ABSTRACT
This study determined the function of thioredoxin and glutaredoxin systems in the phytopathogenic fungus Alternaria alternata via analyzing mutants obtained from the targeted deletion of genes encoding thioredoxin peroxidase (Tsa1), thioredoxin reductase (Trr1), and glutathione reductase (Glr1). Trr1 and Glr1, but not Tsa1, are required for growth and conidiation. The reduced growth and conidiation seen in the Trr1 or Glr1 deletion mutant can be restored by glutathione. Deletion mutants showing growth inhibition by oxidants are defective for H2O2 detoxification and induce smaller lesions on citrus leaves. Trr1 and Glr1, but not Tsa1, also contribute to NaCl resistance. Glr1 is required for sorbitol resistance and is responsible for resistance to mancozeb and boscalid but not chlorothalonil fungicides, a novel phenotype that has not been reported in fungi. Trr1 is required for resistance to boscalid and chlorothalonil fungicides but confers susceptibility to mancozeb. The Tsa1 deletion mutant displays wild-type sensitivity to the tested fungicides. The expression of Tsa1 and Trr1 is regulated by the oxidative stress responsive regulators Yap1, Hog1, and Skn7. The expression of Tsa1, but not Trr1, is also regulated indirectly by the NADPH oxidase. The results indicate that the capability to resist oxidative stress is required for virulence of A. alternata.
IMPORTANCE The thioredoxin and glutaredoxin systems are important thiol antioxidant systems in cells, and knowledge of these two systems in the plant-pathogenic fungus A. alternata is useful for finding new strategies to reduce the virulence of this pathogen. In this study, we demonstrated that thiol antioxidant system-related genes (Tsa1, Trr1, and Glr1) are required for H2O2 detoxification and virulence in A. alternata. Moreover, deletion of Trr1 results in hypersensitivity to the fungicides chlorothalonil and boscalid, and Glr1 deletion mutants are highly sensitive to mancozeb, which is the fungicide mostly used in citrus fields. Therefore, our findings demonstrate that the ability to detoxify reactive oxygen species (ROS) plays a critical role in pathogenesis on citrus and provide novel insights into the physiological functions of thiol-containing systems in fungicide sensitivity for A. alternata.
KEYWORDS: Alternaria alternata, thioredoxin, glutaredoxin, oxidative stress resistance, virulence, fungicide resistance
INTRODUCTION
Experimental evidence indicating that plant pathogens must be able to suppress plant-generated reactive oxygen species (ROS) in order to successfully infect their host cells has mounted (1–3). The thioredoxin and glutaredoxin systems are two main thiol antioxidant systems in cells. The thioredoxin system, containing thioredoxin peroxidase (also known as thiol-specific antioxidant protein [Tsa]), thioredoxin reductase (Trr), and NADPH, plays a key function in cellular defense against oxidative damage (4). The glutaredoxin system, containing glutathione, glutathione reductase (Glr), glutathione peroxidase (Gpx), and NADPH, is also required for cell survival in oxidative stress environments by maintaining high levels of glutathione (5). Both thioredoxin and glutaredoxin are heat-stable oxidoreductases containing two conserved cysteine residues in their active regions (6). Both proteins function in electron transfer supplied from NADPH via the reversible oxidation of two protein sulfhydryl groups (—SH) to a disulfide bridge (S=S). Previous studies indicated that these systems are involved in a wide range of physiological and biochemical functions, including synthesis of deoxyribonucleotides, protein folding, repair of DNA and proteins, sulfur metabolism, and detoxification of ROS (7). In addition, thioredoxin is required for genomic stability, resistance to heat and nitrosative stress, and cell longevity (8–11).
Because of their multifaceted functions and roles, thioredoxin and glutaredoxin systems have been intensively studied in bacteria, the budding yeast Saccharomyces cerevisiae, and mammalian cells; however, these systems have been studied in only a few of the filamentous fungi. Recent data have revealed that their functions in ROS resistance are not completely conserved in different fungi and yeasts. For example, the thioredoxin but not the glutaredoxin system plays a role in resistance to ROS, development, and virulence in the necrotrophic fungal plant pathogen Botrytis cinerea (12). Deletion of any of three thioredoxin-coding genes in Podospora anserine has no effect on H2O2 sensitivity (13), while deletion of a thioredoxin gene in the human pathogen Candida albicans results in a strain that is highly sensitive to H2O2 and attenuated in virulence (14). In the human pathogen Cryptococcus neoformans, thioredoxin peroxidase and glutathione peroxidase are required for virulence (9, 15). Both systems also contribute to ROS resistance and full virulence in the rice blast fungus Magnaporthe oryzae (2, 16, 17).
The plant-pathogenic fungus Alternaria alternata, containing at least seven pathotypes, is a necrotroph; each pathotype produces a unique host-selective toxin to kill its host cells prior to colonization. The tangerine pathotype produces 9,10-epoxy-8-hydroxy-9-methyl-decatrienoic acid (18), which affects tangerines (Citrus reticulate Blanco), grapefruit (C. paradisi Macfad.), and their hybrids and results in necrotic lesions on leaves, twigs, and fruit. Infection with A. alternata on citrus has been shown to induce lipid peroxidation and eventually result in cell death (19). H2O2 is released from plant cells upon cell death (20). To successfully colonize citrus tissues filled with toxic ROS, the pathogen must be able to detoxify them. In the tangerine pathotype of A. alternata, several regulators important for resistance to oxidative stress have been linked to virulence (21). These include genes encoding a Yap1 transcription regulator, a high-osmolarity glycerol 1 (Hog1) mitogen-activated protein (MAP) kinase, a Skn7 response regulator, and NADPH oxidases (Nox). Mutational deletion of any of these genes in A. alternata produces strains showing increased sensitivity to ROS and attenuated virulence on citrus (22–25).
Little is known about the functions of thioredoxin and glutaredoxin systems in relation to survival and virulence of A. alternata. In a previous study, we have demonstrated that the A. alternata glutathione peroxidase 3 (AaGPx3), coordinately regulated by Yap1 and Hog1, contributes to oxidative stress resistance, virulence, melanin biosynthesis, and conidial formation (26). However, AaGPx3 acts as a suppressor for chitin biosynthesis. In the present study, we investigated the importance of thioredoxin and glutaredoxin systems to the virulence of A. alternata. Experiments were performed to characterize mutant strains resulting from the targeted deletion of genes encoding thioredoxin peroxidase (Tsa1), thioredoxin reductase (Trr1), and glutathione reductase (Glr1). The expression of these genes is induced by oxidants and regulated by Yap1, Hog1, and/or Skn7. This work provides novel insights into the physiological functions of thiol-containing systems in fungicide sensitivity and demonstrates that the ability to detoxify ROS plays a critical role in A. alternata pathogenesis in citrus. The study also reports the divergent functions of thioredoxin and glutaredoxin systems in response to different fungicides and environmental stress.
RESULTS
Characterization of thioredoxin- and glutaredoxin-coding genes.
Sequences of both thioredoxin- and glutathione-related genes were retrieved from the complete genome of A. alternata (27). The A. alternata thioredoxin peroxidase-coding gene (AaTsa1) contains a 675-bp open reading frame (ORF) interrupted by two small introns of 58 and 168 bp. AaTsa1 (accession no. MG593564) encodes a 224-amino-acid polypeptide with a predicted molecular mass of 24.3 kDa that shows 65% amino acid similarity to the Saccharomyces cerevisiae Tsa1 (accession no. DAA09870). AaTsa1 contains a conserved domain commonly found in the alkyl hydroperoxide reductase AhpC/thiol-specific antioxidant (TSA) family and a peroxiredoxin domain in the C terminus. AaTsa1 has two conserved cysteine residues (amino acids 57 and 178). The A. alternata thioredoxin reductase-coding gene (AaTrr1) contains a 1,071-bp ORF interrupted by three introns (109, 50, and 50 bp) and encodes a 38.6-kDa polypeptide that shows 60% amino acid similarity to the S. cerevisiae Trr1 (accession no. NP_010640). AaTrr1 (accession no. MG593563) is a flavin adenine dinucleotide (FAD) flavoprotein containing two redox-active cysteine residues and a conserved pyridine nucleotide-disulfide oxidoreductase domain (see Fig. S1 in the supplemental material).
The A. alternata glutathione reductase-coding gene (AaGlr1) has a 1,107-bp ORF interrupted by a 168-bp intron. Similar to the case for AaTrr1, AaGlr1 (accession no. MG593559) contains a pyridine nucleotide-disulfide oxidoreductase domain in the N terminus and a pyridine nucleotide-disulfide oxidoreductase dimerization domain in the C terminus.
Expression of thioredoxin- and glutaredoxin-coding genes in response to oxidative stress.
