ABSTRACT
This study evaluated the geospatial distribution of fecal indicator bacteria (FIB) (i.e., Escherichia coli, Enterococcus spp.) and the alternative fecal indicator pepper mild mottle virus (PMMoV) in tropical freshwater environments under different land use patterns. Results show that the occurrence and concentration of microbial fecal indicators were higher for urban than for parkland-dominated areas, consistent with land use weightage. Significant positive correlations with traditional FIB indicate that PMMoV is a suitable indicator of fecal contamination in tropical catchments waters (0.549 ≤ rho ≤ 0.612; P < 0.01). PMMoV exhibited a strong significant correlation with land use weightage (rho = 0.728; P < 0.01) compared to traditional FIB (rho = 0.583; P < 0.01). In addition, chemical tracers were also added to evaluate the potential relationships with microbial fecal indicators. The relationships between diverse variables (e.g., environmental parameters, land use coverage, and chemical tracers) and the occurrence of FIB and PMMoV were evaluated. By using stepwise multiple linear regression (MLR), the empirical experimental models substantiate the impact of land use patterns and anthropogenic activities on microbial water quality, and the output results of the empirical models may be able to predict the sources and transportation of human fecal pollution or sewage contamination. In addition, the high correlation between PMMoV data obtained from quantitative real-time PCR (qPCR) and viral metagenomics data supports the possibility of using viral metagenomics to relatively quantify specific microbial indicators for monitoring microbial water quality (0.588 ≤ rho ≤ 0.879; P < 0.05).
IMPORTANCE The results of this study may support the hypothesis of using PMMoV as an alternative indicator of human fecal contamination in tropical surface waters from the perspective of land use patterns. The predictive result of the occurrence of human fecal indicators with high accuracy may reflect the source and transportation of human fecal pollution, which are directly related to the risk to human health, and thereafter, steps can be taken to mitigate these risks.
KEYWORDS: fecal indicators, land use, chemical tracers, freshwater environments, regression models
INTRODUCTION
Fecal indicators are used to detect the presence of fecal pathogens in diverse environments. Ideal indicators satisfy the following criteria: presence in the same warm-blooded animal's intestinal tract; presence only in fecally contaminated environments and absence in uncontaminated ones; presence in greater numbers than pathogens; absence of growth and proliferation in the environment; survival patterns similar to those of pathogens; easy detection, cost-effectiveness, and low pathogenic risks (1, 2). To date, numerous studies have correlated the occurrence of traditional fecal indicator bacteria (FIB), most notably Escherichia coli and Enterococcus spp., with fecal pollution in water (3–5). However, the application of traditional FIB in determining recreational water safety has been challenging due to their observed growth and persistence in natural environments (1, 3). In addition, they cannot differentiate the source of fecal contamination between humans and animals (6, 7).
Recently, the pepper mild mottle virus (PMMoV) has been proposed as an alternative indicator for human fecal pollution, as it has been detected at high concentrations in human fecally contaminated surface waters (104 to 107 gene copies/liter) and wastewaters (106 to 1010 gene copies/liter) in Bolivia, Vietnam, Germany, Japan, Singapore, Australia, and the United States (8–14). PMMoV is a plant-pathogenic virus commonly found in human intestinal tracts (8), as it is consumed with peppers in human diets, passing relatively unharmed through the digestive tract, and excreted in feces at concentrations between 105 and 1010 gene copies/gram dry weight feces (8, 10, 15, 16). In terms of specificity, PMMoV has been found in nearly all human samples but was detected in only a minority of chicken, gull, and cow feces at low concentrations (10, 11). Also, this virus has shown great persistence, with a half-life of approximately 1.5 days (11, 17) in seawater and relatively low reduction from conventional and advanced wastewater treatment (8, 10). As such, PMMoV may be used as a conservative marker of sewage contamination (17), though it might overestimate viral pathogens to some extent through wastewater treatment. However, Symonds et al. (18) revealed that PMMoV detection in marine surface waters contaminated with untreated wastewater did not overestimate the risk of gastrointestinal (GI) illness. PMMoV overcomes the false-negative issues caused by reference viral pathogens due to its high concentrations (10, 18).
Most studies related to fecal indicators focus on their detection and occurrence in diverse water environments. However, investigations on the driving factors in the distribution and occurrence of these indicators and associated models are limited. Among the driving factors, environmental (e.g., pH, temperature, salinity, turbidity, UV, etc.) and hydrologic (rainfall runoff) conditions have been well studied, previously using empirical models to predict target microbial concentrations in marine and temperate freshwater systems (6, 19, 20). Predictive models have been applied in the Great Lakes to provide timely information in beach notification programs (21).
Even though the application of site-specific statistical models has recently been promoted by the U.S. Environmental Protection Agency (EPA), many site-specific models have focused on limited predictors and have neglected potential factors (e.g., land use) (21). Land use, due to its difficulty in classification, quantification, and surveillance, has rarely been included in metadata correlation. However, human-induced land use changes are the key drivers in transmitting many infectious diseases and are associated with fecal pollution (22). It is hypothesized that land use change will influence the characteristics of tropical water cycles and would further influence the occurrence of different microbial pathogens and indicators, as well as the distribution patterns of chemical tracers. Previous studies have begun to address the impacts of land cover on the distribution of pathogens in human-impacted water bodies. For example, the occurrence of human viruses showed a distinct pattern determined by land cover for studies conducted in the Milwaukee River watershed, Michigan River, coastal rivers of southern California, and the Great Lakes (23–26). Paule-Mercado et al. (27) revealed that land use and anthropogenic activities influenced intraevent variability of FIB through multiple linear regression (MLR) models in different monitoring sites. However, the studies failed to quantify the land use categories into predictive variables to assess the relationship between indicators and land use (27, 28).
Apart from land cover factors, chemical tracers (e.g., pharmaceuticals and personal care products, fecal sterols, fluorescent whitening agents) are also closely associated with possible wastewater pollution in surface waters (29, 30). A recent study showed that acetaminophen (ACT), salicylic acid (SA), and carbamazepine (CBZ) were suitable chemical markers of raw wastewater contamination in surface water and groundwater for highly urbanized catchment areas (30). These studies have enabled progress in our comprehension of how land cover factors and chemical tracers can influence water quality in the urban water cycle. The challenge moving forward is to derive data-based empirical models between predictors (i.e., environmental parameters, land use cover, and chemical tracers) and the concentration and distribution patterns of fecal indicators to predict fecal contamination in future scenarios. Until now, only one such study has been documented. This study predicted the concentration of fecal coliforms in the Wachusett Reservoir by the combined impacts of temperature, rainfall, and land use characteristics using the approach of Bayesian overdispersed Poisson models (31). However, a single microbial fecal indicator may not be sufficient in identifying the contamination sources in surface waters (32).
To address these gaps, the specific aims of this study were to (i) investigate and compare the spatial variation of E. coli, Enterococcus spp., and PMMoV in tropical surface waters and (ii) evaluate the relationships between land use patterns, chemical markers, and environmental parameters on the distribution of traditional FIB and the alternative fecal indicator, PMMoV, for a tropical urban watershed. Along with molecular (quantitative real-time PCR [qPCR]) and enzyme-based (Idexx Quanti-Tray method) assays for quantifying the concentrations of microbial fecal indicators, viral metagenomics was used to provide insights into the viral community and diversity, which would not have been captured by targeted approaches. Ultimately, the predictive result of the occurrence of human fecal indicators with high accuracy could reflect the source and transportation of human fecal pollution, which are directly related to the risk to human health, and thereafter, steps can be taken to mitigate these risks.
RESULTS AND DISCUSSION
Geospatial analysis of sampling sites.
Table S1 in the supplemental material details the specified land use subcategories under four main land use categories (residential, urban, green, and agriculture). Table S2 in the supplemental material shows a summary of the land use coverage percentage classified under 4 main categories (e.g., residential, urban, green, and agriculture), land use weightage, and general characteristics for each sampling point (i.e., sites A to G). According to the land use assessment, sites A, B, and C were regarded as urban-use-dominated areas, as they included more than 65% of residential and urban areas, and site D was regarded as an agriculturally dominated area, as it was the only sampling location with a reasonable percentage (11 to 67%) of agricultural area. Sites E and G were parkland-dominated areas, as they had more than 95% green areas. Site F had 80% of residential and urban categories; however, there was no drain directly linked to the reservoir at this site, and the chance for possible fecal contamination was relatively small. Hence, site F was regarded as a parkland-dominated area as well. For catchment sampling, only the catchments of sites A, B, and D were sampled. Note that upstream water catchments feed their corresponding water body.
Environmental parameters.
A total of 171 water samples were collected from surface waters in Singapore. The minimum, maximum, and mean of water quality parameters from different sampling locations are summarized in Table S3 in the supplemental material. The water temperature and pH remained relatively stable throughout the sampling period. Other water quality parameters varied widely, especially in catchments, including conductivity (3 to 955 μS), salinity (0 to 1 practical salinity unit [PSU]), dissolved oxygen (DO; 1.04 to 10.3 ppm), total dissolved solids (TDS; 0.15 to 477 ppm), and turbidity (0 to 824 nephelometric turbidity units [NTU]).
