Abstract
Cell mechanics is a multidisciplinary field that bridges cell biology, fundamental mechanics, and micro and nanotechnology, which synergize to help us better understand the intricacies and the complex nature of cells in their native environment. With recent advances in nanotechnology, microfabrication methods and micro-electro-mechanical-systems (MEMS), we are now well situated to tap into the complex micro world of cells. The field that brings biology and MEMS together is known as Biological MEMS (BioMEMS). BioMEMS take advantage of systematic design and fabrication methods to create platforms that allow us to study cells like never before. These new technologies have been rapidly advancing the study of cell mechanics. This review article provides a succinct overview of cell mechanics and comprehensively surveys micro and nano-scale technologies that have been specifically developed for and are relevant to the mechanics of cells.
Here we focus on micro and nano-scale technologies, and their applications in biology and medicine, including imaging, single cell analysis, cancer cell mechanics, organ-on-a-chip systems, pathogen detection, implantable devices, neuroscience and neurophysiology. We also provide a perspective on the future directions and challenges of technologies that relate to the mechanics of cells.
Keywords: Microfabrication, Nanofabrication, Biophysics, Single Cell Analysis, Mechanical Manipulation, Microfluidics
1. Introduction
Cells, similar to most engineering materials, are subject to different types of physical effects including external forces such as compression, tension, fluid shear stress, hydrostatic pressure and internal forces caused by the cytoskeleton. Cells effectively sense the mechanical cues in their microenvironment and respond accordingly by altering their biological, chemical and physical properties [1]. For example, cells can reinforce their cytoskeleton to create stronger surface adhesion [2] or fluidize their cytoskeleton to decrease their structural stiffness in response to changes in their surroundings [3]. It is well known that biochemical signals are important factors that regulate many cellular processes. Mechanical properties and forces are increasingly being recognized as key players in basic cellular processes, and as part of the extracellular signals that regulate the fate and function of cells [4-9]. Cell mechanics influence a wide range of measures such as morphological changes, migration, proliferation, adhesion and differentiation [10]. These measures take place differently in a state of disease, which is generally due to the altered biochemical as well as mechanical microenvironment [11,12]. The study of cell mechanics is a multidisciplinary field that bridges cell biology with fundamental mechanics, and micro and nanotechnology, which synergize to help us better understand the complex nature of cells in their native environment.
To capture a complete picture of all the essential mechanical interactions and physical properties of cells, we need approaches and technologies, which can work and interact with cells. A typical cell body is about 10 micrometres (μm) in diameter, which is approximately one tenth of the thickness of a human hair. The size or resolution of the tools utilized in cellular biophysical studies has to be in the order of size or smaller. Otherwise, numerous theoretical assumptions would have to be employed and measurement errors may cloud our objective examination. With the recent advances in nanotechnology, microfabrication technologies and micro-electro-mechanical-systems (MEMS), we are now well situated to tap into the wondrous micro world of cells. The field that brings biology and MEMS together is currently known as Biological MEMS (BioMEMS). BioMEMS take advantage of systematic design and fabrication methods to create platforms that allow us to study cells like never before. These new technologies have been rapidly advancing the study of cell mechanics. Therefore, in this review we cover micro and nano-scale technologies that have been specifically developed for and are relevant to the mechanics of cells. We present a comprehensive survey of micro and nano technologies relevant to a wide range of cell types, and their applications in biology and medicine, including imaging, cancer research, neuroscience, tissue engineering, regenerative medicine and pathogen detection.
2. Role of cell mechanics in biology and medicine
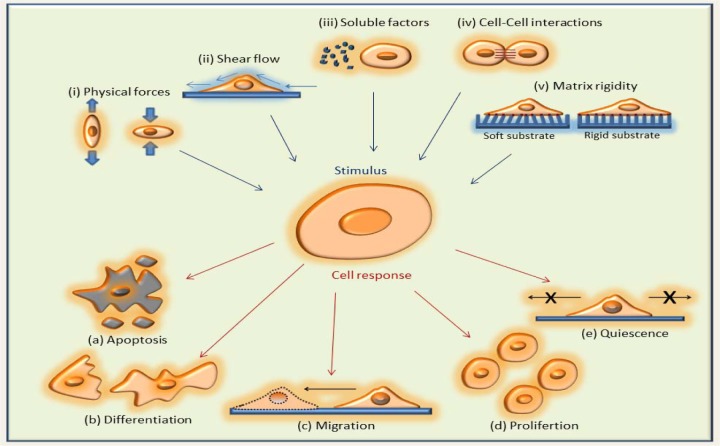
In general, cells consist of a membrane, cytoplasm, nucleus and a cytoskeleton. The cytoskeleton is composed of a network of filamentous proteins, which include microtubules, intermediate filaments, actin filaments and other cellular proteins [13,14]. Cells have a dynamic nature and undergo different types of intracellular and extracellular events to maintain their essential biological functions (Figure 1), including sensing, cell-cell communication, maturation, migration, proliferation, differentiation, apoptosis and quiescence [15-17]. Cells are extraordinarily amenable to adapting to changes in their physiological microenvironment, which is a complex and ever changing medium that is all around them.
