Abstract
Integrated analyses of multiple genomic datatypes are now common in cancer profiling studies. Such data present opportunities for numerous computational experiments, yet analytic pipelines are limited. Tools such as the cBioPortal and Regulome Explorer, while useful, are not easy to access programmatically or implement locally. Here we introduce the MVisAGe R package, which allows users to quantify gene-level associations between two genomic datatypes in order to investigate the effect of genomic alterations (e.g. DNA copy number changes on gene expression). Visualizing Pearson/Spearman correlation coefficients according to the genomic positions of the underlying genes provides a powerful yet novel tool for conducting exploratory analyses. We demonstrate its utility by analyzing three publicly available cancer datasets. Our approach highlights canonical oncogenes in chr11q13 that displayed the strongest associations between expression and copy number, including CCND1 and CTTN, genes not identified by copy number analysis in the primary reports. We demonstrate highly concordant usage of shared oncogenes on chr3q, yet strikingly diverse oncogene usage on chr11q as a function of HPV infection status. Regions of chr19 that display remarkable associations between methylation and gene expression were identified, as were previously unreported miRNA-gene expression associations that may contribute to the epithelial-to-mesenchymal transition.
Keywords: Gene expression, DNA copy number, DNA methylation, integrated analysis, squamous tumors
INTRODUCTION
Technological advances have led to the development of assays, notably microarrays and high-throughput sequencing, which are used to interrogate the genome in cancer profiling studies. Molecular subtypes identified in the seminal gene expression profiling studies of breast cancer (1, 2) provided insight into the underlying biology, including clinically relevant biomarkers, thus illustrating the power of genomic data. Not surprisingly, there has been a corresponding explosion in the development of analysis methods.
Increasingly, cancer studies include data from multiple genomic data types, e.g. gene expression (GE), gene mutation, and DNA copy number (CN), because this approach provides greater insight into the underlying genomic changes, pathways, and biomarkers. Early studies (3–5) showed that somatic CN gains and losses were often accompanied by corresponding changes in GE. A number of methods have been developed to analyze GE and CN data in cancer studies, as reviewed in (6) and (7). Methodology, output, and computational efficiency varies across the procedures, as does the required level of pre-processing, but each approach provides test statistics or measures of statistical significance that can be used to prioritize genes based on the relationship between their GE and CN.
In spite of the availability of methods for modeling the relationship between two genomic data types, Pearson and Spearman correlation coefficients continue to be widely used. For example, the Regulome Explorer (http://explorer.cancerregulome.org/) is a web-based application that allows users to compute correlations between GE and CN data in cohorts from The Cancer Genome Atlas (TCGA). In addition to GE and CN data, the Regulome Explorer includes DNA methylation (ME), microRNA expression (miRNA), and clinical data so that pairwise associations between any two data types can be examined. Although the Regulome Explorer’s broad functionality, high quality graphics, and ease of use make it a valuable tool for mining various TCGA datasets, to the best of our knowledge it cannot be applied to other datasets. This motivated our interest in developing MVisAGe, a tool for Modeling, Visualizing, and Analyzing the cancer Genome.
Here we illustrate MVisAGe’s utility by applying it to GE, CN, ME, and miRNA expression data from the TCGA cervical, head and neck, and lung squamous cell carcinoma cohorts (CESC, n = 191; HNSC, n = 279; LUSC, n = 178). MVisAGe uses matrix operations to compute genome-wide Pearson and Spearman correlation coefficients for two matrices of quantitative genomic data in a matter of seconds. Several plotting functions are available, and these allow users to identify genomic regions where cis-acting alterations, e.g. CN or ME changes, affect GE. Plots of smoothed correlations facilitate the identification of regional effects by reducing noise. By using miRNA-target gene annotation files, MVisAGe can be applied to explore associations between GE and miRNA expression.
Because MVisAGe is available as an R package (https://cran.r-project.org/web/packages/MVisAGe/index.html), researchers can use it to analyze their own datasets, mine publicly available datasets, or customize its capabilities. The package includes a set of “helper functions” that are designed for users without specialized bioinformatics skills who want to analyze TCGA data. These functions facilitate the creation of data matrices with common gene and sample identifiers – items required for MVisAGe’s downstream functions but not available in datasets directly downloaded from web repositories.
Although CESC, HNSC, and LUSC affect different organ systems, they have a number of underlying similarities, including squamous histology and common risk factors – HPV infection in CESC and HNSC; smoking in HNSC and LUSC. These and other categorical factors can be explored with MVisAGe by including optional sample annotation data. This feature facilitates integrated analyses that identify shared or distinct regions of genomic alteration or regulation in separate subcohorts. For example, analysis of the TCGA HNSC cohort highlights regions where CN/GE correlations are markedly different in HPV+ and HPV− patients, suggesting that genes such as EGFR and TRAF3 play different roles in the two groups. To the best of our knowledge separate subcohorts can only be analyzed with the Regulome Explorer by performing independent analyses.
CN/GE correlation coefficients are often accompanied by p-values derived from t statistics in order to prioritize genes where underlying genomic alterations affect gene expression. However, our findings suggest that this approach may not be appropriate, as evidenced by the fact that 11978 of 17442 genes yielded CN/GE Pearson correlation coefficients with false discovery rate (FDR) q-values less than 0.05. For this reason MVisAGE includes two additional methods for assessing statistical significance. The first is a permutation-based approach similar to the DR-Integrator method (8), while the second employs mixtures of normal distributions. Additional approaches are being investigated.
MATERIAL AND METHODS
Datasets
Level 3 mRNA expression, miRNA expression, and DNA methylation data for CESC (n = 191), HNSC (n = 279), and LUSC (n = 178) were downloaded from the Legacy Archive of the Genomic Data Commons (https://portal.gdc.cancer.gov/legacy-archive/search/f), for the following platforms: Illumina HiSeq (mRNA and miRNA) and Illumina 450K (DNA methylation). Gene-level expression measurements for mRNA and miRNA were log2(normalized RSEM + 1) and log2(FPKM + 1), respectively. Quantitative gene-level DNA copy number measurements for the three cohorts were obtained from GISTIC2 output downloaded from the Broad GDAC Firehose (https://gdac.broadinstitute.org/).
