Abstract
Transcriptional activators bind DNA and recruit cofactors to modify chromatin. The extent to which these two events are separable is unclear. Here, using a custom ChIP tiling array to map chromatin modifications, we show that interferon-γ-induced DNA binding of signal transducer and activator of transcription 1 (STAT1), typically associated with the transcription factor interferon regulatory factor 1 (IRF1), causes histone acetylation (H3ac, H4ac). In contrast, among IRF1 sites lacking concomitant STAT1 recruitment, only 25% underwent inducible histone acetylation, 31% exhibited constitutive histone acetylation, and 44% had no histone acetylation. These latter “orphan sites” also lacked other activating modifications (e.g. H3K4me1, H3K4me2) and were typically remote from transcription start sites. In these cases the closest gene was typically an IFNγ-inducible locus that did not respond to IFNγ in this setting. Orphan sites were detected in different cell types, suggesting broad relevance. Despite an atypical downstream response (i.e. no histone modifications), IRF1 binding depended on SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4 or BRG1), as is typical of active IRF1 enhancers. Although SMARCA4 permitted IRF1 access to the orphan sites, there was no corecruitment of the histone acetyltransferases CREB-binding protein (CBP) and p300. Orphan sites were constitutively unacetylated, and several were marked with repressive chromatin modifications (e.g. H3K27me3). In conclusion, although IRF1 can trigger enhanceosome formation independently of STAT1, its ability to do so depends on local chromatin cues.
Keywords: epigenetics, histone modification, interferon regulatory factor (IRF), STAT transcription factor, histone acetylase, interferon, transcription enhancer, gene regulation
Introduction
Gene induction requires recruitment of a large posse of proteins, resulting in the formation of an RNA polymerase preinitiation complex that is subsequently activated to generate a transcript. The multiprotein complex needed to induce transcription is recruited in an orderly cascade that includes activator proteins that bind directly to short DNA motifs, coactivators that bind activators and move or chemically modify nucleosomes, and general transcription factors that associate with activators and coactivators (1). The entire cascade can take minutes, for example at cytokine- or growth factor-induced genes, or days, such as at differentiation-responsive gene targets (2–4).
Chemical modification of histones is a universal theme during gene activation and can include acetylation, methylation, phosphorylation, and other post-translational events (5). Multiple studies of individual or a few genes have suggested that cofactor binding and histone modification inevitably follow activator recruitment. However, it is unclear whether activator binding is always sufficient to induce cofactor recruitment and histone modifications. In vitro work showed that cofactor recruitment is more efficient when multiple activators are cobound to an enhancer, presenting a larger and higher-affinity binding surface (6). Nevertheless, whether individual activators bind to DNA in vivo without concomitant coactivator recruitment and histone modification is uncertain. The estrogen receptor α (ERα)2 induces histone acetylation and methylation at some binding sites, but many binding events are unproductive and not associated with cofactor recruitment or chromatin modification (7, 8). Nuclear receptors can act as silencers or activators, so the extent to which other types of activators bind without modifying chromatin is unclear.
Interferon-γ is an immune cytokine that can induce the expression of hundreds IFN-stimulated genes (ISGs). Receptor binding to the extracellular surface triggers activation of Janus kinase 1 on the intracellular side, which phosphorylates the transcription activator STAT1. Phosphorylated STAT1 forms dimers that translocate to the nucleus, bind to γ-activated sequence motifs, and induce expression of ISGs (9). One of the major early targets of activated STAT1 is the transcription factor interferon regulatory factor 1 (IRF1), which cooperates with STAT1 to induce a second wave of ISGs. One such target of STAT1 and IRF1 is the class II transactivator (CIITA, or MHC2TA), which is the master regulator of major histocompatibility complex class II gene induction (10). This entire cascade of gene induction takes only 5–6 h and represents an ideal system to study events associated with activator binding. Recently, we reported that there are twice as many IRF1 versus STAT1-binding events, and although STAT1 is usually found in close proximity to IRF1, the reverse is not the case (11). Whether there are other fundamental differences in the effect of recruiting these two factors is unknown.
Both STAT1 and IRF1 can recruit cofactors that modify chromatin. Of particular relevance here is their ability to bind to several HATs. In view of their common ability to recruit HATs and their cooperative role in the same pathway, one might expect that histones at the enhancers they interact with would be modified accordingly. However, work on the IFNγ-responsive GBP2 promoter revealed that when STAT1 is mutated to prevent DNA binding, IRF1 is still recruited but does not induce histone acetylation (12). The degree to which this occurs in physiological settings is unknown, as are the distinguishing features of such inactive enhancers.
Our prior genome-scale analysis identified many isolated (STAT1-free) IRF1 sites (11), allowing us to address the degree to which IRF1-binding affects histone modification in vivo. It also allowed us to deduce whether there is any location bias for different classes of IRF1 enhancers, and the chromatin features associated with these subtypes. We identified many isolated IFNγ-induced IRF1-binding events that can induce histone acetylation independent of STAT1, but equally almost half of all such isolated IRF1 sites did not induce histone acetylation or other activating histone modifications. Most of these “orphan sites” were at remote locations, not promoters, but the nearest gene was typically an unresponsive (resistant) ISG. Orphan IRF1 sites did not recruit coactivators and were associated with silencing chromatin marks. Thus, although IRF1 can access repressive chromatin, the local environment appears to define its ability to trigger STAT1-independent enhanceosome formation.
Results
Basal and IFNγ-induced histone acetylation
Using a custom array employed previously to study STAT1 and IRF1 binding (4, 11), we mapped histone H3 and H4 acetylation before and after IFNγ treatment in HeLa cells. The array covers 16 Mb of sequence, representing nonrepetitive DNA on 11 chromosome segments that are heavily enriched for ISGs (4, 11). For ChIP analysis we used antibodies that recognize H3 acetylated on residues Lys9 and Lys14 or H4 acetylated on Lys5, Lys8, Lys12, and Lys16. Detailed acetylation maps are shown in Fig. 1A and Fig. S1 and can be viewed in browser format at http://research.lunenfeld.ca/IFNy.3 ChIP–qPCR was used to validate our ChIP-chip data (Fig. 1B). A detailed summary of the numbers of H3ac and H4ac peaks across studied regions is provided in Table S1. We detected a total of 703 separate positions with acetylated histones (H3ac and/or H4ac) in untreated cells, occupying a total of 788 kb of sequence, with average peak width of 1120 bp (range 479–7210), covering 5% of the studied chromatin. After IFNγ treatment, there were 747 acetylated positions that occupied 852 kb, an average of 1140 bp per peak (range 479–8429). In total, 247 of 747 sites exhibited inducible histone acetylation (a mix of off/on and on/up) (Table S1), which was not associated with a global increase in total H3ac and/or H4ac.4 Only one H3ac site showed a statistically significant drop in acetylation. Thus, as expected, IFNγ treatment triggers chromatin acetylation.