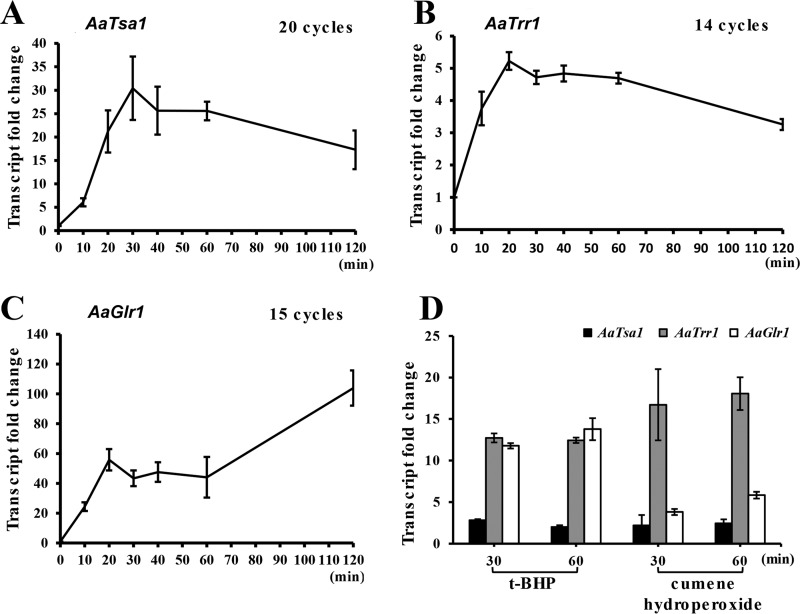
Expression of AaTsa1, AaTrr1, and AaGlr1 was assessed in the Z7 strain after treatment with or without oxidants by quantitative reverse transcriptase PCR (qRT-PCR). Accumulation of both AaTsa1 and AaTrr1 transcripts was elevated in Z7 after exposure to 10 mM H2O2 (Fig. 1). Similarly, expression of AaGlr1 was upregulated to various levels in Z7 in response to H2O2. Upon exposure to 2.6 mM tert-butyl-hydroperoxide (t-BHP) or 1 mM cumene hydroperoxide, significantly increased expression of AaTrr1 and AaGlr1 was also observed (4- to 18-fold increases). However, expression of AaTsa1 was increased only about 2- to 2.8-fold by t-BHP or cumene hydroperoxide.
FIG 1.
Quantitative RT-PCR analysis of gene expression in the wild-type strain of Alternaria alternata. (A) Expression of the thioredoxin peroxidase-coding gene (AaTsa1) in response to H2O2. (B) Expression of the thioredoxin reductase-coding gene (AaTrr1) in response to H2O2. (C) Expression of the glutathione reductase-coding gene (AaGlr1) in response to H2O2. (D) Expression of AaTsa1, AaTrr1, and AaGlr1 in response to tert-butyl-hydroperoxide (t-BHP) or cumene hydroperoxide. The relative expression levels from three independent reactions were calculated using a comparative CT method in relation to the fungal actin gene or comparison of fungal strains grown on medium with or without an oxidant.
The thioredoxin and glutaredoxin systems are required for conidiation, hyphal elongation, and fungal growth.
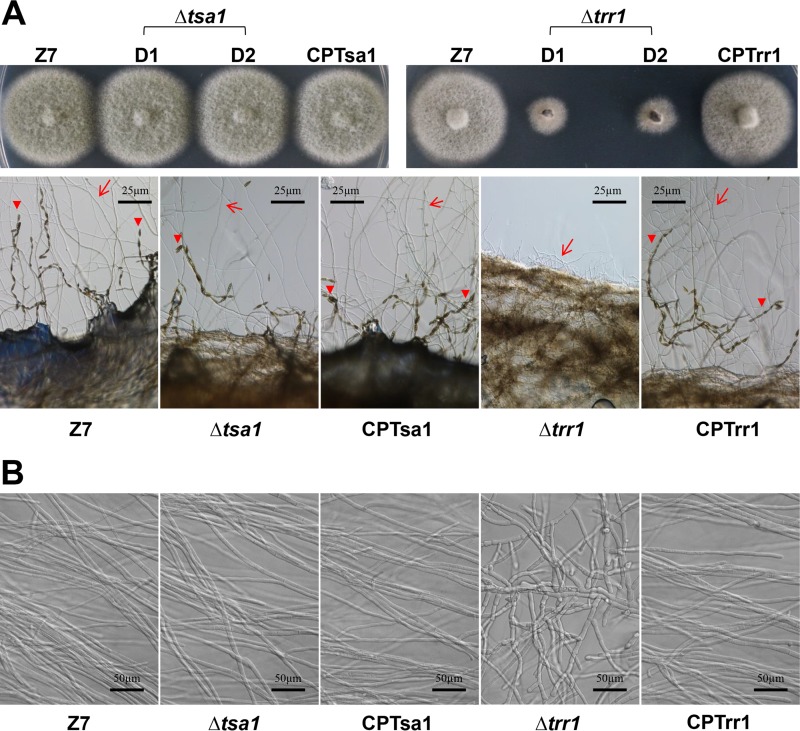
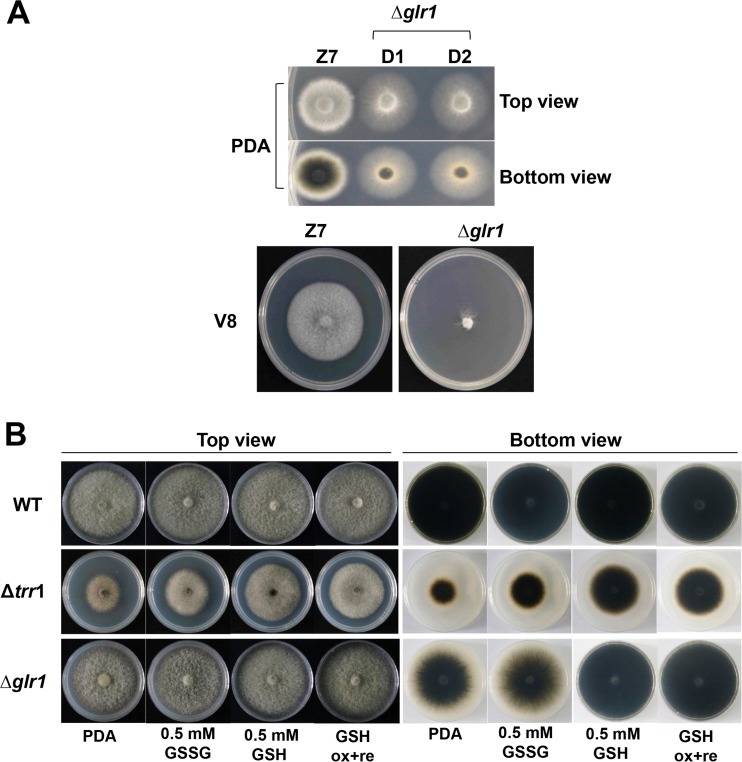
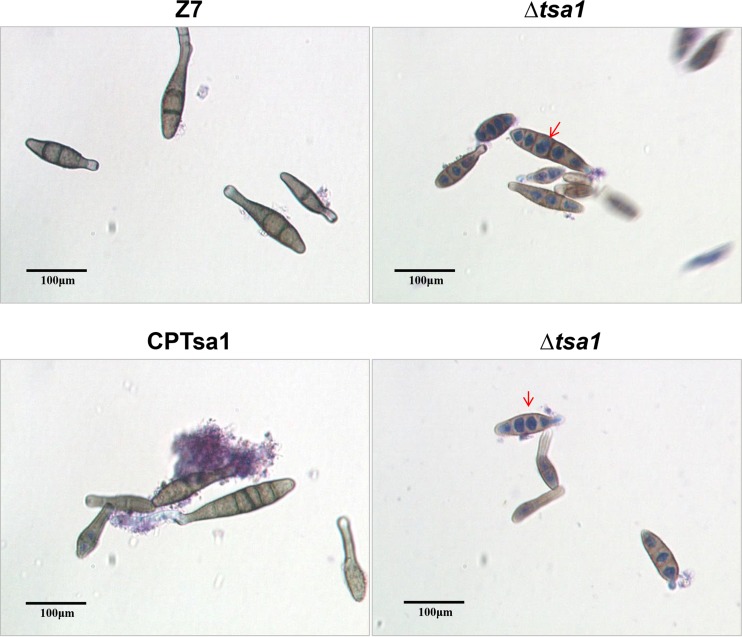
A fungal strain defective for AaTsa1 (Δtsa1) displayed wild-type radial growth on potato dextrose agar (PDA). Both the Δtsa1 and CPTsa1 strains produced ovoid conidia with both cross and longitudinal septae with dark pigmentation similar to those produced by the wild type on PDA. However, radial growth of the Δtrr1 strain was reduced by 50% in relation to that of the wild-type strain (Fig. 2A). The Δtrr1 strain formed less-dense colonies and produced fewer aerial hyphae than the wild type on PDA. Microscopic examination revealed that the Δtrr1 strain failed to produce any conidia and produced stubby and swelling hyphae (Fig. 2B). The complementation strain CPTrr1 expressing AaTrr1::GFP::neo formed fluffy colonies and produced conidia at a rate similar to that for the wild type. Disruption of AaGlr1 in Z7 resulted in the Δglr1 strain, which formed less-dense colonies and produced fewer conidia and less melanin on PDA even though its radial growth was comparable to that of the wild type (Fig. 3A). The Δglr1 strain showed severe growth reduction on V8 agar plates. The growth retardation seen in the Δtrr1 strain could be partially restored by adding 0.5 mM reduced glutathione (GSH), oxidized glutathione (GSSG), or both (Fig. 3B). Addition of GSH to PDA completely restored the colony density, conidiation, and melanin accumulation of the Δglr1 strain.
FIG 2.
The A. alternata thioredoxin reductase 1 gene (AaTrr1), but not the thioredoxin peroxidase 1 gene (AaTsa1), is required for growth and development. (A) Radial growth and formation of hyphae (arrows) and conidia (arrowheads) of the wild-type strain (Z7), two Δtsa1 strains, the CPTsa1 strain expressing a copy of AaTsa1, two Δtrr1 strains, and the CPTrr1 strain expressing a copy of AaTrr1 were cultured on potato dextrose agar or V8 plates for 4 to 7 days. (B) The Δtrr1 strain produces stubby, swelling hyphae, in contrast to those produced by Z7 or other mutants.
FIG 3.
