Abstract
Specialized groups of neurons in the brain are key mediators of circadian rhythms, receiving daily environmental cues and communicating those signals to other tissues in the organism for entrainment and to organize circadian physiology. In Drosophila, the “circadian clock” is housed in seven neuronal clusters, which are defined by their expression of the main circadian proteins, Period, Timeless, Clock, and Cycle. These clusters are distributed across the fly brain and are thereby subject to the respective environments associated with their anatomical locations. While these core components are universally expressed in all neurons of the circadian network, additional regulatory proteins that act on these components are differentially expressed, giving rise to “local clocks” within the network that nonetheless converge to regulate coherent behavioral rhythms. In this review, we describe the communication between the neurons of the circadian network and the molecular differences within neurons of this network. We focus on differences in protein-expression patterns and discuss how such variation can impart functional differences in each local clock. Finally, we summarize our current understanding of how communication within the circadian network intersects with intracellular biochemical mechanisms to ultimately specify behavioral rhythms. We propose that additional efforts are required to identify regulatory mechanisms within each neuronal cluster to understand the molecular basis of circadian behavior.
Circadian rhythms are behavioral and physiological responses that allow anticipation of daily changes in the environment, such as day/night cycles. Indeed, circadian rhythms are entrained by diurnal oscillations in light, temperature, and food availability, each of which can be considered a “zeitgeber,” or “time giver.” Such anticipatory behavior of daily events confers adaptive advantages on individuals that organize physiology around planetary rhythms, as well as within a species as a whole, for purposes of mating, cooperation, protection, etc. In a sense, rhythmic anticipation can be thought of as a primitive mechanism of planning and cognition.
The circadian clock, consisting of transcriptional negative feedback loops, underlies the regulation of a ∼24-h behavioral rhythm. The molecular architecture of this feedback loop is conserved across the majority of multicellular organisms. In animals, the “master clock” is housed in specialized pacemaker neurons in the brain that coordinate various “peripheral clocks” throughout the organism to regulate daily physiological and behavioral changes. Importantly, the various clocks can adapt to changes in daily environmental rhythms, such as the change in light/dark cycles through the seasons, while still maintaining their coordination with each other. Thus, in addition to its role in anticipatory behavior, the master clock ensures that various organs and tissues are physiologically synchronized to respond cooperatively to daily changes in the environment.
In this review, we focus on the circadian system of Drosophila melanogaster. The majority of genes involved in the regulation of circadian rhythms were first discovered in Drosophila, with subsequent analogous or orthologous genes cloned in organisms ranging from fungi to plants to mammals. The genetic tractability of the fly and its simpler circadian neuronal network have allowed us to begin to dissect the biochemical mechanisms involved in regulating behavioral rhythms with molecular resolution. Here we review the latest developments in the biochemistry of the Drosophila circadian clock as it relates to its neuronal network, and fit the known molecular model of the clock into the biochemical restrictions imposed upon each neuron. We will also highlight the molecular pathways operating within the neurons that allow communication between the clocks. Where appropriate, key gaps in our knowledge will be noted.
THE CIRCADIAN CLOCK NEURONAL NETWORK
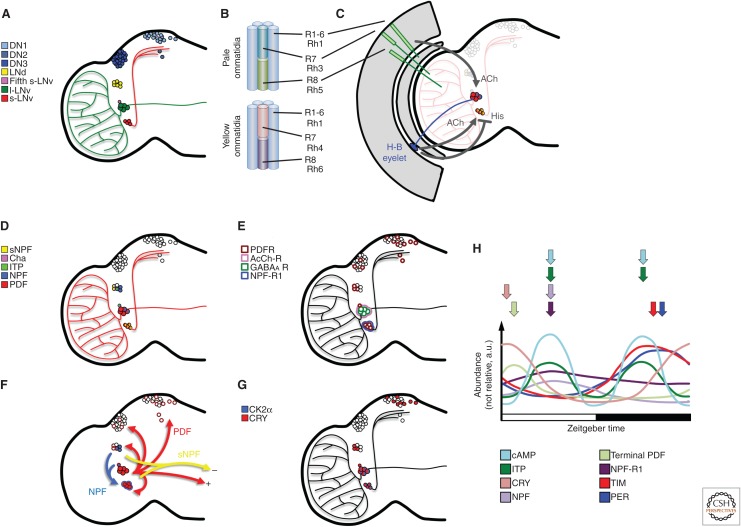
The circadian clock, also referred to as the molecular oscillator in this review, is a transcriptional negative feedback loop that oscillates with a ∼24-h period to regulate rhythmic behavior and physiology. In Drosophila, the master clock is housed in ∼150 neurons that compose the circadian clock neuronal network (CCNN). These neurons are defined by their expression of the core circadian proteins Clock (CLK), Cycle (CYC), Period (PER), and Timeless (TIM) (Houl et al. 2006; Kaneko and Hall 2000). CLK and CYC assemble into a DNA-binding transcriptional activator complex that regulates the transcription of per and tim. After translation, PER and TIM proteins assemble into a transcriptional repressor complex (PER/TIM) that binds to and blocks the activity of CLK/CYC, closing a negative feedback loop that oscillates with ∼24-h periodicity (discussed in detail below). Loss-of-function mutations in any of these core circadian proteins leads to arrhythmic behavior, underscoring their central role within the molecular oscillator. Many clock neurons express the light-sensing protein Cryptochrome (CRY) that communicates light signals to the circadian clock, allowing light-mediated resetting of the oscillator and therefore rhythmic behavior. The retina, which also expresses core clock components, also participates in transmitting light information independently of CRY to the CCNN. Neurons of the CCNN are grouped in seven clusters within the brain, and are named by their anatomical location: small and large ventral lateral neurons (s-LNvs and l-LNvs, respectively), fifth s-LNv, dorsal lateral neurons (LNds), and three clusters of dorsal neurons (DNs 1–3) (Fig. 1A). In this section, we describe signaling between these clusters and how this communication influences their internal biochemistry.
Figure 1.
A schematic of the anatomical structure of the fly brain circadian clock neuronal network. (A) Location and names of the seven neuronal clusters that make up the circadian neuronal network. (B) Structure of the ommatidia that make up the fly retina, displaying the expression of rhodopsins (Rh) in receptor cells (R). Two of the receptor cells within R1-6 are removed from the schematic to reveal R7 and R8 for clarity. (C) The relative position of the receptor cells and H-B eyelet with respect to the LNvs in the brain. The positive and negative effects of acetylcholine (ACh) and histamine (His) are shown in gray. (D) The expression pattern of neuropeptides in each of the circadian neuronal clusters. (E) The expression pattern of receptors in each of the circadian neuronal cluster. (F) The sources and the targets of pigment-dispersing factor (PDF), neuropeptide F (NPF), and short neuropeptide F (sNPF). (+) and (−) symbols represent the functional effect of the neuropeptides in nonspecified motor neurons. (G) The expression pattern of CK2α and CRY in each neuronal cluster. (H) A schematic of peak expression of indicated proteins. Arrows represent the time of peak expression and are color-coded.
The Dual Oscillator Model
The rhythmic behavior pattern of the fly is crepuscular, with peaks of activity during the transitions between light and dark (i.e., sunrise and sunset). These peaks of activity are flanked by periods of low activity reflecting sleep at night or siesta in the day and define the periodicity of rhythmic behavior. Exposure to light for longer periods of time advances morning anticipation while simultaneously delaying evening anticipation. The observation of this divergent anticipatory behavior led to the hypothesis of a dual oscillator model for regulating rhythmic behavior (Pittendrigh and Daan 1976). The dual oscillator model proposed the existence of separate morning and evening oscillators that converge to regulate rhythmic behavior. Studies conducted over 10 years ago provided the first anatomical evidence to support the dual oscillator model. Genetic ablation of the s-LNvs and the l-LNvs leads to a loss of morning anticipation, while ablating the LNds, the fifth s-LNv, and some DNs leads to a loss of evening anticipation (Stoleru et al. 2004). Furthermore, restoring PER expression to LNvs of per mutant flies restores morning but not evening anticipation, whereas restoring PER in both the LNvs and LNds restores both anticipatory behaviors (Grima et al. 2004). These studies established the LNvs as “morning cells” and the remaining cells as “evening cells” (Grima et al. 2004; Stoleru et al. 2004, 2005; Rieger et al. 2006; Guo et al. 2014). More recent genetic manipulations have suggested that four neurons, including three of the six LNds and the fifth s-LNv, are sufficient to serve as evening cells (Guo et al. 2014). However, manipulation of DNs can still alter evening anticipation (Yao et al. 2016; Guo et al. 2016), suggesting that these clusters are nonetheless involved in this behavior. Among morning cells (LNvs), the s-LNvs serve as master pacemaker neurons because they drive rhythmic activity in constant dark conditions (Grima et al. 2004; Stoleru et al. 2005; Rieger et al. 2006); without s-LNv activity, flies become arrhythmic. In the framework of the dual oscillator model, the morning cells (s-LNvs) dominate in the dark to drive coherent behavioral rhythms and also govern the timing of evening anticipation by communicating to the evening cells, placing the morning cells at the top of a neuronal hierarchy.
Since the establishment of the dual oscillator model, the hierarchical organization of the circadian neuronal clusters has been challenged and a more complex understanding of communication within this network has begun to emerge. In constant light conditions that permit rhythmic behavior (i.e., in flies lacking the light-sensing protein CRY; cry0), the LNd/fifth s-LNv cells dominate, driving behavioral rhythmicity when the s-LNvs are either arrhythmic or unable to communicate with the other clusters (Murad et al. 2007; Picot et al. 2007). This exception suggests that s-LNv dominance over the neuronal hierarchy is limited to constant dark conditions. The partitioned roles the different clusters play in morning and evening anticipation become important in seasonal changes of day length, where the evening cells dominate the CCNN in the long days of summer, and the morning cells dominate the CCNN in the long nights of winter, with communication between the two groups maintaining a 24-h behavioral rhythm (Stoleru et al. 2007). In molecular terms, longer light exposure accelerates the LNv clocks, resulting in advanced morning anticipation, and decelerates the LNd/fifth s-LNv/DN clocks to delay evening anticipation, as predicted by the dual oscillator model (Pittendrigh and Daan 1976). Another exception to the dual oscillator model comes from observed morning anticipation in dim light in flies lacking functional morning cells (but possessing functional evening cells) (Bachleitner et al. 2007; Rieger et al. 2009). Thus, the evening cells can compensate for the loss of morning cell activity in dim light conditions and oscillate without being driven by the s-LNvs. As a result, the dual oscillator model is expanded.
The relationships within the CCNN are increasingly revealing higher orders of complexity. In addition to s-LNv (master clock) communication to the DN1s, feedback signals from the DN1s back to the s-LNvs has also been identified, suggesting there is bidirectional flow of information between morning and evening cells (Guo et al. 2016). The LNds also communicate bidirectionally with s-LNvs (discussed in detail below), and it is likely that other groups of neurons within the CCNN communicate similarly. Overexpression of kinases that selectively accelerate or decelerate different clocks within the CCNN lead to split rhythms (a single behavioral rhythm split into behavioral components with short and long periodicity), demonstrating that rhythmic behavior is an emergent property of the communication of the various independent oscillators (Yao and Shafer 2014). Thus, independent molecular oscillators within each cluster are coordinated and synchronized to give rise to coherent behavioral rhythms.
Input from the Eye
Light is an important zeitgeber that helps entrain and reset the circadian clock. Light input to the clock occurs through the deep brain light receptor CRY (Stanewsky et al. 1998; Emery et al. 2000a; Dolezelova et al. 2007) and through the fly retina via rhodopsin activation (Helfrich-Förster et al. 2001). While light-activated CRY induces the degradation of TIM to reset the clock in the brain (Stanewsky et al. 1998; Koh et al. 2006; Peschel et al. 2009), light-activated rhodopsin in the retina triggers Gqα protein to activate no receptor potential A (norpA), a phospholipase that depolarizes the cell and triggers release of the neurotransmitter histamine to influence the CCNN (Bloomquist et al. 1988; Montell 2012; Hardie and Juusola 2015). However, the eye does not contribute to the circadian clock through CRY-dependent light input (Emery et al. 2000b; Bachleitner et al. 2007; Yoshii et al. 2015), despite expressing CRY (Yoshii et al. 2008), suggesting that these two light pathways converge independently to synchronize the circadian clock to the environment. More recently, a deep brain rhodopsin, Rh7, was demonstrated to be involved in light entrainment of pacemaker neurons (Ni et al. 2017). Undoubtedly, future work will uncover the molecular mechanisms by which Rh7 informs the circadian clock.