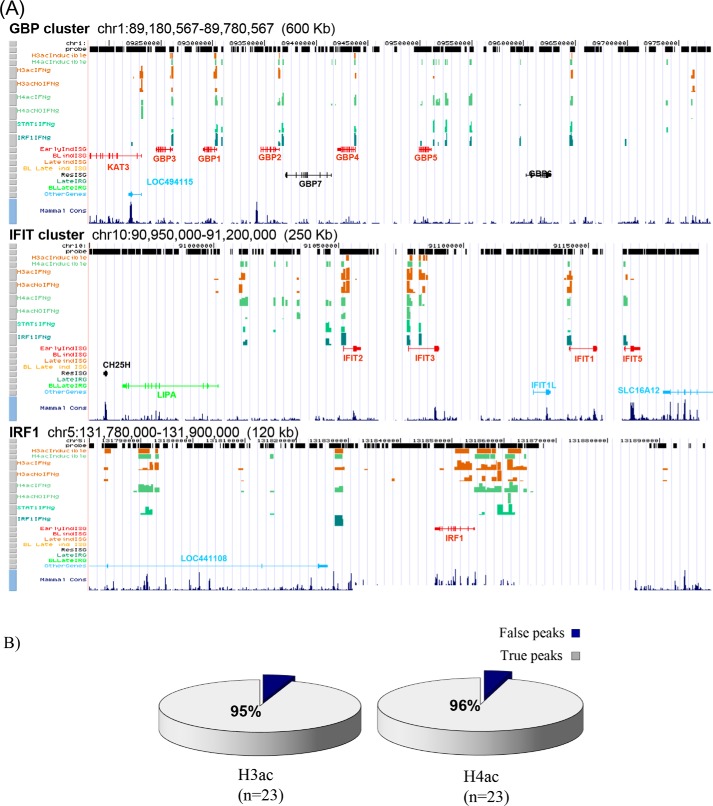
Figure 1.
Mapping constitutive and IFNγ-induced chromatin acetylation at IFNγ-induced STAT1- and IRF1-binding sites. A, chromatin acetylation maps. Tracks are organized in the following order from the top: chromosomal coordinates, probe positions, inducible histone acetylation (H3ac- and H4ac-inducible), plus and minus IFNγ tracks for H3ac (H3acIFNg and H3acNoIFNg) and H4ac (H4acIFNg and H4acNoIFNg), and only the plus IFNg track for STAT1 (STAT1IFNg) and IRF1 (IRF1IFNg) (from Ref. 11). Gene tracks are categorized according to their responses to IFNγ (from Ref. 11) and presented in different colors. Early, 6 h; Late, 24 h; ind, induced; BL, borderline; Res, resistant. Mammal Cons track shows the degree of sequence conservation between 44 vertebrate species. Additional acetylation maps are provided in Fig. S1. B, ChIP–qPCR validation. Arbitrarily selected ChIP-chip H3ac and H4ac sites were examined by ChIP–qPCR on chromatin from HeLa cells with no or 6 h of IFN treatment. ≥95% of peaks were validated in both cases.
The scope of H3ac was similar in untreated versus treated cells (677 peaks covering 714 kb in untreated cells versus 629 covering 686 kb in IFNγ-treated cells). The slight “drop” in H3ac peaks after treatment mainly reflects a reduction in marginal (although significant) basal acetylation. In contrast, there were far fewer basal H4ac than H3ac peaks (192, 3.5-fold less than H3ac, covering 160 kb, 4.5-fold less than H3ac). A prior study also noted stronger and wider H3ac signals compared with H4ac at promoters and enhancers (13). However, we found that H4ac rose considerably after IFNγ treatment (to 394 sites covering 399 kb, now only 1.6- and 1.7-fold less than H3ac, respectively). In IFNγ-treated cells, 91 (14%) H3ac sites versus 156 (40%) H4ac sites were cytokine-induced. Thus, basal acetylation is biased toward H3, whereas IFNγ-induced acetylation is biased toward H4.
Next, we assessed the position of H3ac and H4ac sites relative to gene starts. 64 and 63% of all acetylated sites were within 5 kb of known gene starts in untreated or treated cells, respectively (Table S1). Similar distributions were observed individually for either all H3ac sites (65% −IFNγ, 68% + IFNγ), all H4ac sites (69% −IFNγ, 61% +IFNγ), or constitutive H3ac (67%) and constitutive H4ac (64%) (Fig. 2A, Fig. S2A, and Table S1). However, induced H3ac sites were more likely to be promoter-proximal (73%) compared with induced H4ac sites (56%) (Fig. 2B and Table S1). Randomly generated sites did not show this pattern (Fig. S2B). Thus, in line with previous work (14–16), histone acetylation is mainly promoter-proximal, but still a large fraction is located at remote sites.
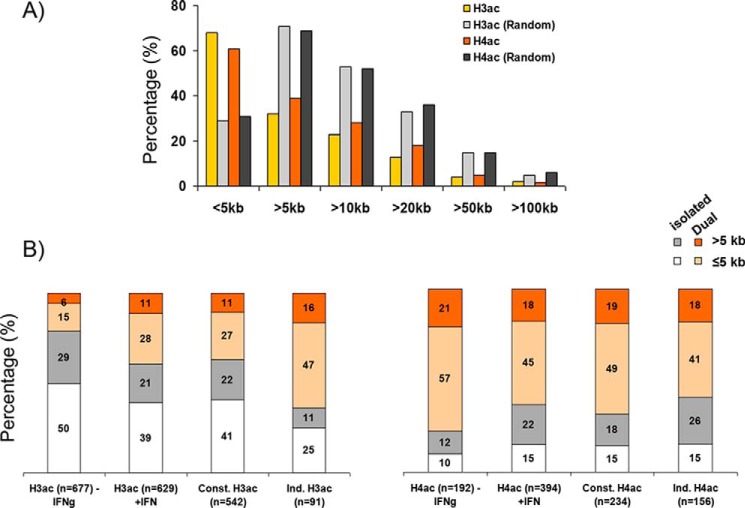
Figure 2.