The A. alternata glutathione reductase 1 gene (AaGlr1) is required for fungal growth and pigmentation. (A) Δglr1 strains form less-dense colonies and produce fewer conidia and less melanin than the wild-type strain Z7 on PDA. (B) The addition of the oxidized (GSSG) or reduced (GSH) form of glutathione, or both (ox+re), to PDA restores radial growth (top view) and pigmentation (bottom view) of the Δglr1 and Δtrr1 strains, which have impaired thioredoxin reductase. All tests were repeated at least twice with three replicates of each treatment. Only representative replicates are shown.
The thioredoxin and glutaredoxin systems are required for resistance to oxidants.
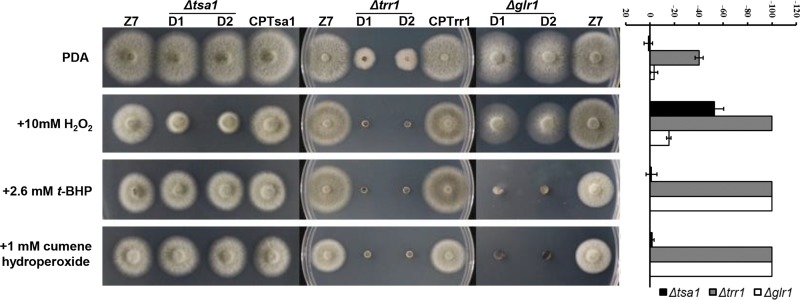
Chemical sensitivity assays performed on PDA revealed that the Δtsa1 strain was more sensitive to H2O2 than the wild type but displayed wild-type resistance to t-BHP and cumene hydroperoxide (Fig. 4). Reintroduction of a functional copy of AaTsa1 into the Δtsa1 strain restored H2O2 resistance to the wild-type level. The Δtrr1 strain displayed increased sensitivity to H2O2, t-BHP, and cumene hydroperoxide. Reintroduction of a functional copy of AaTrr1 into the Δtrr1 strain restored cellular resistance to the three test oxidants at levels comparable to those for the wild type. The Δglr1 strain displayed increased sensitivity to 10 mM H2O2 and failed to grow on PDA amended with 2.6 mM t-BHP or 1 mM cumene hydroperoxide. Fungal cell viability after treatment with H2O2 was determined by staining cells with 1% Evans blue. Live cells could export blue dye and remained light brown, while dead cells were stained blue. The results revealed that more than 97% of Δtsa1 cells were stained blue after exposure to H2O2 (Fig. 5). In contrast, the majority of wild-type and CPTsa1 cells remained light brown after H2O2 treatment. Only 5% of wild-type and 8% of CPTsa1 conidia were stained blue.
FIG 4.
Genes associated with thioredoxin and glutaredoxin are required for resistance to oxidants. Growth of the wild-type strain (Z7), two Δtsa1 strains with impaired thioredoxin peroxidase 1 (AaTsa1), the CPTsa1 strain expressing a copy of AaTsa1, two Δtrr1 strains with impaired thioredoxin reductase 1, the CPTrr1 strain expressing a copy of AaTrr1, and two Δglr1 strains with impaired glutathione reductase 1 cultured on PDA amended with or without H2O2, tert-butyl-hydroperoxide (t-BHP), or cumene hydroperoxide at the indicated concentration is shown. The percentage of growth reduction determined by comparing a cumulative percentage of the growth of the wild type and the deletion mutants grown on the same plate is also shown (far right). All tests were repeated at least twice with three replicates of each treatment. Only representative replicates are shown.
FIG 5.
Viability of Z7, two Δtsa1 strains with impaired AaTsa1, and the CPTsa1 strain expressing a copy of AaTsa1 after being treated with H2O2. Fungal conidia were incubated in 10 mM H2O2 solution for 30 min and stained with Evans blue. Live cells effectively export dye and remain light brown, whereas dead cells fail to export dye and the nucleus is stained blue (arrows).
Mutants defective for thioredoxin- and glutaredoxin-coding genes show changed sensitivity to fungicides.
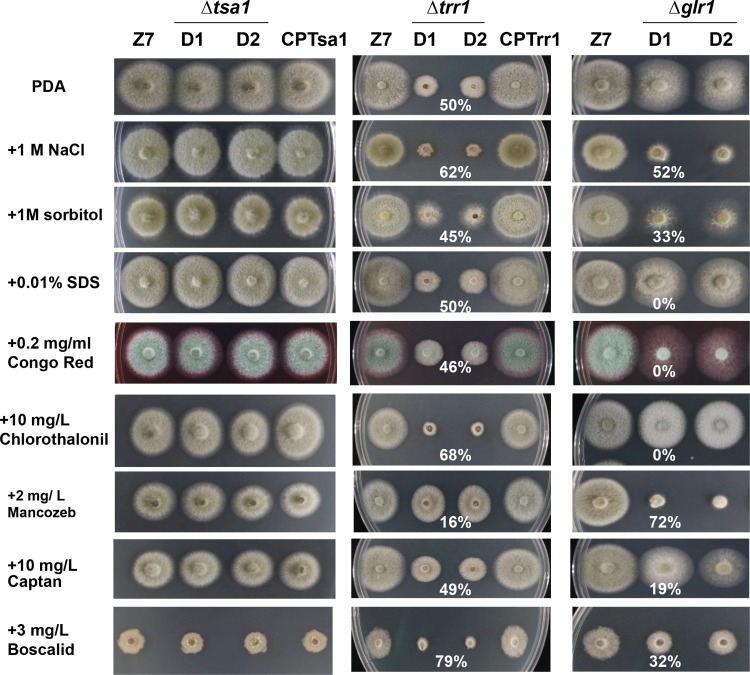
The Δtsa1 strain was not more sensitive to sorbitol, NaCl, or SDS than the Z7 strain (Fig. 6). However, the Δtrr1 and Δglr1 strains had increased sensitivity to NaCl. The Δglr1 but not the Δtrr1 strain had increased sensitivity to sorbitol. None of the Δtsa1, Δtrr1, and Δglr1 strains had increased sensitivity to Congo red. By using PDA as control, assays for fungicide sensitivity indicated that the Δtrr1 strain displayed increased sensitivity to the fungicides chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile) and boscalid [2-chloro-N-(4′-chlorobiphenyl-2-yl)nicotinamide] but displayed decreased sensitivity to mancozeb (Fig. 6). The Δtsa1 strain displayed no increased sensitivity to the test fungicides. The Δglr1 strain displayed increased sensitivity to mancozeb, boscalid, and captan but not chlorothalonil.
FIG 6.
Chemical sensitivity assays. The wild-type strain (Z7), two Δtsa1 strains with impaired thioredoxin peroxidase 1, the CPTsa1 strain expressing a copy of AaTsa1, two Δtrr1 strains with impaired thioredoxin reductase 1, the CPTrr1 strain expressing a copy of AaTrr1, and two Δglr1 strains with impaired for glutathione reductase 1 were grown on PDA with or without the indicated chemicals. All tests were repeated at least twice with three replicates of each treatment. Growth reduction, expressed as a cumulative percentage of the growth of the wild type and the deletion mutants grown on the same plate, is also indicated. Only representative replicates are shown.
The thioredoxin- and glutaredoxin-coding genes are required for fungal virulence.
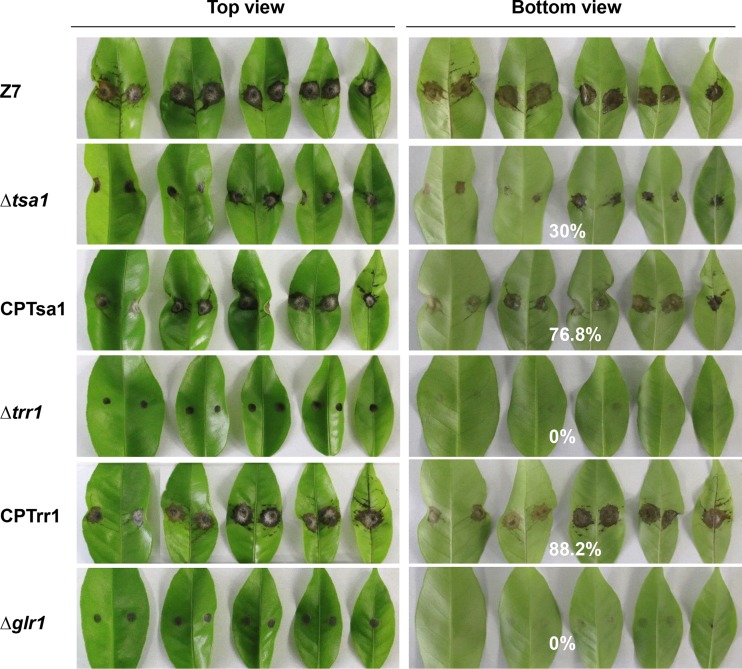
Pathogenicity assays performed on detached Ponkan leaves revealed that the Δtsa1 strain induced smaller necrotic lesions than those induced by the wild type at 2 days postinoculation (dpi) (Fig. 7). The CPTsa1 strain expressing AaTsa1::GFP::neo induced necrotic lesions at a rate and magnitude comparable to those for the wild type on Ponkan leaves at 2 dpi. The Δtrr1 strain failed to induce large spreading lesions as did the wild type. The CPTrr1 strain expressing AaTrr1::GFP::neo produced necrotic lesions similar to those induced by the wild type. Pathogenicity assays revealed that the Δglr1 strain failed to induce large spreading lesions on Ponkan leaves. The impairment of virulence seen in the Δtsa1, Δtrr1, and Δglr1 strains was more drastic when lesions on the bottoms of Ponkan leaves were compared. The Δtsa1 strain induced much smaller lesions on the bottoms of the leaves than the wild type. No necrotic lesions were observed on the bottoms of the leaves inoculated with the Δtrr1 or Δglr1 strain. Quantitative analysis revealed that the sizes of lesions induced by the Δtsa1, CPTsa1, and CPTrr1 strains were about 30%, 76.8%, and 88.2%, respectively, of those induced by the wild type.
