ABSTRACT
The germination of Bacillus spores is triggered by certain amino acids and sugar molecules which permeate the outermost layers of the spore to interact with receptor complexes that reside in the inner membrane. Previous studies have shown that mutations in the hexacistronic gerP locus reduce the rate of spore germination, with experimental evidence indicating that the defect stems from reduced permeability of the spore coat to germinant molecules. Here, we use the ellipsoid localization microscopy technique to reveal that all six Bacillus cereus GerP proteins share proximity with cortex-lytic enzymes within the inner coat. We also reveal that the GerPA protein alone can localize in the absence of all other GerP proteins and that it has an essential role for the localization of all other GerP proteins within the spore. Its essential role is also demonstrated to be dependent on SafA, but not CotE, for localization, which is consistent with an inner coat location. GerP-null spores are shown also to have reduced permeability to fluorescently labeled dextran molecules compared to wild-type spores. Overall, the results support the hypothesis that the GerP proteins have a structural role within the spore associated with coat permeability.
IMPORTANCE The bacterial spore coat comprises a multilayered proteinaceous structure that influences the distribution, survival, and germination properties of spores in the environment. The results from the current study are significant since they increase our understanding of coat assembly and architecture while adding detail to existing models of germination. We demonstrate also that the ellipsoid localization microscopy (ELM) image analysis technique can be used as a novel tool to provide direct quantitative measurements of spore coat permeability. Progress in all of these areas should ultimately facilitate improved methods of spore control in a range of industrial, health care, and environmental sectors.
KEYWORDS: Bacillus, spore, coat, germination, permeability, spore coat, spores
INTRODUCTION
Bacterial endospores (here, spores) are ubiquitous in the environment. They are formed by members of the Bacillales and Clostridiales orders in response to nutrient starvation, with Bacillales being aerobic species and Clostridiales being anaerobes. Their ubiquity results from the protective cellular structure that the spore displays, as shown in Fig. 1. This comprises a central protoplast, or core, which is enveloped, consecutively, by an inner membrane, notable for the reduced fluidity of its lipids, and a thick layer of peptidoglycan, which itself can be subdivided into a structurally distinct germ cell wall and cortex (1). A second membrane, which may be discontinuous, surrounds the cortex, followed by a multilayered coat composed of numerous different proteins. Finally, in some species, the coat itself is surrounded by an outermost structure referred to as the exosporium (2). The various structural features have different primary functions. The coat, for example, protects against degradative enzymes and harmful chemicals (3), whereas the cortex, in conjunction with unique core metabolites, such as dipicolinic acid (DPA), results in a protoplast of sufficiently low water activity to ensure metabolic dormancy (4). In this state, bacterial spores can persist in a dormant state in the environment for extended periods of time.
FIG 1.
Schematic of a bacterial spore. The major cellular structures evident in thin-section transmission electron microscopy images are shown. The spore coat can typically be subdivided into outer and inner coat structures, with the pattern of striation depending on the species. The exosporium is present in some species, including B. cereus, but is absent in others, including B. subtilis.
In order to resume the vegetative cell cycle, the spore must respond to environmental cues that indicate that conditions are once again conducive to growth, all the while being dormant and encased in a multilayered protective shell. The spore-sensing system comprises receptor proteins that are localized to the inner membrane, which respond typically to various amino acids, monosaccharides, and inorganic ions (5). Combinations of these germinant molecules are often required to stimulate efficient germinative responses. In spores of Bacillus species, presumed binding of germinants to receptor proteins results in the release of dipicolinic acid chelated with Ca2+ ions (CaDPA) and other small molecules from the spore core, followed by the activation of specialized lysins that degrade the cortical peptidoglycan. These activities permit hydration of the protoplast, resumption of metabolism, and, concomitant with shedding of the coat, the emergence of a new vegetative cell (5).
A degree of permeability in the spore structure is therefore required to permit the transit of small-molecule germinants through the various integument layers to reach and interact with the germinant receptors. In this context, proteins encoded by the hexacistronic gerP operon, which is present in the genomes of all Bacillus species, have been implicated in having a role in maintaining the permeability of the spore coat. This stems from work conducted initially in Bacillus cereus 569, in which spores with a transposon insertion in the operon had a germination defect that could be relieved by chemical removal of the coat (6). Subsequent mutagenesis analyses with Bacillus subtilis (7) and Bacillus anthracis (8) spores revealed that they too have defective germination phenotypes when the gerP operon is disrupted, or in the case of the B. anthracis, when individual gerP genes are deleted. However, other than being required for efficient spore germination and a suggestion that they are probably coat proteins, little is known of the GerP proteins. Bioinformatic analyses do not reveal any functional clues, only that the various GerP proteins largely resemble orthologous proteins in other Bacillus species. Hence, the purpose of the current study was to use the recently developed ellipsoid localization microscopy (ELM) technique (9) to more precisely determine the location of the various GerP proteins in Bacillus cereus spores. We sought additionally to ascertain whether there is any dependency between the various GerP proteins, and between the GerP and coat morphogenetic proteins, for localization in the spore. Finally, ELM was used to directly assess the permeabilities of gerP spores to fluorescently labeled dextran molecules compared to wild-type spores.
RESULTS
Construction and germination of B. cereus 14579 gerP strains.
Defective spore germination phenotypes associated with the gerP operon were first observed in Bacillus cereus strain 569 bearing a transposon (Tn917) insertion between gerPB and gerPC (6). Our initial attempts at creating markerless deletions of the entire gerP operon and individual genes within the operon in this strain proved to be unsuccessful. Accordingly, the decision was made to switch the work to B. cereus 14579, which appears to be more amenable to genetic manipulation. A markerless allelic exchange system was subsequently used to create a series of strains in the 14579 background where individual genes (start and stop codons aside) from gerPA through gerPF were deleted (Table 1; see also Table S1 and Fig. S1 in the supplemental material). The same markerless approach was used to delete the entire gerP operon, with the exception of the start codon from gerPA and the stop codon of gerPF (strain AG007). In common with B. subtilis and B. anthracis, B. cereus 14579 has two additional gerPF homologues on the chromosome (BC2276 and BC4794; Table S2), although these were not deleted in the course of this work. Analysis of the resultant null mutant spores by transmission electron microscopy revealed no obvious morphological defects compared to wild-type spores (Fig. S2).
TABLE 1.
Bacillus strains used in this study
| Strain | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| PS832 | Wild type | Peter Setlow |
| PS4228 | ΔgerP Tcr | Peter Setlow |
| B. cereus strains | ||
| 14579 | Wild type | Toril Lindbäck |
| AG001 | ΔgerPA | This study |
| AG002 | ΔgerPB | This study |
| AG003 | ΔgerPC | This study |
| AG004 | ΔgerPD | This study |
| AG005 | ΔgerPE | This Study |
| AG006 | ΔgerPF | This study |
| AG007 | ΔgerP | This study |
| AM001 | ΔspoIVA | This study |
| AM002 | ΔsafA | This study |
| AM003 | ΔcotE | This study |
| Mutant transformants | ||
| AG008 | ΔgerPA::pHT315-promoter-gerPA MLSr | This study |
| AG009 | ΔgerPB::pHT315-promoter-gerPB MLSr | This study |
| AG010 | ΔgerPC::pHT315-promoter-gerPC MLSr | This study |
| AG011 | ΔgerPD::pHT315-promoter-gerPD MLSr | This study |
| AG012 | ΔgerPE::pHT315-promoter-gerPE MLSr | This study |
| AG013 | ΔgerPF::pHT315-promoter-gerPF MLSr | This study |
| AG014 | ΔgerP::pHT315-promoter-gerP MLSr | This study |
| GerP localization strains | ||
| AG015 | pHT315-promoter-PA-gfp-PB-PC-PD-PE-PF MLSr | This study |
| AG016 | pHT315-promoter-PA-PB-gfp-PC-PD-PE-PF MLSr | This study |
| AG017 | pHT315-promoter-PA-PB-PC-gfp-PD-PE-PF MLSr | This study |
| AG018 | pHT315-promoter-PA-PB-PC-PD-gfp-PE-PF MLSr | This study |
| AG019 | pHT315-promoter-PA- PB-PC-PD-PE-gfp-PF MLSr | This study |
| AG020 | pHT315-promoter-PA-PB-PC-PD-PE–PF-gfp MLSr | This study |
| Dependency studies | ||
| AM004 | ΔspoIVA::pHT315-promoter-gerPA-gfp MLSr | This study |
| AM005 | ΔsafA::pHT315-promoter-gerPA-gfp MLSr | This study |
| AM006 | ΔcotE::pHT315-promoter-gerPA MLSr | This study |
| AG022 | ΔgerPA::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG023 | ΔgerPA::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG024 | ΔgerPA::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG025 | ΔgerPA::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG026 | ΔgerPA::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG027 | ΔgerPA::pHT315-promoter-gerPF-gfp MLSr | This study |
| AG028 | ΔgerPB::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG029 | ΔgerPB::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG030 | ΔgerPB::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG031 | ΔgerPB::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG032 | ΔgerPB::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG033 | ΔgerPB::pHT315-promoter-gerPF-gfp MLSr | This study |
| AG034 | ΔgerPC::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG035 | ΔgerPC::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG036 | ΔgerPC::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG037 | ΔgerPC::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG038 | ΔgerPC::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG039 | ΔgerPC::pHT315-promoter-gerPF-gfp MLSr | This study |
| AG040 | ΔgerPD::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG041 | ΔgerPD::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG042 | ΔgerPD::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG043 | ΔgerPD::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG044 | ΔgerPD::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG045 | ΔgerPD::pHT315-promoter-gerPF-gfp MLSr | This study |
| AG046 | ΔgerPE::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG047 | ΔgerPE::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG048 | ΔgerPE::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG049 | ΔgerPE::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG050 | ΔgerPE::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG051 | ΔgerPE::pHT315-promoter-gerPF-gfp MLSr | This study |
| AG052 | ΔgerPF::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG053 | ΔgerPF::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG054 | ΔgerPF::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG055 | ΔgerPF::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG056 | ΔgerPF::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG057 | ΔgerPF::pHT315-promoter-gerPF-gfp MLSr | This study |
| AG058 | ΔgerP::pHT315-promoter-gerPA-gfp MLSr | This study |
| AG059 | ΔgerP::pHT315-promoter-gerPB-gfp MLSr | This study |
| AG060 | ΔgerP::pHT315-promoter-gerPC-gfp MLSr | This study |
| AG061 | ΔgerP::pHT315-promoter-gerPD-gfp MLSr | This study |
| AG062 | ΔgerP::pHT315-promoter-gerPE-gfp MLSr | This study |
| AG063 | ΔgerP::pHT315-promoter-gerPF-gfp MLSr | This study |
Tcr, tetracycline resistance; MLSr, macrolide-lincosamide-streptogramin-B resistance.