Histone acetylation is enriched near gene promoters. A, the percentage of IFNγ-induced H3ac or H4ac marked sites versus randomly generated controls at proximal (≤5 kb) or distal (>5 kb) sites relative to the TSS of known genes. B, histograms show the percentages of total, constitutive, and induced H3ac (left) and/or H4ac (right) at proximal (≤5 kb) and distal (>5 kb) sites of ISGs, separated into isolated or dual (overlapping H3ac/H4ac) acetylation. The total number of sites in each category is indicated below the bars. Const., constitutive; Ind., induced.
Next we assessed the degree of overlap of H3ac and H4ac marks. Basal H3ac sites were typically isolated (79%, 524 of 677), whereas the majority of H4ac sites colocalized with H3ac (72%), consistent with the larger number of H3ac sites (677 versus 192) (Table S1). Conceivably, constitutive H3ac sites prime IFNγ-induced histone acetylation because a large fraction of IFNγ-induced H3ac or H4ac sites were marked with basal H3ac (68 or 49%, respectively). Frequently, induced H3ac occurred at dual sites, and 63% (58 of 91) of inducible H3ac sites was observed at any H4 acetylated sites, whereas only 38% (201 of 542) of constitutive H3ac sites (i.e. unchanged − or + IFNγ) were dual (Table S1 and Fig. S2B), increasing total dual H3ac from 21% (139 of 677) before treatment to 39% (239 of 629) after IFNγ treatment (Table S1). In contrast, both constitutive and inducible H4ac sites showed comparable frequencies of overlap with any H3 acetylation (68 and 59%, respectively) (Fig. 2B). Randomly generated sites showed much less overlap with H3ac or H4ac (Fig. S2B). Thus, most basal H3ac is isolated and may prime IFNγ-induced acetylation, preferentially generating dual H3/4ac sites.
Next, we assessed distribution of dual versus isolated marks at gene starts and/or distal locations. Dual H3ac sites were more common at promoter-proximal versus remote regions (2.5–2.9-fold enrichment for all, constitutive or inducible H3ac sites; Fig. 2B). There was a similar proximal bias for most categories of dual H4ac sites (Fig. 2B). Isolated H3ac sites were also skewed to gene promoters albeit to a lesser extent compared with dual sites (1.7–2.3-fold; Fig. 2B). In contrast, isolated (all, constitutive, or induced) H4ac sites were enriched at distal regions (1.2–1.7-fold; Fig. 2B). The preponderance of isolated inducible H4ac at remote sites was evident at multiple loci (e.g. STAT1 and CIITA/SOCS1 regions and GBP, IFI200, and IFIT clusters; Table S1). In summary, whereas isolated H3ac and dual H3ac/H4ac are predominately promoter proximal, isolated H4ac is more often located at distal elements, especially those induced by IFNγ.
Only a subset of STAT1/IRF1-binding sites are preacetylated
Regulatory elements at poised loci can be premarked with active chromatin marks (17, 18). By integrating prior data on STAT1/IRF1 recruitment (11), we assessed whether these TF-binding sites were premarked with histone acetylation in the basal state. Of all 230 distinct TF-binding positions (Fig. 3A, STAT1 and/or IRF1), just under one half were preacetylated before IFNγ treatment. Most (66%) preacetylated TF sites were promoter proximal, which was higher (80%) at dual STAT1/IRF1 sites (Fig. 3A), suggesting a more suitable chromatin state for TF binding. The lower fraction of preacetylated remote TF-binding sites (34%) was similar to the 25% of STAT1 sites that show basal H3K4me (18). Comparison of isolated STAT1 and IRF1 sites showed that the preacetylated fraction was similar for either TF at promoters (70%), but at remote locations 47% of STAT1 versus only 28% of IRF1 sites were preacetylated. Only 21% of remote dual IRF1 + STAT1 sites were preacetylated. In summary, preacetylation is common at ISG promoters, especially those that recruit both STAT1 and IRF1, but although it is seen at approximately half of remote STAT1 sites, most remote IRF1 sites are not preacetylated, including those that show dual IRF1/STAT1 binding.
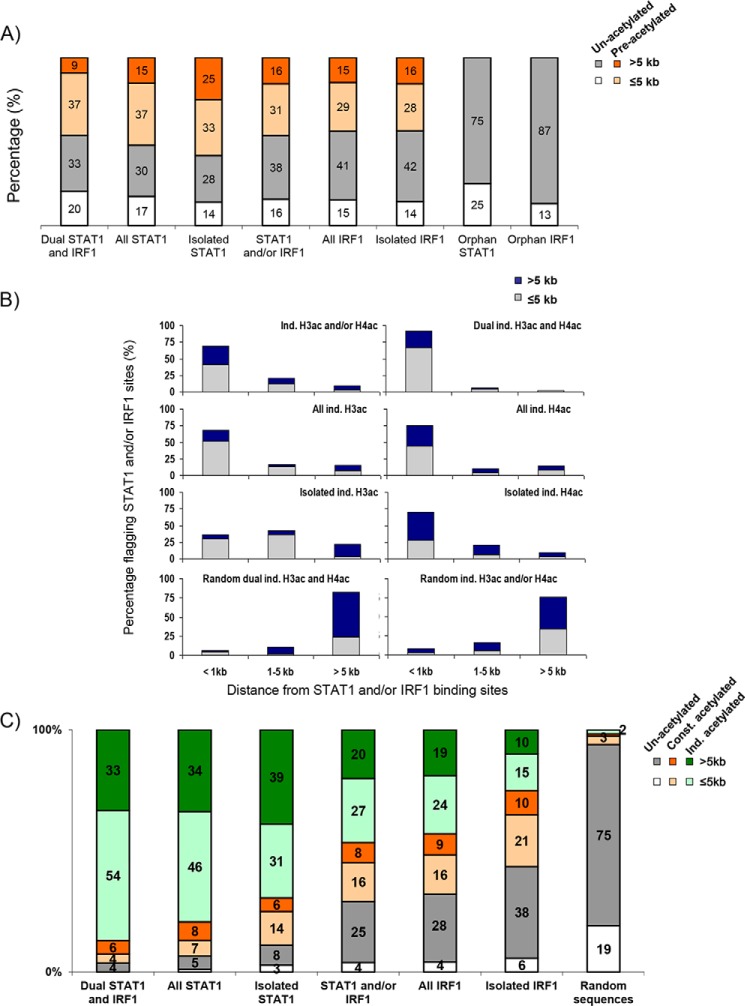
Figure 3.