Different sections of the fly compound eye communicate with circadian neurons to regulate rhythmic behavior. The retina of the compound eye is composed of pale and yellow ommatidia made up of clusters of receptor cells, designated R1-8 (Fig. 1B) (Behnia and Desplan 2015). R7 and R8 in the pale ommatidia express rhodopsin (Rh) 3 and Rh5, respectively, and regulate twilight activity at dawn and dusk (Schlichting et al. 2015). R8 in the yellow ommatidia expresses Rh6 and R1-6 expresses Rh1; collectively they regulate activity in dim light (e.g., moonlight) (Schlichting et al. 2014). Thus, rhodopsins within the receptor cells can communicate dim light signals to the brain to regulate activity. It is likely that these cells can regulate morning anticipation when flies lack functional s-LNvs. R1-6 send projections from the retina into the lamina of the optic lobe, whereas R7 and R8 project further into the brain, to the medulla, where they are in proximity to LNv dendrites (Fig. 1C) (Helfrich-Förster et al. 2002). Despite this anatomical proximity, there is as yet no evidence of synaptic connections between the ommatidia and the l-LNvs despite the influence of R1-6 on moonlight activity (Schlichting et al. 2014). Thus, any R1-6 communication with the CCNN likely occurs through nonsynaptic signaling.
On the other hand, a second structure in the eye, beneath the receptor cells, forms a synaptic connection with the CCNN. The Hofbauer–Buchner (H-B) eyelet, a structure located between the retina and the lamina, communicates with the LNvs (Fig. 1C) (Hofbauer and Buchner 1989; Yasuyama and Meinertzhagen 1999; Malpel et al. 2002). The H-B eyelet is a remnant of the larval Bolwig organ, which plays a developmental role in the normal arborization of the LNvs in the medulla (Malpel et al. 2002). The H-B eyelet expresses Rh6, similarly to R1-6, and is involved in light entrainment (Veleri et al. 2007; Szular et al. 2012; Schlichting et al. 2016). The H-B eyelet makes a synaptic connection with the LNvs, communicating through acetylcholine and histamine release (Helfrich-Förster et al. 2002; Malpel et al. 2002; Schlichting et al. 2014, 2016; Muraro and Ceriani 2015). Blocking synaptic transmission between the H-B eyelet and the LNvs hinders behavioral phase shifts, as well as inhibits neuronal synchrony of TIM/PER oscillations (Veleri et al. 2007), suggesting that the H-B eyelet is important for communicating light signals to the CCNN. Acetylcholine signals likely underlie the transmission of light information to the CCNN because exogenous addition of a cholinergic agonist triggers a spike in Ca2+ and cyclic adenosine monophosphate (cAMP) levels in both LNvs (Wegener et al. 2004; Lelito and Shafer 2012). Indeed, acetylcholine-mediated excitation of the l-LNvs is mediated by the compound eye (including the H-B eyelet) (Muraro and Ceriani 2015; Schlichting et al. 2016), while histamine from the H-B eyelet (and/or other sources) inhibits l-LNv firing (Schlichting et al. 2016). Although no evidence exists of acetylcholine receptor expression in the s-LNvs, excitation of the H-B eyelet also leads to the excitation of the s-LNvs (Schlichting et al. 2016), suggesting that the l-LNvs may relay signals from the H-B eyelet. Together, these findings suggest that the H-B eyelet and photoreceptors communicate to the two LNv clusters through the l-LNvs. In summary, the fly retina communicates with the CCNN to stimulate complex Ca2+ and cAMP responses in the LNvs to influence behavioral rhythmicity.
It is important to note that, although signals from the eye to the CCNN appear to be transmitted through the LNvs, our knowledge is incomplete. For example, flies lacking an oscillator in their morning cells (both LNvs) exhibit rhythmic morning anticipation behavior in dim light (Rieger et al. 2009). Although dim light is thought to be mediated through a CRY-independent mechanism (Bachleitner et al. 2007), suggesting morning anticipation under these conditions arises from retinal activity, recent evidence suggests that CRY may also respond to prolonged exposure to dim light in the brain (Vinayak et al. 2013). The recently discovered Rh7 also responds to dim light (Ni et al. 2017), but how it coordinates with the retinal response is yet to be determined. In a retinal response, either the retina can also communicate to other parts of the CCNN in ways that have not yet been determined, or retinal communication through the LNvs does not require that the LNvs express a functional oscillator. In a CRY response, morning anticipatory behavior may bypass the LNvs altogether and rely on dim light exposure of the evening cells. Future work will determine how these mechanisms converge to regulate morning anticipation.
The Ventral Lateral Neurons, Arousal, and Pigment-Dispersing Factor
The LNvs are the most studied of the circadian neuronal clusters and are easily identifiable by their expression of the neuropeptide pigment-dispersing factor (PDF) (Fig. 1D) (Helfrich-Förster 1995). Elimination of either the LNvs or PDF causes a loss of morning anticipation and leads to arrhythmic behavior in constant dark conditions (Helfrich-Förster 1995, 1998; Renn et al. 1999; Stoleru et al. 2004). Therefore, PDF plays a central role in communication within the CCNN to maintain rhythmic activity in constant (dark) conditions. Although PDF expression does not oscillate in the s-LNv cell bodies, it depends on CLK activity for its expression, yet rhythmically (early morning) accumulates in the s-LNv termini for release during the day (Fig. 1H) (Blau and Young 1999; Park et al. 2000). On the other hand, PDF is not rhythmically released from l-LNvs in constant conditions (Park et al. 2000), highlighting a functional difference in how the two clusters regulate rhythmic behavior through PDF.
The LNvs use PDF to play distinct roles within the CCNN to regulate rhythmic behavior. While the oscillations of the clock in the s-LNvs regulate morning anticipation and drive rhythmic behavior in constant dark conditions (Grima et al. 2004; Stoleru et al. 2005), the l-LNvs are not responsible for either morning activity or evening activity. Instead, exposure to light (sunrise) is communicated to the l-LNv neurons through the eye or by a response mediated by l-LNv CRY, triggering the l-LNvs to fire rapidly and release PDF (Rieger et al. 2006). Neurons that express pdf receptor (pdfr), a G-protein-coupled receptor (GPCR), respond to the PDF released from the l-LNvs by up-regulating cAMP. Initially, identifying the neurons in which pdfr is expressed proved difficult (Hyun et al. 2005; Lear et al. 2005b; Mertens et al. 2005). Later work involving epitope tagging PDFR, measuring pdfr mRNA expression, and critically, testing neuronal response to exogenous PDF injection led to the conclusion that s-LNvs express PDFR and the l-LNvs do not (Fig. 1E) (Shafer et al. 2008; Im and Taghert 2010; Kula-Eversole et al. 2010; Im et al. 2011; Klose et al. 2016). Thus, PDF released from both LNvs impacts only the s-LNvs (and other pdfr-expressing neurons in the CCNN), activating the cellular response leading to light-mediated arousal (Sheeba et al. 2002, 2008a,b; Kula et al. 2006; Parisky et al. 2008; Shang et al. 2008; Im et al. 2011; Guo et al. 2014; Schlichting et al. 2016). Light-induced PDF release from the l-LNvs in turn triggers PDF release from the s-LNvs, and thereby helps to establish a PDF feedback mechanism to reinforce PDF release from the s-LNvs throughout the day (Parisky et al. 2008). Elimination of the l-LNvs reduces the efficacy of a light-pulse response in the late night (Shang et al. 2008), and overexpression of PDF in the l-LNvs induces arrhythmic activity in flies, phenocopying fly behavior in constant light conditions (Rieger et al. 2006; Wülbeck et al. 2008). Thus, the l-LNvs play a critical role in communicating light signals to the CCNN. This role in gating light signals to the CCNN also fits well with their function as a node for signals from the eye structure.
Two processes ensure strong PDF/PDFR-mediated communication in the s-LNvs in the morning. (1) The early morning peak in PDF accumulation in the s-LNv termini is coincident with s-LNv hypersensitivity to PDF, as mediated by the GTPase RalA (Park et al. 2000; Klose et al. 2016). (2) The light-sensitive l-LNvs express dimmed, which encodes a nonoscillating protein that amidates PDF, increasing its activity and half-life (Park et al. 2008). PDF-bound PDFR triggers a cAMP spike in neurons (Mertens et al. 2005; Shafer et al. 2008), triggering further PDF release, strengthening the s-LNv response, and transitioning governance of rhythmic activity from the evening to the morning cells (Choi et al. 2012). The s-LNv clock oscillations dampen in pdf0 flies in constant darkness (Yoshii et al. 2009), underscoring the importance of maintaining the PDF/PDFR feedback. Increased PDF sensitivity that the s-LNvs exhibit in the morning, concurrent with amidated PDF released by the l-LNvs, contribute to a stronger morning arousal mechanism in response to anticipated light. It is also possible that amidated PDF released from the l-LNvs may accentuate arousal at other times (e.g., light pulse; see below). Because the s-LNvs exhibit a stronger response to PDF released by the l-LNvs than PDF released by the s-LNvs themselves, it is likely that the l-LNvs play a critical role in triggering arousal through PDF, contributing to resetting the s-LNv clock in response to the light signal.
Arousal can also be triggered by other neurotransmitters. Dopamine (DA) has also been implicated in stimulating arousal (Kume et al. 2005; Lebestky et al. 2009). Expression of D1-like DA receptor in the l-LNvs provides a molecular basis for the observed DA-mediated arousal in flies (Klose et al. 2016). In the fly, <200 neurons synthesize DA with no evidence of overlap with clock neurons (Mao and Davis 2009). Thus, DA is a means for the CCNN to receive instruction from other circuits and does not appear to be involved in signaling from or within the CCNN. The s-LNv sensitivity to DA is synchronized with PDF sensitivity (Klose et al. 2016), raising the possibility that the neurons either respond directly to DA, or DA sensitivity is communicated to the s-LNvs via PDF-release by the l-LNvs. Given that dopamine receptor expression is low in the s-LNvs (Kula-Eversole et al. 2010), it is more likely that the s-LNvs are responsive to DA stimulation of the l-LNvs. Indeed, dopaminergic neurons make synaptic connections with the LNvs in the fly brain, with a DA response observed in the l-LNvs (Shang et al. 2011). Complicating matters, the cellular response of the s-LNvs to DA is distinct from its response to PDF. Knockdown of adenylyl cyclase AC3 eliminates PDF-mediated cAMP production with no effect on DA-mediated cAMP production in the s-LNvs of explanted brains that are administered either neurotransmitter (Duvall and Taghert 2013; Klose et al. 2016). Knocking down AC13E and ACX-A on the other hand reduces cAMP response to DA but does not alter the response to PDF (Klose et al. 2016). Thus, l-LNv communication of a DA signal to the s-LNvs may be mediated through non-PDF signaling. Distinct adenylyl cyclases for each GPCR not only demonstrates that the PDF and DA signaling pathways are independent of each other, but also underscores how different receptors influence the concentration of intermediary molecules such as cAMP differently, fine tuning a neuronal response.