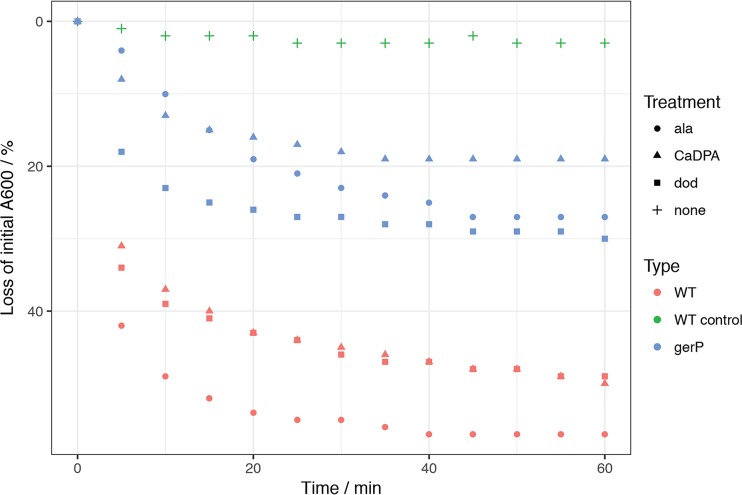
In order to assess whether the deletion of individual or collective gerP genes resulted in defective germination phenotypes in the B. cereus 14579 background, spores of the various null strains were subjected to a heat shock of 70°C for 15 min and cooled on ice, and their germinative responses to l-alanine and inosine were assessed by monitoring the reduction in absorbance associated with the transition of phase-bright dormant spores to phase-dark germinated spores. Spores of all strains, apart from the gerPF null strain, were defective for germination in buffer supplemented with either alanine or inosine with respect to wild-type spores (Fig. 2, S3, and S4). The slight reduction in absorbance recorded after 5 min of germination of mutant spores appeared to be associated with the germination of a small number of spores within the various populations, at least as judged by the light microscope. In contrast, the majority of wild-type spores were phase gray or phase dark after 5 min (data not shown). Even after 60 min, the total loss of absorbance was less than that associated with wild-type spores, with microscopy revealing again the presence of many refractile dormant spores in the mutant populations. In all cases, germination defects were restored by complementation with plasmid-borne copies of the deleted gene(s) (Fig. S4). In contrast to the above-mentioned observations, the gerPF spore germinative responses to alanine and inosine were indistinguishable from those of wild-type spores, indicating that the homologous genes encoded elsewhere on the chromosome can compensate for the loss of gerPF. Additionally, the viability of the various gerP null spores was reduced when incubated overnight on rich solid medium, with colony counts for all mutant spores typically at 20% of those obtained when plating comparable quantities of wild-type spores (data not shown).
FIG 2.
Germination of B. cereus gerP spores in response to nutrient and nonnutrient germinants. Spores at an OD600 of 1 were suspended in buffer supplemented with defined germinants and the resultant changes in absorbance monitored, as described in Materials and Methods. For germinants: circles, 100 mM alanine (ala) (red, wild type; blue, gerP); triangles, 50 mM CaDPA (red, wild type; blue, gerP); squares, 1 mM dodecylamine (dod) (red, wild type; blue, gerP). The + sign represents wild-type spores in buffer without germinant.
Spores can also be stimulated to germinate by various nonphysiological routes, including by exposure to high concentrations of dipicolinic acid chelated with Ca2+ ions (CaDPA) and to the cationic detergent dodecylamine (5). In order to assess the impact of deletion of the various GerP proteins in B. cereus 14579 on germination via these routes, spores of the various strains were prepared and incubated in CaDPA or dodecylamine, and germination was monitored by measuring the absorbance loss. In all cases examined, germination was reduced compared to that observed with wild-type spores, again with the exception of the gerPF spores, which showed essentially wild-type germinative responses (Fig. 2 and S4). Microscopy analyses conducted at the end of each experiment revealed the spore populations to comprise a mixture of predominantly dormant (phase-bright) spores interspersed with lower numbers of germinated (phase-dark) and partially germinated (phase-gray) spores (data not shown). In contrast, wild-type spores were essentially all phase dark. Hence, for both of the nonnutrient germinative pathways examined, a minority of spores can complete germination, whereas most remain dormant.
Localization of GerP proteins.
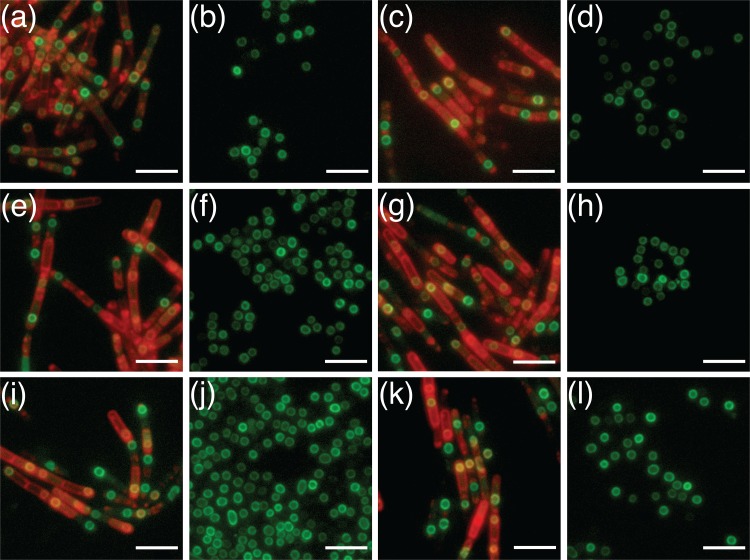
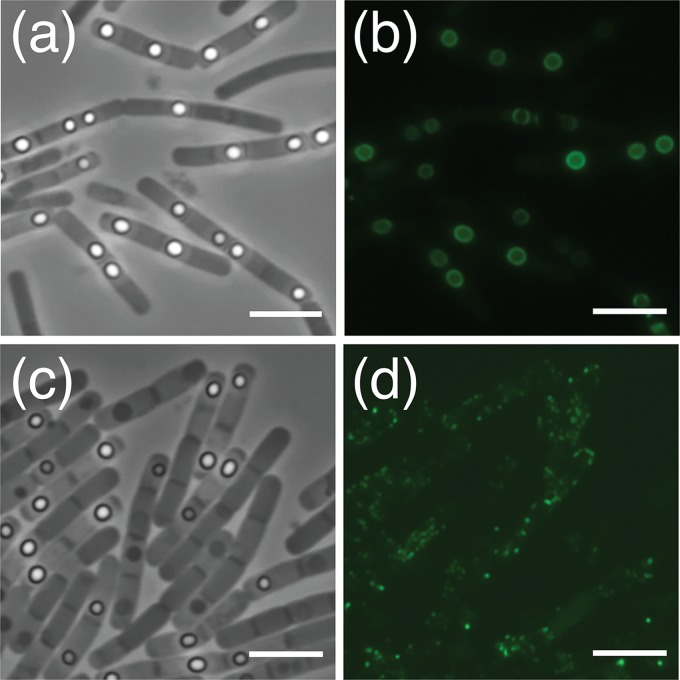
A series of strains designed to individually express each of the respective GerP proteins as C-terminal green fluorescent protein (GFP) fusion proteins were constructed using derivatives of the low-copy-number pHT315 episomal plasmid. The strategy essentially involved cloning the entire gerP operon plus regulatory sequences into pHT315 and then introducing the gfp gene in-frame at the 3′ end of the gene of interest (minus its stop codon). Individual plasmid constructs carrying gerPA-gfp through gerPF-gfp were subsequently introduced to wild-type cells by electroporation (Table 1), meaning that any expressed fusion proteins would be competing for presumed binding sites within the spore with native GerP proteins expressed from the chromosome. Fluorescence microscopy was used to analyze the various gfp-bearing strain cells at intervals throughout sporulation and to examine the final spores in order to observe the pattern of deposition of the various GerP proteins. In all cases, expression was observed first as diffuse fluorescence in the mother cell, followed by a ring of green fluorescence that developed around the still-phase-dark forespore (Fig. 3). In some developing spores, fluorescence was observed as two hemispheres separated by small junctions at either end of the forespore. Fluorescence was retained in mature spores of all strains (Fig. 3), albeit at reduced levels compared to sporulating cells, indicating that the GerP proteins are structural components of the spore rather than being associated purely with the expression or assembly of another component of the spore. Additionally, spores of the various gfp-bearing strains appeared to be free of any germination defects in response to both alanine and inosine (Fig. S4), indicating in all cases that the GFP moiety was not disruptive to function.
