Abstract
Rapid impulse propagation is a defining attribute of the pectinated atrial myocardium and His-Purkinje system (HPS) that safeguards against atrial and ventricular arrhythmias, conduction block, and myocardial dyssynchrony. The complex transcriptional circuitry that dictates rapid conduction remains incompletely understood. Here, we demonstrate that ETV1 (ER81)-dependent gene networks dictate the unique electrophysiological characteristics of atrial and His-Purkinje myocytes. Cardiomyocyte-specific deletion of ETV1 results in cardiac conduction abnormalities, decreased expression of rapid conduction genes (Nkx2–5, Gja5, and Scn5a), HPS hypoplasia, and ventricularization of the unique sodium channel properties that define Purkinje and atrial myocytes in the adult heart. Forced expression of ETV1 in postnatal ventricular myocytes (VMs) reveals that ETV1 promotes a HPS gene signature while diminishing ventricular and nodal gene networks. Remarkably, ETV1 induction in human induced pluripotent stem cell-derived cardiomyocytes increases rapid conduction gene expression and inward sodium currents, converting them towards a HPS phenotype. Our data identify a cardiomyocyte-autonomous, ETV1-dependent pathway that is responsible for specification of rapid conduction zones in the heart and demonstrate that ETV1 is sufficient to promote a HPS transcriptional and functional program upon VMs.
Introduction
Rapidly conducting atrial and Purkinje cells (PC) and slowly conducting, impulse generating nodal cells cooperate to coordinate the heartbeat1. As a result, vertebrate animals establish a primitive conduction hierarchy during embryonic development concurrently with the formation of the early heart tube using highly conserved genetic pathways2–6. Rapid conduction myocytes form in pectinated and trabecular regions during cardiac chamber formation and mature into the pectinated atrial myocardium (PAM) and the His-Purkinje system (HPS), respectively. Rapid cardiac conduction is essential for normal rhythm maintenance7,8 and optimization of synchronous contraction of the cardiac chambers. Consequently, dysregulation of rapid conduction regions produce a large burden of arrhythmic diseases, including atrial fibrillation and Purkinje-mediated ventricular arrhythmias7–10. In addition, conduction block in the HPS is associated with increased mortality in heart failure patients due to myocardial dyssynchrony. Despite their clinical importance, gene regulatory networks (GRNs) that establish rapid conduction in the heart remains poorly defined. In particular, PCs remain the least characterized of all cardiac cell types due in part to their low abundance in the adult heart, comprising less than 1% of the total cell population11.
Seminal work revealed that the His-Purkinje system (HPS) originates from a cardiomyocyte lineage of trabecular myocytes during heart development12–15. Trabecular formation occurs though endocardial-derived neuregulin-1 (NRG1) signaling to cardiomyocytes through its cognate receptor complex ErbB4 and ErbB216. Targeted deletion of Nrg1, Erbb4, or Erbb2 results in the absence of the rapidly conducting trabecular zone in the mouse heart16–18. Moreover, NRG1 is sufficient to convert developing cardiomyocytes towards a rapid conduction cell fate19–21. These studies reinforce that NRG1 dependent signaling engage gene regulatory networks (GRNs) within cardiomyocytes, which are responsible for trabecular and subsequent HPS development.
A mounting body of evidence now shows that cell type-enriched transcription factors (TFs) orchestrate rapid impulse conduction GRNs in these specialized cardiomyocytes. A handful of TFs important for PC specification and function have been identified, including Nkx2-522–25, Tbx526,27, Tbx328–30, Id231, Hopx32, Irx333,34, Irx535,36, and Etv121, however their transcriptional abundance, cardiomyocyte-autonomous molecular function, and requirement for adult cardiac conduction health remain incompletely investigated. What has been clearly demonstrated is these TFs influence PCs to acquire a unique gene signature, compared to surrounding ventricular myocytes (VMs), through the expression of specialized ion channels and gap junction proteins37. Specifically, the enriched expression of the voltage-gated sodium channel protein NaV1.5 (encoded by Scn5a) and the gap junction channel protein Cx40 (encoded by Gja5) enable rapid conduction within the developing and mature heart38–40. Recently, we identified a critical role for the TF ETV1 in NRG1-dependent regulation of fast conduction biology21. ETV1, a member of the Pea3 group of ETS family TFs, is a transcriptional activator regulated through Ras-MAPK-dependent pathways in multiple cells types and functions at the cross roads between cellular proliferation, differentiation, and patterning41–46. In the heart, ETV1 is highly expressed in the developing and mature HPS and pectinated atrial myocardium (PAM)21. Previously, we showed in a germline knockout model that ETV1 is necessary for the expression of fast conduction genes (Nkx2–5, Gja5, Scn5a), determines the unique biophysical properties of the sodium current in atrial and PCs, and promotes proper formation of the HPS in the adolescent mouse heart21. In addition, we found that an ETV1 sequence variant associates with bundle branch and atrioventricular blocks in humans21. Taken together, these data identified ETV1 as a critical regulator of the HPS gene program in cardiomyocytes. However, several questions remain unanswered, including whether the effects of ETV1 knockout reflect a cell-autonomous defect in myocytes, and whether ETV1 is sufficient to reprogram a fast conduction cell fate on rodent and human cardiomyocytes. In this study, we investigated the cardiomyocyte-specific role of ETV1 in regulation of PC specification, rapid conduction GRNs, and electrophysiology in murine and human model systems.
Results
Etv1 defines progressive maturation of rapid conduction zones in the heart
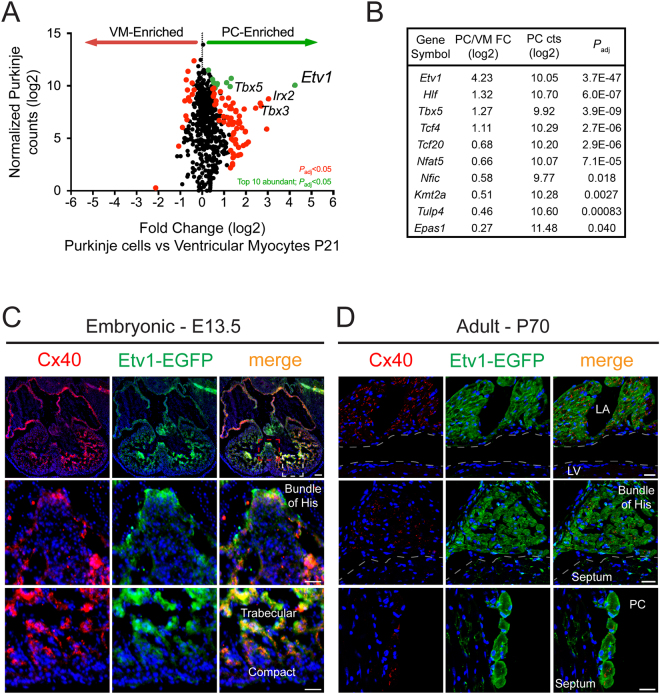
Specific genetic markers of the HPS have facilitated investigation of the molecular circuitry regulating PC specification, patterning, and function48–54. Making use of a Cntn2-EGFP BAC genetic reporter53, we developed and verified a fluorescence-activated cell sorting (FACS)-based purification strategy to isolate PCs and surrounding VMs (Supplemental Fig. 1)47. Leveraging advances in next generation sequencing technology, we performed genome-wide RNA-seq on dissociated ventricles from postnatal day 21 (P21) wild type (WT) mice in a Cntn2-EGFP transgenic background. RNA-seq differential expression analysis between PCs and VMs identified 2,161 differentially expressed protein-coding genes from a total of 12,496 genes with detectible counts in cardiomyocytes (normalized counts > 5). A Kyoto Encyclopedia of Genes and Genomes (KEGG) functional analysis on the 1,336 PC enriched genes revealed an overrepresented number of neuronal gene networks, suggesting TFs and effectors that regulate neuronal development may play a role in cardiac PC development (Supplemental Fig. 2). Of all TFs, Etv1 was the most differently expressed transcript in PCs compared to VMs (Fig. 1A). Tbx3, Tbx5, and Irx2 were also differentially expressed between PCs and VMs in line with previous reports (Fig. 1A)26,30,55. Etv1 was among the most abundant, significantly enriched PC TFs, along with Tbx5 (Fig. 1B).
Figure 1.
RNA Sequencing of fluorescence-activated cell sorted (FACS) postnatal day 21 (P21) mouse Purkinje cells (PC) and ventricular myocytes (VM). (A) Expression of PC (n = 7) and VM (n = 10) transcription factors (TFs) at P21. Differential expression (PC vs VM) on the X-axis and Purkinje transcript levels (normalized counts) are shown on the Y-axis. Significantly enriched (Padj < 0.05) and top ten abundant PC-TFs labeled in green while all remaining significantly different TFs were labeled in red. TFs that were not significantly different labeled in black. (B) Top ten significantly enriched and abundant TFs in PCs. Etv1 was the most enriched and one of the most abundant TFs within PCs. (C,D) Etv1-enhanced green fluorescent protein (EGFP) bacterial artificial chromosome (BAC) transgenic mice were used to confirm regional expression of Etv1 in embryonic (C) and adult (D) hearts. (C) Immunohistochemistry on E13.5 Etv1-EGFP hearts showing overlapping expression of Connexin-40 (Cx40) and GFP in trabecular and atrial myocytes. Bottom two rows are high-magnification views of the Bundle of His (red dashed box) and Trabecular/Compact zone myocytes (white dashed box) outlined in the top row (D) Immunohistochemistry on P70 Etv1-EGFP hearts showing overlapping expression of Cx40 and GFP in atrial myocytes (top row), bundle of His (middle row), and PCs (bottom row). Scale bars: 100 μm (top) 50 μm (middle, bottom) (C); 25 μm (D).
