Traditional fecal indicator bacteria do not distinguish animal from human fecal pollution, which is necessary to evaluate health risks and mitigate pollution sources. Assessing water in urban areas is challenging, since the water can be impacted by sewage, which has a high likelihood of carrying human pathogens, as well as pet and urban wildlife waste. We demonstrate that the Lachno3 and Lachno12 markers are human associated and highly specific for the detection of human fecal pollution from urban sources, offering reliable identification of fecal pollution sources in urban waters.
KEYWORDS: microbial source tracking, 16S rRNA gene, Lachnospiraceae, fecal pollution, urban water, next-generation sequencing
ABSTRACT
The human microbiome contains many organisms that could potentially be used as indicators of human fecal pollution. Here we report the development of two novel human-associated genetic marker assays that target organisms within the family Lachnospiraceae. Next-generation sequencing of the V6 region of the 16S rRNA gene from sewage and animal stool samples identified 40 human-associated marker candidates with a robust signal in sewage and low or no occurrence in samples from nonhuman hosts. Two were chosen for quantitative PCR (qPCR) assay development using longer sequences (the V2 to V9 regions) generated from clone libraries. Validation of these assays with these markers, designated Lachno3 and Lachno12, was performed using fecal samples (n = 55) from cat, dog, pig, cow, deer, and gull sources, and the results were compared with those of established host-associated assays (the Lachno2 marker and two human Bacteroides markers, the HB and HF183/BacR287). Each of the established assays cross-reacted with samples from at least one other animal species, including animals common in urban areas. The Lachno3 and Lachno12 markers were primarily human associated; however, the Lachno12 marker demonstrated low levels of cross-reactivity with samples from select cows and nonspecific amplification with samples from pigs. This limitation may not be problematic when testing urban waters. These novel markers resolved ambiguous results from previous investigations of stormwater-impacted waters, demonstrating their utility. The complexity of the microbiome in humans and animals suggests that no single organism is strictly specific to humans, and the use of multiple complementary markers in combination will provide the highest resolution and specificity for assessing fecal pollution sources.
IMPORTANCE Traditional fecal indicator bacteria do not distinguish animal from human fecal pollution, which is necessary to evaluate health risks and mitigate pollution sources. Assessing water in urban areas is challenging, since the water can be impacted by sewage, which has a high likelihood of carrying human pathogens, as well as pet and urban wildlife waste. We demonstrate that the Lachno3 and Lachno12 markers are human associated and highly specific for the detection of human fecal pollution from urban sources, offering reliable identification of fecal pollution sources in urban waters.
INTRODUCTION
Human fecal pollution enters urban waters through many pathways, such as combined sewage overflows (CSOs), sanitary sewage overflows (SSOs), illicit connections, or failing sanitary sewers that infiltrate systems (1–3). Pathogenic microorganisms from fecal pollution, including bacteria, viruses, and protozoans, pose a risk of waterborne disease for those exposed to the polluted surface waters (4–6). Traditional fecal indicator bacteria (FIB), such as fecal coliforms, Escherichia coli, and enterococci, have historically been used to assess water quality because of the high abundance of microbes in sewage and feces (7, 8); however, some studies have failed to establish direct correlations between FIB levels and pathogens or human health outcomes (9–12). This is likely due to the presence of animal fecal pollution in runoff and non-point sources that contribute to FIB levels without introducing pathogens. Since FIB are common in all warm-blooded animal intestines and do not distinguish human source from animal source fecal pollution (8, 12), there is a need to develop alternative fecal indicators to assess water quality in complex environments where multiple fecal pollution sources contribute to water quality.
Molecular-based methods have demonstrated that fecal anaerobes are useful for tracking fecal pollution sources because they are abundant in the human intestinal tract and specifically associated with one type of host (8, 13, 14). The next-generation sequencing (NGS) technology, which has the ability to yield as many as 1 × 106 sequence reads per sample, provides the opportunity to gain an in-depth inventory of the microbial communities present in a sample, making it possible for similar or identical sequencing reads to be mapped to hosts even if they are present at a low abundance (13, 15, 16). Fecal anaerobes within the Bacteroidales, in particular, members of the genera Bacteroides and Prevotella, have been well studied and successfully used for fecal pollution identification (17–19). However, a large portion of the human microbiome, including members of Clostridiales, which can comprise more than half of the fecal communities, remains untapped for indicator development (20). Additional indicators may be particularly useful in cases where Bacteroidales markers are not abundant in human hosts within particular geographical regions due to diet, culture, or other environmental influences (14, 21–23). In addition, previously reported assays for human-associated Bacteroides have been demonstrated to cross-react with samples from other sources, including cat, dog, chicken, turkey, and raccoon (24–27). Markers from a different bacterial group could add a layer of verification to source tracking studies that are being used to guide mitigation efforts, which are often expensive to implement and require strong stakeholder support.
Previous studies which used the NGS technology to create an inventory of potential new indicators found that about 97% of human fecal community oligotypes were present in sewage samples, with the most abundant ones being matched in these two sources (28), thus demonstrating that the microbial community composition found in sewage comprehensively represents the microbial community composition found in human feces. Members of the family Lachnospiraceae within the order Clostridiales are promising candidates as host-associated genetic markers because of their abundance and high degree of diversity in sewage (29). It has been shown that the host specificity of the family Lachnospiraceae surpasses that of the Ruminococcaceae and Clostridiaceae, two other bacterial families abundant in human feces, when evaluating human-associated markers (30). In particular, members of the genus Blautia within the family Lachnospiraceae demonstrate clear patterns of specificity or preference among sewage and human and animal hosts (13, 22, 31). The human-associated Lachnospiraceae genetic marker Lachno2 (2) was identified on the basis of its presence in sewage but not cows, although the Lachno2 V6 region marker sequence was subsequently found to be present in cats and dogs (13). Despite the noted cross-reactivity, in sewage-contaminated water, the results of the Lachno2 assay and the human Bacteroides (HB) assay, which is modified from the BacHum-UCD marker assay (32) by replacing the forward primer with the HF183 marker (33), are strongly correlated and provide improved confidence for sewage detection (3, 33).
In this study, we examined the population structure of the family Lachnospiraceae in sewage and animal hosts using nearly full-length sequences of the 16S rRNA gene and identified new human-associated Lachnospiraceae genetic markers (i.e., markers preferred for the human host and found only sporadically in other animals). Two novel genetic markers from the family Lachnospiraceae were chosen for quantitative PCR (qPCR) assay development. Assays with these markers, designated Lachno3 and Lachno12, were then validated for their host specificity with stool samples (n = 55) from six animal host species from multiple locations. The results were compared with those of the established human-associated assays with the markers Lachno2, HB, and HF183/BacR287 (2, 26, 33). To demonstrate the applicability of the Lachno3 and Lachno12 genetic markers to improve discrimination of human and animal fecal pollution in an urban area, we further tested environmental samples derived from non-point source pollution with these new assays, as well as the previously described assays with the Lachno2, HB, HF183/BacR287, and DogBact markers (34).
RESULTS
Population structure of Lachnospiraceae in human and animal hosts.