IFNγ-induced STAT1 and IRF1 binding differentially correlate with histone acetylation. A, graph shows the percentages of preacetylated STAT1 and/or IRF1 sites at promoter proximal (≤5 kb) or remote (>5 kb) distances from known genes. B, graphs show the percentages of distribution of IFNγ-induced H3/4ac relative to TF binding within proximal (≤5 kb) or distal (>5 kb) regions of known gene starts. The distribution of equal numbers of randomly generated sites is also shown. C, graph shows the percentages of acetylation of different classes of STAT1- and IRF1-binding sites compared with equal numbers of randomly generated controls. Const., constitutive; Ind. or ind., induced.
Most IFNγ-induced histone acetylation correlates with STAT1 and/or IRF1 binding
Next, we assessed whether IFNγ-induced acetylation correlates with STAT1/IRF1 binding. 69% of all inducible acetylation events occurred within 1 kb of STAT1/IRF1–binding sites and 90% were within 5 kb (Fig. 3B), which accords with previous results showing that STAT1 and IRF1 bind HATs (6, 19–23) and supports the notion that most inducible acetylation is linked to STAT1/IRF1 recruitment. The overlap between acetylation and TF binding was skewed to promoter proximal sites (Fig. 3B). Similar distributions were observed when all inducible H3ac or inducible H4ac sites were considered separately. 91% of dual inducible H3ac and H4ac sites were located within 1 kb of a TF-binding site (Fig. 3B). Although 70% of the isolated inducible H4ac were within 1 kb of a STAT1 or IRF1 binding site, only 36% of isolated inducible H3ac sites showed STAT1 or IRF1 binding. Negligible overlap was observed between STAT1/IRF1 peaks and randomly generated peaks (Fig. 3B). Overall, induction of histone acetylation correlates with STAT1/IRF1 binding, but a significant fraction of isolated H3ac modification could be due to recruitment of (an)other factor(s) or may occur indirectly through looping to one or more of the STAT1/IRF1–binding enhancers.
Distinct effects of isolated IRF1 sites on histone acetylation
STAT1 can recruit HATs (12, 19, 20, 23), and indeed we found that 90% of isolated STAT1 sites were associated with histone acetylation, with most (70%) showing induced H3/4ac and 87% of dual STAT1/IRF1 sites (87%) exhibiting induced histone acetylation (Fig. 3C). IRF1 also binds HATs (6, 21, 22), but at the IFNγ-induced GBP2 promoter, it is insufficient to induce histone acetylation if STAT1 binding is blocked (12). Whether and/or the extent to which such unproductive DNA binding applies at physiological IRF1 sites naturally lacking STAT1 is unclear. Our analysis revealed that 25% of isolated IRF1 sites showed inducible acetylation, 31% exhibited constitutive acetylation, and 44% lacked acetylation. The unacetylated IRF1 sites that lack STAT1 (i.e. unacetylated, isolated IRF1 sites) were labeled orphan sites to discriminate them from other IRF1 sites with concomitant STAT1 binding and/or histone acetylation. These data reveal distinct classes of IRF1 binding sites across the genome. Strikingly, 87% (53 of 61) of orphan sites were remote, whereas the majority (65%) of acetylated IRF1 sites were promoter proximal (Fig. 3C). Indeed, 49% (53 of 109) of all remote IRF1 sites were orphan. Thus, although STAT1 binding is almost always linked to acetylation, the effect of IRF1 recruitment is context-dependent.
Next, we compared the distribution of orphan and nonorphan IRF1 sites in HeLa cells to different classes of IFNγ-target genes. Previously we compared HeLa transcriptome data with that of IFNγ-regulated genes in multiple other cells types and divided HeLa ISGS into those where IFNγ caused induction, no effect (resistant ISGs), or repression and whether the effect was early (detected by 6 h) or late (24 or 48 h) (11). We integrated the HeLa transcriptome and ChIP data sets to compare the distribution of nonorphan versus orphan IRF1 sites relative to the nearest ISG gene class (Fig. 4A) and to calculate their TF-enrichment ratios (TERs), where the fraction of IRF1 sites is normalized to that of randomly generated sites (Fig. 4B) (11). The most proximal gene was an ISG for 60% of productive (nonorphan) IRF1 sites, which reduced to 40% for orphan IRF1 sites, but was still considerably higher than the 20% for randomly generated sites (Fig. 4A). Moreover, TER analysis showed that although nonorphan IRF1 sites were enriched at promoter-proximal regions of early and late ISGs, orphan IRF1 sites were enriched at remote sites linked to either early ISGs or resistant ISGs (Fig. 4B). The latter is consistent with nonproductive IRF1 recruitment, although looping data would be required to conclude unambiguously whether the nearby ISG is the actual target. Irrespective, these analyses demonstrate that orphan IRF1 sites are located almost exclusively at remote elements. These data also suggest that orphan sites are not randomly distributed but are proximal to ISGs. Potentially, therefore, orphan sites are productive enhancers in some cell types.
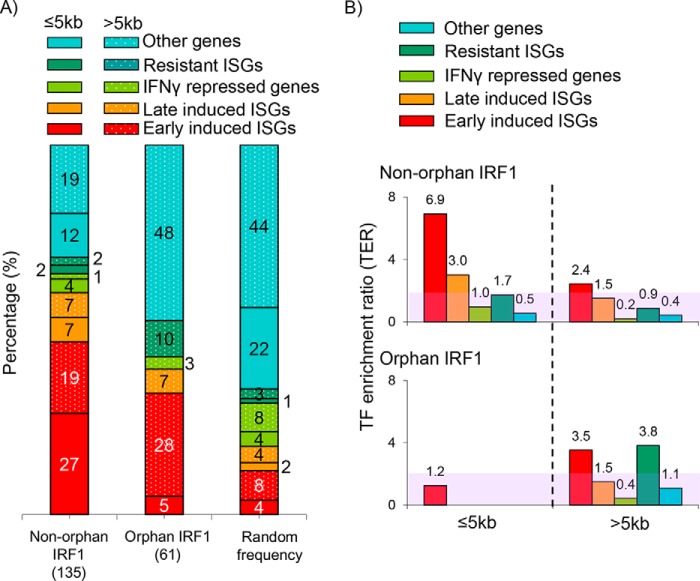
Figure 4.