To verify cell-type specific expression of Etv1 in the heart, we utilized Etv1-EGFP BAC transgenic reporter mice56. Examination of embryonic day 13.5 (E13.5) and P70 Etv1-EGFP sections showed GFP expression in rapid conduction regions of the developing and adult heart in agreement with other Etv1-reporter mice (Fig. 1C,D). At E13.5, Etv1 was expressed specifically in Cx40-positive atrial myocytes (AM), bundle of His, and trabecular myocytes (Fig. 1C). Interestingly, we observed a transmural gradient of Etv1-EGFP expression in the developing ventricles with the highest expression at the trabeculae tips adjacent to the endocardium, the region of terminal PC specification (Fig. 1C, lower panels). In the adult heart, Etv1 expression remained in Cx40-positive AM and the bundle of His (Fig. 1D). Ventricular Etv1 expression was restricted to Cx40-positive mature PCs and absent in VMs (Fig. 1D, lower panels).
Cardiomyocyte-specific ETV1 deletion causes cardiac conduction defects
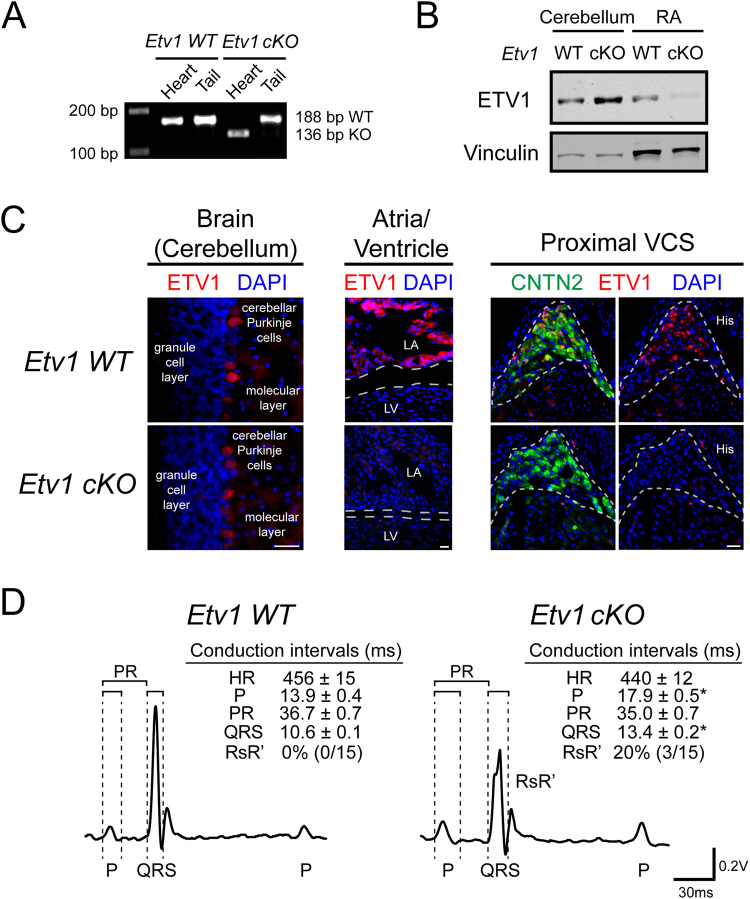
Germline Etv1-deficient mice are runted and die by three-weeks of age due to a progressive neuromuscular defect, limiting cardiac electrophysiological assessments at adult stages43. To circumvent these shortcomings, we generated cardiomyocyte-specific Etv1 null mice by crossing Etv1-floxed mice57 with transgenic Myh6-Cre mice, which express Cre recombinase under the regulatory sequences of the cardiac alpha (α)-myosin heavy chain gene58, hereby referred to as Etv1 cKO mice (Etv1fl/fl; Myh6-Cre). Genomic DNA analysis revealed that Etv1 exon 11 was specifically deleted in the heart of Etv1 cKO mice, but unaltered in non-cardiomyocytes (Fig. 2A). Absence of ETV1 protein was confirmed via immunoblots demonstrating specific loss of right atrial ETV1 expression in Etv1 cKO mouse hearts while ETV1 expression remained preserved in cerebellar brain and WT (Etv1fl/fl) tissues (Fig. 2B). Immunofluorescence of Etv1 WT and cKO heart and brain sections confirmed cell-type specific absence of ETV1 in AM and HPS while expression was unaltered in cerebellar Purkinje cells (Fig. 2C).
Figure 2.
Cardiomyocyte-specific knockout of ETV1 slows atrial and His-Purkinje system conduction. Etv1flox/flox mice crossed with Myh6-Cre transgenic mice enabled generation of cardiac-specific Etv1 mutant mice. (A) Heart and tail RT-PCR analysis of DNA from Etv1 WT (Etv1flox/flox) and Etv1 cKO (Etv1flox/flox, Myh6-Cre) mice demonstrate heart specific deletion of Etv1 exon 11. (B) Immunoblot assessment of Etv1 WT and Etv1 cKO tissue extracts. Protein from cerebellum and right atrial lysates were electrophoresed and immunoblotted to detect ETV1 (top) and Vinculin (bottom). (C) Immunofluorescence evaluation of ETV1 expression in 10-week-old Etv1 WT and Etv1 cKO cerebellar Purkinje cells, atrial myocytes, and His-Purkinje system (HPS). Positive CNTN2 expression identified HPS cells. Nuclei were identified by DAPI (blue). (D) Representative surface ECG traces of 10-week-old Etv1 WT and Etv1 cKO mice. Conduction intervals from 10–12-week-old Etv1 cKO mice compared to Etv1 WT mice showed significantly prolonged P wave and QRS intervals. 20% of Etv1 cKO mice displayed an RsR’ pattern. LA, left atria; LV, left ventricle. Data represent mean ± SEM. Scale bars: 25 um.
In order to examine the effect of Etv1 deletion in the adult heart, echocardiograms and electrocardiograms (ECGs) were performed on anesthetized 10–12-week-old Etv1 WT and cKO mice. Etv1 cKO mice survive to adulthood and maintain normal body weight in contrast to Etv1 germline KO mice (Data not shown). Cardiomyocyte loss of ETV1 resulted in no significant differences in cardiac size or contractility (Supplemental Table 1), however surface ECG of Etv1 cKO mice revealed significant electrophysiological deficits. Etv1 cKO mice had prolonged P wave duration and a widened QRS interval with a 20% prevalence of bundle branch block (RsR’) (Fig. 2D, Supplemental Fig. 3, Supplemental Table 2). RsR’ pattern was not observed in Etv1 WT animals.
Cardiomyocyte-specific ETV1 deletion disrupts expression of genes required for rapid conduction
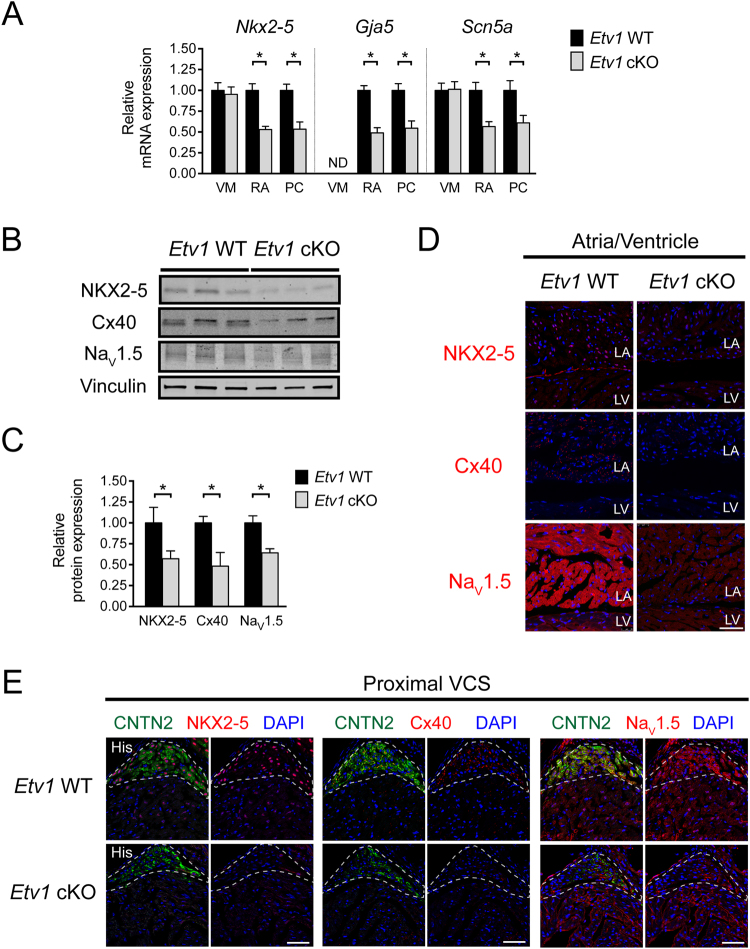
To explore the molecular mechanisms underlying the observed electrophysiological defects in Etv1 cKO mice, we probed whether rapid conduction gene networks were perturbed in these hearts. To test whether these genes were altered due to cardiomyocyte loss of ETV1, we performed quantitative RT-PCR (qPCR) on FACS-purified VMs, AMs, and PCs from 10–12-week-old Etv1 cKO and WT hearts in a Cntn2-EGFP background. Nkx2–5, Gja5, and Scn5a expression was significantly reduced in Etv1 cKO AMs and PCs compared to WT controls while unaltered in VMs where ETV1 is not expressed (Fig. 3A, Supplemental Table 3). In agreement, immunoblots demonstrated reduction of NKX2–5, Cx40, and NaV1.5 protein in Etv1 cKO atrial lysates compared to WT controls (NKX2–5, 57% ± 9%; Cx40, 48% ± 16%; and NaV1.5, 64% ± 5%; P < 0.05; cKO vs. WT) (Fig. 3B,C, Supplemental Table 4). Regional protein expression was further evaluated using immunofluorescence to identify altered expression of fast conduction genes in Etv1 cKO and WT cardiac regions. (Fig. 3D,E) The enrichment of NKX2–5, Cx40, and NaV1.5 in WT AMs (Fig. 3D) and the proximal HPS (Fig. 3E) were reduced to ventricular levels with cardiomyocyte loss of ETV1.