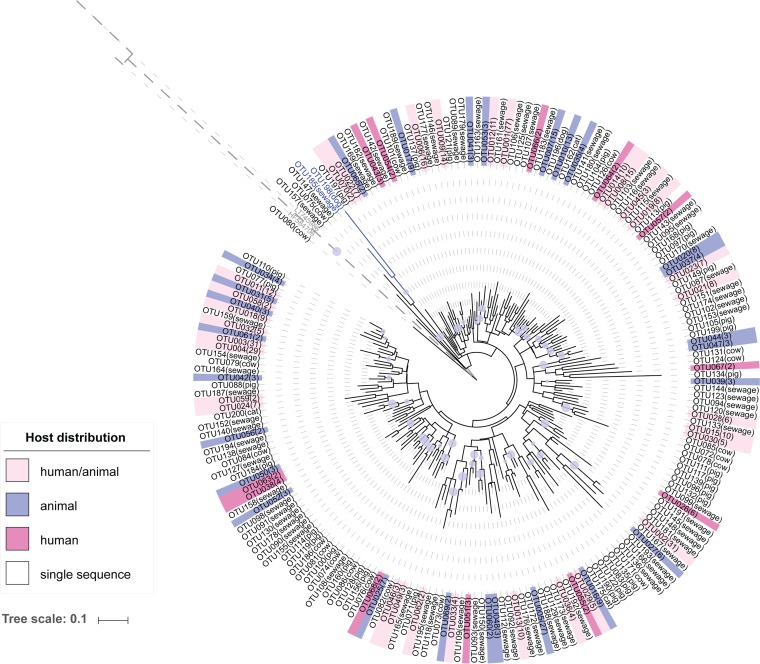
We examined 718 sequences in libraries of Clostridium coccoides sequences from sewage, cat, dog, and pig, as well as previously published Lachnospiraceae sequences from a nearly full-length library comprised of sequences from cows (35). In total, there were 200 operational taxonomic units (OTUs) defined by 97% similarity, of which 70 OTUs contained more than one sequence. A phylogenetic tree was constructed using representative OTU sequences annotated with host information (Fig. 1), which demonstrated that phylogenetically related Lachnospiraceae OTUs did not correspond to the host source. Two OTUs (OTUs 185 and 198) were classified as the family Defluviitaleaceae, which was included in the family Lachnospiraceae in an earlier version of the reference taxonomy, and select members were able to be amplified with the C. coccoides-specific primer. Overall, 31 out of the 70 OTUs with multiple sequences contained sequences from both animals and sewage, suggesting that assessment with the 97% sequence similarity criterion does not resolve these organisms into host groups. A heat map of this distribution is shown in Fig. S1 in the supplemental material.
FIG 1.
Phylogenetic tree comprised of 200 representative OTU sequences from Lachnospiraceae clone libraries. The color range represents OTU host types (i.e., human only, animal only, or human/animal). The number of sequences found in each OTU is in parentheses. The family Defluviitaleaceae clade is in blue. The E. coli outgroup's clade is shown in gray with dashed lines. Bootstrap values larger than 0.7 are indicated by lavender-colored circles, and the values are directly proportional to the circle sizes.
Comparison of host sources using V6 marker regions.
We examined the host distribution patterns of the Lachnospiraceae using the exact sequences of the V6 region of the 16S rRNA gene as markers for organisms uniquely found in a host source. The NGS data set was more extensive than the clone libraries, with 198 samples, 10 host types, and 100,242 unique V6 region sequences (excluding singletons) being recovered. Using exact sequences rather than OTUs, host-associated patterns emerged, with the numbers of highly abundant sequences specifically associated with one host and rarely present in other hosts being determined (Data Set S3).
We used permutation tests with the R package indicspecies with the V6 NGS data set to identify candidates for human-associated markers. In total, 88 indicators with both a sensitivity and a specificity of greater than 90% and P values of 0.001 were selected; 7 out of the 88 indicators were identified with 100% specificity and sensitivity (Fig. S2). The final list of Lachnospiraceae V6 marker candidates in the data set that met our criteria contained 40 candidates, including 10 strictly human-associated candidates (i.e., candidates exclusively found in sewage) and 30 that were human preferred (i.e., candidates that presented at a low relative abundance in one or two other hosts) (Fig. S3).
Comparison of V4V5 and V6 16S rRNA gene regions as markers for human-associated indicators.
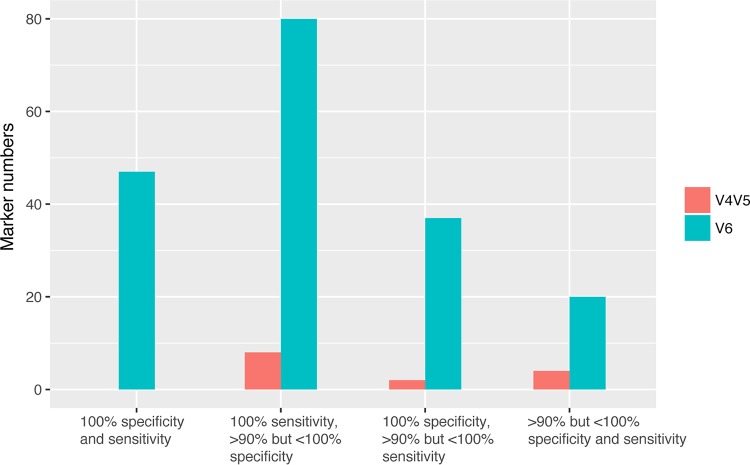
We analyzed a subset of samples that were sequenced for both the V4V5 and V6 regions to determine the region that is more useful for the identification of markers for organisms found in sewage (i.e., human fecal pollution sources). Fifty-two animal samples and 16 sewage samples were utilized for the permutation tests with indicspecies. The results demonstrated that the V6 region had more human markers with a specificity and a sensitivity of over 90% (n = 193) than the V4V5 region (n = 22), and the V6 region showed 49 indicators with a 100% specificity and sensitivity, while the V4V5 region showed none (Fig. 2). In this analysis, a larger number of specific indicators was identified for the V6 region because fewer sequences from animals were included in the data set. Overall, these results suggest that the V6 region is more ideal as a marker region for niche-associated organisms and potentially an ideal target region for Lachnospiraceae assays to discriminate sources of fecal pollution.
FIG 2.
Comparison of results of analysis with the indicspecies program for Lachnospiraceae marker candidates with 90% specificity and sensitivity for the V4V5 and V6 regions.
Continuity of the V4V5 region in closely related organisms with specific V6 markers.