FIG 7.
Thioredoxin- and glutaredoxin-coding genes are required for A. alternata pathogenicity. Pathogenicity was assayed on detached Ponkan leaves by placing a 5- by 5-mm agar plug covering fungal mycelium on the tops of the leaves. The wild-type strain (Z7), the Δtsa1 strain with impaired thioredoxin peroxidase, the CPTsa1 strain expressing a copy of AaTsa1, the Δtrr1 strain with impaired thioredoxin reductase, the CPTrr1 strain expressing a copy of AaTrr1, and the Δglr1 strain with impaired glutathione reductase were grown on PDA, and agar plugs were cut with a sterile toothpick. The leaves were incubated in a plastic box for lesion development. Development of necrotic lesions on detached citrus leaves was observed at 2 days postinoculation (dpi). Only some representative replicates are shown. The percent changes of necrotic lesions appearing on the bottoms of the leaves calculated in relation to those induced by the wild-type strain Z7 are also indicated.
Subcellular localization of AaTsa1 and AaTrr1.
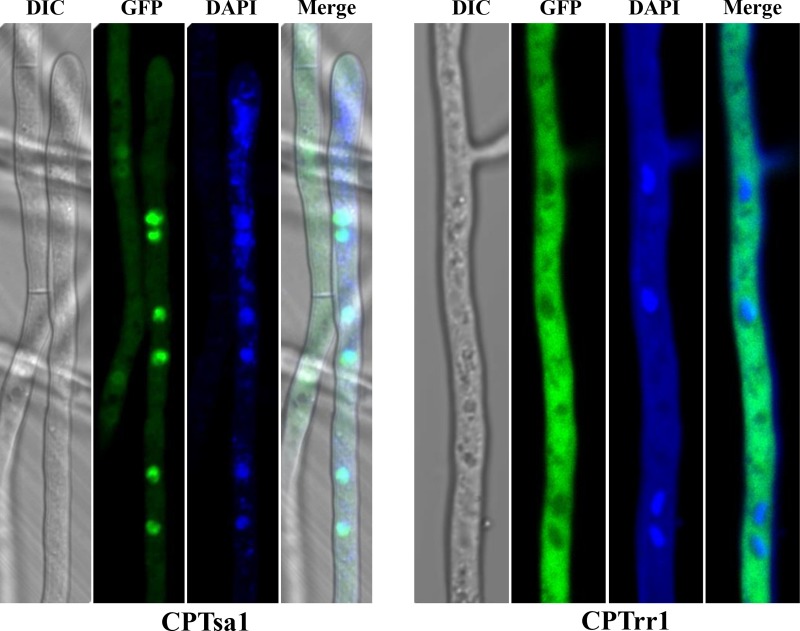
As described above, the Δtsa1 strain had increased sensitivity to H2O2, and the complementation strain (CPTsa1) displayed wild-type resistance to H2O2. CPTsa1 was examined microscopically, revealing green fluorescence in a faint to scattered pattern within fungal hyphae (Fig. 8). Stronger green fluorescence showing distinct foci that closely corresponded to those from the 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclear DNA was observed, indicative of both nuclear and cytoplasmic localization of AaTsa1::GFP. The CPTrr1 strain expressing AaTrr1::GFP::neo displayed wild-type resistance to oxidants. Confocal microscopy examination revealed bright green fluorescence within the CPTrr1 hyphae, indicative of a cytoplasmic location of AaTrr1::GFP. No distinct green foci corresponding to those from DAPI-stained nuclei were observed within the CPTrr1 hyphae.
FIG 8.
Fluorescence microscopy and differential interference contrast (DIC) imaging of the A. alternata CPTsa1 strain expressing an AaTsa1::GFP::neo fragment and the CPTrr1 strain expressing an AaTrr1::GFP::neo fragment. The AaTsa1 (or AaTrr1) fragment carrying its endogenous promoter was translationally fused in frame with a green fluorescent protein (GFP)-coding gene, followed by fusing with a neo gene cassette under the control of the TrpC promoter and terminator, conferring resistance to G418. Fungal nuclei were stained with 4′-6-diamidine-2-phenylindole (DAPI) fluorescent dye.
Expression of AaTsa1 and AaTrr1 is regulated by redox-responsive regulators.
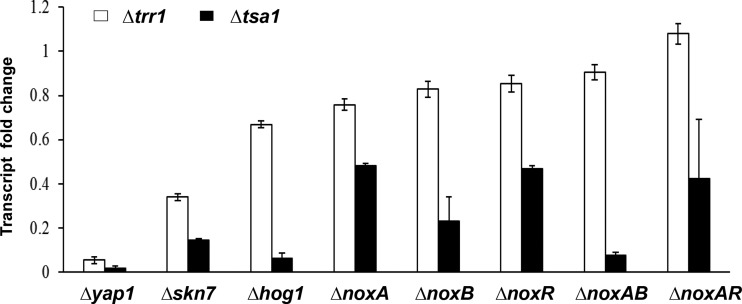
Quantitative RT-PCR analysis revealed that accumulation of the AaTsa1 transcript decreased in the Δyap1 strain, which is defective for the redox-responsive transcription regulator YAP1 in relation to that of the wild type (Fig. 9). Similar reduction of the AaTsa1 transcript was observed in the Δskn7 strain, in which the Skn7 response regulator is impaired, and in the Δhog1 strain, in which the high-osmolarity glycerol 1 (HOG1) MAP kinase is impaired. Expression of AaTsa1 was downregulated considerably in the NoxA, NoxB, and NoxR single and double mutants, in which NADPH oxidases are impaired. Moreover, expression of AaTsa1 was lower in the NoxAB double mutant than in the NoxA and NoxB single mutants, which show an additive effect between NoxA and NoxB. On the other hand, the expression level of AaTsa1 in the NoxAR double mutant was similar to that in the NoxA and NoxR single mutants. This indicates that NoxA and NoxR may functionally complement each other in affecting the expression of AaTsa1. Expression of AaTrr1 was downregulated in the Δyap1 and Δskn7 strains and to a lesser extent in the Δhog1 strain. However, deletion of NoxA, NoxB, and/or NoxR had little or no effect on the expression of AaTrr1.
FIG 9.
qRT-PCR analyses of the expression of the thioredoxin peroxidase 1-coding gene (AaTsa1) and the thioredoxin reductase 1-coding gene (AaTrr1) in A. alternata strains with impaired redox-responsive transcription regulator YAP1 (Δyap1), Skn7 response regulator (Δskn7), high-osmolarity glycerol 1 (HOG1) MAP kinase (Δhog1), and NADPH oxidases (ΔnoxA, ΔnoxB, ΔnoxR, ΔnoxAB, and ΔnoxAR deletion strains). All strains were treated with 10 mM H2O2 for 30 min after being cultured in thermostatic shakers at 25°C. The relative expression level between the wild-type and the mutant strains was calculated from three independent reactions using a comparative CT method in relation to the actin-coding gene.
DISCUSSION
Small thiol-containing proteins play a range of regulatory and metabolic roles in cells. In the current study, we used the targeted gene disruption approach to demonstrate that three genes (AaTsa1, AaTrr1, and AaGlr1) involved in the thioredoxin and glutaredoxin functions are required for resistance to H2O2 and other toxic oxidants and for the growth and development of A. alternata. Genetic analyses also show that some but not all of thiol-containing proteins are associated with osmotic stress resistance and fungicide sensitivity/resistance.
Alignment and comparison of sequences reveal that AaTsa1 is grouped in the alkyl hydroperoxide reductase AhpC/thiol-specific antioxidant (TSA) family. AaTsa1 has two conserved cysteine residues (amino acids 57 and 178) likely involved in the formation of an intermolecular disulfide bond with cysteine residues of other proteins. AaTsa1 is primarily responsible for H2O2 resistance in A. alternata, consistent with the fact that a Tsa1 protein in its reduced dithiol form could reduce H2O2 (28, 29). Both AaTrr1 and AaGlr1 are similar to FAD oxidoreductases that contain two redox-active cysteine residues and a conserved pyridine nucleotide-disulfide domain. FAD-containing oxidoreductase enzymes could transfer reducing equivalents from NADPH to cysteine via FAD, converting disulfide bridges to sulfhydryl groups, and thus detoxifying H2O2 (14). In vitro assays reveal that, unlike Δtsa1, both Δtrr1 and Δglr1 increase sensitivity to H2O2, t-BHP, and cumene hydroperoxide. Previous studies have disclosed that the Δgpx3 strain, defective for a glutathione peroxidase, is hypersensitive to H2O2, t-BHP, cumene hydroperoxide, and superoxide-generating compounds (26). These results indicate that both the thioredoxin and glutaredoxin systems play an important role in cellular resistance to oxidative stress in A. alternata. Moreover, subcellular localization analysis of the AaTsa1::GFP fusion protein revealed that AaTsa1 is localized mainly in the nucleus, suggesting the involvement of AaTsa1 in DNA repair. The Tsa1 homolog in S. cerevisiae is the most important thioredoxin peroxidase for suppressing ROS-induced DNA damage, and loss of Tsa1 may elevate the cellular ROS level and cause genome instability and eventually cell death (10, 30, 31), even though Tsa1 is localized in the cytoplasm (32). Confocal microscopy analysis, however, revealed that AaTrr1 is localized in the cytoplasm of A. alternata hyphae, indicating that AaTrr1 may function to detoxify H2O2 and other toxic ROS in the cytosol.
