Schvartzman et al. review how alterations in the levels of specific metabolites in mammalian cells result in chromatin modifications that influence gene expression.
Abstract
Dynamic regulation of gene expression in response to changing local conditions is critical for the survival of all organisms. In metazoans, coherent regulation of gene expression programs underlies the development of functionally distinct cell lineages. The cooperation between transcription factors and the chromatin landscape enables precise control of gene expression in response to cell-intrinsic and cell-extrinsic signals. Many of the chemical modifications that decorate DNA and histones are adducts derived from intermediates of cellular metabolic pathways. In addition, several of the enzymes that can remove these marks use metabolites as part of their enzymatic reaction. These observations have led to the hypothesis that fluctuations in metabolite levels influence the deposition and removal of chromatin modifications. In this review, we consider the emerging evidence that cellular metabolic activity contributes to gene expression and cell fate decisions through metabolite-dependent effects on chromatin organization.
Introduction
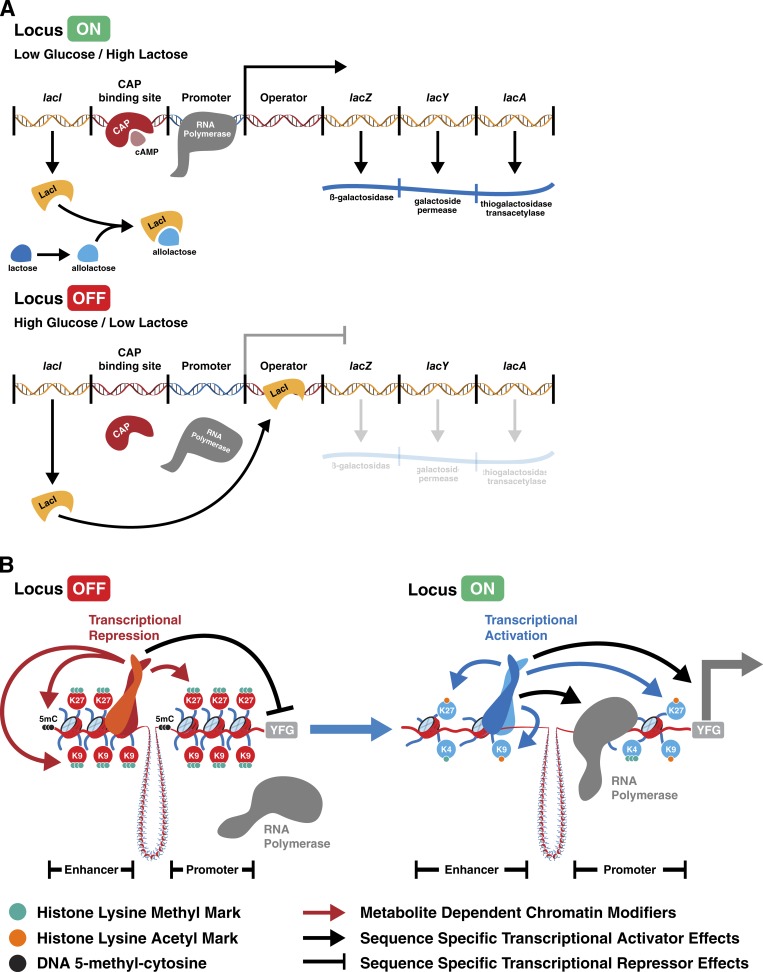
All organisms must adapt to changing environmental conditions to survive and thrive. Therefore, scientists have long studied how changes in nutrient availability influence cellular behaviors. Seminal work by Jacob and Monod (1961) investigating how single-cell organisms adapt to alterations in nutrient supply led to the discovery of the lac operon and laid the groundwork for the modern understanding of gene regulation. After the observation that bacteria could, after a small lag in growth, switch to lactose as a fuel source once glucose was exhausted, Jacob and Monod systematically dissected how bacteria adapt to this metabolic challenge by inducing the expression of genes involved in lactose uptake and catabolism. They proposed a model wherein a metabolite acting as an “inducer” blocks the action of a “repressor” molecule that inhibits expression of a suite of related genes (Fig. 1 A). Subsequent work showed that two metabolic pathways converge to regulate the activity of the lac operon. Allolactose, a product of lactose metabolism, serves as the inducer by binding the repressor, thereby reducing the fraction of repressor that can bind and repress the operon. Cyclic AMP (cAMP), which increases dramatically in the absence of glucose, positively increases transcription of the operon by promoting the binding of a coactivator that recruits RNA polymerase (Fig. 1 A; Lewis, 2005). Thus, the lac operon serves as an AND gate that integrates multiple metabolic inputs to coordinate appropriate gene expression in response to environmental fluctuations. This model, whereby sequence-specific DNA binding proteins regulate the transcription of genes that contain their cognate sequence (Ptashne, 1988) in direct proportion to the ability to bind and recruit RNA polymerase, serves as a basis for how specific gene regulation is thought to be effected.
Figure 1.
Paradigms of metabolic regulation of gene expression. (A) Summarized model of the E. coli lac operon as outlined by Jacob and Monod (1961). In low glucose/high lactose conditions, the lac repressor (LacI) binds allolactose and RNA polymerase is able to activate transcription of genes required for lactose metabolism. Conversely, in high glucose/low lactose conditions, LacI is not bound to allolactose and can bind to the operator sequence, repressing the ability of RNA polymerase to transcribe operon genes. CAP, catabolite activator protein. (B) Schematic representation of how sequence-specific DNA binding proteins recruit chromatin modifying enzymes that serve to deposit inhibitory (left) or activating (right) marks. In this model, transcription factors recruit local chromatin modifying enzymes. YFG, your favorite gene; 5mC, 5-methyl-cytosine; K9, histone H3 lysine 9; K27, histone H3 lysine 27; K4, histone H3 lysine 4.
Nutrient signaling in metazoan organisms is more complex than in prokaryotes. Multicellular organisms have evolved signaling pathways that respond to specific nutrients as well as hormones that reflect organismal metabolic status (Chantranupong et al., 2015). The response of an individual cell (e.g., whether to rewire metabolic pathways to favor an anabolic vs. catabolic state) to such extracellular signals depends in turn on a variety of intracellular nutrient and bioenergetic sensors including AMP-activated protein kinase (AMPK), mammalian target of rapamycin (mTOR), and GCN2. These enzymes sense changes in intracellular metabolites and convert these variations into an output, substrate phosphorylation, which is able to be effected at all ratios of ATP/ADP that exist in viable cells. Collectively, these signaling pathways enable cells to coordinate organismal metabolic status (through extracellular signaling pathways) with intracellular metabolic status. Furthermore, these kinases allow metazoan organisms to enact changes in gene expression over a wide range of variation in the substrates used to maintain bioenergetics.
However, metazoan cells also retain features of direct nutrient sensing within their nuclear organization. All organisms harbor variable levels of chemical modification on their DNA and DNA-associated proteins (Yung and Elsässer, 2017). The deposition and removal of these marks require metabolites that are intermediates of distinct metabolic pathways. This has led to the hypothesis that these chromatin modifications respond to fluctuations in nutrient availability to modulate gene expression. In contrast to the basic model of transcription proposed by Jacob and Monod (1961) in Escherichia coli, it appears gene regulation in more complex organisms has evolved to also be regulated by chemical modifications of DNA and its attendant proteins (Fig. 1 B). In this review, we discuss examples of how metabolite availability shapes gene expression in mammalian cells through effects on chromatin modifications. In particular, we focus on the hypothesis that levels of specific metabolites provide signals that can be integrated in chromatin modifications to influence gene expression.
Theories and limitations
Regulation of gene expression
Viewed from an enzymatic perspective, gene expression involves the recruitment of RNA polymerase to a specific DNA template and the activation of its transcribing ability (Sainsbury et al., 2015). However, in more complex organisms, it appears a prerequisite to this reaction is that the chromatin regions to which RNA polymerase is recruited have previously been rendered accessible (Levine et al., 2014). Accessibility is largely a result of the combined activity of transcription factors, nucleosome remodeling complexes, and chromatin-modifying enzymes that add or remove marks that favor recruitment of specific coactivators. Key to the links between metabolism and gene expression is the fact that these marks are derived from intermediary metabolites.
This hypothesis does not challenge the importance of transcription factors in directing gene expression. However, transcription factors act in concert with a number of chromatin-modifying activities that may sense the metabolic state of a cell. Although controversy still exists as to the causative or correlative nature of chromatin modifications with respect to gene expression, it is clear that metazoan transcription factors do not survey the entire genome landscape (Kolomeisky, 2011) and that chromatin accessibility can be increased or decreased by various chromatin modifications. As demonstrated by the inefficiency of the inducible pluripotent stem cell reprogramming process (Takahashi and Yamanaka, 2006), transcription factors are not free to bind and/or activate any genomic region. In contrast to the deterministic effect of metabolites on gene expression in E. coli as demonstrated by the lac operon model (Fig. 1 A), metazoan cells engage a model in which transcription factors, chromatin remodelers, and metabolic state cofactors act in concert to influence whether specific gene loci are activated or repressed (Fig. 1 B).
