMCM species with an acetyl group in the inner leaflet of the IM (16). This protein is highly conserved across all Corynebacterineae, and TMCM acetylation appears to be required for efficient transport of TMCM to the periplasmic space and the subsequent transfer of corynomycolyl chains to form other cell envelope components, such as TDCM and the mAGP complex (16). Using a global lipidomic approach, we confirm that loss of TmaT leads to loss of its product, trehalose monoacetylcorynomycolate (AcTMCM), and accumulation of the TMCM precursors, trehalose monohydroxylcorynomycolate (hTMCM) and trehalose monoketocorynomycolate (ketoTMCM). Interestingly, we also detected the loss and accumulation of two novel acylated species (Acyl-AcTMCM and Acyl-hTMCM species, respectively) in the IM. Importantly, we also show that loss of TmaT leads to broader changes in synthesis or loss of other lipid subclasses, including PG, acylated PG (Acyl-PG)-like species, and alanylated-lipids, as well as the accumulation of triacylglycerols (TAGs) in the IM fraction. These results highlight an unanticipated connectivity between different pathways of C. glutamicum cell envelope synthesis and/or compensatory mechanisms for maintaining cell envelope integrity and the importance of global lipidomics profiling approaches for defining the biochemical phenotype of cell envelope mutants.
METHODS
Growth conditions and manipulation of Escherichia coli and C. glutamicum
Escherichia coli DH5α was cultured in Luria-Bertani medium at 37°C. C. glutamicum ATCC 13032 was grown in brain heart infusion (BHI) medium (Oxoid or Becton, Dickinson and Co.) at 30°C. Fifteen grams of agar per liter was used for preparation of solid medium, and 10% sucrose (wt/vol) was added when required. C. glutamicum electrocompetent cells were prepared by growing at 18°C for 3–4 days until an OD600 of 0.6 was reached. Pelleted cells were washed four times with 10% (vol/vol) glycerol and then resuspended in 1 ml of 10% (vol/vol) glycerol and aliquoted in 100 μl volumes and stored at −80°C until further use. Electroporation was performed at the following settings: 2.5 kV, 200 Ω, and 25 µF. Kanamycin (30 µg/ml) and ampicillin (100 µg/ml) were added to the medium when needed.
DNA manipulations
All restriction endonucleases and DNA modification enzymes were obtained from Sigma-Aldrich and New England Biolabs. Genomic DNA was extracted from C. glutamicum as described (26), using the NucleonTM kit for cell cultures and blood (Amersham Biosciences). PCR was performed using Taq polymerase (Sigma-Aldrich) using the following conditions: 95°C for 5 min followed by 25 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min, then incubation at 72°C for 4 min. PCR products were separated by agarose gel (Amersham Biosciences) electrophoresis. Probes for Southern blot hybridizations were digoxygenin (DIG)-labeled using a DIG labeling kit (Sigma-Aldrich). DNA sequences were determined using an Applied Biosystems 3730S capillary sequencer (Micromon, Monash University).
Construction of a CL synthase (NCgl2646) mutant
Deletion of the NCgl2646 gene was performed using gBlocks® Gene Fragments (Integrated DNA Technologies). A 500 bp gene fragment was synthesized, consisting of 220 bp upstream and downstream of NCgl2646, with a central region of 48 bp comprising the 5′ and 3′ ends of the gene. The termini of the synthetic DNA included XbaI and HindIII sites to facilitate cloning into pK18mobsacB, a suicide vector for C. glutamicum (27). After sequence verification, the plasmid was electroporated into C. glutamicum ATCC 13032. Kanamycin-resistant colonies, resulting from integration of the plasmid via a single crossover event were confirmed by PCR. A clone was cultured on BHI plates containing 10% sucrose to derive double crossover strains, representing potential NCgl2646 knockouts. Gene deletion was confirmed by PCR and Southern blot hybridization. The complementation construct and the probe for the Southern blot were prepared by PCR amplifying a 2.1 kb fragment consisting of the NCgl2646 gene plus 500 bp upstream and 100 bp downstream sequences using primer pair NCgl2646F (5′-GCGATATCCCATCGGCGCCCGCTGCG-3′) and NCgl2646R (5′-GCGATATCCACAAACGCGTGGCCCGAC-3′), containing EcoRV restriction sites (underlined). The product was cloned into the PvuII site of pSM22 (28) to create pSM22:NCgl2646. Construction of the NCgl2759 (tmaT) mutant was described in a previous study (16).
Extraction of IM and OM lipids
WT and mutant C. glutamicum strains were grown to exponential phase (OD600nm = 2.5–3) in 100 ml of BHI medium (six replicate flasks for each strain), and cell suspensions were split in two and harvested by centrifugation (1,800 g, 10 min, 4°C). The two cell pellets were washed twice in PBS (10 ml) and then extracted separately. The first cell pellet was extracted in 20 volumes of chloroform/methanol (2:1, vol/vol) and then chloroform/methanol/water (CMW; 1:2:0.8, vol/vol/vol) (29). After removal of insoluble material by centrifugation (450 g, 5 min, room temperature), the extracts were dried under nitrogen and subjected to biphasic partitioning in 1-butanol and water (2:1, vol/vol). The organic phase was dried, and lipids were resuspended in 40 μl/100 mg (pellet wet weight) water-saturated 1-butanol [total lipid (TL) fraction]. The second cell pellet was extracted in water-saturated 1-butanol (RT for 30 min) to selectively remove OM lipids (30). After centrifugation (450 g, 5 min, 4°C), the pellets were sequentially extracted in 20 volumes of chloroform/methanol (2:1, vol/vol) and then CMW (1:2:0.8, vol/vol/vol) to extract the remaining lipids (IM lipids). Lipid fractions were analyzed by high-performance TLC (HPTLC) using aluminum-backed silica gel sheets (Merck). One-dimensional HPTLCs were developed in chloroform/methanol/13 M ammonium solution/1 M ammonium acetate/water (180:140:9:9:23, vol/vol/vol/vol/vol). Glycolipids were stained and visualized with orcinol/H2SO4. Phospholipids (and glycerolipids) were visualized by baking the silica gel sheets for a few minutes at 160°C. The silica gel sheets were scanned, and a color restoration tool was applied (Adobe Photoshop), by which glycolipid bands appear blue whereas pure phospholipid bands appear gray.
LC/MS analyses
Lipid extracts (1 μl of the lipid extracts was used for MS1 and 2 μl for MS/MS analyses) were separated on an Agilent 1290 Infinity Quaternary LC System (Agilent Technologies) using a C18 column (Phenomenex Kinetex, 2.6 μm EVO C18 100Å) eluted with an isopropanol (IPA) mobile phase binary solvent system at a flow rate of 260 µl/min (31). The mobile phase A consisted of acetonitrile (ACN):H2O (60:40, vol/vol) with 10 mM ammonium formate and mobile phase B of IPA:ACN (90:10, vol/vol), with 10 mM ammonium formate. The 30 min gradient program was used: 0–5 min 68% A, 1.5–4 min 55% A, 4–5 min 48% A, 5–8 min 42% A, 8–11 min 34% A, 11–14 min 30% A, 14–18 min 25% A, 18–21 min 3% A, 21–25 min 3% A, 25–25.1 min 68% A, and 25.1–30 min 68% A. Eluted lipids were detected using a 6550 iFunnel Q-TOF LC/MS system (Agilent Technologies). The scan range was adjusted to 100–3,200 Da (high mass) at the acquisition rate of 2 spectra/s for MS1 and 4 spectra/s for MS/MS. Data acquisition was performed in negative and positive ionization mode. The gas temperature was set to 225°C and the gas flow to 13 l/min. The nebulizer was set to 20 pounds per square inch gage with a detection window of 100 parts per million (ppm) and a minimum height of 1,000 counts. The sheath gas flow was set to 12 l/min, with a temperature of 275°C. The capillary voltage was set at 4,000 V with a nozzle voltage of 1,500 V. The voltages of the Fragmentor, Skimmer 1, and OctopoleRFPeak were 175/360; 65; and 750 V, respectively. For product ion scans, collision energies were set to 10, 20, 30, and 40 (CE10-40). After tuning, the resolving power of the 6550 iFunnel Q-TOF LC/MS system varied between 19,000 and 24,000 for masses between 600 and 1,500 Da. As reference ions, purine ([M+H]+, 121.050873) and HP-921 ([M+H]+, 922.009798) were used.
Software analysis tools
The custom-made and LipidBlast libraries were imported in MS-DIAL (25) (Version 2.06; MS/MS data), and the following parameters were used for identification: retention time window 1–24 min, mass range window 300–2500 Da, minimum peak height 10,000. MetaboAnalyst 3.0 (32) was used for the analysis of unpaired peak intensity tables [MS1 datasets, derived after XCMS was applied to find peaks in the raw MS1 data files using the centwave method (33)], with the following parameters: selected missing value imputation, interquantile range filtering, median normalization (total ion current) and log transformation of datasets, and no scaling. Univariate and multivariate analyses were also performed, including principal component analysis, dendrogram and heatmap comparisons, t-test verification, or volcano plot data visualization. For a comparison of a lipid species which was, i.e., highly abundant in the IM and missing in the OM, a value of 0 was filled in. The structural formulas were designed with the JChem Excel Add-In from ChemAxon (34).