How do the LNvs respond to a range of signals while maintaining coherent molecular oscillators that regulate rhythmic behavior? Although narrowing a large variety of signals down to a few intermediary molecules (such as Ca2+ and cAMP) may seem like an overly simple mechanism for communicating complex information, small differences in the concentrations and ratios of cAMP and Ca2+ can activate different cellular processes to generate a range of complex responses (Siso-Nadal et al. 2009). The internal environment of the LNvs will vary depending on the signals the neurons respond to. PDF released from the l-LNvs induces an increase in cAMP in the s-LNvs, without a corresponding increase in cytosolic Ca2+ (Yao et al. 2012; Schlichting et al. 2016). In contrast, H-B eyelet signaling to the s-LNvs induces an increase in both cAMP and cytosolic Ca2+ (Schlichting et al. 2016). However, Ca2+ levels oscillate in the s-LNvs in a light-independent manner in constant dark conditions (Liang et al. 2016, 2017), indicating that Ca2+ is also subject to regulation by the s-LNv clock. Thus, depending on the time of day, PDF input, amidated PDF input, light input through the retina and H-B eyelet, and signals from other sources, the cytosol will exhibit variations in cytoplasmic cAMP and Ca2+ that trigger a variety of responses. The s-LNvs (and l-LNvs) may “know” where a signal is coming from and respond appropriately by sensing the variation in the intermediary molecules. This phenomenon is important when considering the different GPCRs that participate in regulating rhythmic behavior.
The Dorsal Lateral Neurons and the Fifth s-LNV
Evening anticipation is regulated by non-PDF (PDF−) neuronal clusters within the CCNN, as discussed above. Ablation of three CRY+ LNds out of six, the fifth s-LNv and some DNs eliminate evening anticipation, defining the evening cells as CRY+ PDF− neurons (Stoleru et al. 2004). Later work narrowed the definition further by demonstrating that the CRY+ LNds and the fifth s-LNv are sufficient to drive evening anticipation (Rieger et al. 2006; Picot et al. 2007; Guo et al. 2014).
The response of the evening cells to PDF released by the morning cells facilitates morning anticipation. Restoring PDFR in both morning and evening cells in a pdfr− genetic background restores wild-type activity in flies, while restoring PDFR only in the LNvs results in loss of morning anticipation similar to that of pdfr− or pdf01 flies (Renn et al. 1999; Lear et al. 2009; Im and Taghert 2010). Because there is a reported lack of synaptic connections between the LNvs and the LNds (Kim et al. 2013), the LNd response is dependent on PDF diffusion, which is sufficient for signaling (Jan and Jan 1982; Nässel and Wegener 2011). Expression of a membrane-tethered (nondiffusing) PDF peptide is sufficient to restore morning anticipation when expressed in the evening cells (LNds), but not when expressed in the morning cells (Choi et al. 2012), suggesting that morning anticipation is regulated by the response of evening cells to PDF. Split behavioral rhythms arise from kinase overexpression in LNvs that correspond to the speed of the LNv (morning) clocks and the LNd (evening) clocks (Yao and Shafer 2014). Under the same conditions that lack PDFR, wild-type behavioral rhythms are restored, suggesting that instructions from the morning cells are communicated through evening cells (Yao and Shafer 2014). Others have similarly observed that overexpression of the CK2 mutant Timekeeper in the morning cells is masked in a pdfr− genetic background (Lear et al. 2005b). Finally, ablation of the evening cells in flies leads to a loss of rhythmic behavior in constant dark conditions, similar to flies with an arrhythmic s-LNv (Stoleru et al. 2004). Together, these data suggest that the LNv clocks (at least in part) regulate rhythmic behavior by communicating through the evening cells through clock-regulated PDF release. Investigation of rhythmic behavior of mutants in the ion channel narrow abdomen (na), an output gene of the circadian clock that regulates locomotion activity (Lear et al. 2005a), points to a similar conclusion. Restoring a wild-type copy in the morning neurons of na mutant flies does not restore wild-type rhythmic behavior, while restoring na in the evening neurons does (Lear et al. 2005a). Finally, interfering with the neuronal clocks through the overexpression of a dominant-negative CLK mutant in the evening neurons severely weakens behavioral rhythmicity in a light/dark cycle and results in arrhythmic flies in constant dark conditions (Dissel et al. 2014). Therefore, the LNds, while retaining some autonomy of their own clock, are responsible for communicating instructions from the s-LNvs (by way of PDF) to downstream targets to regulate rhythmic behavior.
The intracellular response of neurons to signals such as PDF is cell-dependent. The LNds respond to PDF with an increase in intracellular cAMP (but not Ca2+) (Yao et al. 2012). However, the adenylyl cyclase that mediates PDF-dependent cAMP production in the LNds is different than that in the LNvs (Duvall and Taghert 2013). Therefore, the same GPCR responds to PDF to generate different cAMP responses in the two clusters. This important distinction suggests that a PDF signal can induce different cAMP responses depending on cell type. Because both groups of neurons express the same core clock components, the different adenylyl cyclases may therefore tune the speed of each local clock differently.
Other Neuropeptides and Evening Anticipation
The GPCRs and neuropeptides discussed so far regulate morning anticipation and arousal. Neuropeptide F (NPF) and its receptor NPFR1, help regulate evening anticipation. NPF is produced in two to three l-LNvs, three LNds, and the fifth s-LNv (Hermann et al. 2012; Kim et al. 2013), with one study reporting production in all four s-LNvs (He et al. 2013), although not consistently observed by others (Fig. 1D) (Hermann et al. 2012; Kim et al. 2013). The s-LNvs do express NPFR1 however, which associates with Gi to inhibit cAMP production (Garczynski et al. 2002; Kim et al. 2013) that may antagonize PDFR or D1-like DA receptor production of cAMP in these neurons (Fig. 1E). Therefore, PDF produced in the s- and l-LNvs communicate to the LNds, fifth s-LNv, some DNs, as well as feeding back to the s-LNvs, while NPF produced in the LNds, fifth s-LNv, and some LNvs communicate back to the s-LNvs in a neuronal feedback loop (Fig. 1F). Flies expressing RNAi that targets npf or npfR1 exhibit a loss of evening anticipation (He et al. 2013), mirroring the loss of morning anticipation in pdf0 or pdfr− flies (Renn et al. 1999; Hyun et al. 2005; Lear et al. 2005b; Mertens et al. 2005). NPF expression peaks in the late day (Fig. 1H), correlating well with its regulatory role in evening anticipation and contrasting with the PDF peak in s-LNv termini in the early day (Park et al. 2000; He et al. 2013). NPF and PDF also differently regulate delays and advances in the morning and evening neurons. Loss of pdf modestly shortens rhythmic behavior in constant darkness, while loss of npf modestly lengthens it, suggesting an antagonistic relationship between the morning and evening clocks (Fig. 1F) (Renn et al. 1999; Shafer and Taghert 2009; Hermann et al. 2012). The proposed antagonistic mechanism mirrors the advances and delays exhibited by morning and evening anticipation to light, discussed above (Pittendrigh and Daan 1976). Finally, such a feedback between the morning and evening neurons suggests that while morning anticipation is the response of evening cells to PDF, evening anticipation may be an s-LNv response to NPF, although not necessarily exclusively by these neurons.
NPF/PDF responses may regulate expression of PER in the circadian clock by influencing cellular cAMP. Unlike PDF, NPF and NPFR1 production is rhythmic, peaking in the mid- to late day in an apparent reverse phase with TIM and PER protein production (Fig. 1H) (Hardin et al. 1992; Park et al. 2000; He et al. 2013; Hermann-Luibl et al. 2014). If NPF/NPFR1 is dominant over PDF signaling in a target neuron such as the s-LNvs, it would drive cAMP levels down. Conversely, if PDF signaling is dominant over NPF in a target neuron, cAMP levels would rise. It is not known whether cAMP regulates CLK activity, but cAMP does influence the activity of HDAC4, which is required for rhythmic per transcription (Fogg et al. 2014). An increase in cAMP levels induces the phosphorylation and nuclear entry of HDAC4, which promotes per expression at ZT16, with low cAMP levels and low per expression at ZT8 (Liu and Schneider 2013; Fogg et al. 2014; Luan et al. 2014). Expression of per remains low in flies expressing a nonfunctional form of HDAC4 (Fogg et al. 2014). Assuming rhythmic bimodal dominance of either PDFR or NPFR1 GPCRs in s-LNvs, or simply the expression of one of the receptors in a given cluster, PER oscillations in a neuron can be regulated or reinforced by signals from other circadian neurons. Another cAMP-responsive protein, protein kinase A (PKA), stabilizes PER and TIM in high cAMP conditions (Li et al. 2014; Seluzicki et al. 2014). Thus, a model for PER accumulation that incorporates cAMP activity can be proposed. Throughout the day, increased PDF release allows cAMP to accumulate, which increases signals to phosphorylate HDAC4, shifting it into the nucleus. An accumulation of nuclear HDAC4 thus results in increased gene (per) expression in the late day and into the night. An accumulation in cAMP also activates PKA, which stabilizes PER and TIM proteins, contributing to the accumulation of the repressor complex at night. NPF/NPFR1 production peaks in the late day, permitting a drop in cAMP, reversing the process for the oncoming morning. This proposed mechanism relies on delays in signals that begin with neuropeptide release and culminates in gene regulation.
Two other less-studied neuropeptides also help regulate rhythmic behavior in Drosophila. Ion transport protein (ITP) is a neurotransmitter peptide that helps regulate evening anticipation (Hermann-Luibl et al. 2014). Similar to NPF, it peaks during the day and is CLK regulated (Fig. 1H) (Hermann-Luibl et al. 2014). ITP activity also stabilizes PER in the s-LNvs at night (Hermann-Luibl et al. 2014). The short-NPF (sNPF) neuropeptide is expressed in the two LNds lacking NPF, as well as in the s-LNvs (Fig. 1D) (Johard et al. 2009; Kula-Eversole et al. 2010). Although similar in name, the sNPF and NPF neurotransmitters are very different in amino acid sequence and are encoded by different genes. Less is known about sNPF involvement in rhythmic behavior, but is likely to regulate other behaviors under circadian control, such as feeding, locomotion, learning, and olfaction (Nässel and Wegener 2011). sNPF is also involved in regulating sleep, partly by inhibiting the arousal-promoting LNvs (Shang et al. 2013). PDF produced by the LNvs and sNPF produced by the s-LNvs and LNds, respectively, oppose each other in excitation and inhibition of motor neurons (Vecsey et al. 2014), which may form the basis of rhythmic locomotion output from the CCNN (Fig. 1F). It has recently been shown that sNPF also communicates to the DN1s to regulate DN1 Ca2+ oscillations (Liang et al. 2017). sNPF is likely to prove to be a critical link between the CCNN and other circuits to regulate rhythmic behavior.
The Intersection of CRY and PDF Function
Although the hypomorphic cryb mutant flies exhibit wild-type behavioral rhythmicity, luciferase reporters driven by tim and per promoters show a lack of transcription oscillation in whole flies (Stanewsky et al. 1998; Ivanchenko et al. 2001). This loss of luciferase signal is reportedly caused by the nonoscillating luciferase reporter expressed in the eyes, masking detection of any signal from luciferase that is oscillating in the CCNN (Stanewsky et al. 1998). This conclusion suggests that CRY is necessary for normal clock oscillations in the eye, but not necessary for oscillations in the pacemaker neurons. Similarly, flies lacking cry expression exhibit no tim oscillations in the l-LNv, suggesting that CRY is critical to maintaining the l-LNv clock as well (Yoshii et al. 2015), although it is unclear whether lack of CRY expression is necessary for the l-LNv clock, or for signals from a functional clock in the eye. Thus, CRY is necessary to maintain a functional clock in the eye and possibly the l-LNvs.
In other neuronal clusters in the CCNN, CRY participates in maintaining the phase of the local clocks. CRY− LNd clocks oscillate in antiphase with respect to the CRY+ s-LNvs and the CRY+ LNd clocks in constant darkness (Yoshii et al. 2009). Although the authors suggest that this antiphasic oscillation may be a consequence of the limitations of the 12-h time resolution in the study (Yoshii et al. 2009), the speed with which the phase reversal occurs (within a day in the CRY− LNds) and an increasing amplitude in reverse phase through constant darkness may suggest otherwise (Fig. 3 in Yoshii et al. 2009). The phase-reversal phenomenon in the CRY− LNds is further complicated by the resynchronization of the CRY− LNd phase with the s-LNv and CRY+ LNd phases in a pdf0 background, suggesting that PDF plays a role in keeping the CRY− neurons in antiphase. It is not known whether exogenous CRY or PDFR expression in the CRY− LNds would synchronize their clocks with the CRY+ LNds. Echoing these data, all LNds in a pdfr−;;cryb genetic background display antiphasic PER cycling when compared to cryb flies (Zhang et al. 2009), as does PER in the CRY+ LNds of cryb,pdf0 flies compared to cryb or pdf0 flies alone (Cusumano et al. 2009). In other words, antiphase oscillations within the LNds depend both on CRY and PDF. This suggests that CRY and PDF converge to direct the CRY− PDFR− LNds to oscillate in reverse phase in constant conditions. It remains to be seen how CRY and PDF converge to regulate the clocks in the two LNd subgroups.
