Abstract
Objectives
Genome-wide association studies (GWASs) have discovered associations of numerous SNPs and genes with obesity. However, the underlying molecular mechanisms through which these SNPs and genes affect the predisposition to obesity remain not fully understood. Aims of our study are to comprehensively characterize obesity GWAS SNPs and genes through computational approaches.
Methods
For obesity GWAS identified SNPs, functional annotation, effects on miRNAs binding and impact on protein phosphorylation were performed via RegulomeDB and 3DSNP, miRNASNP, and the PhosSNP 1.0 database, respectively. For obesity associated genes, protein-protein interaction network construction, gene ontology and pathway enrichment analyses were performed by STRING, PANTHER and STRING, respectively.
Results
A total of 445 SNPs are significantly associated with obesity related phenotypes at threshold P < 5×10−8. A number of SNPs were eQTLs for obesity associated genes, some SNPs located at binding sites of obesity related transcription factors. SNPs that might affect miRNAs binding and protein phosphorylation were identified. Protein-protein interaction network analysis identified the highly-interconnected “hub” genes. Obesity associated genes mainly involved in metabolic process and catalytic activity, and significantly enriched in 15 signal pathways.
Conclusions
Our results provided the targets for follow-up experimental testing and further shed new light on obesity pathophysiology.
Introduction
Obesity is a worldwide epidemic with increasing global morbidity and mortality [1]. Obesity rates in adults aged 18+ have risen from around 9% in 1975 to 26% in 2014 with a marked increase over the past 4 decades [2]. In 2013, the American Medical Association (AMA) called obesity a disease, highlighting the importance of obesity in public health concern. Obesity results from a prolonged energy imbalance owing to intake exceeds expenditure, which may be defined as a condition in which there is an excessive accumulation of body fat and abnormally high body fat percentage caused by an increase in the number and volume of adipocytes at the cellular level. Obesity is a complex and multi-factorial disorder, defined by body mass index (BMI). Clinically, obesity is often accompanied by diversely serious chronic diseases, such as metabolic syndrome, type 2 diabetes (T2D), cardiovascular disease and certain forms of cancer, which become the leading causes of morbidity and mortality.
In the genome era, genome-wide association study (GWAS) is a major and effective approach to discover genes and SNPs that contribute to complex diseases. To date, 46 obesity GWASs and 12 meta-analyses had reported 445 SNPs involved in 389 genes with a significant threshold P < 5×10−8. The future challenge of obesity genetic study is to elucidate functional mechanisms through which these GWAS associated loci modulate obesity risk.
Computational approaches are powerful and essential means for post-GWAS studies, which can screen the potential and promising SNPs that deserve experimental testing for follow-up functional assays within amounts of candidate variants. Computational analyses represent a starting point to guide the functional research. Our group has previously characterized the GWAS SNPs and genes by computational approaches for osteoporosis [3], T2D [4] and Alzheimer's disease [5]. Here, we used computational approaches to comprehensively analyze obesity GWAS identified SNPs and genes, including functional annotation using RegulomeDB and 3DSNP, effects on miRNAs binding and protein phosphorylation for obesity associated SNPs, and protein-protein interaction network, gene ontology and pathway enrichment analyses for obesity associated genes, in order to identify functional SNPs for follow-up experimental assays, and to provide guidance for future study with regard to the pathogenesis and etiology of obesity.
Methods
Search strategy and data collection
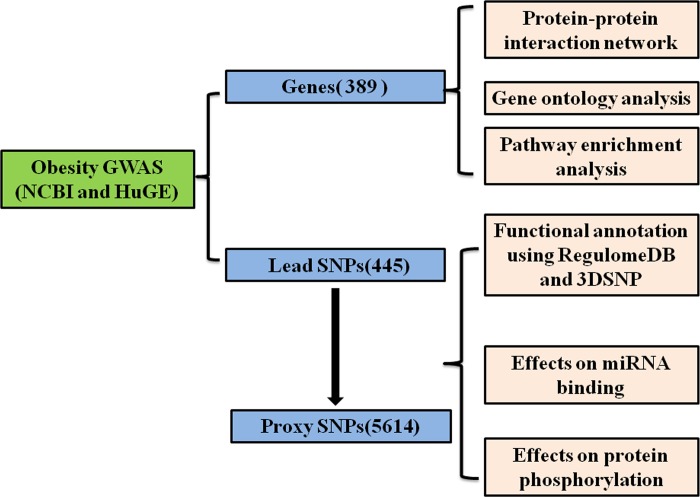
NCBI Association Results Browser and HuGE Navigator were used to extract obesity GWAS SNPs. “Obesity”, “BMI”, “body weight”, “overweight” and “FBM” were used as keywords and P < 5×10−8 as a significant threshold (updated to September, 2017). We first mapped these GWAS SNPs to the genome (hg19: GRCh37) by dbSNP (https://www.ncbi.nlm.nih.gov/snp/). Genes within a distance limit of 1Mb from GWAS SNPs were considered to be obesity GWAS genes, which were used to perform protein-protein interaction network, GO and KEGG pathway enrichment analyses. SNP Annotation and Proxy Search (SNAP, http://www.broadinstitute.org/mpg/snap/ldsearch.php) was used to identify proxy SNPs that were in strong linkage disequilibrium (LD) with obesity GWAS identified lead SNPs, based on genotype data from the 1000 Genomes Pilot 1 Project and the International HapMap Project (v3) with the CEU population panel. A distance limit of 500kb from the query lead SNP and r2 ≥ 0.8 from pairwise LD calculations were used as search and inclusion criteria. All loci were organized into an excel.file (S1 Table). A detailed workflow overview is illustrated in Fig 1.