Lipid library development, adaptation, and functional evaluation
The LC-MS/MS ESI QTOF run was performed in positive and negative ionization mode with collision energies of 10, 20, 30, and 40 eV (CE10-40) on C. glutamicum TL extracts. Totals of 1,582 and 1,308 features were detected in positive and negative ion modes, respectively, and CE30 gave the best fragmentation pattern results, with the lowest background and high abundance of fragment ions. Manual curation of peaks detected in positive ion mode led to the identification of ∼300 lipid species (including isomers) representing 28 lipid (sub)classes. The 6550 iFunnel Q-TOF LC/MS system has a resolving power of 19–24,000, which is sufficient to clearly resolve isotopomers having the same nominal mass (i.e., M0 and M+2 isotopologues of PG18:1/18:1 and PG18:0/18:1). A number of trehalose mycolates (i.e., hTMCM and ketoTMCM) could not be resolved with the 2 ppm mass accuracy in MS1 mode, but were clearly resolved by LC (a ketoTMCM elutes later from the column compared with its hTMCM mass-counterpart). While the libraries contained a large number of isobaric lipids, the majority of these were clearly resolved by the high-quality MS/MS fragmentation data (with the exception of some species of TDCM and CL). After deconvolution and removal of homodimers (protonated species that contained two identical lipid species with the same LC retention time as monomeric species), isotopologues, and isomers, 160 unique lipid species were identified in the positive ion analysis based on accurate mass and their MS/MS fragmentation pattern. This information was coupled with literature searches (to include all known lipid classes of C. glutamicum in our libraries with MS/MS fragmentation pattern) to create 37 lipid libraries containing 90,260 potential lipid species plus an additional ∼600,000 CL species, including many isobars. Each of these libraries contains a list of all possible molecular species within the class, including their chemical formulae, exact mass, and diagnostic fragments generated in positive ionization mode using a collision energy of 30 eV. Libraries were constructed using a range of FA chain lengths (C12-22), which is slightly broader than previously determined for C. glutamicum (C12-19) (22, 35, 36), as well as a range of saturated, monounsaturated, diunsaturated, triunsaturated, and tetraunsaturated species. Lipid libraries were generated (containing ∼3,000 theoretical species each) for the following lipid (sub)classes: diacylglycerol (DAG), alanylated DAG (Ala-DAG), lysinylated DAG (Lys-DAG), cytidine diphosphate DAG (CDP-DAG), PG, phosphatidylglycerol phosphate (PGP), alanylated PG (Ala-PG), lysinylated phosphatidylglycerol (Lys-PG), Acyl-PG, Acyl-PG-like, PI, phosphatidylinositol phosphate (PIP), PIM1-4, Gl-A (glucopyranosyluronic acid DAG) and Gl-X to Gl-Z (1-3mannose-α1–4-glucuronic acid-α1-diacylglycerol). Existing LipidBlast libraries for monoacylglycerol (MAG), TAG, phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), PS phosphatidylserine (PS), and SM were also added to complete lipid coverage and as negative controls. The libraries for acetylated PIMs 2-4 (AcPIM2-4) and diacetylated PIM2 (diAcPIM2) were slightly smaller (approximately 540 species), as the mannose-linked acyl chain was restricted to just FA16:0 and FA18:1. On the other hand, the CL library contains 665,500 theoretical species reflecting the large number of fatty acyl combinations on the two different glycerol backbone moieties (for summary, see supplemental Table S1). For corynomycolic acid-containing lipid species, we also extended the theoretical length to MA20-42, compared with empirically determined MA22-38 (22, 37) as well as including saturated, monounsaturated, diunsaturated, triunsaturated, and tetraunsaturated species. The libraries for the hTMCM, AcTMCM, ketoTMCM, Acyl-hTMCM (C16:0 and C18:1), Acyl-AcTMCM (C16:0 and C18:1), glucose monohydroxylcorynomycolate (hGMM), glucose monoacetylcorynomycolate (AcGMM), acyl-glucose monohydroxylcorynomycolate, and glycerol monohydroxylcorynomycolate (hGroMM) lipid classes contained 115 species each. The trehalose di-hydroxylcorynomycolate (h2TDCM) and Ac1-hTDCM libraries (both containing two corynomycolic acids) contained 13,225 entities (for summary, see supplemental Table S1). The master library of all 37 lipid (sub)classes can be downloaded as an Excel data file from http://bio21.unimelb.edu.au/.
The lipid libraries created de novo or taken from LipidBlast were subsequently adapted for the use in MS-DIAL (for MS/MS-derived data). The LipidBlast Excel template contains a Visual Basic macro program that automatically creates the MS/MS libraries in a mass spectral export format (MSP format). Libraries in the MSP format can be uploaded in MS-DIAL. Therefore, the newly created lipid libraries were adapted to the LipidBlast template and its macro program. In a last step, the LC-MS/MS output files (“.D” format from Agilent Technologies) were converted to an Abf file and uploaded to MS-DIAL together with each adapted lipid library or with a master library containing the lipids from all lipid (sub)classes.
The functionality of the lipid libraries was initially evaluated using a mixture of lipid standards that are not synthesized by C. glutamicum [Avanti Polar Lipids, 40 μM final concentration; 4 μM for PI8:0/8:0 (603.64Da)]: DAG14:0/14:0 (512.81Da), PG14:0/14:0 (688.85Da), PE14:0/14:0 (635.86Da), PS14:0/14:0 (701.85Da), PC14:0/14:0 (677.94Da) and CL28:0/28:0 (1275.71Da). LC-ESI-QTOF-MS/MS analysis of this mixture in positive ionization mode with a collision energy of 30 (CE30) resulted in the detection of all species except PI8:0/8:0, possibly due to its low concentration (data not shown).
Statistical analyses and development of a targeted lipid screen
LC-MS1 datasets (positive ionization mode, six replicates each) were converted from MS1.D output files (MassHunter) with the ProteoWizard tool into the .mzXML format (38). XCMS was then used to find peaks in the data files using the centwave method (33). These peaks were aligned and missing peaks reintegrated, resulting into large data tables containing the masses, the retention times, and the different abundances of each peak/lipid. Manual inspection of the peaks was performed, and peaks with a poor peak shape were removed from the matrix. We created three lipid matrices to compare the relative ion intensities of peaks/lipids from the TL fraction (3,123 peaks), IM fraction (1,864 peaks), and OM fraction (1,124 peaks). As the initial MS1 datasets contained ∼1,000–3,000 peaks, we developed a targeted screen whereby only the previously MS/MS identified lipids were included. Of the 233 identified lipids (Table 1), 185 were also identified in the MS1 datasets. We selected 10 hGMM, 31 hTMCM, 6 ketoTMCM, 7 AcTMCM, 7 Acyl-hTMCM, 3 Ac1-hTDCM, 35 TDCM, 7 DAG, 12 Ala-DAG, 1 PA, 1 CDP-DAG, 8 TAG, 9 PG, 2 Ala-PG, 3 Acyl-PG, 4 Acyl-PG-like, 5 PI, 1 PIM1, 1 PIM2, 5 AcPIM2, 1 AcPIM3, 1 AcPIM4, 6 Gl-A, 4 Gl-X, 1 Gl-Y, 1 Gl-Z, and 13 CL species. This limited dataset required less computational time and was used to compare the IM/OM of the WT with the IM/OM of the TmaT KO mutant (MS1 based). The datasets were uploaded to MetaboAnalyst and analyzed as described in the Software Analysis Tools section.
TABLE 1.
Detected lipid (sub)classes and lipid species of the TL, IM, and OM fraction of the C. glutamicum WT, TmaT mutant, and PBQC via LC-MS/MS ESI TOF in positive ionization mode (CE30)
| Number of Lipid Species | ||||||||||||
| WT | TmaT KO Mutant | PBQC | ||||||||||
| Lipid (Sub)Class | RT (min) | TL | IM | OM | TL | IM | OM | TL | IM | OM | Total Number | |
| 1 | hTMCM | 4.6–12.6 | 19 | 18 | 20 | 26 | 26 | 15 | 26 | 22 | 19 | 33 |
| 2 | AcTMCM | 6.6–11.5 | 4 | 3 | 8 | 0 | 0 | 0 | 6 | 5 | 7 | 8 |
| 3 | keto-TMCM | 6.0–9.0 | 4 | 3 | 0 | 10 | 10 | 3 | 9 | 7 | 2 | 11 |
| 4 | Acyl-hTMCM | 15–19 | 10 | 7 | 5 | 12 | 12 | 5 | 11 | 11 | 6 | 14 |
| 5 | Acyl-AcTMCM | 16.9–19.3 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 |
| 6 | h2TDCM | 20.0–22.0 | 30 | 28 | 28 | 22 | 30 | 25 | 29 | 29 | 23 | 43 |
| 7 | Ac1-hTDCM | 21.3–21.6 | 3 | 3 | 3 | 2 | 3 | 1 | 3 | 3 | 3 | 3 |
| 8 | hGMM | 6.5–13.0 | 9 | 5 | 11 | 6 | 8 | 7 | 8 | 7 | 7 | 12 |
| 9 | DAG | 11–14.5 | 7 | 7 | 4 | 8 | 8 | 4 | 7 | 8 | 4 | 8 |
| 10 | Ala-DAG | 10–13.5 | 13 | 11 | 5 | 9 | 9 | 2 | 12 | 12 | 4 | 13 |
| 11 | CDP-DAG | 6–7 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| 12 | TAG | 20.0–22.0 | 9 | 10 | 10 | 10 | 11 | 11 | 10 | 9 | 10 | 13 |