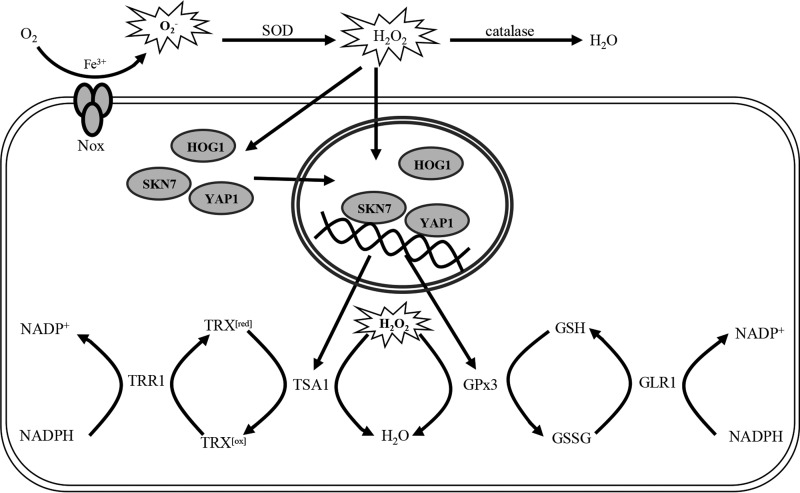
Experiments further demonstrated that the thioredoxin and glutaredoxin systems play a critical role in fungal colonization and lesion formation on a citrus host. qRT-PCR analyses and previous studies showed that AaTsa1 and AaGpx3 are regulated by Yap1, Skn7, Hog1, and Nox, which have previously been shown to be required for ROS resistance and fungal virulence (21, 26). As with those ROS-responsive regulators, Δtsa1, Δtrr1, and Δglr1 strains are all more sensitive to ROS and incite less-severe necrosis than the wild type. Previous research also has shown that Nox can produce low-level H2O2 (25). Thus, we speculate that the low-level H2O2 produced by Nox triggers the activity of YAP1, SKN7, and HOG1, whose products in turn regulate genes (AaTsa1 and AaGpx3) in the thioredoxin and glutaredoxin systems leading to H2O2 detoxification (Fig. 10). AaTrr1 may be regulated by YAP1, SKN7, and HOG1 and not via Nox, since the Nox-related knockout mutants have no significant effect on AaTrr1 expression, which suggests that the regulation of AaTrr1 may be more complex. In addition, the mutant strains have no defects in Alternaria citri toxin (ACT) production as evidenced by in vitro bioassays (data not shown). Because A. alternata strains showing increased sensitivity to ROS are also defective for lesion development, the results confirm further that A. alternata must deal with toxic ROS during infection and colonization and that the ability to detoxify ROS is required for fungal virulence.
FIG 10.
Proposed network of transcriptional regulation that involves YAP1, SKN7, HOG1, NOX, GPx3, GLR1, TSA1, and TRR1 and leads to ROS detoxification in A. alternata. Low-level H2O2, generated by superoxide dismutases (SOD) from superoxide (O2−) produced by NOX, may serve as a signal that upregulates the expression of YAP1, SKN7, and HOG1, and nuclear localization of these proteins transcriptionally upregulates TSA1, TRR1, GPx3, and GLR1. TSA1 can detoxify H2O2 in conjunction with thioredoxin reductase (TRR1) and thioredoxin cycling between reduced thioredoxin (TRX[red]) and oxidized thioredoxin (TRX[ox]). GPx3 can detoxify H2O2 in conjunction with glutathione reductase (GLR1) and glutathione cycling between reduced glutathione (GSH) and oxidized glutathione (GSSG).
One of the most significant findings of this study is the observation of the involvement of the protective oxidative stress genes in fungicide sensitivity. Depending on the type of fungicides tested, deletion mutants of A. alternata with impaired thioredoxin or glutaredoxin have very different reactions. Δtrr1 increases sensitivity to chlorothalonil and boscalid but not captan, indicating that the AaTrr1 product is required for cellular resistance to chlorothalonil and boscalid. However, the Δtrr1 strain displays elevated resistance to mancozeb. Δglr1 increases sensitivity to mancozeb, boscalid, and captan but not chlorothalonil, indicating that the AaGlr1 product is required for cellular resistance to mancozeb and boscalid. Previous studies also have found that Δgpx3 increases resistance to fluioxonil and vinclozolin fungicides (26). Unlike AaTrr1 and AaGlr1 deletion mutants, the Δtsa1 strain displays wild-type sensitivity to all tested fungicides, indicating that AaTsa1 plays no role in the response to fungicides. Alternatively, the function of AaTsa1 could be compensated for by other antioxidant enzymes in response to fungicides. It appears that the thioredoxin and glutaredoxin systems have independent and overlapping functions, as their components are not all involved in fungicide sensitivity or resistance. Although these fungicides have been widely used in agricultural production, the observed results indicate that they have more complex modes of action than previously thought.
Chlorothalonil is a broad-spectrum fungicide and was shown to be an inhibitor of specific NAD thiol-dependent glycolytic and respiratory enzymes in a previous study (33). The reason why chlorothalonil is widely used is due to its high efficacy as a multisite fungicide, which is attributed to thiol moiety inactivation in proteins such as glutathione and coenzyme A (34, 35). Chlorothalonil has no impact on AaTsa1 and AaGlr1, as disruption of either coding gene does not affect fungal sensitivity to this fungicide. In contrast, AaTrr1 is required for resistance to chlorothalonil. A previous report revealed that the lack of regeneration of the reduced thiol could be an important feature in chlorothalonil-treated cells (33). Our results indicate that thiol-containing proteins in the thioredoxin system maybe be involved in the tolerance to chlorothalonil. Boscalid is a carboxamide fungicide which has been known to inhibit proteins associated with the mitochondrial complex II system, leading to respiration malfunction and affecting ATP production (36). Chemical sensitivity assays reveal that both Δtrr1 and Δglr1 mutants have increased sensitivity to boscalid. It appears that A. alternata itself could resist low levels of boscalid through the thioredoxin and glutaredoxin systems. We speculate that boscalid may cause ROS accumulation in A. alternata and therefore require the detoxification of these ROS by the thioredoxin and glutaredoxin systems, since boscalid resulted in oxidative stress and lipid peroxidation in zebrafish (37). Captan, belonging to the dicarboximide family, is a chloroalkylthiol fungicide. Although captan has been known to inhibit fungal respiration by nonspecifically interacting with proteins containing a thiol moiety (38), deletion of AaTrr1 or AaTsa1 has no impact on captan sensitivity in A. alternata. However, AaGlr1 plays a role in resistance to captan. As is well known, glutathione reductase is indispensable for maintaining a proper concentration balance of reduced glutathione (GSH) (39). We speculate that GSH is important for A. alternata to withstand captan based on the facts that ROS accumulation is increased in captan-treated S. cerevisiae and that the yeast cell viability is increased when yeast cells are previously treated with GSH (40).
Interestingly, sensitivity assays revealed that A. alternata strains with impaired AaTrr1, AaTsa1, or AaGlr1 display different levels of sensitivity to the mancozeb fungicide. Δtrr1 increases resistance to mancozeb, while Δglr1 increases sensitivity to mancozeb and the Δtsa1 strain displays wild-type sensitivity. Mancozeb, belonging to the class of ethylene bisdithiocarbamates (EHBDCs) is not toxic until it is degraded to release ethylene bisiothiocyanate sulfide (EBIS) in water (41). EBIS could be converted further to highly toxic ethylene bisiothiocyanates (EBI) upon exposure to UV light. Mancozeb has been known to interact with and to inhibit metal- and/or sulfhydryl-containing enzymes involved in respiration, ATP production, and lipid metabolism (42, 43). Recent research shows that mancozeb causes stoichiometric loss of protein thiols, reversibly dimerizes the protein, and inhibits its enzymatic activity (44). A. alternata, at least in part, depends on the function of glutathione reductase to deal with the toxicity of mancozeb, because deletion of AaGlr1 results in strains that are highly sensitive to mancozeb. On the other hand, in contrast to the case for AaGlr1, thioredoxin reductase likely has the reverse function, as disruption of AaTrr1 in A. alternata increases resistance to mancozeb. Although Δtrr1 grows better in the presence of mancozeb, it still has a worse growth phenotype than the wild type, which may be due to its severe growth inhibition. The involvement of the redox responsive transcription factor Yap1 in response to mancozeb treatment has also been observed in S. cerevisiae (45). The expression of AaTrr1 is also regulated by Yap1. Nevertheless, the contradictory functions of AaTrr1 and AaGlr1 in response to mancozeb highlight the complexity of cellular resistance to any given fungicide. Cellular resistance to both oxidative stress and fluconazole mediated by the bZip transcription regulator Yap1 has also been reported in animal fungal pathogens (46, 47).
The Δgpx3 strain, carrying a glutathione peroxidase defect, reduces conidial formation and vegetative growth (26). In this study, we showed that AaTrr1 but not AaTsa1 is required for normal hyphal elongation, fungal growth, and conidiation. The Δtrr1 strain produces very few conidia and produces stubby, swelling hyphae. The Δglr1 strain is also defective for vegetative growth and conidial production. The deficiencies of hyphal growth and conidiation seen in either the Δtrr1 or Δglr1 strain could be remedied by the addition of oxidized or reduced glutathione to the medium, strengthening the link between sensitivity to oxidative stress and the thioredoxin and glutaredoxin systems in the context of fungal growth and conidial formation.