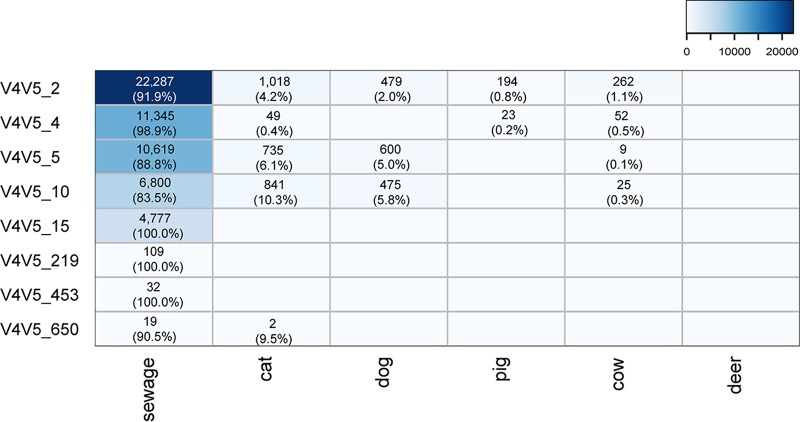
We chose two candidates to be explored further and with which to develop qPCR assays. The clone libraries allowed us to identify the upstream V4V5 region sequences that matched the V4V5 regions in the organisms represented by the Lachno3 and Lachno12 markers. We were then able to use our NGS data set of the much more deeply sequenced V4V5 region to examine if these matching V4V5 regions were unique to sewage samples or if they were also found in animals. Among the clones that contained the Lachno3 marker (n = 79), 18 were matched with the V4V5 NGS data set with eight unique sequence types (Fig. 3). All of the V4V5 types showed dominance in sewage, but several V4V5 types were also found at a lower abundance in animals, suggesting that the V4V5 marker region of the organisms represented by Lachno3 is not as specific as the V6 region. For the Lachno12-related V6 marker sequence, only one V4V5 type (i.e., V4V5_15) which was exclusive to sewage was found. However, it is possible that there are other Lachno12-related V4V5 types that cross over with animals since the depth of the sewage clone library limits the identification of more Lachno12-related V4V5 types.
FIG 3.
Abundances of Lachno3-associated V4V5 sequence types in sewage and five animal hosts. The abundance shown in each cell is normalized to the median sequence count for all samples and is converted to a percentage according to each V4V5 type's total abundance. The values increase from white to blue.
Development of qPCR assays for Lachno3 and Lachno12.
We designed two human-associated Lachnospiraceae assays based on marker candidates Lachno3 and Lachno12 by using our library of C. coccoides sequences from sewage and animals to find regions of specificity. The Lachno3 and Lachno12 clones with V4V5 region sequences that did not cross over into animals were considered targets, and the animal clone library sequences were used for exclusion of nontargets. The forward and reverse Lachno3 and Lachno12 primer sequences had at least 1 mismatch with the animal sequences, and the probes had several mismatches. We purposefully choose one marker (i.e., Lachno3) that was strictly specific, based on the results obtained with our NGS data set of sequences from animals, and one marker that was more abundant in sewage but that had low levels in other hosts (i.e., Lachno12) as a means to benchmark the performance of these assays when these organisms may occur at very low levels in nontarget animal sources. The primer and probe positions are shown in Fig. S4.
Lachno3 and Lachno12 assay validation.
The relative abundance levels of Lachno3 and Lachno12 in sequenced sewage samples (n = 28) indicated that the Lachno3 marker was generally present at levels about 3.0-fold ± 1.3-fold the levels of the Lachno12 marker. The qPCR results of the Lachno3 and Lachno12 assays with untreated Milwaukee, WI, sewage influent samples (n = 8) indicated that we could expect the Lachno3 marker copy number (CN) (1.24 × 105 ± 7.74 × 104) to be generally 2-fold the Lachno12 marker CN (6.61 × 104 ± 4.47 × 104).
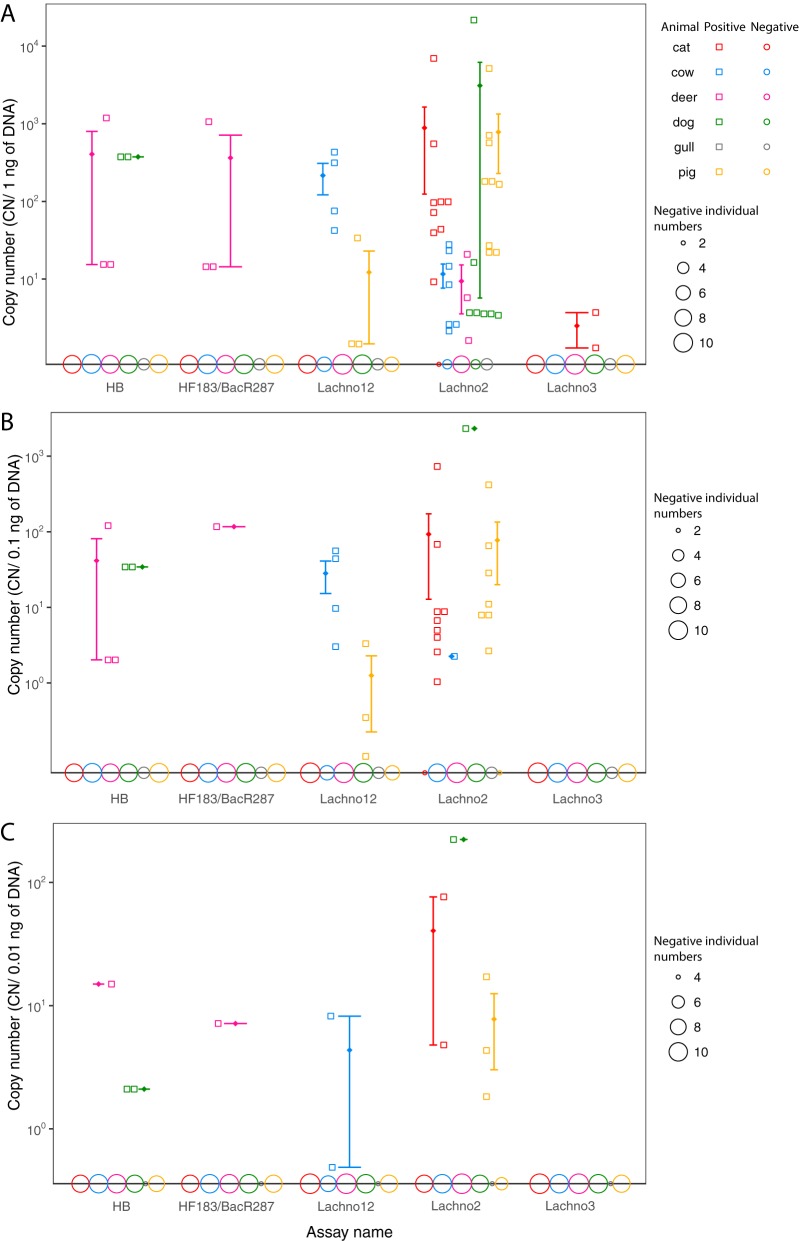
Both assays were tested with animal fecal samples (Fig. 4). The Lachno3 assay demonstrated a very low level of amplification with samples from two cats with a 1-ng μl−1 fecal sample dilution; however, the Lachno3 sequence was not found in the NGS data set of sequences from these animals, suggesting that there may have been nonspecific amplification in these samples. All other samples were negative. Lachno12 cross-reacted with samples from four cows (25% of cows) (Fig. 4A; Table S2), with the average copy number being 2.16 × 102, which is equivalent to a 1:300 dilution of sewage DNA. Lachno12 also showed positive qPCR results with samples from three pigs (33.3% of pigs) with an average CN of 12; however, the V6 sequencing results for these pig samples were negative for the Lachno12 marker, indicating nonspecific amplifications. In addition, a low occurrence of Lachno12 (a 1:280 relative abundance compared to that in sewage) was observed in the sample from one dog in the NGS data set, while this marker was not detected by qPCR of that dog sample or any other samples tested. This demonstrates that sequencing approaches may be more sensitive than amplification methods because of what can practically be amplified in a sample. Lachno12 was considered human preferred with cross-reaction with samples from cows, while Lachno3 was considered human specific in our results.