Quantitative detection of microbial targets and chemical tracers in freshwater samples.
E. coli and Enterococcus spp. were detected in approximately 90% of the samples, with geometric mean counts of 79 most probable number (MPN)/100 ml and 41 MPN/100 ml, respectively (Table 1). In Singapore, the microbial water quality guideline for recreational freshwater is 200 MPN/100 ml Enterococcus spp. or lower for 95% of the time. All the reservoir sites met the Singapore recreational water criteria. qPCR results for PMMoV showed a lower detection frequency (69%) than those of the bacteria, with a geometric mean concentration of 763 log of standard plasmid concentration (GC)/100 ml of all the samples. This result is similar to that of a previous study conducted in Japan, in which PMMoV had a 76% detection frequency in surface water with a mean concentration of 550 GC/100 ml (16). Elsewhere, PMMoV had a 100% detection frequency (5 × 105 to 1.07 × 105 GC/100 ml) in river waters containing wastewater treatment plant (WWTP) effluent in Germany, a 94% detection frequency in surface water in Vietnam, and an 85% detection frequency in groundwater in Mexico (1.79 × 103 to 1.04 × 103 GC/100 ml) (10, 33). Another study conducted in Florida, an area with a climate similar to that of Singapore, showed a 60% detection frequency in coastal waters, with the highest concentrations reaching 8.73 × 104 GC/100 ml (18). In Singapore, there is no pepper farming or harvesting in the vicinity of the sampling sites, indicating that our samples containing PMMoV were likely impacted by fecal contamination from possible sewage or anthropogenic activities. Nonetheless, there is a possibility that improper disposal of processed food containing peppers could be a contributor as well, since such food products containing peppers are known to contain PMMoV (9).
TABLE 1.
Frequencies and occurrence of fecal indicators (E. coli, Enterococcus spp., and PMMoV) and chemical tracersa
| Microorganism or chemical tracer (unit for geometric mean or SD or for detection limit) | No. of samples | Detection frequency (%) | Geometric mean | Geometric SD | Detection limit |
|---|---|---|---|---|---|
| E. coli (MPN/100 ml) | 171 | 90 | 79 | 34.44 | 1 |
| Enterococcus spp. (MPN/100 ml) | 171 | 89 | 41 | 24.64 | 1 |
| PMMoV (GC/100 ml) | 171 | 69 | 763 | 42.86 | 16 |
| ACE (ng/liter) | 119 | 100 | 100 | 2.90 | 1 |
| ACT (ng/liter) | 118 | 49 | 11 | 29.26 | 1 |
| CBZ (ng/liter) | 119 | 95 | 0.90 | 4.20 | 0.01 |
| CYC (ng/liter) | 118 | 98 | 44 | 3.60 | 0.5 |
Abbreviations: ACE, acesulfame; ACT, acetaminophen; CYC, cyclamate; CBZ, carbamazepine.
Three of the four selected chemical tracers were detected in more than 95% of samples, while the chemical ACT was detected in only 49% of the samples. Their geometric mean concentrations varied widely, with acesulfame (ACE) and cyclamate (CYC) having higher concentrations (100 ng/liter and 44 ng/liter, respectively) than did CBZ, which had a concentration of 0.9 ng/liter, and ACT, which had a concentration of 11 ng/liter. The relatively low detection frequency of ACT in collected samples indicated low raw wastewater contamination in urban surface waters, as ACT is biodegradable and has been utilized for detecting raw wastewater contamination in urban surface waters (30).
Distribution of microbial fecal indicators and chemical tracers at different sampling sites.
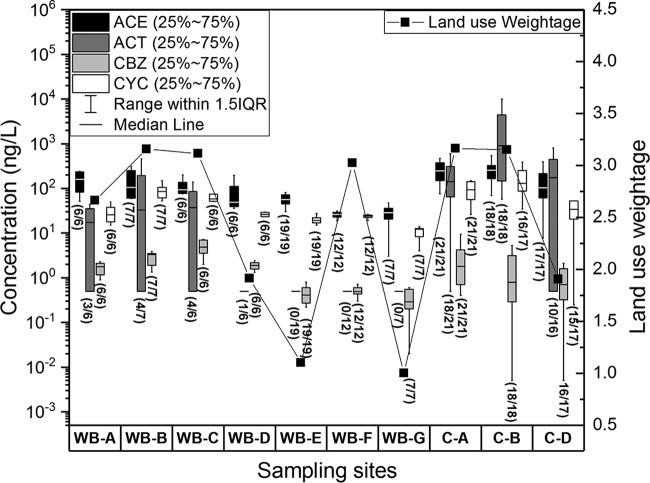
The concentrations of microbial fecal indicators and chemical tracers were generally lower in water bodies (sites A, B, and D) than those in their corresponding catchments (sites A, B, and D) (Mann-Whitney U test, P value < 0.05), suggesting that the contamination occurs in the upstream catchments for these sites (Fig. 1 and 2), as upstream water catchments feed their corresponding water bodies. For the other sites, the concentrations of microbial fecal indicators and chemical tracers were lower in water bodies E, F, and G than in water bodies A, B, and C (Mann-Whitney U test, P < 0.05). From the land use assessment, sites A, B, and C were classified as urban-use-dominated areas, while E, F, and G were classified as parkland-dominated areas. These results confirm the fecal contamination on water quality by anthropogenic activities or sewage contamination possibly caused by sewage at the urban-use-dominated sites A, B, and C. In addition, concentrations of microbial fecal indicators in water body D (an agriculture-dominated area) were higher than the concentrations in parkland-dominated areas but lower than the concentrations in urban-use-dominated areas, indicating that agricultural runoff may have a moderate impact on human fecal contamination due possibly to poultry, aquaculture, and vegetable farming. In general, the concentrations of selected targets were consistent with land use weightage, except for sampling site WB-F, where there were no direct drain connections and relatively small chances for fecal contamination from the surrounding environment (Fig. 1 and 2). These results substantiate the high correlations observed between the selected targets and land use weightage, implying that land use patterns and anthropogenic activities greatly influence the distribution patterns of microbial fecal indicators and chemical markers.
FIG 1.
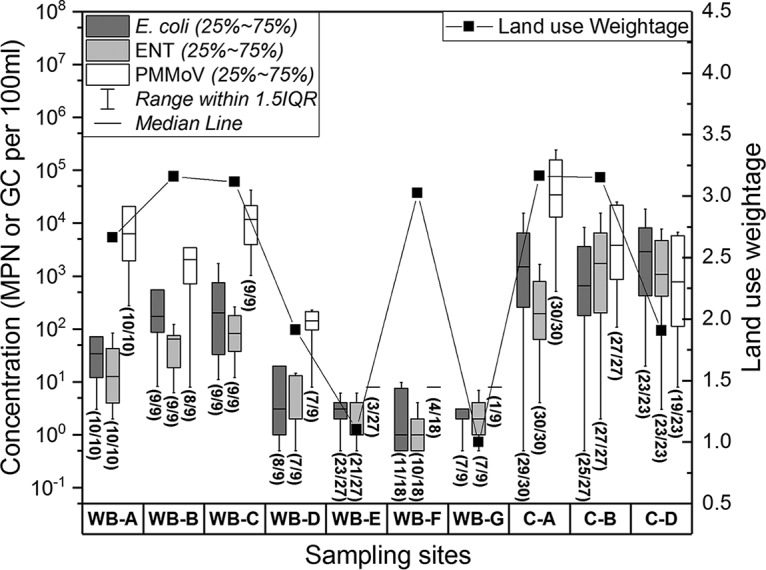
Box plot concentrations of E. coli, Enterococcus spp. (ENT), and PMMoV in different water bodies and catchments (different sampling points within the same location are merged for simplicity). The median value is represented by the line inside the box, which indicates the range (25% to 75%). The solid line (■) shows the land use weightage at different sampling points. The higher weightage of land use category means that the areas are more impacted by anthropogenic activities. All the samples, including the nondetect samples with reactive oxygen species (ROS) imputed values, are shown in the plots. Numbers in parentheses show the detection frequency of different targets at different sampling sites as the number of positive samples over the total number of samples at each site. IQR, interquartile range.
FIG 2.
Box plot concentrations of selected chemical tracers in different water bodies and catchments (different sampling points within the same location are merged for simplicity). The median value is represented by the line inside the box, which indicates the range (25% to 75%). The solid line (■) shows the land use weightage at different sampling points. The higher weightage of land use category means that the areas are more impacted by anthropogenic activities. All the samples, including the nondetect samples with ROS imputed values, are shown in the plots. Numbers in parentheses show the detection frequency of different selected chemicals at different sampling sites as the number of positive samples over the total number of samples at each site.
In terms of detection frequency, there was a statistically significant association between the different sampling locations from different land use categories for E. coli [χ2(9) = 26.13; P = 0.002], Enterococcus spp. [χ2(9) = 39.91; P < 0.0001], PMMoV [χ2(9) = 112.95; P < 0.0001], and ACT [χ2(9) = 65.33; P < 0.0001] (see Table S4 in the supplemental material). In particular, a robust strength of association was observed between the different sampling locations from different land use categories and detection frequency of PMMoV (Φ value = 0.813; P < 0.0001) and ACT (Φ value = 0.771; P < 0.0001). This indicates that the detection frequencies of PMMoV and ACT were largely influenced by different sampling locations from different land use categories.