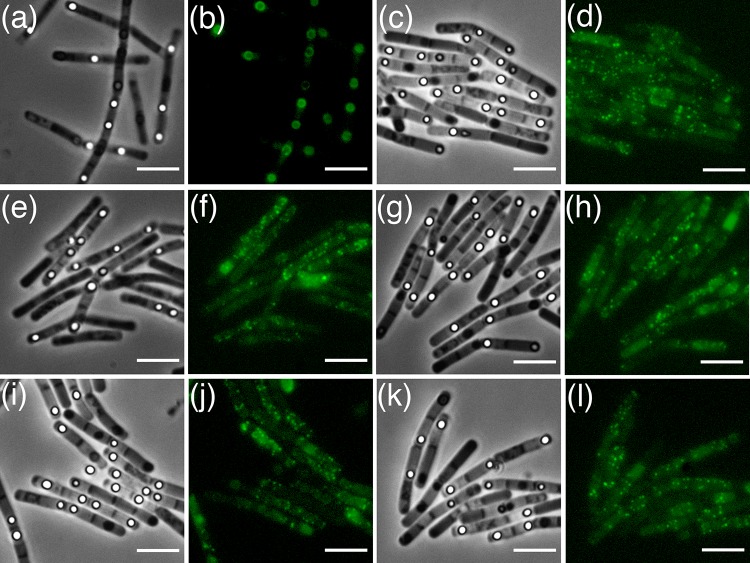
FIG 3.
Fluorescence microscopy of B. cereus 14579 sporulating cells and spores with plasmid-borne copies of gerPA-gfp (a and b), gerPB-gfp (c and d), gerPC-gfp (e and f), gerPD-gfp (g and h), gerPE-gfp (i and j), and gerPF-gfp (k and l). The expression of various genes was controlled by native gerP operon regulatory sequences. Red fluorescence is associated with the lipophilic FM4-64 dye, which was used to visualize cell membranes. Scale bar = 5 μm.
Ellipsoid localization microscopy was then used to more precisely locate the various GerP proteins in B. cereus spores. Information about the location and distribution of proteins in the coat and exosporium of B. cereus is sparse in comparison to that available for B. subtilis spores. However, previous studies have revealed that the BclA protein forms the fiber-like nap on the exterior surface of the exosporium (10, 11), whereas CotD has been identified as a component of the spore coat (12). Accordingly, strains bearing gfp fusions to the 3′ ends of these genes were constructed to serve as benchmarks for the exosporium and coat locations. Strains designed to express C-terminal GFP fusions to SleL and CwlJ were also constructed, since at least in B. subtilis, these cortex-lytic enzymes have been localized to the inner coat (13). Spores of additional strains, including those with bxpB-gfp (BxpB is an exosporium protein [11, 14]), were prepared but failed to show any fluorescence, whereas the irregular distribution of fluorescent foci evident in cotE-gfp spores precluded ELM analysis. CotE is important in the assembly of spore outer coat proteins (13), and it may be that the GFP moiety disrupted its key morphogenetic role in our study.
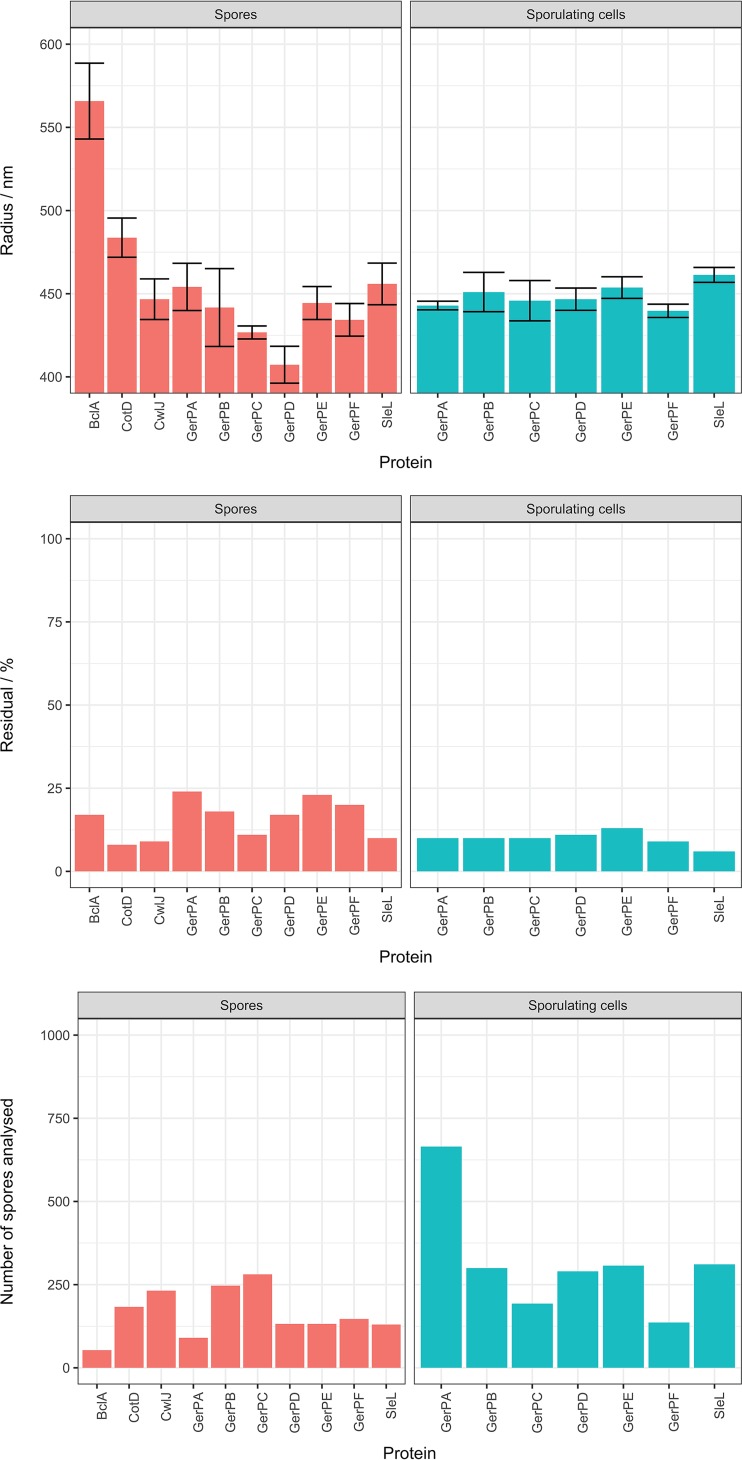
The radial locations of the GerP proteins with respect to the spore center are shown in Fig. 4, together with the numbers of individual spores analyzed and the residual fitting error of the ELM measurements. We infer the radial location of GerPA, for example, as 454 nm ± 14 nm in mature spores, where the ±14 nm is the standard deviation of radial locations found in repeated measurements. The radial locations are as follows: GerPB, 442 nm (±23 nm); GerPC, 427 nm (±4 nm); GerPD, 407 nm (±11 nm); GerPE, 444 nm (±10 nm); and GerPF, 434 nm (±10 nm) (Fig. 4). As expected, the BclA-GFP protein was measured as having the largest equivalent radius at 566 nm (±23 nm), which is consistent with its position on the exterior of the spore. Similarly, ELM-derived measurements indicate that the CotD-GFP protein, with an equivalent radius of 484 nm (±12 nm), is located to the exterior of both SleL-GFP (456 nm ± 12 nm) and CwlJ-GFP (447 nm ± 12 nm), with the two cortex-lytic enzymes (CLEs) apparently occupying similar locations within the spore. We can infer from these data that the GerP proteins are distributed within the inner spore coat, using the CLEs as indicators of this location. However, the apparent spread of the distribution, coupled with relatively high residuals associated with measurements of some of the GerP proteins (stemming from the low brightness of the samples examined), led us to conduct additional measurements with sporulating cells. Brighter fluorescence and improved separation of cells enabled a greater number of individual cells to be included in the analyses, samples for which were drawn from cultures a few hours prior to release of mature spores from mother cells. In this case, the radial locations of all six GerP proteins was more uniform, averaging 445 nm ± 10 nm. The reduced residual values associated with these measurements increase the level of confidence with which we can state that all six GerP proteins reside in a similar location within the inner spore coat. Additionally, none of the GerP-GFP spores cross-reacted with anti-GFP antisera, which is consistent with an inner coat location (data not shown).
FIG 4.
The locations of several GFP-fusion proteins with respect to the spore center were estimated using the ELM image analysis technique. All of the GerP proteins were located near the CLEs SleL and CwlJ, in the inner coat. The number of analyzed spores and the residual error of the model fit are shown and indicate that more accurate estimates were obtained within sporulating cells, just prior to mother cell lysis, than with mature spores, which produced larger residuals attributed to clumping.