| 13 | PG | 5.9–10.1 | 11 | 11 | 5 | 5 | 5 | 2 | 10 | 10 | 5 | 11 |
| 14 | Acyl-PG | 14–15 | 3 | 3 | 0 | 1 | 1 | 0 | 2 | 2 | 0 | 3 |
| 15 | Ala-PG | 9–10 | 2 | 2 | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 2 |
| 16 | PA | 8.6–9.2 | 2 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 |
| 17 | CL | 17–21 | 16 | 16 | 3 | 14 | 14 | 4 | 14 | 14 | 3 | 18 |
| 18 | PI | 6.7–9.8 | 4 | 4 | 3 | 2 | 3 | 2 | 4 | 4 | 2 | 6 |
| 19 | PIM1 | 5.9–10.2 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 |
| 20 | PIM2 | 7.0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 21 | AcPIM2 | 5 | 4 | 1 | 6 | 5 | 1 | 4 | 5 | 1 | 7 | |
| 22 | AcPIM3 | 12–14.5 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 |
| 23 | AcPIM4 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | |
| 24 | Gl-A | 5.6–10 | 7 | 6 | 2 | 6 | 5 | 1 | 7 | 5 | 1 | 8 |
| 25 | Gl-X | 5.7–8.4 | 6 | 6 | 1 | 4 | 4 | 1 | 4 | 5 | 1 | 6 |
| 26 | Gl-Y | 8.1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| 27 | Gl-Z | 7.9 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 |
| 28 | Acyl-PG-like | 14.0–17.0 | 4 | 4 | 3 | 0 | 0 | 0 | 3 | 3 | 3 | 4 |
| Sum: | 174 | 161 | 114 | 151 | 162 | 86 | 178 | 171 | 103 | 233 | ||
RESULTS
Generation of a new pipeline for profiling C. glutamicum lipids
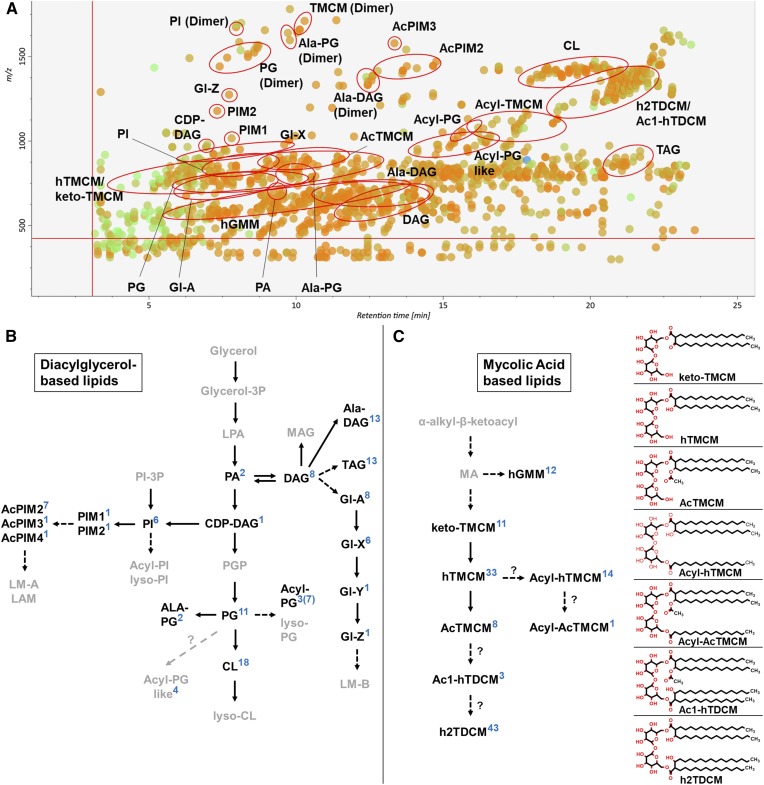
LC-ESI-QTOF-MS/MS analysis of total C. glutamicum lipids in positive and negative ion modes resulted in the detection of 1582 and 1308 peaks, respectively. Manual curation of these peaks led to the identification of 28 different lipid (sub)classes, comprising ∼300 molecular species, most of which were detected with greater sensitivity in positive ionization mode (the exception being Acyl-PG), as NH4+ and/or H+ ion adducts. Therefore, all subsequent analyses were performed in positive ionization mode. Interestingly, a number of abundant lipid species (i.e., hTMCM, PI, PG, Ala-PG, and Ala-DAG) were also detected as protonated dimeric forms, which had identical LC retention times as the monomers and are likely generated in-source (Fig. 1A and supplemental Tables S2–S8). After removal of isotopologues, dimer species, isomers, and isobars, 160 molecular species were identified with high confidence. These species included glycerolipids (DAG, TAG, Ala-DAG, and CDP-DAG), CLs, Gl-A and Gl-X to Gl-Z, glycerophospholipids (PG, Ala-PG, Acyl-PG, Acyl-PG-like, PA, and PI), PIMs (PIM1-4, and AcPIM2-4), and corynomycolic acid-containing lipids (hTMCM, AcTMCM, ketoTMCM, Acyl-hTMCM, Acyl-AcTMCM, h2TDCM, and Ac1-hTDCM). Each of these MS/MS libraries were imported into MS-DIAL and used to process MS/MS fragmentation data. Lipid libraries were also created for AcGMM, Acyl-GMM, hGroMM, Lys-DAG, PGP, Lys-PG, PIP, and diAcPIM2 species or directly imported from LipidBlast (MAG, TAG, PA, PC, PE, PS, and SM). Some of the latter lipids/lipid classes were not identified (i.e., PC, PE, PS, and SM) but were included to serve as negative controls to exclude lipid contaminations of foreign origin. In summary, our lipid libraries contain the in silico MS/MS spectra for 37 lipid (sub)classes comprising 90,260 lipid species and 665,500 CL species. Each lipid library contains the accurate mass, the chemical formula, and the MS/MS fragmentation data (predicted) for each molecular species. The LC retention time information for each lipid subclass is given in Table 1. The size of each library varied between 100 and ∼3,000 species, reflecting the degree of acylation and/or other modifications within each class. The high mass resolution of the 6550 iFunnel Q-TOF-MS (between 19 and 24,000) was sufficient to resolve isotopologues of related lipid species. However, some lipids with the same accurate mass were resolved by LC, indicating the occurrence of isobaric species which share the same mass-to-charge ratio and MS/MS fragmentation pattern, but can differ in the position of the acyl chain, the position of the double bond, or the double bond geometry (39). Identification of the structure of these species will require more detailed analysis and was not attempted here. Interrogation of LC-MS/MS output files with MS-DIAL routinely resulted in the identification of 78–100% of the C. glutamicum lipids that had been previously identified by manual curation (Fig. 1A). In a limited number of cases, manual interrogation was required to reliably identify specific isobaric lipid species. For example, keto-TMCM species were commonly annotated as hTMCMs, as both classes can have the same molecular mass and very similar MS/MS spectra, but can be readily distinguished following manual curation based on the relative abundance of a fragment ion corresponding to loss of trehalose headgroup. Other overlaps between different libraries included Acyl(16:0)-hTMCM and Acyl(18:1)-hTMCM, between Ac1-hTDCM and h2TDCM, and between CL and PG. In the latter case, the CL library detects dimers of PG, although these species are easily distinguished based on their different retention times (PG/PG dimers: 5.9–10.1 min; CL: 17–21 min).
Fig. 1.
LC-MS analysis of C. glutamicum lipids. A: MS/MS peak spot viewer of the C. glutamicum WT tTL fraction in MS-DIAL. The retention time (x axis) is plotted against the mass/charge ratio (y axis). Blue spots represent peaks of lower abundance based on peak area, green of intermediate abundance, and red of high abundance. A total of 174 lipid species (26 subclasses) were detected in the TL fraction of WT C. glutamicum. Each lipid (sub)class is highlighted with a red circle in the peak spot viewer of the MS-DIAL software. Note that not every spot within a circle belongs to the lipid (sub)class. B, C: Overview of pathways involved in DAG-based (B) and MA-based (C) lipid biosynthesis. Lipid (sub)classes identified via LC-MS/MS ESI QTOF in positive ionization mode using the established lipid libraries in MS-DIAL are in bold. The number of identified species in each lipid (sub)class are indicated in blue. Altogether, 233 lipids across 28 lipid (sub)classes were identified in the different C. glutamicum strains analyzed (WT, ΔtmaT, and the ΔtmaT complementation strain) and in the pooled biological quality control (PBQC); 108 were DAG-based and 125 MA-based lipids. The chemical structures of trehalose-containing MAs are shown.
Overview of the major lipid classes in WT C. glutamicum
The abundance of different lipid classes in C. glutamicum is broadly reflected by the number of identified molecular species in each class. Our results represent the relative ion intensity of individual species Trehalose corynomycolates are the most abundant lipids in this bacterium and are represented by 125 TMCM/TDCM and related molecular species (Fig. 1C and supplemental Tables S2–S4). The majority of these lipids contained saturated (45 = 36%) and monounsaturated (38 = 30%) corynomycolates, although species containing diunsaturated (27 = 22%) and triunsaturated (15 = 12%) corynomycolates were also detected. The predominant h2TDCM species contained MA32:0 (15x; where 15x means that 15 different h2TDCM lipids contain the MA residue MA32:0), MA34:1 (10x), MA34:0 (8x), MA36:2 (6x), and MA36:1/MA26:0 (5x each) (supplemental Table S3).
DAG-based lipids constitute the second major class of lipids, with 108 DAG-based lipid species detected. These species included, i.e., DAG (8), Ala-DAG (13), TAG (13), PG (11), Ala-PG (2), and Acyl-PG (3). The majority of these lipids contained saturated (42.4%) or monounsaturated (51.2%) FAs, whereas diunsaturated or triunsaturated FA species constituted less than 7% (Fig. 1B and supplemental Tables S5–S8). The most abundant FAs were FA18:1 (43%), FA16:0 (26%), FA18:0 (8.5%), and FA16:1 (4.5%). The remaining 18% of the FAs were based on FA12:0, FA14:0, FA14:1, FA15:0, FA16:2, FA17:0, and FA18:2. CLs are not included here, because the CL MS/MS library only contains DAG fragments. The most commonly identified CL DAG moieties were DAG34:1 (13x), DAG36:2 (7x), and DAG32:1 (4x). Further DAG-based lipids detected in these analyses included Gl-A, X, and Z that contain glucuronic acid linked to DAG and form membrane anchors for the subclass of LMs that are anchored in the IM but can extend to the OM and the bacterial cell surface.
Acyl-PG species constitute a minor class of DAG-based lipids. Unlike most other lipid classes, acyl-PG was detected with greater sensitivity in negative ionization mode (six molecular species) compared with positive ion mode (three molecular species detected).