As mentioned before, ACT is used to indicate raw wastewater contamination and has been reported to be present in groundwater and surface waters near sewage systems (30). PMMoV has also been observed at high concentrations in untreated and/or inadequately treated wastewaters (9, 34). Within our sampling areas, used water from both domestic and nondomestic sources is collected through a combination of gravity sewers and pumping stations leading to four water reclamation plants. The final effluent is also further treated using advanced membrane and reverse osmosis technologies to high-grade water (35). Hence, it is likely that urban-use-dominated areas received wastewater contamination (possibly from sewage), as they showed the presence of both ACT and PMMoV.
Correlations between molecular methods and metagenomics analysis.
In the parallel project with the same batch of samples, significant Spearman's rank correlations were observed between molecular methods (qPCR) and metagenomics analysis for the occurrence of PMMoV (36). The results showed a strong significant correlation between PMMoV data (continuous, not binary variables) from qPCR and metagenomics (i.e., PMMoV contig 1-6, PMMoV-sum, and PMMoV-mapped reads to reference genome) (0.588 < rho < 0.879; P < 0.01), indicating that the relative abundance of PMMoV from metagenomics was aligned with the absolute abundance from qPCR (36). With the widespread application of next-generation sequencing, the NCBI SRA databases have recently expanded. In the future, with access to shared viral metagenomics data sets in such databases, the relative abundance of the alternative fecal indicator PMMoV may be obtained by viral metagenomics across different samples from diverse water environments. However, metagenomics is still unable to provide absolute quantification and as such cannot be used for quantitative microbial risk assessment (QMRA). In QMRA, risks are estimated based on the absolute concentrations of specific pathogens in the environment and the infectivity of those pathogens to humans (37). Furthermore, due to high sequencing costs, relatively long sequencing time, and extensive bioinformatics analysis of metagenomics, qPCR is still the preferable approach to detect the presence of fecal contamination. In addition, the relative abundance of E. coli and Enterococcus metagenomics data did not show a significant correlation with results from the Idexx Quanti-Tray method (P > 0.05) (see Table S5 in the supplemental material). This lack of correlation could be due to disparities in the detection methods used and the uneven biodiversity across all the samples. The Idexx Quanti-Tray method is based on the presence of enzymes from E. coli (β-glucuronidase) and Enterococcus spp. (glucosidase) (38, 39), while the metagenomics data rely on sequence detection assays. Further examination of bacterial samples shows that our samples are dominated by other bacterial classes (i.e., Betaproteobacteria, Actinobacteria, Cyanobacteria, and Acidimicrobiia), suggesting a poor coverage for E. coli and Enterococcus spp. Rarefaction curves also showed that the samples from catchments and urbanized reservoirs did not reach the plateau on genus and species levels (see Fig. S1 and S2 in the supplemental material). More sequencing reads were needed to sufficiently represent the environment from which the samples were taken. Therefore, E. coli and Enterococcus metagenomics data were not included in our later analysis. This also indicates the intrinsic limitation of metagenomics: the relative abundance data might be skewed if the sequencing depth is not sufficient to cover the low relative abundance of targets in each library.
Correlations between fecal indicators (E. coli, Enterococcus spp., and PMMoV), environmental parameters, and chemicals.
Spearman's rho correlation analysis was performed to assess the empirical relationships between traditional FIB (E. coli, Enterococcus spp.) and PMMoV as an alternative human fecal indicator. The results showed that there were moderate to strong significant positive correlations between all of the microbial fecal indicators (0.549 < rho < 0.888; P < 0.01) (Table 2). The highest correlation (rho = 0.888; P < 0.01) was observed between the PMMoV metagenomics data (i.e., PMMoV_reads mapped to reference genome) and the PMMoV qPCR data. In addition, a strong significant positive correlation was observed between E. coli and Enterococcus spp. from the culture-based method (rho = 0.859; P < 0.01). The significant positive correlations between PMMoV and traditional indicators (0.549 < rho < 0.612; P < 0.01) are in agreement with the results of Haramoto et al. (16) (P < 0.05) but contrast with other results indicating that the PMMoV concentration did not correlate well with FIB in groundwater from a karst aquifer in Mexico (P > 0.05) (33) and from the Ruhr and Rhine rivers in Germany (P > 0.01) (10). In our study, PMMoV had positive correlations with traditional FIB E. coli and Enterococcus spp., suggesting that the virus might be used as a fecal indicator, especially if there is no pepper farming or harvesting in the surrounding sampling environment, which is the case in Singapore. However, past studies have shown that FIB are not always correlated with enteric viruses (3, 6, 34), causing false-positive signals, especially in tropical regions due to the longer survival time and natural occurrence of FIB (40). Therefore, these results support PMMoV as an indicator in tropical regions.
TABLE 2.
Spearman's rank correlation between microbial indicators and environmental parameters, chemical tracers, and land use categoriesa
| Correlation parameter | Correlation coefficient |
||||
|---|---|---|---|---|---|
| E. coli (MPN/100 ml) | Enterococcus spp. (MPN/100 ml) | PMMoV by qPCR (GC/100 ml) | PMMoV _contig1 | PMMoV_ reads mapped to reference genome | |
| Microbial indicators | |||||
| E. coli (MPN/100 ml) | 0.554 | 0.599 | |||
| Enterococcus spp. (MPN/100 ml) | 0.859 | 0.588 | 0.541 | ||
| PMMoV (GC/100 ml) | 0.612 | 0.549 | 0.788 | 0.888 | |
| Environmental parametersb | |||||
| Temp (°C) | −0.344 | −0.426 | −0.209 | ||
| pH | 0.432 | 0.394 | |||
| EC | 0.521 | 0.588 | 0.394 | 0.565 | 0.543 |
| TDS (ppm) | 0.507 | 0.585 | 0.358 | 0.516 | 0.495 |
| Salinity (PSU) | 0.515 | 0.595 | 0.371 | 0.525 | 0.518 |
| DO (ppm) | −0.345 | −0.333 | −0.440 | −0.356 | |
| Turbidity (PFU) | 0.503 | 0.572 | 0.379 | ||
| Chemical tracers | |||||
| ACE (ng/liter) | 0.590 | 0.638 | 0.658 | 0.721 | 0.549 |
| ACT (ng/liter) | 0.631 | 0.727 | 0.473 | 0.638 | 0.642 |
| CBZ (ng/liter) | 0.530 | 0.717 | 0.712 | ||
| CYC (ng/liter) | 0.564 | 0.590 | 0.609 | 0.811 | 0.792 |
| Land use categories | |||||
| Residentialb | 0.421 | 0.412 | 0.678 | 0.728 | 0.608 |
| Urban | 0.497 | 0.503 | 0.473 | ||
| Green | −0.588 | −0.585 | −0.702 | −0.757 | −0.705 |
| Weightage | 0.583 | 0.583 | 0.728 | 0.766 | 0.713 |
Only significant correlations are shown (with Bonferroni correction, 2-tailed t test). All correlations were significant at P values of <0.01, except for values in boldface, for which P values were <0.05. Abbreviations: TDS, total dissolved solids; DO, dissolved oxygen; PSU, practical salinity units; ACE, acesulfame; ACT, acetaminophen; CYC, cyclamate; CBZ, carbamazepine.
For environmental parameters oxidation-reduction potential [ORP] (in millivolts), 24-h rainfall and 5-day rainfall (in millimeters), and antecedent dry period (in days) and for land use category “Agriculture,” no significant correlation was found; therefore, these parameters are not included in the table.
Among all the environmental variables, the microbial fecal indicators had moderate to strong significant positive correlations (0.358 < rho < 0.595) with conductivity, TDS, and salinity. The traditional FIB had higher correlation coefficients (0.507 < rho < 0.595; P < 0.05) with conductivity, TDS, and salinity than with PMMoV (0.358 < rho < 0.394; P < 0.05). This is similar to the results reported by Hamza et al. (10), which showed that PMMoV had moderate positive correlation with conductivity in a river in Germany, with a correlation coefficient, rho, of 0.33 (P = 0.0053, n = 72). Galdino Pereira et al. (41) also reported a possible association between conductivity and FIB in surface water in Brazil and concluded that the higher value of conductivity, indicating large amounts of ions, was associated with organic matter decomposition (rho = 0.42; P < 0.05). The significant positive correlation between TDS and traditional fecal indicators could be due to the fact that TDS provides the suitable medium for microbial fecal indicators to grow, carrying supportive nutrients (27, 42). A previous study conducted by Lian et al. in tropical surface waters in Singapore in 2015 showed that there was no significant correlation between microbial fecal indicators, conductivity, and salinity (6). The difference could be due to different types of catchments studied. In our study, samples were collected from many different environments, including urbanized, agricultural, and parkland areas, whereas the samples analyzed by Liang et al. (6) originated from highly urbanized catchments. The positive correlation between turbidity and all the microbial fecal indicators (0.333 < rho < 0.440), except PMMoV metagenomics data, was expected, and a similar correlation was also found in the earlier study by Liang et al. (6). The exception of PMMoV metagenomics data could be caused by the limited metagenomics sample size (n = 38) in our study compared to the total sample size (n = 171).