GerPA-dependent localization.
Having established that the various GerP proteins appear to localize to broadly the same vicinity within the spore, we then sought to identify whether the localization of any individual GerP protein is dependent upon presumed interactions with other protein(s) encoded within the operon. In the first instance, this was achieved by introducing variant pHT315 plasmids containing the gerP promoter sequence plus an open reading frame (ORF) encoding the GerP protein of interest with a C-terminal GFP fusion to strains bearing markerless chromosomal deletions in single gerP genes. For example, six plasmids encoding GerPA-GFP through GerPF-GFP were introduced individually to the gerPA background strain (AG001), and then to the gerPB background, and so on, creating a total of 36 new strains. Each of these strains was then sporulated by nutrient exhaustion and analyzed at intervals by fluorescence microscopy until mature spores were released. The results of these analyses reveal that in all backgrounds, with the exception of gerPA, the various GerP-GFP fusion proteins are expressed and localize around the developing forespore in a manner reminiscent of that observed for GFP fusion proteins in the wild-type background (Fig. S5). Hence, in the absence of GerPB, for example, all six GFP fusion proteins were observed to localize during sporulation and to persist in the mature spore. The exception to this occurred in the gerPA background, where in the absence of GerPA, none of the GerPB-GFP to GerPF-GFP fusion proteins localized around the developing forespore, although diffuse fluorescence interspersed with bright fluorescent foci was observed in the mother cell in each case (Fig. 5). Similar observations were made when the entire gerP operon, modified with individual ORFs containing in-frame gfp fusions, was expressed in trans from a series of plasmids introduced to the gerP null background, reducing the potential for differences in expression levels between chromosomal and plasmid-borne genes being responsible for the apparent dependency on GerPA for localization of all other GerP proteins. Similarly, when individual gerP genes were introduced on pHT315-derived plasmids to the gerP null background, only the GerPA-GFP protein was observed to localize and to produce fluorescent mature spores, conferring further evidence that this protein is key to the localization of the other GerP proteins (Fig. S6).
FIG 5.
Phase-contrast and fluorescence microscopy of sporulating B. cereus gerPA cells with plasmid-borne copies of gerPA-gfp (a and b), gerPB-gfp (c and d), gerPC-gfp (e and f), gerPD-gfp (g and h), gerPE-gfp (i and j), and gerPF-gfp (k and l). None of the remaining GerP proteins can localize around the developing forespore in the absence of GerPA. Scale bar = 5 μm.
GerP dependence on coat morphogenetic proteins.
A small number of morphogenetic proteins have been identified from genetic and microscopy-based studies that appear to function as interaction hubs for the recruitment and localization of defined subsets of proteins during the assembly of the B. subtilis spore coat (13). The SpoIVA protein, for example, is required for the localization of proteins that comprise the basement layer of the coat (15), whereas SafA and CotE are responsible for localizing the inner and outer coat proteins, respectively (16, 17). Although details of the structural hierarchy in terms of morphogenetic protein dependency are sparse in species other than in B. subtilis, the conserved presence of genes encoding orthologues of these proteins indicates that SpoIVA, SafA, and CotE probably fulfill related functions in most, if not all, Bacillus species (1, 13). With this in mind, and with a view to investigate which, if any, of these proteins are crucial to GerP localization, spoIVA, safA, and cotE null mutant strains were prepared in the B. cereus 14579 background. Plasmid-borne copies of gerPA-gfp were introduced subsequently to each mutant strain and sporulation allowed to proceed by nutrient starvation. Microscopy analyses revealed that the deletion of spoIVA results in early lysis of the developing forespore; hence, this strain was not examined further. In contrast, fluorescence associated with GerPA-GFP was observed in both the safA and cotE backgrounds (Fig. 6). Localization of the protein, visible as a fluorescent ring encircling the developing forespore, was evident only in the cotE background, indicating that GerPA, and presumably by extension all GerP proteins, are SafA-dependent proteins.
FIG 6.
Establishing the localization hierarchy for GerP proteins. Phase-contrast and fluorescence microscopy images of sporulating B. cereus cotE cells (a and b) and safA cells (c and d), both carrying plasmid-borne gerPA-gfp under the control of its native promoter. The GerPA-GFP protein is observed to localize around the developing forespore in the cotE background. The same protein is expressed in the safA strain but fails to localize, indicating that GerPA, and by extension all GerP proteins, are SafA dependent. These observations are consistent with an inner coat location for the GerP proteins. Scale bar = 5 μm.
Measuring the permeability of GerP spores.
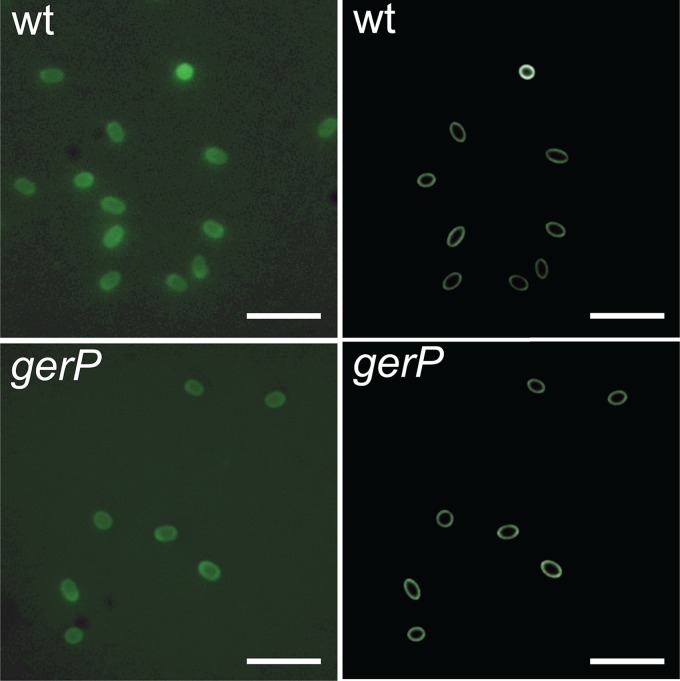
The seemingly hindered passage of germinant molecules through the spore coats of various species to interact with the inner membrane-located receptors provides an indirect indication of a permeability defect in gerP spores. In an attempt to provide a more direct measurement of the permeability of the coats of wild-type and mutant spores, we examined the possibility of using ELM to measure the diffusion of fluorescein isothiocyanate (FITC)-labeled dextrans of differing sizes through the spore coat. Accordingly, both B. cereus and B. subtilis wild-type and gerP spores were incubated in solutions of FITC-labeled dextrans ranging in size from 3 kDa to 70 kDa, before examination by fluorescence microscopy, as described in Materials and Methods. The best results in terms of amenability to ELM analysis were obtained with B. subtilis spores, for which the fluorescence micrographs showed many well-separated spores with bright ring-like images, indicating that an outer portion of the spore had been fluorescently stained (Fig. 7).
FIG 7.
Fluorescence microscopy and superresolved reconstructions of B. subtilis wild-type (wt) spores (top panels) and gerP spores (bottom panels), stained with FITC-dextrans, with an average molecular mass of 10 kDa. The superresolved reconstructions are generated by feeding precise structure parameters from the fluorescence images back into the ellipsoid model for its image, while decreasing the inferred point spread function to remove instrumental blurring (9). Scale bar = 5 μm.
Figure 8 presents the midpoint radial locations inferred by ELM analysis for the fluorescent FITC-dextran stains. For every size of FITC-dextran tested, the midpoint of the stained layer was located about 20 nm closer to the spore center in the wild-type spores than in the gerP mutants. This is consistent with the notion of impaired coat permeability in the gerP mutants that is implied by the germination data; however, the measurement shown in Fig. 8, on its own, would also be consistent with the gerP mutants simply being larger than the wild-type spores, so it is essential to consider this result together with the germination data. Additionally, in both the wild type and the gerP mutants, the smallest dextran molecules (3 to 5 kDa) were located on average about 15 nm closer to the spore center than the heavier dextrans. The larger dextrans (10, 20, and 70 kDa) were located in similar regions to one another.
FIG 8.