Identification of new lipid subclasses in C. glutamicum
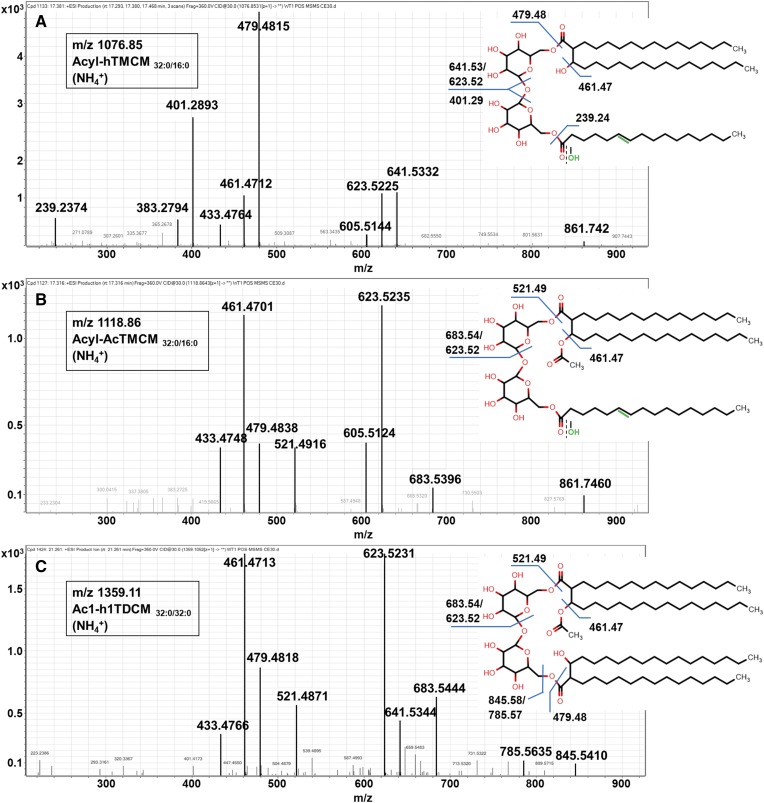
While the major subclass of TMCMs in C. glutamicum contains a hydroxylated corynomycolate (hTMCM), two additional subclasses have also been identified in which the corynomycolate is modified with a carbonyl group (ketoTMCM) or an acetyl group (AcTMCM) (16). KetoTMCM species are low-abundance precursors for mature hTMCM, while acetylation of hTMCM is a transient modification that facilitates TMCM transport across the IM (16). In the course of this study, we identified two additional subclasses of TMCM in which the second nonmycolylated glucose in the trehalose headgroup is modified with an acyl chain (Fig. 2A, B). Acylated hTMCM species were initially identified based on the accurate mass of their [M+NH4]+ molecular ions that differed from major hTMCM species by either m/z 238.23 or 264.24, corresponding to the predicted masses of FA16:0 and FA18:1 or the corresponding monounsaturated fatty alcohols. Collision-induced dissociation of the putative Acyl-hTMCM32:0/16:0 (Fig. 2A) yielded fragments of m/z 479.5/461.5 and m/z 641.5/623.5, consistent with canonical hTMCM32:0/16:0 (16), while additional diagnostic fragments at m/z 401.29 and 239.24 indicated the presence of palmitoyl (16:0) fatty acyl modification (or the corresponding fatty alcohol) of the C(O)6 hydroxyl of the nonmycolylated glucose unit in the trehalose headgroup. This new subclass of lipids is referred to as Acyl-hTMCM.
Fig. 2.
Putative MS/MS fragmentation pattern of the three newly identified trehalose MAs. Fragmentation pattern of Acyl-hTMCMC32:0/16:0 (A), Acyl-AcTMCMC32:0/16:0 (B), and Ac1-hTDCMC32:0/C32:0 (C). Altogether, 18 novel lipid species, comprising 14 Acyl-hTMCM, 1 Acyl-AcTMCM, and 3 Ac1-hTDCM species, were detected. The FA moiety of Acyl-hTMCM (A) and Acyl-AcTMCM (B) could either carry a keto group or a hydroxyl group. In the case of a hydroxyl group, the FA moiety is monounsaturated (highlighted in green).
Molecular ions corresponding to an acylated form of AcTMCMC32:0/C16:0 were also identified. This species/subclass differed from AcTMCM by m/z 238.23, but otherwise had identical MS/MS spectra (Fig. 2B) (16).
A third series of previously unidentified TMCM-related lipids corresponded to a modified form of h2TDCM (TDCM with two hydroxy corynomycolic acids). These species contained characteristic fragment ions at m/z 521.49/683.54 and m/z 479.48/641.53, corresponding to acetylated and hydroxylated corynomycolic acids, respectively, and are referred to as Ac1-hTDCM (Fig. 2C).
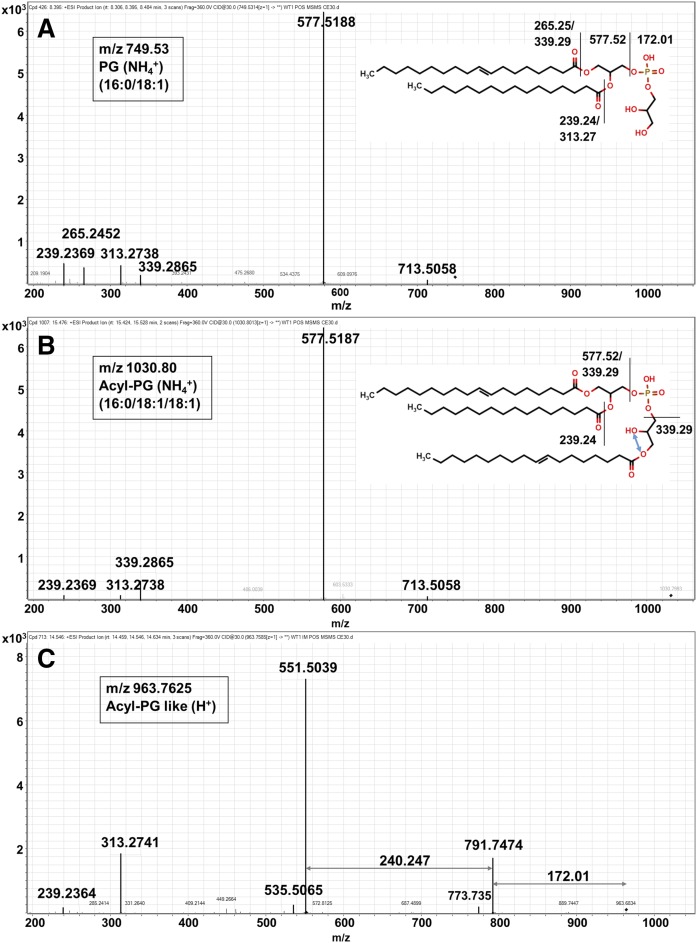
A fourth lipid (sub)class that could not be matched to known lipids was provisionally characterized as a putative Acyl-PG (Acyl-PG-like). The mass spectra of PG16:0/18:1 (m/z 749.53), Acyl-PG16:0/18:1/18:1 (m/z 1030.80), and Acyl-PG-like?/C32:0 (m/z 963.76) are shown in Fig. 3. In common with other Acyl-PG species, the MS/MS fragmentation spectrum of the Acyl-PG like species contained fragment ions corresponding to DAG and the PG headgroup (m/z 172.01). However, the mass spectrum of Acyl-PG-like species contained an additional fragment ion at m/z 240.247 (or m/z 266.26 and 268.276), which may correspond to a putative fatty alcohol. These species had the same LC elution time as acyl-PG species consistent with the presence of three acyl chains. Acyl-PG species were only detected in the IM fraction, whereas Acyl-PG-like species were detected in the IM and OM fraction. Altogether, 11 PG, 3 Acyl-PG, and 4 Acyl-PG-like species were detected.
Fig. 3.
MS/MS fragmentation pattern of three different PG-based lipids in positive ionization mode. Fragmentation pattern of PG16:0/18:1 (m/z 749.53) (A), AcylPG16:0/18:1/18:1 (m/z 1,030.80) (B), and Acyl-PG-like?/C32:0 (m/z 963.76) (C). Acyl-PG-like species are related to PG and Acyl-PG, in having the same LC-retention time and loss of a fragment ion corresponding to a PG headgroup (172.01 Da). These lipids also characteristically lose additional fragment ions (240.247; 266.26; or 268.276 Da) that may correspond to loss of fatty alcohol. Acyl-PG species were only detected in the IM fraction, whereas Acyl-PG-like species were detected in the IM and OM fractions. Acyl-PG-like species were completely missing in the ΔtmaT mutant. Altogether, 11 PG, 3 Acyl-PG, and 4 Acyl-PG-like species were detected. While the additional fatty acyl chain is shown linked to sn-3 hydroxyl group of the glycerol headgroup, attachment could also occur on the sn-2 hydroxyl (blue arrow).
Compartmentalization of C. glutamicum lipids across the IM and OM
We have previously shown that OM lipids can be selectively extracted from live C. glutamicum with water-saturated 1-butanol and IM lipids subsequently extracted from OM-depleted cell pellets with CMW (16). LC-ESI-QTOF-MS/MS analysis of the IM and OM fraction of WT C. glutamicum demonstrated that the OM and IM fractions had markedly different lipid compositions. Targeted analysis of 185 lipid species (diagnostic of all major corynebacterial lipid classes) indicated that the IM contained DAGs, Ala-DAGs, PGs, Gl-As, hTMCMs, and ketoTMCMs as major lipids. The IM was also highly enriched for intermediates in de novo phospholipid synthesis (i.e., CDP-DAG), the newly characterized acyl-PG and acyl-TMCM lipids and glycolipid precursors for the cell envelope lipoglycans, LM and LAM (i.e., Gl-A-Z and PIM2-4), consistent with the IM being the major site of lipid synthesis (Figs. 4A–F and 5B and supplemental Table S4). In contrast, the major lipids in the OM fraction comprised hTMCMs, h2TDCMs, Gl-As, PGs, and CLs (Fig. 4A–F and supplemental Table S4). This membrane was also enriched for minor lipids such as Ac1-hTDCM, Acyl-PG-like, AcTMCM, hGMM, and specific molecular species of Ala-DAG (Fig. 5B). Interestingly, nonbilayer generating lipids, such as TAGs and abundant CL species, were found to be equally distributed across the OM/IM fractions. C. glutamicum lacks the capacity to synthesize TAGs de novo, although these bacteria express a number of TAG lipases. C. glutamicum may therefore scavenge TAGs from the medium and degrade them in the cytoplasm. Whether these IM TAG remain associated with the plasma membrane and/or form lipid bodies in the cytoplasm remains to be determined.