In each tumor type methylation beta values were used to produce binary gene-level methylation calls, as described previously (9). Briefly, we began by restricting to probes in transcription start sites, as designated by TSS200 or TSS1500 in the Illumina 450K annotation data. For each tumor sample the gene-level beta value for gene g was computed by finding the mean beta value over all probes corresponding to gene g. These gene-level beta values were then compared to a gene-specific threshold defined below in order to make binary methylated calls. The threshold for gene g was defined based on the mean and standard deviation of the beta values for all probes corresponding to gene g in the normal samples. More specifically, the gene-specific threshold is three standard deviations above the mean, i.e.
where beta_ng is the set of beta values from normal samples for gene g. The value 1 is included here because beta values lie between 0 and 1. Each tumor sample was classified as methylated at gene g if its gene-level beta value was greater than threshg, otherwise it was classified as unmethylated.
Gene positions (hg38 RefGene) were downloaded from Galaxy (https://galaxyproject.org/). Genes not in chr1 – chr22, chrX, or chrY were removed, as were genes having names that appeared in multiple chromosomes. Start and stop positions for each gene were defined by taking the mean across multiple start and stop positions, if necessary.
Matrix-based computation of Pearson and Spearman correlation coefficients
Let X and Y be n × m matrices with numeric entries, where rows are indexed by genes and columns are indexed by samples. The n Pearson correlation coefficients for each row of X and each row of Y were computed as follows: First, create n × m matrices X1 and Y1 by standardizing each row of X and Y to have mean 0 and Euclidean norm 1. Then define the n × m matrix Z = X1 * Y1 by multiplying the corresponding entries of each matrix. Finally, the vector of length n corresponding to the row sums of Z gives the n Pearson correlation coefficients. Spearman correlations were computed using the above approach after first converting entries in X and Y to row-specific ranks.
Smoothing
Smoothing reduces local variability and thus facilitates the identification of regions with large correlation coefficients produced by somatic alterations in DNA copy number or methylation level. Loess smoothing was applied to the gene-level correlation coefficients using the gene positions and the loess() R function. The size of the smoothing window is an input parameter, and smoothing is performed separately for each chromosome. The effect of smoothing at each telomere is minimized by artificially extending the chromosome, smoothing the extended chromosome, and then restricting to the smoothed correlation coefficients from the original chromosomal markers. No adjustment was made at the centromeres.
Statistical significance
Traditionally the significance of an observed Pearson correlation coefficient ρ = ρobs is assessed with the test statistic . Positive correlation coefficients are of interest when working with CN and GE data, so MVisAGe computes the p-value p(tn−2 > tobs), the area under the curve in the right tail of the t distribution with n − 2 degrees of freedom. In the permutation-based approach the correlation coefficients ρperm are computed after randomly permuting the sample identifiers in the expression data N times. Each resulting gene-specific null distribution is then used to assess the significance of ρobs for a given gene using the formula , where #(ρperm > ρobs) is the number of ρperm values larger than ρobs. Our final method for assessing statistical significance was motivated by the observation that the CN/GE ρ values in CESC, HNSC, and LUSC all have bimodal distributions (Supplementary Figure 1A–C). This suggests that genes fall into two broad categories: genes whose expression values are driven by underlying copy number alterations and genes whose expression values are not driven by underlying copy number alterations. The mixtools R package (10) was used to obtain parameter estimates (μ1, μ2, σ1, σ2) for a mixture of two normal distributions N(μ1, σ1) and N(μ2, σ2), and here we assume μ2 > μ1. One would expect correlation coefficients to be larger when copy number alterations drive changes in gene expression. For this reason, the N(μ2, σ2) distribution is used to assess significance or obtain thresholds for identifying genes of interest. Additional details are provided in the Supplementary Methods.
Computing
R 3.4.1 (11) was used to create all figures and perform all data analyses.
RESULTS
Somatic copy number alterations and gene expression in HNSC
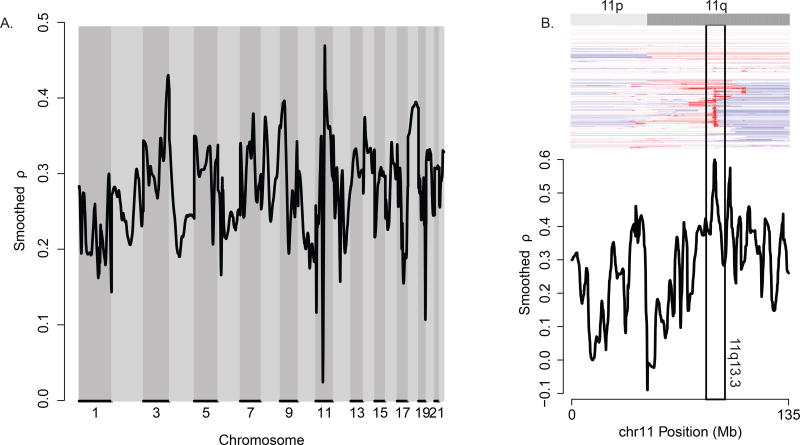
We began by analyzing the GE and gene-level CN data from the TCGA HNSC cohort. After restricting to the genes present on both platforms, MVisAGe was used to compute and plot smoothed Pearson correlation coefficients (CN/GE ρ values) across the genome (Figure 1A). Details are presented in the Methods. Several regions with large smoothed CN/GE ρ values were observed in Figure 1A, including 3q26-qter, 9p21, and 11q13. Prior studies (12, 13) have shown that each of these regions exhibit recurrent copy number aberrations in HNSC – amplifications in 3q and 11q13, deletions in 9p21. Although these regions were identified by GISTIC in the TCGA HNSC study (14), in each case the GISTIC region contained only a small number of genes (n = 7 in 3q26; n = 3 in 11q13, one of which is a miRNA; and n = 2 in 9p21). Our results suggest that, broadly speaking, these copy number alterations lead to corresponding changes in expression of regional genes. Moreover, it seems likely that the affected genes extend beyond the GISTIC regions because of the number of large unsmoothed CN/GE ρ values. In the discussion that follows, we focus primarily on 11q13 because the focal gains in this region produce extreme CN/GE ρ values for a number of genes. This region includes PPFIA1 and FADD, which were identified by GISTIC (Figure 1B), as well as known oncogenes CCND1 and CTTN not highlighted by GISTIC.
Figure 1.