The microbial fecal indicators measured by culture-based or molecular methods had weak to strong significant negative correlations (−0.440 < rho <−0.202) with temperature and DO, similar to results from Liang et al. (6). This negative correlation between microbial fecal indicators and temperature is different from results from previous studies in temperate countries, where the temperature range is very large, spanning winter to summer temperatures (27, 43). Although warmer temperature does lead to higher growth rates (27), in equatorial Singapore the temperature range is very small (27 to 29.4°C; mean value, 28.6°C), and the warmer temperature could also result in higher decay rates of bacteria due to increased solar UV irradiation (44–46). The low DO levels may be caused by a fecal contamination event, since an increased fecal indicator concentration would increase oxygen demand and result in decreased DO levels (27).
To assess the potential of chemical tracers as markers of fecal contamination, correlations with microbial fecal indicators (E. coli, Enterococcus spp., PMMoV, PMMoV-contig1, and PMMoV-mapped reads) were evaluated for pooled data across all seven sampling sites. Coefficients of correlations between chemical tracers and microbial fecal indicators are shown in Table 2. It can be seen that all four chemical tracers showed a moderate to strong significant positive correlation to microbial fecal indicators, where two human wastewater-associated chemical markers (i.e., ACE and ACT) exhibited a moderate to strong correlation to microbial fecal indicators (0.473 < rho < 0.727; P < 0.01). This indicates that human wastewater was likely an important source of fecal indicator microorganisms (E. coli, Enterococcus spp., PMMoV, PMMoV-contig1, and PMMoV-mapped reads) in the study area, as ACE and ACT are known to be specific chemical markers of human fecal wastewater sources (29, 30, 47, 48).
Geospatial distribution of fecal indicators (E. coli, Enterococcus spp., and PMMoV) and chemicals.
To evaluate the influence of land use categories on microbial fecal indicators/chemical tracers, Spearman's rank correlations were performed among all the variables (Tables 2 and S6). There were moderate to strong significant positive correlations between all of the microbial fecal indicators/chemical tracers and residential/urban land use categories, while there were significant negative correlations with green land use categories. This suggests that microbial fecal indicators/chemical tracer sources are related to urban/residential runoff and anthropogenic activities. This finding is in agreement with those of Paule-Mercado et al. (27), who reported a positive association between FIB and urban land use in Korea. Among all of the microbial fecal indicators, PMMoV had a higher correlation with different land use categories than with traditional indicators. The strong correlations between PMMoV and residential and green categories (∣rho∣ > 0.6; P < 0.01) suggest that PMMoV may be a potential indicator of anthropogenic impacts on the distribution of pathogens in such freshwater environments. This could be expected, as FIB have their intrinsic limitations. First, FIB are able to proliferate and/or occur naturally in freshwater environments (1, 40). However, PMMoV is generally not able to replicate in aquatic environments in Singapore due to the absence of specific hosts (i.e., peppers) needed for proliferation, as there is no farming or harvesting of peppers. Second, in terms of specificity, E. coli and Enterococcus spp. cannot differentiate between human and animal feces (49–52), whereas PMMoV is more human specific (8, 10, 11). In our study, PMMoV has overcome the limitations of regrowth and specificity associated with traditional FIB; thus, it has the potential to be a more-suitable indicator of human fecal contamination. In addition, all of the chemical tracers had moderate to strong positive correlations with residential and urban land use categories, while exhibiting negative significant correlations with green land use categories (see Table S6 in the supplemental material).
Regression analysis.
In this study, multiple regression models between microbial fecal indicators and environmental parameters, land use categories, and chemical tracers were examined, followed by the stepwise variables selection approach (Table 3). These models will facilitate the prediction of microbial fecal indicators, thus indicating the potential of fecal contamination and hence, human pathogens. Note that these models used pooled data from all sampling locations.
TABLE 3.
MLR models of E. coli, Enterococcus spp., and PMMoV concentration and PMMoV metagenome relative abundancea
| Model (unit) | r2 | Model significance | Independent parameter | Coefficient | 95% confidence interval |
Significance | Constant | |
|---|---|---|---|---|---|---|---|---|
| Lower bound | Upper bound | |||||||
| E. coli (MPN/100 ml) | ||||||||
| 1 | 0.406 | 0 | Green | −0.028 | −0.037 | −0.02 | <0.0001 | 3.445 |
| 2 | 0.552 | 0 | Green | −0.027 | −0.035 | −0.019 | <0.0001 | 3.124 |
| Agriculture | 0.032 | 0.017 | 0.046 | <0.0001 | ||||
| 3 | 0.758 | 0 | Green | −0.017 | −0.023 | −0.01 | <0.0001 | 2.029 |
| Agriculture | 0.039 | 0.028 | 0.049 | <0.0001 | ||||
| ACE | 0.002 | 0.002 | 0.003 | <0.0001 | ||||
| Enterococcus spp. (MPN/100 ml) | ||||||||
| 1 | 0.37 | 0 | ACE | 0.002 | 0.002 | 0.003 | <0.0001 | 1.096 |
| 2 | 0.63 | 0 | ACE | 0.002 | 0.002 | 0.003 | <0.0001 | −0.282 |
| EC | 0.004 | 0.003 | 0.006 | <0.0001 | ||||
| 3 | 0.69 | 0 | ACE | 0.002 | 0.001 | 0.003 | <0.0001 | 10.166 |
| EC | 0.004 | 0.003 | 0.005 | <0.0001 | ||||
| Temp (°C) | −0.347 | −0.551 | −0.142 | 0.001 | ||||
| PMMoV (GC/100 ml) | ||||||||
| 1 | 0.65 | 0 | Green | −0.042 | −0.05 | −0.034 | <0.0001 | 5.273 |
| 2 | 0.68 | 0 | Green | −0.038 | −0.047 | −0.029 | <0.0001 | 4.848 |
| ACE | 0.001 | 0 | 0.002 | 0.03 | ||||
| 3 | 0.72 | 0 | Green | −0.038 | −0.046 | −0.029 | <0.0001 | 4.759 |
| ACE | 0.002 | 0.001 | 0.003 | 0.001 | ||||
| ACT | −6.87E−05 | 0 | 0 | 0.005 | ||||
| 4 | 0.74 | 0 | Green | −0.039 | −0.047 | −0.031 | <0.0001 | 5.055 |
| ACE | 0.002 | 0.001 | 0.003 | <0.0001 | ||||
| ACT | −5.91E−05 | 0 | 0 | 0.014 | ||||
| Antecedent dry period (days) | −0.099 | −0.186 | −0.012 | 0.027 | ||||
| PMMoV_contig1 (relative abundance) | ||||||||
| 1 | 0.641 | 0 | ACE | 0.001 | 0.001 | 0.001 | <0.0001 | 0.035 |
| 2 | 0.733 | 0 | ACE | 0.001 | 0 | 0.001 | 0.001 | −0.129 |
| Residential | 0.009 | 0.001 | 0.017 | 0.038 | ||||
| 3 | 0.809 | 0 | ACE | 0.001 | 0 | 0.001 | <0.0001 | −0.083 |
| Residential | 0.013 | 0.005 | 0.021 | 0.004 | ||||
| Antecedent dry period (days) | −0.041 | −0.079 | −0.004 | 0.033 | ||||
| PMMoV_reads mapped to reference genome | ||||||||
| 1 | 0.558 | 0 | Green | −0.027 | −0.036 | −0.018 | <0.0001 | −4.774 |
| 2 | 0.617 | 0 | Green | −0.025 | −0.034 | −0.016 | <0.0001 | −8.832 |
| pH | 0.529 | 0.004 | 1.054 | 0.048 | ||||
Dependent variables of the MLR models have been log10 transformed; the best-fit model under each category is highlighted in bold.
The first few models in each microbial fecal indicator group included only either land use categories or the chemical tracers as variables, with r2 values ranging from 0.37 to 0.65. When land use categories, environmental parameters, and the chemical tracers were combined in the analysis, the resulting models (i.e., E. coli in model 3, Enterococcus spp. in model 3, PMMoV in models 3 and 4, PMMoV_contig1 in models 2 and 3, and PMMoV_reads in model 2) were improved significantly (r2 values > 0.617). In model 4 of PMMoV, 74% of the data variation was explained by green land use, ACE, ACT, and antecedent dry period (days). In model 3 of the PMMoV_contig1, the relative abundance of PMMoV_contig1 had 80.9% of the data variation explained by ACE, residential land use, and antecedent dry period (days). These results indicated that the multiple-linear-regression models had a much better prediction ability (with r2 values of >0.617) when land use categories (i.e., green, residential, agriculture), environmental parameters (i.e., electrical conductivity [EC], temperature), and/or chemical tracers (i.e., ACE and ACT) were combined than when the variables were used alone. In particular, PMMoV_contig1 exhibited the highest r2 value, 0.809 (see Fig. S3 in the supplemental material), suggesting that metagenomics data could be applied in multiple linear regressions to predict the relative abundance of PMMoV in freshwater environments.