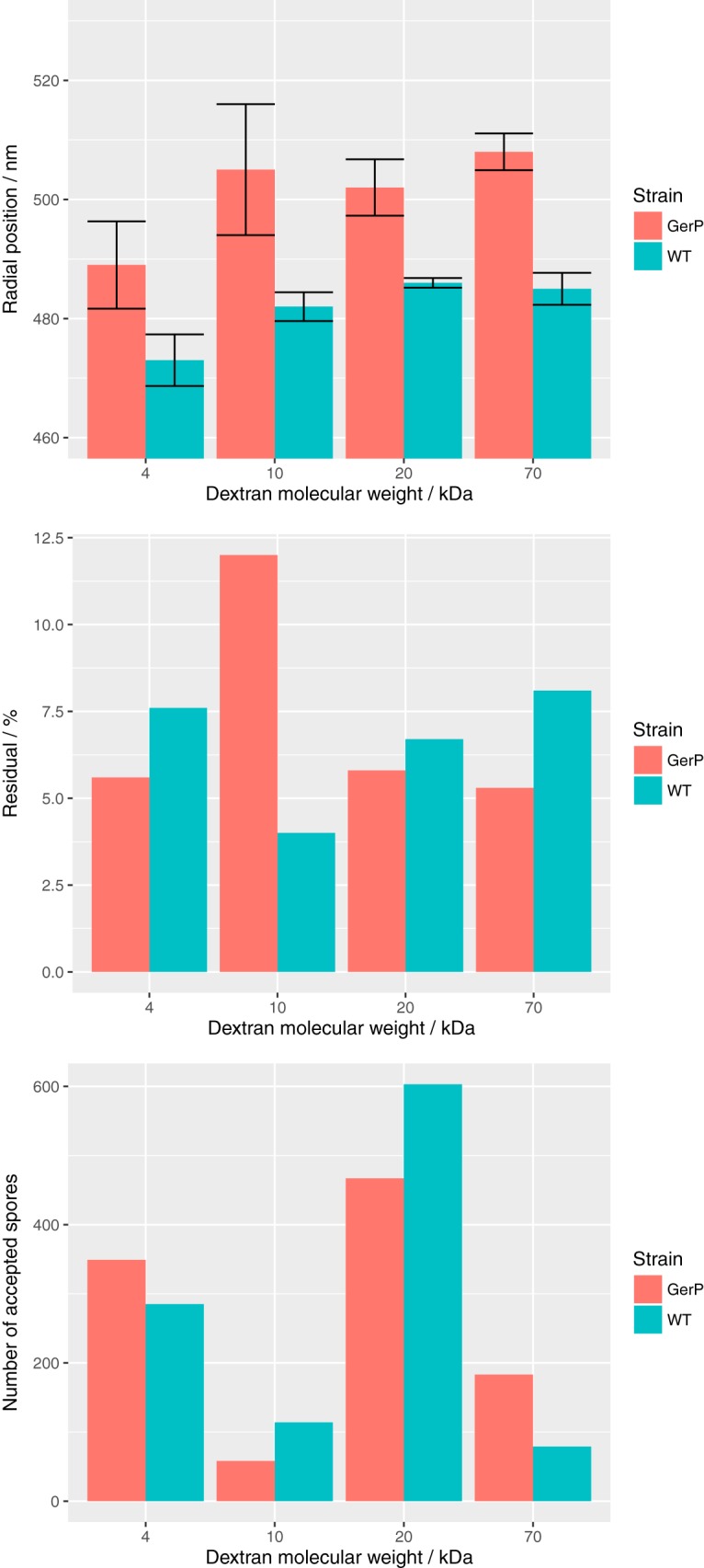
The midpoint radial locations of regions of B. subtilis spores fluorescently stained with FITC-dextran molecules were inferred using the ELM image analysis technique. The number of analyzed spores and average residual error of the model fits are also shown for each case.
DISCUSSION
Bacterial spores of all species are presented with a dilemma in that their protective structures must prevent ingress of potentially damaging molecules or chemicals to the cellular protoplast within while permitting access of small-molecule germinant molecules to the inner membrane-bound germinant receptors. Additionally, they must be capable of rapidly releasing solutes from the spore core, including CaDPA, to the environment upon spore germination. This is achieved, at least in part, by the presence of a multilayered coat and, in some cases, exosporium structures, one function of which is to serve collectively as molecular sieves (1). The sieving properties vary between species, but in general, it seems that moderately sized proteins and other molecules can breach the outermost layers of spores, exemplified by recent work showing transit of the 26-kDa red fluorescent protein through the exosporium of Bacillus megaterium spores and apparently through the outer, but not inner, coat of B. subtilis spores (18, 19). These observations are consistent with those of earlier studies (20–22), which imply that permeability decreases with progression toward the interior of the spore.
The results of the germination experiments conducted in this study with B. cereus 14579 spores that are null for the entire gerP operon, or individual genes therein, are similarly consistent with previous studies and with the hypothesis that the GerP proteins influence the permeability of the spore coat to small hydrophilic molecules. Defective germinative responses were observed, for example, whether induction was via stimulation of the nutrient germinant receptors by alanine or inosine, or in response to exogenous CaDPA or dodecylamine. The result with dodecylamine is in contrast to results observed in B. subtilis, where dodecylamine stimulated a faster germinative response in gerP spores than in wild-type spores (7). Dodecylamine is known to trigger germination by stimulating the opening of inner membrane-located DPA channels that are present in both B. subtilis and B. cereus (5); hence, it is not clear why the deletion of gerP should cause differing germinative responses between species. The exception to the above-mentioned concerns is B. cereus gerPF null spores, which displayed essentially wild-type germinative responses when stimulated via nutrient or nonnutrient pathways. Presumably, the loss of gerPF in this case was compensated by either of the two additional gerPF homologues encoded elsewhere on the chromosome, as demonstrated previously in closely related B. anthracis spores (8).
A primary objective of this study was to ascertain whether the GerP proteins are structural components of the spore or whether they are involved only in spore assembly. Fluorescence microscopy of strains expressing GerP-GFP fusion proteins supports the notion that GerP proteins are indeed located in the spore coat, and the images of smooth fluorescent rings appeared suitable for ELM image analysis using a spherical shell model as an approximation to the GerP-GFP location. We then used ELM to more precisely locate the GerP proteins with respect to some other coat and exosporium proteins. These included the BclA exosporial nap protein, coat-localized CotD, and the CLEs CwlJ and SleL. The radial location of each of these benchmark proteins was established using ELM; BclA, as expected, was outermost, followed by CotD, and then SleL and CwlJ. The SleL and CwlJ proteins occupied a similar radial location around 450 nm, which we identify as the position of the inner coat. Overall, we found that all the GerP proteins were also located in the inner coat. Our analysis of GerP locations in mature spores was, unfortunately, complicated by the fact that the spores tended to clump, and hence, the number of well-separated spores available for analysis was limited. Additionally, even the available images were often poorly fitted due to adjacent fluorescent material. The high residual fitting errors shown in Fig. 4 indicate that the radial locations found for the GerP proteins in mature spores (407 to 454 nm) have limited accuracy and may be biased by image analysis limitations. In order to obtain more accurate estimates of GerP location, additional fluorescence microscopy was conducted with sporulating GerP-GFP cells in which the fluorescent proteins were observed to have completely localized to rings around phase-bright forespores but prior to mother cell lysis. We obtained superior fluorescence brightness with these samples, and much more importantly, we found that this removed many of the difficulties due to spore clumping and enabled a greater number of cells to be analyzed. Subject to the assumption that the GerP protein locations in these specimens are consistent with their locations in mature spores, we believe this method more accurately located the GerP proteins. These results, in which the GerP protein radial locations span a narrower range from 440 nm to 454 nm, indicate that all six proteins remain in the inner coat but now seem more likely to share proximity with CwlJ (447 nm) and SleL (456 nm) within the spore (SleL was also observed to occupy a similar location in both sporulating cells and in mature spores). This is an important distinction, since the location of CLEs in the inner spore coat could pose a mechanistic problem as to how they access the cortical substrate during germination, especially if the GerP proteins form a proteinaceous layer that lies between the CLEs and the cortex/outer membrane boundary. A future objective will be to improve the resolution of these measurements to definitively ascertain the location of CLEs with respect to the cortex.
A further objective of this work was to establish the localization hierarchy for the GerP proteins, both in terms of morphogenetic protein dependency and to identify any dependency between the individual GerP proteins. With regard to morphogenetic protein dependency, fluorescence microscopy analyses conducted with B. cereus cotE and safA spores revealed GerPA, and therefore presumably all GerP proteins, to be SafA dependent. Hence, the SafA protein's role as the interaction hub for inner coat localization in B. subtilis spores appears to be conserved in B. cereus spores, and again, presumably across all Bacillus species. Similar fluorescence microscopy-based analyses conducted with a series of null mutants and appropriate GerP-GFP-expressing strains indicate that only the GerPA protein is required for each of the remaining GerP proteins to localize in the developing spore. We cannot rule out, however, that one or both of the additional GerPF homologues encoded on the chromosome permit localization of GerPA-GerPE in the gerPF null mutant; this is something that will have to be tested in the future. Regardless, we can infer from these analyses that the GerPB, GerPC, GerPD, and GerPE proteins each have an essential role in maintaining the permeability of the coat to permit ingress of germinants to their sites of interaction with receptors at the inner membrane. GerPA is also essential, although whether this goes beyond its requirement for localization of the remaining GerP proteins or whether it has an additional function relating to spore permeability has yet to be established. Unfortunately, preliminary experiments aimed at identifying physical interactions that may occur between the GerP proteins using a bacterial 2-hybrid system were unsuccessful, owing in part to the relative insolubility of at least some of the proteins when expressed in Escherichia coli (data not shown).
Finally, the ELM technique was applied to compare the permeabilities of wild-type and gerP spores, since it offered the prospect of making direct quantitative measurements using fluorescent dextran stains as opposed to inferring permeability defects based on germination kinetics. Measurements with wild-type B. subtilis spores revealed that the smallest FITC-dextran tested (3 to 5 kDa) permeated to a depth that is comparable in location to the CotG protein when measured by the same method (radial locations of 473 nm versus 475 nm [9]), whereas the larger dextrans localized to a position comparable to the location of the CotZ outer crust protein (485 nm versus 480 nm [9]). In contrast, midpoint radial locations for all dextran-stained gerP spores measured outside the location of the wild-type CotZ protein (489 nm for 3- to 5-kDa dextrans and ∼505 nm for the larger dextrans). Since CotZ is located on the surface of B. subtilis spores (3), these data indicate that none of the dextran stains significantly permeated the gerP spores, instead adhering and accumulating on the spore surface. This may be too simplistic a view, however, since the layer order of proteins in wild-type spores may not apply to gerP spores, and indeed, a previous study includes a transmission electron microscopy (TEM) image of a single B. subtilis gerP spore that appears to lack the spore crust (23). It is intriguing, therefore, that despite being located in the inner coat, the GerP proteins appear to exert an influence on the permeability of the coat that extends to the surface of the spore. One possible explanation for this is that absence of GerP proteins in the inner coat results in relatively subtle structural perturbations that ripple through to the layers on top, perhaps even leading to loss of the crust. This hypothesis will be tested in future by establishing the layer order and precise locations of defined coat proteins in gerP spores with respect to those in wild-type spores.