Fig 1. The workflow of obesity associated loci derived from GWAS.
Data were obtained from the NCBI Association Results Browser (http://www.ncbi.nlm.nih.gov/projects/gapplusprev/sgap_plus.htm) and HuGE Navigator (http://64.29.163.162:8080/HuGENavigator/startPagePhenoPedia.do) and analyzed by many tools.
Functional annotation of obesity GWAS identified lead SNPs and their proxy SNPs via RegulomeDB and 3DSNP
RegulomeDB is an online database that provides functional interpretation of SNPs outside the coding region using high-throughput data from the ENCODE Project (ENCODE Project Consortium, 2012) and other resources like expression Quantitative Trait Loci (eQTL) and NCBI Sequence Read Archive. It is also a valuable tool for investigation of potential regulatory functions of SNPs on gene expression and disease phenotype. In RegulomeDB, SNPs are classified into classes based on the combinatorial presence/absence status of functional categories including protein binding, motifs, chromatin structure, eQTLs, histone modifications, and related data. And each SNP was assigned to an annotation score (range 1–7) to indicate the potential function [6]. 3DSNP is an integrated database for comprehensively annotating the regulatory function of human non-coding SNPs by exploring their three-dimensional (3D) interactions with genes and other genetically associated SNPs mediated by chromatin loops. Unlike the scoring scheme of RegulomeDB, 3DSNP integrates six different functional categories, including 3D interacting genes, enhancer state, promoter state, transcription factor binding sites, sequence motifs altered and conservation, in a scoring system to quantitatively evaluate the functionality of a SNP [7], and the sum of scores of the six functional categories is the total functionality score of a SNP. The higher 3DSNP score indicates the more likely functionality of a SNP. Here, we used RegulomeDB (v1.1) and 3DSNP (v1.0) to score each SNP.
Effects of obesity GWAS identified lead SNPs and their proxy SNPs on miRNA binding
MiRNAs can modulate gene expression by predominantly decreasing mRNA stability through base pairing with the 3'UTR of target mRNAs. A small complementary sequence from 2 to 7 nucleotides long usually is involved in the recognition of target mRNA by miRNA. Thus, only one nucleotide alteration in recognition sequence by SNPs can either create or destroy miRNA binding sites, and increase or decrease the miRNA binding affinity with target mRNAs. Several online web-based databases, such as SNPinfo (http://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi), miRNASNP (http://bioinfo.life.hust.edu.cn/miRNASNP2/), mrSNP (http://mrsnp.osu.edu/), MirSNP (http://202.38.126.151/hmdd/mirsnp/search/), and PolymiRTS (http://compbio.uthsc.edu/miRSNP/), can provide a resource for identifying the miRNA-related SNPs. We used miRNASNP (v2.0) to predict the “gain or loss” effects of miRNA binding sites by SNPs in 3'UTR of target mRNAs.
Effects of obesity GWAS lead SNPs and their proxy SNPs on protein phosphorylation
Protein phosphorylation is one of the most important reversible and intensively studied post-translational modification types, related to diverse signaling pathways and performing essential roles in modulating almost all kinds of biological processes and normal cellular functions. Protein phosphorylation is catalyzed by protein kinases (PKs) and principally targeting on serine (S), threonine (T), and tyrosine (Y) residues. In human genome, approximately 70% of nonsynonymous SNPs are potential phosphorylation-related SNPs (phosSNPs) that might influence protein phosphorylation status. Here, the PhosSNP 1.0 database was applied to identify phosSNPs for obesity GWAS lead SNPs and their proxy SNPs.
Protein-protein interaction network analyses
Search Tool for the Retrieval of Interacting Genes (STRING, v10.0, http://string-db.org) is a database that covers known and predicted protein interactions, including direct (physical) and indirect (functional) associations. The database quantitatively integrates interaction data derived from four sources: genomic context, high-throughput experiments, co-expression (conserved) and previous knowledge. Currently, STRING covers 5,214,234 proteins from 1133 organisms. In this study, STRING was applied to build protein-protein interaction network.
GO and pathway enrichment analyses
Gene ontology (GO, http://geneontology.org) is a widely utilized source of gene functional annotation including biological process, molecular function and cellular component. The Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.8) (https://david.ncifcrf.gov/) was used to identify significantly enriched GO terms. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding high-level functions and utilities of the biological system. In our work, STRING was used to investigate the enriched KEGG pathways for obesity GWAS genes. KEGG pathways with FDR < 0.05 were considered to be significant.