Paule-Mercado et al. (27) predicted FIB concentrations by using various environmental parameters in different land use categories. Even though higher R2 (0.83 to 0.91) values were obtained in the MLR models, the results were unable to quantify land use factors and monitored only FIB that might be limited for fecal source tracking. To date, there have been no studies on the relationship between microbial fecal indicator data (both traditional FIB and the alternative, PMMoV) and the addition of land use categories and chemical tracers. Our study showed that with the addition of land use categories (green, agriculture, and residential) and chemical tracers (i.e., ACE and ACT), the model performance significantly improved for the prediction of microbial fecal indicators (r2 values > 0.69). This analysis has evaluated the impacts of different natural and human-induced changes (i.e., land use patterns) on microbial water quality, which is of significance to devise urban development planning to protect human health (53). The models of E. coli, Enterococcus spp., and PMMoV in terms of absolute quantification might be applied to assess human health risks in recreational waters.
Evaluation of fecal indicators in terms of their correlation with different enteric viruses.
The empirical relationships between fecal indicators and selected microbial pathogens were evaluated in other studies. Significant associations between PMMoV and viruses (i.e., human adenovirus [HAdV] and human polyomavirus) were observed by Hamza et al. (10) in river water in Germany, where PMMoV had a moderately significant correlation coefficient with HAdV (rho = 0.42; P < 0.0001; n = 102). Significant cross-correlations were also observed between PMMoV and all human-associated MST markers in coastal waters from southeastern Florida (rho was not given) (18).
In terms of PMMoV performance, although PMMoV shares with enteric viruses similar viral replication modes, fates (i.e., adsorption, sedimentation, resuspension, and inactivation), and detection methods (i.e., qPCR) (54), the moderately weak significant correlations could have been caused by differences in morphological properties and origins. First, the capsid structure of PMMoV is more robust than that of enteric viruses (9, 10). The log10 reduction of PMMoV was 1.1, whereas the log10 reduction of HAdV was 3.7 after a 21-day incubation experiment in river water (25°C) (10). Second, PMMoV is of dietary origin and is not well correlated to human enteric infection (9, 10). The high prevalence of PMMoV (106 to 109 viruses/g dry weight of human feces) was observed in 12 of 18 healthy human feces samples by metagenomics RNA sequencing in the United States and Singapore (8) and in 19 of 20 healthy human feces samples in Germany by reverse transcription (RT)-qPCR (10). One exception was noted in a study conducted in France, which showed that only 22 of 304 (7.2%) stool samples tested positive for PMMoV; however, the samples were from adult hospitalized patients instead of healthy adults (15) and these disparities could have been caused by dietary habits in hospital patients (10).
Singapore cuisine typically contains fresh peppers (Capsicum spp.) or manufactured pepper-containing products (e.g., chili sauces or curry), indicating a higher chance of PMMoV presence in the healthy human gut. In other countries with a high prevalence of PMMoV in surface waters, such as Germany (100%, n = 111), Japan (76%, n = 140), and Vietnam (94%, n = 17), fresh and processed peppers might make contributions to high occurrences as well (10, 13, 16). High concentrations of PMMoV indicating high levels of human fecal pollution might not imply the presence or high concentrations of enteric viruses, as the gastrointestinal tract of healthy people does not necessarily contain a high density of enteric viruses. In addition to enteric viruses, a few more other pathogens could be present in human stool samples, such as Campylobacter jejuni, Giardia lamblia, Shigella, and toxic strains of E. coli (55). To further evaluate the human feces specificity of PMMoV, the investigation of cooccurrence between PMMoV and those pathogens should be conducted in the future. Nonetheless, in our study of tropical urban and rural catchments, PMMoV appeared to be sensitive to land use patterns and anthropogenic activities, thus making it appropriate for tracking human fecal pollution. Likewise, previous studies reported the high human feces specificity of PMMoV in the United States, Germany, and Singapore (8, 10, 11). This study evaluates the land use impacts on the occurrence and distribution of traditional FIB and PMMoV in tropical surface waters. To summarize, this finding would support the hypothesis of using PMMoV as an alternative indicator of human fecal contamination in tropical surface waters from the perspective of land use patterns. However, due to patterns similar to those of traditional FIB in terms of variable correlations with enteric viruses, PMMoV may not be suitable as an indicator of viral contamination (3, 6).
Conclusion.
This work characterized the geospatial distribution of traditional FIB and an alternative fecal indicator (PMMoV) at seven sampling locations with varied land use categories. Diverse land use in different water bodies across Singapore is one of the key factors impacting the occurrence and concentration of fecal indicators, i.e., concentrations were found to be higher in an urban-use-dominated catchment than a parkland-dominated one. Correlations between traditional FIB and the alternative fecal indicator suggest that PMMoV can be used as a suitable indicator of fecal pollution in tropical surface waters. The strong correlations between the occurrence and concentration of PMMoV and residential and green categories (∣rho∣ > 0.6; P < 0.01) suggest that PMMoV has potential as an indicator of human impact and hence, the possible incidence of pathogens in urban freshwater environments. The empirical models to predict the concentrations of traditional and alternative fecal indicators were improved with the inclusion of land use and chemical tracers as independent variables compared to the use of environmental parameters alone. These models can be applied to identify the impacts of anthropogenic activities and land use patterns in surface water quality. They also can be used to reflect sources and transportation of human fecal pollution or sewage contamination, offering valuable information for devising management strategies for public health.
MATERIALS AND METHODS
Sampling sites and land use studies.
This study was conducted in Singapore, which has a land area of 719.1 km2 and a population of 5.61 million people as of 2016 (56). Sampling sites and land use categories were mapped with ArcGIS version 10.3.1 software (ESRI, Redlands, CA, USA). Land use shape files were obtained from the Singapore Land Authority (SLA) and PUB (Singapore's National Water Agency) where the land use subcategories were defined by the Urban Redevelopment Authority (URA). However, to reduce the number of parameters, subcategories were merged into 5 main categories: (i) residential, (ii) urban, (iii) green, (iv) agricultural, and (v) water body.
Four different layers of maps were used for the analysis: (i) catchment land use shape files, (ii) river shape files, (iii) subcatchment shape files, and (iv) drain line maps. WGS_1984_Web_Mercator_Auxiliary_Sphere was used as the projection coordinate system, while GCS_WGS_1984/SVY21 Singapore was used as the geographic coordinate system.
The percentage area of land cover around each sampling point was calculated based on the drains into the sampling sites. However, when there were no direct drains into the specific water body, the effect of land use was calculated based on a 2-km buffer around the site. In order to reduce the number of parameters, the weightage of land use (which is the combination of different land use categories) was calculated. For the calculation of weightage, numbers 4, 3, 2, and 1 were assigned to residential, urban, agriculture, and green, respectively. A final weightage number was calculated according to the contribution of each land use category to each sampling site. The formula for calculating final weightage was as follows: weightage = (residential PA × 4) + (urban PA × 3) + (agriculture PA × 2) + (green PA × 1), where PA (as a percentage) is the fraction of area devoted to a particular use.
Sample collection and measurement of physical-chemical parameters.
A total of 171 water samples were collected from 7 different sampling zones in Singapore (A to G) with 10 sampling sites in 7 different water bodies (WB-A to WB-G) and 9 sampling sites in 3 different upstream catchment zones (C-A, C-B, and C-D) over a period of 14 months (November 2014 to January 2016) (see Table S7 in the supplemental material). Water samples were collected during the Northeast Monsoon (sample size [n] = 61, collected in November 2014, December 2014, January 2015, December 2015), Southwest Monsoon (n = 36; July 2015 and August 2015), pre-Northeast Monsoon (n = 18; October 2015), and pre-Southwest Monsoon (n = 56; March 2015, April 2015, January 2016).
Water samples from each site were collected in a 5-liter sterilized carboy for quantification of traditional FIB (e.g., E. coli and Enterococcus spp.) and target chemical tracers and in three 10-liter sterilized carboys for concentration of viral particles (e.g., PMMoV). Physiochemical parameters, including temperature, conductivity, turbidity, salinity, pH, dissolved oxygen (DO), and total dissolved solids (TDS), were measured in situ using the HI9828 multiparameter meter (Hanna Instruments). All the physiochemical environmental parameters were measured according to the American Public Health Association (APHA) standard methods (57), and rainfall information (i.e., 24-hour rainfall, 5-day rainfall, and the antecedent dry period) for each sampling site was obtained from a Singapore government-linked website (58).
Sample concentration and analysis.