In addition to increased sensitivity to ROS, Δglr1 increases sensitivity to NaCl and sorbitol, suggesting the involvement of glutathione reductase in osmotic stress resistance. The Δtrr1 strain displays increased sensitivity to NaCl but not sorbitol, while the Δtsa1 strain displays wild-type sensitivity to NaCl and sorbitol. In the rice blast fungus Magnaporthe oryzae, deletion of a Tsa1 or Trr1 homolog but not a Glr1 homolog results in fungi that display cellular sensitivity to osmolytes (16). Another interesting feature is that both Δtsa1 and Δtrr1 strains display wild-type sensitivity to the cell wall-disturbing compound Congo red, while the Δgpx3 strain is more resistant to this compound (26). Although thioredoxin and glutaredoxin systems are conserved in all cells, their actual functions could be different between species. The roles of these systems in maintaining cellular redox homeostasis, conferring oxidative stress resistance, and preventing programmed cell death are well known. In the present study, we have found additional roles in fungicide sensitivity and resistance. Some of the thiol-containing proteins could also play a role in resistance to osmotic stress, even though the exact mechanisms contributing to this phenotype remain elusive. In conclusion, this study provides genetic evidence to determine the important role of the thioredoxin and glutaredoxin systems in the diverse biological and physiological functions of A. alternata. The study also demonstrates that both thioredoxin and glutaredoxin systems contribute to A. alternata virulence on citrus, supporting the requirement of ROS detoxification for successful infection by this phytopathogenic fungus.
MATERIALS AND METHODS
Fungal strains and growth conditions.
The wild-type Z7 strain of Alternaria alternata (Fr.) Keissler was single spore cultured from diseased Ougan (Citrus suavissima Hort. ex Tanaka) leaves in Zhejiang, China (27) and used for transformation and mutagenesis. Fungal strains defective for a Skn7 response regulator (Δskn7), a high-osmolarity glycerol (Δhog1) mitogen-activated protein kinase, a Yap1 regulator (Δyap1), and NADPH oxidases (ΔnoxA, ΔnoxB, ΔnoxR, ΔnoxAB, and ΔnoxAR) were created from the EV-MIL31 strain in previous studies (22–25, 48). The A. alternata strains used in this study and their characteristics are listed in Table 1. All fungal strains were grown on potato dextrose agar (PDA) at 26°C unless otherwise stated. Conidia were harvested from fungal cultures grown on V8 juice agar for 7 to 14 days by immersing, scraping with sterile water, and passing through two layers of sterile cheesecloth. Fungal mycelium was harvested from 2-day liquid cultures by passing through cheesecloth and was used for purification of DNA or RNA. A regeneration medium (RMM) containing sucrose (49) was used to recover fungal transformants. Assays for sensitivity to chemicals were performed on PDA plates amended with a test compound. Fungal hyphae and conidia from 2- to 4-day-old cultures were transferred with sterile toothpicks onto the test medium. Each treatment contained three replicates, and experiments were repeated at least two times. The colony radius of the disruption mutants relative to that of the wild type grown on the same plate was measured. Percent growth inhibition was calculated by dividing the relative difference in the growth by the wild-type growth and then multiplying by 100. Mancozeb (M-45; 80% wettable powder) was purchased from Taoshiyinong Agriculture Technology (Nantong, Jiangsu, China), chlorothalonil (98.5%) was from Suli Chem (Wuxi, Jiangsu, China), boscalid (98%) was from Jiahe Chemicals (Nantong, Jiangsu, China), and captan (99.15%) was from Aladdin Industrial Corp. (Shanghai, China).
TABLE 1.
Phenotypic characterization of Alternaria alternata strains used in this study
| Strain | Phenotype | H2O2 sensitivity | Pathogenicity | Vegetative growth | Reference |
|---|---|---|---|---|---|
| Z7 | Wild type | Resistant | Aggressive | Normal | 27 |
| Δtsa1 mutant | Impaired thioredoxin peroxidase | Sensitive | Reduced | Normal | This study |
| CPTsa1 | AaTsa1 complementation | Resistant | Aggressive | Normal | This study |
| Δtrr1 mutant | Impaired thioredoxin reductase | Sensitive | Reduced substantially | Inhibited | This study |
| CPTrr1 | AaTrr1 complementation | Resistant | Aggressive | Normal | This study |
| Δglr1 mutant | Impaired glutathione reductase | Sensitive | Reduced | Inhibited | This study |
| ΔnoxA mutant | Impaired NADPH oxidase NoxA | Sensitive | Reduced | Inhibited | 48 |
| ΔnoxB mutant | Impaired NADPH oxidase NoxB | Sensitive | Reduced | Inhibited | 25 |
| ΔnoxR mutant | Impaired NADPH oxidase NoxR | Sensitive | Reduced | Inhibited | 25 |
| ΔnoxAB mutant | Impaired NADPH oxidases NoxA and NoxB | Sensitive | Not determined | Inhibited | 25 |
| ΔnoxAR mutant | Impaired NADPH oxidases NoxA and NoxR | Sensitive | Not determined | Inhibited | 25 |
| Δhog1 mutant | Impaired HOG1 MAP kinase | Sensitive | Reduced substantially | Inhibited | 23 |
| Δyap1 mutant | Impaired transcriptional regulator YAP1 | Sensitive | Reduced substantially | Inhibited | 22 |
| Δskn7 mutant | Impaired SKN7 response regulator | Sensitive | Reduced | Inhibited | 24 |
Targeted gene disruption.
The genome of A. alternata strain Z7 has been sequenced and annotated (27). The A. alternata thioredoxin peroxidase (AaTsa1)- and thioredoxin reductase (AaTrr1)-coding genes were identified from the genome sequence and amplified by PCR with gene-specific primers from genomic DNA of Z7. Fungal DNA was purified from mycelium according to the protocol described by Raeder and Broda (53). Open reading frame (ORF) and exon/intron positions in genes were identified using the Softberry gene-finding software. Both AaTsa1 and AaTrr1 were mutated by integrating a bacterial phosphotransferase B gene (HYG) cassette under the control of the Aspergillus nidulans trpC gene promoter and terminator, resulting in resistance to hygromycin in the Z7 genome using a split-marker approach (50). Truncated but overlapping HYG fragments (HY/g and h/YG) were amplified by PCR with different primer sets from plasmid pTFcm (51) and joined with truncated AaTsa1 fragments by two-round PCR according to the principles described by Lin et al. (54). The oligonucleotide primers used in this study and their sequences are reported in Table 2.
TABLE 2.
Oligonucleotide primers used in this studya
| Forward primers |
Reverse primers |
||
|---|---|---|---|
| Primer (gene) | Nucleotide sequence (5′→3′) | Primer (gene) | Nucleotide sequence (5′→3′) |
| 1F (AaTsa1) | GCTTATTGGCGGTACTTAT | 1R (AaTsa1) | TAGGCATTCATTGTTGACCTCCGTTTGGCTGGACCCTTG |
| 2F (AaTsa1) | CAAGGGTCCAGCCAAACGGAGGTCAACAATGAATGCCTA | 2R (AaTsa1) | GCTCCATACAAGCCAAACC |
| 3F (AaTsa1) | CAGCGTCTCCGACCTGA | 3R (AaTsa1) | GAGACAACACGGCTGCGGGATTACCTCTAAACAAGTGTACC |
| 4F (AaTsa1) | GGTACACTTGTTTAGAGGTAATCCCGCAGCCGTGTTGTCTC | 4R (AaTsa1) | TTCCTTTCGTCTTGGGTT |
| 5F (AaTsa1) | GACCGCCGATAGCAACT | 5R (AaTsa1) | CGTAACGCAATACCCTCC |
| 6F (AaTsa1) | GGGGATGGGCATACTTG | 6R (AaTsa1) | CAGATGATAATAATGTCCTCGT |
| 7F (AaTsa1) | TTATGTTTATCGGCACTTTG | 7R (AaTsa1) | TGGACTGGGAGGATTTGA |
| 8F (AaTsa1) | GTTATGCGGATACAGGCG | 8R (AaTsa1) | AGCTCCTCGCCCTTGCTCACCATAAGGATTACCTCTAAACAAGTG |
| 9F (AaTsa1) | CACTTGTTTAGAGGTAATCCTTATGGTGAGCAAGGGCGAGGAGCT | 9R (AaTsa1) | GCCTCTTCGCTATTACGC |
| 10F (AaTrr1) | GAAGGGACCAGCGAAAT | 10R (AaTrr1) | TAGGCATTCATTGTTGACCTCCTGGGTACGGAGAATAAGGA |
| 11F (AaTrr1) | TCCTTATTCTCCGTACCCAGGAGGTCAACAATGAATGCCTA | 11R (AaTrr1) | GCTCCATACAAGCCAAACC |
| 12F (AaTrr1) | CAGCGTCTCCGACCTGA | 12R (AaTrr1) | TTTGTTTGCGACTTCTCAGGGATTACCTCTAAACAAGTGTACC |
| 13F (AaTrr1) | GGTACACTTGTTTAGAGGTAATCCCTGAGAAGTCGCAAACAAA | 13R (AaTrr1) | CTCCACTTACCCACAGCA |
| 14F (AaTrr1) | GCACCGAAATCATCTCCC | 14R (AaTrr1) | CCTGACGATACCGCTTGT |
| 15F (AaTrr1) | TTGGCATAACTGGTGAAGA | 15R (AaTrr1) | GAGGTAAACCCGAAACG |
| 16F (AaTrr1) | GTGGTTTGGCTTGTATGG | 16R (AaTrr1) | TTGGCGAGTGGTGCCTATGT |
| 17F (AaTrr1) | GTTCTGATTTTCGGTAGGC | 17R (AaTrr1) | AGCTCCTCGCCCTTGCTCACCATTAGAAGAGGGTTGGAGCGGTA |
| 18F (AaTrr1) | TACCGCTCCAACCCTCTTCTAATGGTGAGCAAGGGCGAGGAGCT | 18R (AaTrr1) | GCCTCTTCGCTATTACGC |
| 19F (AaTsa1) | AGCAAGAAGGACGGAGGC | 19R (AaTsa1) | GGTGTCGCTTTGATGGTTT |
| 20F (AaTrr1) | GCACCGAAATCATCTCCC | 20R (AaTrr1) | CACCACCTCCGATAACGAC |
| 21F (AaGlr1) | CTGCGTGCCGAAGAAAGT | 22R (AaGlr1) | TCAACCTCGTCCTTAGCG |
| 22F (AaGlr1) | ATCAGCGTCTTCACATACC | 22R (AaGlr1) | TAGGCATTCATTGTTGACCTCCGGACAACGGCAGATACTT |
| 23F (AaGlr1) | AAGTATCTGCCGTTGTCCGGAGGTCAACAATGAATGCCTA | 23R (AaGlr1) | GCTCCATACAAGCCAAACC |
| 24F (AaGlr1) | CAGCGTCTCCGACCTGA | 24R (AaGlr1) | CTCTGGGTGGGCGAAAAGGATTACCTCTAAACAAGTGTACC |
| 25F (AaGlr1) | GGTACACTTGTTTAGAGGTAATCCTTTTCGCCCACCCAGAG | 25R (AaGlr1) | CAGCCAATCAAGCAATAAGG |
| 26F (AaGlr1) | ATTGGTAGTGGAGGTATTGC | 26R (AaGlr1) | GACGGTTGATTCATTCTTTGT |
| 27F (AaGlr1) | GCCACGGAAGGATGAAG | 27R (AaGlr1) | GCTATTTACCCGCAGGAC |
| 28F (AaGlr1) | CTTCCTCCCTTTATTTCAGA | 28R (AaGlr1) | GCCGCACTACAGCACAG |
Primer pairings 19F plus 19R (AaTsa1), 20F plus 20R (AaTrr1), and 21F plus 21R (AaGlr1) were used for quantitative RT-PCR. Descriptions of other primers are given in Results and in Materials and Methods.