Orphan IRF1 sites are skewed to remote sites of ISGs. A, histogram shows the percentages of nonorphan (productive) and orphan IRF1 sites or an equal number of randomly generated sites at proximal (≤5 kb) or distal (>5 kb) sites of the indicated classes of IFNγ affected genes or other genes unaffected by IFNγ. The number of sites in each category is indicated in parentheses. Random frequency indicates the distribution of 135 randomly generated sites. B, distribution of IRF1 sites normalized to random controls. The number of IRF1 sites from ChIP-chip data, or random computer-generated sites near to each gene class are determined, and the ratio of the former over the latter provides the TER. The unshaded region indicates TER >2-fold.
Generality of orphan IRF1 sites
To validate and extend the ChIP-chip data, we selected 12 of the 61 orphan IRF1 sites (Or1–12) for ChIP–qPCR analysis (browser shots are provided in Fig. S3). These sites were compared with seven locations at the CIITA and SOCS1 loci that were positive for IRF1 or STAT1 binding and H3/4 acetylation and four negative controls at these loci that lacked TF binding or histone acetylation. The assays were performed at 0 or 6 h after IFNγ treatment, as in the ChIP-chip study, and also at 24 h to assess late histone acetylation, as well as other histone modifications.
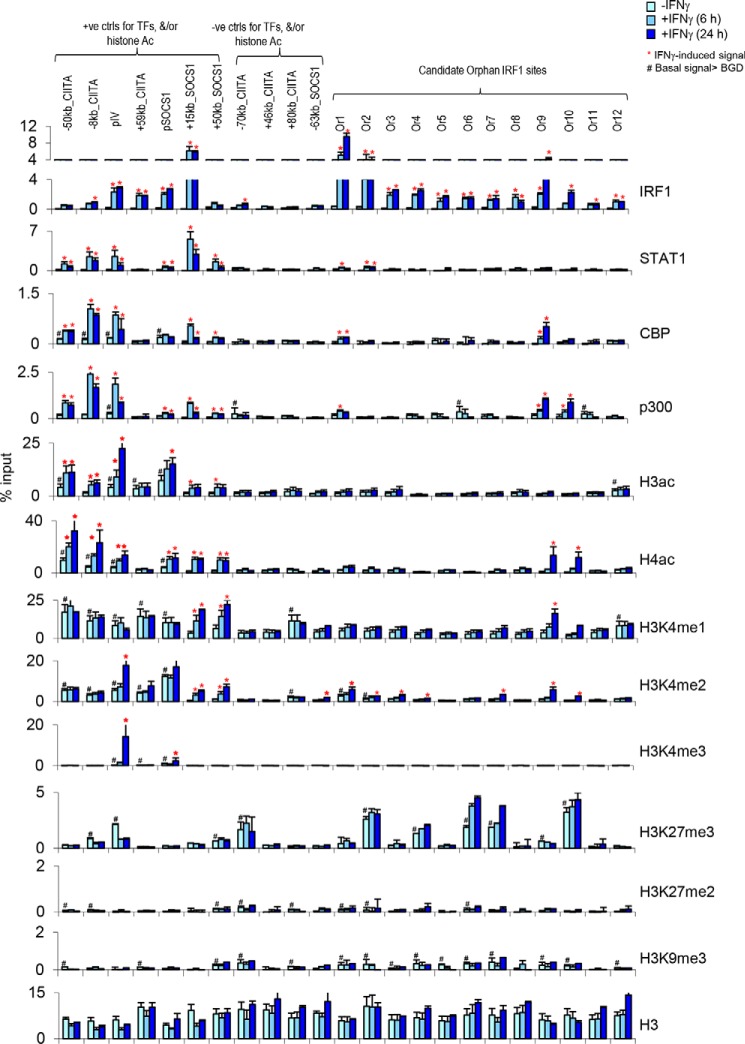
6 h after IFNγ treatment, IRF1 binding was confirmed at 83% (10/12) of orphan sites, rising to 100% by 24 h (Fig. 5). In contrast, STAT1 and H3ac signals were negligible at 10 of 12 sites and very low at the remaining 2 (cf. positive controls at CIITA and SOCS1 loci; Fig. 5). There was no H3ac or H4ac induction at any site after 6 h, still no H3ac at 24 h, and only 2 of 12 sites exhibited H4ac at 24 h. These analyses validate the ChIP-chip data, and further reveal that, apart from rare cases, acetylation is absent rather than delayed at orphan sites.
Figure 5.
A dearth of activating chromatin marks at orphan IRF1 sites in HeLa cells. ChIP–qPCR was performed to detect STAT1, IRF1, and the indicated coactivators or epigenetic marks in HeLa cells 0, 6, and 24 h after IFNγ treatment. Tested sites included 7 positive controls (ctrls), 4 negative controls at the CIITA and SOCS1 loci, and 12 randomly selected orphan IRF1 sites (Or1–Or12). *, site at which the IFNγ-induced signal is >2-fold above basal levels at the same site and also >2-fold above the signal at the negative control +46 kb site near SOCS1 after treatment. #, site at which basal levels are >2-fold above basal levels at the +46 kb negative control site. The data are expressed as percentages of input and are from at least two independent experiments, and error bars indicate the range.
We also examined histone methylation in these follow up studies. H3K4me3 marks promoters, and consistent with the location of orphan sites outside annotated TSSs (Figs. 3A and 4), none exhibited basal or induced levels of this mark (Fig. 5). H3K4me1 and H3K4me2 mark enhancers, and similar to acetylation, these modifications were not induced at 6 h (Fig. 5). At 24 h, H3K4me1 was induced at one of the two sites also marked with H4ac, whereas H3K4me2 showed low-level induction at several orphan sites, including both those marked with H4ac (Fig. 5). Thus, there is some low-level IFNγ-induced methylation at orphan sites, consistent with a potential enhancer function in some contexts, but it is delayed and below the basal or induced levels at active enhancers (cf. positive controls in Fig. 5). These data reveal a dearth of activating histone modifications linked to IRF1 binding at orphan sites.