Results
Obesity GWAS SNPs/genes
A total of 445 SNPs were identified to be associated with obesity, BMI, body weight, overweight or FBM at a significant threshold P < 5×10−8, and 389 genes were considered as obesity GWAS genes. Of these SNPs, 263 mapped to intronic regions, 63 mapped to intergenic regions, 64 located in gene upstream and 58 in gene downstream, 19 located in 3'UTR and 8 in 5'UTR, 15 missense variant, 10 nc transcript variants, and 3 synonymous variant. A total of 5,614 proxy SNPs were considered to be in strong LD with obesity GWAS lead SNPs by SNAP (r2 ≥ 0.80). The detailed information of obesity GWAS loci was showed in S1 Table.
Functional annotation of obesity GWAS lead SNPs and proxy SNPs
Of the 6,059 SNPs, 36 SNPs returned error and 1,739 SNPs had score of ‘7’ that means no data available for these SNPs in RegulomeDB database, 4,284 SNPs returned with scores of 1–6, in which 680 SNPs (55 lead SNPs and 625 proxy SNPs) had score less than or equal to 3. The detailed regulatory functions of all GWAS SNPs and their proxy SNPs were given in S1 Table. Of particular note, four proxy SNPs which were in strong LD with obesity GWAS lead SNP rs977747 (located at 1p32 region) had scores of 1, were eQTLs for PDZK1IP1. Twenty-two proxy SNPs located at 1q21-q22 region had scores of 1, were eQTLs for CTSS. Four lead SNPs and multiple proxy SNPs located in 2p23.3 region were eQTLs for ADCY3. Additionally, some SNPs were located at the binding sites of obesity related transcription factors CEBPB, TCF7L2, STAT3, SPI1, GATA2, CREB1 and MEF2C (Table 1 and S2 Table). For 3DSNP, eleven GWAS lead SNPs had scores more than 100, and twelve GWAS lead SNPs were in the range of 60–100 (S1 and S3 Tables). GWAS lead SNP rs823114 had the highest 3DSNP score of 205.28 and a RegulomeDB score of 4, which located in 2kb upstream of the NUCKS1 gene. RegulomeDB suggested that rs823114 influence the binding of 59 different proteins (S3 Table). GWAS lead SNP rs329120 had the second highest 3DSNP score of 204.84 and a RegulomeDB score of 4, which located in the intronic region of the JADE2 gene and 500kb downstream of the LOC107986451 gene. RegulomeDB uncovered that rs329120 affect the binding of 34 different proteins (S3 Table). The functions of NUCKS1, JADE2 and LOC107986451 in obesity are unknown, and need to explore in the future.
Table 1. SNPs located in the binding sites of obesity related transcription factors.
| Transcription factor | SNPs |
|---|---|
| CEBPB | rs2815752,rs4836133,rs17150703,rs11191580,rs7132908,rs9925964,rs329120,rs2270204,rs11257655 |
| TCF7L2 | rs11208659,rs6548238,rs13201877,rs7132908,rs329120,rs823114,rs2357760,rs427943 |
| STAT3 | rs4836133,rs7132908,rs9925964,rs9940128,rs1861866,rs11671664,rs329120,rs2281727, rs4430979 |
| SPI1 | rs13191362,rs1121980,rs11671664,rs329120,rs633715,rs11257655,rs3783890 |
| GATA2 | rs2272903,rs9400239,rs7132908,rs4357030,rs11257655 |
| CREB1 | rs823114 |
Effects of obesity GWAS lead SNPs and proxy SNPs on miRNA binding and protein phosphorylation
Using miRNASNP, nine obesity GWAS lead SNPs and 35 proxy SNPs might influence the recognition and targeting of miRNAs (S4 Table). Among these miRNAs, miR-326, let-7, miR-31, miR-342, miR-181a, miR-148, miR-196a and miR-548 have been reported to relate with adipogenesis and lipid metabolism. The target genes of these miRNAs were further identified using Target Scan Human 7.1 and literature mining through NCBI PubMed (Table 2).
Table 2. The target genes of the miRNAs reported to relate with adipogenesis and lipid metabolism.
| miRNA | Predicted and validated target genes |
|---|---|
| miR-326 | RASSF1, AAK1, FAIM2, DGKG, ARAP1, TCF4, |
| miR-31 | PIK3C2A, C/EBPα, IDE, FTO, SLC2A4 |
| miR-548d-5p | PPARγ, ADAM30, NFKB1, LIN7C, CDKN2B, KRAS, TP53INP1, ABCB5, HHEX, PIK3C2A, SLC30A8, PTEN, EXOC4, JAZF1, VPS26A, ADIPOQ, IRS1, CCDC171, TMEM18, G6PC2, |
| miR-342 | CTBP2, CCDC171, EP300, SH2B1 |
| let-7 | HMGA2, ADCY9, IGF2BP2, KCNJ11, KCTD15, SEC16B |
| miR-196a | HOXC8, ACACB, ADIPOQ, CCDC171, G6PC2, NEGR1 |
| miR-181a | TNF-a, Smad7,Tcf7l2, IDH1,sirtuin1 (SIRT1), CDKN2B, FAM120A, SLC30A8 |
| miR-148 | WNT1, ADCY9, ASB4, CREB1, FAM120A, FAM120AOS, IRS1, KCTD15, LIN7C, LPL, NEGR1, NOTCH2, SLC30A8, KLF9 |
Note: Bold genes represent validated target genes of miRNAs.