Fig. 4.
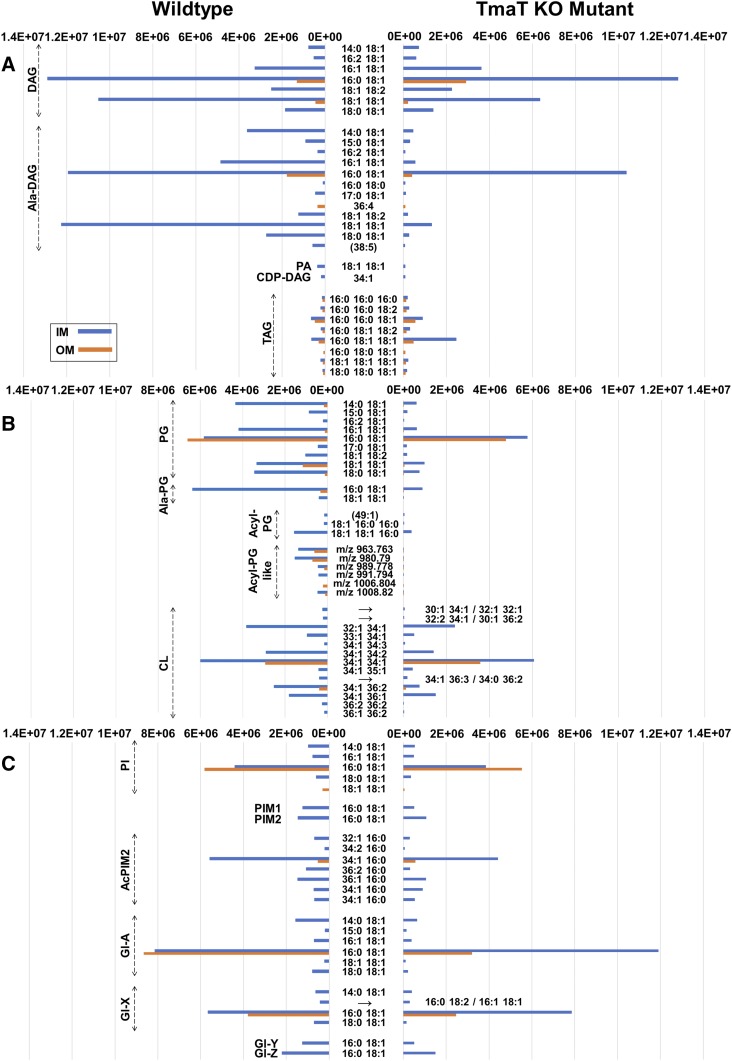
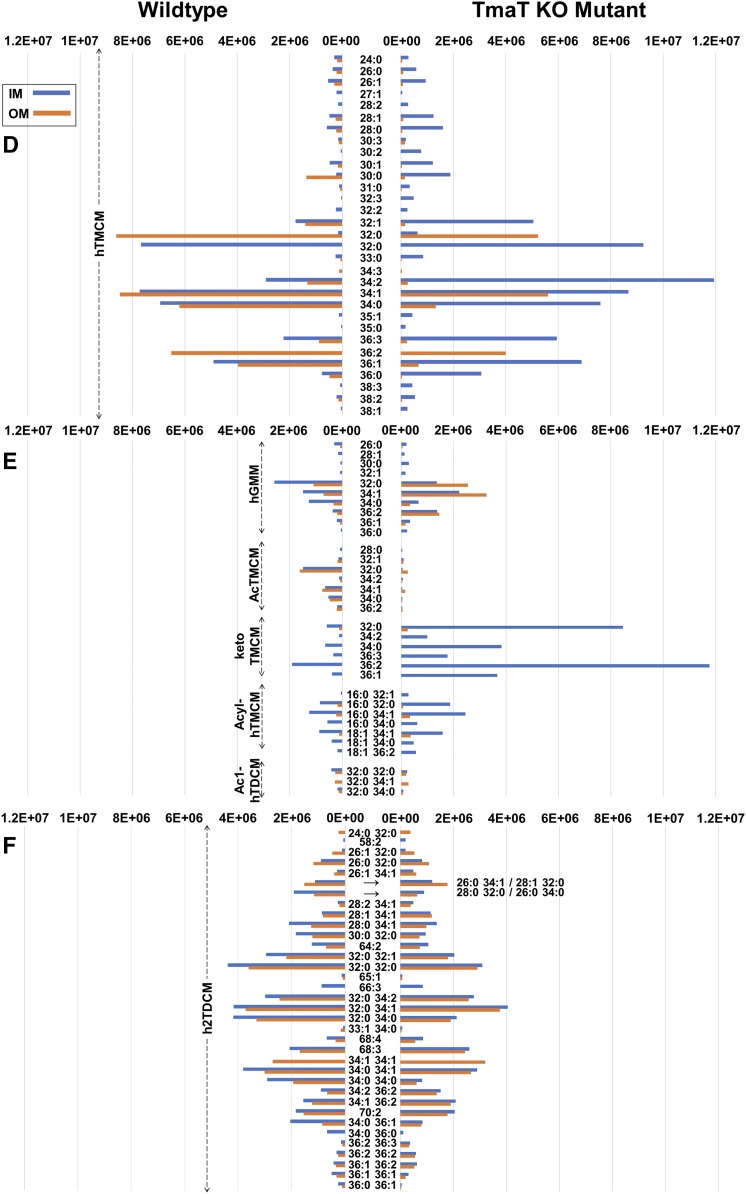
Comparison of the relative abundances of IM and OM lipids in C. glutamicum WT and the Δtmat mutant bacteria. The relative abundance of 185 species, identified by LC-MS/MS profiling, in the IM and OM fractions of C. glutamicum WT and the ΔtmaT mutant bacteria. Lipid abundances (based on ion intensities) represent the mean value of six replicates. The relative abundance of DAG and TAG based lipid classes (A), PG and CL-based lipid classes (B), PI/PIM and Gl glycolipid classes (C), hTMCM species (D), other trehalose and glucose corynemycolates (E) and h2TDCM species (F) are shown. The most abundant lipids of WT IM were the DAGs, Ala-DAGs, PGs, CLs, Gl-As, hTMCMs, and ketoTMCMs, and for the WT OM, the hTMCMs, h2TDCMs, Gl-As, PGs, and CLs. Compared with WT bacteria, the IM fraction of the ΔtmaT mutant was deficient in AcTMCM, Acyl-AcTMCM, and Acyl-PG-like species and had decreased amounts of PGs, Acyl-PGs, Ala-DAGs, and h2TDCMs, while the OM fraction was deficient in Acyl-PG, decreased in PG, hTMCM, AcTMCM, and h2TDCM and increased in hGMM and Acyl-hTMCM.
Fig. 5.
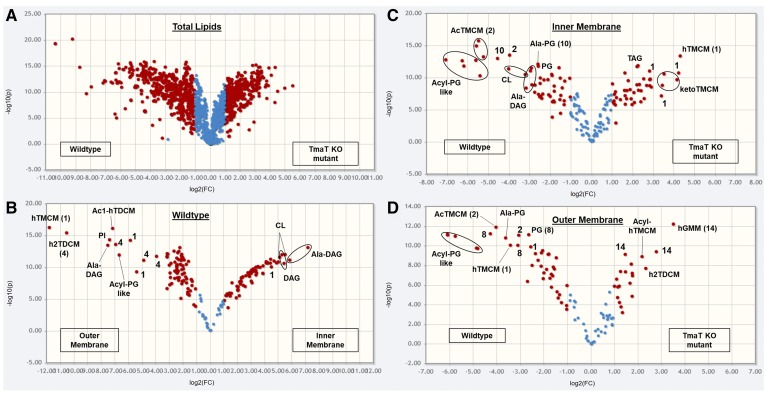
Comparison of total, IM and OM lipid composition of C. glutamicum WT versus the Δtmat mutant. Volcano plots of TLs (A), WT IM and OM lipids (B), WT versus ΔtmaT IM lipids (C), and WT versus ΔtmaT OM lipids (D). The x axis represents the fold change (threshold of 1) and the y axis the t-test (threshold of 0.05). Both values are median-normalized and log-transformed. The most highly enriched lipids of the IM of the WT (compared with its OM) were Ala-DAG, DAG, CL, PG, ketoTMCM, Gl-A to GLZ, and PIM2 to AcPIM2-4.