FIG 4.
qPCR validation of the results for animal fecal samples obtained with the Lachno3, Lachno12, Lachno2, HB, and HF183/BacR287 assays. The y axis indicates the copy numbers, and the x axis shows the assay names. The results obtained with 1 ng μl−1 (A), 0.1 ng μl−1 (B), and 0.01 ng μl−1 (C) animal fecal DNA are shown. The results for different animals are shown in different colors, with those for pools being represented as two adjacent data points. The result for each positive sample is shown as a hollow square, and the results for all negative samples are shown as open circles on the x axis, with the sizes of the circles being directly proportional to the CN for each animal. The error bars represent the mean CN (shown as a rhombus) with the standard error.
Samples from the same animals used to validate the specificity of the Lachno3 and Lachno12 assays were used to test established assays for human Bacteroides as defined by the presence of the HF183 sequence, which is the forward primer of the HB assay. The HB assay was positive for samples from one dog pooled sample (20% of dog samples tested) and one individual deer as well as one pooled deer sample (27% of deer samples tested); the HF183/BacR287 assay was positive for the same deer samples but not for any dog samples. The Lachno2 assay was run at an annealing temperature of 61°C rather than the previously reported 60°C and showed a cross-reaction with samples from cats (82% of cat samples tested), dogs (80% of dog samples tested), and pigs (100% of pig samples tested) at the highest concentration of fecal material tested. Some samples from cats and pigs were also positive when lower concentrations of fecal material were used. The Lachno2 qPCR results also showed low levels of amplification for samples from three deer (27% of deer samples tested) and seven cows (70% of cows samples tested), while the Lachno2-related V6 sequence was found by sequencing in only one of the samples from cows (i.e., sample PU73), supporting the conclusion that most Lachno2 signals in cow samples were from nonspecific qPCR amplifications; the three positive samples from deer were not sequenced, but all had decreased CN when the temperature was increased from 60°C to 61°C (data not shown), indicating that they could also be nonspecific amplifications. The gull samples were negative by all five assays.
Application of Lachno3 and Lachno12 assays for urban water samples.
We tested several environmental water samples that demonstrated inconsistent results with the HB and Lachno2 assays (i.e., a high CN in one human marker assay but a low CN in another or results that were not of the same magnitude). The HB assay detects HF183-positive organisms that have also been found in dogs and deer, and the Lachno2 assay is sensitive for detecting sewage but sporadically cross-reacts with samples from dogs, cats, and other nonurban animals. Samples were tested by the Lachno3 and Lachno12 assays and a combination of other available assays (Table 1). Samples in which organisms were not detected (or that had results below the detection limit) by the DogBact assay could potentially be excluded as being from a dog source. Samples with positive Lachno3 CN were considered contaminated with human fecal pollution. Because the Lachno12 marker is less specific and sensitive than the Lachno3 marker, the ratios of these markers that were not typical of those found in sewage or the presence of only Lachno12 was considered a suspicion of nonhuman sources. In addition, because the BacHum-UCD assay cross-reacted with samples from raccoons (27), dogs (32), and deer, urban water samples that were positive only by the HB assay were interpreted as containing contamination from raccoon when the DogBact assay was negative (e.g., sample FT15268). Deer are not expected to be in this highly urbanized area. The possibility of contamination from a limited number of humans that had atypical microbiome compositions could not be ruled out as an explanation for the inconstancies in human-associated indicator results.
TABLE 1.
Applications of Lachno3 and Lachno12 assays for testing environmental samples that had inconsistent results by the HB and Lachno2 assays
| Sample name | Sample type | Site | Sampling date (mo/day/yr) | CN/100 ml |
Interpretation of presumptive sourcesa | ||||
|---|---|---|---|---|---|---|---|---|---|
| HB | Lachno2 | Lachno3 | Lachno12 | DogBact | |||||
| FT12431 | Stormwater | Honey Creek 05 | 7/24/12 | 672 | 318 | 151 | 162 | 0 | Human |
| FT17171 | River water | Kinnickinnic River | 7/22/14 | 375 | 6,450 | 821 | 1,610 | 0 | Human |
| FT18040 | Stormwater | Wilson Park Creek outfall 18 | 10/14/14 | BLDb | 9,620 | 1,890 | 185 | 0 | Human |
| FT19920 | River water | Menomonee River | 7/9/15 | 0 | 675 | 265 | 161 | 0 | Human |
| FT21217 | River water | Kinnickinnic River grab | 5/3/16 | 801 | 27,300 | 6,510 | 4,450 | 0 | Human |
| FT21380 | Stormwater | Kinnickinnic River grab | 6/7/16 | 7,500 | 548,000 | 173,000 | 40,500 | 0 | Human |
| FT14569 | Beach | South Shore Old Beach 001 | 7/9/13 | BLD | 1,760 | 394 | 391 | 276 | Human/dog |
| FT14570 | Beach | South Shore Old Beach 002 | 7/9/13 | 166 | 3,460 | 1,000 | 1,430 | 1,060 | Human/dog |
| FT14571 | Beach | South Shore Old Beach 003 | 7/9/13 | 0 | 18,100 | 985 | 765 | 27,900 | Human/dog |
| FT15280 | Stormwater | Kinnickinnic River outfall, new | 11/6/13 | 225 | 33,700 | 249 | 196 | 8,710 | Human/dog |
| FT17167 | River water | Kinnickinnic River | 7/22/14 | 1,381 | 34,000 | 6,730 | 8,900 | 944 | Human/dog |
| FT17708 | Stormwater | Wilson Park Creek outfall 07 | 8/25/14 | BLD | 9,020 | 1,630 | 466 | 839 | Human/dog |
| FT17713 | Stormwater | Wilson Park Creek outfall 15 | 8/25/14 | 0 | 6,150 | 107 | 193 | 408 | Human/dog |
| FT20193 | Beach | South Shore Old Beach 001 | 8/10/15 | 0 | 1,320 | 132 | 0 | 320 | Human/dog |
| FT20574 | River water | Kinnickinnic River autosampler | 9/8/15 | 39,700 | 188,000 | 75,400 | 37,300 | 15,800 | Human/dog |
| FT21332 | Stormwater | Kinnickinnic River manhole | 5/10/16 | 0 | 1,350 | 0 | 170 | 19,200 | Dog |
| FT12198 | Stormwater | Wilson Park Creek outfall 25 | 6/21/12 | 566 | 0 | 0 | 132 | 0 | Raccoon |
| FT15268 | Stormwater | Kinnickinnic River outfall 47 | 10/31/13 | 3,540 | 0 | 0 | 0 | 0 | Raccoon |
| FT20724 | Stormwater | Russell Avenue manhole | 10/28/15 | 8,560 | 0 | 45 | 256 | 0 | Raccoon |
Cow, pig, and deer feces are not expected in these urban water samples.