Figure 3.
Cardiomyocyte deletion of ETV1 resulted in decreased expression of fast conduction genes in atrial and His-Purkinje system (HPS) myocytes. (A) Quantitative RT-PCR of fast conduction gene RNA levels (normalized to Gapdh) comparing 10–12-week-old Etv1 WT (Etv1flox/flox) and Etv1 cKO (Etv1flox/flox, Myh6-Cre) FACS-purified ventricular, atrial, and Purkinje myocytes. Relative Nkx2–5, Gja5, and Scn5a expression displayed versus control, Etv1 WT (n = 4). (B) Immunoblot assessment of Etv1 WT and Etv1 cKO atrial tissue lysates detecting NKX2–5, Cx40, NaV1.5, and Vinculin (loading control). (C) Protein level densitometric quantification (normalized to vinculin), displayed relative to Etv1 WT (n = 5). (D) Immunofluorescence evaluation of NKX2–5, Cx40, and NaV1.5 expression in 10-week-old Etv1 WT and Etv1 cKO atria/ventricular sections. (E) Immunofluorescence evaluation of NKX2–5, Cx40, and NaV1.5 expression in 10-week-old Etv1 WT and Etv1 cKO HPS sections. Positive CNTN2 expression identified HPS cells. Nuclei were identified by DAPI (blue). LA, left atria; LV, left ventricle. Data represent mean ± SEM. *P < 0.05, 2-tailed Student’s t test. Scale bars: 50 um.
Hypoplasia of the His-Purkinje system in cardiomyocyte-specific Etv1 knockout mice
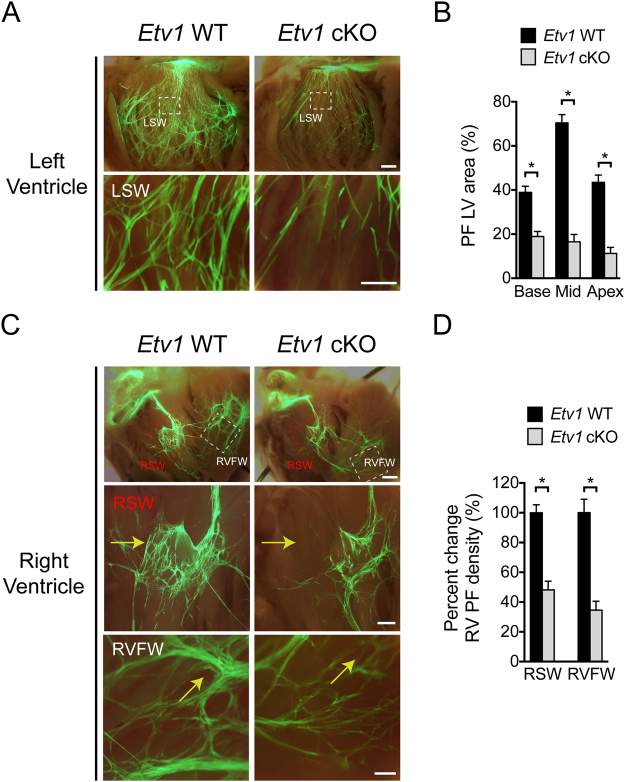
Cardiomyocyte loss of ETV1 resulted in diminished expression of Nkx2–5 in AM and PCs, suggesting that Etv1 cKO mice might have an underlying HPS structural deficit that phenocopies defects in Nkx2–5 heterozygous mice. To further explore the mechanism underlying conduction deficits in Etv1 cKO, we used the Cntn2-EGFP reporter line to visualize the morphology of the adult left and right HPS using whole mount techniques. In these mice, cardiomyocyte loss of ETV1 resulted in a hypomorphic left and right HPS mirroring the defects observed in Nkx2–5 heterozygous mice (Fig. 4A,C)22. Regional analysis of the left and right HPS showed a significant reduction of PC density in all regions of the heart (Fig. 4B,D). Taken together, we conclude that ETV1-dependent signaling pathways in developing cardiomyocytes play a critical role in the development of the mature HPS.
Figure 4.
ETV1 is required for normal development of the His-Purkinje system (HPS). Etv1flox/flox mice backcrossed with Cntn2-EGFP were used to visualize the left and right HPS. (A) Representative images of the left ventricle (LV) HPS of 10-week-old Etv1 WT (Etv1flox/flox) Cntn2-EGFP and Etv1 cKO (Etv1flox/flox, Myh6-Cre) Cntn2-EGFP hearts. Lower row is a high-magnification image of the white dashed boxes in the top row. (B) Quantification of EGFP+ Purkinje cell density (base, mid and apex) in Etv1 WT and Etv1 cKO hearts. Percentage of EGFP+ Purkinje cell area was significantly reduced on the left ventricular septal wall (LSW) in Etv1 cKO hearts versus control (n = 3). (C) Representative images of the right ventricle (RV) HPS of 10-week-old Etv1 WT Cntn2-EGFP and Etv1 cKO Cntn2-EGFP hearts. Middle and bottom rows are high-magnification views of red and white dashed boxes outlined in the top row. Yellow arrows highlight regional loss of HPS cells. (D) Percentage of EGFP+ Purkinje cell area was significantly reduced on the RV septum wall (RSW) and RV free wall (RVFW) in Etv1 cKO Cntn2-EGFP hearts versus control (n = 3). Data represent mean ± SEM. *P < 0.05, 2-tailed Student’s t test. Scale bars: 1 mm (top) (A,B), 200um (middle, bottom) (A,B).
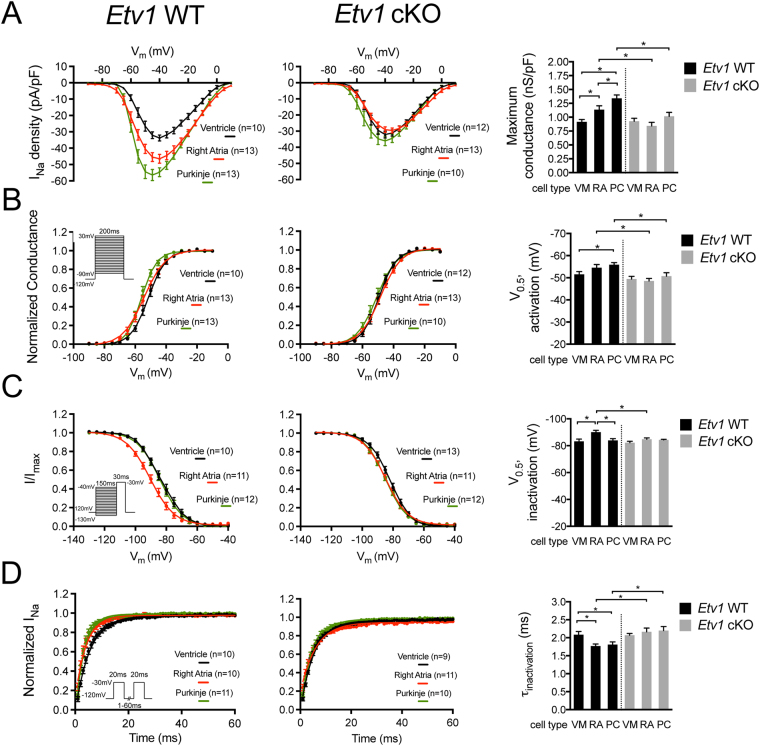
Single-cell electrophysiology analysis reveals homogenization of sodium channel properties between Etv1-null ventricular, atrial, and Purkinje cardiomyocytes
Given that Etv1 cKO hearts show a significant reduction of Scn5a in AMs and PCs, we investigated whether cardiomyocyte-specific loss of ETV1 would influence sodium current properties in dissociated adult myocytes. We recorded sodium currents from voltage clamped 10–12-week-old Etv1 WT and cKO dissociated VMs, AMs, and PCs. In agreement with previous studies59,60, voltage step commands elicited the greatest sodium current densities in WT PCs followed by AM and lastly VMs (maximum conductance: 1.34 ± 0.06 nS/pF, 1.14 ± 0.07 nS/pF, 0.92 ± 0.04 nS/pF, respectively) (Fig. 5A, Supplemental Tables 5 and 6). Sodium channels in WT right AMs and PCs underwent steady-state activation at more negative potentials compared to VMs (Fig. 5B, Supplemental Tables 5 and 7). Sodium channels in WT right AMs underwent steady-state inactivation at more negative potentials compared to VMs and PCs (Fig. 5C, Supplemental Tables 5 and 8). Sodium channel recovery from inactivation was faster in right AMs and PCs compared to VMs (Fig. 5D, Supplemental Tables 5 and 9). Cardiomyocyte loss of ETV1 altered the sodium current biophysical properties of AMs and PCs to resemble the sodium current identity of VMs (Fig. 5A–D, Supplemental Tables 5–8).
Figure 5.