Ten obesity GWAS lead SNPs and thirteen proxy SNPs may affect protein phosphorylation (Table 3). Among them, SNP rs6265 within BDNF influencing BDNF phosphorylation has been verified [8].
Table 3. Effect of obesity lead SNPs and proxy SNPs on protein phosphorylation.
| SNPs | Mapped genes | Types | |
|---|---|---|---|
| Lead SNPs | rs11676272 | ADCY3 | Type I(−), Type II(−) |
| rs6265 | BDNF | Type II(−) | |
| rs591120 | SEC16B | Type III(+)/(−) | |
| rs671 | ALDH2 | Type II(+),Type III(−) | |
| rs7498665 | SH2B1 | Type I(+) | |
| rs2230061 | CTSS | Type III(+) | |
| rs1190736 | GPR101 | Type II (+)/(−),Type III(+)/(−) | |
| rs2228213 | HIVEP1 | Type II(−),Type III(+)/(−) | |
| rs5215 | KCNJ11 | Type III(−) | |
| rs17826219 | ATAD5 | Type III(+)/(−) | |
| Proxy SNPs | rs749670 | ZNF646 | Type III(+) |
| rs12102203 | DMXL2 | Type I(−), Type II(+), Type III(+) | |
| rs61750814 | NUP54 | Type I(+), Type III(+)/(−) | |
| rs2277598 | BBS4 | Type I(+), Type III(+) | |
| rs1344642 | STK36 | Type II(−), Type III(−) | |
| rs2230115 | ZNF142 | Type I(+), Type III(+) | |
| rs3770213 | ZNF142 | Type III(+)/(−) | |
| rs3770214 | ZNF142 | Type I(−) | |
| rs2228209 | HIVEP1 | Type II(+),Type III(+)/(−) | |
| rs3816780 | ATAD5 | Type I(+),Type III(+)/(−) | |
| rs11657270 | ATAD5 | Type I(−),Type II(−), | |
| rs9910051 | ATAD5 | Type III(+)/(−),Type IV(−) | |
| rs1336900 | HORMAD1 | Type I(−),Type III(+) | |
Note: Type I (+)/(−): change of an amino acid with S/T/Y residue or vice versa to create a new or remove an original phosphorylation site.
Type II (+)/(−): variations to add or remove adjacent phosphorylation sites.
Type III (+)/(−): mutations to change PK types of adjacent phosphorylation sites.
Type IV(+)/(−): an amino acid substitution among S, T, or Y that could change the PK types in the phosphorylated position.
Protein-protein interaction network
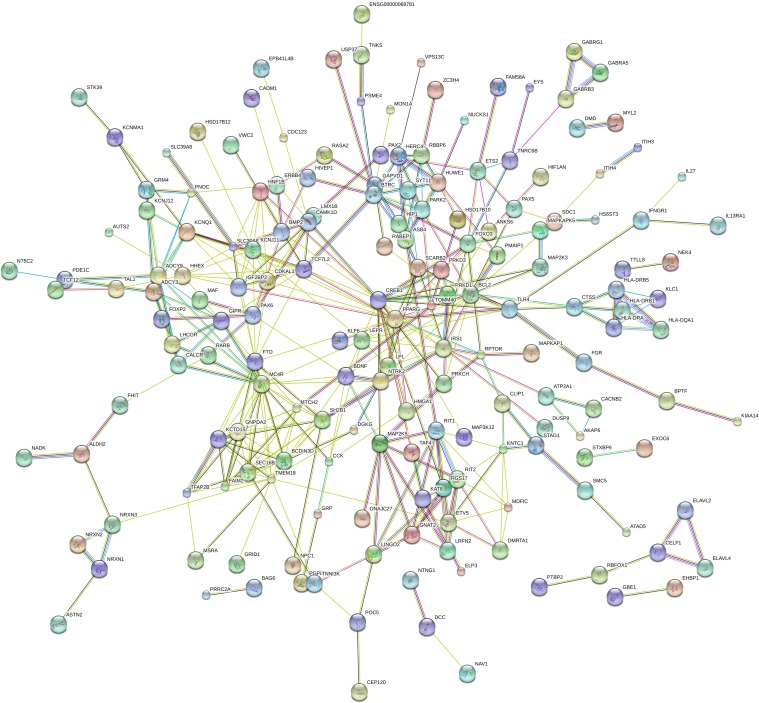
Fig 2 showed protein-protein interaction network of obesity GWAS genes coding proteins. Proteins with the strong connections were considered as being ‘Hub proteins’, such as CREB1, MC4R, PPARG, TMEM18, FTO, BCL2, TCF7L2, IRS1, SH2B1, KCNJ11, SEC16B, SLC30A8, BCDIN3D, BTRC, CDKAL1, FOXO3, GNPDA2, IGF2BP2, KCTD15, MTCH2 and RIT2 (S5 Table). These hub proteins were mainly involved in AMPK signaling pathway (CREB1, FOXO3, IRS1 and PPARG), Neurotrophin signaling pathway (BCL2, FOXO3, IRS1 and SH2B1), PI3K-Akt signaling pathway (BCL2, CREB1, FOXO3 and IRS1), Circadian rhythm (BTRC and CREB1), T2D (IRS1 and KCNJ11), and several cancer associated pathways. These hub proteins encoding genes were defined as “hub” genes.