Figure 1.
Cells respond to different microenvironmental stimulus in vivo. A schematic showing the different factors, (i) physical forces, (ii) shear flow, (iii) soluable factors, (iv) cell-cell interactions and (v) matrix rigity that trigger the cells to undergo changes in their behaviors and functions such as (a) apoptosis, (b) differentiation, (c) migration, (d) proliferation and (e) quiescene.
Cells generally respond to the mechanical forces and mechanical properties of their microenvironment in two different ways: with a physical response (e.g., alignment of cell shape and cytoskeleton on anisotropic surfaces) [18-21], or a biochemical response (e.g., activation intracellular or extracellular signaling cascades) [22-24]. These responses lead to the emergence of many cellular events including stiffening, softening, maturation, calcium influx, morphological changes, generation of tractions forces or focal adhesions [25], as well as disease states such as cancer [26,27], osteoporosis [28], osteoarthritis [29], asthma [30], glaucoma [31], malaria [32], atherosclerosis [33] and sickle cell anaemia [34], as a result of complex cellular mechanisms. Therefore, the main objective in studies related to cell mechanics is to understand these interactions, events, and their biological and functional consequences, which are covered in the following sections.
2.1 Mechanical properties of cells
The intracellular components of cells such as cytoskeletal proteins, cytoplasm and membrane contribute to the mechanical properties of cells and tissues. For example, the physical properties of the cytoskeleton play an important role in cellular functions such as spreading, crawling, adhesion and polarity. Furthermore, the cytoskeleton is important in maintaining cell shape by providing structural stiffness [13,35,36]. The cell nucleus provides a degree of structural stiffness and plasticity [37,38]. Maintaining the cell shape is crucial to performing biological functions. Cell shape can be determined and controlled by cellular attachments to the surrounding extracellular matrix [39]. The tensegrity (tension integrity) approach states that the combination of tension and compression elements provides a stable form that maintains the cell shape through the balancing of cytoplasmic pressure [14]. According to this approach, cells withstand shape distortion through pre-tension or prestress in their structural elements [14], in which actin filaments and microtubules are the dominant structures for determining cell stiffness [40,41]. Cell stiffness has been extensively studied and quantitative cell stiffness values have been reported in literature (Table 1). Reported cell stiffness values cover a wide range, which mainly depend on the cell type and measurement method. The stiffness of diseased cells can be dramatically different [11,42,43]. For instance, cancer cells are known to be significantly softer than normal cells [11] and sickle red blood cells are known to be significantly stiffer than healthy blood cells [34,43] (Table 1).
Table 1.
Mechanical properties of cells reported in the literature
| Aspects of cell mechanics | Cell type | Magnitude | Tool/Technique | References |
|---|---|---|---|---|
| Stiffness | Fibroblasts Fibroblasts Vascular endothelial cells Vascular smooth muscle cells Rat ASM(airway smooth muscle)cells |
0.02 N/m 0.02 N/m 0.03–0.04 N/m 0.09–0.88 N/m 0.099 N/m |
Mcropipette Magnetic twisting cytometer |
[231] [232] |
| Elastic modulus | Cancer MCF-7 cell Osteoblasts Skeletal muscle cells Cardiocytes Erythrocytes Leukocytes Fibroblasts Endothelial cells Outer hair cells |
0.95 – 1.19 kPa 0.3–20 kPa 8–45 kPa 90–110 kPa 14–33 kPa 0.2–1.4 kPa 0.6–12 kPa 0.2–2 kPa 2–4 kPa |
Atomic force microscopy |
[233] [234] [235,236] [237] [238,239] [240] [241,242] [243] [244] |
| Viscoelasticity | Cytoplasm | 210 Pa s 2000 Pa s |
Magnetic bead microrheology | [245] [246] |
| Cell adhesion force | Human cervical carcinoma cell Epithelial cells Murine fibroblast cells Rat cardiac fibroblast |
19–204 nN 100 nN 300–400 nN 10 nN |
Atomic force microscopy High-speed centrifugation technique Manipulation force microscope Traction force microscopy |
[247] [248] [249] [250] |
| Cell traction forces | Fibroblasts Fish keratocytes |
100 nN 20 nN |
Microcantilevers micro pads Flexible substrate |
[251] [58] |
| Shear stress | Endothelial cells Bovine aortic endothelial cells Human umbilical vein endothelial cells |
1–15 dyn/cm2 10 dyn/cm2 1–3 dyn/cm2 |
Microfluidics | [252-254] [255] [256] |
The deformation of certain cell types is indispensable for performing their essential biological functions. For instance, the typical diameter of a red blood cell is about 7.0–8.5 μm and these cells can undergo up to 100% elastic deformation, when they flow through tiny capillaries with inner diameters as little as 3 μm [15]. In sickle cell disease(SCD), due to the intracellular haemoglobin polymerization, red blood cells lose their elasticity. Because of this stiffening, sickled red blood cells cannot deform to pass through capillaries and cause blockages which lead to pain in patients [43]. On the other hand, for some cell types, excessive or repeated mechanical deformation can trigger harmful signaling pathways that can result in physiological disorders or diseases. For example, hypertension and other cardiovascular diseases are associated with an altered compliance of blood vessel walls normally regulated by the deformability of smooth muscle cells [15].