ETV1 regulates the diversity of sodium channel biophysical properties between ventricular, atrial, and Purkinje myocytes. Whole-cell patch clamp data from dissociated cardiomyocytes (ventricular, right atrial, Purkinje myocytes) using 10–12 week-old Etv1 WT (Etv1flox/flox) and Etv1 cKO (Etv1flox/flox, Myh6-Cre) mice in a Cntn2-EGFP background (n = 4). (A) Comparison of sodium current–voltage (I–V) relationship. Maximum conductance was calculated to assess significant differences among experimental groups. (B) Voltage dependence of steady-state activation. Voltage at half activation (V0.5, activation) was calculated to assess significant differences among experimental groups. (C) Voltage dependence of steady-state inactivation. Voltage at half inactivation (V0.5, inactivation) was calculated to assess significant differences among experimental groups. (D) Time course of recovery from inactivation. Tau of recovery (τrecovery) was calculated to assess significant differences among experimental groups. Number of cells analyzed per cell type (ventricle, right atria, Purkinje) included in each graph legend. Patch clamp protocol diagrams are included for each endpoint. Data represent mean ± SEM. *P < 0.05, 1-way ANOVA.
ETV1 overexpression reprograms neonatal ventricular cardiomyocytes towards a His-Purkinje cell gene signature
Next, we examined whether forced expression of ETV1 in P1 neonatal rat ventricular myocytes (NRVMs), which normally lack ETV1, was sufficient to enrich for rapid conduction gene networks. NRVMs monolayers were infected with an adenovirus that co-expresses mouse ETV1 and GFP (Ad-Etv1-EGFP), or GFP alone (Ad-EGFP). Four days post-transduction, cells were collected for either RNA or protein level analysis (Supplemental Fig. 4A). NRVM transduced with Ad-Etv1-EGFP results in a ~49 fold increase in Etv1 quantified by qPCR (Supplemental Fig. 4B) and robust nuclear expression of ETV1 visualized by Immunofluorescence (Supplemental Fig. 4C).
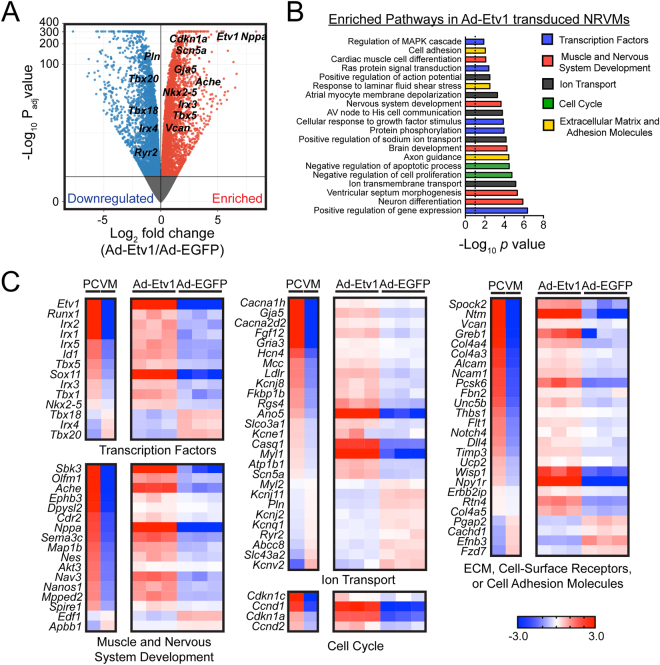
To comprehensively explore the GRNs controlled by ETV1, we performed RNA-seq transcriptional profiling on NRVMs transduced with either Ad-Etv1-EGFP or Ad-EGFP. We found a majority of genes were significantly different (9,236 genes, 62%) from a total of 14,813 genes tested with detectible counts (normalized counts > 5). Indeed, rapid conduction genes Nkx2–5 (1.5 fold, Padj = 5.4E-27), Gja5 (2.6 fold, Padj = 1.8E-87), and Scn5a (3 fold, Padj = 5.0E-140) were all significantly enriched with ectopic ETV1 expression along with numerous other genes identified as critical markers of trabecular/PCs identity, including Nppa, Cdkn1a, Ache, Irx3, Tbx5, Kcne1, and Vcan (Fig. 6A)51,61–64. In addition, genes associated with nodal or VM identity including Irx465, Tbx1866, Tbx2067, Ryr268, and Pln69 were all significantly diminished with ectopic ETV1 expression (Fig. 6A). To classify the GRNs altered with ETV1 overexpression, we performed KEGG functional network analysis on the 4,696 upregulated genes and found that many of these genes belonged to nervous system development pathways (Fig. 6B). A majority of these functional pathways including neuron differentiation, axon guidance, neuron development, and cell adhesion were consistent with known transcriptional targets of ETV1 in nervous system development70. Indeed, many of these functional networks were enriched in PCs, suggesting that ETV1 plays a key role in endowing cardiac PCs with their rapidly conducting, neuronal-like identity (Fig. 6B, Supplemental Fig. 2B)47.
Figure 6.
ETV1-transduced neonatal rat ventricular myocytes (NRVMs) upregulates a His-Purkinje system gene signature. (A) Volcano plot of relative transcript expression from NRVMs transduced with either Ad-Etv1-EGFP or Ad-EGFP. RNA-sequencing (RNA-seq) comparison revealed a total of 9,236 differentially expressed genes (normalized counts ≥ 5, padj < 0.05). All significantly different genes (padj < 0.05) are labeled blue (downregulated) or red (enriched) and all nonsignificantly different transcripts labeled in gray. Of these there were 4,696 upregulated and 4,540 downregulated genes in Ad-Etv1-EGFP versus Ad-EGFP transduced NRVMs. (B) Functional clustering of upregulated genes in Ad-Etv1 transduced NRVMs highlighted significantly enriched ETV1-dependent cellular processes (top 20 non-redundant categories are shown). Pathways are color coded to represent genes clustered into functional classes for heat maps in C. (C) Comparative RNA-seq between 21-day-old (P21) wild-type mouse FACS-purified Purkinje cell (PC)/ventricular myocytes (VM) and Ad-Etv1-EGFP/Ad-EGFP transduced NRVMs. Heat map representation of 88 genes differentially expressed in Ad-Etv1-EGFP versus Ad-EGFP transduced NRVMs (n = 3) plotted adjacent to average fold change expression in PCs and VMs. Genes clustered into functional groups demonstrate that ETV1 regulates a PC transcriptome in neonatal cardiomyocytes.
To compare the gene expression profile between endogenous PCs and VMs (Supplemental Fig. 2A) with NRVMs transduced with either Ad-Etv1-EGFP or Ad-GFP (Fig. 6A), heat maps were populated with genes identified via KEGG analysis and clustered into functional groups including TFs, muscle and nervous system development, ion transport, cell cycle, and ECM, cell-surface receptor, or cell adhesion molecules. Heat maps of average log2 fold change values were plotted for PCs and VMs while individual log2 fold change values were plotted for Ad-Etv1-EGFP or Ad-GFP transduced NRVMs demonstrating reproducibility between replicates (Fig. 6C). The gene expression profile of these 88 transcripts revealed a high level of similarity in PC gene expression patterns compared to ETV1-transduced NRVMs. This data establishes that ETV1-directed GRNs are an important component of the PC transcriptome.
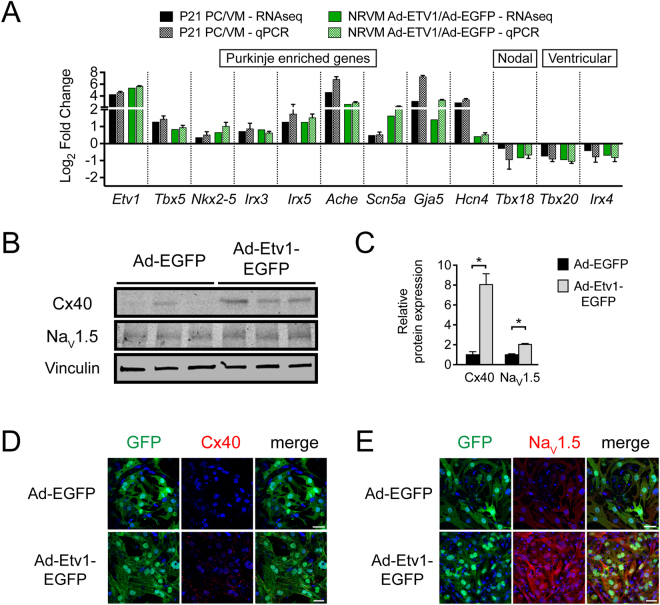
We next confirmed select ETV1-regulated genes critical for cardiac conduction by qPCR, including PC-enriched Etv1, Tbx5, Nkx2–5, Irx3, Irx5, Ache, Scn5a, Gja5, Hcn4; Nodal Tbx18; and Ventricular Tbx20, Irx4 (Fig. 7A, Supplemental Table 10). Forced expression of ETV1 was sufficient to promote a PC identity in postnatal VMs. We further confirmed that ETV1 was sufficient for protein level enrichment of the gap junction Cx40 and the cardiac sodium channel NaV1.5, both necessary for rapid conduction. Immunoblots of NRVMs expressing ETV1 demonstrated a substantial increase in Cx40 protein expression (~8 fold increase) as well as an increase in NaV1.5 expression (~2 fold increase) compared to control (Fig. 7B,C). Protein expression changes were further evaluated using immunofluorescence to detect expression of Cx40 and NaV1.5 in Ad-Etv1-EGFP or Ad-EGFP transduced NRVMs (Fig. 7D,E). Cx40 was ectopically detected in VMs with transduction of ETV1 (Fig. 7D). Moreover, the expression of NaV1.5 was markedly enriched in VMs expressing ETV1 (Fig. 7E).