Viral particles in 30-liter water samples were concentrated using hollow fiber ultrafiltration (Hemoflow HF80S; Fresenius Medical Care, Gad Homburg, Germany) to obtain a final volume of 600 ml (59). Samples spiked with bacteriophage MS2 indicated a recovery efficiency of 70 to 80%. Briefly, the water sample was recirculated until the final volume reached approximately 200 to 250 ml. Elution was carried out by recirculating 300 ml of elution buffer (0.1 g of sodium polyphosphate [NaPP], 5 ml of Tween 80, and 10 μl of Antifoam in 1 liter of Nanopure water) for 5 min. Both retentate and eluent were combined to a final volume of 600 ml. Meanwhile, bacterial nucleic acids were extracted from 10 ml of primary concentrate according to the PowerSoil DNA isolation kit instructions (Mo Bio Laboratories, Inc., Carlsbad, CA). A secondary concentration of virus particles was performed using polyethylene glycol (PEG) precipitation, followed by Ultra-15 Amicon ultrafiltration (60, 61). Briefly, the pH of primary concentrate was adjusted to 7.2, followed by the addition of 0.3 M NaCl and 0.8% (wt/vol) of PEG powder to the sample. The mixture was then incubated at 4°C with 100-rpm agitation for about 24 h. After the incubation, the sample was centrifuged at 14,000 × g for 45 min at 4°C to pellet the virus particles. The virus particles were resuspended by adding 10 ml of phosphate-buffered saline (PBS). Subsequently, the virus particles were separated from the suspended solids by adding an equal volume (∼10 ml) of chloroform, followed by 45 min of centrifugation at 3,000 × g at 4°C. The aqueous phase (containing virus particles) was collected and filtered through a 0.22-μm filter. An Ultra-15 centrifugal tube with a molecular weight cutoff of 30 kDa (Amicon Merck, Germany) was used to further concentrate the sample to a final volume of 1 ml (centrifuged at 4,000 × g at 4°C). Samples were stored at −80°C until further analysis. After primary and secondary concentrations, viral nucleic acids (DNA and RNA) were extracted from 140 μl of the final concentrate using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) and then stored at −80°C (62).
Quantification of PMMoV.
The extracted viral nucleic acids were reverse transcribed using the ImProm-II reverse transcription system (Promega) following the manufacturer's instructions. qPCRs for the samples were performed in 20 μl of reaction mixture containing a 2.5-μl aliquot of cDNA mixed with 17.5 μl of qPCR mixture containing 10 μl of FastStart Universal Probe Master (Rox; Roche Diagnostics, Basel, Switzerland) and 10 pmol forward and reverse primers and TaqMan probe (16, 63). Duplicate qPCR amplification was performed in a StepOnePlus real-time PCR system under the following thermal conditions: 95°C for 30 s and 45 cycles of 95°C for 5 s and 60°C for 60 s. Both positive-control (kindly provided by the Charles Gerba lab at the University of Arizona) and negative-control (nuclease-free reagent grade water and cDNA negative control) assays were performed for quality control. Serial dilutions of plasmid DNA (101 to 107 gene copies per reaction mixture) containing the PMMoV target sequence (68 bp) were used to generate the standard curve. The standard curve equation with R2 of 0.9 to 0.99 was obtained by plotting the cycle threshold (CT) value against the log of standard plasmid concentration (GC/reaction) (see Table S8 in the supplemental material). The detection limit of PMMoV was 16 GC/100 ml, which was calculated based on 1 GC per reaction mixture.
To avoid false negatives or underestimates due to inhibitions in qPCR, TaqMan exogenous internal positive control (IPC) reagents were used to assess the level of inhibition in each environmental sample (64). A sample was considered to be inhibited when the difference between the CT values of IPC with and without cDNA sample was more than 3 CT units. In this case, cDNA samples were diluted with double-distilled H2O (ddH2O; sterile ultrapure water) to achieve the difference below 3 CT units (65). In the current study, the highest dilution factor required to decrease the inhibition level was 4.
Quantification of Enterococcus spp. and E. coli.
Colilert-18 and Enterolert (Idexx Laboratories Inc., USA) were used to quantify E. coli and Enterococcus spp. in the water samples. Medium powder was mixed with a volume of 100 ml of water sample and 10-fold serially diluted in the provided sterile bags before the mixture was poured into a multiwell tray (Quanti-Tray/2000). This enables a wider coverage of detection range due to high concentrations of FIB at certain locations. Colilert-18 and Enterolert samples were incubated at 35 ± 0.5°C for 18 h and 41 ± 0.5°C for 24 h, respectively. After incubation, the Quanti-Tray was visualized under UV light using a 6-watt fluorescent UV lamp to identify and count the positive wells. With reference to the most probable number (MPN) table provided, the MPN of that particular sample was obtained (MPN/100 ml).
Detection and quantification of chemical markers.
This study targeted compounds acesulfame (ACE), acetaminophen (ACT), cyclamate (CYC), and carbamazepine (CBZ), widely used as chemical markers of human fecal sources in surface waters and groundwater, as previously described (29, 30, 47, 66, 67).
The detection and quantification of the chemical markers were carried out using solid-phase extraction (SPE) coupled with ultrahigh-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) and isotope dilution as discussed by Tran et al. (68), with slight modification. The method detection limits (MDLs) for the target chemical markers were 1.0 ng/liter for ACE and ACT, 0.5 ng/liter for CYC, and 0.01 ng/liter for CBZ (Table 1).
Metagenomics sequencing for bacterial and viral samples.
Bacterial nucleic acids were extracted from a total of 36 samples from August and October 2015, and viral nucleic acids were extracted from a total of 38 samples from January and April 2015. Downstream reverse transcription and amplification for viral nucleic acids were performed to obtain sufficient quantities of cDNA for sequencing (69–71). Products were sent to The Singapore Centre on Environmental Life Sciences Engineering (SCELSE), where the nucleic acids were fragmented to 250-bp lengths and sequenced on two lanes of Illumina Hiseq 2500 platform in a rapid mode run. The Illumina TruSeq Nano DNA library kit was used in library construction.
Processing and analysis of Illumina reads.
The sequencing data were trimmed by BBtools (http://jgi.doe.gov/data-and-tools/bbtools/) to remove adaptors, low-quality reads, reads smaller than 50 bp, and PhiX reads. PhiX reads serve as a quality control and a calibration control in sequencing runs (72). “Primer B” sequences, which were used in viral random amplification, were also removed in viral sequencing samples.
Bacterial metagenomes.
All the reference genomes of E. coli and Enterococcus spp. were downloaded from NCBI (373 E. coli complete genomes and 51 Enterococcus complete genomes as of 8 October 2017). High-quality reads were mapped to the reference genomes in order to obtain the relative abundance of E. coli and Enterococcus spp. in each library. Mapping was performed using the CLC Genomics Workbench 8.0.3 based on the default setting (mismatch gap = 2, with the linear gap cost, length fraction of 0.5, similarity fraction is 99%). The proportion of mapping reads equals the number of mapping reads in each library divided by the total reads in the library. Metaxa2 2.2 beta 10 was used to extract 16S rRNA sequences from metagenomics reads and assign bacterial taxonomy with default parameters (73). A rarefaction curve was obtained using Metaxa2 Diversity Tools based on the generated bacterial rarefaction tables at level 6 (genera) and level 7 (species) with default parameters (74).
Viral metagenomes.
CLC Genomics Workbench version 8.0.3 (CLC bio, Boston, MA, USA) was used for de novo assembly. The assembly setting was as follows: minimum contig length, 1,000 bp; similarity fraction, 0.95. All the contigs from the pooled 38 libraries were then uploaded into MetaVir2 pipeline for taxonomy assignment (75, 76). In order to quantify the relative abundance of PMMoV in different libraries, reads were remapped to contigs using Novoalign. RPKM (reads per kilobase of transcript per million mapped reads) was generated by standardizing and quantifying every contig's relative abundance throughout the 38 libraries (77). RPKM was calculated using the following equation: RPKM = (raw reads mapped to one contig in one library)/{[total reads mapped to the library (millions)] × [length of the contig (kbp)]}.
In addition, high-quality reads were mapped to the reference genome of PMMoV (NC_003630.1), observing the same mapping method as the one stated above.
Statistical analysis.
In this study, ProUCL version 5.1.02, the software recommended by the U.S. Environmental Protection Agency, was used to estimate data for nondetects by using the regression on order statistics method (78). The method was used to generate imputed values for nondetects. All statistical tests were performed using IBM SPSS 23 (IBM, Portsmouth, UK). Spearman's rank correlation was performed among 25 factors, including all the microbial fecal indicators, environmental parameters, and chemical tracers. To avoid multiple-comparison problems, the Bonferroni correction was performed to mitigate false-positive associations. The P value was adjusted to α/n (where n is the number of tests in the matrix). Different levels of correlation were defined (i.e., strong, rho ≥ 0.7; moderately strong, 0.5 ≤ rho < 0.7; moderately weak, 0.3 ≤ rho < 0.5; and weak, rho < 0.3). A Pearson's chi-squared test of independence and a nominal association with Φ value was performed to find the significance and coefficient of association between different sampling locations with different land use categories and the detection frequency of microbial fecal indicators. MLR analyses, including stepwise variable selection approaches, were performed to find the significant explanatory variables in fitting the observed microbial fecal indicators. Spearman's rank correlation of PMMoV, E. coli, and Enterococcus spp. was performed between qPCR and metagenomics data (relative abundance) for viral (n = 38) and bacterial (n = 36) metagenomics samples.
Data availability.
Bacterial and viral data sets were deposited in NCBI Sequence Read Archive (SRA) under accession numbers SRR6423776 to SRR6423811 and SRR5995660 to SRR5995697, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This research grant is supported by the Singapore National Research Foundation under its Environment and Water Research Programme and administered by PUB, Singapore's national water agency (Ref: 1301-IRIS-37 [IDD 90301/1/65]).