Associations between DNA copy number and gene expression in head and neck squamous cell carcinoma (HNSC). (A) Genome-wide plot of smoothed gene-level Pearson correlation coefficients between quantitative measurements of DNA copy number and gene expression for n = 279 HNSC samples. (B) Plot of smoothed gene-level Pearson correlation coefficients between quantitative measurements of DNA copy number and gene expression for n = 279 HNSC samples in chromosome 11. Extreme correlation coefficients are observed in 11q13.3, the site of recurrent, high-level DNA copy number gains seen in the heatmap display of gene-level DNA copy number measurements. Genes are ordered by genomic position in the heatmap display; hierarchically clustered samples appear in rows.
Several genes have been proposed as the target of the 11q13 amplicon, including CCND1, FADD, and CTTN (15 – 17). CCND1 promotes progression through the cell cycle by dimerizing with CDK4/6 and regulating the G1/S transition (15). Although FADD is a component of the death-inducing signaling complex that mediates apoptosis, it can also act as an oncogene by regulating members of the NFκB and MAPK pathways (16). Cortactin is an actin-associated scaffolding protein, and high expression of CTTN has been shown to contribute to cell motility and tumor invasion through degradation of the extracellular matrix (17). Therefore, the observed copy number gains result in increased expression of genes with biological and therapeutic relevance.
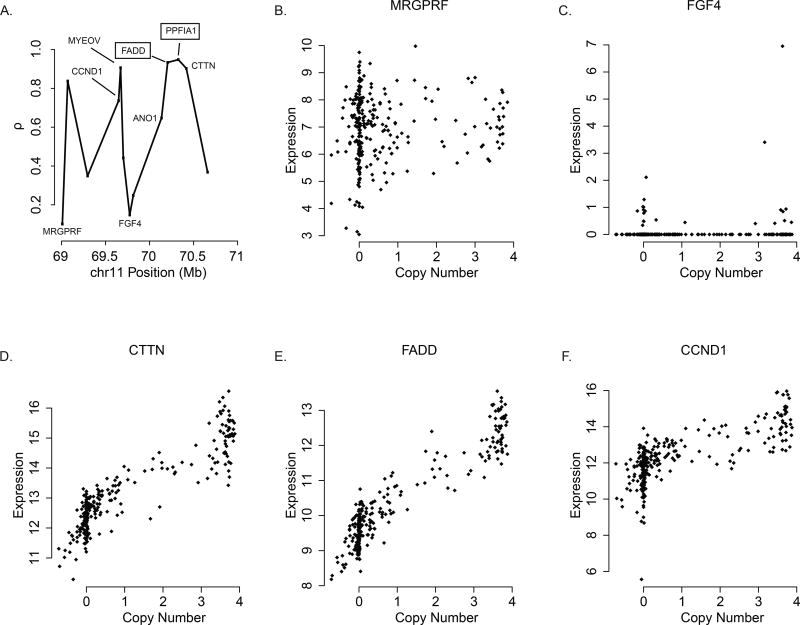
As shown in Figure 2A, the unsmoothed CN/GE ρ values in 11q13 exhibit surprising levels of variability even though the underlying CN values for these genes are very similar (Supplementary Data). This suggests that at the gene level there are exceptions to the broad association between regions of somatic copy number alteration and corresponding changes in gene expression as depicted in Figures 1A/B. Gene-specific scatterplots of CN and GE are shown in Figures 2B–F. Interestingly, even though both MRGPRF and FGF4 have low CN/GE ρ values (ρ = 0.10 and 0.02, respectively), the relationship between the CN and GE values for these two genes is quite different. Unexpectedly, the scatterplots for FADD and CTTN show that both gains and losses are accompanied by corresponding changes in expression (ρ = 0.93 and 0.90, respectively), whereas the effect is much less pronounced for CCND1 (ρ = 0.74). As noted above, FADD can induce apoptosis and activate the NFκB pathway, so both increases and decreases in expression may be beneficial to the tumor. At present, it is not clear why CTTN would exhibit such a duality. Future work is needed in order to determine whether the decrease in expression that accompanies copy number loss is functional or merely a passenger event. CCND1 plays a fundamental role in regulation of the cell cycle, and Klein et al. (18) describe the effect of numerous transcriptional inducers and repressors. Indeed, it may be the case that the complexity of the transcriptional regulation of CCND1 contribute to the observation that copy number changes have less of an effect on expression than other regional genes such as FADD and CTTN.
Figure 2.
Gene-level DNA copy number and gene expression in the 11q amplicon in head and neck squamous cell carcinoma. (A) Plot of gene-level Pearson correlation coefficients for select genes in 11q13.3. The correlation coefficients exhibit considerable variability even though the underlying DNA copy number values are very similar. (B – F) Scatterplots of quantitative DNA copy number and gene expression measurements for select genes in 11q13.3.
Integrated analyses of subcohorts
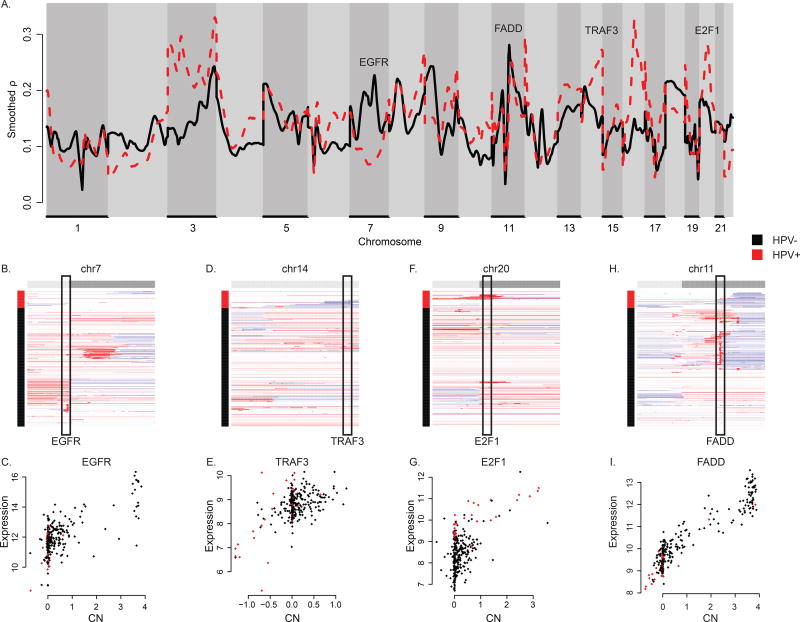
Sample annotation data can be incorporated into MVisAGe so the gene-specific correlations are simultaneously computed for two or more subcohorts. As an illustration, we repeated the above analysis using subcohorts defined by HPV status in HNSC. Figure 3A shows genome-wide plots of smoothed CN/GE ρ values in HNSC for HPV negative (HPV−, n = 243) and HPV positive (HPV+, n = 36) subjects. There are a number of striking differences, including 7p11 (EGFR), 11q13 (FADD), 14q32 (TRAF3), and 20q11 (E2F1). Figure 3B shows that broad copy number gains in chr7p and focal amplification of EGFR are seen almost exclusively in HPV− subjects (19). In general these gains lead to increased expression of EGFR in the HPV− subjects (Figure 3C), resulting in large CN/GE ρ values. On the other hand, HPV+ samples have large CN/GE ρ values in chr14 that appear to be driven by copy number losses and decreased expression of regional genes, including TRAF3 (Figure 3D/E). This could result in dysregulated NFκB signaling (20). On chr20, copy number gains and increased expression of E2F1 are seen in HPV+ subjects (Figure 3F/G). Figure 3G also shows that among E2F1 copy neutral samples, expression levels are markedly higher in HPV+ samples than in HPV− samples, perhaps because of E2F1 transcription autoregulation in the absence of negative control by pRb (21).