For AaTsa1 disruption, a 5′ AaTsa1 fragment (1,301 bp) was amplified with primer set 1F/1R and fused with a 1,049-bp HY/g fragment amplified with primers 2F/2R to produce a 5′-AaTsa1::HY/g fragment. A 3′ AaTsa1 fragment (1,395 bp) was amplified with primers 4F/4R and fused with a 1,701-bp h/YG fragment amplified with primers 3F/3R to produce an h/YG::AaTsa1-3′ fragment. The two fragments 5′-AaTsa1::HY/g and h/YG::AaTsa1-3′ were combined and transformed into protoplasts prepared from the Z7 strain using CaCl2 and polyethylene glycol as previously described (49) to disrupt AaTsa1. Fungal transformants were recovered from a regeneration medium (RMM) containing 100 μg/ml hygromycin and examined for gene deletion by PCR with an AaTsa1 primer that was not localized in the split-marker fragments pairing with a Hyg-specific primer. Primer set 6F/6R amplified a 1,995-bp fragment and 7F/7R amplified a 3,101-bp fragment from genomic DNA of transformants whose AaTsa1 allele was replaced with the Hyg cassette. Those primers failed to amplify fragments from genomic DNA of the wild type. Of 27 transformants recovered from hygromycin-containing medium, 22 had correct integration of Hyg within AaTsa1 based on PCR screening with an AaTsa1-specific primer whose sequence was not located in the split-marker fragments pairing with a Hyg-specific primer (see Fig. S2 in the supplemental material).
To disrupt AaTrr1, the two overlapping fragments 5′-AaTrr1::HY/g and h/YG::AaTrr1-3′ (see Fig. S3 in the supplemental material) were generated by two-round PCR and introduced into Z7 as described above. Primer set 15F/15R amplified a 3,730-bp fragment and 16F/16R amplified a 2,739-bp fragment from transformants whose AaTrr1 allele was replaced with the Hyg cassette. Primer set 15F/15R or 16F/16R failed to amplify any fragments from genomic DNA of the wild type. Diagnostic PCR analyses indicated that an AaTrr1-specific primer whose sequence was not located in the split-marker fragments amplified the expected DNA fragment when pairing with a Hyg primer from genomic DNA prepared from 15 transformants but not from the wild type and the remaining 4 transformants (Fig. S3 and data not shown).
The split-marker approach was also used to disrupt AaGlr in Z7 as illustrated in Fig. S4 in the supplemental material. Primers used to generate split Hyg marker fragments and for diagnostic PCR were also included (Table 2).
To exclude that other integration events caused the phenotypes of the gene mutants we describe below, two mutants from each gene disruption were selected for further analyses.
Genetic complementation.
For genetic complementation, an AaTsa1 (or AaTrr1) fragment carrying its endogenous promoter but without a stop codon was amplified from Z7 genomic DNA and translationally fused in frame with a green fluorescent protein (GFP)-coding gene with a TrpC terminator by fusion PCR (Fig. S2 and S3). The resultant AaTsa1 (or AaTrr1)::GFP fragment was fused with a neo gene cassette under the control of the TrpC promoter and terminator, conferring resistance to G418 from plasmid pNEO1300. The AaTsa1 (or AaTrr1)::GFP::neo fragment was transformed into protoplasts prepared from an AaTsa1 (or AaTrr1) mutant. Fungal transformants were recovered from RMM supplemented with 100 mg/ml G418, screened by PCR with primers 5F/5R (for AaTsa1) and 14F/14R (for AaTrr1), and examined for H2O2 sensitivity and the emission of green fluorescence. We could not obtain the Δglr1 complementation strain because we failed to generate sufficient protoplasts of this mutant.
Gene expression.
Relative expression of the genes in A. alternata strains treated with or without chemicals was quantified by quantitative RT-PCR in a 7300 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). Fungal RNA was extracted with an Axygen RNA purification kit (Capital Scientific, Union City, CA, USA). First-strand DNA was synthesized from RNA using a Prime Script RT reagent kit (TaKaRa) and used for PCR amplification with gene-specific primers. Expression of the A. alternata actin-coding gene was included as a control. The expression levels relative to that of the wild type from three independent reactions were calculated using a comparative threshold cycle (CT) method as previously described (52).
Microscopy.
Fungal conidia and hyphae were examined with a Nikon microscope equipped with an LV100ND image system (Nikon, Japan). Nuclei in fungal hyphae grown in liquid medium for 2 days were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, USA). GFP fluorescence was examined with a Zeiss LSM780 confocal microscope (Gottingen, Niedersachsen, Germany).
Pathogenicity assay.
Fungal virulence was assessed on detached Ponkan (also known as Chinese honey orange) (Citrus poonensis Yu Tanaka; syn. C. reticulata Blanco) leaves inoculated by placing a 5- by 5-mm agar plug covering fungal mycelium on each spot. The inoculated leaves were kept in a plastic box at 26°C for 2 to 4 days for lesion development. Each fungal strain was tested on at least 10 leaves, and experiments were repeated twice.
Statistical analysis.
The significance of treatments was determined by analysis of variance and means separated by Duncan's test (P < 0.05).
Accession number(s).
Newly determined sequence data have been deposited in GenBank under accession numbers MG593559, MG593563, and MG593564.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Foundation of Natural Science of China (31571948) and the earmarked fund for China Agriculture Research System (CARS-27) to H.L.
We thank our colleagues in the Institute of Biotechnology, Zhejiang University, for insightful discussions.
We declare no competing financial interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00086-18.