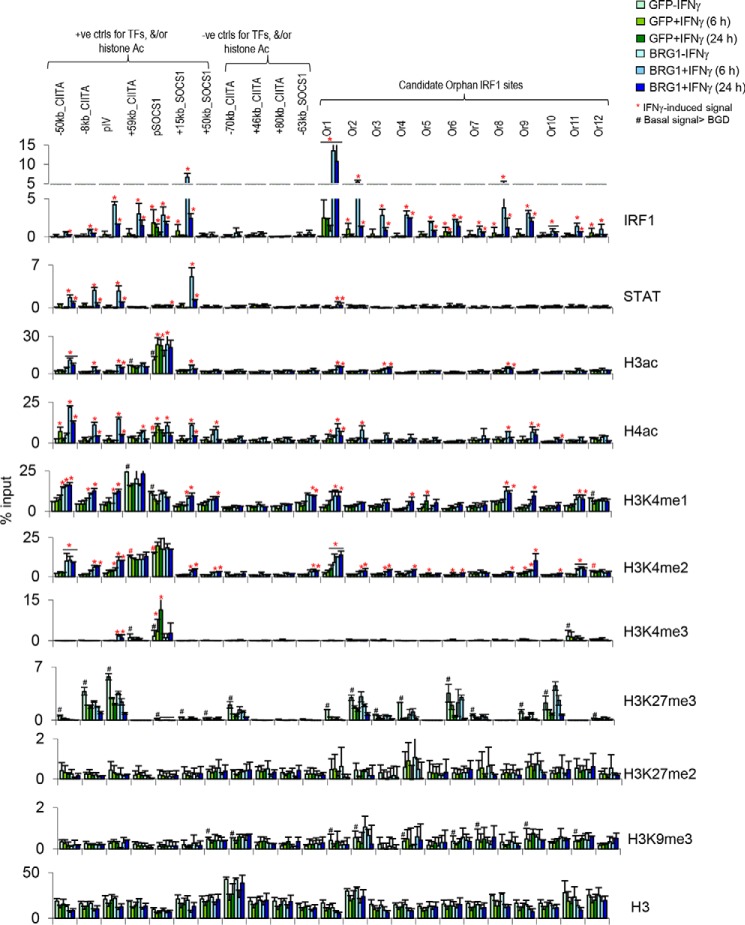
To address the generality of these findings, we assessed acetylation at orphan sites in the SW13 human adrenal carcinoma cell line. These cells lack BRG1, a chromatin remodeling enzyme critical to prime IFNγ targets for recruitment of IRF1, other TFs, and subsequent chromatin modifications (4, 24–26). Therefore, this approach also allowed us to assess whether orphan sites, like other IFNγ regulated elements, exhibit BRG1 dependence. We repeated the above ChIP–qPCR experiments in SW13 cells transduced with adenovirus vectors expressing either GFP or BRG1, as described (25). One day after viral transduction, the cells were stimulated with IFNγ for 0, 6, or 24 h. Strong STAT1, IRF1 binding, and H3/4ac induction was observed at positive control sites, which was BRG1-dependent as reported before (Fig. 6) (4). No TF recruitment or H3/4ac was seen at any of the negative controls. All 12 of the orphan IRF1 sites found originally in HeLa cells showed BRG1-dependent IRF1 binding in SW13 cells (Fig. 6). STAT1 was absent in 10 of 11 of the sites and very weakly induced at the remaining site (Fig. 6). H3ac and H4ac were unaffected at 9 of 12 and 8 of 12 sites, respectively, and positive sites exhibited only weak induction (Fig. 6). Similarly, H3K4me1 was induced at only 3 of 12 sites by 6 h, rising to 6 of 12 at 24 h, but again, induction was typically lower than basal or induced levels at positive control enhancers (Fig. 6). These data indicate that orphan sites are bona fide IRF1 targets in different cell types and show that despite the atypical downstream response, the upstream regulation of recruitment matches that of productive IRF1 enhancers. Moreover, although SWI/SNF permits IRF1 binding, it is insufficient to ensure subsequent coactivator recruitment, invoking the role of other factors.
Figure 6.
Absence of activating chromatin marks and BRG1 dependence of orphan sites in SW13 adrenocortical cells. ChIP–qPCR was performed to detect STAT1, IRF1, and the indicated coactivators or epigenetic marks in SW13 cells 0, 6, and 24 h after IFNγ treatment. The cells were transduced with adenovirus expressing GFP or BRG1 24 h before IFNγ treatment. Tested sites, abbreviations, and analysis are as in Fig. 5. ctrl, control.
Repressive chromatin and coactivator absence characterize orphan sites
The inability of IRF1 to induce acetylation at orphan sites could reflect inactivation of recruited HATs or failure of IRF1 to recruit HATs. To distinguish between these possibilities, we assessed p300 and CBP binding in HeLa cells, because IFNγ-induced recruitment of these HATs occurs at multiple CIITA enhancers (4, 25). p300/CBP binding was detected at several of the acetylated positive control enhancers but was absent at orphan sites, with the notable exception of the two orphan sites that also showed delayed induction of H4ac (Fig. 5 and Fig. S4A).
IRF1 is insufficient to recruit HATs at the GBP2 promoter unless STAT1 is also present (12). However, 25% of physiologically relevant IRF1-binding events induce acetylation without corecruitment of STAT1 (Fig. 3C). Potentially, chromatin context interferes with coactivator recruitment at orphan sites. Thus, we next assessed whether repressive histone methylation is associated with orphan sites, including H3K9 and H3K27 methylation. H3K9me3 was present at orphan sites, but at very low levels (Fig. 6). However, high levels of H3K27me3 marked half of the orphan sites (Fig. 6). Thus, the inability of orphan sites to recruit HATs may be linked to a repressive chromatin environment.
Discussion
Recruitment of activators to DNA is associated with histone modifications. Here, we used H3 and H4 acetylation as proxies to indicate productive STAT1 and IRF1 recruitment to DNA in vivo. STAT1 binding was virtually always associated with histone acetylation, primarily of histone H4. Prior work revealed that IRF1 binding to the GBP2 promoter is insufficient for histone acetylation in the absence of STAT1 (12). The extent to which such unproductive IRF1 binding occurs naturally was unknown. By integrating prior STAT1/IRF1 ChIP-chip data (11) with new H3ac and H4ac ChIP-chip data, our results show that at isolated IRF1 sites lacking STAT1, 25% undergo inducible histone acetylation, 31% exhibit constitutive histone acetylation, and 44% show no histone acetylation (orphan sites). Thus, IRF1 does promote histone acetylation independent of STAT1 but in a context-specific manner.