Figure 7.
ETV1 overexpression leads to marked enrichment of fast conduction genes in postnatal ventricular myocytes (VMs). (A) Quantitative RT-PCR (qPCR) validation of RNA sequencing (RNA-seq) gene expression data comparing known cardiac conduction associated genes from endogenous Purkinje cells (PCs) and VMs versus Ad-Etv1-EGFP or Ad-EGFP transduced neonatal rat ventricular myocytes (NRVMs). Log2 fold change displayed relative to VM levels (P21 PC/VM, n = 7,10 respectively) or Ad-EGFP levels (NRVM Ad-Etv1-EGFP/Ad-EGFP, n = 3) (Supplemental Table 10). (B) Western blots of Ad-EGFP and Ad-Etv1-EGFP transduced NRVM lysates detecting Cx40, NaV1.5, and Vinculin (loading control). (C) Protein level densitometric quantification (normalized to vinculin), displayed relative to Ad-EGFP (n = 5). (D,E) Immunofluorescence staining of (D) Cx40 and (E) NaV1.5 in Ad-Etv1-EGFP and Ad-EGFP transduced NRVMs. ETV1-transduced cells displayed ectopic expression of Cx40 and marked enrichment of NaV1.5 compared to Ad-GFP-transduced NRVMs. Nuclei were co-stained with DAPI. Data represent mean ± SEM. *P < 0.05, 2-tailed Student’s t test. Scale bars: 50 um.
ETV1 induces expression of rapid conduction genes and Purkinje-like sodium currents on human induced pluripotent stem cell-derived cardiomyocytes
To assess whether ETV1 has a conserved function in human HPS specification, we investigated whether ectopic ETV1 induction in hiPSC-CMs would convert these cells toward a HPS lineage. Using a protocol previously validated for the generation of purified and mature hiPSC-CMs71, we infected hiPSC-CM monolayers with Ad-Etv1-EGFP or Ad-EGFP. Two weeks post-transduction, cells were collected for either RNA or cellular electrophysiological analysis (Fig. 8A). hiPSC-CMs transduced with Ad-Etv1-EGFP results in a ~70 fold increase in Etv1 quantified by qPCR (Fig. 8B). Similar to results observed in NRVMs, hiPSC-CMs expressing Etv1 demonstrated a substantial increase in NKX2–5 expression (3.0 fold, P = 4.6E-3), GJA5 expression (5.3 fold, P = 9.5E-4), and SCN5A expression (3.4 fold, P = 9.9E-4) compared to control (Fig. 8B, Supplemental Table 11). ETV1 was able to dictate rapid conduction GRNs in human cardiomyocytes.
Figure 8.
Activation of ETV1 in human induced pluripotent stem cells-derived cardiomyocytes (hiPSC-CMs) leads to increased expression of rapid conduction genes and sodium current. (A) Schematic representation of hiPSC-CM generation and maturation (day 0–21), transduction of Ad-Etv1-EGFP or Ad-EGFP (day 24), and timepoint for experimentation (day 38–40). (B) Quantitative RT-PCR analysis of Etv1, NKX2–5, GJA5, SCN5A, and MYL2 in hiPSC-CM transduced with either Ad-Etv1-EGFP or Ad-EGFP (n = 4). (C) Whole-cell patch clamp was performed on Ad-Etv1-EGFP (n = 12) or Ad-EGFP (n = 9) transduced hiPSC-CMs. Sodium current–voltage (I–V) relationship comparison. (D) hiPSC-CM NaV peak conductance (gNaV-peak). gNaV-peak following −120 mV to −35 mV depolarization step was measured for Ad-Etv1-EGFP (n = 12) or Ad-EGFP (n = 9) transduced hiPSC-CMs. Data represent mean ± SEM. *P < 0.05, 2-tailed Student’s t test.
To probe whether these changes in gene expression altered sodium current properties, we recorded sodium currents from voltage clamped transduced hiPSC-CMs. Voltage step commands elicited larger transient sodium current densities in Etv1 transduced hiPSC-CMs (n = 12) versus GFP transduced hiPSC-CMs (n = 9) (Fig. 8C). Peak currents in Etv1 transduced hiPSC-CMs were double compared to controls (maximum conductance: 2.52 ± 0.33 nS/pF, 1.26 ± 0.014 nS/pF, respectively) (Fig. 8C,D, Supplemental Table 12). Sodium channels in Etv1 transduced hiPSC-CMs trended to undergo steady-state activation at more negative potentials compared to control (Supplemental Fig. 5A, Supplemental Table 12). No difference was observed in sodium channel steady-state inactivation in transduced hiPSC-CMs (Supplemental Fig. 5B, Supplemental Table 12). Taken together, Etv1 overexpression was able to transform the sodium current properties of hiPSC-CMs towards a PC-like phenotype.
Discussion
Rapid conduction through the atria and specialized HPS enables coordinated activation and contraction of the four-chamber heart. Our studies reinforce ETV1 as a key regulatory factor that controls rapid conduction cardiac GRNs. Using cardiomyocyte-specific loss of function and viral transduction strategies, we found that ETV1 positively regulated gene networks necessary for active (Scn5a) and passive (Gja5) conductance along with structural (Nkx2–5) determinants of the mature HPS. As a consequence, cardiomyocyte loss of ETV1 significantly slows conduction in atrial and His-Purkinje myocytes and produces severe specification defects of the adult HPS. When placed in the context of previous work19–21, endocardially-derived NRG1 signaling establishes progenitor HPS cells and simultaneously activates ETV1 signaling networks to create rapid conduction regions. Our study now links this established upstream signaling network with downstream cardiomyocyte-specific nuclear responses that occur to create distinct electrical networks in the adult heart.
Purkinje and VMs share a common ventricular progenitor12. Their transcriptional signature begins to diverge with the inception of NRG1 signaling gradients creating trabecular and compact zones within the embryonic heart20. To define the transcriptional landscape of rapidly conducting PCs, we probed transcriptional differences between PCs and VMs by RNA-seq. We found that Etv1 is the most differentially expressed TF between PC and VMs. In addition, we identified that Etv1 is one of the most abundant expressed TFs in PCs. Confirming this approach, we also show Tbx5, another established HPS regulator required for maintenance of normal cardiac rhythm, to be highly abundant and enriched within HPS26,27,72. We believe this newest dataset is the benchmark for defining the transcriptional identity of endogenous murine cardiac PCs for comparative investigation.
To verify the specificity of Etv1 expression within the heart, we analyzed a BAC transgenic mouse, Etv1-EGFP. This reporter mouse was originally developed to identify and study neuronal cell types;56,73 we speculated it would also be an effective tool to study the HPS with the added benefit of not altering Etv1 gene dosage. In line with our previous description of the Etv1-nlz reporter in the heart21, Etv1-EGFP expression was observed throughout the HPS at all stages of cardiogenesis and the mature heart. GFP expression was also detected in the PAM in the developing and mature heart. This suggests that ETV1-dependent GRNs establish early in development and are engaged in atrial and trabecular myocytes. During the transition from precursor trabecular myocytes to a Purkinje lineage, ETV1 continues to be highly expressed in cells destined to populate the post mitotic HPS, and persistent ETV1 activity may be required to promote conduction health. Moreover, the commonality of ETV1 expression in PAM and HPS cells reveal a mechanistic understanding of the shared GRNs and electrical properties that govern these fast conducting, heterogeneous cell types.
ETV1 is expressed within many neuronal cell types and plays an important role in their specification41,43,45,57,70,74. Our initial germline knockout studies were not able to address whether neuronal influences contributed to the observed cardiac conduction phenotype. It is well recognized that neuronal cell types, particularly neural crest cells, are required for normal heart development75. Cardiomyocyte loss of ETV1 circumvents the adolescent lethality observed in Etv1 germline KO mice, enabling electrophysiological assessment of the adult heart. We found that Etv1 cKO mutant hearts resemble Etv1 germline null hearts in virtually all aspects of functional, molecular, and cellular electrophysiological phenotype. Conduction defects including P wave and QRS interval prolongation in Etv1 cKO hearts arose from a combination of hypoplastic HPS creating current-to-load mismatch, decrease in junctional conductance, and alterations in sodium channel density and gating; all present in Etv1 germline KO heart. A key point of distinction between Etv1 germline KO hearts and Etv1 cKO hearts is that Etv1 cKO hearts had a normal PR duration, suggesting normal AV nodal function21. This difference could suggest a neuronal component to AV nodal development, an effect of ETV1 on autonomic innvervation of the AVN, or a lack of Etv1 deletion in E9.0 cardiomyocytes that populate the developing AVN. This observation supports a need for future experiments focused on elucidating the role of ETV1 in AV nodal development and/or autonomic innervation. Neuronal-restricted gene targeting will prove important to elucidate these potential mechanisms.
Our ETV1 transduction study demonstrates that this single TF is sufficient to impart a PC genetic program onto postnatal VMs. Indeed, ETV1 is an established orchestrator of neuronal GRNs and appears to convey a neuronal-like gene signature to cardiac PCs. When compared to our Purkinje genome wide expression profile the transition of ETV1 transduced VMs to a PC identity appears to be of high fidelity. Genes required for rapid conduction and HPS specification were significantly enriched including Gja5, Scn5a, Nkx2–5, Tbx5, Irx3, and Irx5. Moreover, classic markers of the HPS49 were also induced in ETV1 transduced VMs including Ache, Kcne1 (MinK), Hcn4, and Cacna1h. Of note, ectopic ETV1 expression in VMs was not able to enrich mature PC markers including Pcp447 and Cntn253,76. Since NRVM cultures are only viable up to one week, it will be interesting to see whether in vivo transductions that facilitate more prolonged expression of ETV1 could mitigate these shortcomings or if other unknown TFs are required for expression of these terminal markers. Similar to our work, a recent study using IRX3 overexpression in NRVMs demonstrated that IRX3 was able to promote expression of 12 HPS-enriched genes33. In their model, IRX3 interacts with NKX2–5/TBX5 in a post-translational TF complex required for proper HPS development and function33. Our work extends their findings by demonstrating that ETV1 transcriptionally activates these TFs for appropriate specification of the HPS.