MATERIALS AND METHODS
Bacterial strains and spore preparation.
The Bacillus cereus strains employed in this study (Table 1) were all isogenic with the ATCC 14579 strain, which was a gift from Toril Lindbäck (Norwegian School of Veterinary Science, Norway). Bacillus cereus strains were routinely cultured in LB medium at 30°C, supplemented with 1 μg/ml erythromycin and 25 μg/ml lincomycin where appropriate (Table 1). Competent Escherichia coli DH5α cells (NEB UK) were used for cloning procedures and for the propagation of plasmids. Bacillus cereus spores were prepared by nutrient exhaustion in casein-hydrolysate yeast-extract (CCY) liquid medium (24). Efficient sporulation was achieved using 200-ml cultures in 2-liter baffled flasks that were subject to orbital agitation at 30°C for 48 h. The resultant spores were purified from cellular debris by centrifuging and resuspending the spore pellets initially in phosphate-buffered saline (PBS) supplemented with 0.1% (wt/vol) Tween 20, followed by several rounds of washing and resuspending spores in ice-cold deionized water. Spores of B. subtilis strains were prepared by sporulation at 37°C on 2× Schaeffer's glucose agar plates without antibiotics, as described previously (7). Purified spores of both species were stored as suspensions (optical density at 600 nm [OD600], 50) in water at 4°C.
Mutant construction.
A series of gerP null mutant strains in the B. cereus 14579 background were constructed using a markerless allelic exchange methodology (19, 25). To delete gerPA (BC1145), for example, PCR was used to prepare an amplicon comprising approximately 500 bp of sequence upstream and inclusive of the gerPA start codon. A second amplicon comprising 500 bp of downstream sequence starting from, and including the gerPA stop codon, was also prepared by PCR. Primers were designed to include approximately 15 bp of overlapping sequence between the 3′ end of the upstream amplicon and the 5′ end of the downstream amplicon (sequences of primers used in this work are shown in Tables 2 to 5). Additionally, the 5′ end of the upstream amplicon contained 15 bp of overlapping sequence with the 5′ end of EcoRI- and BamHI-digested pMAD vector (26), as did the 3′ end of the downstream amplicon with the 3′ end of pMAD. A variant pMAD vector containing a single I-SceI restriction site was used in this work (a gift from Toril Lindbäck, Norwegian School of Veterinary Science, Oslo, Norway). The fragments were assembled using a Klenow-based ligation independent cloning technique (https://openwetware.org/wiki/Klenow_Assembly_Method:_Seamless_cloning) and then used to transform E. coli to carbenicillin resistance. Transformant E. coli strains were screened by colony PCR to identify clones with the correct construct, and plasmids were subsequently purified and validated by DNA sequencing. Similar procedures were used to prepare plasmids designed to individually delete the remaining gerP genes (gerPB [BC1144], gerPC [BC1143], gerPD [BC1142], gerPE [BC1141], and gerPF [BC1140]) and to delete the entire operon.
TABLE 2.
Oligonucleotide primers used to create null mutant strains
| Strain mutant type | Primer | Sequence (5′ to 3′) |
|---|---|---|
| gerPA | pMAD gerPA up For | CCATGGTACCCGGGAGCTCGAATTCCTCATACCCTTTAATTGTATTCGCA |
| ATGTAA gerPA up Rev | AAAAATTCAATGCAATTGCCTCCTTTACATACAAAACACCCTTCGTTTTT | |
| ATGTAA gerPA down For | AAAAACGAAGGGTGTTTTGTATGTAAAGGAGGCAATTGCATTGAATTTTT | |
| pMAD gerPA down Rev | GCCTCGCGTCGGGCGATATCGGATCCAAACCAATATTTAAAGTACCATTT | |
| gerPB | pMAD gerPB up For | GCCATGGTACCCGGGAGCTCGAATTCGCGATTGAAAAAAAACGTGAAAAA |
| ATGTAA gerPB up Rev | ATATTTATGACAAGGATGTCTTACATTGCAATTGCCTCCTTTAAGTTGAG | |
| ATGTAA gerPB down For | GCTCAACTTAAAGGAGGCAATTGCAATGTAAGACATCCTTGTCATAAATA | |
| pMAD gerPB down Rev | GCCTCGCGTCGGGCGATATCGGATCCCATCCACCATCATTTGTCGATACA | |
| gerPC | pMAD gerPC up For | GCATGCCATGGTACCCGGGAGCTCGAATTCTGGTCGGACATATTCGTATC |
| ATGTAA gerPC up Rev | CGTTAAGGTTCATACATACTTACATTTGTGAAACCTCCTTTTCAATAGGG | |
| ATGTAA gerPC down For | CCCTATTGAAAAGGAGGTTTCACAAATGTAAGTATGTATGAACCTTAACG | |
| pMAD gerPC down Rev | GCCTCGCGTCGGGCGATATCGGATCCAAAAGTGTGCGCAATTCAACGTTA | |
| gerPD | pMAD gerPD up For | GCATGCCATGGTACCCGGGAGCTCGAATTCAAAATCGCCCCTCCTCTTCT |
| ATGTAA gerPD up Rev | ATAATTTTTCAGTCTCCTTACATACATACTTACTCCTTTCGGAAATTTC | |
| ATGTAA gerPD down For | GAAATTTCCGAAAGGAGTAAGTATGTATGTAAGGAGACTGAAAAATTAT | |
| pMAD gerPD down Rev | CCTCGCGTCGGGCGATATCGGATCCAATTGAAAGAACCGTTACTGTTTTG | |
| gerPE | pMAD gerPE up For | TGCATGCCATGGTACCCGGGAGCTCGAATTCAAAATCGCCCCTCCTCTTC |
| ATGTAA gerPE up Rev | CATAATTTTTCAGTCTCCTTACATACATACTTACTCCTTTCGGAAATTTC | |
| ATGTAA gerPE down For | GAAATTTCCGAAAGGAGTAAGTATGTATGTAAGGAGACTGAAAAATTATG | |
| pMAD gerPE down Rev | GCCTCGCGTCGGGCGATATCGGATCCAATTGAAAGAACCGTTACTGTTTTG | |
| gerPF | pMAD gerPF up For | TGCATGCCATGGTACCCGGGAGCTCGAATTCAACAGTTACAGAGGGTCCA |
| ATGTAA gerPF up Rev | AGAATATCTCCTACTTTTTTTACATCCACCTACTTTACTAAACTTTGTAA | |
| ATGTAA gerPF down For | TTACAAAGTTTAGTAAAGTAGGTGGATGTAAAAAAAGTAGGAGATATTCT | |
| pMAD gerPF down Rev | CCTCGCGTCGGGCGATATCGGATCCAACATCCTGTAAAAGAGAAATGTTT | |
| gerP (operon) | pMAD gerP up For | GCCATGGTACCCGGGAGCTCGAATTCTCTCCATCCTAAAACT |
| ATGTAA gerP up Rev | CTTGTTTATACCACTATTACATTTATTGCGTGTGTGTTTTGAACG | |
| ATGTAA gerP down For | CGTTCAAAACACACACGCAATAAATGTAATAGTGGTATAAACAAGTAAG | |
| pMAD gerP down Rev | GCCTCGCGTCGGGCGATATCGGATCCAGAAAAGCTTACAACATCCTGT | |
| cotE | pMAD cotE up For | CCATGGTACCCGGGAGCTCGAATTCATTTCTGAAACAGAAGAAGTTGATT |
| ATGTAA cotE up Rev | ACTTCTCCCTAGCTTTCTATTACATTCGTAACCCTCCTCAATCACTATTC | |
| ATGTAA cotE down For | TAGAAAGCTAGGGAGAAGTTCTTCC | |
| pMAD cotE down Rev | CCTCGCGTCGGGCGATATCGGATCCAATTGTACTACTTCACGACGTACTA | |
| safA | pMAD safA up For | CCATGGTACCCGGGAGCTCGAATTCATATGATTGAATCAGCGCCACCTGG |
| ATGTAA safA up Rev | CGGGAATACTCCCGCCTTTTTACATATTTTCCCCCTCCTGTATAACTTAT | |
| ATGTAA safA down For | AAATATGTAAAAAGGCGGGAGTATTCCCGCCTTTT | |
| pMAD safA down Rev | CCTCGCGTCGGGCGATATCGGATCCCTGTTTTACCATCATTGTTAATGTA |
TABLE 3.