Global changes in cell envelope lipid composition in the C. glutamicum TmaT mutant
We have previously shown that deletion of the C. glutamicum gene NCgl2759, encoding a TmaT, leads to the accumulation of hTMCM and ketoTMCM in the IM and to defects in the transport of hTMCM to the OM with concomitant defects in the synthesis of h2TDCM (16). To investigate whether disruption of TMCM transport resulted in other pleiotropic changes in lipid composition, we analyzed IM and OM lipid extracts of the C. glutamicum ΔtmaT mutant and a complemented strain expressing the plasmid pSM22:tmaT (encoding the native acetyltransferase). Consistent with our previous analyses, the ΔtmaT mutant lacked detectable amounts of AcTMCM, the product of the TmaT enzyme, and contained elevated levels of the AcTMCM precursors, hTMCM and ketoTMCM, in the IM fraction (Figs. 4D,E, 5C). Levels of hGMM and the newly identified minor subclass Acyl-hTMCM were also found to be elevated in both the IM and OM of the ΔtmaT mutant (Figs. 4E, 5D). Unexpectedly, levels of h2TDCM, the downstream product of hTMCM and ketoTMCM, were also elevated in the OM fraction of the mutant (Fig. 4F). It is possible that the accumulated hTMCM or ketoTMCM species are transported across the IM by the same transporter that mediates the transport as AcTMCM. Alternatively, the transbilayer movement of these species may be mediated by separate transporters. In this respect, C. glutamicum has been shown to express two TMCM transporters, NCgl2769/CmpL1 and NCgl0228/CmpL4, which could potentially mediate transport of these different species. Complementation of the ΔtmaT mutant with a plasmid expressing the tmat gene completely reversed these changes in trehalose lipid composition and IM/OM distribution (Fig. 6A).
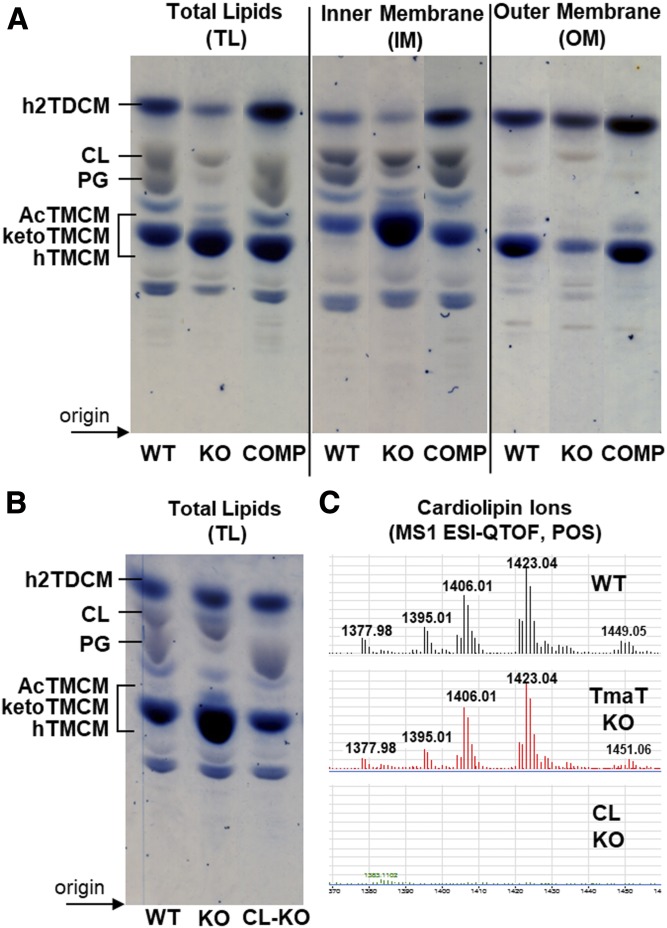
Fig. 6.
HPTLC analysis of lipids from C. glutamicum WT and the Δtmat mutant. A: TL extracts, as well as lipids derived from IM and OM fractions from C. glutamicum WT, the Δtmat mutant (KO), and the complementation strain (COMP) were analyzed by HPTLC. Glycolipids (blue) and phospholipid (gray) were detected by orcinol-sulfuric acid staining. Select TLC lanes are shown from replicate analyses (supplemental Fig. S1). B: A C. glutamicum mutant lacking CL synthase (NCgl2646) was generated, and TLs were analyzed by HPTLC. C: LC-MS of lipids from C. glutamicum WT bacteria, the ΔtmaT, and the ΔNCgl2646 mutants. Only the region of the LC chromatogram showing CL species is shown.
Loss of TmaT was also associated with major changes in the synthesis and membrane distribution of several other lipid classes thought to be unrelated to trehalose corynomycolates. Specifically, the C. glutamicum ΔtmaT mutant was highly depleted in a number of PG molecular species (55–60% fewer PG species were identified in the IM and OM fraction compared with the WT), Acyl-PG, alanylated-PG, and Ala-DAG (Fig. 4A, B). Moreover, Acyl-PG-like species were completely absent in ΔtmaT, while TAG species were highly increased in the CMW fraction of the mutant (the relative abundance, P values, and fold change for 185 IM and OM lipid species are summarized in supplemental Table S9). The depletion of PGs in the mutant was confirmed by HPTLC analysis of the TL extracts of WT, ΔtmaT, and the complemented line (Fig. 6A and supplemental Fig. S1). PG is both a major membrane component and precursor for CL. Unexpectedly, no differences were observed in expression of CL in the mutant, suggesting that the rate of PG synthesis is reduced in the mutant, but the pool of PG precursors is still sufficient to sustain CL synthesis. Genetic disruption of the gene encoding CL synthase (gene NCgl2646) and HPTLC and LC-MS1 analysis of the TLs of the derived mutant confirmed that CL species detected in the WT and ΔtmaT mutant were the major products of this gene (Fig. 6B, C). These results reveal an unanticipated degree of interdependency between different pathways of lipid synthesis and/or potential mechanisms for compensating for loss of specific lipids in these bacteria.
DISCUSSION
The Corynebacterineae comprise a number of pathogens of medical and veterinary significance, as well as bacteria with important biotechnology applications. A detailed understanding of the composition and biogenesis of the complex IM and OM of these bacteria is expected to lead to the development of new antibiotics, as well as understanding the metabolic and bioenergetic demands of cell envelope synthesis on bacterial growth. Here, we describe a new lipidomics pipeline for characterizing the IM/OM fractions of the model Corynebacterineae, C. glutamicum. IM/OM lipids were extracted using differential solvent extraction and analyzed by LC-ESI-QTOF-MS in positive ion mode. Lipid species were identified using curated in silico generated MS/MS libraries covering 37 distinct lipid classes and 90,260 lipid species (with an additional 665,500 CL molecular species). Each of these libraries comprises all known and theoretical glycero- and trehalose lipids containing FA (C12-22) and MA (MA20-42) chain lengths, as well as a range of saturated, monounsaturated, diunsaturated, triunsaturated, and tetraunsaturated species. Our master library includes seven additional lipid libraries that were directly imported from LipidBlast (of MAG, TAG, PA, PC, PE, PS, and SM). Inclusion of these libraries allowed detection of lipid classes that are not synthesized by C. glutamicum but may be acquired from the growth medium. For example, while many corynebacteria lack enzymes required for synthesis of TAGs (40), we identified significant levels of TAGs in both the IM and OM fractions, which are likely derived from the BHI growth medium. Interestingly, C. glutamicum contains a putative TAG lipase (gene KIQ_011745; UniProt: A0A072Z966), suggesting that they may be able to catabolize these acquired lipids. Using this new pipeline, we were able to rapidly identify and semiquantitate 28 lipid classes/subclasses, comprising >233 molecular species in total or IM/OM lipid fractions (Table 1).
Consistent with previous studies, we have shown that the major glycerolipids of C. glutamicum contain predominantly C18:1 (43%), C16:0 (26%), C18:0 (8.5%), and C16:1 (4.5%) FAs (22, 35, 36) (Fig. 1B and supplemental Tables S5–S8). Similarly, we identified 125 glucose/trehalose corynomycolic acids containing 34 different MA moieties, with chain length ranging from MA22:0 to MA38:3 (Fig. 1C and supplementary Tables S2–S4) (22, 37). These glycolipids contained predominantly saturated (45 = 36%) and monounsaturated (38 = 30%) MA chains, while diunsaturated (27 = 22%) and triunsaturated (15 = 12%) MAs were less abundant. As an example, we identified 43 different molecular species of the trehalose corynomycolate, h2TDCM (supplemental Table S3), with the most abundant of these species containing MA32:0 (15x), MA34:1 (10x), MA34:0 (8x), MA36:2 (6x), and MA36:1/MA26:0 (5x each). These results are in line with the studies of Yang et al. (2012), where the MAs 32:0, 34:1, and 36:2 were identified as the major MA species in C. glutamicum (37).
Our analyses led to the identification of three new TMCM-related lipid subclasses, comprising Acyl-hTMCM, Acyl-AcTMCM, and Ac1-hTDCM. These species have not been reported previously in C. glutamicum and comprise acylated forms of hTMCM and AcTMCM and a monoacetylated form of TDCM, respectively. Based on their MS/MS fragmentation pattern (Fig. 2A, B), we propose that the additional FA or fatty alcohol is located on the nonmycolylated glucose residue in the trehalose headgroup. These species are present at low levels and are predominantly located in the IM fraction. While the function of TMCM acylation is currently unknown, this modification could play a similar role to MA acetylation in promoting transbilayer flipping of TMCM intermediates across the IM and/or transport to the OM.
We have previously shown that differential solvent extraction of live C. glutamicum cells with water-saturated 1-butanol, followed by CMW, results in the selective extraction of OM and IM lipids, respectively (16). Detailed lipidomic analysis of these fractions provides further support for the selectivity of this procedure as well as further information on the lipid composition of the IM and OM. In particular, the IM fraction was highly enriched in key intermediates of several lipid pathways (PA, CDP-DAG, PIM1, Gl-A, keto-TMCM, AcTMCM, and the acylated hTMCM) as well as other lipids that may function as bulk constituents of the IM (hTMCM, DAGs, Ala-DAGs, PGs, and CLs) (Fig. 4 and Table 1). In contrast, the OM was primarily composed of the trehalose lipids, hTMCMs, and h2TDCMs, and the negatively charged phospholipids Gl-A, PG, and CLs. Minor lipid species in this layer included Ac1-hTDCM, AcTMCM, hGMM, Acyl-PG-like, and TAG. It has previously been proposed that CL may be tightly linked to the AG-peptidoglycan complex in the periplasmic space, based on the inaccessibility of this lipid to a reverse micellar extraction protocol designed to extract OM lipids (14). We recovered approximately 30% of the total CL fraction in the initial 1-butanol extraction (OM fraction), with the remainder being extracted in CMW. These data would suggest that approximately 70% of the CL is either associated with the IM or the AG-peptidoglycan complex.