THE ARCHITECTURE OF THE Drosophila melanogaster CIRCADIAN CLOCK
The Negative Feedback Loop Model
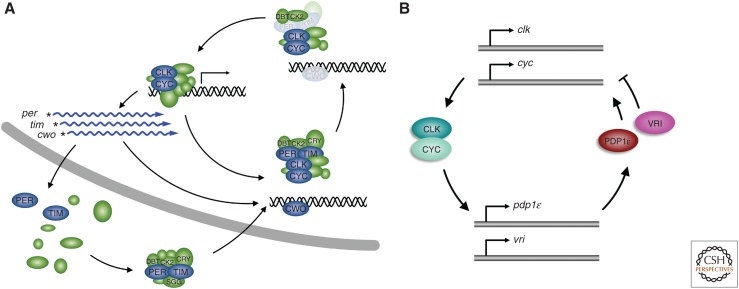
The circadian clock is a collection of proteins assembled into a delayed negative feedback loop that maintains a 24-h rhythm of transcriptional regulation. Four feedback loops are centered around the CLK/CYC activator complex (Fig. 2). In the primary feedback loop, the CLK/CYC complex mediates the transcription of per and tim by binding to an E-box sequence in their promoters (Fig. 2A) (Hao et al. 1997, 1999; Allada et al. 1998; Rutila et al. 1998; Dushay et al. 1990; Darlington et al. 2000; Menet et al. 2010). After a delay, the transcripts are translated and the PER and TIM proteins are assembled to form the repressor complex in the cytoplasm. After a second delay, the PER/TIM complex is imported into the nucleus under regulation by the Shaggy (SGG) and casein kinase II (CK2) kinases (Martinek et al. 2001; Smith et al. 2008; Top et al. 2016). In the nucleus, the repressor complex binds to the activator complex leading to CLK phosphorylation by as-yet-unknown mechanisms, followed by the removal of both complexes from the DNA, ending circadian transcriptional activity of the CLK/CYC target genes, including tim and per (Hardin et al. 1990; Lee et al. 1999; Kim and Edery 2006; Yu et al. 2006; Abruzzi et al. 2011). After a third delay, the repressor complex is degraded through the activity of CK2, casein kinase I/doubletime (DBT) and Nemo (NMO), releasing the CLK/CYC complex to initiate the next round of transcription, and closing the negative feedback loop (Kloss et al. 1998; Price et al. 1998; Glossop et al. 1999; Lin et al. 2002, 2005; Meissner et al. 2008; Smith et al. 2008; Chiu et al. 2011; Yu et al. 2011). The delays in the feedback ensure a clock oscillation with a ∼24-h frequency. The second loop involves CLK/CYC-mediated expression of Clockwork Orange (CWO) that reinforces PER/TIM repressor activity. CWO cycles similarly to PER and binds to the CLK E-box sequences, competing with the activator complex and reinforcing TIM/PER-mediated repression of transcription (Fig. 1H) (Kadener et al. 2007; Lim et al. 2007; Matsumoto et al. 2007; Zhou et al. 2016). In two other secondary feedback loops, the CLK/CYC activator complex mediates the transcription of vrille (vri) and par domain protein 1 (pdp1ε), which are translated in the cytoplasm (Fig. 2B) (Blau and Young 1999; Cyran et al. 2003). VRI represses CLK/CYC activity, whereas PDP1ε promotes it, helping tune the 24-h oscillations of the circadian clock. The function of the positive feedback loop is to either moderate the negative feedback, or to increase the rate of CLK/CYC recovery as the repressor complex is removed. Further details of the circadian clock, the secondary and tertiary structures of the main proteins, and their interaction motifs that mediate protein complex assembly are reviewed elsewhere (Crane and Young 2014).
Figure 2.
Model of the negative feedback loop of Drosophila melanogaster. (A) The CLK/CYC activator complex binds to target promoters to activate transcription of target genes, including per, tim, and cwo. PER and TIM assemble in the cytoplasm to form the repressor complex, which is transported into the nucleus through the activity of SGG and CK2 and binds to the activator complex on the DNA. CWO binding to CLK-binding regions on the DNA coincides with repressor complex-mediated removal of the activator complex, serving as a competitive inhibitor. CK2-mediated TIM degradation leads to DBT-mediated PER degradation and release of the activator complex, closing the negative feedback loop. (B) The CLK/CYC activator complex promotes the expression of PDP1ε and VRI. PDP1ε promotes the expression of clk and cyc, while VRI inhibits it.
The Oscillating Transcriptome
The circadian clock regulates the transcription of a large number of genes in Drosophila. CLK binds to about 1500 sites in the Drosophila genome, with more than 800 of these sites exhibiting rhythmic CLK binding (Abruzzi et al. 2011). About 500 of these sites also display oscillating RNA Pol II binding (Abruzzi et al. 2011), which is in the range of the number of transcripts previously estimated to fluctuate over the course of the day (150–750) (Wijnen et al. 2006). CLK/CYC binding to DNA peaks at night, between ZT14 and ZT18 with exceptions, but always remains bound at lower levels throughout the day (Abruzzi et al. 2011). This residual level of CLK/CYC E-box occupancy indicates that PER/TIM-mediated removal of CLK/CYC from the DNA is incomplete, but is still sufficient to ensure oscillations of the target transcription. Residual occupancy also indicates that small variations in protein concentration, or critically, variations in protein affinity as mediated by different posttranslational modifications or protein partners, may differently tune gene expression. In support of this idea, microarray data reveal variations in the onset of oscillating transcripts (Boothroyd et al. 2007; Nagoshi et al. 2010), suggesting that the activator and repressor complexes do not necessarily act at the same time for each gene. Similarly, there are tissue-specific differences in CLK-mediated gene expression. For example, CLK binds to two regions of the pdp1ε promoter in wild-type fly heads at ZT14, but binds to only one of these regions in eyeless flies (Abruzzi et al. 2011), suggesting that the second site is eye-specific. More generally, a need for differences in the regulation of CLK–DNA interactions at different times and different cells is supported by microarray data that reveal a wide range of gene expression differences across time and tissues (Nagoshi et al. 2010). Thus, although expression levels of activator and repressor are critical in maintaining the timing of gene expression, they are not necessarily sufficient. It is very likely that the circadian clock uses a wide range of regulatory proteins to tightly regulate circadian protein activity and gene expression across the tissues in the organism.
Regulatory Kinases of the Circadian Clock and Cryptochrome
The activity of the different core clock components is primarily regulated by protein interactions and posttranslational modifications. Different accessory proteins interact with and influence how the repressor and activator complexes are regulated. Here we will review the current knowledge of some of these modifying proteins that regulate timed activity, before discussing their roles in specific tissues.
Doubletime/Casein Kinase I
Doubletime (Dbt) encodes casein kinase I (DBT), a serine/threonine kinase that phosphorylates PER to regulate PER stability and nuclear accumulation (Kloss et al. 1998; Price et al. 1998; Cyran et al. 2005). Two mutant forms of DBT are named for the manner in which they alter behavioral rhythmicity, dbt short (dbtS) and dbt long (dbtL) (Kloss et al. 1998). Overexpression of either form of DBT in vivo results in a dominant change in behavior, while overexpression of the wild-type form has a modestly longer (∼1 h) period (Muskus et al. 2007; Venkatesan et al. 2015). The lack of a meaningful change in phenotype with DBT overexpression suggests that DBT activity is not a rate-limiting step in the circadian clock. Of the DBT mutants, surprisingly, both long and short forms exhibit reduced kinase activity (Preuss et al. 2004; Kivimäe et al. 2008), suggesting that DBT may serve two opposing roles in regulating rhythmicity. Indeed while DBTL and a DBT variant lacking ATPase activity (DBTK38R) change the kinetics of PER degradation that reflect changes in behavioral rhythmicity (i.e., delayed PER degradation, longer behavioral periodicity), DBTS does not appear to alter the kinetics of PER degradation at all (Preuss et al. 2004; Syed et al. 2011). If DBTS is not impaired in PER degradation, but can still impose a short behavioral rhythm on the fly, then DBT must play an additional role in regulating behavioral rhythmicity. One possibility is that DBTS leads to early termination of PER transcription inhibition (Bao et al. 2001; Syed et al. 2011). The presence of DBT, but not its kinase activity, is required to hyperphosphorylate and inactivate CLK to release it from DNA (Yu et al. 2009; Menet et al. 2010), which may be how dbtS flies exhibit a short behavioral rhythm. Because DBT cannot phosphorylate CLK in vitro or interact with it directly, DBT likely mediates its effect on CLK through PER (Kim and Edery 2006; Yu et al. 2006; Kivimäe et al. 2008; Menet et al. 2010) by either being recruited to phosphorylate CLK, or through phosphorylation of PER to influence its repressor function. Thus, while mutants like DBTL are defective in triggering timely degradation of PER protein (Syed et al. 2011), DBTS may be defective in regulating the activity of CLK protein.
DBT triggers and mediates a phosphorylation cascade on PER, beginning at the PER short downstream region (PERSD), includes the classic PER short (PERS) site, and culminates with modification of S47 at the amino terminus (Konopka and Benzer 1971; Kivimäe et al. 2008; Chiu et al. 2011; Garbe et al. 2013). Phosphorylation of S47 triggers physical association between PER and Slimb, an F-Box/WD-40 repeat protein that ubiquitinates PER to mediate its degradation (Grima et al. 2002; Ko et al. 2002; Chiu et al. 2008). Mutations that block phosphorylation of S47 or the PERSD region stabilize PER and lengthen the periodicity of behavioral rhythms (Chiu et al. 2008; Kivimäe et al. 2008; Garbe et al. 2013). On the other hand, mutation of the PERS site, also involved in the phosphorylation cascade, destabilizes PER primarily in the late night, resulting in a short behavioral rhythm (Konopka and Benzer 1971; Edery et al. 1994), suggesting that modification of this site is required for PER stabilization. These data suggest that PER may be alternatively stabilized and destabilized in different phases of the circadian cycle. DBT physically associates with the amino terminus of PER, and possibly a second, internal region spanning ∼50 residues in the latter third of PER, remaining associated with the repressor complex until PER is degraded (Kloss et al. 1998; Kim et al. 2007). PER is not immediately degraded however, because of its association with TIM, despite accumulating phosphorylation events over time (Edery et al. 1994; Rothenfluh et al. 2000; Kloss et al. 2001), which may mean that TIM influences how PER is phosphorylated. Therefore, DBT activity in the nucleus and nuclear TIM degradation form the basis of the temporal regulation of PER-mediated transcription repression (Kloss et al. 2001; Venkatesan et al. 2015).
Casein Kinase II
Timekeeper (Tik) and Andante, respectively, encode the α (catalytic) and β (regulatory) subunits that make up the heterotetrameric casein kinase II (CK2), a kinase that phosphorylates PER and TIM to regulate protein stability and nuclear entry (Lin et al. 2002, 2005; Akten et al. 2003; Top et al. 2016). RNAi against α or β subunits causes a long behavioral rhythm (Szabó et al. 2013; Top et al. 2016). Overexpression of each individual subunit gives rise to a similar long rhythm phenotype (Lin et al. 2005; Szabó et al. 2013; Top et al. 2016), an initially paradoxical phenotype that has been more recently suggested to reflect a dominant-negative effect arising from altered CK2 stoichiometry and titration of the functional complex (Top et al. 2016). Consistent with this notion, overexpression of both subunits yields a short behavioral rhythm suggestive of a CK2 gain-of-function (Top et al. 2016). CK2 physically associates with and phosphorylates both PER and TIM to regulate rhythmic behavior (Lin et al. 2002, 2005; Szabó et al. 2013; Top et al. 2016). Thus, CK2-mediated phosphorylation is a rate-limiting step in the circadian clock (Top et al. 2016).