The application of this pathway to hiPSC-CMs suggests a conserved role of ETV1 in defining rapid conduction GRNs across vertebrate species. Human and mouse ETV1 are both encoded by 13 exons, share a highly conserved amino acid sequence (98%), and have identical sequence homology in the DNA binding ETS domain (Supplemental Fig. 6)77. Indeed, the conserved function of ETV1 has been probed in dopaminergic neurons where mouse ETV1 was able to rescue the ast-1 (Etv1 ortholog) mutant phenotype when expressed in transgenic worms74. Our study suggests that mouse ETV1 is able to induce rapid conduction genes and modify sodium current properties resembling a PC-like signature in hiPSC-CM reinforcing a shared mechanism. To our knowledge, ETV1 is the first TF sufficient to modify the transcriptome and sodium current properties of human cardiomyocytes towards a PC-like phenotype. Investigating whether ETV1 alone or combined with other TFs and/or signaling pathways, such as Notch78 or NRG119, are able to differentiate adult VMs or pluripotent stem cells into mature His-PCs will be of significant interest.
Current treatment options to improve conduction in failing hearts are limited to artificial pacemakers. While these implantable devices have proved to be clinically effective, they do not address the causal elements of impaired electrical function. Our findings shed light on the cell-type specific molecular mechanisms that orchestrate HPS development and rapid conduction physiology. Moreover, our data suggest that the ETV1-dependent rapid conduction GRN can be modulated for the management of selected human conduction disorders.
Methods
Experimental animals and Study design
All procedures and protocols were approved by the Institutional Animal Care and Use Committee of the New York University Langone Health, protocol IA16–01599, and complied with the standards for the care and use of animal subjects as stated in the Guide for the Care and Use of Laboratory Animals.
This study was preformed using Etv1flox/flox mice46,57 (provided by Dr. Ping Chi, Memorial Sloan-Kettering Cancer Center NY, NY, USA), Myh6-Cre58, Cntn2-EGFP BAC transgenic53, and Etv1-EGFP BAC transgenic56 mutant mice that have been previously characterized. Cntn2-EGFP and Etv1-EGFP mice were maintained in the CD1 genetic background. Etv1flox/flox and Myh6-Cre mice were maintained in the C57BL/6 genetic background. For Purkinje cell FACS, HPS morphology imaging/quantification, and cellular electrophysiology, Etv1flox/flox; Myh6-Cre mouse line was bred into the Cntn2-EGFP background. Etv1flox/flox; Myh6-Cre; Cntn2-EGFP compound mutant mouse lines were backcrossed greater than 5 generations into the C57BL/6 genetic background. Etv1flox/flox mice were used as WT controls. For NRVM isolations, P1 wild type litters in a Sprague-Dawley background were purchased from Charles River Laboratories.
NRVM isolation
NRVMs were isolated from P1 wild type male and female rats using the Miltenyi Biotec neonatal heart dissociation kit (Miltenyi Biotec, 130–098–373) with a gentleMACS octo dissociator (Miltenyi Biotec, 130–096–427) according to the manufacture’s protocol. P1 NRVM heart lysates were purified using Miltenyi Biotec rat neonatal cardiomyocyte isolation kit (Miltenyi Biotec, 130–105–420) according to the manufacture’s protocol. The final cell suspension was collected, counted, and plated as confluent monolayers on fibronectin-coated (25 μg/mL) plastic dishes. 24 hours post-plating in DMEM plus 10% FBS plus supplements (3 mM pyruvic acid, 2 g/l BSA, 0.5 mg/ml primocin, 15 mM HEPES, 4 μg/ml transferrin, 0.7 ng/ml sodium selenite, 5 μg/ml linoleic acid, 10 μM ascorbic acid), cells were cultured in serum-free DMEM with supplements as previously described21. The final myocyte cultures contained purified cardiomyocytes at a density of 3 × 106 cells per 35-mm dish. Viral transductions were performed during the media change at 24 hours after plating. Cardiomyocytes were used for either transcript or protein level analysis four days post-transduction.
Human induced pluripotent stem cell lines
NCRM1 iPSC line from Codex BioSolutions Inc. (MD, USA) were plated on Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Gibco, A1413202) coated plates and cultured with Essential 8 Medium (Gibco, A1517001). The differentiation and maturation protocol has been previously published71. Briefly, day one hiPSC were treated with small molecule CHIR99021 (Tocris, 4423, final concentration 10 μM) in the RPMI-BSA medium [RPMI 1640 Medium (HyClone, SH30027.01) supplemented with 213 μg/ml AA2P (l-ascorbic acid 2-phosphate magnesium) (A8960, Sigma) and 0.1% bovine serum albumin (BSA) (A1470, Sigma)] for 24 h followed by RPMI-BSA medium replacement. On differentiation day four, cells were treated with IWP2 (Tocris, 3533, final concentration 5 μM) in RPMI-BSA medium. After 48 h, media was replaced with RPMI-BSA medium. During maturation and purification, differentiated hiPSC-cardiomyocytes were treated with metabolic selection medium (fatty acid + T3). Fatty acid medium was prepared as DMEM Medium (No Glucose) supplemented with 0.1% BSA (Sigma, A1470) and 1 × Linoleic Acid-Oleic Acid-Albumin (Sigma, L9655). Fatty acid + T3 medium was fatty acid medium supplemented with T3 (Acros Organics, 437260010, final concentration 10 nM) for 8 days. RPMI 1640 Medium supplemented with 3% KnockOut Serum Replacement (Gibco, 10828–028, the routine medium) was used to culture differentiated cardiomyocytes. Medium was changed every two days unless specified.
Adenovirus reagents
A monolayer of NRVMs or iPSC-CMs was transduced with either Ad-Etv1-EGFP (Etv1 vector; multiplicity of infection 5:1; Vector Biolabs) or Ad-EGFP (control vector; multiplicity of infection 5:1; Vector Biolabs) 24 hours post cell isolation (NRVMs) or 24 days post hiPSC differentiation.
Antibody reagents
Immunofluorescent antibodies used in this study were all previously described21. The antibody target, dilution, species, company, and product number are the following: NKX2–5, 1:100 (rabbit, Abcam, ab91196); ETV1, 1:100 (rabbit, Abcam, ab36788); NaV1.5, 1:50 (rabbit, Alomone Labs, ASC-005); Cx40, 1:250 (rabbit, Alpha Diagnostic, Cx40A); CNTN2, 1:40 (goat, R&D Systems, AF4439); GFP, 1:100 (rabbit, Abcam, ab290). Secondary antibodies were donkey anti-rabbit, 1:500 (Santa Cruz Biotechnology, sc-2784); donkey anti-goat, 1:500 (Santa Cruz Biotechnology, sc-2024); and donkey anti-mouse, 1:500 (Santa Cruz Biotechnology, sc-2099). Western blot primary antibodies were [target, dilution (species, company, product number)] Nkx2–5, 1:1,000 (mouse, Abcam, ab91196); ETV1, 1:200 (goat, Santa Cruz Biotechnology, sc-1953); NaV1.5, 1:500 (rabbit, Alomone Labs, ASC-005); Cx40, 1:1,000 (rabbit, Alpha Diagnostic, Cx40A); and vinculin, 1:5,000 (mouse, Abcam, ab11194). Secondary antibodies were goat anti-rabbit, 1:15,000 (LI-COR, 926–32211); goat anti-mouse, 1:15,000 (LI-COR, 926–32220); and donkey anti-goat, 1:15,000 (LI-COR, 926–32214).
Immunohistochemistry
Embryonic and adult hearts were removed and either emersion or retrograde perfusion-fixed, respectively, and stored in 4% paraformaldehyde overnight (adult) or 2 hours (embryonic) at 4 °C. Samples were then processed as previously described21.
Electrocardiograms
Surface ECGs were acquired in anesthetized mice (2% isoflurane) by using subcutaneous needle electrodes attached in a 3-limb configuration, as previously described21,79. Parameters measured from the ECG data included P wave duration, PR interval, QRS duration, and QT interval. The P wave duration included both the first positive and negative deflections from baseline. QRS duration was measured from the initial deflection from baseline after the P wave until the return to baseline prior to the T wave peak. PR interval was measured from the start of the P wave to the beginning of the QRS complex. RSR’ was defined as two upward deflections above baseline resulting from a single ventricular depolarization. The R wave is the first upward deflection in the QRS complex. The S component is the first downward deflection. The R’ wave is a second upward deflection in the QRS complex prior to the T wave peak.