We thank National University of Singapore and Public Utilities Board (PUB) for supporting this research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00287-18.
REFERENCES
- 1.Rochelle-Newall E, Nguyen TMH, Le TPQ, Sengtaheuanghoung O, Ribolzi O. 2015. A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Front Microbiol 6:308. doi: 10.3389/fmicb.2015.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed W, Sidhu J, Toze S. 2012. Evaluation of the nifH gene marker of Methanobrevibacter smithii for the detection of sewage pollution in environmental waters in Southeast Queensland, Australia. Environ Sci Technol 46:543–550. doi: 10.1021/es203372u. [DOI] [PubMed] [Google Scholar]
- 3.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith JF, Cao Y, McGee CD, Weisberg SB. 2009. Evaluation of rapid methods and novel indicators for assessing microbiological beach water quality. Water Res 43:4900–4907. doi: 10.1016/j.watres.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Tallon P, Magajna B, Lofranco C, Leung KT. 2005. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Pollut 166:139–166. doi: 10.1007/s11270-005-7905-4. [DOI] [Google Scholar]
- 6.Liang L, Goh S, Vergara G, Fang H, Rezaeinejad S, Chang S, Bayen S, Lee W, Sobsey MD, Rose J. 2015. Alternative fecal indicators and their empirical relationships with enteric viruses, Salmonella enterica, and Pseudomonas aeruginosa in surface waters of a tropical urban catchment. Appl Environ Microbiol 81:850–860. doi: 10.1128/AEM.02670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed W, Sritharan T, Palmer A, Sidhu J, Toze S. 2013. Evaluation of bovine feces-associated microbial source tracking markers and their correlations with fecal indicators and zoonotic pathogens in a Brisbane, Australia, reservoir. Appl Environ Microbiol 79:2682–2691. doi: 10.1128/AEM.03234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T, Breitbart M, Lee WH, Run J-Q, Wei CL, Soh SWL, Hibberd ML, Liu ET, Rohwer F, Ruan Y. 2005. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitajima M, Iker BC, Pepper IL, Gerba CP. 2014. Relative abundance and treatment reduction of viruses during wastewater treatment processes-identification of potential viral indicators. Sci Total Environ 488:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 10.Hamza IA, Jurzik L, Überla K, Wilhelm M. 2011. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res 45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 11.Rosario K, Symonds EM, Sinigalliano C, Stewart J, Breitbart M. 2009. Pepper mild mottle virus as an indicator of fecal pollution. Appl Environ Microbiol 75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Symonds E, Verbyla M, Lukasik J, Kafle R, Breitbart M, Mihelcic J. 2014. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res 65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda K, Nakada N, Hanamoto S, Inaba M, Katayama H, Do AT, Nga TTV, Oguma K, Hayashi T, Takizawa S. 2015. Pepper mild mottle virus as an indicator and a tracer of fecal pollution in water environments: comparative evaluation with wastewater-tracer pharmaceuticals in Hanoi, Vietnam. Sci Total Environ 506:287–298. doi: 10.1016/j.scitotenv.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Hughes B, Beale D, Dennis P, Cook S, Ahmed W. 2017. Cross-comparison of human wastewater-associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Appl Environ Microbiol 83:e00028-17. doi: 10.1128/AEM.00028-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colson P, Richet H, Desnues C, Balique F, Moal V, Grob J-J, Berbis P, Lecoq H, Harlé J-R, Berland Y. 2010. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One 5:e10041. doi: 10.1371/journal.pone.0010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haramoto E, Kitajima M, Kishida N, Konno Y, Katayama H, Asami M, Akiba M. 2013. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl Environ Microbiol 79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 18.Symonds E, Sinigalliano C, Gidley M, Ahmed W, McQuaig-Ulrich S, Breitbart M. 2016. Faecal pollution along the southeastern coast of Florida and insight into the use of pepper mild mottle virus as an indicator. J Appl Microbiol 121:1469–1481. doi: 10.1111/jam.13252. [DOI] [PubMed] [Google Scholar]
- 19.Fauvel B, Cauchie H-M, Gantzer C, Ogorzaly L. 2016. Contribution of hydrological data to the understanding of the spatio-temporal dynamics of F-specific RNA bacteriophages in river water during rainfall-runoff events. Water Res 94:328–340. doi: 10.1016/j.watres.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 20.Safaie A, Wendzel A, Ge Z, Nevers MB, Whitman RL, Corsi SR, Phanikumar MS. 2016. Comparative evaluation of statistical and mechanistic models of Escherichia coli at beaches in southern lake Michigan. Environ Sci Technol 50:2442–2449. doi: 10.1021/acs.est.5b05378. [DOI] [PubMed] [Google Scholar]
- 21.US Environmental Protection Agency. 2012. Recreational water quality criteria (EPA publication no. 820-F-12-058). US Environmental Protection Agency, Washington, DC. [Google Scholar]
- 22.Patz JA, Daszak P, Tabor GM, Aguirre AA, Pearl M, Epstein J, Wolfe ND, Kilpatrick AM, Foufopoulos J, Molyneux D. 2004. Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. Environ Health Perspect 112:1092. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenaker PL, Corsi S, Borchardt MA, Spencer SK, Baldwin AK, Lutz MA. 2017. Hydrologic, land cover, and seasonal patterns of waterborne pathogens in Great Lakes tributaries. Water Res 113:11–21. doi: 10.1016/j.watres.2017.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corsi SR, Borchardt M, Spencer S, Hughes PE, Baldwin AK. 2014. Human and bovine viruses in the Milwaukee River watershed: hydrologically relevant representation and relations with environmental variables. Sci Total Environ 490:849–860. doi: 10.1016/j.scitotenv.2014.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins TM, Scott TM, Morgan MR, Rose JB. 2005. Occurrence of alternative fecal indicators and enteric viruses in Michigan rivers. J Great Lakes Res 31:22–31. doi: 10.1016/S0380-1330(05)70235-5. [DOI] [Google Scholar]
- 26.Jiang S, Noble R, Chu W. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl Environ Microbiol 67:179–184. doi: 10.1128/AEM.67.1.179-184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paule-Mercado M, Ventura J, Memon S, Jahng D, Kang J-H, Lee C-H. 2016. Monitoring and predicting the fecal indicator bacteria concentrations from agricultural, mixed land use and urban stormwater runoff. Sci Total Environ 550:1171–1181. doi: 10.1016/j.scitotenv.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 28.Tiefenthaler L, Stein ED, Schiff KC. 2011. Levels and patterns of fecal indicator bacteria in stormwater runoff from homogenous land use sites and urban watersheds. J Water Health 9:279–290. doi: 10.2166/wh.2010.056. [DOI] [PubMed] [Google Scholar]
- 29.Tran NH, Gin KY-H, Ngo HH. 2015. Fecal pollution source tracking toolbox for identification, evaluation and characterization of fecal contamination in receiving urban surface waters and groundwater. Sci Total Environ 538:38–57. doi: 10.1016/j.scitotenv.2015.07.155. [DOI] [PubMed] [Google Scholar]
- 30.Tran NH, Li J, Hu J, Ong SL. 2014. Occurrence and suitability of pharmaceuticals and personal care products as molecular markers for raw wastewater contamination in surface water and groundwater. Environ Sci Pollution Res 21:4727–4740. doi: 10.1007/s11356-013-2428-9. [DOI] [PubMed] [Google Scholar]
- 31.Cha Y, Park M-H, Lee S-H, Kim JH, Cho KH. 2016. Modeling spatiotemporal bacterial variability with meteorological and watershed land-use characteristics. Water Res 100:306–315. doi: 10.1016/j.watres.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Wilkes G, Edge T, Gannon V, Jokinen C, Lyautey E, Medeiros D, Neumann N, Ruecker N, Topp E, Lapen DR. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res 43:2209–2223. doi: 10.1016/j.watres.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 33.Rosiles-González G, Ávila-Torres G, Moreno-Valenzuela OA, Acosta-González G, Leal-Bautista RM, Grimaldo-Hernández CD, Brown JK, Chaidez-Quiroz C, Betancourt WQ, Gerba CP. 2017. Occurrence of pepper mild mottle virus (PMMoV) in groundwater from a karst aquifer system in the Yucatan Peninsula, Mexico. Food Environ Virol 9:487–497. doi: 10.1007/s12560-017-9309-1. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz BW, Kitajima M, Campillo ME, Gerba CP, Pepper IL. 2016. Virus reduction during advanced Bardenpho and conventional wastewater treatment processes. Environ Sci Technol 50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- 35.Singapore's National Water Agency (PUB). 2017. The overview of used water. Singapore's National Water Agency (PUB), Singapore: https://www.pub.gov.sg/usedwater Accessed 20 October 2017. [Google Scholar]
- 36.Gu X, Tay QXM, Te SH, Saeidi N, Goh SG, Kushmaro A, Thompson JR, Gin KY-H. 2018. Geospatial distribution of viromes in tropical freshwater ecosystems. Water Res 137:220–232. doi: 10.1016/j.watres.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization. 2003. Assessing microbial safety of drinking water improving approaches and methods: improving approaches and methods. OECD Publishing, Paris, France. [Google Scholar]