CK2 modifies PER and TIM proteins to regulate their stability. Overexpression of the CK2α mutant Tik in vivo results in a behavioral rhythm of ∼31 h, and correlates with the stabilization of PER and TIM proteins (Smith et al. 2008). Accumulation of both PER and TIM is also seen with the CK2β Andante mutation (Akten et al. 2003). RNAi depletion of CK2 in S2 cells stabilizes nuclear PER/TIM levels, while lengthening behavioral period in flies to ∼33 h (Szabó et al. 2013; Top et al. 2016). Together, these data suggest that the activity of CK2 is required for normal oscillations of PER and TIM proteins. Mutation of CK2 target sites in PER do not lead to an accumulation of PER protein above wild-type levels despite exhibiting longer behavioral periods in flies (Lin et al. 2005), but mutation of CK2 sites in TIM does (Top et al. 2016). Another allele of tim, tim ultralong (timUL), carries a mutation at a serine site that is predicted to block CK2-mediated phosphorylation (Meissner et al. 2008), which causes TIM to persist in the nucleus and delay PER degradation (Rothenfluh et al. 2000). Thus, the evidence suggests that the main role for CK2 is likely to regulate the stability of TIM. In addition to its role in stabilizing the repressor complex, CK2 activity also contributes to nuclear accumulation of the repressor complex (Smith et al. 2008; Top et al. 2016), and phosphorylates CLK in a PER-dependent manner to stabilize it, contrasting DBT-mediated destabilization of CLK (Szabó et al. 2013). The involvement of CK2 in the stability and nuclear accumulation of the repressor complex and the stability of the activator complex is likely to ensure that these different regulatory processes are biochemically and chronologically linked.
Shaggy/Glycogen Synthase Kinase 3
Shaggy encodes glycogen synthase kinase 3 (SGG), a key regulator of nuclear accumulation of the repressor complex through modification of PER and TIM (Martinek et al. 2001; Ko et al. 2010; Top et al. 2016). In contrast to DBT and CK2, kinases that regulate stability of the repressor complex, SGG has no effect on the stability of PER or TIM (Top et al. 2016). Instead, SGG physically associates with the repressor complex, with a preference for binding TIM over PER, phosphorylating TIM and triggering a CK2 phosphorylation cascade to regulate PER/TIM nuclear accumulation (Top et al. 2016). Mutations that block SGG phosphorylation sites on TIM delay PER/TIM nuclear entry and lead to a high accumulation of cytoplasmic PER protein (Top et al. 2016), suggesting a link between the mechanism that regulates nuclear entry and nuclear degradation. It is likely that SGG-mediated triggering of nuclear entry of the TIM/PER complex is followed by CK2-mediated destabilization of TIM (Top et al. 2016), and then by DBT-mediated phosphorylation and destabilization of PER.
SGG is known to be regulated by various extracellular signals. SGG is a target of signaling cascades that are activated by Wnt or Hedgehog, reducing SGG activity (Harwood 2001; Doble 2003; Zhang et al. 2005; Hur and Zhou 2010) and offering a potential biochemical link between developmental signals and the circadian clock. SGG is also a target of signaling cascades that are activated by insulin or by growth factor, reducing SGG activity through the Akt/TOR pathway (Harwood 2001; Zhang et al. 2006; Zheng and Sehgal 2010), offering another potential biochemical link between metabolic signals and the circadian clock. In these cascades, SGG activity is modulated by phosphorylation as a function of cAMP levels (Fang et al. 2000). Thus, it is likely that the cAMP levels adjusted by PDFR, NPFR, and D1-like DA receptor modulates SGG activity through the signaling between neurons of the CCNN. Indeed, GSK3/SGG activity in the mouse has been shown to be regulated by activation of dopamine receptors (Beaulieu et al. 2011). Thus, SGG represents a potential communication hub for extracellular signals, and the biochemical state of the cell (e.g., cAMP levels) to influence processes such as nuclear entry of the repressor complex to regulate phase and synchrony of the different clocks.
Other Kinases
Nemo (NMO) is a proline-directed kinase that physically associates with and helps regulate PER and CLK stability (Yu et al. 2011). NMO is required for S596 phosphorylation on PER as part of the DBT phosphorylation cascade that regulates PER stability (Chiu et al. 2011; Garbe et al. 2013). S596 phosphorylation antagonizes DBT activity and helps stabilize PER, possibly by delaying degradation (Chiu et al. 2011). Surprisingly, RNAi directed against nmo in flies does not alter PER or TIM levels (Yu et al. 2011), despite shortening behavioral rhythmicity (Chiu et al. 2011; Yu et al. 2011), conflicting with the purported stabilizing role for S596 phosphorylation. Because NMO interacts with CLK (Yu et al. 2011), NMO may destabilize CLK and shorten behavioral rhythmicity through a mechanism of transcription termination. Whereas NMO stabilizes PER and destabilizes CLK, CK2 has the opposite effect on these proteins (Lin et al. 2005; Chiu et al. 2011; Yu et al. 2011), suggesting that the antagonistic action of these kinases on the repressor and activator complexes may generate a more robust transcriptional negative feedback loop.
PKA is a kinase that communicates extracellular signals to the cell by phosphorylating target proteins, activating, or inactivating them. In circadian rhythms, PKA deficiencies lead to a loss of locomotor rhythmicity in flies (∼80% arrhythmic), while per RNA oscillation and eclosion rhythmicity remains intact (Majercak et al. 1997), indicating that PKA is involved in output mechanisms that specifically regulate locomotor rhythmicity. PKA activity is required to stabilize TIM in some clock neurons (LNds and DN1s) and stabilize PER in other clock neurons (LNvs) (Li et al. 2014; Seluzicki et al. 2014), offering a basis for observed behavioral arrhythmicity, but without a satisfactory explanation for how eclosion remains rhythmic. The molecular basis for cell-type-specific differences by which PKA stabilizes TIM and PER are not understood. While PKA is known to be activated by cAMP levels, mechanisms by which extracellular signals may contribute to differential PKA activity in various circadian neurons are poorly defined. Like SGG, PKA may transduce extracellular signals of the CCNN to influence the stability of the repressor complex to regulate clock phase or synchrony. Whereas extracellular signaling through SGG affects nuclear entry of the TIM/PER repressor complex and alters the onset of transcriptional repression (Martinek et al. 2001; Top et al. 2016), PKA signaling alters TIM and PER stability, impacting the termination of transcriptional repression (Li et al. 2014; Seluzicki et al. 2014), suggesting that these pathways may regulate target genes at different times of day.
Cryptochrome
CRY is the primary photoreceptor for entraining fly circadian rhythms (Stanewsky et al. 1998; Cashmore 2003). Flies that lack cry (cry01) or express the cry hypomorph (cryb) adjust to changes in light regiments over a span of several days, instead of a single day (Stanewsky et al. 1998; Dolezelova et al. 2007). In the presence of light, CRY binds to TIM and both proteins are ubiquitinated for degradation by complexes involving Ramshackle (BRWD3)/JET/Cullin4 and JET/Cullin3, respectively (Ceriani et al. 1999; Koh et al. 2006; Peschel et al. 2009; Ozturk et al. 2011, 2013). Several studies suggest that light triggers the reduction of the FAD coenzyme bound within CRY, inducing a conformational change that releases the CRY carboxy-terminal tail (CTT) (Dissel et al. 2004; Hoang et al. 2008; Ozturk et al. 2011, 2008; Vaidya et al. 2013; Ganguly et al. 2016). However, other experiments indicate that light causes CTT release through a process that does not depend on flavin reduction (Ozturk et al. 2014). The release of the CRY CTT allows a TIM region of similar sequence to bind to the exposed pocket in CRY, communicating the light signal to TIM (Vaidya et al. 2013). Consistent with this model, removal of the CRY CTT promotes CRY-TIM binding without the need for light activation, leading to weaker entrainment in flies lacking the CTT (Busza et al. 2004). Despite a weakened ability to entrain to light, cry mutants lacking the CTT can be entrained under low light conditions (Dissel et al. 2004), likely through the CRY-independent visual system, or the deep brain photoreceptor Rh7 as described above (Rieger et al. 2003; Schlichting et al. 2014; Ni et al. 2017).
RELATING THE CIRCADIAN CLOCK MODEL TO THE CCNN AND CONFLICTS
Recent studies using luciferase and calcium reporters in wild-type and mutant flies have challenged the notion that the molecular architecture, rhythmic dynamics, and oscillatory phase of core clock components are regulated the same in all pacemaker neurons (Roberts et al. 2015; Liang et al. 2016, 2017). The molecular model of the circadian clock, however, is presented as a stereotyped model, often described as a finely tuned mechanism that maintains a 24-h rhythm. If the model is correct, how does the circadian clock oscillate with different frequencies in different neurons of the circadian system? To understand rhythmic behavior, our knowledge of intracellular mechanisms that regulate the circadian clock must be integrated within the context of extracellular signaling and synaptic transmission between pacemaker neurons. In this section, we will discuss “speeds” of clocks to refer to the period of the circadian clock within a given cluster of neurons, to distinguish an “advance” or “delay” in anticipatory behavior in morning/evening cells.
The Effects of Different Kinases on the s-LNv and LNd Clocks
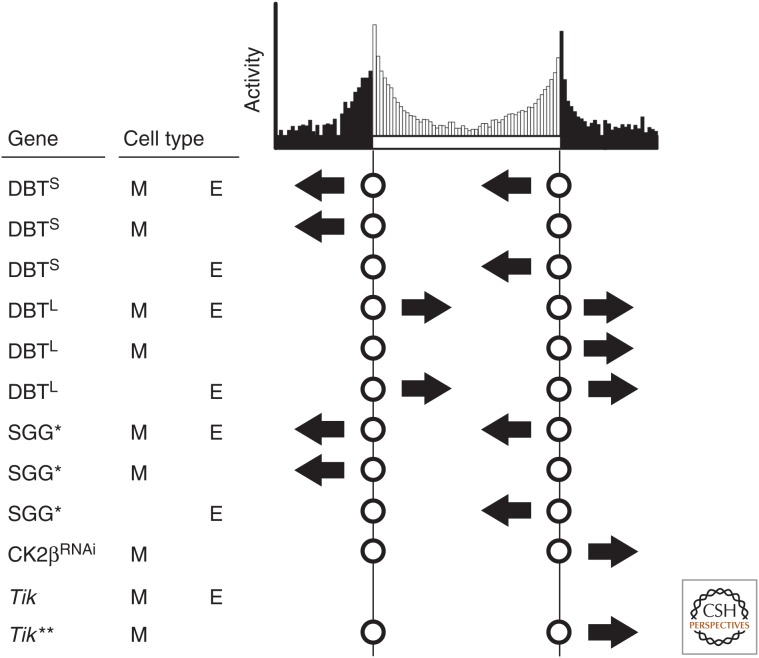
The s-LNvs have been described as master pacemaker cells that dominate behavior in constant dark conditions through the release of PDF (Renn et al. 1999; Grima et al. 2004; Stoleru et al. 2004, 2005). Speeding up the LNv clock through overexpression of SGG also speeds up the LNd clock over a few days in constant darkness, suggesting communication between the two clusters (Stoleru et al. 2005). However, speeding up the LNv clock through overexpression of DBTS speeds up the LNv clock, but fails to speed up the LNd/fifth s-LNv clocks. LNvs that are slowed by DBTL overexpression fails to slow the fifth s-LNv (LNd clocks were not measured), contrasting the effect of DBTS overexpression (Yao et al. 2016). Focusing on the contrasting results between SGG and DBTS overexpression, it appears that the method by which the LNv clocks are sped up is important in instructing the LNd clock speed. We speculate that because DBT activity is required for releasing transcriptional repression and SGG activity is required for nuclear entry of PER/TIM to activate transcriptional repression, it is possible that the differences observed in LNd clock speeds lie in the differences in the beginning or the ending of circadian transcriptional regulation.