BLD, below the limit of detection.
DISCUSSION
Organisms with a specific host niche offer an opportunity to discover new indicators of fecal pollution.
The gut microbiome of humans and animals is largely shaped by diet and host physiology (20, 36), and organisms specifically adapted to fill a niche within a specific host are promising candidates for developing new indicators of fecal pollution sources. The gut microbiomes of humans and animals have a limited number of bacterial families and genera but have extensive species and strain diversity that could indicate diversification among heterogeneous hosts (37). Our work to examine the population structure within the family Lachnospiraceae found that clustering of OTUs at 97% similarity is not sufficient to distinguish patterns of host specificity (Fig. 1) (30). This suggests that genetic traits that determine a host association do not map to the overall phylogeny within a group.
While overall phylogenetic patterns were not detected, we found that finer-scale methods could track host-associated organisms within the family Lachnospiraceae. Our previous work within the genus Blautia showed that use of a 60-bp region within the 16S rRNA gene as a marker region was sufficient to reveal ecologically relevant distribution patterns among hosts (31). Here, we expanded this work to include all Lachnospiraceae and demonstrate that this family is rich in potential indicators, with 88 V6 region sequences being identified by the biomarker identification program indicspecies (38). Analysis of the V4V5 regions in clone libraries demonstrated that organisms tracked by use of a particular V6 sequence can be further discriminated into subpopulations by their V4V5 sequences (i.e., one sequence type according to the sequence of the V6 region could have multiple associated V4V5 sequence types), with some of the V4V5 sequences from organisms positive for the Lachno3 marker being found in other animals. The analysis performed with indicspecies (38) identified far fewer markers in the V4V5 region than in the V6 region, demonstrating that V6 was more discriminatory of host patterns. Analysis of two regions at the same sequence depth verified that sequencing depth could not account for these results. Overall, while the V4V5 region is longer, it offered less resolution for tracking populations associated with particular hosts. These results are consistent with the V6 region showing the highest variability (40). Our findings support the hypothesis that marker gene distribution patterns may reflect differences in the genome that account for a presence in different host niches, but we reiterate that analysis of only a portion of the 16S rRNA gene cannot be used to indicate the exact organism that it comes from and mapping of the genetic markers to longer sequence reads could improve the tracking of specific organisms that are uniquely adapted to a host.
Dietary, geographic, and environmental factors may affect the presence of fecal genetic markers.
We found that the most abundant Lachnospiraceae V6 region sequence in sewage in our NGS data set (designated Lachno1) was not found in dairy cows in this study, but we have recovered this marker in the steer population in previous studies (2). Lachno1 was also found in the cow clone library, which consisted of organisms from beef cattle feces. This could be attributed to the different diets of these cow populations, as it has been reported that beef cattle fecal microbial communities are very likely shaped by feeding operations (41). In addition, beef cattle and dairy cows have been found to have different patterns of abundance of major and minor gut bacterial groups (35). Our qPCR results demonstrated that three out of the four cows positive for Lachno12 were from the same farm in Racine, WI, and all six of the negative cows came from different farms but in the same city of Brodhead, WI. Considering the possibility of different diets in cattle populations, there may be trade-offs in sensitivity and specificity when choosing markers, and it might be necessary to develop markers that are directed toward certain types of animal operations or feeding regimens.
The most abundant markers are stable in sewage.
The ranks of marker abundance differed slightly across various sewage samples, but within most of our sewage samples or sewage-contaminated water samples (n = 38), the Lachno1, Lachno2, and Lachno3 markers were within the top four most abundant Lachnospiraceae sequences. The stability of these markers was also found over a 3-year period at two wastewater treatment plants in a single city (30). The initial taxonomy of the NGS data set was based on release 102 of the SILVA database and was later updated so that it was based on release 119 of the SILVA database, and previously annotated Lachnospiraceae were annotated to Christensenellaceae and Defluviitaleaceae within the order Clostridiales. We included sequences annotated as Christensenellaceae, recently described in a study of the human fecal microbial community (42), as members of this family appear to be preferentially found in humans. We excluded Defluviitaleaceae sequences because organisms in this family within sewer systems appear to have a nonfecal origin (43). However, these sequences might be good candidates for tracing sewage release into the environment, since they were not found in any of the animals tested; they may ultimately demonstrate the presence of sewage more specifically than any of the human-derived markers that are found to cross over with other nontarget hosts.
Lachno3 is highly human specific.
Deep sequencing has revealed that only on rare occasions are marker sequences exclusive to a host, and even in these cases, further sequencing may reveal that they are shared between two or more hosts. Rather, certain community members appear to prefer one host and occur at low levels or only sporadically in other hosts (15). For human-associated marker assays, including the Lachno2 assay (2), the Lachno12 assay, and the previously published human Bacteroides assays (17, 18, 26), cross-reactivity was found, but usually for a low number of animals (Fig. 4). The use of these assays synergistically could ultimately improve specificity. Current fecal identification is often based on the usage of single human-associated alternative fecal indicators; however, there are several factors that can influence the sharing of organisms by human and animal microbiomes (e.g., similar diets or cohabitation), and the use of a combination of human-associated assays can exclude false-positive detection of human sources (32).
True animal cross-reaction needs to be differentiated from nonspecific amplification by assay primers. In the qPCR validation portion of this study, the Lachno3 qPCR results showed very low copy numbers in two cats, but the V6 marker sequences were absent according to the sequencing results for these cats. The Lachno2 assay validation results also included nonspecific amplification. These signals could be caused by primers amplifying targets that are very close to the marker V6 sequences. High levels of similar but nontarget DNA could account for nonspecific amplification in other studies (44, 45). Increasing the temperature could reduce nonspecific amplification but may negatively impact assay efficiency. This was observed in the case of optimization of the temperature for the Lachno3 assay, as well as validation of the Lachno2 assay; the slight increase of the temperature eliminated false-positive results by the Lachno2 assay with the highest level of fecal material from six deer, one cat, and one pig. This complication further highlights the usefulness of using two unrelated assays to detect human fecal pollution.
We also developed an assay with the Lachno12 marker, which was primarily human associated but which was found at low levels in samples from dogs and sporadically present in samples from certain cows. We found that despite a very low occurrence in one dog sample according to the sequencing results, the marker was not detected by qPCR. This finding illustrates that while sequencing may reveal a low level of an organism, it may not be relevant in practical applications, such as detection in water samples, where fecal material is already diluted. Further, these results helped confirm that the low levels of amplification in cat samples by the Lachno3 assay were most likely nonspecific, as the cat samples were sequenced to a similar depth and the Lachno3 sequence was absent.
Assessments for the host specificity and sensitivity of markers should be ongoing, given the high degree of diversity of the microbiomes of animals; mechanisms like cohabitation, which gives rise to a shared gut microbiome; and diet and geographic differences among individuals within a host type. For example, Lachno2 was originally chosen because of its high sensitivity for the detection of organisms in sewage and its absence in cows (2). With the inclusion of samples from dairy cows in this study, we observed crossover reactions with this target. Additionally, we found the sporadic presence of Lachno2 in samples from cats, dogs, and pigs, demonstrating the high sensitivity but low specificity of this marker. Similarly, the HF183 assay was later found to amplify signals in cat and dog samples; however, redesign of the reverse primer and probe improved its specificity in subsequent work (26).