Links between metabolites and chromatin modifications
The repertoire of annotated chromatin modifications is large and continually expanding. To date, the most common modifications include methylation of nucleic acids and acetylation, methylation, phosphorylation, and ubiquitination of histones. Additional histone acylations, such as crotonylation, succinylation, and propionylation, arise in the presence of high levels of the specific acyl-donor (Sabari et al., 2017). Here, we focus on the common histone and DNA modifications that are most affected by the metabolic state of the cell.
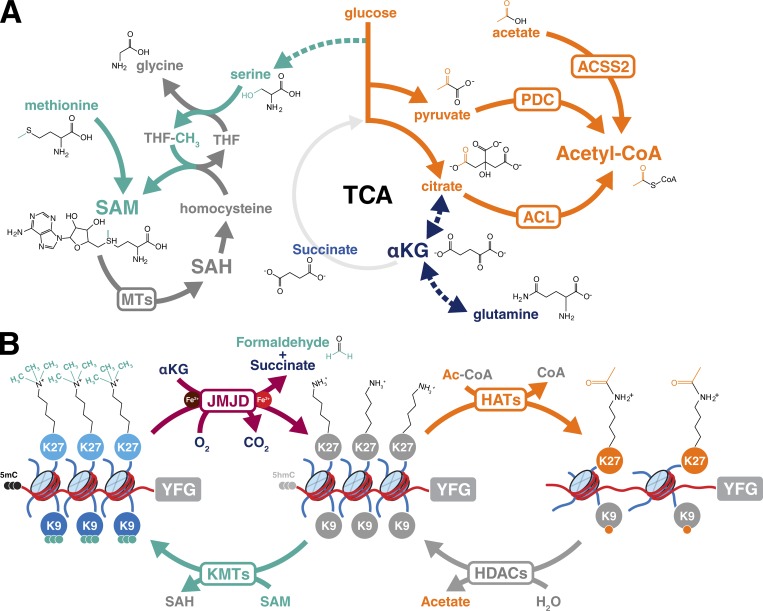
All methylation reactions require the metabolite S-adenosylmethionine (SAM) as a methyl donor. This metabolite is directly derived from the combined activities of serine-glycine-one carbon metabolism and the methionine cycle (Fig. 2 A). Cytosine DNA methylation is catalyzed by the three DNA methyltransferases, DNMT1, DNMT3a, and DNMT3b (Lyko, 2018). DNMT1, the maintenance methyltransferase, recognizes hemimethylated DNA to produce a symmetrically modified DNA duplex (Gruenbaum et al., 1982). DNMT3a and 3b, in contrast, are de novo methyltransferases that deposit methyl marks on previously unmodified cytosines (Okano et al., 1999). A wide variety of histone methyltransferases, including lysine methyltransferases and arginine methyltransferases, similarly use SAM as a methyl donor. This can result in mono- or dimethylation of arginine residues or mono-, di-, or trimethylation of the amide nitrogen of lysine (Fig. 2 B). In all cases, the reaction results in the donation of a methyl group from SAM to the substrate and the release of S-adenosylhomocysteine, which can serve as a competitive inhibitor of these enzymes or be restored to SAM via intracellular metabolic pathways (Fig. 2 A).
Figure 2.
Metabolic pathways provide substrates for enzymes that modify chromatin. (A) Metabolic pathways implicated in the generation of carbon groups required for methylation (turquoise), acetylation (orange), or demethylation (dark blue) of chromatin. Note that TCA cycle metabolites serve to provide carbon units for both acetylation and demethylation via αKG. THF, tetrahydrofolate; SAH, S-adenosylhomocysteine; MTs, methyltransferases; PDC, pyruvate dehydrogenase complex. Details in the text. (B) Chromatin modification reactions that require intermediary metabolites. Repressed chromatin is shown on the left; open chromatin is shown on the right; and a poised state is indicated in the center. Color coding is as in A, with methyl marks in turquoise and acetylation in orange. 5-Methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) are indicated, but their reactions are omitted for clarity. HATs, histone acetyltransferases; JMJD, Jumonji-domain containing histone demethylases; KMTs, histone lysine methyltransferases; YFG, your favorite gene. For simplicity, the flavin-dependent LSD1 family of histone demethylases and the sirtuin family NAD-dependent HDACs are omitted but are discussed in the text.
Although DNA and histone methylation can be lost passively through replication or nucleosome eviction, the active removal of methylated moieties can proceed through one of two major chemical mechanisms. The LSD1 family of histone demethylases uses oxygen and a flavin-dependent reaction mechanism to remove methyl moieties from mono- or dimethylated histone residues (Shi et al., 2004). A second, more general mechanism of methyl removal involves the family of α-ketoglutarate (αKG)–dependent dioxygenases (Gao et al., 2017), which encompasses a diverse array of enzymes responsible for histone demethylation (JmjC family of histone lysine demethylases), DNA 5-methyl-cytosine hydroxylation (Tet family of DNA modifying enzymes), RNA N6-methyladenosine (m6A) demethylation (FTO, fat mass and obesity-associated protein, and ALKBH5), and EglN prolyl-4-hydroxylation, among others. All αKG-dependent dioxygenases use oxygen and αKG as substrates and release CO2 and succinate as products. This reaction requires ferrous iron (Fe(II)) as a cofactor and, like LSD1-dependent demethylation, results in hydroxylation of the enzymatic substrate (Fig. 2 B). In the case of lysine or arginine demethylation, this hydroxyl-methyl intermediate is released as formaldehyde, which can ultimately be recycled into formate to fuel nucleotide biosynthesis (Burgos-Barragan et al., 2017).
DNA methylation and several histone lysine methylation states are traditionally considered to exert repressive effects on gene expression (Mozzetta et al., 2015; Rao et al., 2017; Schuettengruber et al., 2017). In contrast, histone acetylation is primarily associated with actively transcribed loci (Xu et al., 2005; Tessarz and Kouzarides, 2014; Fig. 2 B). The acetylation of histone lysines is performed by acetyl-transferases that use acetyl-CoA to modify target proteins, which may not always be histones. Acetyl-CoA can be generated either through the mitochondrial oxidation of carbon sources including glucose, amino acids, and fatty acids or the recycling of acetate groups (Fig. 2 A). Three classes of histone deacetylases (HDAC classes I, II, and IV) remove acyl moieties from histone lysine residues though simple amide hydrolysis (Seto and Yoshida, 2014). Class III HDACs, also known as sirtuins, use a more complex method of deacylation in which the cosubstrate NAD+ is cleaved to form acyl-ADP-ribose and nicotinamide.
Enzymatic considerations for the metabolic regulation of gene expression
Thus, metabolites are integral components of the reactions that regulate chromatin modifications. A multitude of factors can influence the efficiency of these enzymatic reactions. As has been reviewed previously (Fan et al., 2015; Su et al., 2016; Reid et al., 2017), the KM of the enzyme must be equal to, or higher than, the range of substrate concentrations for physiological fluctuations in substrate concentration to affect the rate of the reaction. Therefore, if substrate concentrations are well above enzymatic KM, small variations in substrate concentration will not influence enzymatic rate. For example, cellular ATP concentrations (∼10 mM) are far higher than the levels needed to saturate kinase activity. Furthermore, in viable cells, the ADP and Pi levels are much lower than the ATP levels and therefore play essentially no role in product inhibition. Thus, kinases are unlikely to be influenced by fluctuations in substrate or product concentrations. In the case of enzymes that regulate methylation or acetylation, however, the measured KM is frequently in the range of observed substrate concentrations in cells (Su et al., 2016; Reid et al., 2017).
Therefore, a key question in the field is whether or not substrate concentrations are limiting or product levels are too high to allow constitutive activity of the suite of enzymes that regulate chromatin modifications. This is not a trivial question. First, several additional factors influence enzymatic rate, including cofactor availability, pH, and allosteric regulation by metabolites or enzymatic partners. As an example, high acetyl-CoA levels in the mitochondria (0.1–1.5 mM) coupled with high mitochondrial pH (7.9–8.0) enable nonenzymatic acetylation of mitochondrial proteins to proceed as an energetically favorable event (Wagner and Payne, 2013). At the lower levels of cytoplasmic acetyl-CoA (2–13 µM; Lee et al., 2014) coupled with a lower pH, acetylation of proteins occurs only if catalyzed by cytoplasmic and nuclear acetyltransferases (Wagner and Payne, 2013).