Figure 3.
Associations between DNA copy number and gene expression in head and neck squamous cell carcinoma by human papillomavirus (HPV) infection status. (A) Genome-wide plot of smoothed gene-level Pearson correlation coefficients between quantitative measurements of DNA copy number and gene expression for samples with (HPV+, n = 36) and without (HPV−, n = 243) HPV. Pronounced differences are observed at select genes. (B, D, F, H) Heatmap displays of gene-level quantitative DNA copy number measurements in select chromosomes. Genes are ordered by genomic position (columns); samples appear in rows and are hierarchically clustered according to HPV status. Rectangles highlight select regions containing driver genes with recurrent copy number changes. (C, E, G, I) Scatterplots of quantitative measurements of DNA copy number and gene expression for select genes of interest by HPV status.
The heatmap in Figure 3H shows that focal gains of 11q13.3 are found almost exclusively in the HPV− samples. In spite of this observation, the CN/GE ρ values for FADD are nearly the same among HPV+ and HPV− subjects (Figure 3I). This appears to result from the fact that CN losses and gains of FADD, which are observed primarily in HPV+ and HPV− samples, respectively, both lead to corresponding decreases and increases in gene expression as a function of HPV status. On the other hand, CN changes have a less pronounced effect on CCND1 expression, particularly in HPV+ subjects (Supplementary Figure 2A–C). Although the reason for this behavior is not clear, the hypothesis that it may be connected to inactivation of RB1 in HPV+ samples is intriguing.
Arm-level alterations of 3p (loss) and 3q (gain) are observed in the majority of the HNSC samples (Supplementary Figure 3A), but some differences are seen between HPV+ and HPV− samples. For example, a subset of HPV− samples is essentially copy neutral throughout chr3, and two HPV+ samples have gains on both arms. Even though HPV+ samples have larger CN/GE ρ values across most of chr3, manual review suggests that this distinction arises because of the differences in CN noted above, not because of gene targets that are present in HPV+ and absent in HPV−. Although there is no clear target of the deletion in 3p, it is noteworthy that known tumor suppressors MLH1 and BAP1 produce two of the ten largest CN/GE ρ values in 3p among the HPV+ samples. MLH1 is a mismatch repair gene, and a previous study found an association between promoter methylation and decreased expression (22). Although we did not observe altered methylation in the TCGA cohort, the effect of copy number losses suggests an alternate method for regulating MLH1 expression in HNSC. Supplementary Figure 3B shows that both HPV+ and HPV− samples produce a focal peak in CN/GE ρ values in 3q. Strikingly, DVL3, which plays a role in Wnt signaling, has the first and second largest CN/GE ρ values in 3q for HPV+ and HPV− samples, respectively. Wnt signaling is known to be important in HPV+ HNSC because it regulates cellular differentiation and thereby contributes to viral replication (23). Kwan et al. (24) found that although Dvl3 expression was positively associated with Wnt-βcatenin activity in cervical cancer, they also observed a decrease in DVL3 expression and cell proliferation when cervical cancer cell lines were treated with the AMPK activator metformin. This suggests a possible therapeutic approach in HNSC.
Having considered the role of coordinated gene expression and copy number within a single anatomic site and across subsets of patients within that tumor as a function of viral infection, we turned our attention to similar relationships in squamous tumors across anatomic sites. We extended the dataset by obtaining from TCGA smoothed CN/GE ρ values for cohorts of cervical (CESC) and lung squamous cell carcinoma (LUSC). This combined cohort allowed investigation of those shared genomic correlations across squamous tumors of three anatomic sites in which two tumors are primarily associated with tobacco as an etiology (HPV− HNSCC and LUSC) and two others are driven primarily by a viral etiology (CESC and HPV+ HNSCC). The genome-wide plot of smoothed CN/GE ρ values in Supplementary Figure 4A shows that LUSC has pronounced peaks in 3q and 9p that are shared with HNSC, as well as a peak in 19p (KEAP1, TYK2) that is unique among these three tumor types. Although PIK3CA, SOX2, and TP63 are often mentioned as the proposed targets of the 3q amplicon in LUSC (25), it is important to note that other regional genes may be relevant as well. Supplementary Figure 4B shows that DCUN1D1, DVL3, and SENP2 exhibit larger CN/GE ρ values than PIK3CA, SOX2, or TP63 in LUSC. The importance of these genes in LUSC is supported by previous findings (26). In the previous paragraph, we discussed the potential relevance of DVL3 in HNSC, and the fact that DCUN1D1 and SENP2 also have large values of ρ in CESC and HNSC (Supplementary Figure 4C/D) suggests that they may be important in these tumor types as well. HNSC and LUSC also have a common peak at 11q13, although the association between GE and CN is stronger in HNSC. CESC has a distinct peak in 11q22/23 (YAP1, BIRC2), and large smoothed CN/GE ρ values are observed in HNSC but not LUSC. In particular, the CN/GE ρ values for CESC and HNSC display a surprising level of similarity at the YAP1, BIRC2, and DCUN1D5 loci (Supplementary Figure 4E/F). Prior studies (27, 28) illustrate the importance of elevated YAP1 expression in both cervical carcinoma and HNSC cell lines, suggesting a common role for this gene in pathways that are dysregulated in these tumor types. In LUSC the CN/GE ρ values have peaks at BIRC2 and DCUN1D5 (Supplementary Figure 4G), but here ρ is smaller than the corresponding values in CESC and HNSC. This combined with the fact that the CN/GE ρ value for YAP1 is markedly lower in LUSC than CESC and HNSC suggests that copy number alterations in 11q22 may be less relevant in this tumor type.