Cardiomyocyte enzymatic dissociation for whole-cell patch clamp and FACS purification
Cardiomyocytes were isolated from P21 Cntn2-EGFP hearts for RNA-seq and 10–12 week old Etv1fl/fl Cntn2-EGFP hearts for qPCR studies. Hearts were Langendorff-perfused and enzymatically digested as previously described80. Myocyte FACS-based purification was performed as previously described21,81. Cardiomyocytes were purified based on their mitochondrial content with the incubation of the mitochondrial dye tetramethylrhodamine methyl ester perchlorate (TMRM; Invitrogen). 50 nM TMRM was added to the enzymatic digestion buffer for the Langendorff-perfusion. Cell suspensions were purified by FACS (Beckman Coulter MoFlo, 100 µm nozzle). Purkinje (TMRMhiGFP+) and ventricular myocytes (TMRMhiGFP–) fractions were collected for RNA isolation (Supplemental Fig. 1). VMs and PCs for patch clamp experiments were not stained for TMRM. PCs were identified visually via endogenous GFP expression.
RNA-sequencing and quantitative RT-PCR
Purkinje and Ventricular Myocyte RNA-seq: RNA was isolated from FACS purified Purkinje and Ventricular myocytes by the PicoPure RNA Isolation Kit (Arcturus Engineering). Isolated RNA was amplified and sequencing libraries were prepared using the Trio Low Input RNA Kit (Nugen). NRVM RNA-seq: Total RNAs were extracted using the RNeasy kit (Qiagen). Sequencing libraries were prepared using the TruSeq RNA Library Prep Kit v2 (Illumina). For both sequencing experiments, samples were sequenced 50 bp paired-ended at 10 million to 20 million reads per replicate on an Illumina HiSeq. 2500 instrument. Library preparation and sequencing were performed at New York University School of Medicine Genome Technology Center. All the reads were mapped to the mouse or rat (based on samples genome) reference genome (mm10/Rnor6) using the STAR aligner (v2.5.0c)82. Alignments were guided by a Gene Transfer File (GTF, version GRCm38.74) and the mean read insert sizes and their standard deviations were calculated using Picard tools (v.1.126) (http://broadinstitute.github.io/picard/). The read count tables were generated using HTSeq (v0.6.0)83 and normalized based on their library size factors using DESeq. 2 (v3.0)84 and differential expression analysis was performed. The Read Per Million (RPM) normalized BigWig files were generated using BEDTools (v2.17.0)85 and bedGraphToBigWig tool (v4), and downstream statistical analyses and generating plots were performed in R environment (v3.1.1) (http://www.r-project.org/). Gene ontology analysis was performed with DAVID (http://david.abcc.ncifcrf.gov/). Candidate targets were validated with qPCR using primers specific to the gene of interest. (Supplemental Table 13) RNA was reverse-transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). qPCR was performed with a SYBR qPCR Kit (Qiagen) on a StepOne™ Real-Time PCR System (Applied Biosystems). qPCR probes, if available, were purchased from Origene and used according to the manufacturer’s instructions. Remaining qPCR probes were designed or identified and validated from prior literature.
Whole-cell sodium current (INa) recordings
INa recordings in isolated cardiomyocytes and hiPSC-CM were conducted in whole-cell configuration at room temperature with voltage step protocols as previously described21. As described in published methods21, recording pipettes were filled with a solution containing (in mM) NaCl 5, CsF 135, EGTA 10, MgATP 5, HEPES 15, pH 7.2, with CsOH. Cells were maintained in a solution containing (in mM) NaCl 5, CsCl 112.5, TEACl 20, CdCl2 0.1, MgCl2 1, CaCl2 1, HEPES 20, glucose 11, pH 7.4, with CsOH. All recordings were obtained three times to verify reproducible recordings and within 15 minutes after establishing whole-cell configuration using an Axon multiclamp 700B Amplifier coupled to a pClamp system (version 10.2, Axon Instruments).
Transthoracic echocardiography
Echocardiography was performed on anesthetized (2% isoflurane) mice as previously described21,86 using the Vevo 2100 high-resolution ultrasound imaging system (VisualSonics). VisualSonics Vevo 2100 V1.5.0 software was used for all data analysis. Short axis M-mode parameters were measured that include: diastolic and systolic left ventricular anterior and posterior wall thickness. From long axis B-mode measurements, left ventricular end diastolic volume, end systolic volume, stroke volume and ejection fraction were calculated from semi-automated LV area traces
HPS whole-mount quantification
His-Purkinje system imaging and quantification were conducted using Etv1flox/flox; Myh6-Cre; Cntn2-EGFP reporter mice as previously described21. Hearts were excised, washed in 1% PBS, and retrograde perfusion-fixed with 4% paraformaldehyde and stored overnight. For imaging of the left and right HPS, the left and right free walls were cut open as previously described21. Littermate controls (Etv1flox/flox; Cntn2-EGFP) were imaged on the same day at comparable magnification, exposure, and light intensity. GFP positive area normalized to total area (as specified) was measured using ImageJ software for the left ventricular septum, right ventricular septum, and right ventricular free wall.
Statistics
Comparisons were tested using 1-way ANOVA or 2-tailed Student’s t test where appropriate. P < 0.05 were considered statistically significant. All results were expressed as Mean ± SEM for each group. Mouse ECGs and ECHO data analysis were conducted by 2 operators. Genotypes were blinded until the endpoints were analyzed. Mouse studies were performed prior to genotyping, ensuring blinded observations.
Data availability
The mouse lines will be made publicly available through a mouse repository. RNA-seq data has been deposited with the NCBI under the Gene Expression Omnibus (GEO) accession number: GSE115061. All other data and materials are available upon corresponding author request.
Electronic supplementary material
Acknowledgements
This work was funded by grants from the National Institutes of Health (NIH) to A.S. (T32 GM066704, F31 HL132438), D.S.P. (R01 HL132073), and G.I.F. (R01 HL105983). Additional support for D.S.P. from the American Heart Association Scientist Development Grant (17SDG33411201) and generous support from the Saperstein family (Saperstein Scholar Award). We are grateful to Ping Chi (Memorial Sloan-Kettering Cancer Center, NY) for providing the Etv1fl/fl mouse line and Thomas Jessell (Columbia University, NY) and Silvia Arber (Friedrich Miescher Institute (FMI) for Biomedical Research, Switzerland) for producing the Etv1flox allele; Michael Gregory and Kamilah Ryan of the NYU Cytometry and Cell Sorting Core; Adriana Heguy and Karina Ray of the NYU Genome Technology Center, supported in part by the NYU CTSA grant UL1TR000038 from the National Center for Advancing Translational Sciences (NCATS), NIH.
Author Contributions
A.S., G.I.F. and D.S.P. conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. X.L. performed cell electrophysiological experiments. B.L. carried out human induced pluripotent stem cell experiments, under the advice and supervision of L.B. A.K.-J. performed RNA sequencing analysis, under the advice and supervision of A.T. A.S., F.L. and J.Z. performed all other experimental work.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28239-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Glenn I. Fishman, Email: glenn.fishman@nyumc.org
David S. Park, Email: david.park@nyumc.org
References
- 1.Legato MJ. Ultrastructure of the atrial, ventricular, and Purkinje cell, with special reference to the genesis of arrhythmias. Circulation. 1973;47:178–189. doi: 10.1161/01.CIR.47.1.178. [DOI] [PubMed] [Google Scholar]
- 2.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Weerd JH, Christoffels VM. The formation and function of the cardiac conduction system. Development. 2016;143:197–210. doi: 10.1242/dev.124883. [DOI] [PubMed] [Google Scholar]
- 4.Lin Q, Srivastava D, Olson EN. A transcriptional pathway for cardiac development. Cold Spring Harb Symp Quant Biol. 1997;62:405–411. doi: 10.1101/SQB.1997.062.01.048. [DOI] [PubMed] [Google Scholar]
- 5.Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272:671–676. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- 6.Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90:509–519. doi: 10.1161/01.RES.0000013072.51957.B7. [DOI] [PubMed] [Google Scholar]
- 7.Jongbloed MR, et al. Embryonic conduction tissue: a spatial correlation with adult arrhythmogenic areas. J Cardiovasc Electrophysiol. 2004;15:349–355. doi: 10.1046/j.1540-8167.2004.03487.x. [DOI] [PubMed] [Google Scholar]
- 8.Wakili R, Voigt N, Kaab S, Dobrev D, Nattel S. Recent advances in the molecular pathophysiology of atrial fibrillation. J Clin Invest. 2011;121:2955–2968. doi: 10.1172/JCI46315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King JH, Huang CL, Fraser JA. Determinants of myocardial conduction velocity: implications for arrhythmogenesis. Front Physiol. 2013;4:154. doi: 10.3389/fphys.2013.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haissaguerre M, Vigmond E, Stuyvers B, Hocini M, Bernus O. Ventricular arrhythmias and the His-Purkinje system. Nat Rev Cardiol. 2016;13:155–166. doi: 10.1038/nrcardio.2015.193. [DOI] [PubMed] [Google Scholar]
- 11.Boyden PA, Hirose M, Dun W. Cardiac Purkinje cells. Heart Rhythm. 2010;7:127–135. doi: 10.1016/j.hrthm.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 13.Mikawa T, Cohen-Gould L, Fischman DA. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus. III: Polyclonal origin of adjacent ventricular myocytes. Dev Dyn. 1992;195:133–141. doi: 10.1002/aja.1001950208. [DOI] [PubMed] [Google Scholar]
- 14.Mikawa T, Borisov A, Brown AM, Fischman DA. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus: I. Formation of the ventricular myocardium. Dev Dyn. 1992;193:11–23. doi: 10.1002/aja.1001930104. [DOI] [PubMed] [Google Scholar]
- 15.Meilhac SM, et al. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development. 2003;130:3877–3889. doi: 10.1242/dev.00580. [DOI] [PubMed] [Google Scholar]
- 16.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 17.Gassmann M, et al. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee KF, et al. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 19.Rentschler S, et al. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci USA. 2002;99:10464–10469. doi: 10.1073/pnas.162301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai D, et al. Neuregulin 1 sustains the gene regulatory network in both trabecular and nontrabecular myocardium. Circ Res. 2010;107:715–727. doi: 10.1161/CIRCRESAHA.110.218693. [DOI] [PubMed] [Google Scholar]