Kinase overexpression also affects morning and evening anticipation behaviors differently. Overexpressing DBTS in the LNvs advances morning anticipation with no effect on evening anticipation, while overexpressing SGG in the same neurons advances both morning and evening anticipation (Stoleru et al. 2005; Yao et al. 2016), reflecting their effect on speeds of the individual molecular oscillators. Conversely, overexpressing DBTL in the LNvs slows the LNv clocks, yet delays evening anticipation with no effect on morning anticipation (Fig. 3) (Yao et al. 2016). It is suggested that this delay in evening anticipation is possibly a result of the slowing effect of the LNvs on the DN clocks (Yao et al. 2016). An effect similar to that of DBTL expression is also true of overexpression of the CK2 mutant Timekeeper, or RNAi directed against CK2β in the LNvs (Lear et al. 2005b; Smith et al. 2008; Zhang et al. 2010). Because both DBT and CK2 are involved in triggering degradation of either PER or TIM (Price et al. 1998; Meissner et al. 2008; Smith et al. 2008; Syed et al. 2011), we can speculate that the prolonged transcriptional repression in the LNvs caused by delayed PER/TIM degradation has the effect of sending signals that slow down the evening cells.
Figure 3.
Summary of morning and evening anticipation advances and delays. The indicated gene is overexpressed in morning cells (LNvs; M), evening cells (LNds, fifth s-LNv, DNs; E), or both. The advance or delay of the anticipation peaks in the first 24 h of constant darkness is depicted by the arrow. Presence of absence of the circles indicates the presence or absence of an anticipation peak. The DBT data (Yao et al. 2016), SGG data (Stoleru et al. 2005), CK2β RNAi data (Lear et al. 2005b; Zhang et al. 2010), and Timekeeper (Tik) data (Smith et al. 2008) are summarized from the indicated sources. *Changes in anticipation were inferred from data in the indicated source. **Lack of change in morning anticipation is inferred from incomplete data that coincides with wild-type flies in the indicated source.
Differences in CK2 Expression
CK2 is a circadian kinase that is expressed differentially in circadian neurons (Fig. 1G). CK2α is expressed only in the LNvs, and is more abundant in l-LNvs than s-LNvs (Lin et al. 2002). Other circadian neurons apparently lack CK2 expression (Top et al. 2016). Because ectopic expression of CK2 in circadian neurons can change clock periodicity (Top et al. 2016), it should be assumed that the specific concentration of CK2 in each of the LNvs and apparent lack of CK2 in other clock neurons is appropriately titrated in establishing local oscillators with ∼24-h periods. Thus, the different clusters that express the same core protein components (i.e., PER, TIM, etc.) must have degradation and nuclear entry mechanisms that compensate for the differences in CK2 expression. However, if CK2 is necessary to regulate nuclear entry and nuclear degradation of TIM and PER, how is TIM/PER nuclear entry and degradation regulated in pacemaker neurons such as the LNds, if CK2 is not expressed there? The CK2 phosphorylation sites identified on PER (Lin et al. 2005) and TIM (Top et al. 2016) may either be phosphorylated by another kinase in the LNds, or PER and TIM degradation may be regulated through an entirely different mechanism. It is also possible that CK2 does not participate in regulating a mechanism of the circadian clock despite being expressed in a neuronal cluster. A mutant form of TIM that delays CK2-mediated nuclear entry in s-LNvs does not delay nuclear entry in the l-LNvs despite a higher CK2α expression in the l-LNvs (Lin et al. 2002; Top et al. 2016). In summary, the LNds that express no CK2 must use another mechanism to regulate PER/TIM nuclear entry and degradation, and the l-LNvs that express abundant CK2 but do not exhibit delayed nuclear entry when TIM phosphorylation sites are mutated likely use yet another mechanism to regulate PER/TIM nuclear entry. The differences LNvs exhibit in CK2 levels and the efficacy of mutations that block CK2 activity indicate that different mechanisms are involved in regulating local clocks in the CCNN.
Differences in NMO Expression
In the fly brain, NMO is expressed in two of four l-LNvs, all s-LNvs, a subset of DN1s, at low levels in some LNds, and is not observed at all in DN2s or DN3s (Yu et al. 2011). An RNAi-mediated decrease of NMO shortens rhythmic behavior in constant dark conditions, advancing both morning and evening anticipation (Chiu et al. 2011). The effect an nmo knockdown has on both anticipation events is consistent with its expression in morning and evening cells. However, the variability of NMO expression in the different circadian neurons raises questions similar to those in the case of CK2. Is PER regulated by the same phosphorylation cascade in the neurons lacking NMO? Do the clocks in these neurons oscillate slightly longer or is there a different mechanism (i.e., another kinase) that substitutes for the absence of NMO in these neurons? Importantly, the absence of NMO in some neurons indicate that the model for regulating PER stability in the different neurons is not universally applicable.
Differences in CRY Expression and Light Response
Clock neurons differ in their expression of CRY and also in their responsiveness to light (Benito et al. 2008; Yoshii et al. 2008). For example, TIM in the s-LNv clock is nonresponsive to a light pulse at ZT 15, likely because of the low levels of CRY in the cells at that time (Tang et al. 2010). Increasing CRY expression in these cells increases the degradation of TIM in response to light and causes behavioral phase shifts (Tang et al. 2010). To speculate, this molecular and behavioral response may be extrapolated to circadian neurons that express low levels of CRY, suggesting a weakened or even absent light response in such low expressers. Indeed, a light pulse differentially shifts the phase of each local clock (e.g., s-LNvs, LNds, and DN1s), but each local clock returns to synchrony with each other in less than 3 days, presumably through communication between these neurons (Roberts et al. 2015). Responsiveness to light by the molecular oscillator also depends on the protein Jetlag (JET). Increasing JET levels in s-LNvs augments TIM degradation in response to a light pulse, similar to CRY overexpression (Tang et al. 2010). Yet, JET overexpression does not elicit a behavioral response, in contrast to the observations made with CRY overexpression (Tang et al. 2010). Thus, a single external signal can induce different phase shifts in each clock that may manifest differently in rhythmic behavior of the fly. Additionally, while these findings demonstrate that TIM degradation is important to reset the molecular oscillator, it appears that a behavioral phase response may be regulated by CRY, not TIM degradation.
Monitoring clocks in different clusters when analyzing cry0 flies reveals that non s-LNvs phase shift rapidly to accommodate a phase change in light regimen, while s-LNvs are slow to respond (Yoshii et al. 2015). This suggests that CRY-mediated light responsiveness is critical in the s-LNvs and that non s-LNvs possibly depend more on signals from the eye. These conclusions are supported by analysis of light-mediated TIM degradation in jet knockdown flies at ZT15 and ZT21, before and after TIM nuclear entry (Lamba et al. 2014). A reduction of JET in the LNds at either time point has no effect on light-mediated TIM degradation in either LNds or LNvs, nor does it induce a phase response as compared to wild-type. JET reduction in the LNvs has no effect on LNv TIM degradation in response to light, but alters behavioral phase shifts. These data reaffirm that behavioral phase response is likely mediated through CRY, and whether light-mediated TIM degradation will occur depends on the neuronal cluster in which TIM is expressed. Therefore, the sensitivity of s-LNvs to CRY-mediated light detection, the different changes of clock phase in the s-LNvs and LNds with a light pulse, and the differences in CRY expression (and light sensitivity) in CRY-expressing neurons result in the different light responses by each neuronal cluster.
Effects of PER on Different Clocks
The period gene is expressed in all clock neurons, in addition to many other tissues in the fly. PER is the key repressor of transcription in the circadian clock, with its expression directly correlated with behavioral rhythmicity (i.e., lower per expression leads to longer rhythms) (Baylies et al. 1992). Thus, any differences in PER concentration in the different clusters would be expected to affect the frequency of the local oscillator. Consistent with this model, the comparison of perS with wild-type flies yields a range of differences in calcium oscillations in the different clusters (Liang et al. 2016). This observation suggests that altering the stability of PER has different consequences on the oscillators in the different clusters. This is also consistent with long PER protein oscillations that the l-LNvs exhibit relative to the s-LNvs in wild-type flies (Fig. 4A in Roberts et al. 2015) that correlate with lower per expression in the l-LNvs compared to the s-LNvs (Kula-Eversole et al. 2010).
The regulation of PER function in specific neuronal clusters also relies on the spatial organization of functional domains within the PER protein. In the current model of PER function, DBT binds to the amino terminus of PER to regulate protein degradation (Kloss et al. 1998; Chiu et al. 2008), while CLK appears to interact with PER at the carboxy-terminal end (Chang and Reppert 2003), seemingly spatially separating the degradation and repression activities of PER. Consistent with this, deletions within the last third of PER alter PER repressor activity (Chang and Reppert 2003; Sun et al. 2010) and a carboxy-terminal truncation of the last third of PER (BG) cannot rescue the behavioral rhythmicity of a per0 fly (Stanewsky et al. 1997). However, despite demonstrating arrhythmic behavior, oscillation of PER in BG fly LNvs is indistinguishable from wild-type (Stanewsky et al. 1997). Therefore, the last third of PER that encodes the “CLK-binding domain” is important for regulating behavioral rhythmicity, but has no effect on the oscillation of the LNv clocks. Thus, it is tempting to speculate that a functional CLK-binding domain is necessary for mediating rhythmic behavior, but dispensable for the function of the LNv clock. Undoubtedly, the resolution of this conflict will uncover interesting mechanistic insight into the function of the LNv clocks.
Activity of Promoters
The mechanisms that regulate nuclear accumulation and nuclear stability of the repressor complex effectively titrate the repressor to regulate gene expression. Screens of RNA expression in whole heads or brains reveal a wide range of peaks and phases of oscillating transcripts (Claridge-Chang et al. 2001; Ceriani et al. 2002; Ueda et al. 2002; Wijnen et al. 2006; Abruzzi et al. 2011; Hughes et al. 2012). Genome-wide ChIP experiments also reveal that CLK association with circadian-regulated promoters itself occurs with a circadian rhythm, with peak DNA-binding phases spread throughout the circadian cycle (Abruzzi et al. 2011). Therefore, small changes in the accumulation of the activator or repressor complex will have significant consequences for some genes, while leaving other genes unaffected. Slight differences in SGG/CK2 expression, for example, could alter the timing and the extent of nuclear accumulation of the repressor complex, effectively shifting the CLK-PER-binding equilibrium to alter the expression of genes in a promoter-dependent manner. In this respect, gene expression may be tuned with modifications that alter binding affinity of proteins, which would, in turn, alter transcription activity. Additional mechanisms that regulate protein stability, translation rates and protein complex formation may give rise to functionally important differences in the dynamics of circadian gene expression.
Understanding the Local Mechanisms that Regulate Local Clocks
Together, the studies discussed here demonstrate that an understanding of specific mechanisms that regulate the circadian clock in individual neuronal clusters is required to understand the principles that govern behavior. Traditional overexpression studies have revealed a lot about circadian rhythms. However, manipulation of entire clusters by overexpressing accessory components such as kinases makes two assumptions: (1) that the intended target of the kinase is being manipulated, and (2) that the kinase is endogenous in that cluster and is being supplemented through overexpression. Neither assumption is necessarily correct. For example, CK2 is not endogenously expressed in the LNds (Lin et al. 2002; Top et al. 2016). Therefore, a behavioral change as a result of CK2 overexpression in the LNds may not necessarily reflect an effect of phosphorylation of the intended targets, because CK2 is involved a wide range of other cellular processes that may alter various cellular pathways and neuronal activity.
Cell-autonomous, loss-of-function manipulation of endogenous clock mechanism may provide a useful method to understand the properties of individual clock neurons as well as that of the network. Mutation of TIM that block SGG/CK2 phosphorylation of the protein delays nuclear accumulation of the repressor complex in the s-LNvs (Top et al. 2016). In the LNds, the delay is less severe, while in the l-LNvs there is no delay. Thus, nuclear entry is regulated by a different mechanism in the l-LNvs neurons (Top et al. 2016). It is not clear, however, whether the TIM mutations delay nuclear entry of the LNd clock by blocking the nuclear entry mechanism, or whether nuclear entry is delayed because of the slower s-LNv clock. Additionally, the absence of a nuclear entry mechanism involving CK2 in the l-LNvs does not preclude other possible functions of CK2, such as degradation of TIM or PER.