Fig 2. Protein-protein interaction network of obesity GWAS associated genes.
The nodes and edges represent the proteins (genes) and their interactions, respectively. Colored nodes represent query proteins and first shell of interactors, white nodes represent second shell of interactors, empty nodes represent proteins of unknown 3D structure, filled nodes represent some 3D structure is known or predicted. Purple edges (experimentally determined) and light blue edges (from curated databases) represent known interactions, green edges (gene neighborhood), red edges (gene fusions) and dark blue edges (gene co-occurrence) represent predicted interactions, yellow-green edges (textmining), black edges (co-expression) and light blue edges (protein homology) represent others.
GO and pathway enrichment analyses of obesity GWAS genes
GO and pathway enrichment analyses found that obesity GWAS genes significantly enriched in 104 GO functional categories with P-value < 0.05 (S6 Table) and 15 KEGG signaling pathways with FDR < 0.05 (S7 Table). Notably, of 104 GO functional categories, two GO terms “glucose homeostasis” and “response to glucose” were also enriched by T2D GWAS genes (such as TCF7L2 and FTO), indicating that obesity associated genes might confer T2D risk through its primary effect on adiposity.
Discussion
One of the molecular mechanisms by which SNPs mediate the onset of a disease is to regulate gene expression by affecting the binding of transcription factors, and increase disease susceptibility [9]. For example, SNP rs4684847, in LD with obesity GWAS lead SNP rs1801282, is located in the upstream 6.5 kb of PPARG2 promoter. The risk C allele of rs4684847 could enhance the binding to PRRX1 and inhibit PPARG2 mRNA expression, thereby leading to abnormal regulation of free fatty acids turnover and glucose homeostasis [10]. In this study, we identified some obesity GWAS SNPs that were located at the binding sites of adipogenic differentiation and lipid metabolism related transcription factors CEBPB, TCF7L2, STAT3, SPI1, GATA2, CREB1 and MEF2C, and were eQTLs of obesity or lipid metabolism associated genes, such as ADCY3, CTSS, KCTD15, MTCH2 and SPI1. Genetic and epigenetic studies have demonstrated that ADCY3 is involved in the pathogenesis of obesity [11]. ADCY3 haploinsufficiency resulted in increased expression of genes involved in adipogenesis in peripheral tissues of mice [12]. CTSS gene expression increases with obesity in the adipose tissue of obese rodent and human. Compared with normal-weight subjects, obese subjects had a 2-fold increase in CTSS mRNA in adipose tissue and 30% increase in circulating CTSS levels [13]. Variants of the KCTD15 were associated with risk for obesity. In adipose tissues of 2 obesity mice models, KCTD15 exhibited concomitant obesity-dependent alterations in DNA methylation and gene expression [14]. MTCH2 is a conserved regulator of lipid homeostasis. Knockdown of MTCH2/mtch2 reduced lipid accumulation (in adipocyte-like cells, C. elegans and mice) [15] and number of adipocytes (in zebrafish) [16], while overexpression of MTCH2 increased fat accumulation (in adipocyte-like cells, C. elegans and mice) [15,17]. Loss of muscle MTCH2 protected mice from diet-induced obesity and hyperinsulinemia and increased energy expenditure [18]. The transcription factor SPI1 is expressed in mature adipocytes of white adipose tissue, whose mRNA and protein levels is increased markedly in mouse models of genetic or diet-induced obesity [19].
Another molecular mechanism by which SNPs mediate disease susceptibility is to affect the binding of miRNAs [9]. In this study, 9 lead SNPs and 35 proxy SNPs were predicted to affect the binding of miRNAs (S2 Table). Among them, of particular note, SNP rs7132908 located in the 3'UTR of FAIM2 had a 3DSNP score of 162.69. The rs7132908 A allele was predicted to destroy the binding sites for hsa-miR-330-5p and hsa-miR-326. SNP rs2650492 located in the 3'UTR of SBK1, had a 3DSNP score of 90.2 and a RegulomeDB score of 2c. The rs2650492 A allele was predicted to destroy the binding sites for hsa-miR-331-3p. SNP rs2531995 located in the 3'UTR of ADCY9, had a 3DSNP score of 43.01 and a RegulomeDB score of 1f. The rs2531995 A allele was predicted to create the binding sites for hsa-miR-632 and hsa-miR-654-3p. Therefore, it is very likely that these SNPs play regulatory function by affecting miRNA binding, which needs to be confirmed by experimental methods in the future.