Statistical significance in HNSC
As noted in the Introduction, in HNSC approximately two-thirds of the genes produced CN/GE Pearson correlation coefficients whose FDR q-values were less than 0.05 when the significance of the correlation coefficients was assessed using t-distributions. Interestingly, highly concordant results were observed when the permutation-based approach was applied (Supplementary Figure 5). In fact, when significance was assessed using 1000 permutations, a total of 11943 genes had q-values less than 0.05. Moreover, 11891 genes had q-values less than 0.05 using both approaches. Supplementary Figure 1B shows a histogram of the CN/GE ρ values in HNSC along with normal densities N(μ1, σ1) and N(μ2, σ2) produced by the mixtools R package, and here we assume μ2 > μ1. As noted in the Methods, we use the N(μ2, σ2) distribution to identify genes of interest. In HNSC a total of 1622 genes have CN/GE ρ values larger than μ2 + σ2 = 0.432 + 0.164 = 0.594. Although this set of genes includes known tumor suppressors and oncogenes such as CDKN2A, EGFR, and CCND1, others such as TP63 do not achieve this threshold. These results suggest that the use of mixture models provides a flexible, data-driven approach for identifying genes having CN/GE ρ values of interest in a given cohort. Results for each of the three methods appear in the Supplementary Data.
DNA methylation- and microRNA-based regulation of gene expression
Using a similar approach we computed gene-specific values of ρ based on (a) GE and binary ME calls (0 = unmethylated, 1 = methylated) or (b) expression levels of miRNAs and predicted target genes (29). Binary methylation measurements were chosen because similar values were computed in the TCGA LUSC study (9) and because the study of Singhal et al. (29) suggests that beta values are not amenable to correlation-based analyses. The miRNA analysis involved the computation of ρ for all predicted miRNA/target gene combinations (30). However, in an effort to focus on the miRNA/target gene combinations that produced the strongest associations, we filtered the results by allowing each gene to appear exactly once in the final output and with the miRNA that produced the smallest (i.e. largest negative) value of ρ.
Methylation-based regulation of expression of zinc finger genes
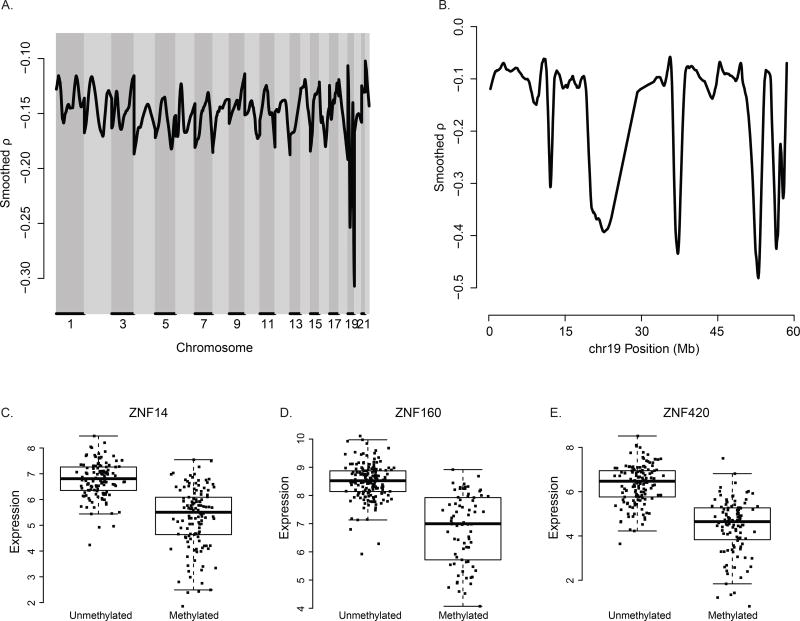
Like somatic copy number alterations, gene methylation changes have a cis effect on the expression of regional genes. Thus, it is natural to plot the smoothed ρ values from GE and ME in genomic order, as was done earlier for GE and CN. Figure 4A shows the results for HNSC, and here we observe a pronounced negative peak on chr19 that to our knowledge has not been previously reported and which is unmatched across the remainder of the genome. Closer inspection (Figure 4B) shows the presence of three distinct negative peaks near 22Mb, 37Mb, and 52Mb, as well as smaller peaks near 10Mb and 57Mb. In their analysis of two independent HNSC cohorts, Gaykalova et al. (31) noted that expression of ZNF14 (19.8Mb), ZNF420 (37.5Mb), and ZNF160 (53.5Mb) were all strongly affected by gene methylation levels. We observed similar results in the TCGA HNSC data (Figure 4C – E).
Figure 4.
Associations between DNA methylation and gene expression in head and neck squamous cell carcinoma (HNSC). (A) Genome-wide plot of smoothed gene-level Pearson correlation coefficients between binary DNA methylation measurements and gene expression for n = 250 HNSC samples. (B) Plot of smoothed gene-level Pearson correlation coefficients between binary DNA methylation measurements and gene expression for n = 250 HNSC samples in chromosome 19. (C – E) Boxplot displays of expression values of select zinc finger genes by DNA methylation status illustrate strong regulation of gene expression by DNA methylation.