- 38.Edberg SC, Allen MJ, Smith DB. 1989. National field evaluation of a defined substrate method for the simultaneous detection of total coliforms and Escherichia coli from drinking water: comparison with presence-absence techniques. Appl Environ Microbiol 55:1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edberg SC, Allen MJ, Smith DB. 1988. National field evaluation of a defined substrate method for the simultaneous enumeration of total coliforms and Escherichia coli from drinking water: comparison with the standard multiple tube fermentation method. Appl Environ Microbiol 54:1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekklesia E, Shanahan P, Chua L, Eikaas H. 2015. Associations of chemical tracers and faecal indicator bacteria in a tropical urban catchment. Water Res 75:270–281. doi: 10.1016/j.watres.2015.02.037. [DOI] [PubMed] [Google Scholar]
- 41.Galdino Pereira VM, Bemvindo Gomes R, Vinícius do Carmo e Sá M. 2011. Correlation between Escherichia coli and limnological variables in water samples of the Lagoa da Maraponga, Fortaleza, Ceará State, Brazil. Acta Sci Biol Sci 33:145–151. [Google Scholar]
- 42.Narkis N, Armon R, Offer R, Orshansky F, Friedland E. 1995. Effect of suspended solids on wastewater disinfection efficiency by chlorine dioxide. Water Res 29:227–236. doi: 10.1016/0043-1354(94)E0117-O. [DOI] [Google Scholar]
- 43.Selvakumar A, Borst M. 2006. Variation of microorganism concentrations in urban stormwater runoff with land use and seasons. J Water Health 4:109–124. [PubMed] [Google Scholar]
- 44.Oladeinde A, Bohrmann T, Wong K, Purucker S, Bradshaw K, Brown R, Snyder B, Molina M. 2014. Decay of fecal indicator bacterial populations and bovine-associated source-tracking markers in freshly deposited cow pats. Appl Environ Microbiol 80:110–118. doi: 10.1128/AEM.02203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitman RL, Nevers MB, Korinek GC, Byappanahalli MN. 2004. Solar and temporal effects on Escherichia coli concentration at a Lake Michigan swimming beach. Appl Environ Microbiol 70:4276–4285. doi: 10.1128/AEM.70.7.4276-4285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran NH, Hu J, Li J, Ong SL. 2014. Suitability of artificial sweeteners as indicators of raw wastewater contamination in surface water and groundwater. Water Res 48:443–456. doi: 10.1016/j.watres.2013.09.053. [DOI] [PubMed] [Google Scholar]
- 48.Van Stempvoort DR, Roy JW, Grabuski J, Brown SJ, Bickerton G, Sverko E. 2013. An artificial sweetener and pharmaceutical compounds as co-tracers of urban wastewater in groundwater. Sci Total Environ 461–462:348–359. doi: 10.1016/j.scitotenv.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. 2012. Enterococci in the environment. Microbiol Mol Biol Rev 76:685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernhard AE, Goyard T, Simonich MT, Field KG. 2003. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res 37:909–913. doi: 10.1016/S0043-1354(02)00384-6. [DOI] [PubMed] [Google Scholar]
- 51.Nshimyimana JP, Freedman AJE, Shanahan P, Chua LC, Thompson JR. 2017. Variation of bacterial communities with water quality in an urban tropical catchment. Environ Sci Technol 51:5591–5601. doi: 10.1021/acs.est.6b04737. [DOI] [PubMed] [Google Scholar]
- 52.Staley C, Dunny GM, Sadowsky MJ. 2014. Environmental and animal-associated enterococci. Adv Appl Microbiol 87:147–186. doi: 10.1016/B978-0-12-800261-2.00004-9. [DOI] [PubMed] [Google Scholar]
- 53.de Brauwere A, Ouattara NK, Servais P. 2014. Modeling fecal indicator bacteria concentrations in natural surface waters: a review. Crit Rev Environ Sci Technol 44:2380–2453. doi: 10.1080/10643389.2013.829978. [DOI] [Google Scholar]
- 54.Brookes JD, Antenucci J, Hipsey M, Burch MD, Ashbolt NJ, Ferguson C. 2004. Fate and transport of pathogens in lakes and reservoirs. Environ Int 30:741–759. doi: 10.1016/j.envint.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Leclerc H, Schwartzbrod L, Dei-Cas E. 2002. Microbial agents associated with waterborne diseases. Crit Rev Microbiol 28:371–409. doi: 10.1080/1040-840291046768. [DOI] [PubMed] [Google Scholar]
- 56.Department of Statistics Singapore. 2016. Population in brief 2016. https://www.nptd.gov.sg/PORTALS/0/HOMEPAGE/HIGHLIGHTS/population-in-brief-2016.pdf. Accessed 18 September 2017.
- 57.American Public Health Association. 2005. Standard methods for the examination of water and wastewater, 21st ed American Public Health Association, Washington, DC. [Google Scholar]
- 58.The National Environment Agency Singapore. 2017. Climate historical daily records. http://www.weather.gov.sg/climate-historical-daily/ The National Environment Agency Singapore, Singapore: Accessed 19 January 2017. [Google Scholar]
- 59.Liu P, Hill VR, Hahn D, Johnson TB, Pan Y, Jothikumar N, Moe CL. 2012. Hollow-fiber ultrafiltration for simultaneous recovery of viruses, bacteria and parasites from reclaimed water. J Microbiol Methods 88:155–161. doi: 10.1016/j.mimet.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 60.Arnone R, Walling J. 2007. Waterborne pathogens in urban watersheds. J Water Health 5:149–162. doi: 10.2166/wh.2006.001. [DOI] [PubMed] [Google Scholar]
- 61.Gibson KE, Schwab KJ. 2011. Tangential-flow ultrafiltration with integrated inhibition detection for recovery of surrogates and human pathogens from large-volume source water and finished drinking water. Appl Environ Microbiol 77:385–391. doi: 10.1128/AEM.01164-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saeidi N, Gu X, Goh SG, Xin CLY, Gin KY-H. 2017. Evaluating the efficacy of commercial kits for viral DNA/RNA extraction. Water Practice Technol 12:80–86. doi: 10.2166/wpt.2017.015. [DOI] [Google Scholar]
- 63.Department of Statistics Singapore. 2016. Statistics Singapore—latest data. http://www.singstat.gov.sg/statistics/latest-data-16 Accessed 19 January 2017.
- 64.Hartman LJ, Coyne SR, Norwood DA. 2005. Development of a novel internal positive control for Taqman® based assays. Mol Cell Probes 19:51–59. doi: 10.1016/j.mcp.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 65.Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res 39:559–568. doi: 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 66.Kuroda K, Murakami M, Oguma K, Muramatsu Y, Takada H, Takizawa S. 2012. Assessment of groundwater pollution in Tokyo using PPCPs as sewage markers. Environ Sci Technol 46:1455–1464. doi: 10.1021/es202059g. [DOI] [PubMed] [Google Scholar]
- 67.Nakada N, Kiri K, Shinohara H, Harada A, Kuroda K, Takizawa S, Takada H. 2008. Evaluation of pharmaceuticals and personal care products as water-soluble molecular markers of sewage. Environ Sci Technol 42:6347–6353. doi: 10.1021/es7030856. [DOI] [PubMed] [Google Scholar]
- 68.Tran NH, Hu J, Ong SL. 2013. Simultaneous determination of PPCPs, EDCs, and artificial sweeteners in environmental water samples using a single-step SPE coupled with HPLC–MS/MS and isotope dilution. Talanta 113:82–92. doi: 10.1016/j.talanta.2013.03.072. [DOI] [PubMed] [Google Scholar]
- 69.Bibby K, Peccia J. 2013. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ Sci Technol 47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim Y, Aw TG, Teal TK, Rose JB. 2015. Metagenomic investigation of viral communities in ballast water. Environ Sci Technol 49:8396–8407. doi: 10.1021/acs.est.5b01633. [DOI] [PubMed] [Google Scholar]
- 71.Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. 2002. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A 99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Illumina. 2016. Using a PhiX control for HiSeq® sequencing runs. Technical note: sequencing. Illumina, Inc, San Diego, CA. [Google Scholar]
- 73.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH. 2015. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Res 15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 74.Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjöling Å, Nilsson RH. 2016. Metaxa2 diversity tools: easing microbial community analysis with Metaxa2. Ecol Inform 33:45–50. doi: 10.1016/j.ecoinf.2016.04.004. [DOI] [Google Scholar]
- 75.Roux S, Tournayre J, Mahul A, Debroas D, Enault F. 2014. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinform 15:1. doi: 10.1186/1471-2105-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 77.Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, Smith MI, Wang D, Virgin HW, Rohwer F. 2015. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U S A 112:11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Office of Research and Development, US Environmental Protection Agency. 2015. ProUCL version 5.1 user guide. Statistical software for environmental applications for data sets with and without nondetect observations. Office of Research and Development, US Environmental Protection Agency, Washington, DC: https://www.epa.gov/sites/production/files/2016-05/documents/proucl_5.1_user-guide.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bacterial and viral data sets were deposited in NCBI Sequence Read Archive (SRA) under accession numbers SRR6423776 to SRR6423811 and SRR5995660 to SRR5995697, respectively.