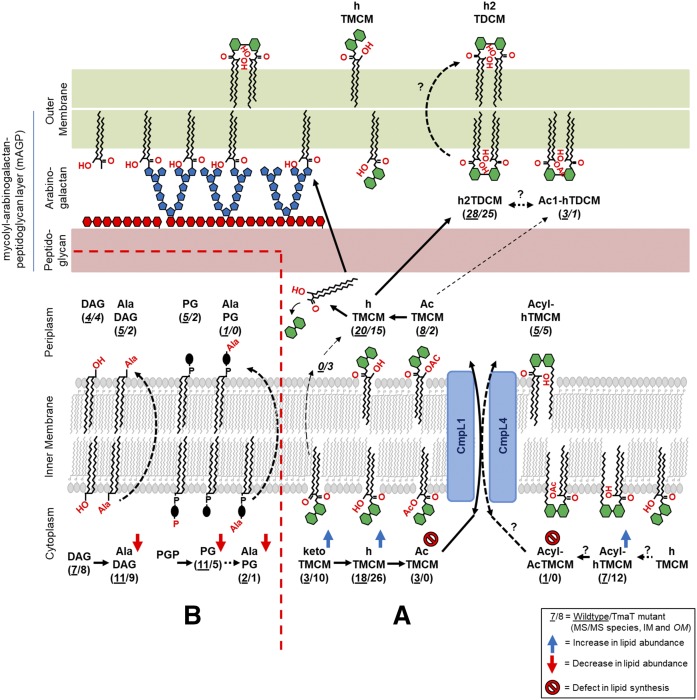
While considerable progress has been made in dissecting individual pathways of lipid synthesis in Corynebacterineae, comparatively little is known about how these pathways are regulated to generate the mature cell envelopes and/or the extent to which perturbations in one pathway lead to altered fluxes in other pathways. We have previously shown that loss of the C. glutamicum IM acetyltransferase TmaT/ΔNCgl2759 leads to disruption in the transport of hTMCM from the inner leaflet of the IM to the OM and reduced synthesis of h2TDCM (16). To investigate whether loss of TmaT leads to broader changes in the synthesis of other trehalose lipids or biosynthetically unrelated lipids, we undertook global lipid analysis of the ΔNCgl2759 mutant. Consistent with previous analyses, we show that loss of TmaT is associated with loss of synthesis of AcTMCM and accumulation of the lipid species, hTMCM, ketoTMCM, and Acyl-hTMCM in the IM fraction. The accumulation of ketoTMCM and hTMCM in the mutant is consistent with both lipids being precursors to AcTMCM and the acetylation of the MA chain of hTMCM being a critical step in the transport of this species across the IM. The enhanced accumulation of ketoTMCM relative to hTMCM could also indicate physical interactions between the enzymes, CmrA (which converts ketoTMCM to hTMCM), and TmaT (which converts hTMCM to AcTMCM) and substrate channeling of ketoTMCM to form downstream transport-competent intermediates. Interestingly, the novel lipid Acyl-hTMCM also accumulated in the IM of the ΔNCgl2759 mutant, supporting the notion that alternative acylation of the second glucose moiety in the trehalose headgroup may constitute a second mechanism for facilitating the transbilayer movement of TMCM species from the inner to the outer leaflet of the IM. It remains to be determined whether acylation and acetylation are independent and possibly redundant mechanisms for facilitating transbilayer movement of TMCM species or whether both modifications are required for efficient transport across the IM or transport from the IM to OM (Fig. 7A). The possibility that acylation enhances the transbilayer movement of hTMCM [although insufficient to sustain normal flux of TMCM to the periplasmic space/OM in the absence of TmaT acetylation (9, 17, 18)] is indicated by the continued synthesis of low levels of h2TDCM in the OM fraction of the TmaT mutant. Together, these analyses confirm that TmaT-mediated acetylation of TMCM is important for the IM to OM transport of TMCM, but also raises the possibility that additional modifications, such as acylation of hTMCM, AcTMCM, or ketoTMCM, could also facilitate this step (Fig. 7A).
Fig. 7.
Revised scheme for synthesis of TMCM/TDCM and other C. glutamicum lipids. A: ketoTMCM is converted to hTMCM on the inner leaflet of the IM and subsequently acetylated (on the corynomycolic acid) or acylated (on the second glucose moiety of the trehalose headgroup), facilitating transport by the two Cmpl transporters (Cmpl1 + 4) across the IM. It is proposed that the acetyl/acyl groups are removed in the periplasm prior to the synthesis of TDCM and the transfer of MAs to the mAGP complex. The identification of Ac1-hTDCM species in this study suggests that not all of the AcTMCM is deacylated and that some of these “transport” intermediates can be converted to the Ac1-hTDCM pool. B: Several PGs, alanylated-PG, and Ala-DAG lipids were highly enriched in the IM and their levels decreased in the Δtmat mutant.
Strikingly, loss of TmaT was also associated with marked changes in the cellular levels of several classes of biosynthetically unrelated lipids. In particular, IM levels of PG, Acyl-PG-like species, and alanylated-PG and -DAG were dramatically downregulated in the ΔtmaT mutant. PG functions as both a bulk lipid in the IM, as well as a precursor for alanylated PG and CL. Interestingly, depletion of PG pools did not result in concomitant decrease in levels of CL, suggesting that the IM pool of PG may function as a buffer for CL synthesis and that limiting levels of PG are preferentially directed to CL synthesis at the expense of other PG-related lipids. It is possible that TmaT-mediated changes in h/ketoTMCM levels in the IM directly impact the activities of PG synthase and/or the extent to which DAG, PG, or Acyl-PG-like species are flipped across the IM for synthesis of CL in the periplasmic space. Alternatively, loss of TmaT activity may result in global changes in cellular physiology that lead to downregulation of other pathways of lipid biosynthesis. However, it is notable that not all pathways of lipid synthesis were affected in the TmaT null mutant, indicating that this phenotype is not the result of loss of viability.
Some of the changes observed in the lipid composition of the IM fraction in the ΔtmaT mutant could also represent compensatory responses to accumulation of hTMCM in this membrane. In particular, previous studies have suggested that aminoacylation of lipids reduces the overall net negative charge of cytoplasmic membranes in other bacteria and increases their resistance against cationic antimicrobial peptides (41–43). The loss of Ala-DAG, as well as Ala-PG, in the IM fraction could therefore represent a broader compensatory change to maintain the physiochemical properties of the IM. In contrast to the PG levels, levels of TAGs were increased in the TmaT mutant. Previous studies have indicated a link between TAG degradation in mycobacteria and MA biosynthesis (44). As the loss of TmaT leads to accumulation of hTMCM and precursors in the IM, it is possible that reduced requirement for MA synthesis may lead to reduced need for TAG degradation and buildup of these scavenged lipids. Finally, disruption of the mycobacterial TMCM transporter, MmpL3 (CmpL1+4 in C. glutamicum), results in similar changes in lipid composition as in the ΔtmaT mutant (45). Analysis of changes in the transcriptional responses of the ΔmmpL3 mutant indicated the induction of genes related to osmoprotection and metal homeostasis, suggesting that the inability to synthesize a correct OM leads to changes in cellular permeability and a metabolic downshift.
In conclusion, we have provided a high-resolution analysis of the lipid composition of C. glutamicum IM and OM fractions using a new LC-QTOF-MS/MS lipidomics platform. These analyses have highlighted a number of new lipid species/subclasses in these bacteria that may have important roles in TMCM synthesis and/or transport across the IM/OM. They have also revealed an unexpected level of interconnection of trehalose-mycolate and PG/CL synthesis in these bacteria that may elevate enzymes involved in their synthesis as high-priority targets for new generations of antimycobacterial/corynebacterial drugs.