Similarly, overexpressing or down-regulating SGG in evening cells (LNds/fifth s-LNv/DNs) does not change behavioral rhythmicity, while the same manipulation in morning cells (LNvs), respectively, shortens or lengthens behavioral rhythms (Stoleru et al. 2005; Fischer et al. 2016; Top et al. 2016). These data indicate that SGG activity is limited to the morning cells, specifically the s-LNvs. However, manipulation of the morning cells by TIM mutants that block SGG phosphorylation delay nuclear entry in evening cells (Top et al. 2016). Given that SGG overexpression or down-regulation in evening cells does not alter rhythmic behavior (Fischer et al. 2016), either the evening cells express a different kinase that targets the SGG sites on TIM or mutations at these sites are silent, and nuclear entry is regulated through another mechanism that is likely dependent on signaling from s-LNvs. As shown by SGG and CK2, mutations alone are not predictive of behavioral phenotypes but rather by the combination of the mutation and the regulatory proteins that may be differentially expressed or regulated within the CCNN.
In the example of another point mutation, the period short mutant (perS) alters a DBT phosphorylation site that regulates PER stability (Chiu et al. 2008, 2011; Kivimäe et al. 2008; Garbe et al. 2013). As the name suggests, perS flies exhibit shortened rhythms in DD, and an advanced evening anticipation peak in LD with no effect on morning anticipation (Stanewsky et al. 1998; Renn et al. 1999; Emery et al. 2000b; Li et al. 2014). Also, a perS, cryb double-mutant fly exhibits the same 19-h behavioral rhythm in constant light as a perS fly, conditions in which the LNds (i.e., evening cells) dominate rhythmic behavior (Emery et al. 2000a). This suggests that the “short” activity of perS affects evening cells. Therefore, it is possible that phosphorylation of the perS site is critical for the regulation of evening anticipation in the LNds. This does not mean that perS has no effect on the LNvs, however. Calcium peaks in each neuronal cluster are different, but critically, perS affects the peak in each cluster differently (Liang et al. 2016). The DN1s and DN3s are advanced by ∼2 h, while s-LNvs and LNds are advanced by 6 and 10 h, respectively, in LD conditions. These data suggest that while perS does not alter morning anticipation behavior, the neurons that regulate morning anticipation (s-LNvs) can still exhibit a large phase shift in peak cytoplasmic calcium, possibly as a result of changes in the timing of signals being received, albeit without altering their influence on morning anticipation. Future work will reveal the relationship of Ca2+ oscillations, clock speeds, timed anticipation, and rhythmic behavior.
CONCLUSIONS
The Drosophila circadian rhythm field is increasingly becoming more molecular and attention should increasingly be focused on cell-type-specific properties of neurons in the circadian network. We now have a growing appreciation that differences in the intracellular biochemical pathways within these neurons alter the activity of the entire network and its behavioral outputs, yet many important questions remain unanswered. What are the components of the activator and repressor complexes in each of the clusters? If a regulatory kinase is absent, what is the substitute mechanism that ensures that the local clock runs smoothly? How does a local clock regulate the release of neurotransmitters to communicate to other neuronal clusters? What mechanism of the clock is changed in response to a neurotransmitter? Understanding the complex interplay between cytoplasmic secondary messengers, core clock proteins, and their regulators that are differentially expressed within the circadian neuron network is likely to provide new insights into the relationships between biochemistry, neurobiology, and behavior.
Footnotes
Editors: Paolo Sassone-Corsi, Michael W. Young, and Akhilesh B. Reddy
Additional Perspectives on Circadian Rhythms available at www.cshperspectives.org
REFERENCES
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. 2011. Drosophila CLOCK target gene characterization: Implications for circadian tissue-specific gene expression. Genes Dev 25: 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. 2003. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6: 251–257. [DOI] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. 1998. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93: 791–804. [DOI] [PubMed] [Google Scholar]
- Bachleitner W, Kempinger L, Wülbeck C, Rieger D, Helfrich-Förster C. 2007. Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc Natl Acad Sci 104: 3538–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Rihel J, Bjes E, Fan JY, Price JL. 2001. The Drosophila double-time S mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci 21: 7117–7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies MK, Vosshall LB, Sehgal A, Young MW. 1992. New short period mutations of the Drosophila clock gene per. Neuron 9: 575–581. [DOI] [PubMed] [Google Scholar]
- Beaulieu J-M, Del’guidice T, Sotnikova TD, Lemasson M, Gainetdinov RR. 2011. Beyond cAMP: The regulation of Akt and GSK3 by dopamine receptors. Front Mol Neurosci 4: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Desplan C. 2015. Visual circuits in flies: Beginning to see the whole picture. Curr Opin Neurobiol 34: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Houl JH, Roman GW, Hardin PE. 2008. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms 23: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW. 1999. Cycling vrille expression is required for a functional Drosophila clock. Cell 99: 661–671. [DOI] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. 1988. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54: 723–733. [DOI] [PubMed] [Google Scholar]
- Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. 2007. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet 3: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. 2004. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304: 1503–1506. [DOI] [PubMed] [Google Scholar]
- Cashmore AR. 2003. Cryptochromes. Cell 114: 537–543. [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Más P, Petti AA, Weitz CJ, Kay SA. 1999. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285: 553–556. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. 2002. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 22: 9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DC, Reppert SM. 2003. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr Biol 13: 758–762. [DOI] [PubMed] [Google Scholar]
- Chiu JC, Vanselow JT, Kramer A, Edery I. 2008. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev 22: 1758–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I. 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Cao G, Tanenhaus AK, McCarthy EV, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JCP, Nitabach MN. 2012. Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in Drosophila. Cell Rep 2: 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. 2001. Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657–671. [DOI] [PubMed] [Google Scholar]
- Crane BR, Young MW. 2014. Interactive features of proteins composing eukaryotic circadian clocks. Annu Rev Biochem 83: 191–219. [DOI] [PubMed] [Google Scholar]
- Cusumano P, Klarsfeld A, Chélot E, Picot M, Richier B, Rouyer F. 2009. PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci 12: 1431–1437. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NRJ, Hardin PE, Young MW, Storti RV, Blau J. 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. 2005. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci 25: 5430–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Lyons LC, Hardin PE, Kay SA. 2000. The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J Biol Rhythms 15: 462–471. [DOI] [PubMed] [Google Scholar]
- Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. 2004. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci 7: 834–840. [DOI] [PubMed] [Google Scholar]
- Dissel S, Hansen CN, Özkaya Ö, Hemsley M, Kyriacou CP, Rosato E. 2014. The logic of circadian organization in Drosophila. Curr Biol 24: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble BW. 2003. GSK-3: Tricks of the trade for a multi-tasking kinase. J Cell Sci 116: 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezelova E, Dolezel D, Hall JC. 2007. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics 177: 329–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushay MS, Konopka RJ, Orr D, Greenacre ML, Kyriacou CP, Rosbash M, Hall JC. 1990. Phenotypic and genetic analysis of Clock, a new circadian rhythm mutant in Drosophila melanogaster. Genetics 125: 557–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall LB, Taghert PH. 2013. E and M circadian pacemaker neurons use different PDF receptor signalosome components in Drosophila. J Biol Rhythms 28: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. 1994. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci 91: 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Hall JC, Rosbash M. 2000a. A unique circadian-rhythm photoreceptor. Nature 404: 456–457. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Förster C, Emery-Le M, Hall JC, Rosbash M. 2000b. Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26: 493–504. [DOI] [PubMed] [Google Scholar]
- Fang X, Yu SX, Lu Y, Bast RC, Woodgett JR, Mills GB. 2000. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc Natl Acad Sci 97: 11960–11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Helfrich-Förster C, Peschel N. 2016. GSK-3 β does not stabilize cryptochrome in the circadian clock of Drosophila. PLoS ONE 11: e0146571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg PCM, O’Neill JS, Dobrzycki T, Calvert S, Lord EC, McIntosh RLL, Elliott CJH, Sweeney ST, Hastings MH, Chawla S. 2014. Class IIa histone deacetylases are conserved regulators of circadian function. J Biol Chem 289: 34341–34348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Manahan CC, Top D, Yee EF, Lin C, Young MW, Thiel W, Crane BR. 2016. Changes in active site histidine hydrogen bonding trigger cryptochrome activation. Proc Natl Acad Sci 113: 10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe DS, Fang Y, Zheng X, Sowcik M, Anjum R, Gygi SP, Sehgal A. 2013. Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS Genet 9: e1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. 2002. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides 23: 773–780. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Lyons LC, Hardin PE. 1999. Interlocked feedback loops within the Drosophila circadian oscillator. Science 286: 766–768. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chélot E, Papin C, Limbourg-Bouchon B, Rouyer F. 2002. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420: 178–182. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. 2004. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431: 869–873. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. 2014. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife 3: e02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M. 2016. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Allen DL, Hardin PE. 1997. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol 17: 3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Glossop NR, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Förster C, Hardin P. 1999. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci 19: 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC, Juusola M. 2015. Phototransduction in Drosophila. Curr Opin Neurobiol 34: 37–45. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. 1992. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci 89: 11711–11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ. 2001. Regulation of GSK-3: A cellular multiprocessor. Cell 105: 821–824. [DOI] [PubMed] [Google Scholar]
- He C, Cong X, Zhang R, Wu D, An C, Zhao Z. 2013. Regulation of circadian locomotor rhythm by neuropeptide Y-like system in Drosophila melanogaster. Insect Mol Biol 22: 376–388. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. 1995. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci 92: 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. 1998. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: A brain-behavioral study of disconnected mutants. J Comp Physiol A 182: 435–453. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. 2001. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30: 249–261. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Edwards T, Yasuyama K, Wisotzki B, Schneuwly S, Stanewsky R, Meinertzhagen IA, Hofbauer A. 2002. The extraretinal eyelet of Drosophila: Development, ultrastructure, and putative circadian function. J Neurosci 22: 9255–9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann C, Yoshii T, Dusik V, Helfrich-Förster C. 2012. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol 520: 970–987. [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, Helfrich-Förster C. 2014. The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J Neurosci 34: 9522–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, Wu W, Berndt A, Wolf E, Bittl R, Ahmad M. 2008. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol 6: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A, Buchner E. 1989. Does Drosophila have seven eyes? Naturwissenschaften 76: 335–336. [Google Scholar]
- Houl JH, Yu W, Dudek SM, Hardin PE. 2006. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J Biol Rhythms 21: 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. 2012. Brain-specific rescue of clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet 8: e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EM, Zhou FQ. 2010. GSK3 signalling in neural development. Nat Rev Neurosci 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. 2005. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron 48: 267–278. [DOI] [PubMed] [Google Scholar]
- Im SH, Taghert PH. 2010. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol 518: 1925–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Li W, Taghert PH. 2011. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS ONE 6: e18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko M, Stanewsky R, Giebultowicz JM. 2001. Circadian photoreception in Drosophila: Functions of cryptochrome in peripheral and central clocks. J Biol Rhythms 16: 205–215. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. 1982. Peptidergic transmission in sympathetic ganglia of the frog. J Physiol (Lond) 327: 219–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HAD, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, Nässel DR. 2009. Peptidergic clock neurons in Drosophila: Ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol 516: 59–73. [DOI] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21: 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. 2000. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422: 66–94. [DOI] [PubMed] [Google Scholar]
- Kim EY, Edery I. 2006. Balance between DBT/CKIε kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci 103: 6178–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I. 2007. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol 27: 5014–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Jan LY, Jan YN. 2013. A PDF/NPF neuropeptide signaling circuitry of male Drosophila melanogaster controls rival-induced prolonged mating. Neuron 80: 1190–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimäe S, Saez L, Young MW. 2008. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol 6: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose M, Duvall LB, Li W, Liang X, Ren C, Steinbach JH, Taghert PH. 2016. Functional PDF signaling in the Drosophila circadian neural circuit is gated by Ral A-dependent modulation. Neuron 90: 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L. 2001. Phosphorylation of period is influenced by cycling physical associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila clock. Neuron 30: 699–706. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678. [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. 2010. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3β/SGG in circadian clocks. J Neurosci 30: 12664–12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A. 2006. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312: 1809–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. 1971. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula E, Levitan ES, Pyza E, Rosbash M. 2006. PDF cycling in the dorsal protocerebrum of the Drosophila brain is not necessary for circadian clock function. J Biol Rhythms 21: 104–117. [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. 2010. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci 107: 13497–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. 2005. Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba P, Bilodeau-Wentworth D, Emery P, Zhang Y. 2014. Morning and evening oscillators cooperate to reset circadian behavior in response to light input. Cell Rep 7: 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Lin J-M, Keath JR, McGill JJ, Raman IM, Allada R. 2005a. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48: 965–976. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin J-M, Schroeder A, Zhang L, Allada R. 2005b. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron 48: 221–227. [DOI] [PubMed] [Google Scholar]