Comparing the effect of copy number, methylation, and miRNA expression changes on gene expression
We then examined the results from the different platforms to determine if changes in CN, ME, and miRNA expression produced expression changes in different classes of genes across all three tumor types. For each platform – CN, ME, and miRNA – we began by identifying the genes that produced the 1000 largest (CN) or smallest (ME and miRNA) values of ρ. We then used the DAVID annotation database (32, 33) to identify pathways that are enriched for the three gene sets, as described by Gene Ontology (GO) terms for biological processes. The results are presented in the Supplementary Data. Pathways for genes strongly regulated by copy number changes included protein transport/localization, protein catabolic process, RNA processing, and other related pathways that contribute to cell proliferation. In contrast, genes controlled by methylation were involved in developmental pathways including anterior/posterior pattern formation, as well as neuron development/differentiation, urogenital system development, and epithelium development. miRNA genes were regulatory in nature, including regulation of cell motion/migration, regulation of cell proliferation, and regulation of phosphorylation. Highly concordant results were observed in the TCGA CESC data (Supplementary Data). If we assume that the strong associations (positive or negative) arose because of underlying changes in DNA copy number, methylation, or miRNA expression that contributed to the tumor phenotype, these results suggest a level of diversity and complementarity that may not have been previously appreciated.
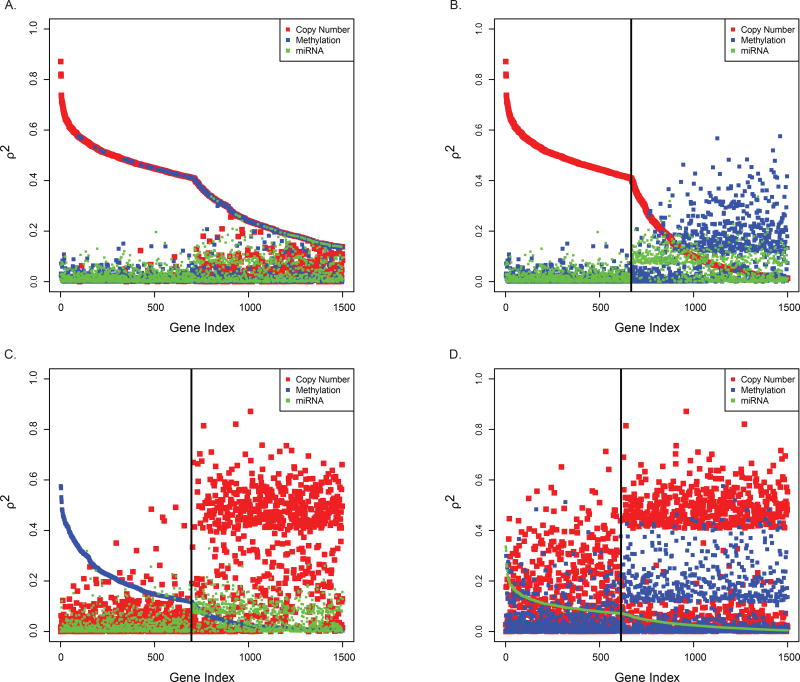
In addition, we were interested in the degree to which gene expression is regulated by changes in DNA copy number, methylation, or miRNA expression. Although an approach based on multiple linear regression models could be used to assess the joint effect of the three genomic datatypes on gene expression, for ease of interpretation we chose to compare the univariate associations. As above, we began by identifying the genes that produced the 1000 largest (CN) or smallest (ME, miRNA) values of ρ in HNSC. This yielded a set of 2831 unique genes, 1871 of which had ρ values for all three datatypes. These 1871 genes were then separated by whether they belonged to the list of 1000 CN genes (n = 668), the list of 1000 ME genes (n = 696), or the list of 1000 miRNA genes (n = 614). We then squared the ρ values, which eliminates differences in sign and thus allows a direct comparison of the results across the platforms. Each of the 1871 genes had three corresponding values of ρ2, one from each platform. The three values of ρ2 for each gene were plotted on a common axis based on different orderings of the genes so that each × coordinate (gene) has three y coordinates corresponding to the values of ρ2 for the three platforms. Figure 5A shows results based on ordering the genes according to the maximum value of ρ2 across the three platforms. In Figure 5B the genes are ordered according to the value ρ2 of in CN. Panels C and D of Figure 5 are similar to panel B, only now the genes are ordered according to the values of ρ2 ME and miRNA, respectively. The strongest associations are observed between GE and CN. Moreover, the vertical line at n = 668 in Figure 5B shows that genes with larger values of ρ2 from CN had considerably smaller values of ρ2 from either ME or miRNA. Although a similar result is seen in Figure 5C for the n = 696 genes with large ρ2 values from ME, the difference between the ME ρ2 values and those from either CN or miRNA is much less pronounced. In contrast, Figure 5D shows that many of the genes with the largest ρ2 values from miRNA have expression values that are in fact more strongly influenced by either CN or ME. This suggests that while many genes have expression patterns that are driven by copy number changes, very few genes are regulated to the same extent by targeting miRNAs.
Figure 5.
Associations between gene expression, DNA copy number, DNA methylation, and microRNA expression in head and neck squamous cell carcinoma. Gene-level Pearson correlation coefficients were computed based on expression values, quantitative DNA copy number measurements, binary DNA methylation values, and expression levels of targeting microRNAs (miRNAs). For each platform the largest 1000 (DNA copy number) or smallest 1000 (DNA methylation, miRNA expression) Pearson correlation coefficients were identified. 1871 of these genes yielded Pearson correlation coefficients for all three platforms. The three squared correlation coefficients (ρ2) for each gene are plotted after ordering the genes according to (A) largest overall gene-level value of ρ2, (B) gene-level value of ρ2 for DNA copy number, (C) gene-level value of ρ2 for DNA methylation, or (D) gene-level value of ρ2 for miRNA expression.