Supplementary Material
Footnotes
Abbreviations:
- Ac1-hTDCM
- trehalose acetyl-hydroxylcorynomycolate
- AcPIM
- acetylated PIM
- AcTMCM
- trehalose monoacetylcorynomycolate
- Acyl-AcTMCM
- acyl-trehalose monoacetylcorynomycolate
- Acyl-hTMCM
- acyl-trehalose monohydroxylcorynomycolate
- Acyl-PG
- acylated phosphatidylglycerol
- AG
- arabinogalactan
- Ala-DAG
- alanylated diacylglycerol
- Ala-PG
- alanylated phosphatidylglycerol
- BHI
- brain heart infusion
- CDP-DAG
- cytidine diphosphate diacylglycerol
- CL
- cardiolipin
- CMW
- chloroform/methanol/water
- DAG
- diacylglycerol
- Gl
- glucuronic acid diacylglycerol
- GMM
- glucosylmonocorynomycolate
- h2TDCM
- trehalose di-hydroxylcorynomycolate
- hGMM
- glucose monohydroxylcorynomycolate
- HPTLC
- high-performance TLC
- hTMCM
- trehalose monohydroxylcorynomycolate
- IM
- inner membrane
- ketoTMCM
- trehalose monoketocorynomycolate
- LAM
- lipoarabinomannan
- LM
- lipomannan
- MA
- mycolic acid
- MAG
- monoacylglycerol
- MS/MS
- tandem mass spectrometry
- OM
- outer membrane
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PIM
- phosphatidylinositol mannoside
- PS
- phosphatidylserine
- QTOF
- quadrupole TOF
- TAG
- triacylglycerol
- TDCM
- trehalose dicorynomycolate
- TL
- total lipid
- TmaT
- TMCM mycolyl acetyltransferase
- TMCM
- trehalose monocorynomycolate
This work was supported by the Australian National Health and Medical Research Council (NHMRC), the National Collaborative Research Infrastructure Strategy, and Bioplatforms Australia. MJM is an NHMRC Principal Research Fellow.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Franco-Paredes C., and Rodriguez-Morales A. J.. 2016. Unsolved matters in leprosy: a descriptive review and call for further research. Ann. Clin. Microbiol. Antimicrob. 15: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokrousov I. 2009. Corynebacterium diphtheriae: genome diversity, population structure and genotyping perspectives. Infect. Genet. Evol. 9: 1–15. [DOI] [PubMed] [Google Scholar]
- 3.Murray C. J., Ortblad K. F., Guinovart C., Lim S. S., Wolock T. M., Roberts D. A., Dansereau E. A., Graetz N., Barber R. M., Brown J. C., et al. . 2014. Global, regional, and national incidence and mortality for HIV, tuberculosis, and malaria during 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 384: 1005–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 2015. Global Tuberculosis Report 2015. 20th edition Geneva: World Health Organization. [Google Scholar]
- 5.De La Iglesia A. I., and Morbidoni H. R.. 2006. [Mechanisms of action of and resistance to rifampicin and isoniazid in Mycobacterium tuberculosis: new information on old friends] Rev. Argent. Microbiol. 38: 97–109. [PubMed] [Google Scholar]
- 6.Stehr M., Elamin A. A., and Singh M.. 2014. Filling the pipeline—new drugs for an old disease. Curr. Top. Med. Chem. 14: 110–129. [DOI] [PubMed] [Google Scholar]
- 7.Lanéelle M. A., Tropis M., and Daffé M.. 2013. Current knowledge on mycolic acids in Corynebacterium glutamicum and their relevance for biotechnological processes. Appl. Microbiol. Biotechnol. 97: 9923–9930. [DOI] [PubMed] [Google Scholar]
- 8.Lee J. Y., Na Y. A., Kim E., Lee H. S., and Kim P.. 2016. The actinobacterium Corynebacterium glutamicum, an industrial workhorse. J. Microbiol. Biotechnol. 26: 807–822. [DOI] [PubMed] [Google Scholar]
- 9.Lea-Smith D. J., Pyke J. S., Tull D., McConville M. J., Coppel R. L., and Crellin P. K.. 2007. The reductase that catalyzes mycolic motif synthesis is required for efficient attachment of mycolic acids to arabinogalactan. J. Biol. Chem. 282: 11000–11008. [DOI] [PubMed] [Google Scholar]
- 10.Alderwick L. J., Lloyd G. S., Ghadbane H., May J. W., Bhatt A., Eggeling L., Futterer K., and Besra G. S.. 2011. The C-terminal domain of the Arabinosyltransferase Mycobacterium tuberculosis EmbC is a lectin-like carbohydrate binding module. PLoS Pathog. 7: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rainczuk A. K., Yamaryo-Botte Y., Brammananth R., Stinear T. P., Seemann T., Coppel R. L., McConville M. J., and Crellin P. K.. 2012. The lipoprotein LpqW is essential for the mannosylation of periplasmic glycolipids in Corynebacteria. J. Biol. Chem. 287: 42726–42738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Layre E., Sweet L., Hong S., Madigan C. A., Desjardins D., Young D. C., Cheng T. Y., Annand J. W., Kim K., Shamputa I. C., et al. . 2011. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol. 18: 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartain M. J., Dick D. L., Rithner C. D., Crick D. C., and Belisle J. T.. 2011. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J. Lipid Res. 52: 861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bansal-Mutalik R., and Nikaido H.. 2011. Quantitative lipid composition of cell envelopes of Corynebacterium glutamicum elucidated through reverse micelle extraction. Proc. Natl. Acad. Sci. USA. 108: 15360–15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchand C. H., Salmeron C., Bou Raad R., Meniche X., Chami M., Masi M., Blanot D., Daffe M., Tropis M., Huc E., et al. . 2012. Biochemical disclosure of the mycolate outer membrane of Corynebacterium glutamicum. J. Bacteriol. 194: 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaryo-Botte Y., Rainczuk A. K., Lea-Smith D. J., Brammananth R., van der Peet P. L., Meikle P., Ralton J. E., Rupasinghe T. W., Williams S. J., Coppel R. L., et al. . 2015. Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem. Biol. 10: 734–736. [DOI] [PubMed] [Google Scholar]
- 17.Takayama K., Wang C., and Besra G. S.. 2005. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18: 81–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tropis M., Meniche X., Wolf A., Gebhardt H., Strelkov S., Chami M., Schomburg D., Kramer R., Morbach S., and Daffe M.. 2005. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae. J. Biol. Chem. 280: 26573–26585. [DOI] [PubMed] [Google Scholar]
- 19.Yagüe G., Segovia M., and Valero-Guillén P. L.. 2003. Phospholipid composition of several clinically relevant Corynebacterium species as determined by mass spectrometry: an unusual fatty acyl moiety is present in inositol-containing phospholipids of Corynebacterium urealyticum. Microbiology. 149: 1675–1685. [DOI] [PubMed] [Google Scholar]
- 20.Layre E., Al-Mubarak R., Belisle J. T., and Branch Moody D.. 2014. Mycobacterial lipidomics. Microbiol. Spectr. 2: 1–19. [DOI] [PubMed] [Google Scholar]
- 21.Crick P. J., and Guan X. L.. 2016. Lipid metabolism in mycobacteria—Insights using mass spectrometry-based lipidomics. Biochim. Biophys. Acta. 1861: 60–67. [DOI] [PubMed] [Google Scholar]
- 22.Radmacher E., Alderwick L. J., Besra G. S., Brown A. K., Gibson K. J., Sahm H., and Eggeling L.. 2005. Two functional FAS-I type fatty acid synthases in Corynebacterium glutamicum. Microbiology. 151: 2421–2427. [DOI] [PubMed] [Google Scholar]
- 23.Kind T., Liu K. H., Lee D. Y., DeFelice B., Meissen J. K., and Fiehn O.. 2013. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 10: 755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., Spener F., van Meer G., Wakelam M. J., and Dennis E. A.. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50(Suppl): S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., and Arita M.. 2015. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 12: 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderberg R. J., Strachan J. A., and Cangelosi G. A.. 1995. Purification of DNA from Mycobacterium species without sonication or phenol. Biotechniques. 18: 217–219. [PubMed] [Google Scholar]
- 27.Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., and Pühler A.. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 145: 69–73. [DOI] [PubMed] [Google Scholar]
- 28.McKean S., Davies J., and Moore R.. 2005. Identification of macrophage induced genes of Corynebacterium pseudotuberculosis by differential fluorescence induction. Microbes Infect. 7: 1352–1363. [DOI] [PubMed] [Google Scholar]
- 29.Billman-Jacobe H., McConville M. J., Haites R. E., Kovacevic S., and Coppel R. L.. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33: 1244–1253. [DOI] [PubMed] [Google Scholar]
- 30.Morita Y. S., Velasquez R., Taig E., Waller R. F., Patterson J. H., Tull D., Williams S. J., Billman-Jacobe H., and McConville M. J.. 2005. Compartmentalization of lipid biosynthesis in mycobacteria. J. Biol. Chem. 280: 21645–21652. [DOI] [PubMed] [Google Scholar]
- 31.Bird S. S., Marur V. R., Sniatynski M. J., Greenberg H. K., and Kristal B. S.. 2011. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal. Chem. 83: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia J., Sinelnikov I. V., Han B., and Wishart D. S.. 2015. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 43: W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith C. A., Want E. J., O’Maille G., Abagyan R., and Siuzdak G.. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78: 779–787. [DOI] [PubMed] [Google Scholar]
- 34.Csizmadia F. 2000. JChem: Java applets and modules supporting chemical database handling from web browsers. J. Chem. Inf. Comput. Sci. 40: 323–324. [DOI] [PubMed] [Google Scholar]
- 35.Brennan P. J., and Lehane D. P.. 1971. The phospholipids of corynebacteria. Lipids. 6: 401–409. [DOI] [PubMed] [Google Scholar]
- 36.Collins M. D., Goodfellow M., and Minnikin D. E.. 1982. Fatty acid composition of some mycolic acid-containing coryneform bacteria. J. Gen. Microbiol. 128: 2503–2509. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y., Shi F., Tao G., and Wang X.. 2012. Purification and structure analysis of mycolic acids in Corynebacterium glutamicum. J. Microbiol. 50: 235–240. [DOI] [PubMed] [Google Scholar]
- 38.Kessner D., Chambers M., Burke R., Agus D., and Mallick P.. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 24: 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groessl M., Graf S., and Knochenmuss R.. 2015. High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst. 140: 6904–6911. [DOI] [PubMed] [Google Scholar]
- 40.Plassmeier J., Li Y., Rueckert C., and Sinskey A. J.. 2016. Metabolic engineering Corynebacterium glutamicum to produce triacylglycerols. Metab. Eng. 33: 86–97. [DOI] [PubMed] [Google Scholar]
- 41.Slavetinsky C. J., Peschel A., and Ernst C. M.. 2012. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 56: 3492–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maloney E., Lun S., Stankowska D., Guo H., Rajagoapalan M., Bishai W. R., and Madiraju M. V.. 2011. Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front. Microbiol. 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maloney E., Stankowska D., Zhang J., Fol M., Cheng Q. J., Lun S., Bishai W. R., Rajagopalan M., Chatterjee D., and Madiraju M. V.. 2009. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 5: e1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabruja M., Mondino S., Tsai Y. T., Lara J., Gramajo H., and Gago G.. 2017. A conditional mutant of the fatty acid synthase unveils unexpected cross talks in mycobacterial lipid metabolism. Open Biol. 7: 160277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degiacomi G., Benjak A., Madacki J., Boldrin F., Provvedi R., Palu G., Kordulakova J., Cole S. T., and Manganelli R.. 2017. Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci. Rep. 7: 43495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.