- 21.Shekhar A, et al. Transcription factor ETV1 is essential for rapid conduction in the heart. J Clin Invest. 2016;126:4444–4459. doi: 10.1172/JCI87968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jay PY, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meysen S, et al. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev Biol. 2007;303:740–753. doi: 10.1016/j.ydbio.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 24.Harris BS, et al. Differentiation of cardiac Purkinje fibers requires precise spatiotemporal regulation of Nkx2-5 expression. Dev Dyn. 2006;235:38–49. doi: 10.1002/dvdy.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pashmforoush M, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/S0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 26.Moskowitz IP, et al. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131:4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- 27.Arnolds DE, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakker ML, et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- 29.Frank DU, et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci USA. 2012;109:E154–163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoogaars WM, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 31.Moskowitz IP, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 32.Ismat FA, et al. Homeobox protein Hop functions in the adult cardiac conduction system. Circ Res. 2005;96:898–903. doi: 10.1161/01.RES.0000163108.47258.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim KH, et al. Irx3 is required for postnatal maturation of the mouse ventricular conduction system. Sci Rep. 2016;6:19197. doi: 10.1038/srep19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SS, et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci USA. 2011;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costantini DL, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaborit N, et al. Cooperative and antagonistic roles for Irx3 and Irx5 in cardiac morphogenesis and postnatal physiology. Development. 2012;139:4007–4019. doi: 10.1242/dev.081703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatcher CJ, Basson CT. Specification of the cardiac conduction system by transcription factors. Circ Res. 2009;105:620–630. doi: 10.1161/CIRCRESAHA.109.204123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamaddon, H. S. et al. High-resolution optical mapping of the right bundle branch in connexin40 knockout mice reveals slow conduction in the specialized conduction system. Circ Res87, 929-936 (2000). [DOI] [PubMed]
- 39.Remme CA, et al. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res Cardiol. 2009;104:511–522. doi: 10.1007/s00395-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miquerol L, et al. Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc Res. 2004;63:77–86. doi: 10.1016/j.cardiores.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Willardsen M, Hutcheson DA, Moore KB, Vetter ML. The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mech Dev. 2014;131:57–67. doi: 10.1016/j.mod.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Janknecht R. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J Biol Chem. 2002;277:42669–42679. doi: 10.1074/jbc.M205501200. [DOI] [PubMed] [Google Scholar]
- 43.Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/S0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 44.Yoneshima H, et al. Er81 is expressed in a subpopulation of layer 5 neurons in rodent and primate neocortices. Neuroscience. 2006;137:401–412. doi: 10.1016/j.neuroscience.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 45.Okazawa M, Abe H, Nakanishi S. The Etv1 transcription factor activity-dependently downregulates a set of genes controlling cell growth and differentiation in maturing cerebellar granule cells. Biochem Biophys Res Commun. 2016;473:1071–1077. doi: 10.1016/j.bbrc.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 46.Ran L, et al. Combined inhibition of MAP kinase and KIT signaling synergistically destabilizes ETV1 and suppresses GIST tumor growth. Cancer Discov. 2015;5:304–315. doi: 10.1158/2159-8290.CD-14-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim EE, et al. PCP4 regulates Purkinje cell excitability and cardiac rhythmicity. J Clin Invest. 2014;124:5027–5036. doi: 10.1172/JCI77495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aanhaanen WT, et al. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ Res. 2010;107:728–736. doi: 10.1161/CIRCRESAHA.110.222992. [DOI] [PubMed] [Google Scholar]
- 49.Liang X, Evans SM, Sun Y. Insights into cardiac conduction system formation provided by HCN4 expression. Trends Cardiovasc Med. 2015;25:1–9. doi: 10.1016/j.tcm.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miquerol L, et al. Resolving cell lineage contributions to the ventricular conduction system with a Cx40-GFP allele: a dual contribution of the first and second heart fields. Dev Dyn. 2013;242:665–677. doi: 10.1002/dvdy.23964. [DOI] [PubMed] [Google Scholar]
- 51.Kondo RP, Anderson RH, Kupershmidt S, Roden DM, Evans SM. Development of the cardiac conduction system as delineated by minK-lacZ. J Cardiovasc Electrophysiol. 2003;14:383–391. doi: 10.1046/j.1540-8167.2003.02467.x. [DOI] [PubMed] [Google Scholar]
- 52.Rentschler S, et al. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pallante BA, et al. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhattacharyya S, Bhakta M, Munshi NV. Phenotypically silent Cre recombination within the postnatal ventricular conduction system. PLoS One. 2017;12:e0174517. doi: 10.1371/journal.pone.0174517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lebel M, et al. The Iroquois homeobox gene Irx2 is not essential for normal development of the heart and midbrain-hindbrain boundary in mice. Mol Cell Biol. 2003;23:8216–8225. doi: 10.1128/MCB.23.22.8216-8225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 57.Patel TD, et al. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003;38:403–416. doi: 10.1016/S0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- 58.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li GR, Lau CP, Shrier A. Heterogeneity of sodium current in atrial vs epicardial ventricular myocytes of adult guinea pig hearts. J Mol Cell Cardiol. 2002;34:1185–1194. doi: 10.1006/jmcc.2002.2053. [DOI] [PubMed] [Google Scholar]
- 60.Vaidyanathan R, et al. The ionic bases of the action potential in isolated mouse cardiac Purkinje cell. Heart Rhythm. 2013;10:80–87. doi: 10.1016/j.hrthm.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- 62.Park, D. S. & Fishman, G. I. Development and Function of the Cardiac Conduction System in Health and Disease. J Cardiovasc Dev Dis4, 10.3390/jcdd4020007 (2017). [DOI] [PMC free article] [PubMed]
- 63.Munshi NV. Gene regulatory networks in cardiac conduction system development. Circ Res. 2012;110:1525–1537. doi: 10.1161/CIRCRESAHA.111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christoffels VM, Moorman AF. Development of the cardiac conduction system: why are some regions of the heart more arrhythmogenic than others? Circ Arrhythm Electrophysiol. 2009;2:195–207. doi: 10.1161/CIRCEP.108.829341. [DOI] [PubMed] [Google Scholar]
- 65.Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- 66.Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stennard FA, et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262:206–224. doi: 10.1016/S0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 68.Kushnir A, Marks AR. The ryanodine receptor in cardiac physiology and disease. Adv Pharmacol. 2010;59:1–30. doi: 10.1016/S1054-3589(10)59001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taur Y, Frishman WH. The cardiac ryanodine receptor (RyR2) and its role in heart disease. Cardiol Rev. 2005;13:142–146. doi: 10.1097/01.crd.0000128709.84812.86. [DOI] [PubMed] [Google Scholar]
- 70.Abe H, Okazawa M, Nakanishi S. The Etv1/Er81 transcription factor orchestrates activity-dependent gene regulation in the terminal maturation program of cerebellar granule cells. Proc Natl Acad Sci USA. 2011;108:12497–12502. doi: 10.1073/pnas.1109940108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin B, et al. Culture in Glucose-Depleted Medium Supplemented with Fatty Acid and 3,3′,5-Triiodo-l-Thyronine Facilitates Purification and Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Front Endocrinol (Lausanne) 2017;8:253. doi: 10.3389/fendo.2017.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nadadur RD, et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci Transl Med. 2016;8:354ra115. doi: 10.1126/scitranslmed.aaf4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Groh A, et al. Cell-type specific properties of pyramidal neurons in neocortex underlying a layout that is modifiable depending on the cortical area. Cereb Cortex. 2010;20:826–836. doi: 10.1093/cercor/bhp152. [DOI] [PubMed] [Google Scholar]
- 74.Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Plein A, Fantin A, Ruhrberg C. Neural crest cells in cardiovascular development. Curr Top Dev Biol. 2015;111:183–200. doi: 10.1016/bs.ctdb.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 76.Maass K, et al. Isolation and characterization of embryonic stem cell-derived cardiac Purkinje cells. Stem Cells. 2015;33:1102–1112. doi: 10.1002/stem.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coutte L, et al. Characterization of the human and mouse ETV1/ER81 transcription factor genes: role of the two alternatively spliced isoforms in the human. Oncogene. 1999;18:6278–6286. doi: 10.1038/sj.onc.1203020. [DOI] [PubMed] [Google Scholar]
- 78.Rentschler S, et al. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation. 2012;126:1058–1066. doi: 10.1161/CIRCULATIONAHA.112.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutstein DE, Danik SB, Sereysky JB, Morley GE, Fishman GI. Subdiaphragmatic murine electrophysiological studies: sequential determination of ventricular refractoriness and arrhythmia induction. Am J Physiol Heart Circ Physiol. 2003;285:H1091–1096. doi: 10.1152/ajpheart.00100.2003. [DOI] [PubMed] [Google Scholar]
- 80.Kang G, et al. Purkinje cells from RyR2 mutant mice are highly arrhythmogenic but responsive to targeted therapy. Circ Res. 2010;107:512–519. doi: 10.1161/CIRCRESAHA.110.221481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hattori F, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 82.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan J, et al. Generation of a tamoxifen inducible Tnnt2MerCreMer knock-in mouse model for cardiac studies. Genesis. 2015;53:377–386. doi: 10.1002/dvg.22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse lines will be made publicly available through a mouse repository. RNA-seq data has been deposited with the NCBI under the Gene Expression Omnibus (GEO) accession number: GSE115061. All other data and materials are available upon corresponding author request.