Abnormal protein phosphorylation has been reported in a number of diseases, such as Parkinson's disease, Alzheimer's disease, and other degenerative disorders. Deng et al. [8] identified and characterized phosSNPs significant for bone mineral density in humans, and successfully dissected the functions of phosSNPs. Niu et al. [20] showed that phosSNPs rs3755955 and rs6831280 of IDUA, rs2707466 of WNT16 associate with bone mineral density phenotypes, and in silico analyses revealed that phosSNP rs2707466 of WNT16 directly destroyed a phosphorylation site, which could have a deleterious effect on WNT16 protein, but phosSNPs rs3755955 and rs6831280 of IDUA might play indirect effects on nearby phosphorylation sites. In this study, 10 GWAS lead SNPs and 13 proxy SNPs were identified as being phosSNPs that might affect the protein phosphorylation status, indicating that similar mechanism might also exist in obesity.
Protein-protein interaction network identified a number of key “hub” genes including FTO, TMEM18, MC4R, CREB1, PPARG and TCF7L2. FTO was the first obesity susceptibility gene identified by GWASs and continued to be the locus with the largest effect on obesity risk and BMI, most widely replicated with variety of obesity traits across diverse ancestries and throughout the life course [21]. Intriguingly, in human brains, obesity associated SNPs within FTO are functionally connected, at megabase distances, with regulation of the homeobox gene IRX3 expression, but not FTO, and in vivo studies in mice demonstrated that the expression levels of IRX3 affect body mass and composition phenotypes, suggesting that although the obesity associated SNPs reside in the first intron of FTO gene, they may not only influence FTO but mediate their obesity effects through long-range interaction with nearby genes (notably IRX3 and RPGRIP1L) [22]. Of note, as the first mRNA demethylase that has been identified, the demethylase activity of FTO is required for adipogenesis [23]. TMEM18 is the second largest effect size among all loci identified so far via GWASs or GWAS meta-analyses [24]. TMEM18 seems to influence energy levels via insulin and glucagon signaling, and significantly inhibited adipocyte maturation in human adipogenesis [25]. MC4R is widely expressed in the central nervous system, which plays an important role in the leptin-melanocortin pathway in modulating energy homeostasis, affecting both energy intake and expenditure [26]. In both MC4R knock-out mice and humans, mutations in MC4R could cause decreased energy expenditure and increased food intake [26,27]. CREB1 is a transcription factor that can drive the expression of a number of genes involved in the regulation of food intake and energy expenditure. Expression of constitutively active CREB induced expression of endogenous C/EBP β, and caused adipogenesis [28]. Mice that lack CREB1 in SIM1-positive neurons developed an obese phenotype as a result of reduced energy expenditure, not because of excessive energy intake [29]. PPARG is a member of the nuclear receptor superfamily of ligand-dependent transcription factors and that functions as a master regulator of adipocyte differentiation and metabolism [30]. TCF7L2 is an important transcription factor in the canonical Wnt signaling pathway, and Wnt signaling via TCF7L2 can inhibit adipogenesis [31].
Pathway enrichment analysis identified 15 signaling pathways. Neurotrophins are a family of trophic factors, consisting of BDNF, nerve growth factor, neurotrophin 3, and neurotrophin 4, which exert their opposite functional outcomes through engagement of p75 neurotrophin receptor (p75NTR) or Trk tyrosine kinase receptors. Neurotrophin receptor signaling could affect how the central nervous system control body weight change and energy intake [32–34]. In the hypothalamus, BDNF signals through TrkB could suppress appetite and reduce body weight. Mice conditionally-depleted of BDNF in neurons [33] or BDNF+/- mice [32] overeat and become obese on a normal diet. Adipocyte-specific deficiency of p75NTR or transplantation of p75NTR-null white adipose tissue into wild-type could protect mice from high-fat diet-induced obesity and insulin resistance [35]. GWAS have identified associations between BMI and two loci close to cell adhesion molecule 1 and 2 (CADM1 and CADM2), risk variants within them associate with elevated CADM1 and CADM2 expression in the hypothalamus of human subjects, respectively. In obese mice, expression of CADM1 and CADM2 increased, and Cadm1 loss protected mice from obesity. In excitatory neurons, induction of Cadm1 could facilitate weight gain while exacerbate energy expenditure. In the hypothalamus and hippocampus, decreased Cadm1 expression promoted a negative energy balance and weight loss [36]. Approximately 15–40% of inflammatory bowel disease (IBD) patients are obese. IBD and obesity share environmental link and mechanistic connection, obesity can contribute to the development of IBD and response to therapy in IBD patients [37]. It is suggested that toxoplasmosis associate with obesity by alteration of inflammatory fat distribution as organisms change and reside in fatty tissues [38]. Men and women with cutaneous leishmaniasis presented higher body mass index than the controls [39]. Association between overweight and asthma have been found among females, persistent asthma associated with high BMI throughout childhood [40]. Also, BMI has been shown to be associated with the risk of immune-related infectious diseases such as tuberculosis [41]. Besides, high-fat diet-induced obesity can enhance allograft rejection [42]. Obesity is considered as a risk factor for posttransplantation complications including acute graft-versus-host disease [43].
Our computational analyses have some strengths and weaknesses. Compared with labour-intensive and time-consuming wet experiments, computational approach could quickly identify the promising and potential causal SNPs from a large amount of GWAS variants, and the validity has been proved by previous work [8,9]. On the other hand, each computational tool has its own unique features, and a single approach might not correctly find true causal SNP. Application of multiple methods would reduce false discovery rate. An appropriate follow-up to our study would be to validate computational prediction by wet experiments. This two-step process of identifying potential functional SNPs using computational tools followed by conventional experimentation would be a good strategy to reveal functional mechanisms at obesity GWAS loci.