Oligonucleotide primers used to validate null mutant strains by diagnostic PCR
| Strain mutant type | Primer | Sequence (5′ to 3′) |
|---|---|---|
| gerPA | gerPA Out For | GTCATGTACATACGGATTTACACCCTAATCAAACGTTCAAAACACACACG |
| gerPA Out Rev | GATAGGGCTTTAATACTTCCGGCAGTACCAATTTGGAACACAGAAGAGG | |
| gerPB | gerPB Out For | CCGGCTCTTTCAACGTTGGAGATAATGTTTCTGTCTACAATTATCA |
| gerPB Out Rev | GCTCTTCTTGGAGTTGGCGCACTTGATCTTCTAAATTTAGGATGGCTGC | |
| gerPC | gerPC Out For | GGTACTGCCGGAAGTATTAAAGCCCTATCTAAATTTTCAAATACGGGCGG |
| gerPC Out Rev | CGGACGACGATACACCGTTCATTTTAATCTGTCCGACTTTTAACTCACGG | |
| gerPD | gerPD Out For | GCATCGACTCCCTTATTATTTATCACAAGCGCAATCATATGAAGGTA |
| gerPD Out Rev | CTCCCCGTATATAACAAGGAATTTCACGGTGGACGGCAATAGCTCTAC | |
| gerPE | gerPE Out For | GCCTTTCTCTCCTTTATTCAGCACATACCGGGAAATTTCCGAAAGGAG |
| gerPE Out Rev | GATCTGCCACATCAGAATCAAATGTATTCGTTGCACTAACACCGTTAAAC | |
| gerPF | gerPF Out For | GCGCACACTTTTAAACTCTGGTGGATTTCAAATTGGAAATGTTGATTATG |
| gerPF Out Rev | TAGTGCCTCTTCACTACTTACTTGTTTATACCACTATTTGTGGAC | |
| gerP | gerP Out For | CGGCGCTAATATTTTCACCTCTGTTAACGGATAATACGCCGCTTCCCCG |
| gerP Out Rev | GATGCTTGCAGCAGAAGAAGCATATCAAGATTGGCAAGGGAAGAGTG |
TABLE 4.
Oligonucleotide primers used to create GFP fusion strains
| Primer | Sequence (5′ to 3′) | Overlap |
|---|---|---|
| gfp For | AGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCCAATTCTTGTTGAATT | |
| gfp Rev | TTATTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCTGTTACAA | |
| pHT315 Prom gerPA For | CAGCTATGACCATGATTACGCCAAGCTTTCATTTGGCATAAAATGTAG | pHT315 |
| pHT315 gerPF Rev | GTTGTAAAACGACGGCCAGTGAATTCTTACGCTGTTCCAATCTGATCTTG | pHT315 |
| gfp gerPA Rev | CCAGTGAAAAGTTCTTCTCCTTTACTAGTTGAGCCAATTATCGCTTGATC | gfp |
| gfp gerPB For | GCATGGATGAACTATACAAATAAAGGAGGCAATTGCATTGAATTTTTA | gfp |
| gfp gerPB Rev | GTGAAAAGTTCTTCTCCTTTACTAGAAGAACTAGTTGATGGTTTGATAG | gfp |
| gfp gerPC For | GGCATGGATGAACTATACAAATAAGACATCCTTGTCATAAATATATAGCA | gfp |
| gfp gerPC Rev | CCAGTGAAAAGTTCTTCTCCTTTACTCTCCTTTCGGAAATTTCCCGGTAT | gfp |
| gfp gerPD For | GGCATGGATGAACTATACAAATAAGTATGTATGAACCTTAACGTTGTAAA | gfp |
| gfp gerPD Rev | CTCCAGTGAAAAGTTCTTCTCCTTTACTACCTGGAGTAGGAGGGACATCT | gfp |
| gfp gerPE For | GGCATGGATGAACTATACAAATAAGGAGACTGAAAAATTATGTTGCATCA | gfp |
| gfp gerPE Rev | CCAGTGAAAAGTTCTTCTCCTTTACTTTGTGCGGAAGGTTCATCTGTAAT | gfp |
| gfp gerPF For | CATGGCATGGATGAACTATACAAATAATTCATATAGTAATTACAAAGTTT | gfp |
| gfp gerPF Rev | GTGAAAAGTTCTTCTCCTTTACTCGCTGTTCCAATCTGATCTTGATCTG | gfp |
| pHT315 gfp Rev | GTTGTAAAACGACGGCCAGTGAATTCTTATTTGTATAGTTCATCCATGCC | gfp |
TABLE 5.
Oligonucleotide primers used to create gfp fusions to defined spore coat proteins
| Strain genotype | Primer | Sequence (5′ to 3′) |
|---|---|---|
| bclA-gfp | pHT315 bclA For | GCTATGACCATGATTACGCCAAGCTTCTTCCAAATCAATCATATGTTATA |
| bclA gfp Rev | CAGTGAAAAGTTCTTCTCCTTTACTAGCGATTTTTTCAATAATAATAGAT | |
| bclA gfp For | ATCTATTATTATTGAAAAAATCGCTAGTAAAGGAGAAGAACTTTTCACTG | |
| pHT315 gfp Rev | GTTGTAAAACGACGGCCAGTGAATTCTTATTTGTATAGTTCATCCATGCC | |
| cotD-gfp | pHT315 cotD For | GCTATGACCATGATTACGCCAAGCTTTGCCACCACCATCTCCGTATGC |
| cotD gfp Rev | CCAGTGAAAAGTTCTTCTCCTTTACTCTTTTTAAATATTCCACCAACG | |
| cotD gfp For | CGTTGGTGGAATATTTAAAAAGAGTAAAGGAGAAGAACTTTTCACTGG | |
| sleL-gfp | pHT315 sleL For | CTATGACCATGATTACGCCAAGCTTCATACCACTCAGGATATAACTTTTT |
| sleL gfp Rev | CAGTGAAAAGTTCTTCTCCTTTACTGCCCTTTTTCGTAATCGTAAAGTTT | |
| sleL gfp For | AAACTTTACGATTACGAAAAAGGGCAGTAAAGGAGAAGAACTTTTCACTG | |
| cwlJ-gfp | pHT315 cwlJ For | GCTATGACCATGATTACGCCAAGCTTAGATACCAAAGTAGGCTCAATTAC |
| gfp cwlJ Down | CCAGTGAAAAGTTCTTCTCCTTTACTATATACGCTAGGGCAGTCTTCGCC | |
| cwlJ gfp Up | GGCGAAGACTGCCCTAGCGTATATAGTAAAGGAGAAGAACTTTTCACTGG | |
| cwlJ gfp Down | TCCCTCCTCATTTTCATCTCTTATTTGTATAGTTCATCCATGCC | |
| gfp gerQ Up | GGCATGGATGAACTATACAAATAAGAGATGAAAATGAGGAGGGA | |
| pHT315 gerQ down | CGACGTTGTAAAACGACGGCCAGTGAATTCTTATGGTCTTGGAGTATAAG |
pMAD-derived plasmids were subsequently introduced to B. cereus by electroporation, with transformants being selected at 30°C on LB plates supplemented with 1 μg/ml erythromycin, 5 μg/ml lincomycin, and 90 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Electroporation was conducted using a Gene Pulsar instrument (Bio-Rad), operating at 200 Ω, 2 kV, and 25 μF, and cuvettes that contained 500 ng of plasmid DNA plus 50 μl of thawed electrocompetent cells. Clones that had integrated plasmids by homologous recombination were selected by incubating blue colonies at 37°C on fresh LB plates and restreaking after 24 and 48 h. Plasmid pBKJ223, which encodes the I-SceI restriction enzyme, was introduced by electroporation to single-crossover cells, which were recovered on LB medium containing 10 μg/ml tetracycline. Transformant colonies were subsequently passaged every 24 h in fresh LB medium containing tetracycline at 37°C, with aliquots being plated and screened on LB agar containing tetracycline and X-Gal for white colonies that were sensitive to erythromycin and lincomycin. Candidate colonies that had undergone a second recombination event to excise the integrated plasmid, leaving behind the truncated gene, were validated by PCR and sequencing and then passaged on LB medium minus antibiotics to promote excision of the pBKJ223 plasmid. The same markerless deletion strategy was used to create B. cereus 14579-derived strains that were null for the coat morphogenetic proteins SpoIVA (BC1509), SafA (BC4420), and CotE (BC3770).
The low-copy-number episomal plasmid pHT315 (27) was used as the basis to construct a series of GerP-GFP fusion strains. Two different strategies were employed. First, overlap PCR was used to construct six variant gerP operons, each of which contained the native promoter sequence followed by gerPA through gerPF. In each variant plasmid, a single gerP gene, minus the stop codon, was fused at the 3′ end with an amplicon encoding the gene for GFP. Fragments containing the gerP promoter sequence, variant gerP operons, and linearized pHT315 were assembled using the Klenow assembly technique and used to transform E. coli to carbenicillin resistance. A similar approach was used to construct pHT315-derived plasmids containing the gerP promoter sequence and single gerP genes fused at the 3′ end with gfp. Constructs containing gfp fusions to bclA (BC1207), cotD (BC1560), sleL (BC3607), and cwlJ (BC5390), each under the control of their native promoter sequences, were prepared similarly. In all cases, the resultant plasmids were purified from positive E. coli clones, validated for the intended construction by DNA sequencing, and introduced to B. cereus wild-type and gerP null strains by electroporation.
Spore germination assays.