Cells exhibit viscoelastic behaviour which gives them the characteristics of both solids and fluids [44]. Due to their viscoelastic properties, cells deform in a time dependent manner, whereby mechanical stresses relax under constant deformation, or deformation increases over time as a result of a constant load [44,45]. Viscoelasticity plays an important role in cellular processes, such as in the regulation of cell shape and in the regulation of genetic expression through viscoelastic coupling between the plasma membrane and the nucleus [46,47]. The viscoelastic properties of cells have been of interest and they have been studied using the available micro/nano-scale tools (Table 1).
2.2 Mechanical interactions of cells
Cells-matrix and cell-cell interactions are involved in numerous signaling pathways and they are required for maintaining the functional and structural integrity of cells. Cells are able to pull and push on their microenvironment through cytoskeletal contractility to sense and assess their microenvironment or other cells [48]. Any miscommunication between cells and the surrounding matrix, or in cell-cell interactions, may lead to the emergence of a disease state [5]. While cell-matrix interactions are primarily mediated by integrins, cell-cell interactions involve the secretion of signaling molecules, gap junctions, neurotransmission, and intercellular nanotubes [5].
Cellular adhesion is a critical process, which governs migration, immobilization and the attachment to the extracellular matrix (ECM). Cellular adhesion is regulated by a combination of several factors including biochemical stimuli, internal and external forces, and the mechanical properties of the extracellular environment [49,50]. For instance, cell adhesion strength increases under mechanical stress, which results in the upregulation of adhesion molecules [48,51]. Furthermore, the presence of mechanical forces can directly impact the size, shape and composition of focal adhesions, implying a direct relationship between the applied forces and the generation of biochemical signals [52]. For example, leucocytes exhibit different adhesive states during an inflammatory response: fast rolling, slow rolling and firm adhesion [53].
In addition to being subjected to external forces, cells can generate their own mechanical forces during migration, contraction and cytoskeletal activity. For instance, all muscle cells have a molecular motor, which is composed of actin and myosin with a well-defined structural arrangement, and used to generate active contraction [54]. Cell traction forces were first observed as distortions on a flexible substrate due to fibroblast locomotion [55]. Traction forces generated by faster migrating cells, such as leukocytes, could not be detected in the same manner as fibroblasts [56], implying that slow moving cells have a stronger adhesion whereas fast moving cells have a weaker adhesion [57]. Therefore, it was determined that slow moving cells have greater cytoskeletal contractility and fast cell migration/motion requires both weak adhesion forces and cytoskeletal contractility [58]. In this sense, traction forces are often measured in slow moving cells, and magnitudes vary depending on the cell type and assessment state (Table 1).
2.3 Behaviour of cells in fluid flow
Endothelial cells are the innermost layer in vascular walls and they interact strongly with the blood flow. The essential functions of endothelial cells include the maintenance of the anticoagulant properties of blood vessel walls and the regulation of vascular permeability [59]. When blood flows through the vessels, this generates haemodynamic forces that are essentially a combination of two fluid forces: shear stress and hydrostatic pressure [60]. Even though the entire vascular wall experiences the hydrostatic pressure, only the inner lining endothelial cells undergo blood flow induced shear stress [61,62]. Therefore, the morphology of endothelial cells is affected by shear stress and they align parallel to flow [63,64]. Flow disturbances, separation and vortexes negatively influence the endothelial cell morphology due to cellular mis-orientation [60].
Shear stress on the cell surface leads to intracellular stress generation, cytoskeletal reorganization, and hence the balancing of internal and external forces [59,60,64,65]. Alterations in shear stress may also contribute to vascular diseases as in the example of atherosclerosis [33,66]. Therefore, measurement of the shear stress is an essential aspect of cell mechanics studies. Shear stresses experienced by different cell types cover a wide range as presented in Table 1.