REFERENCES
- 1.Heller J, Tudzynski P. 2011. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol 49:369–390. doi: 10.1146/annurev-phyto-072910-095355. [DOI] [PubMed] [Google Scholar]
- 2.Huang K, Czymmek KJ, Caplan JL, Sweigard JA, Donofrio NM. 2011. HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog 7:e1001335. doi: 10.1371/journal.ppat.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samalova M, Meyer AJ, Gurr SJ, Fricker MD. 2014. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol 201:556–573. doi: 10.1111/nph.12530. [DOI] [PubMed] [Google Scholar]
- 4.Holmgren A, Johansson C, Berndt C, Lönn ME, Hudemann C, Lillig CH. 2005. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans 33:1375–1377. [DOI] [PubMed] [Google Scholar]
- 5.Blokhina O, Virolainen E, Fagerstedt KV. 2003. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot 91(Spec No):179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmgren A. 1989. Thioredoxin and glutaredoxin systems. J Biol Chem 264:13963–19366. [PubMed] [Google Scholar]
- 7.Grant CM. 2001. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol Microbiol 39:533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- 8.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW. 2004. Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Missall TA, Pusateri ME, Lodge JK. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol Microbiol 51:1447–1458. doi: 10.1111/j.1365-2958.2004.03921.x. [DOI] [PubMed] [Google Scholar]
- 10.Iraqui I, Kienda G, Soeur J, Faye G, Baldacci G, Kolodner RD, Huang ME. 2009. Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet 5:e1000524. doi: 10.1371/journal.pgen.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molin M, Yang J, Hanzén S, Toledano MB, Labarre J, Nyström T. 2011. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell 43:823–833. doi: 10.1016/j.molcel.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Viefhues A, Heller J, Temme N, Tudzynski P. 2014. Redox systems in Botrytis cinerea: impact on development and virulence. Mol Plant Microbe Interact 27:858–874. doi: 10.1094/MPMI-01-14-0012-R. [DOI] [PubMed] [Google Scholar]
- 13.Malagnac F, Klapholz B, Silar P. 2007. PaTrx1 and PaTrx3, two cytosolic thioredoxins of the filamentous ascomycete Podospora anserina involved in sexual development and cell degeneration. Eukaryot Cell 6:2323. doi: 10.1128/EC.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dantas ADS, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, Quinn J. 2010. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol Cell Biol 30:4550–4563. doi: 10.1128/MCB.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Missall TA, Cherryharris JF, Lodge JK. 2005. Two glutathione peroxidases in the fungal pathogen Cryptococcus neoformans are expressed in the presence of specific substrates. Microbiology 151:2573. doi: 10.1099/mic.0.28132-0. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez J, Wilson RA. 2014. Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS One 9:e87300. doi: 10.1371/journal.pone.0087300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Yin Z, Tang W, Cai X, Gao C, Zhang H, Zheng X, Wang P, Zhang Z. 2017. The thioredoxin MoTrx2 protein mediates ROS balance and controls pathogenicity as a target of the transcription factor MoAP1 in Magnaporthe oryzae. Mol Plant Pathol 18:1199–1209. doi: 10.1111/mpp.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohmoto K, Akimitus K, Otani H. 1991. Correlation of resistance and susceptibility of citrus to Alternaria alternata with sensitivity to host-specific toxins. Phytopathology 81:719–722. doi: 10.1094/Phyto-81-719. [DOI] [Google Scholar]
- 19.Lin CH, Yang SL, Chung KR. 2011. Cellular responses required for oxidative stress tolerance, colonization, and lesion formation by the necrotrophic fungus Alternaria alternata in citrus. Curr Microbiol 62:807–815. doi: 10.1007/s00284-010-9795-y. [DOI] [PubMed] [Google Scholar]
- 20.Zandalinas SI, Mittler R. 2017. ROS-induced ROS release in plant and animal cells. Free Radic Biol Med doi: 10.1016/j.freeradbiomed.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 21.Chung KR. 2014. Reactive oxygen species in the citrus fungal pathogen Alternaria alternata: the roles of NADPH-dependent oxidase. Physiol Mol Plant Pathol 88:10–17. doi: 10.1016/j.pmpp.2014.08.001. [DOI] [Google Scholar]
- 22.Lin CH, Yang SL, Chung KR. 2009. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol Plant Microbe Interact 22:942. doi: 10.1094/MPMI-22-8-0942. [DOI] [PubMed] [Google Scholar]
- 23.Lin CH, Chung KR. 2010. Specialized and shared functions of the histidine kinase- and HOG1 MAP kinase-mediated signaling pathways in Alternaria alternata, a filamentous fungal pathogen of citrus. Fungal Genet Biol 47:818–827. doi: 10.1016/j.fgb.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Chen LH, Lin CH, Chung KR. 2012. Roles for SKN7 response regulator in stress resistance, conidiation and virulence in the citrus pathogen Alternaria alternata. Fungal Genet Biol 49:802–813. doi: 10.1016/j.fgb.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Yang SL, Chung KR. 2013. Similar and distinct roles of NADPH oxidase components in the tangerine pathotype of Alternaria alternata. Mol Plant Pathol 14:543–556. doi: 10.1111/mpp.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang SL, Yu PL, Chung KR. 2016. The glutathione peroxidase-mediated reactive oxygen species resistance, fungicide sensitivity and cell wall construction in the citrus fungal pathogen Alternaria alternata. Environ Microbiol 18:923–935. doi: 10.1111/1462-2920.13125. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Sun X, Yu D, Xu J, Chung K, Li H. 2016. Genomic and transcriptomic analyses of the tangerine pathotype of Alternaria alternata in response to oxidative stress. Sci Rep 6:32437. doi: 10.1038/srep32437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carmel-Harel O, Storz G. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54:439. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 29.Wood ZA, Schröder E, Harris JR, Poole LB. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28:32. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 30.Huang ME, Rio AG, Nicolas A, Kolodner RD. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci U S A 100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pannunzio NR, Lieber MR. 2017. AID and reactive oxygen species can induce DNA breaks within human chromosomal translocation fragile zones. Mol Cell 68:901. doi: 10.1016/j.molcel.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SG, Cha MK, Jeong W, Kim IH. 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J Biol Chem 275:5723–5732. doi: 10.1074/jbc.275.8.5723. [DOI] [PubMed] [Google Scholar]
- 33.Tillman RW, Siegel MR, Long JW. 1973. Mechanism of action and fate of the fungicide chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile) in biological systems. I. Reactions with cells and subcellular components of Saccharomyces pastorianus. Pesticide Biochem Physiol 3:160–167. [Google Scholar]
- 34.Gisi U, Sierotzki H. 2008. Fungicide modes of action and resistance in downy mildews. Eur J Plant Pathol 122:157–167. doi: 10.1007/s10658-008-9290-5. [DOI] [Google Scholar]
- 35.Bessi H, Cossuleguille C, Zaïd A, Vasseur P. 1999. Effects of chlorothalonil on glutathione and glutathione-dependent enzyme activities in Syrian hamster embryo cells. Bull Environ Contam Toxicol 63:582–589. doi: 10.1007/s001289901020. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn PJ. 1984. Mode of action of carboximides. Br Mycol Soc Sym Ser 9:155–183. [Google Scholar]
- 37.Zang X, Wang M, Zhang XY, Tian XH. 2017. Effects of boscalid on the antioxidant enzyme system of adult zebrafish (Danio rerio). Agric Sci Technol 18:287–293. [Google Scholar]
- 38.Gordon EB. 2001. Captan and folpet, p 1711–1742. In Krieger RI, Krieger WC (ed), Handbook of pesticide toxicology, 2nd ed Elsevier Inc., Amsterdam, The Netherlands. [Google Scholar]
- 39.Muller EG. 1996. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol Biol Cell 7:1805–1813. doi: 10.1091/mbc.7.11.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scariot FJ, Jahn L, Apl D, Echeverrigaray S. 2017. Necrotic and apoptotic cell death induced by captan on Saccharomyces cerevisiae. World J Microbiol Biotechnol 33:159. doi: 10.1007/s11274-017-2325-3. [DOI] [PubMed] [Google Scholar]
- 41.Marrs TT, Ballantyne B (ed). 2004. Pesticide toxicology and international regulation. John Wiley & Sons, New York, NY. [Google Scholar]
- 42.Domico LM, Cooper KR, Bernard LP, Zeevalk GD. 2007. Reactive oxygen species generation by the ethylene-bis-dithiocarbamate (EBDC) fungicide mancozeb and its contribution to neuronal toxicity in mesencephalic cells. Neurotoxicology 28:1079–1091. doi: 10.1016/j.neuro.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dias PJ, Teixeira MC, Telo JP, Sá-Correia I. 2010. Insights into the mechanisms of toxicity and tolerance to the agricultural fungicide mancozeb in yeast, as suggested by a chemogenomic approach. OMICS 14:211–227. doi: 10.1089/omi.2009.0134. [DOI] [PubMed] [Google Scholar]
- 44.Roede JR, Jones DP. 2014. Thiol-reactivity of the fungicide maneb. Redox Biol 2:651–655. doi: 10.1016/j.redox.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teixeira MC, Dias PJ, Simões T, Sácorreia I. 2008. Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem Biophys Res Commun 367:249–255. doi: 10.1016/j.bbrc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 46.Dantas AS, Day A, Ikeh M, Kos I, Achan B, Quinn J. 2015. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5:142. doi: 10.3390/biom5010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul S, Doering TL, Moyerowley WS. 2015. Cryptococcus neoformans Yap1 is required for normal fluconazole and oxidative stress resistance. Fungal Genet Biol 74:1–9. doi: 10.1016/j.fgb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang SL, Chung KR. 2012. The NADPH oxidase-mediated production of hydrogen peroxide (H2O2) and resistance to oxidative stress in the necrotrophic pathogen Alternaria alternata of citrus. Mol Plant Pathol 13:900–914. doi: 10.1111/j.1364-3703.2012.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung KR, Shilts T, Li W, Timmer LW. 2002. Engineering a genetic transformation system for Colletotrichum acutatum, the causal fungus of lime anthracnose and postbloom fruit drop of citrus. FEMS Microbiology Lett 213:33–39. doi: 10.1111/j.1574-6968.2002.tb11282.x. [DOI] [PubMed] [Google Scholar]
- 50.Chung KR, Lee MH. 2015. Split-marker-mediated transformation and targeted gene disruption in filamentous fungi, p 175–180. In van den Berg M, Maruthachalam K (ed), Genetic transformation systems in fungi, vol 2 Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 51.Wang JY, Li HY. 2008. Agrobacterium tumefaciens-mediated genetic transformation of the phytopathogenic fungus Penicillium digitatum. J Zhejiang Univ Sci B 9:823–828. doi: 10.1631/jzus.B0860006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun X, Wang J, Feng D, Ma Z, Li H. 2011. PdCYP51B, a new putative sterol 14α-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl Microbiol Biotechnol 91:1107–1119. doi: 10.1007/s00253-011-3355-7. [DOI] [PubMed] [Google Scholar]
- 53.Raeder U, Broda P. 1985. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol 1(1):17–20. [Google Scholar]
- 54.Lin C-H, Yang SL, Wang N-Y, Chung K-R. 2010. The FUS3 MAPK signaling pathway of the citrus pathogen Alternaria alternata functions independently or cooperatively with the fungal redox-responsive AP1 regulator for diverse developmental, physiological and pathogenic processes. Fungal Genet Biol 47:381–391. doi: 10.1016/j.fgb.2009.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.