Second, the determination of enzyme KM in vitro does not necessarily provide a reliable measure of enzyme function in vivo. It is impossible to recapitulate in a test tube the complex microenvironment an enzyme would experience in cells, including the presence of multiple metabolites that could compete for the substrate binding site and thus raise the effective KM of the enzyme for its normal substrate. Similarly, measures of in vitro KM do not account for the inclusion of the enzyme in a multiprotein complex as found for the majority of chromatin-modifying enzymes (e.g., histone H3K4 methyltransferases as part of Set1/COMPASS [complex of proteins associated with Set1; Miller et al., 2001] or polycomb repressive complex 2 [PRC2] histone H3K27 methyltransferases as part of PRC2 [Klymenko et al., 2006] methyltransferase complexes). Moreover, local concentrations of enzyme cofactors, allosteric regulators, cosubstrates, end products, and any existing posttranslational modifications of the chromatin-modifying enzymes may influence in vivo KM.
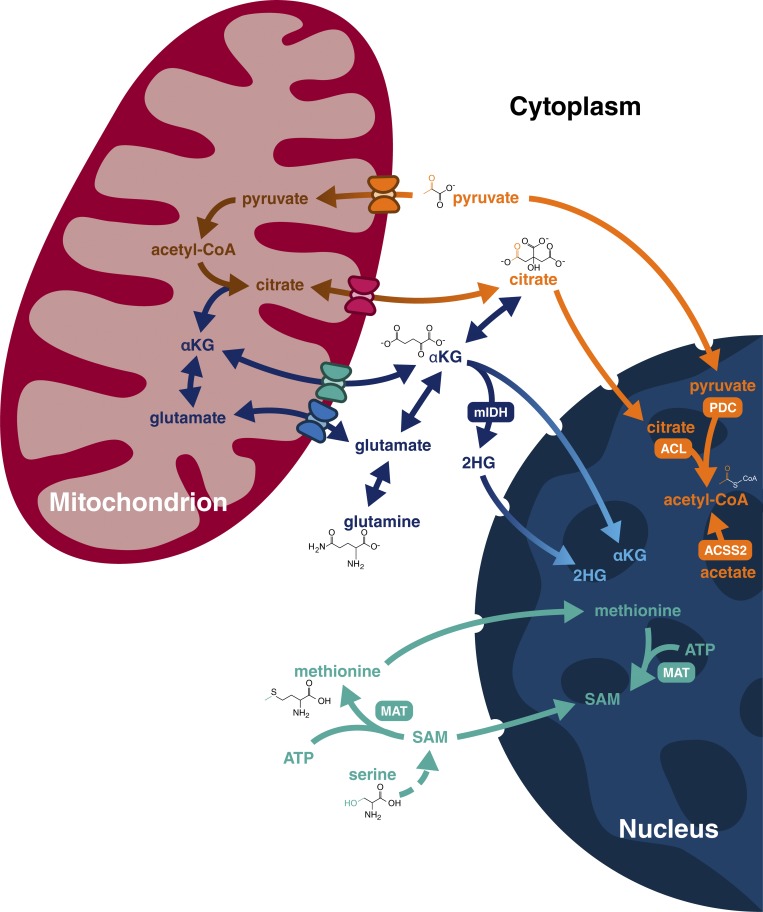
Third, it is almost impossible with current technology to estimate the local concentration of individual metabolites. To date, most estimates of metabolite availability are based on whole-cell analyses and thus scramble different pools with vastly different substrate concentrations (Chen et al., 2016). However, localization of enzymes that produce key metabolites can provide a clue as to the mechanisms cells use to generate local pools of substrates for chromatin modifications (Fig. 3). For example, the pyruvate dehydrogenase complex, traditionally found in the mitochondrial matrix adjacent to the inner mitochondrial membrane, can transit to the nucleus where it generates locally high concentrations of acetyl-CoA to fuel histone acetylation required for transcription of genes involved in S phase (Sutendra et al., 2014) or lipid biosynthesis (Chen et al., 2018). Similarly, nuclear ATP-citrate lyase generates acetyl-CoA pools upon DNA damage, facilitating the histone acetylation required for efficient double-strand break repair by homologous recombination (Sivanand et al., 2017). Another example that involves local nuclear production of acetyl-CoA depends on the function of acetyl-CoA synthetase 2 (ACSS2). Under normoxic conditions, cytoplasmically localized ACSS2 drives the use of acetate for lipogenesis (Kamphorst et al., 2014; Schug et al., 2015). Under hypoxia, however, ACSS2 translocates to the nucleus where it allows the recycling of acetate produced by histone deacetylation reactions for histone reacetylation (Bulusu et al., 2017).
Figure 3.
Subcellular localization of selected metabolic pathways. A simplified schematic depicting the interplay between mitochondrial, cytoplasmic, and nuclear metabolism. Metabolic enzymes found to localize to the nucleus: methionine adenosyl-transferase (MAT), ATP-citrate lyase (ACL), pyruvate dehydrogenase complex (PDC), and ACSS2. Metabolite transport across the inner mitochondrial membrane requires transporters that are shown in simplified form. Note: complete enzymatic reactions are not depicted; we highlight the metabolites directly relevant to chromatin regulation for simplicity.
SAM-generating enzymes can also localize to the nucleus. Methionine-adenosyltransferase complex II associates with the large protein complex associated with transcriptional repression of the COX2 locus mediated by methylation of H3K9 by the methyltransferase SETDB1 (Katoh et al., 2011; Kera et al., 2013). Interestingly, this mechanism is not restricted to targeting SAM for repressive functions. The serine-responsive SAM-containing metabolic enzyme (SESAME) complex interacts with the Set1 methyltransferase complex to coordinate H3K4 methylation and H3T11 phosphorylation at the pyruvate kinase M2 locus (Li et al., 2015), leading to its transcriptional activation. These examples suggest that despite the free passage of metabolites from cytoplasm to nucleus through the large nuclear pore complex, local generation of metabolite pools can have biologically relevant functions. Nonetheless, a great deal of work remains to explore how regional pools of metabolites might affect chromatin modifications. A combination of in nucleo chemical assays to probe enzyme responsiveness to variations in metabolites (Shrimp et al., 2015) and chemoproteomic approaches to quantitate metabolite/enzyme interactions in native contexts (Montgomery and Meier, 2016) may begin to answer this question.
Regulation of gene expression by metabolites
Metabolites as substrates
Despite the challenge of estimating the biochemical milieu of chromatin-modifying enzymes in living cells, increasing evidence suggests that changes in metabolite availability do influence chromatin modifications and ultimately gene expression in mammalian cells. In cultured cells, nucleocytosolic pools of acetyl-CoA for histone acetylation are derived primarily through the action of ATP-citrate lyase, which converts mitochondrial-derived citrate to oxaloacetate and acetyl-CoA. Signaling-induced changes in glucose oxidation and ATP-citrate lyase activity increase acetyl-CoA availability and total histone acetylation (Wellen et al., 2009; Lee et al., 2014). Outside idealized cell culture conditions, acetate may provide a major source of carbon for histone acetylation: in hypoxia and in tumors, conversion of acetate to acetyl-CoA by ACSS2 provides an additional source of nucleocytosolic acetyl-CoA and could represent an important recycling mechanism for recapture of acetate after histone deacetylation (Comerford et al., 2014; Schug et al., 2015; Bulusu et al., 2017).
Perturbations in SAM availability likewise influence total levels of histone methylation. In contrast to human cells or differentiated mouse cells, some strains of mouse embryonic stem cells (ESCs) can generate SAM from threonine catabolism; consequently, threonine depletion reduces H3K4me3 levels in these ESCs (Shyh-Chang et al., 2013). In both stem cells and differentiated cells, methionine restriction similarly reduces SAM pools and compromises histone methylation (Shiraki et al., 2014). Human pluripotent cells regulate SAM pools through the action of nicotinamide N-methyltransferase, an enzyme that consumes SAM and buffers low levels of H3K27me3 at key developmental genes (Sperber et al., 2015). In all cases, a major experimental hurdle is to uncouple direct effects on chromatin from secondary changes driven by reduced viability or proliferation that may result from depletion of critical metabolites. This is especially true for studies attempting to manipulate αKG levels, as cultured cells largely derive αKG from glutamine, a metabolite that is critical for proliferation and survival of cultured mammalian cells (Wise et al., 2008). However, mouse ESCs in the ground state of naive pluripotency exhibit the unusual ability to proliferate in the absence of exogenous glutamine. Restricting glutamine in these cells triggered accumulation of several trimethylated histone lysine residues, consistent with impaired αKG-dependent demethylation (Carey et al., 2015). Together, these cell-based studies have begun to lay the groundwork for our understanding of how metabolic networks provide the substrates that regulate chromatin modifications.