Future application of Lachnospiraceae assays for detection of fecal sources in urban water.
Humans and animals in urban areas contribute fecal pollution to waterways, including recreational beaches. It is not practical or perhaps even feasible to develop assays for every possible source in a complex watershed comprised of urban land use; however, the use of multiple assays and interpretation of results in a tiered approach may provide insight into possible sources. For example, use of the Lachno2 assay with highly specific assays like the Lachno3 and HF183/BacR287 assays could help identify when nonhuman sources are present, without running separate assays for organisms from dogs, cats, or raccoons. Further, human-associated indicators target organisms that are generally present and the most abundant in the human population (28), but when fecal pollution is derived from a smaller number of individuals, for example, as a result of a broken lateral sewage line from a home or a cross connection between a sanitary sewer pipe and a stormwater pipe, results may be atypical. Multiple assays may be necessary when investigating small-scale contamination, like locating failures in sanitary sewer systems. Future work to increase the types and numbers of animals tested and increase the geographic coverage will provide more comprehensive assessments of specificity. Stormwater with fecal contamination from urban wildlife in particular lacks characterization, and it is difficult to distinguish fecal contamination from urban wildlife from contributions from a limited number of humans. Shared resources, such as fecal sample banks, may be useful for researchers so that they may validate the assays used in their watershed and compare their assays with those used in other areas. Overall, the use of a combination of human-associated fecal marker assays with known cross-reaction potentials, as well as animal marker assays, will improve the resolution of fecal pollution source identification. This information is crucial for assessing the possible risk from co-occurring pathogens and for remediation of pollution sources in urban water environments.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Two sets of animal fecal samples were used in this study; the first (set 1) was used for clone library construction and included samples from 5 cats, 5 dogs, and 10 pigs. The second (set 2) was used for qPCR validation and included samples from 11 cats, 10 dogs, 9 pigs, 11 deer, 10 cows, and 4 gulls. The animals providing the samples for set 1 were different from the individuals providing the samples for set 2. Sequencing was also performed for the majority of samples (n = 44) but was not performed in cases in which not enough material was available. Collection locations are detailed in Data Set S1 in the supplemental material. Animal stool samples were transported to University of Wisconsin—Milwaukee (Milwaukee, WI, USA) on ice within 24 h of collection and stored at −80°C upon arrival. Fecal sample preparation and DNA extraction followed the protocol for pathogen detection described in the instructions in the QIAamp DNA stool minikit (Qiagen Inc., Valencia, CA), which increases the yield of nonhost fecal genomic DNA. In some cases, extracted DNA was sent directly from the originating laboratory. All DNA samples were stored at −20°C.
Fecal clone libraries.
Clone libraries were generated from set 1 animal fecal samples using Clostridium coccoides-specific primers, which amplified a portion of the 16S rRNA gene from Lachnospiraceae (primers Ccoc-F/1492R) (2, 30, 46). These amplicons were then analyzed by Sanger sequencing (ABI Prism 3700xi genetic analyzer; Applied Biosystems, Foster City, CA). Sequences from two previously published data sets were also used: (i) C. coccoides clone libraries from four different sewage samples (GenBank accession numbers JX228967 to JX230954) (30) and (ii) full-length whole-community 16S rRNA gene libraries from cows (GenBank accession numbers FJ672948 to FJ674268 and FJ675665 to FJ685516) (35). Only Lachnospiraceae sequences from the 16S rRNA gene libraries from cows were used; for both of the previously published sequence data sets, libraries were subsampled to 200 sequences. The cloning and sequencing methods used, including PCR, ligation, transformation, plasmid preparation, and sequencing reactions, were previously described (2).
Sequence processing and analysis.
Three primers (primers Ccoc-F, 331F, and 1492R) were used to sequence the animal clone libraries. Sequences were assembled using the SeqMan Pro program (Lasergene, v12; DNAStar, Madison, WI). Sequences of less than 900 bp were discarded, and chimeras were subsequently removed using the Chimera Vsearch tool (47) in the mothur program (48). A total of 718 sequences, including 200 sewage, 80 cat, 85 dog, 153 pig, and 200 cow sequences, were used. Operational taxonomic units (OTUs) were created at the 97% similarity level using the nearest-neighbor method in mothur and were based on release 119 of the SILVA taxonomic reference database.
A phylogenetic tree of all representative OTU sequences was constructed to examine the phylogenetic relationships among the Lachnospiraceae sequences from different hosts. The host source was annotated for each OTU (i.e., human only, animal only, or human/animal). The representative OTU sequences, along with two Escherichia coli 16S rRNA gene sequences that were used as an outgroup (GenBank accession numbers HF584706 and LT745986), were aligned using the MUSCLE program (49) and trimmed to the same length with the MEGA7 program (50). The tree was constructed using the maximum-likelihood method in the Kimura 2-parameter (K2) model with gamma distribution rates and invariant sites (G+I), bootstrapped for 1,000 replicates, and visualized in the Interactive Tree of Life (iTOL; http://itol.embl.de) (51). Representative sequences and their host annotation for each OTU are shown in Data Set S2. A heatmap was generated using the R packages RColorBrewer and ggplot2 to display the relative abundance of members of the Lachnospiraceae in different hosts based on the clone libraries (Fig. S1). To better visualize the distribution of clones in different hosts, the relative abundance was normalized to 100% for each of the 70 most abundant OTUs.
NGS data sets.
Sequences were generated using the Illumina MiSeq and HiSeq platforms at the Marine Biological Laboratory (MBL), University of Chicago. Whole-community data sets of partial 16S rRNA gene sequences were generated from the V4V5 (primers 518F/926R) (52) and V6 (primers 967F/1064R) (53) regions and stored in the Visualization and Analysis of Microbial Populations (VAMPs) platform (54). Sequence counts were normalized to the median abundance for all bacterial sequences from all samples, and singleton sequences were removed. Lachnospiraceae sequences were parsed out of whole microbial communities using taxonomy assignments in GAST software (55). Sequences from the newly described family Christensenellaceae (42), which were previously designated to belong to the Lachnospiraceae and which were very likely also host adapted, were added into the data sets. In all, the data set included 20,587 unique V4V5 sequences with 741,927 reads and 100,242 unique V6 sequences with 17,143,353 reads. The Lachnospiraceae sequences were enumerated according to their rank abundance in the composite data set of sequences from sewage samples. The second and third most abundant Lachnospiraceae in this data set had appeared in the inverse order in a previous analysis and had been designated Lachno3 and Lachno2, respectively. Likewise, the 10th most abundant member of the Lachnospiraceae in the data set had previously been designated Lachno12. Since the exact order is somewhat dependent on the sewage samples used in the analysis, we chose to keep the original designations for Lachno3 and Lachno2. Therefore, Lachno3 in this study is the second most abundant Lachnospiraceae in the data set, and Lachno2 is the third most abundant. The designations Lachno12 and Lachno10 correspond to the 10th and 12th most abundant Lachnospiraceae, respectively. The 100 most abundant sequences for each animal and sewage source are detailed in Data Set S3.