miRNA-based regulation of EMT genes
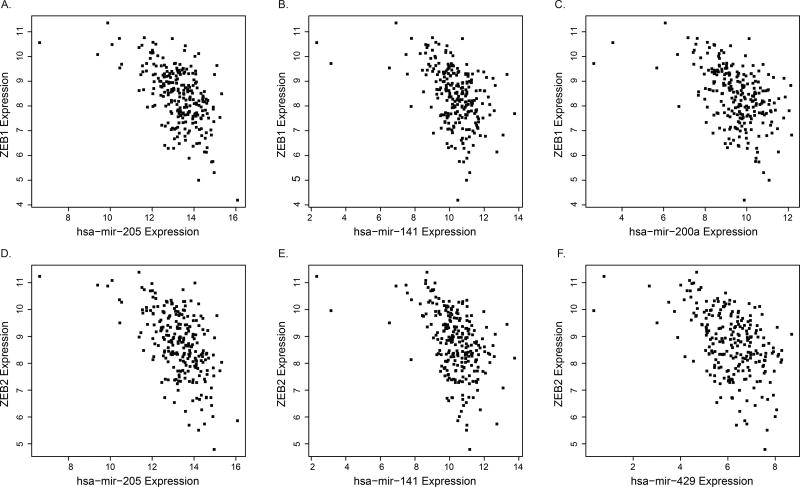
The epithelial to mesenchymal transition (EMT) is a biological process in which epithelial cells undergo changes that lead to the development of a mesenchymal phenotype. Although EMT occurs naturally during development, in tumor cells it can result in increased metastatic potential (34). Earlier we discussed genes whose expression patterns in HNSC were strongly influenced by miRNA expression, and we noted that the DAVID analysis identified enrichment for GO terms such as regulation of cell motion/migration. Closer inspection of the genes and miRNAs that produced large negative ρ values revealed several miRNAs and target genes that are known to play a role in EMT, including members of the miR-200 family, miR-205, and ZEB1/2 (Figure 6A – F). Both ZEB1 and ZEB2 are transcription factors that contribute to EMT by inhibiting the expression of CDH1 (34), and ZEB1/2 in turn are regulated by miR-205 and the miR-200 family (35). These associations were observed for genes and miRNAs on multiple chromosomes, suggesting that a single underlying genomic event – e.g. CN or ME change – involving miRNAs is unlikely. In their study of immortalized human mammary epithelial cells, Lim et al. (36) noted that different mechanisms of epigenetic regulation of mir-200 family members contributed to a mesenchymal phenotype (histone modification for miR-429/200a/b and methylation for miR-141/200c). miR-205 and the members of the miR-200 family do not appear to be the targets of copy number loss. Additional study will be required in order to determine if epigenetic regulation plays a role in HNSC.
Figure 6.
Expression of epithelial to mesenchymal transition (EMT) genes and microRNAs in the miR-200 family in head and neck squamous cell carcinoma. Scatterplots show expression levels of EMT-related transcription factors ZEB1/2 and select members of the miR-200 family (n = 244). Strong negative correlations suggest transcriptional regulation of ZEB1/2 by the miR-200 family.
DISCUSSION
Current cancer profiling studies regularly examine data from multiple high throughput technologies in an effort to better characterize genes and genomic alterations that are associated with disease. Although many methods have been developed for analyzing a single type of genomic data – e.g. gene expression, DNA copy number, or miRNA expression – considerably fewer methods exist for jointly analyzing multiple datatypes. Here we introduce MVisAGe, a tool for exploring associations between two quantitative genomic datatypes, and we illustrate its capabilities by analyzing three publicly available datasets from The Cancer Genome Atlas.
Like the Regulome Explorer, MVisAGe uses Pearson and Spearman correlation coefficients to quantify associations between gene expression and other genomic variables. Matrix multiplication is used for computational efficiency, and built-in plotting functions allow users to visualize the correlation coefficients according to the genomic position of the underlying genes in order to easily assess the cis effect of DNA copy number or methylation effects on gene expression. Regions with extreme correlation coefficients (large positive or large negative values) are of interest because it is believed they arise from somatic changes in DNA copy number or methylation level. Graphical output may not be appropriate for some genomic data types – e.g. miRNA and target gene expression – so output datasets can also be produced. In either case, MVisAGe can be used to perform exploratory analyses that highlight genes or regions of interest for later study.
By incorporating optional sample annotation data, MVisAGe can be applied to compare and contrast associations within two or more groups of subjects. Here we illustrate the utility of performing such integrated analyses by examining HPV+ and HPV− subjects in the TCGA HNSC cohort. 11q13 is commonly amplified in HNSC, and the region contains oncogenes such as CCND1, FADD, and CTTN that have been shown to play an important role in the disease. Interestingly, local copy number changes were largely defined by HPV status, with gains and losses primarily appearing in HPV− and HPV+ subjects, respectively. Moreover, the overall effect of these events on expression, as quantified by the Pearson correlation coefficient, was similar for some genes (e.g. FADD and CTTN) but not others (CCND1). Future studies may provide additional insight, but these are the sort of hypothesis-generating analyses that MVisAGe was designed to perform.
Gene expression is known to be regulated by the underlying DNA copy number, promoter methylation level, and the expression of targeting miRNAs. Therefore we applied MVisAGe using all of the above genomic data types. In general, DNA copy number changes had the strongest effect on gene expression, followed by changes in DNA methylation levels and miRNA expression. In addition, different classes of genes were most strongly regulated by changes in DNA copy number, DNA methylation level, or miRNA expression in both HNSC and CESC. At the moment it is unclear whether the concordant results observed in HNSC and CESC extend to other tumor types.
Cancer profiling studies involving multiple genomic datatypes are becoming increasingly common, and methods for performing integrated analyses are needed to fully leverage the power of these datasets. Although the CBioPortal and Regulome Explorer can be used to analyze pre-loaded datasets from high-profile studies like those conducted by the TCGA, considerable infrastructure is required to apply these methods to other datasets. The MVisAGe R package is a flexible yet powerful tool that researchers can apply to their own data or use to mine publicly available datasets. MVisAGe allows users to visualize gene-level associations between two genomic datatypes in order to explore the effect of underlying genomic aberrations (e.g. DNA copy number changes on gene expression). Genome-wide analysis of DNA copy number data and gene expression data from the TCGA HNSC project illustrated remarkably strong associations in 11q13. However, MVisAGe highlighted genes in this region that were not identified in the original analyses of copy number or expression alone. Moreover, we identified differential use of canonical oncogenes in these regions depending on HPV status. Taken together, these and other examples illustrate the types of exploratory analyses that can be performed with MVisAGe.
Supplementary Material
Significance.
This study presents an important bioinformatics tool that will enable integrated analyses of multiple genomic datatypes.