Metabolites as inhibitors
An additional determinant of enzymatic rate is the inhibitory effect of enzymatic products and other closely related metabolites. The biological importance of enzyme inhibition by small molecules is underscored by the observation that mutations in metabolic enzymes occur in a variety of human malignancies (Kaelin and McKnight, 2013). For example, loss-of-function mutations in the metabolic enzymes succinate dehydrogenase (SDH; Baysal et al., 2000) and fumarate hydratase (FH; Tomlinson et al., 2002) were originally associated with cancer syndromes including hereditary paraganglioma and pheochromocytomas and hereditary leiomyomatosis and renal cell carcinoma, respectively. Subsequent large scale sequencing studies found FH and SDH mutations in a number of other malignancies (Carvajal-Carmona et al., 2006; Bardella et al., 2011). Mechanistically, high levels of succinate and fumarate driven by loss of SDH or FH function, respectively, can act as competitive inhibitors of αKG-dependent enzymes. In particular, SDH and FH mutations are associated with impaired DNA and histone demethylation and the accumulation of repressive chromatin modifications that contribute to malignant progression (Laukka et al., 2016). In addition, high levels of succinate can inhibit the activity of EglN prolyl-4 hydroxylases, leading to stabilization of the heterodimeric transcription factors HIF1α and HIF2α, which may represent possible mediators of the oncogenic effects of succinate and fumarate accumulation seen in SDH and FH mutant tumors, respectively (Koivunen et al., 2012; Laukka et al., 2016).
The discovery of neomorphic point mutations in the genes coding for isocitrate dehydrogenase 1 and 2 in a number of different cancers was a key finding in expanding the understanding of the connections between metabolism and gene expression. Point mutations in IDH1 (R132) and IDH2 (R172K or R140Q) are seen in tumors including acute myelogenous leukemia (AML), grade II–III glioblastoma, chondrosarcoma, cholangiocarcinoma, and many other tumors at low frequencies. Wild-type IDH1 and IDH2 catalyze the interconversion of isocitrate and αKG, with concurrent reduction of NAD(P)+ to NAD(P)H and release of CO2 in the forward reaction. The neomorphic mutant enzymes catalyze the irreversible partial reduction of αKG to d-2-hydroxyglutarate (D-2HG), a metabolite not normally present in physiological conditions (Dang et al., 2009; Ward et al., 2010). D-2HG is in turn a competitive inhibitor of αKG-dependent enzymes (Xu et al., 2011).
As a result of D-2HG–mediated inhibition, tumors harboring IDH1 or IDH2 mutations show various degrees of hypermethylation of DNA and histone marks, which are thought to mediate the transcriptional changes that block terminal differentiation (Figueroa et al., 2010; Chowdhury et al., 2011; Lu et al., 2012). Interestingly, not all D-2HG effects are pro-oncogenic. In non-IDH mutant leukemias, D-2HG leads to inhibition of the m6A demethylase FTO and consequent hypermethylation and destabilization of Myc transcripts (Su et al., 2018).
Metabolites that accumulate in physiological conditions can also inhibit chromatin-modifying enzymes. For example, fasting-induced increases in circulating levels of the ketone body β-hydroxybutyrate inhibit class I HDACs in multiple tissues and induce expression of genes associated with resistance to oxidative stress (Shimazu et al., 2013). Similarly, short-chain fatty acids such as propionate and butyrate derived from commensal bacterial populations increase histone acetylation and promote extrathymic maturation of regulatory T cells (Arpaia et al., 2013). Low oxygen or acidic pH induce chromatin hypermethylation in part by stimulating the production of L-2HG (the enantiomer of D-2HG) by wild-type forms of malate dehydrogenase or lactate dehydrogenase (Intlekofer et al., 2015, 2017; Oldham et al., 2015). These selected examples illustrate how cell-autonomous and circulating metabolites serve as signals that transmit information about environmental changes to individual cells to induce appropriate adjustments in gene expression.
Metabolic modulation of enzyme activity
To maintain their activity, αKG-dependent dioxygenases use ascorbate (vitamin C) to facilitate regeneration of the required Fe(II) cofactor (Myllylä et al., 1978). Providing exogenous ascorbic acid potently enhances αKG-dependent dioxygenase activity (Knowles et al., 2003). This effect can have biological relevance. Defective collagen production—a process mediated by the αKG-dependent prolyl hydroxylases—is a major driver of the pathogenesis of scurvy. The therapeutic effect of vitamin C–rich fruits in patients with scurvy provides a dramatic example of the relevance of this cofactor for organismal homeostasis (Kuiper and Vissers, 2014).
Tet enzymes are known to be particularly sensitive to ascorbate levels, and multiple studies have shown that variations in ascorbate levels can affect stem cell biology via Tet enzymatic activity (Blaschke et al., 2013; Chen et al., 2013b; Minor et al., 2013; Yin et al., 2013; Cimmino et al., 2017). Metabolite profiling of isolated hematopoietic progenitors revealed 2–20-fold higher levels of ascorbate in hematopoietic stem cells (HSCs) and multipotent progenitors compared with other hematopoietic precursors (Agathocleous et al., 2017). Reducing ascorbate availability in vivo increased HSC frequency and function as a result of impaired Tet2 function. Consequently, ascorbate depletion results in increased leukemogenesis in the setting of a cooperating oncogene (Agathocleous et al., 2017). These collective findings suggest that HSC expansion is favored by the inhibition of Tet2 that results from decreased ascorbate. As such, conditions of ascorbate depletion would mimic loss of Tet2 and favor leukemogenesis in the setting of additional oncogenic events.
Integrating metabolism and gene expression
Transcription factors are required but may not be sufficient for cell fate decisions
Although metabolites may contribute to gene regulation, the ability of a cell to make state or fate changes relies on the expression of specific transcriptional programs. One explanation for how specific biological events lead to activation of appropriate transcriptional programs is through the DNA binding specificity of transcription factors. Transcription factors are hence an essential force determining cell identity, as demonstrated by the pivotal role of MyoD in directing muscle differentiation and factors such as Oct4, Sox2, and Klf4 in directing the acquisition of the pluripotent state (Davis et al., 1987; Takahashi and Yamanaka, 2006). At a first approximation, it is difficult to see how the metabolic changes discussed above can lead to specific transcriptional programs that effect cell specification. The modifications affected by intermediary metabolite levels are often present genome-wide, and the metabolites themselves are not localized to specific genomic loci.
However, gene expression is not simply a binary output driven by the presence or absence of transcription factor binding. Rather, gene expression is the result of multiple inputs including the recruitment of chromatin-modifying activities that deposit activating marks (e.g., acetylation at H3K9 and H3K27 and methylation at H3K4) and remove inhibitory marks (e.g., methylation of H3K9 and H3K27). These marks can in turn enhance gene expression by altering charge-driven nucleosomal contacts or promoting recruitment of additional coactivators. Gene expression therefore represents the integrated output of multiple disparate inputs, including metabolite availability. Taken together, metabolic alterations may influence the probability of expressing a given gene, and, therefore, in aggregate may contribute to the induction or repression of specific gene expression programs.
Modification-specific effects
A major challenge to the hypothesis that metabolites provide important information that links gene expression programs to environmental conditions is the problem of specificity. First, the same chemical modification on different residues can exert vastly different biological effects. For example, H3K9me3 and H3K27me3 are associated with gene repression (Mozzetta et al., 2015), H3K4me1 with gene enhancers, and H3K4me3 with gene expression (Shilatifard, 2012; Rao et al., 2017; Schuettengruber et al., 2017), and yet all of these modifications are derived from SAM. Likewise, the enzymatic removal of the trimethyllysine modification always requires αKG, regardless of the residue being modified. Second, to exert coherent effects on gene expression, marks must be deposited on specific regulatory regions and not distributed randomly genome-wide. How, then, do cells translate global shifts in metabolite availability into specific gene expression programs?
One answer to the first problem may be hardwired in the biochemistry of the enzymes themselves. The KM of each enzyme for its substrate can vary even between closely related members of a protein family, thus providing a hierarchy of enzymes that will be more or less sensitive to fluctuations in the availability of a specific substrate. For example, PCAF and GCN5, which acetylate H3K9 and H3K14 residues, exhibit a KM of ∼0.6 µM for acetyl-CoA, whereas p300, which acetylates H3K27 residues, has a 10-fold higher KM for acetyl-CoA (Jin et al., 2011; Fan et al., 2015). Acetyltransferases also exhibit diversity in their susceptibility to inhibition by free CoA (Montgomery et al., 2015).