Design of host-specific molecular assays for humans.
Initially, animal and sewage samples that were both sequenced for V4V5 and V6 regions were compared using the R package indicspecies (38) with 999 permutation tests to identify the region that would provide the most specific and sensitive Lachnospiraceae marker candidates. The human-associated marker candidates were first chosen by use of the criteria that they had sensitivities and specificities of greater than 90% and that they were among the top 95% most abundant Lachnospiraceae in sewage. Candidates were retained if they were present at lower levels in two other animal hosts or less (i.e., cat, dog, pig, cow, deer, chicken, and raccoon). We chose the V6 region as the most promising marker region and then compared the V6 NGS data set sequences to the sequences in the sewage clone library using analysis with the local BLAST program (56) to find clones that represented longer sequences and contained each V6 marker sequence for primer design.
Two qPCR assays were developed to target Lachno3 and Lachno12 (Table 2). Primers and probes were designed using the alignments of sequences from animals and sewage in the MegAlign Pro program in DNAStar software. Alignments included each respective V6 marker sequence, a Lachnospiraceae full-length 16S rRNA gene reference sequence (GenBank accession number EF036467), and the marker's exact matches with the sewage clone library sequences. Animal sequences that had >97% similarity with the genetic marker by analysis with the BLAST program and representative sequences from the top 10 OTUs of all animal sequences were also added to the alignment to design primers that would discriminate animal sources. Primers specific for the V6 sequence region were designed, as that region provided the largest amount of discrimination for the assays.
TABLE 2.
Lachno3 and Lachno12 marker primer and probe sequences
| Marker | Sequence |
||
|---|---|---|---|
| Forward primer | Probea | Reverse primer | |
| Lachno3 | 5′-CAACGCGAAGAACCTTACCAAA-3′ | FAM-5′-CTCTGACCGGTCTTTAATCGGA-3′-MGB | 5′-CCCAGAGTGCCCACCTTAAAT-3′ |
| Lachno12 | 5′-ATCTTGACATCCCTCTGACCGGGA-3′ | FAM-5′-CGTCCCTTTCCTTCGGGACAGG-3′-MGB | 5′-CTCAGAGTGCCCACCACTACGT-3′ |
FAM, 6-carboxyfluorescein; MGB, TaqMan minor groove binder.
The Lachno3 and Lachno12 genetic markers were mapped into longer sequence reads that included the V4V5 region to evaluate whether the related V4V5 sequences had the same specificities. We examined the V4V5 sequence types associated with each V6 marker in sewage clone library sequences since both regions spanned the clone sequences. These V4V5 sequence types were then compared to the larger V4V5 region sequence NGS data set for multiple animals and sewage samples by using the BLAST program. The NGS data sets for the V4V5 region were named numerically according to the rank abundance of the sequences in sewage.
Quantitative PCR analyses.
All qPCR experiments were performed on an Applied Biosystems StepOne Plus real-time PCR system thermal cycling block (Applied Biosystems, Foster City, CA). To validate the Lachno3 and Lachno12 assays, animal stool DNA extracts (set 2) were tested in the formats of at least six individual samples and one pooled sample (made from samples from 2 individuals), except for gull stool samples, which were run only as single samples. Each animal stool DNA was diluted to 1 ng μl−1, 0.1 ng μl−1, and 0.01 ng μl−1, with 5 μl being used in each qPCR mixture. Sewage samples were diluted 1:100, and environmental samples were run without dilution. All standard curves were run in triplicate with DNA from sewage clones whose sequences matched those of the Lachno3 and Lachno12 markers, and samples were serially diluted to contain from 1.5 × 106 to 1.5 copies per reaction mixture. For each validation run, a sewage control and a blank control were used. The qPCR setting was as described by Templar et al. (33). To optimize the annealing temperature for these two assays, we tested diluted sewage DNA samples (diluted at ratios of 1:100, 1:500, 1:1,000, 1:2,000, 1:4,000, and 1:8,000) from 60°C to 64°C to determine if any amplification efficiency was lost. The amplification program included 1 cycle at 50°C for 2 min, followed by 1 cycle at 95°C for 10 min and then 40 cycles of 95°C for 15 s, followed by 1 min at 64°C for Lachno3 or 61°C for Lachno12; the Lachno2 assay was run at 61°C in this study. The qPCR assay slopes, y intercepts, and efficiencies are shown in Table S1. For validation result output, each animal's qPCR copy number (CN) was converted to the CN per nanogram of DNA, the CN per 0.1 ng of DNA, and the CN per 0.01 ng of DNA, and the result for each sewage sample was converted to the CN per nanogram of DNA.
Accession number(s).
The partial 16S rRNA gene clone library sequences were deposited in the GenBank database under accession numbers MG702648 to MG702965. A portion of the NGS data used in this study was from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA; SRA accession number SRP041262) (V6 sequences) and BioProject (BioProject accession number PRJNA261344) (V4V5 sequences). The NGS data generated for this study are stored in SRA under accession numbers SRP132402 (V6 region sequences) and SRP132403 (V4V5 region sequences).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Marine Biological Laboratory (MBL), University of Chicago, and the Great Lakes Genomics Center (GLGC), University of Wisconsin—Milwaukee, for providing expertise in NGS and Sanger sequencing. We thank Adelaide Roguet for lending bioinformatics expertise to the project, Keri A. Lydon for her insightful discussion, and Katherine Halmo for assistance with DNA extractions.
Funding for this work was provided by grant number R01 AI091829 from the National Institutes of Health (NIH).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00309-18.