Spore germination assays were conducted using spores that were synchronized to germinate by incubating at 70°C for 15 min and then cooling on ice. Spores (300 μl) were resuspended at an optical density at 600 nm (OD600) of 1 (approximately 108 spores · ml−1) in 10 mM Tris-HCl buffer (pH 7.4), supplemented with 10 to 100 mM l-alanine or inosine, and the resultant changes in absorbance were monitored at 30°C using a PerkinElmer EnVision Xcite multilabel plate reader fitted with a 600-nm photometric filter. Germination experiments conducted with nonphysiological germinants were conducted by resuspending spores at an OD600 of 1 in 2.5 mM Tris-HCl (pH 7.4) supplemented with 50 mM CaDPA at 30°C, or in 25 mM KPO4 (pH 7.5), supplemented with 1 mM dodecylamine at 45°C. Changes in absorbance of the spore suspensions were monitored as described above. The presented data are from single experiments, which are representative of multiple analyses conducted with at least two batches of spores. Spore viability was assessed by plating serially diluted spore suspensions on LB agar plates, followed by colony enumeration after 24 h of incubation at 30°C.
Microscopy.
Three microliters of spore or cellular suspensions was dispersed onto poly-l-lysine-coated microscope slides and sealed under a coverslip. The samples were imaged on an Olympus BX53 microscope with a 100× 1.30 numerical aperture (NA) oil objective lens, illumination from a mercury lamp, and filters for GFP and red fluorescence (i.e., FM4-64 dye was added to some samples to enable visualization of membranes). Images were captured with a Retiga-2000R charge-coupled-device (CCD) camera, giving a pixel width of 74 nm on the specimen, and 12-bit gray levels. The image data were recorded as 1,600 by 1,200 pixel Tiffs.
Immunolabeling of spores.
Spores (1 ml; OD600, 10) were incubated with gentle agitation at room temperature in phosphate-buffered saline (PBS) containing 2% (wt/vol) bovine serum albumin (BSA) for 1 h. Spores were harvested by centrifugation (13,500 × g for 1 min), resuspended in 400 μl PBS-BSA, and incubated with 100 μl of 500-fold diluted anti-GFP antibody (ab290; Abcam, Cambridge, UK) for 30 min, followed by three washes in PBS-BSA. Resuspended spores (400 μl) were then incubated with 100 μl of 500-fold diluted Dylight594-conjugated anti-rabbit IgG antibody (ab96885; Abcam) for 30 min. Antibody-labeled spores were washed three times with PBS-BSA and then analyzed by fluorescence microscopy.
Dextran permeability experiments.
A series of fluorescein isothiocyanate (FITC)-labeled dextrans (Sigma-Aldrich, Dorset, UK), with average molecular masses of 3 to 5 kDa, 10 kDa, 20 kDa, and 70 kDa, were used to investigate the permeabilities of B. subtilis wild-type and gerP spores. Essentially, 50 μl of the respective FITC-dextran solutions (25 mg/ml) were added to the same volume of spores (OD600, 10), which were then incubated for 24 h at 4°C. In order to reduce background fluorescence, the dextran-spore suspensions were pelleted by centrifugation (13,500 × g for 1 min), enabling removal of the unbound dextran-containing supernatant, before resuspending the spores in 1 ml of sterile deionized (DI) water. The spores were centrifuged again and resuspended in 100 μl of water, and then 3 μl of the suspension were transferred to poly-l-lysine-coated microscope slides for imaging by fluorescence microscopy.
Ellipsoid localization microscopy.
The quantitative fluorescence ELM technique was used to measure the location of GFP fusion proteins in mature spores and in sporulating cells, as well as to measure the extent to which fluorescently labeled dextran molecules had permeated spores. The ELM analysis was previously reported in detail (9). Briefly, several independent fields of GFP- or FITC-dextran-labeled spores or sporulating cells were imaged with fluorescence microscopy. Each field contained about 50 spores or cells on a dark background. Automated image segmentation was used to identify single spores, and the image of each candidate was used to fit the parameters of a model that describes the image of a spheroidal fluorescent shell. For B. cereus, an equation describing the image of a spherical fluorescent layer was fitted to the image data; for B. subtilis, a model for an ellipsoidal shell was fitted because this spore cannot be well approximated by a sphere. A filter was applied to exclude fits from overlapping spores and fragments of debris. The average radius parameter fitted to the spores provides an estimate of the midpoint radial position of the GFP fusion or FITC-dextran layer with respect to the spore center. (The equivalent radius of a sphere of equal volume is reported for FITC-dextran layers that were analyzed with the ellipsoid model.) The mean and standard deviation of the estimates from several fields of spores or cells with each protein are presented in Fig. 4 and 8. Sample data and ELM software are provided in supporting data (https://doi.org/10.17863/CAM.23124).
Supplementary Material
ACKNOWLEDGMENTS
We declare no competing financial interests.
A.G. was the recipient of a Cambridge Nehru Scholarship and received support from the Raymond and Beverly Sackler Foundation. We gratefully acknowledge support from MedImmune through the Beacon collaboration, and from the Engineering and Physical Sciences Research Council Centre for Doctoral Training in Sensor Technologies and Applications (grant EP/L015889/1).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00760-18.
REFERENCES
- 1.Henriques AO, Moran CP Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 2.Stewart GC. 2015. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol Mol Biol Rev 79:437–457. doi: 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popham DL, Bernhards CB. 2015. Spore peptidoglycan. Microbiol Spectr 3:157–177. doi: 10.1128/microbiolspec.TBS-0005-2012. [DOI] [PubMed] [Google Scholar]
- 5.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behravan J, Chirakkal H, Masson A, Moir A. 2000. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J Bacteriol 182:1987–1994. doi: 10.1128/JB.182.7.1987-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butzin XY, Troiano AJ, Coleman WH, Griffiths KK, Doona CJ, Feeherry FE, Wang G, Li YQ, Setlow P. 2012. Analysis of the effects of a gerP mutation on the germination of spores of Bacillus subtilis. J Bacteriol 194:5749–5758. doi: 10.1128/JB.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr KA, Janes BK, Hanna PC. 2010. Role of the gerP operon in germination and outgrowth of Bacillus anthracis spores. PLoS One 5:e9128. doi: 10.1371/journal.pone.0009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manetsberger J, Manton JD, Erdelyi MJ, Lin H, Rees D, Christie G, Rees EJ. 2015. Ellipsoid localization microscopy infers the size and order of protein layers in Bacillus spore coats. Biophys J 109:2058–2066. doi: 10.1016/j.bpj.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sylvestre P, Couture-Tosi E, Mock M. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol 45:169–178. doi: 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- 11.Todd S, Moir A, Johnson M, Moir A. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J Bacteriol 185:3373–3378. doi: 10.1128/JB.185.11.3373-3378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eijlander RT, Kuipers OP. 2013. Live-cell imaging tool optimization to study gene expression levels and dynamics in single cells of Bacillus cereus. Appl Environ Microbiol 79:5643–5651. doi: 10.1128/AEM.01347-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driks A, Eichenberger P. 2016. The spore coat. Microbiol Spectr 4:179–200. doi: 10.1128/microbiolspec.TBS-0023-2016. [DOI] [PubMed] [Google Scholar]
- 14.Steichen CT, Kearney JF, Turnbough CL Jr. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J Bacteriol 187:5868–5876. doi: 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramamurthi KS, Losick R. 2008. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell 31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little S, Driks A. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol Microbiol 42:1107–1120. doi: 10.1046/j.1365-2958.2001.02708.x. [DOI] [PubMed] [Google Scholar]
- 17.Ozin AJ, Henriques AO, Yi H, Moran CP Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J Bacteriol 182:1828–1833. doi: 10.1128/JB.182.7.1828-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donadio G, Lanzilli M, Sirec T, Ricca E, Isticato R. 2016. Localization of a red fluorescence protein adsorbed on wild type and mutant spores of Bacillus subtilis. Microb Cell Fact 15:153. doi: 10.1186/s12934-016-0551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindbäck T, Mols M, Basset C, Granum PE, Kuipers OP, Kovacs AT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ Microbiol 14:2233–2246. doi: 10.1111/j.1462-2920.2012.02766.x. [DOI] [PubMed] [Google Scholar]
- 20.Gerhardt P, Black SH. 1961. Permeability of bacterial spores. II. Molecular variables affecting solute permeation. J Bacteriol 82:750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshikawa T, Beaman TC, Pankratz HS, Nakashio S, Corner TR, Gerhardt P. 1984. Resistance, germination, and permeability correlates of Bacillus megaterium spores successively divested of integument layers. J Bacteriol 159:624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishihara T, Takubo Y, Kawamata E, Koshikawa T, Ogaki J, Kondo M. 1989. Role of outer coat in resistance of Bacillus megaterium spore. J Biochem 106:270–273. doi: 10.1093/oxfordjournals.jbchem.a122843. [DOI] [PubMed] [Google Scholar]
- 23.Nagler K, Setlow P, Reineke K, Driks A, Moeller R. 2015. Involvement of coat proteins in Bacillus subtilis spore germination in high-salinity environments. Appl Environ Microbiol 81:6725–6735. doi: 10.1128/AEM.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Zhou K, Wisnivesky F, Wilson DI, Christie G. 2017. Effects of culture conditions on the size, morphology and wet density of spores of Bacillus cereus 569 and Bacillus megaterium QM B1551. Lett Appl Microbiol 65:50–56. doi: 10.1111/lam.12745. [DOI] [PubMed] [Google Scholar]
- 25.Janes BK, Stibitz S. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119. doi: 10.1016/0378-1119(91)90495-W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.