In yeast, the H3K4 methyltransferase, Set1, has a comparatively high KM for SAM (Sadhu et al., 2013). Consequently, H3K4me3 is most sensitive to conditions that restrict SAM availability (Shyh-Chang et al., 2013; Shiraki et al., 2014; Mentch et al., 2015). Conversely, whereas most histone trimethyllysine modifications were increased by restricting intracellular αKG pools, H3K4me3 was comparatively unaffected (Carey et al., 2015). These findings lead to the intriguing hypothesis that activating histone modifications (acetylation, H3K4me3) may be sensitive to an ON rate driven by acetyl-CoA and SAM availability, whereas repressive modifications may be sensitive to an OFF rate mediated by αKG pools. In this manner, gene activation may require disparate metabolic inputs that signal nutrient sufficiency. Alternatively, metabolic regulation of individual marks might have context specificity: in addition to H3K4 methylation, H3K9 and H3K27 methylation were responsive to methionine restriction in vitro, although in vivo this could only be seen for H3K4 (Dann et al., 2015; Mentch et al., 2015).
Locus-specific effects
Even more challenging is the question of how metabolites could affect a particular chromatin modification in a site-specific manner. One possibility is that DNA sequence-specific transcription factors enable recruitment of chromatin-modifying complexes that respond to fluctuations in metabolite levels. Similarly, enzymes that produce or consume metabolites may also be directly recruited to specific chromatin loci to influence local substrate concentrations. Alternatively, the local landscape of transcription factors, chromatin-modifying enzymes, and chromatin readers may collectively determine the likelihood of specific marks being deposited or removed.
Although most examples of differential expression programs are a consequence of local changes in chromatin, some biological processes are driven by global alterations in the chromatin landscape. Global erasure of parental methylation patterns occurs early in vertebrate embryonal development and is followed by a transient pluripotent period marked by globally accessible chromatin. This has been observed in a number of species by both electron microscopy and the pattern of chromatin modifications (Ahmed et al., 2010; Borsos and Torres-Padilla, 2016).
Metabolites may play particularly important roles regulating the global chromatin landscape in stem cells, which are characterized by dynamic chromatin regulation. Mouse ESCs exhibit global chromatin hypomethylation (Ficz et al., 2013; Habibi et al., 2013; Hackett et al., 2013; Leitch et al., 2013; Pedersen et al., 2016; von Meyenn et al., 2016), and interventions that elevate αKG levels enhance demethylation and promote self-renewal of mouse ESCs (Carey et al., 2015; Hwang et al., 2016). In contrast, mouse epiblast stem cells and human primed pluripotent cells must undergo demethylation to differentiate. As a result, αKG accelerates differentiation of human ESCs (TeSlaa et al., 2016). This is also the metabolic node that has been coopted by tumor cells harboring IDH, SDH, and FH mutations. By preventing activation of genes associated with differentiation, these mutations lock malignant cells into a dedifferentiated state (Figueroa et al., 2010; Lu et al., 2012, 2013). AML stem cells can promote a similar hypermethylated state in the absence of IDH mutations by elevating activity of branched-chain amino acid transaminase 1, thus restricting intracellular levels of αKG (Raffel et al., 2017). Together, these findings suggest that αKG levels are limiting for cells that must demethylate regions of repressed chromatin to differentiate. Thus, pathological restriction of αKG or inhibition of αKG-dependent enzymes locks cells in a malignant state.
Changes in availability of acetyl-CoA donors for histone acetylation can likewise induce global chromatin alterations defined by alterations in bulk histone acetylation. Both mouse and human pluripotent cells consume high levels of glucose, which can be used to generate nucleocytosolic acetyl-CoA by the combined activities of the tricarboxylic acid (TCA) cycle and ATP-citrate lyase (Fig. 2 B; Zhou et al., 2012; Gu et al., 2016; Flores et al., 2017). Declines in glucose metabolism that accompany differentiation are associated with decreased acetyl-CoA availability and reduced histone acetylation (Moussaieff et al., 2015). Similarly, restricting acetyl-CoA availability via loss of ATP-citrate lyase has dramatic effects on broad gene expression programs, including the ability to respond to growth factors or differentiate (Wellen et al., 2009). Nevertheless, locus-specific regulation still occurs, as not all transcription is shut off when acetyl-CoA is limited. Bromodomain-containing proteins such as Brd4 that recognize acetylated lysines provide one major mechanism through which changes in histone acetylation can influence gene expression (Dey et al., 2003; Jang et al., 2005). The identification of recurrent translocations involving Brd4 in human cancer led to the development of JQ1, a small molecule that impairs the ability of Brd4 to bind acetylated lysine (Filippakopoulos et al., 2010). Although JQ1 has potent inhibitory effects on highly expressed genes such as Myc, many chromatin loci remain active despite the impaired ability of Brd4 to read acetylated histones in the presence of JQ1. Although the mechanisms responsible for this locus-specific JQ1 resistance are unclear, certain genetic loci may have an advantage in recruiting transcription factors and coactivators that mediate specific transcriptional programs (Finley et al., 2018). In support of this hypothesis, tumors driven by mutant IDH1 exhibit globally repressed chromatin and enhanced sensitivity to JQ1 (Chen et al., 2013a), suggesting that metabolic alterations that shape the chromatin landscape may determine sensitivity to perturbations in gene expression networks. This exemplifies the combinatorial effect of global metabolically mediated expression changes and locus-specific recruitment of transcriptional coactivators that collectively define gene expression programs.
Dysregulation in cancer
As previously discussed, mutations in metabolic enzymes result in accumulation of pathological levels of metabolites that can interfere with normal regulation of histone and DNA methylation (Figueroa et al., 2010; Chowdhury et al., 2011; Lu et al., 2012). Even in the absence of such mutations, cancer-associated changes in metabolic networks can induce alterations in the chromatin landscape (Sciacovelli et al., 2016). A recent study of pancreatic adenocarcinoma (PDAC) highlights the global effects of metabolic changes on gene expression in the context of metastatic progression (McDonald et al., 2017). Comparison of primary PDAC samples with liver and lung metastatic samples as well as primary cell lines derived from these tumors revealed widespread reprogramming of chromatin modifications at the level of regions rich in H3K9me3 and DNA methylation (large heterochromatin domains [LOCKs]). These changes occurred in the absence of metastasis-specific driver mutations, arguing for clonal chromatin drivers of metastatic progression. The decrease in repressive marks in LOCKs correlated with increased expression of genes contained in these regions in metastatic lines. Moreover, metastatic lines showed increased activity in the oxidative pentose phosphate pathway at the level of 6-phosphogluconate dehydrogenase (PGD) in glucose tracing experiments. Genetic or chemical inhibition of PGD, the enzymatic step that showed the most increased flux in metastatic lines compared with primary lines, was sufficient to revert the chromatin changes of metastatic lines to those seen in primary tumors. PGD inhibition also led to impaired metastatic ability in metastatic lines but not growth inhibition of the primary lines. These results argue that a clonal metabolic adaptation at the level of PGD leads to chromatin alterations that drive prometastatic behaviors conducive to dissemination and/or growth in highly vascularized organs such as the lung or liver where glucose concentrations are high. Although the direct link between higher PGD activity and loss of heterochromatic marks has not been explored, it is tempting to speculate that metabolic effects like the ones outlined in this review are directly responsible for the observed phenotype.
Another notable example of how master regulators of cellular metabolism can affect gene expression globally was recently outlined in the context of LKB1 mutant pancreatic adenocarcinoma (Kottakis et al., 2016). LKB1, a kinase that activates AMPK, is a tumor suppressor inactivated in a variety of malignancies, including pancreatic adenocarcinoma (Hawley et al., 2003; Woods et al., 2003; Shaw et al., 2004). Using genetically engineered mouse models harboring a KrasG12D and/or LKB1 homozygous loss, Kottakis et al. (2016) noted that a combination of KrasG12D mutation and LKB1 loss led to an mTOR-dependent rewiring of metabolism that resulted in increased activity through the serine-glycine-one carbon network. This in turn resulted in higher levels of SAM, increased global DNA methylation, and transcriptional silencing of retrotransposons. The effect was specific to KrasG12D;LKB1−/− cells and was validated in human pancreatic adenocarcinoma cell lines with this genetic makeup. Moreover, these cells were uniquely sensitive to DNA methyltransferase inhibitors, outlining a potential therapeutic vulnerability. This study exemplifies how changes in the metabolic state of a cell can have global effects on gene expression that are mediated through a change in availability of substrates of chromatin-modifying enzymes.