REFERENCES
- 1.Marsalek J, Rochfort Q. 2004. Urban wet-weather flows: sources of fecal contamination impacting on recreational waters and threatening drinking-water sources. J Toxicol Environ Health A 67:1765–1777. doi: 10.1080/15287390490492430. [DOI] [PubMed] [Google Scholar]
- 2.Newton RJ, VandeWalle JL, Borchardt MA, Gorelick MH, McLellan SL. 2011. Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Appl Environ Microbiol 77:6972–6981. doi: 10.1128/AEM.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer EP, VandeWalle JL, Bootsma MJ, McLellan SL. 2011. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res 45:4081–4091. doi: 10.1016/j.watres.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 4.Donovan E, Unice K, Roberts JD, Harris M, Finley B. 2008. Risk of gastrointestinal disease associated with exposure to pathogens in the water of the Lower Passaic River. Appl Environ Microbiol 74:994–1003. doi: 10.1128/AEM.00601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffin DW, Donaldson KA, Paul JH, Rose JB. 2003. Pathogenic human viruses in coastal waters. Clin Microbiol Rev 16:129–143. doi: 10.1128/CMR.16.1.129-143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brokamp C, Beck AF, Muglia L, Ryan P. 2017. Combined sewer overflow events and childhood emergency department visits: a case-crossover study. Sci Total Environ 607-608:1180–1187. doi: 10.1016/j.scitotenv.2017.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.U.S. Environmental Protection Agency. 2012. Recreational water quality criteria. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 8.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 9.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemarchand K, Lebaron P. 2003. Occurrence of Salmonella spp. and Cryptosporidium spp. in a French coastal watershed: relationship with fecal indicators. FEMS Microbiol Lett 218:203–209. doi: 10.1111/j.1574-6968.2003.tb11519.x. [DOI] [PubMed] [Google Scholar]
- 11.Colford JM, Wade TJ, Schiff KC, Wright CC, Griffith JF, Sandhu SK, Burns S, Sobsey M, Lovelace G, Weisberg SB. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18:27–35. doi: 10.1097/01.ede.0000249425.32990.b9. [DOI] [PubMed] [Google Scholar]
- 12.Ercumen A, Pickering AJ, Kwong LH, Arnold B, Parvez SM, Alam M, Sen D, Islam S, Kullmann C, Chase C, Ahmed R, Unicomb L, Luby S, Colford JM. 2017. Animal feces contribute to domestic fecal contamination: evidence from E. coli measured in water, hands, food, flies and soil in Bangladesh. Environ Sci Technol 51:8725–8734. doi: 10.1021/acs.est.7b01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher JC, Eren AM, Green HC, Shanks OC, Morrison HG, Vineis JH, Sogin ML, McLellan SL. 2015. Comparison of sewage and animal fecal microbiomes by using oligotyping reveals potential human fecal indicators in multiple taxonomic groups. Appl Environ Microbiol 81:7023–7033. doi: 10.1128/AEM.01524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed W, Hughes B, Harwood VJ. 2016. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231. doi: 10.3390/w8060231. [DOI] [Google Scholar]
- 15.McLellan SL, Eren AM. 2014. Discovering new indicators of fecal pollution. Trends Microbiol 22:697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan B, Ng C, Nshimyimana JP, Loh LL, Gin KYH, Thompson JR. 2015. Next-generation sequencing (NGS) for assessment of microbial water quality: current progress, challenges, and future opportunities. Front Microbiol 6:1027. doi: 10.3389/fmicb.2015.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ Microbiol 7:249–259. doi: 10.1111/j.1462-2920.2004.00702.x. [DOI] [PubMed] [Google Scholar]
- 19.Fremaux B, Gritzfeld J, Boa T, Yost CK. 2009. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res 43:4838–4849. doi: 10.1016/j.watres.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 20.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okabe S, Shimazu Y. 2007. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl Microbiol Biotechnol 76:935–944. doi: 10.1007/s00253-007-1048-z. [DOI] [PubMed] [Google Scholar]
- 22.Koskey AM, Fisher JC, Eren AM, Ponce-Terashima R, Reis MG, Blanton RE, McLellan SL. 2014. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep 6:696–704. doi: 10.1111/1758-2229.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorvitovskaia A, Holmes SP, Huse SM. 2016. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 4:15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed W, Yusuf R, Hasan I, Goonetilleke A, Gardner T. 2010. Quantitative PCR assay of sewage-associated Bacteroides markers to assess sewage pollution in an urban lake in Dhaka, Bangladesh. Can J Microbiol 56:838–845. doi: 10.1139/W10-070. [DOI] [PubMed] [Google Scholar]
- 25.Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst Appl Microbiol 33:348–357. doi: 10.1016/j.syapm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, Shanks OC. 2014. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl Environ Microbiol 80:3086–3094. doi: 10.1128/AEM.04137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van De Werfhorst LC, Sercu B, Holden PA. 2011. Comparison of the host specificities of two Bacteroidales quantitative PCR assays used for tracking human fecal contamination. Appl Environ Microbiol 77:6258–6260. doi: 10.1128/AEM.00239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. 2015. Sewage reflects the microbiomes of human populations. mBio 6:e02574. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML. 2010. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12:378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLellan SL, Newton RJ, Vandewalle JL, Shanks OC, Huse SM, Eren AM, Sogin ML. 2013. Sewage reflects the distribution of human faecal Lachnospiraceae. Environ Microbiol 15:2213–2227. doi: 10.1111/1462-2920.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL. 2015. A single genus in the gut microbiome reflects host preference and specificity. ISME J 9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res 41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 33.Templar HA, Dila DK, Bootsma MJ, Corsi SR, McLellan SL. 2016. Quantification of human-associated fecal indicators reveal sewage from urban watersheds as a source of pollution to Lake Michigan. Water Res 100:556–567. doi: 10.1016/j.watres.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 34.Sinigalliano CD, Fleisher JM, Gidley ML, Solo-Gabriele HM, Shibata T, Plano LRW, Elmir SM, Wanless D, Bartkowiak J, Boiteau R, Withum K, Abdelzaher AM, He G, Ortega C, Zhu X, Wright ME, Kish J, Hollenbeck J, Scott T, Backer LC, Fleming LE. 2010. Traditional and molecular analyses for fecal indicator bacteria in non-point source subtropical recreational marine waters. Water Res 44:3763–3772. doi: 10.1016/j.watres.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durso LM, Harhay GP, Smith TPL, Bono JL, DeSantis TZ, Harhay DM, Andersen GL, Keen JE, Laegreid WW, Clawson ML. 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl Environ Microbiol 76:4858–4862. doi: 10.1128/AEM.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dethlefsen L, McFall-Ngai M, Relman DA. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jabari L, Gannoun H, Cayol JL, Hamdi M, Fauque G, Ollivier B, Fardeau ML. 2012. Characterization of Defluviitalea saccharophila gen. nov., sp. nov., a thermophilic bacterium isolated from an upflow anaerobic filter treating abattoir wastewaters, and proposal of Defluviitaleaceae fam. nov. Int J Syst Evol Microbiol 62:550–555. doi: 10.1099/ijs.0.030700-0. [DOI] [PubMed] [Google Scholar]
- 44.Shanks OC, White K, Kelty CA, Sivaganesan M, Blannon J, Meckes M, Varma M, Haugland RA. 2010. Performance of PCR-based assays targeting Bacteroidales genetic markers of human fecal pollution in sewage and fecal samples. Environ Sci Technol 44:6281–6288. doi: 10.1021/es100311n. [DOI] [PubMed] [Google Scholar]
- 45.Layton BA, Cao Y, Ebentier DL, Hanley K, Ballesté E, Brandão J, Byappanahalli M, Converse R, Farnleitner AH, Gentry-Shields J, Gidley ML, Gourmelon M, Lee CS, Lee J, Lozach S, Madi T, Meijer WG, Noble R, Peed L, Reischer GH, Rodrigues R, Rose JB, Schriewer A, Sinigalliano C, Srinivasan S, Stewart J, Van De Werfhorst LC, Wang D, Whitman R, Wuertz S, Jay J, Holden PA, Boehm AB, Shanks O, Griffith JF. 2013. Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res 47:6897–6908. doi: 10.1016/j.watres.2013.05.060. [DOI] [PubMed] [Google Scholar]
- 46.Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, Oyaizu H, Tanaka R. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol 68:5445–5451. doi: 10.1128/AEM.68.11.5445-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks, p 361–362. Abstr Third Int AAAI Conf Weblogs Soc Media. [Google Scholar]
- 49.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. 2016. Interactive Tree Of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. 2014. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS One 9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huse SM, Welch DBM, Voorhis A, Shipunova A, Morrison HG, Eren AM, Sogin ML. 2014. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics 15:41. doi: 10.1186/1471-2105-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.