In addition to acetylation, IRF1 binding to orphan sites also did not induce activating histone methylation marks. Thus, multiple features argue that IRF1 recruitment is indeed unproductive at these locations. Similar results were observed for multiple test sites in both cervical epithelia (HeLa) and adrenocortical (SW13) cell types, suggesting broad relevance. Binding of IRF1 to orphan sites was BRG1-dependent, exactly like acetylated IRF1 sites; thus the upstream regulation of activator recruitment is typical, even though the downstream result is unproductive.
Acetylation at IRF1 sites correlated precisely with corecruitment of coactivators like p300 and CBP. Such productive enhanceosome formation may depend on cooperation with other DNA-bound activators that together produce an ideal binding surface for coactivators (6). In line with this idea, STAT1 and IRF1 binding together almost always induced acetylation. However, STAT1 is not a prerequisite for productive enhanceosome formation, because we identified many isolated IRF1 binding events that induced histone acetylation. Our genome-scale analyses reveals key genomic and biochemical features that distinguish contexts in which IRF1 does or does not induce acetylation. First, most isolated IRF1 sites that are acetylated inducibly or constitutively are promoter-proximal, whereas the vast majority of orphan sites are remote (Fig. 3C). In addition, the latter sites are often associated with repressive chromatin that is hypoacetylated and/or marked with repressive histone methylation (e.g. H3K27me3). Thus, genomic location and chromatin context appear to be key features that influence the ability of IRF1, in the absence of STAT1, to activate an enhancer. Limited accessibility could interfere with the recruitment of other TFs to nearby sites and thus impede construction of a full enhanceosome. H3K27me3 is deposited by polycomb repressive complex 2 (PRC2), which BRG1 combats to ensure ISGs remain IFN responsive (26, 27). In BRG1-deficient cells, PRC2/H3K27me3 completely blocks IRF1 (or STAT1) binding, but at orphan sites marked with H3K27me3, IRF1 could bind, providing BRG1 was present. It would be informative in the future to use chromatin accessibility assays to compare orphan versus nonorphan sites and to define whether PRC2 has any role in blocking HAT recruitment at orphan sites.
Recent work showed that IRF1 competes with IRF4 at the interleukin 9 locus in T helper 9 cells (28). This intriguing property is distinct from the orphan sites we identified here, because whereas IRF1 reduces histone acetylation at the IL9 locus, the orphan sites in our study are already completely deacetylated. Thus, at the IL9 locus in T helper 9 cells, IRF1 actively represses transcription, whereas at orphan sites in HeLa and SW13 cells, multiple criteria indicate that binding is simply unproductive rather than actively repressive.
The function of nonproductive IRF1 recruitment is unclear. Our data indicate that the most proximal gene is usually an ISG, arguing that they could be functional in certain contexts. Conceivably, orphan IRF1 sites may increase responsiveness to a second signal or so-called “priming.” This appears to be the case for the tumor necrosis factor locus in monocytes and macrophages where IFNγ induces IRF1 recruitment to an upstream enhancer without affecting H3K27 acetylation but promotes subsequent responsiveness to lipopolysaccharide (29). Orphan IRF1 sites may thus prime enhancers to respond to this or other foreign agents.
The absence of chromatin activity at a large subset of IRF1 sites resembles observations at a subset of ERα targets (7, 8). Nuclear receptors can act as silencers or activators, so it was unclear whether the absence of positive chromatin activity might apply to activators other than ERα. Our work suggests that unproductive activator binding is a general phenomenon. Presumably, orphan sites may be functional in a distinct cell type or in response to another signal. Irrespective, this discovery suggests that interpretation of binding data requires insight into the functional consequences of TF binding.
Experimental procedures
Cell lines and adenovirus transduction
HeLa–ini1–11 cells (HeLa) and SW13 cells were grown as described (24). IFNγ treatment and transduction with adenovirus was also as described (25).
Custom oligonucleotide ChIP tiling array design
A custom oligonucleotide tiling array was designed to cover 11 genomic regions spanning a total of 16 Mb of human genomic DNA in 8 chromosomes (11). Briefly, regions covered from 1 to 5 Mb genomic sequences. Arrays consisted of 50-mers, in quadruplicate, with median probe spacing of 80 bp within nonrepetitive DNA regions.
ChIP ChIP-chip and ChIP-quantitative PCR
ChIP-chip assays were performed and analyzed as described (24). Data are shown in browser format at http://research.lunenfeld.ca/IFNy3 and also were deposited in GEO under code GSE11131. ChIP–qPCR assays were performed as described (24). Antibodies and primers are listed in Tables S2 and S3, respectively.
Author contributions
M. A. E. H., Z. X., and R. B. conceptualization; M. A. E. H., Z. X., and T. Y. data curation; M. A. E. H., K. H., Z. X., T. Y., and R. B. formal analysis; M. A. E. H. validation; M. A. E. H., Z. X., and T. Y. investigation; M. A. E. H. and Z. X. visualization; M. A. E. H., Z. X., and T. Y. methodology; M. A. E. H. and R. B. writing-original draft; M. A. E. H. and R. B. writing-review and editing; R. B. supervision; R. B. funding acquisition; R. B. project administration.
Supplementary Material
This work was supported by funding from the Krembil Foundation (to R. B.) and from the Canadian Cancer Society Research Institute (to R. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S4.
Data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession number GSE11131.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
M. Abou El Hassan and R. Bremner, unpublished data.
- ERα
- estrogen receptor α
- STAT1
- signal transducer and activator of transcription 1
- IRF1
- interferon regulatory factor 1
- IFN
- interferon
- ISG
- IFN-stimulated gene
- CREB
- cAMP-responsive element-binding protein
- CBP
- CREB-binding protein
- qPCR
- quantitative PCR
- TER
- transcription factor enrichment ratio
- PRC2
- polycomb repressive complex 2.