The interplay between metabolites and gene expression programs that sustain malignancy may provide opportunities for therapeutic intervention. For example, glutamine, which provides many critical metabolic functions that fuel tumor growth (DeBerardinis et al., 2007), also generates the αKG used by mutant IDH to produce 2HG. Consequently, IDH mutant cells are more sensitive than wild-type counterparts to glutaminase inhibitors (Elhammali et al., 2014; Emadi et al., 2014). Glutamine metabolism may affect chromatin even in tumors lacking IDH mutations. A number of studies have reported histone methylation changes after treatment with glutaminase inhibitors, although whether these effects are direct or a downstream consequence of cellular adaptations to the drug remains unknown (Simpson et al., 2012a,b). Reductive carboxylation of glutamine-derived αKG has been shown to contribute to acetyl-CoA pools (Metallo et al., 2011; Mullen et al., 2011; Wise et al., 2011). Therefore, glutaminase inhibitors may also affect gene expression by decreasing citrate pools that maintain acetyl-CoA levels. Hence, the glutamine dependence of cancer cells may reflect its contributions to carbon pools used for both demethylation and acetylation reactions.
Alterations in glutamine levels, a common feature of human tumors (Roberts and Frankel, 1949; Roberts and Tanaka, 1956; Kamphorst et al., 2015), may allow tumor cells to change their chromatin environment in ways that allow escape from oncogene addiction. Studies in a panel of xenograft tumors revealed that glutamine and αKG levels were decreased in the ischemic central areas of tumors compared with the tumor periphery, and these metabolic alterations contributed to increased histone methylation in tumor cores (Pan et al., 2016). Decreased αKG impaired activity of the histone demethylase KDM6B and resulted in H3K27 hypermethylation. In BRAFV600E mutant melanoma cells, low glutamine promoted features of dedifferentiation and resistance to BRAFV600E inhibitors driven in part by H3K27 methylation. Consequently, inhibiting the H3K27 methyltransferase EZH2 restored sensitivity to BRAFV600E inhibition (Pan et al., 2016). Thus, extracellular nutrient changes, in this case as a result of ischemia in poorly vascularized tumors, may have effects on tumor biology that are a result of changes to the chromatin landscape.
Conclusion
Across evolution, fluctuations in nutrient availability influence gene expression programs. Increasing evidence supports the hypothesis that the chromatin landscape of metazoan cells integrates nutrient sensing with transcriptional control to effectively regulate transcriptional programs. These effects of metabolites on gene expression provide a supplemental layer of regulation on top of the more canonical regulators of gene expression, including sequence-specific binding proteins.
Fundamental questions regarding how metabolites affect gene expression programs remain. What enzymes preferentially respond to fluctuations in metabolite availability? Can physiological changes in extracellular nutrient availability alter levels of intracellular metabolites to a degree sufficient to affect chromatin modifications? What are the factors that control local generation of metabolites that serve as substrates or inhibitors of chromatin modifying enzymes? How do specific chromatin regions become sensitive (or resistant) to effects of metabolism when similar chromatin marks are present genome wide? Can the chromatin-modifying enzymes that link intermediary metabolites with the recruitment of RNA polymerase be pharmacologically targeted for therapeutic effect in cancer or other disease states? Continued technological developments will aid the pursuit of these questions. Measures of in vivo metabolic activity of specific cell populations, determination of metabolite concentrations in subcellular compartments, acute genetic manipulation of developing tissues, and sequence-specific targeting of chromatin-modifying enzymes will collectively enable direct assessment of the extent to which changes in metabolite availability influence chromatin modifications, gene expression, and cell fate decisions in mammalian cells.
Acknowledgments
We thank members of the Thompson and Finley laboratories for discussion.
J.M. Schvartzman is supported by a Hope Funds for Cancer Research Postdoctoral Fellowship. L.W.S. Finley is a Dale F. Frey-William Raveis Charitable Fund Scientist supported by the Damon Runyon Cancer Research Foundation (DFS-23-17). This work was additionally supported by the Memorial Sloan Kettering Cancer Center Support Grant P30 CA008748.
C.B. Thompson is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He also serves on the board of directors of Merck and Charles River Laboratories.
The authors declare no further conflict of interest.
J.M. Schvartzman, C.B. Thompson, and L.W.S. Finley wrote the manuscript.
References
- Agathocleous M., Meacham C.E., Burgess R.J., Piskounova E., Zhao Z., Crane G.M., Cowin B.L., Bruner E., Murphy M.M., Chen W., et al. 2017. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 549:476–481. 10.1038/nature23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K., Dehghani H., Rugg-Gunn P., Fussner E., Rossant J., and Bazett-Jones D.P.. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 5:e10531 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N., Campbell C., Fan X., Dikiy S., van der Veeken J., deRoos P., Liu H., Cross J.R., Pfeffer K., Coffer P.J., and Rudensky A.Y.. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 504:451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardella C., Pollard P.J., and Tomlinson I.. 2011. SDH mutations in cancer. Biochim. Biophys. Acta. 1807:1432–1443. 10.1016/j.bbabio.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., et al. 2000. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 287:848–851. 10.1126/science.287.5454.848 [DOI] [PubMed] [Google Scholar]
- Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martínez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A., et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 500:222–226. 10.1038/nature12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsos M., and Torres-Padilla M.-E.. 2016. Building up the nucleus: Nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 30:611–621. 10.1101/gad.273805.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulusu V., Tumanov S., Michalopoulou E., van den Broek N.J., MacKay G., Nixon C., Dhayade S., Schug Z.T., Vande Voorde J., Blyth K., et al. 2017. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Reports. 18:647–658. 10.1016/j.celrep.2016.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Barragan G., Wit N., Meiser J., Dingler F.A., Pietzke M., Mulderrig L., Pontel L.B., Rosado I.V., Brewer T.F., Cordell R.L., et al. 2017. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature. 548:549–554. 10.1038/nature23481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Finley L.W.S., Cross J.R., Allis C.D., and Thompson C.B.. 2015. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 518:413–416. 10.1038/nature13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Carmona L.G., Alam N.A., Pollard P.J., Jones A.M., Barclay E., Wortham N., Pignatelli M., Freeman A., Pomplun S., Ellis I., et al. 2006. Adult leydig cell tumors of the testis caused by germline fumarate hydratase mutations. J. Clin. Endocrinol. Metab. 91:3071–3075. 10.1210/jc.2006-0183 [DOI] [PubMed] [Google Scholar]
- Chantranupong L., Wolfson R.L., and Sabatini D.M.. 2015. Nutrient-sensing mechanisms across evolution. Cell. 161:67–83. 10.1016/j.cell.2015.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Y., Lu C., Cross J.R., Morris J.P. IV, Shroff A.S., Ward P.S., Bradner J.E., Thompson C., and Lowe S.W.. 2013a Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 27:1974–1985. 10.1101/gad.226613.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z., et al. 2013b Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45:1504–1509. 10.1038/ng.2807 [DOI] [PubMed] [Google Scholar]
- Chen J., Guccini I., Mitri D.D., Brina D., Revandkar A., Sarti M., Pasquini E., Alajati A., Pinton S., Losa M., et al. 2018. Compartmentalized activities of the pyruvate dehydrogenase complex sustain lipogenesis in prostate cancer. Nat. Genet. 50:219–228. 10.1038/s41588-017-0026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.W., Freinkman E., Wang T., Birsoy K., and Sabatini D.M.. 2016. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 166:1324–1337.e11. 10.1016/j.cell.2016.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Yeoh K.K., Tian Y.-M., Hillringhaus L., Bagg E.A., Rose N.R., Leung I.K.H., Li X.S., Woon E.C.Y., Yang M., et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12:463–469. 10.1038/embor.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L., Dolgalev I., Wang Y., Yoshimi A., Martin G.H., Wang J., Ng V., Xia B., Witkowski M.T., Mitchell-Flack M., et al. 2017. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 170:1079–1095.e20. 10.1016/j.cell.2017.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford S.A., Huang Z., Du X., Wang Y., Cai L., Witkiewicz A.K., Walters H., Tantawy M.N., Fu A., Manning H.C., et al. 2014. Acetate dependence of tumors. Cell. 159:1591–1602. 10.1016/j.cell.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C., et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 462:739–744. 10.1038/nature08617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann S.G., Ryskin M., Barsotti A.M., Golas J., Shi C., Miranda M., Hosselet C., Lemon L., Lucas J., Karnoub M., et al. 2015. Reciprocal regulation of amino acid import and epigenetic state through Lat1 and EZH2. EMBO J. 34:1773–1785. 10.15252/embj.201488166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., and Lassar A.B.. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 51:987–1000. 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- DeBerardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S., and Thompson C.B.. 2007. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 104:19345–19350. 10.1073/pnas.0709747104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Chitsaz F., Abbasi A., Misteli T., and Ozato K.. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA. 100:8758–8763. 