Acknowledgments
This work was supported by the National Cancer Institute [P30 CA006973 to L. Danilova, 5U10CA181009 to D.N. Hayes] and the National Institutes of Health [U24 CA126544, U24 CA143848-02S1 to D.N. Hayes]
Footnotes
Conflict of Interest
None
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Nat. Acad. Sci. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu W, Choi H, Ghosh D. Global associations between copy number and transcript mRNA microarray data: an empirical study. Cancer Inform. 2008;6:17–23. doi: 10.4137/cin.s342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myllykangas S, Junnila S, Kokkola A, Autio R, Scheinin I, Kiviluoto T, et al. Intergrated gene copy number and expression microarray analysis of gastric cancer highlights potential target genes. Int. J. Cancer. 2008;123(4):817–825. doi: 10.1002/ijc.23574. [DOI] [PubMed] [Google Scholar]
- 5.Ortiz-Estevez M, De Las Rivas J, Fontanillo C, Rubio A. Segmentation of genomic and transcriptomic microarrays data reveals major correlation between DNA copy number aberrations and gene-loci expression. Genomics. 2011;97(2):86–93. doi: 10.1016/j.ygeno.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Lahti L, Schafer M, Klein H-U, Bicciato S, Dugas M. Cancer gene prioritization by integrative analysis of mRNA expression and DNA copy number data: a comparative review. Brief. Bioinform. 2013;14(1):27–35. doi: 10.1093/bib/bbs005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang N, Shah PK, Li C. Lessons from a decade of integrating cancer copy number alterations with gene expression profiles. Brief. Bioinform. 2012;13(3):305–316. doi: 10.1093/bib/bbr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salari K, Tibshirani R, Pollack JR. DR-Integrator: a new analytic tool for integrating DNA copy number and gene expression data. Bioinformatics. 2010;26(3):414–416. doi: 10.1093/bioinformatics/btp702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;491(7423):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benaglia R, Chauveau D, Hunter DR, Young D. mixtools: an R package for analyzing finite mixture models. J. Stat. Soft. 2009;32(6):1–29. [Google Scholar]
- 11.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. http://R-project.org. [Google Scholar]
- 12.Lin M, Smith LT, Smiraglia DJ, Kazhiyur-Mannar R, Lang JC, Schuller DE, et al. DNA copy number gains in head and neck squamous cell carcinoma. Oncogene. 2006;25(9):1424–1433. doi: 10.1038/sj.onc.1209166. [DOI] [PubMed] [Google Scholar]
- 13.Smeets SJ, Braakhuis BJM, Abbas S, Snijders PJF, Ylstra B, van de Wiel MA, et al. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with and without oncogene-expressing human papillomavirus. Oncogene. 2006;25:2558–2564. doi: 10.1038/sj.onc.1209275. [DOI] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasamny JJ, Allak A, Krook KA, Jo VY, Policarpio-Nicolas ML, Sumner HM, et al. Cyclin D1 and FADD as biomarkers in head and neck squamous cell carcinoma. Otolaryngol Head Neck Surg. 2012;146(6):923–931. doi: 10.1177/0194599811435052. [DOI] [PubMed] [Google Scholar]
- 16.Dent P. FADD the bad. Cancer Biol. Ther. 2013;14(9):780–781. doi: 10.4161/cbt.26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigo JP, Garcia-Carracedo D, Garcia LA, Menendez ST, Allonca E, Gonzalez MV, et al. Distinctive clinicopathological associations of amplification of the cortactin gene at 11q13 in head and neck squamous cell carcinoma. J. Pathol. 2009;217:516–523. doi: 10.1002/path.2462. [DOI] [PubMed] [Google Scholar]
- 18.Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J. Cell. Sci. 2008;121(Pt23):3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter V, Yin, Wilkerson MD, Cabanski CR, Zhao N, Du Y, et al. Molecular subtypes of head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE. 2013;8(2):e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajek M, Sewell A, Kaech S, Burtness B, Yarbrough WG, Issaeva N. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck cancer. Cancer. 2017;123:1778–1790. doi: 10.1002/cncr.30570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8(13):1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 22.Koutsimpelas D, Pongsapich W, Heinrich U, Mann S, Mann WF, Brieger J. Promoter methylation of MGMT, MLH1, and RASSF1A tumor suppressor genes in head and neck squamous cell carcinoma: pharmalogical genome demethylation reduces proliferation of head and neck squamous carcinoma cells. Oncology Rep. 2012;27:1135–1141. doi: 10.3892/or.2012.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rampias T, Psyrri A. Human papillomavirus (HPV)-positive head and neck cancer and the Wnt signaling pathway. In: Burtness B, Golemis EA, editors. Molecular Determinants of Head and Neck Cancer. New York: Springer-Verlag; 2014. pp. 215–225. [Google Scholar]
- 24.Kwan HT, Chan DW, Cai PCH, Mak CSL, Yung MMH, Leung THY, et al. AMPK activators suppress cervical cancer cell growth through inhibition of DVL3 mediated Wnt/β-catenin signaling activity. PLoS ONE. 2013;8(1):e53597. doi: 10.1371/journal.pone.0053597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature Genet. 2009;21(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Qian J, Hoeksema MD, Zou Y, Espinosa AV, Rahman SM, et al. Integrative genomics analysis identifies candidate drivers at 3q26–29 amplicon in squamous cell carcinoma of the lung. Clin Cancer Res. 2013;19(20):5580–5590. doi: 10.1158/1078-0432.CCR-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenzetto E, Brenca M, Boeri M, Verri C, Piccinin E, Gasparini P, et al. YAP1 acts as oncogenic target of 11q22 amplification in multiple cancer subtypes. Oncotarget. 2014;5(9):2608–2621. doi: 10.18632/oncotarget.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge L, Smail M, Meng W, Shyr Y, Ye F, Fan KH, et al. Yes-associated protein expression in head and neck squamous cell carcinoma nodal metastasis. PLoS ONE. 2011;6(11):e27529. doi: 10.1371/journal.pone.0027529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singhal SK, Usmani N, Michiels S, Metzger-Filho O, Saini KS, Kovalchuk O, Parliament M. Towards understanding the breast cancer epigenome: a comparison of genome-wide DNA methylation and gene expression data. Oncotarget. 2016;7(3):3002–3017. doi: 10.18632/oncotarget.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaykalova DA, Vatapalli R, Wei Y, Tsai H-L, Wang H, Zhang C, et al. Outlier analysis defined zinc finger gene family DNA methylation in tumors and saliva of head and neck cancer patients. PLoS ONE. 2015;10(11):e0142148. doi: 10.1371/journal.pone.0142148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichments tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 36.Lim Y-Y, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci. 2013;126:2256–2266. doi: 10.1242/jcs.122275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.