- Lear BC, Zhang L, Allada R. 2009. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol 7: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang JSC, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. 2009. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron 64: 522–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. 1999. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol Cell Biol 19: 5316–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelito KR, Shafer OT. 2012. Reciprocal cholinergic and GABAergic modulation of the small ventrolateral pacemaker neurons of Drosophila’s circadian clock neuron network. J Neurophysiol 107: 2096–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guo F, Shen J, Rosbash M. 2014. PDF and cAMP enhance PER stability in Drosophila clock neurons. Proc Natl Acad Sci 111: E1284–E1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. 2016. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351: 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. 2017. A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron 94: 1173–1189.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. 2007. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol 17: 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. 2002. A role for casein kinase 2α in the Drosophila circadian clock. Nature 420: 816–820. [DOI] [PubMed] [Google Scholar]
- Lin J-M, Schroeder A, Allada R. 2005. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J Neurosci 25: 11175–11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schneider MF. 2013. Opposing HDAC4 nuclear fluxes due to phosphorylation by β-adrenergic activated protein kinase A or by activity or Epac activated CaMKII in skeletal muscle fibres. J Physiol 591: 3605–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B, Goodarzi MO, Phillips NG, Guo X, Chen YDI, Yao J, Allison M, Rotter JI, Shaw R, Montminy M. 2014. Leptin-mediated increases in catecholamine signaling reduce adipose tissue inflammation via activation of macrophage HDAC4. Cell Metab 19: 1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Kalderon D, Edery I. 1997. Drosophila melanogaster deficient in protein kinase A manifests behavior-specific arrhythmia but normal clock function. Mol Cell Biol 17: 5915–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpel S, Klarsfeld A, Rouyer F. 2002. Larval optic nerve and adult extra-retinal photoreceptors sequentially associate with clock neurons during Drosophila brain development. Development 129: 1443–1453. [DOI] [PubMed] [Google Scholar]
- Mao Z, Davis RL. 2009. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: Anatomical and physiological heterogeneity. Front Neural Circuits 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769–779. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, et al. 2007. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev 21: 1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner RA, Kilman VL, Lin JM, Allada R. 2008. TIMELESS is an important mediator of CK2 effects on circadian clock function in vivo. J Neurosci 28: 9732–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. 2010. Dynamic PER repression mechanisms in the Drosophila circadian clock: From on-DNA to off-DNA. Genes Dev 24: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. 2005. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48: 213–219. [DOI] [PubMed] [Google Scholar]
- Montell C. 2012. Drosophila visual transduction. Trends Neurosci 35: 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. 2007. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro NI, Ceriani MF. 2015. Acetylcholine from visual circuits modulates the activity of arousal neurons in Drosophila. J Neurosci 35: 16315–16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. 2007. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol 27: 8049–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. 2010. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci 13: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR, Wegener C. 2011. A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides 32: 1335–1355. [DOI] [PubMed] [Google Scholar]
- Ni JD, Baik LS, Holmes TC, Montell C. 2017. A rhodopsin in the brain functions in circadian photoentrainment in Drosophila. Nature 545: 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, Song SH, Selby CP, Sancar A. 2008. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem 283: 3256–3263. [DOI] [PubMed] [Google Scholar]
- Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. 2011. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci 108: 516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, VanVickle-Chavez SJ, Akileswaran L, Van Gelder RN, Sancar A. 2013. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1-CUL4-ROC1 E3 ligase complex. Proc Natl Acad Sci 110: 4980–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, Selby CP, Zhong D, Sancar A. 2014. Mechanism of photosignaling by Drosophila cryptochrome: Role of the redox status of the flavin chromophore. J Biol Chem 289: 4634–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJL, Kang K, Liu X, Garrity PA, Rosbash M, et al. 2008. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. 2000. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci 97: 3608–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Veenstra JA, Park JH, Taghert PH. 2008. Mapping peptidergic cells in Drosophila: Where DIMM fits in. PLoS ONE 3: e1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R. 2009. Light-dependent interactions between the Drosophila circadian clock factors Cryptochrome, Jetlag, and Timeless. Curr Biol 19: 241–247. [DOI] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. 2007. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol 5: e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S. 1976. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol 106: 223–252. [Google Scholar]
- Preuss F, Fan JY, Kalive M, Bao S, Schuenemann E, Bjes ES, Price JL. 2004. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol 24: 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- Rieger D, Stanewsky R, Helfrich-Förster C. 2003. Cryptochrome, compound eyes, Hofbauer–Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms 18: 377–391. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, Helfrich-Förster C. 2006. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci 26: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger D, Wülbeck C, Rouyer F, Helfrich-Förster C. 2009. Period gene expression in four neurons is sufficient for rhythmic activity of Drosophila melanogaster under dim light conditions. J Biol Rhythms 24: 271–282. [DOI] [PubMed] [Google Scholar]
- Roberts L, Leise TL, Noguchi T, Galschiodt AM, Houl JH, Welsh DK, Holmes TC. 2015. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol 25: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Young MW, Saez L. 2000. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26: 505–514. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93: 805–814. [DOI] [PubMed] [Google Scholar]
- Schlichting M, Grebler R, Peschel N, Yoshii T, Helfrich-Förster C. 2014. Moonlight detection by Drosophila’s endogenous clock depends on multiple photopigments in the compound eyes. J Biol Rhythms 29: 75–86. [DOI] [PubMed] [Google Scholar]
- Schlichting M, Grebler R, Menegazzi P, Helfrich-Förster C. 2015. Twilight dominates over moonlight in adjusting Drosophila’s activity pattern. J Biol Rhythms 30: 117–128. [DOI] [PubMed] [Google Scholar]
- Schlichting M, Menegazzi P, Lelito KR, Yao Z, Buhl E, Dalla Benetta E, Bahle A, Denike J, Hodge JJ, Helfrich-Förster C, et al. 2016. A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila. J Neurosci 36: 9084–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R. 2014. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PloS Biol 12: e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. 2009. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: The anatomical basis of a neuropeptide’s circadian functions. PLoS ONE 4: e8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. 2008. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. 2008. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci 105: 19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Haynes P, Pírez N, Harrington KI, Guo F, Pollack J, Hong P, Griffith LC, Rosbash M. 2011. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. 2013. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 80: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Chandrashekaran M, Joshi A, Sharma V. 2002. Locomotor activity rhythm in Drosophila melanogaster after 600 generations in an aperiodic environment. Naturwissenschaften 89: 512–514. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. 2008a. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. 2008b. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol 99: 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siso-Nadal F, Fox JJ, Laporte SA, Hébert TE, Swain PS. 2009. Cross-talk between signaling pathways can generate robust oscillations in calcium and cAMP. PLoS ONE 4: e7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Lin JM, Meissner RA, Allada R. 2008. Dominant-negative CK2α induces potent effects on circadian rhythmicity. PLoS Genet 4: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC. 1997. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: Mapping elements of the PER protein involved in circadian cycling. J Neurosci 17: 676–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431: 862–868. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. 2005. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438: 238–242. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernández M de LP, Menet JS, Ceriani MF, Rosbash M. 2007. The Drosophila circadian network is a seasonal timer. Cell 129: 207–219. [DOI] [PubMed] [Google Scholar]
- Sun WC, Jeong EH, Jeong HJ, Ko HW, Edery I, Kim EY. 2010. Two distinct modes of PERIOD recruitment onto dCLOCK reveal a novel role for TIMELESS in circadian transcription. J Neurosci 30: 14458–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed S, Saez L, Young MW. 2011. Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. J Biol Chem 286: 27654–27662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó Á, Papin C, Zorn D, Ponien P, Weber F, Raabe T, Rouyer F. 2013. The CK2 kinase stabilizes CLOCK and represses its activity in the Drosophila circadian oscillator. PLoS Biol 11: e1001645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szular J, Sehadova H, Gentile C, Szabo G, Chou WH, Britt SG, Stanewsky R. 2012. Rhodopsin 5- and Rhodopsin 6-mediated clock synchronization in Drosophila melanogaster is independent of retinal phospholipase C-β signaling. J Biol Rhythms 27: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C-HA, Hinteregger E, Shang Y, Rosbash M. 2010. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron 66: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, Harms E, Syed S, Adams EL, Saez L. 2016. GSK-3 and CK2 kinases converge on Timeless to regulate the master clock. Cell Rep 16: 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. 2002. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem 277: 14048–14052. [DOI] [PubMed] [Google Scholar]
- Vaidya AT, Top D, Manahan CC, Tokuda JM, Zhang S, Pollack L, Young MW, Crane BR. 2013. Flavin reduction activates Drosophila cryptochrome. Proc Natl Acad Sci 110: 20455–20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Pírez N, Griffith LC. 2014. The Drosophila neuropeptides PDF and sNPF have opposing electrophysiological and molecular effects on central neurons. J Neurophysiol 111: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Rieger D, Helfrich-Förster C, Stanewsky R. 2007. Hofbauer–Buchner eyelet affects circadian photosensitivity and coordinates TIM and PER expression in Drosophila clock neurons. J Biol Rhythms 22: 29–42. [DOI] [PubMed] [Google Scholar]
- Venkatesan A, Fan JY, Nauman C, Price JL. 2015. A doubletime nuclear localization signal mediates an interaction with bride of Doubletime to promote circadian function. J Biol Rhythms 30: 302–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak P, Coupar J, Hughes SE, Fozdar P, Kilby J, Garren E, Yoshii T, Hirsh J. 2013. Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genet 9: e1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener C, Hamasaka Y, Nässel DR. 2004. Acetylcholine increases intracellular Ca2+ via nicotinic receptors in cultured PDF-containing clock neurons of Drosophila. J Neurophysiol 91: 912–923. [DOI] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. 2006. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet 2: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wülbeck C, Grieshaber E, Helfrich-Förster C. 2008. Pigment-dispersing factor (PDF) has different effects on Drosophila’s circadian clocks in the accessory medulla and in the dorsal brain. J Biol Rhythms 23: 409–424. [DOI] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. 2014. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343: 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. 2012. Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol 108: 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Bennett AJ, Clem JL, Shafer OT. 2016. The Drosophila clock neuron network features diverse coupling modes and requires network-wide coherence for robust circadian rhythms. Cell Rep 17: 2873–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA. 1999. Extraretinal photoreceptors at the compound eye’s posterior margin in Drosophila melanogaster. J Comp Neurol 412: 193–202. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. 2008. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol 508: 952–966. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. 2009. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci 29: 2597–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Hermann-Luibl C, Kistenpfennig C, Schmid B, Tomioka K, Helfrich-Förster C. 2015. Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J Neurosci 35: 6131–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. 2006. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Price JL, Hardin PE. 2009. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol 29: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Houl JH, Hardin PE. 2011. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol 21: 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J. 2005. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell 8: 267–278. [DOI] [PubMed] [Google Scholar]
- Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell 24: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lear BC, Seluzicki A, Allada R. 2009. The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr Biol 19: 2050–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. 2010. DN1p circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol 20: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. 2010. AKT and TOR signaling set the pace of the circadian pacemaker. Curr Biol 20: 1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu W, Hardin PE. 2016. CLOCKWORK ORANGE enhances PERIOD mediated rhythms in transcriptional repression by antagonizing E-box binding by CLOCK-CYCLE. PLoS Genet 12: e1006430. [DOI] [PMC free article] [PubMed] [Google Scholar]