References
- 1. Cosma M. P. (2002) Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10, 227–236 10.1016/S1097-2765(02)00604-4 [DOI] [PubMed] [Google Scholar]
- 2. Cosma M. P., Tanaka T., and Nasmyth K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97, 299–311 10.1016/S0092-8674(00)80740-0 [DOI] [PubMed] [Google Scholar]
- 3. Soutoglou E., and Talianidis I. (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295, 1901–1904 10.1126/science.1068356 [DOI] [PubMed] [Google Scholar]
- 4. Ni Z., Abou El Hassan M., Xu Z., Yu T., and Bremner R. (2008) The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat. Immunol. 9, 785–793 10.1038/ni.1619 [DOI] [PubMed] [Google Scholar]
- 5. Bemer M. (2018) Unraveling the complex epigenetic mechanisms that regulate gene activity. Methods Mol. Biol. 1675, 205–231 10.1007/978-1-4939-7318-7_13 [DOI] [PubMed] [Google Scholar]
- 6. Merika M., Williams A. J., Chen G., Collins T., and Thanos D. (1998) Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol. Cell 1, 277–287 10.1016/S1097-2765(00)80028-3 [DOI] [PubMed] [Google Scholar]
- 7. Lupien M., Eeckhoute J., Meyer C. A., Krum S. A., Rhodes D. R., Liu X. S., and Brown M. (2009) Coactivator function defines the active estrogen receptor α cistrome. Mol. Cell. Biol. 29, 3413–3423 10.1128/MCB.00020-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lupien M., Meyer C. A., Bailey S. T., Eeckhoute J., Cook J., Westerling T., Zhang X., Carroll J. S., Rhodes D. R., Liu X. S., and Brown M. (2010) Growth factor stimulation induces a distinct ERα cistrome underlying breast cancer endocrine resistance. Genes Dev. 24, 2219–2227 10.1101/gad.1944810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Negishi H., Taniguchi T., and Yanai H. (2017) The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb. Perspect. Biol. pii, a028423 10.1101/cshperspect.a028423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suzuki K., and Luo Y. (2017) Histone acetylation and the regulation of major histocompatibility class II gene expression. Adv. Protein Chem. Struct. Biol. 106, 71–111 10.1016/bs.apcsb.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 11. Abou El Hassan M., Huang K., Eswara M. B., Xu Z., Yu T., Aubry A., Ni Z., Livne-Bar I., Sangwan M., Ahmad M., and Bremner R. (2017) Properties of STAT1 and IRF1 enhancers and the influence of SNPs. BMC Mol. Biol. 18, 6 10.1186/s12867-017-0084-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramsauer K., Farlik M., Zupkovitz G., Seiser C., Kröger A., Hauser H., and Decker T. (2007) Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-γ-inducible gbp2 gene. Proc. Natl. Acad. Sci. U.S.A. 104, 2849–2854 10.1073/pnas.0610944104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., and Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 14. Roh T. Y., Cuddapah S., and Zhao K. (2005) Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19, 542–552 10.1101/gad.1272505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., Afzal V., Ren B., Rubin E. M., and Pennacchio L. A. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., and Jaenisch R. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robertson A. G., Bilenky M., Tam A., Zhao Y., Zeng T., Thiessen N., Cezard T., Fejes A. P., Wederell E. D., Cullum R., Euskirchen G., Krzywinski M., Birol I., Snyder M., Hoodless P. A., et al. (2008) Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 18, 1906–1917 10.1101/gr.078519.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varinou L., Ramsauer K., Karaghiosoff M., Kolbe T., Pfeffer K., Müller M., and Decker T. (2003) Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-γ-dependent innate immunity. Immunity 19, 793–802 10.1016/S1074-7613(03)00322-4 [DOI] [PubMed] [Google Scholar]
- 20. Zakharova N., Lymar E. S., Yang E., Malik S., Zhang J. J., Roeder R. G., and Darnell J. E. (2003) Distinct transcriptional activation functions of STAT1α and STAT1β on DNA and chromatin templates. J. Biol. Chem. 278, 43067–43073 10.1074/jbc.M308166200 [DOI] [PubMed] [Google Scholar]
- 21. Eklund E. A., and Kakar R. (1999) Recruitment of CREB-binding protein by PU.1, IFN-regulatory factor-1, and the IFN consensus sequence-binding protein is necessary for IFN-γ-induced p67phox and gp91phox expression. J. Immunol. 163, 6095–6105 [PubMed] [Google Scholar]
- 22. Leung Y. T., Shi L., Maurer K., Song L., Zhang Z., Petri M., and Sullivan K. E. (2015) Interferon regulatory factor 1 and histone H4 acetylation in systemic lupus erythematosus. Epigenetics 10, 191–199 10.1080/15592294.2015.1009764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J. J., Vinkemeier U., Gu W., Chakravarti D., Horvath C. M., and Darnell J. E. (1996) Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc. Natl. Acad. Sci. U.S.A. 93, 15092–15096 10.1073/pnas.93.26.15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pattenden S. G., Klose R., Karaskov E., and Bremner R. (2002) Interferon-γ-induced chromatin remodeling at the CIITA locus is BRG1 dependent. EMBO J. 21, 1978–1986 10.1093/emboj/21.8.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ni Z., Karaskov E., Yu T., Callaghan S. M., Der S., Park D. S., Xu Z., Pattenden S. G., and Bremner R. (2005) Apical role for BRG1 in cytokine-induced promoter assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 14611–14616 10.1073/pnas.0503070102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abou El Hassan M., Huang K., Eswara M. B., Zhao M., Song L., Yu T., Liu Y., Liu J. C., McCurdy S., Ma A., Wither J., Jin J., Zacksenhaus E., Wrana J. L., and Bremner R. (2015) Cancer cells hijack PRC2 to modify multiple cytokine pathways. PLoS One 10, e0126466 10.1371/journal.pone.0126466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abou El Hassan M., Yu T., Song L., and Bremner R. (2015) Polycomb repressive complex 2 confers BRG1 dependency on the CIITA locus. J. Immunol. 194, 5007–5013 10.4049/jimmunol.1403247 [DOI] [PubMed] [Google Scholar]
- 28. Campos Carrascosa L., Klein M., Kitagawa Y., Lückel C., Marini F., König A., Guralnik A., Raifer H., Hagner-Benes S., Rädler D., Böck A., Kang C., Lohoff M., Garn H., Schaub B., et al. (2017) Reciprocal regulation of the Il9 locus by counteracting activities of transcription factors IRF1 and IRF4. Nat. Commun. 8, 15366 10.1038/ncomms15366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chow N. A., Jasenosky L. D., and Goldfeld A. E. (2014) A distal locus element mediates IFN-γ priming of lipopolysaccharide-stimulated TNF gene expression. Cell Rep. 9, 1718–1728 10.1016/j.celrep.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.