Conclusions
Our computational analysis identified a number of SNPs that located at binding sites of obesity related transcription factors, and were eQTLs for obesity associated genes, and might affect miRNAs binding and protein phosphorylation. Protein-protein interaction network analysis identified the highly-interconnected “hub” genes. Obesity associated genes significantly enriched in 104 GO categories and 15 signaling pathways. Taken together, our study uncovered potential functional mechanisms of obesity GWAS SNPs and genes, and provided targets and clues for future functional analysis with regard to the etiology and pathogenesis of obesity.
Supporting information
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Basic Research Program of China (973 Program, No. 2011CB504004), the National Natural Science Foundation of China (Nos. 31371275 and 30971635) and self-determined research funds of CCNU from the colleges’ basic research and operation of MOE (No. CCNU14Z01003) to QH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2014;384: 766–781. doi: 10.1016/S0140-6736(14)60460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Health Observatory (GHO) data. Overweight and obesity Web site. http://www.who.int/gho/ncd/risk_factors/overweight/en/ (last accessed June 6, 2017)
- 3.Qin L, Liu Y, Wang Y, Wu G, Chen J, Ye W, et al. Computational characterization of osteoporosis associated SNPs and genes identified by genome-wide association studies. PLoS One. 2016;11: e0150070 https://doi.org/10.1371/journal.pone.0150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng M, Liu X, Yang M, Han L, Xu A, Huang Q. Computational analyses of type 2 diabetes-associated loci identified by genome-wide association studies. J Diabetes. 2017;9: 362–377. doi: 10.1111/1753-0407.12421 [DOI] [PubMed] [Google Scholar]
- 5.Han Z, Huang H, Gao Y, Huang Q. Functional annotation of Alzheimer's disease associated loci revealed by GWASs. PLoS One. 2017;12: e0179677 https://doi.org/10.1371/journal.pone.0179677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22: 1790–1797. doi: 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Quan C, Chen H, Bo X, Zhang C. 3DSNP: a database for linking human noncoding SNPs to their three-dimensional interacting genes. Nucleic Acids Res. 2017;45: D643–D649. doi: 10.1093/nar/gkw1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng FY, Tan LJ, Shen H, Liu YJ, Liu YZ, Li J, et al. SNP rs6265 regulates protein phosphorylation and osteoblast differentiation and influences BMD in humans. J Bone Miner Res. 2013;28: 2498–2507. doi: 10.1002/jbmr.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Q. Genetic study of complex diseases in the post-GWAS era. J Genet Genomics. 2015;42: 87–98. doi: 10.1016/j.jgg.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 10.Claussnitzer M, Dankel SN, Klocke B, Grallert H, Glunk V, Berulava T, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156: 343–358. doi: 10.1016/j.cell.2013.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, Shen C, Seed Ahmed M, Östenson CG, Gu HF. Adenylate cyclase 3: a new target for anti-obesity drug development. Obes Rev. 2016;17: 907–914. doi: 10.1111/obr.12430 [DOI] [PubMed] [Google Scholar]
- 12.Tong T, Shen Y, Lee HW, Yu R, Park T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci Rep. 2016;6: 34179 doi: 10.1038/srep34179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naour N, Rouault C, Fellahi S, Lavoie ME, Poitou C, Keophiphath M, et al. Cathepsins in human obesity: changes in energy balance predominantly affect cathepsin s in adipose tissue and in circulation. J Clin Endocrinol Metab. 2010;95: 1861–1868. doi: 10.1210/jc.2009-1894 [DOI] [PubMed] [Google Scholar]
- 14.Sonne SB, Yadav R, Yin G, Dalgaard MD, Myrmel LS, Gupta R, et al. Obesity is associated with depot-specific alterations in adipocyte DNA methylation and gene expression. Adipocyte. 2017;6: 124–133. doi: 10.1080/21623945.2017.1320002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rottiers V, Francisco A, Platov M, Zaltsman Y, Ruggiero A, Lee SS, et al. MTCH2 is a conserved regulator of lipid homeostasis. Obesity (Silver Spring). 2017;25: 616–625. [DOI] [PubMed] [Google Scholar]
- 16.Landgraf K, Strobach A, Kiess W, Körner A. Loss of mtch2 function impairs early development of liver, intestine and visceral adipocytes in zebrafish larvae. FEBS Lett. 2016;590: 2852–2861. doi: 10.1002/1873-3468.12330 [DOI] [PubMed] [Google Scholar]
- 17.Bar-Lev Y, Moshitch-Moshkovitz S, Tsarfaty G, Kaufman D, Horev J, Resau JH, et al. Mimp/Mtch2, an obesity susceptibility gene, induces alteration of fatty acid metabolism in transgenic mice. PLoS One. 2016;11: e0157850 https://doi.org/10.1371/journal.pone.0157850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzaglo-Azriel L, Kuperman Y, Tsoory M, Zaltsman Y, Shachnai L, Zaidman SL, et al. Loss of muscle MTCH2 increases whole-body energy utilization and protects from diet-induced obesity. Cell Rep. 2016;14: 1602–1610. doi: 10.1016/j.celrep.2016.01.046 [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Tong Q. Transcription factor PU.1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol. 2008;295: C213–C220. doi: 10.1152/ajpcell.00422.2007 [DOI] [PubMed] [Google Scholar]