10.1073/pnas.1433065100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhammali A., Ippolito J.E., Collins L., Crowley J., Marasa J., and Piwnica-Worms D.. 2014. A high-throughput fluorimetric assay for 2-hydroxyglutarate identifies Zaprinast as a glutaminase inhibitor. Cancer Discov. 4:828–839. 10.1158/2159-8290.CD-13-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emadi A., Jun S.A., Tsukamoto T., Fathi A.T., Minden M.D., and Dang C.V.. 2014. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp. Hematol. 42:247–251. 10.1016/j.exphem.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Fan J., Krautkramer K.A., Feldman J.L., and Denu J.M.. 2015. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 10:95–108. 10.1021/cb500846u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.-L., Walter J., et al. 2013. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 13:351–359. 10.1016/j.stem.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 18:553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. 2010. Selective inhibition of BET bromodomains. Nature. 468:1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley L.W.S., Vardhana S.A., Carey B.W., Alonso-Curbelo D., Koche R., Chen Y., Wen D., King B., Radler M.R., Rafii S., et al. 2018. Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat. Cell Biol. 20:565–574. 10.1038/s41556-018-0086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A., Schell J., Krall A.S., Jelinek D., Miranda M., Grigorian M., Braas D., White A.C., Zhou J.L., Graham N.A., et al. 2017. Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat. Cell Biol. 19:1017–1026. 10.1038/ncb3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Ji M., Fang X., Liu Y., Yu Z., Cao Y., Sun A., Zhao L., and Zhang Y.. 2017. Capillary electrophoresis—Mass spectrometry metabolomics analysis revealed enrichment of hypotaurine in rat glioma tissues. Anal. Biochem. 537:1–7. 10.1016/j.ab.2017.08.012 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., and Razin A.. 1982. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 295:620–622. 10.1038/295620a0 [DOI] [PubMed] [Google Scholar]
- Gu W., Gaeta X., Sahakyan A., Chan A.B., Hong C.S., Kim R., Braas D., Plath K., Lowry W.E., and Christofk H.R.. 2016. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell. 19:476–490. 10.1016/j.stem.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H.D., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C., et al. 2013. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 13:360–369. 10.1016/j.stem.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., and Surani M.A.. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 339:448–452. 10.1126/science.1229277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Boudeau J., Reid J.L., Mustard K.J., Udd L., Mäkelä T.P., Alessi D.R., and Hardie D.G.. 2003. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28 10.1186/1475-4924-2-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.-Y., Kwak S., Lee S., Kim H., Lee S.E., Kim J.-H., Kim Y.A., Jeon Y.K., Chung D.H., Jin X., et al. 2016. Psat1-dependent fluctuations in α-ketoglutarate affect the timing of ESC differentiation. Cell Metab. 24:494–501. 10.1016/j.cmet.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Intlekofer A.M., Dematteo R.G., Venneti S., Finley L.W.S., Lu C., Judkins A.R., Rustenburg A.S., Grinaway P.B., Chodera J.D., Cross J.R., and Thompson C.B.. 2015. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 22:304–311. 10.1016/j.cmet.2015.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Wang B., Liu H., Shah H., Carmona-Fontaine C., Rustenburg A.S., Salah S., Gunner M.R., Chodera J.D., Cross J.R., and Thompson C.B.. 2017. L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat. Chem. Biol. 13:494–500. 10.1038/nchembio.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., and Monod J.. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318–356. 10.1016/S0022-2836(61)80072-7 [DOI] [PubMed] [Google Scholar]
- Jang M.K., Mochizuki K., Zhou M., Jeong H.-S., Brady J.N., and Ozato K.. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 19:523–534. 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Jin Q., Yu L.-R., Wang L., Zhang Z., Kasper L.H., Lee J.-E., Wang C., Brindle P.K., Dent S.Y.R., and Ge K.. 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30:249–262. 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G. Jr., and McKnight S.L.. 2013. Influence of metabolism on epigenetics and disease. Cell. 153:56–69. 10.1016/j.cell.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst J.J., Chung M.K., Fan J., and Rabinowitz J.D.. 2014. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2:23 10.1186/2049-3002-2-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst J.J., Nofal M., Commisso C., Hackett S.R., Lu W., Grabocka E., Vander Heiden M.G., Miller G., Drebin J.A., Bar-Sagi D., et al. 2015. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75:544–553. 10.1158/0008-5472.CAN-14-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Ikura T., Hoshikawa Y., Tashiro S., Ito T., Ohta M., Kera Y., Noda T., and Igarashi K.. 2011. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell. 41:554–566. 10.1016/j.molcel.2011.02.018 [DOI] [PubMed] [Google Scholar]
- Kera Y., Katoh Y., Ohta M., Matsumoto M., Takano-Yamamoto T., and Igarashi K.. 2013. Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J. Biol. Chem. 288:13592–13601. 10.1074/jbc.M112.429738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T., Papp B., Fischle W., Köcher T., Schelder M., Fritsch C., Wild B., Wilm M., and Müller J.. 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20:1110–1122. 10.1101/gad.377406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles H.J., Raval R.R., Harris A.L., and Ratcliffe P.J.. 2003. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 63:1764–1768. [PubMed] [Google Scholar]
- Koivunen P., Lee S., Duncan C.G., Lopez G., Lu G., Ramkissoon S., Losman J.A., Joensuu P., Bergmann U., Gross S., et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 483:484–488. 10.1038/nature10898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomeisky A.B. 2011. Physics of protein-DNA interactions: Mechanisms of facilitated target search. Phys. Chem. Chem. Phys. 13:2088–2095. 10.1039/C0CP01966F [DOI] [PubMed] [Google Scholar]
- Kottakis F., Nicolay B.N., Roumane A., Karnik R., Gu H., Nagle J.M., Boukhali M., Hayward M.C., Li Y.Y., Chen T., et al. 2016. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 539:390–395. 10.1038/nature20132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper C., and Vissers M.C.M.. 2014. Ascorbate as a co-factor for Fe- and 2-oxoglutarate dependent dioxygenases: Physiological activity in tumor growth and progression. Front. Oncol. 4:359 10.3389/fonc.2014.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukka T., Mariani C.J., Ihantola T., Cao J.Z., Hokkanen J., Kaelin W.G. Jr., Godley L.A., and Koivunen P.. 2016. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 291:4256–4265. 10.1074/jbc.M115.688762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.V., Carrer A., Shah S., Snyder N.W., Wei S., Venneti S., Worth A.J., Yuan Z.-F., Lim H.-W., Liu S., et al. 2014. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20:306–319. 10.1016/j.cmet.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A., et al. 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20:311–316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Cattoglio C., and Tjian R.. 2014. Looping back to leap forward: Transcription enters a new era. Cell. 157:13–25. 10.1016/j.cell.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. 2005. The lac repressor. C. R. Biol. 328:521–548. 10.1016/j.crvi.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Li S., Swanson S.K., Gogol M., Florens L., Washburn M.P., Workman J.L., and Suganuma T.. 2015. Serine and SAM responsive complex SESAME regulates histone modification crosstalk by sensing cellular metabolism. Mol. Cell. 60:408–421. 10.1016/j.molcel.2015.09.024 [DOI] [PubMed] [Google Scholar]
- Lu C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C.R., Khanin R., Figueroa M.E., Melnick A., et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 483:474–478. 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Venneti S., Akalin A., Fang F., Ward P.S., Dematteo R.G., Intlekofer A.M., Chen C., Ye J., Hameed M., et al. 2013. Induction of sarcomas by mutant IDH2. Genes Dev. 27:1986–1998. 10.1101/gad.226753.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F. 2018. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19:81–92. 10.1038/nrg.2017.80 [DOI] [PubMed] [Google Scholar]
- McDonald O.G., Li X., Saunders T., Tryggvadottir R., Mentch S.J., Warmoes M.O., Word A.E., Carrer A., Salz T.H., Natsume S., et al. 2017. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 49:367–376. 10.1038/ng.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch S.J., Mehrmohamadi M., Huang L., Liu X., Gupta D., Mattocks D., Gómez Padilla P., Ables G., Bamman M.M., Thalacker-Mercer A.E., et al. 2015. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22:861–873. 10.1016/j.cmet.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., et al. 2011. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 481:380–384. 10.1038/nature10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T., Krogan N.J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J.F., and Shilatifard A.. 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA. 98:12902–12907. 10.1073/pnas.231473398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor E.A., Court B.L., Young J.I., and Wang G.. 2013. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288:13669–13674. 10.1074/jbc.C113.464800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D.C., and Meier J.L.. 2016. Mapping Lysine Acetyltransferase–Ligand Interactions by Activity-Based Capture. In Enzymes of Epigenetics, Part B. New York, Elsevier; 105–123. 10.1016/bs.mie.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Montgomery D.C., Sorum A.W., Guasch L., Nicklaus M.C., and Meier J.L.. 2015. Metabolic regulation of histone acetyltransferases by endogenous acyl-CoA cofactors. Chem. Biol. 22:1030–1039. 10.1016/j.chembiol.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M., et al. 2015. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 21:392–402. 10.1016/j.cmet.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Mozzetta C., Boyarchuk E., Pontis J., and Ait-Si-Ali S.. 2015. Sound of silence: The properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 16:499–513. 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- Mullen A.R., Wheaton W.W., Jin E.S., Chen P.-H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., and DeBerardinis R.J.. 2011. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 481:385–388. 10.1038/nature10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllylä R., Kuutti-Savolainen E.-R., and Kivirikko K.I.. 1978. The role of ascorbate in the prolyl hydroxylase reaction. Biochem. Biophys. Res. Commun. 83:441–448. 10.1016/0006-291X(78)91010-0 [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., and Li E.. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Oldham W.M., Clish C.B., Yang Y., and Loscalzo J.. 2015. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 22:291–303. 10.1016/j.cmet.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Reid M.A., Lowman X.H., Kulkarni R.P., Tran T.Q., Liu X., Yang Y., Hernandez-Davies J.E., Rosales K.K., Li H., et al. 2016. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 18:1090–1101. 10.1038/ncb3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M.T., Kooistra S.M., Radzisheuskaya A., Laugesen A., Johansen J.V., Hayward D.G., Nilsson J., Agger K., and Helin K.. 2016. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 35:1550–1564. 10.15252/embj.201593317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. 1988. How eukaryotic transcriptional activators work. Nature. 335:683–689. 10.1038/335683a0 [DOI] [PubMed] [Google Scholar]
- Raffel S., Falcone M., Kneisel N., Hansson J., Wang W., Lutz C., Bullinger L., Poschet G., Nonnenmacher Y., Barnert A., et al. 2017. BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature. 551:384–388. 10.1038/nature24294 [DOI] [PubMed] [Google Scholar]
- Rao V.K., Pal A., and Taneja R.. 2017. A drive in SUVs: From development to disease. Epigenetics. 12:177–186. 10.1080/15592294.2017.1281502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid M.A., Dai Z., and Locasale J.W.. 2017. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 19:1298–1306. 10.1038/ncb3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E., and Frankel S.. 1949. Free amino acids in normal and neoplastic tissues of mice as studied by paper chromatography. Cancer Res. 9:645–648. [PubMed] [Google Scholar]
- Roberts E., and Tanaka T.. 1956. Free amino acids of the Yoshida ascites tumor. Cancer Res. 16:204–210. [PubMed] [Google Scholar]
- Sabari B.R., Zhang D., Allis C.D., and Zhao Y.. 2017. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18:90–101. 10.1038/nrm.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhu M.J., Guan Q., Li F., Sales-Lee J., Iavarone A.T., Hammond M.C., Cande W.Z., and Rine J.. 2013. Nutritional control of epigenetic processes in yeast and human cells. Genetics. 195:831–844. 10.1534/genetics.113.153981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S., Bernecky C., and Cramer P.. 2015. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16:129–143. 10.1038/nrm3952 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.-M., Di Croce L., and Cavalli G.. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 171:34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Schug Z.T., Peck B., Jones D.T., Zhang Q., Grosskurth S., Alam I.S., Goodwin L.M., Smethurst E., Mason S., Blyth K., et al. 2015. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 27:57–71. 10.1016/j.ccell.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacovelli M., Gonçalves E., Johnson T.I., Zecchini V.R., da Costa A.S.H., Gaude E., Drubbel A.V., Theobald S.J., Abbo S.R., Tran M.G.B., et al. 2016. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 537:544–547. 10.1038/nature19353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., and Yoshida M.. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6:a018713 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.J., Kosmatka M., Bardeesy N., Hurley R.L., Witters L.A., DePinho R.A., and Cantley L.C.. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA. 101:3329–3335. 10.1073/pnas.0308061100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., and Shi Y.. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 119:941–953. 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81:65–95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T., Hirschey M.D., Newman J., He W., Shirakawa K., Le Moan N., Grueter C.A., Lim H., Saunders L.R., Stevens R.D., et al. 2013. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 339:211–214. 10.1126/science.1227166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F., and Kume S.. 2014. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19:780–794. 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Shrimp J.H., Sorum A.W., Garlick J.M., Guasch L., Nicklaus M.C., and Meier J.L.. 2015. Characterizing the covalent targets of a small molecule inhibitor of the lysine acetyltransferase P300. ACS Med. Chem. Lett. 7:151–155. 10.1021/acsmedchemlett.5b00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J.J., Zhu H., et al. 2013. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 339:222–226. 10.1126/science.1226603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson N.E., Tryndyak V.P., Beland F.A., and Pogribny I.P.. 2012a An in vitro investigation of metabolically sensitive biomarkers in breast cancer progression. Breast Cancer Res. Treat. 133:959–968. 10.1007/s10549-011-1871-x [DOI] [PubMed] [Google Scholar]
- Simpson N.E., Tryndyak V.P., Pogribna M., Beland F.A., and Pogribny I.P.. 2012b Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics. 7:1413–1420. 10.4161/epi.22713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand S., Rhoades S., Jiang Q., Lee J.V., Benci J., Zhang J., Yuan S., Viney I., Zhao S., Carrer A., et al. 2017. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell. 67:252–265.e6. 10.1016/j.molcel.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K.A., Devi A., Detraux D., Gu H., et al. 2015. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 17:1523–1535. 10.1038/ncb3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., Deng X., Wang Y., Weng X., Hu C., et al. 2018. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 172:90–105.e23. 10.1016/j.cell.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Wellen K.E., and Rabinowitz J.D.. 2016. Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 30:52–60. 10.1016/j.cbpa.2015.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T.H., Haromy A., Hashimoto K., Zhang N., Flaim E., and Michelakis E.D.. 2014. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 158:84–97. 10.1016/j.cell.2014.04.046 [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S.. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- TeSlaa T., Chaikovsky A.C., Lipchina I., Escobar S.L., Hochedlinger K., Huang J., Graeber T.G., Braas D., and Teitell M.A.. 2016. α-Ketoglutarate accelerates the initial differentiation of primed human pluripotent stem cells. Cell Metab. 24:485–493. 10.1016/j.cmet.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P., and Kouzarides T.. 2014. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 15:703–708. 10.1038/nrm3890 [DOI] [PubMed] [Google Scholar]
- Tomlinson I.P.M., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E.M., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., et al. Multiple Leiomyoma Consortium . 2002. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30:406–410. 10.1038/ng849 [DOI] [PubMed] [Google Scholar]
- von Meyenn F., Iurlaro M., Habibi E., Liu N.Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z., et al. 2016. Impairment of dna methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell. 62:848–861. 10.1016/j.molcel.2016.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G.R., and Payne R.M.. 2013. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288:29036–29045. 10.1074/jbc.M113.486753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P.S., Patel J., Wise D.R., Abdel-Wahab O., Bennett B.D., Coller H.A., Cross J.R., Fantin V.R., Hedvat C.V., Perl A.E., et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 17:225–234. 10.1016/j.ccr.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., and Thompson C.B.. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 324:1076–1080. 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D.R., DeBerardinis R.J., Mancuso A., Sayed N., Zhang X.-Y., Pfeiffer H.K., Nissim I., Daikhin E., Yudkoff M., McMahon S.B., and Thompson C.B.. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 105:18782–18787. 10.1073/pnas.0810199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D.R., Ward P.S., Shay J.E.S., Cross J.R., Gruber J.J., Sachdeva U.M., Platt J.M., DeMatteo R.G., Simon M.C., and Thompson C.B.. 2011. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA. 108:19611–19616. 10.1073/pnas.1117773108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Johnstone S.R., Dickerson K., Leiper F.C., Fryer L.G.D., Neumann D., Schlattner U., Wallimann T., Carlson M., and Carling D.. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13:2004–2008. 10.1016/j.cub.2003.10.031 [DOI] [PubMed] [Google Scholar]
- Xu F., Zhang K., and Grunstein M.. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 121:375–385. 10.1016/j.cell.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.-H., Ito S., Yang C., Wang P., Xiao M.-T., et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 19:17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R., Mao S.-Q., Zhao B., Chong Z., Yang Y., Zhao C., Zhang D., Huang H., Gao J., Li Z., et al. 2013. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135:10396–10403. 10.1021/ja4028346 [DOI] [PubMed] [Google Scholar]
- Yung P.Y.K., and Elsässer S.J.. 2017. Evolution of epigenetic chromatin states. Curr. Opin. Chem. Biol. 41:36–42. 10.1016/j.cbpa.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C., and Ruohola-Baker H.. 2012. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 31:2103–2116. 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]