- 20.Niu T, Liu N, Yu X, Zhao M, Choi HJ, Leo PJ, et al. Identification of IDUA and WNT16 phosphorylation-related non-synonymous polymorphisms for bone mineral density in meta-analyses of genome-wide association studies. J Bone Miner Res. 2016;31: 358–368. doi: 10.1002/jbmr.2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10: 51–61. doi: 10.1038/nrendo.2013.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507: 371–375. doi: 10.1038/nature13138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, Sanderson J, et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun. 2015;6: 6792 doi: 10.1038/ncomms7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518: 197–206. doi: 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernhard F, Landgraf K, Klöting N, Berthold A, Büttner P, Friebe D, et al. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia. 2013;56: 311–322. doi: 10.1007/s00125-012-2773-0 [DOI] [PubMed] [Google Scholar]
- 26.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88: 131–141. [DOI] [PubMed] [Google Scholar]
- 27.Ste Marie L, Miura GI, Marsh DJ, Yagaloff K, Palmiter RD. A metabolic defect promotes obesity in mice lacking melanocortin-4 receptors. Proc Natl Acad Sci USA. 2000;97: 12339–12344. doi: 10.1073/pnas.220409497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang JW, Klemm DJ, Vinson C, Lane MD. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J Biol Chem. 2004;279: 4471–4478. doi: 10.1074/jbc.M311327200 [DOI] [PubMed] [Google Scholar]
- 29.Chiappini F, Cunha LL, Harris JC, Hollenberg AN. Lack of cAMP-response element-binding protein 1 in the hypothalamus causes obesity. J Biol Chem. 2011;286: 8094–8105. doi: 10.1074/jbc.M110.178186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefterova MI, Haakonsson AK, Lazar MA, Mandrup S. PPARγ and the global map of adipogenesis and beyond. Trends Endocrinol Metab. 2014;25: 293–302. doi: 10.1016/j.tem.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289: 950–953. [DOI] [PubMed] [Google Scholar]
- 32.Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, et al. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci USA. 1999;96: 15239–15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu B, Goulding EH, Zang K, Cepoi D, Cone RD, Jones KR, et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003;6: 736–742. doi: 10.1038/nn1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu B, Xie X. Neurotrophic factor control of satiety and body weight. Nat Rev Neurosci. 2016;17: 282–292. doi: 10.1038/nrn.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baeza-Raja B, Sachs BD, Li P, Christian F, Vagena E, Davalos D, et al. P75 neurotrophin receptor regulates energy balance in obesity. Cell Rep. 2016;14: 255–268. doi: 10.1016/j.celrep.2015.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathjen T, Yan X, Kononenko NL, Ku MC, Song K, Ferrarese L, et al. Regulation of body weight and energy homeostasis by neuronal cell adhesion molecule 1. Nat Neurosci. 2017;20: 1096–1103. doi: 10.1038/nn.4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh S, Dulai PS, Zarrinpar A, Ramamoorthy S, Sandborn WJ. Obesity in IBD: epidemiology, pathogenesis, disease course and treatment outcomes. Nat Rev Gastroenterol Hepatol. 2017;14: 110–121. doi: 10.1038/nrgastro.2016.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter CJ. Toxoplasmosis and polygenic disease susceptibility genes: extensive toxoplasma gondii host/pathogen interactome enrichment in nine psychiatric or neurological disorders. J Pathog. 2013;2013: 965046 doi: 10.1155/2013/965046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunha DF, Cunha SF, Nunes AG, Silva-Vergara ML. Is an increased body mass index associated with a risk of cutaneous leishmaniasis? Rev Soc Bras Med Trop. 2009;42: 494–495. [DOI] [PubMed] [Google Scholar]
- 40.Ekström S, Magnusson J, Kull I, Andersson N, Bottai M, Besharat Pour M, et al. Body mass index development and asthma throughout childhood. Am J Epidemiol. 2017;186: 255–263. doi: 10.1093/aje/kwx081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Li X, Xin H, Li H, Li M, Lu W, et al. Association of body mass index with the tuberculosis Infection: a population-based study among 17796 adults in rural China. Sci Rep. 2017;7: 41933 doi: 10.1038/srep41933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molinero LL, Yin D, Lei YM, Chen L, Wang Y, Chong AS, et al. High-fat diet-induced obesity enhances allograft rejection. Transplantation. 2016;100: 1015–1021. doi: 10.1097/TP.0000000000001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuji S, Kim SW, Yoshimura K, Akiyama H, Okamoto S, Sao H, et al. Possible association between obesity and posttransplantation complications including infectious diseases and acute graft-versus-host disease. Biol Blood Marrow Transplant. 2009;15: 73–82. doi: 10.1016/j.bbmt.2008.10.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.