3. Conventional analysis and imaging methods for studying cell mechanics
The most commonly used conventional techniques in cell mechanics are: (a) atomic force microscopy (AFM), (b) optical tweezers, (c) micropipettes, (d) flow chambers, and (e) microscopy imaging including confocal and fluorescence microscopy.
3.1 Atomic force microscopy
AFM is a type of scanning probe microscopy for imaging, manipulating and quantifying the sample surface at a nano-scale resolution. The basic components of an AFM are a cantilever with a sharp tip controlled by piezoelectric actuators, a laser and a detector. When the tip of the AFM is scanning the sample surface, the cantilever is deflected as a result of the forces between the tip and the surface. This deflection can then be quantified with a detector (photodiodes) by determining the position of the laser beam reflected by the cantilever. AFM has been widely utilized in cell mechanics studies in the literature [67,68]. The cytoskeleton structure was investigated and the mechanical properties including the elasticity, viscoelasticity and plasticity of L929 cells were quantified using AFM force measurements [69]. Elastic modulus and viscosity can be used as indicators of cellular differentiation or can be utilized in observing the effects of external stimuli. Furthermore, AFM can be used as a manipulation tool at the single cell level.
3.2 Optical tweezers
Optical tweezers, also known as laser tweezers, have the ability to manipulate dielectric particles by focusing a laser to a diffraction-limited point through a microscope objective that has a high numerical aperture [70]. Particles near the focused laser are trapped because of the restoring force towards the focus. The size of the particles that can be trapped in optical tweezers range from 20 nm to several micrometres, such as organelles, cells and polystyrene or silica microspheres. Furthermore, forces ranging from 0.1 to 100 pN can be exerted using optical tweezers [70]. In a typical study, the mechanical properties of the red blood cell membrane spectrin network were characterized using optical tweezers. The isolated spectrin skeleton was deformed by applying forces to silica beads bound to the membrane [71].
3.3 Micropipettes
Micropipettes are used for the mechanical analysis of cells by applying a suction to a small portion of a cell, while measuring the deformation of the cell membrane and the suction pressure of the micropipette [72]. A novel micromanipulation technique was presented to determine alterations in cellular rheology during cell spreading [73]. Chick fibroblasts were allowed to spread on the surface of a glass microplate and micropipette aspiration was applied to cells at controlled pressure levels. The internal pipette radius, cell radius outside the pipette and the length of the aspired portion of the cells were quantified [73]. The automated micropipette aspiration was utilized in conjunction with a video microscopy system. Membrane deformation, membrane area, and cell volume were measured and tracked at a nano-scale resolution.
3.4 Flow chambers
Flow chambers simulate fluid shear stresses on cells to mimic their physiological environment. A typical flow chamber includes inlet-outlet ports, a vacuum slot, a gasket that specifies the height of the chamber and a glass coverslip that encloses the chamber, the glass coverslip can be coated with different cell layers or proteins [74]. A mouse endothelial blood-brain barrier (BBB) model including dynamic interactions between T cells and spinal cord microvessels was developed [75]. Flow chambers are widely used in the literature to mimic blood cell-endothelial wall interactions to understand disease pathophysiology such as in SCD. Post-capillary venules, where blood cell-endothelium interactions occur in vivo in SCD, were modelled in vitro with cultured endothelium on the chamber walls to study abnormal red blood cell adhesion on the endothelium [76,77].
3.5 Optical microscopy
Optical microscopy tools have been commonly used in studies of cell mechanics. High resolution imaging and 3D volume construction are invaluable for cell deformation and strain measurements. Modern fluorescent and confocal microscopes offer these properties with live cell imaging functions, which have enabled recent advances in the study of cell mechanics. The confocal microscopy allows point-by-point illumination of the samples using a focused laser beam resulting in higher resolution and 3D information. Fluorescence microscopy is based on obtaining images of fluorophore-labelled samples illuminated with a specific wavelength. Furthermore, a novel confocal microscopy-based indentation system was presented for studying chondrocyte mechanics [78]. 3D reconstructions of the cells were obtained and cellular deformations at different controlled loading conditions were evaluated. A fluorescence microscopy-based 3D particle tracking system was developed for motion tracking within a 100 micrometre range [79]. The viscoelastic mechanical response of kidney cells was analyzed using this technique.
4. Micro and nano technologies in cell mechanics
Conventional tools with high sensitivity and accuracy, such as AFM and laser tweezers, have been used extensively for mechanical characterization and the manipulation of cells as described above. While these tools have played an essential role in understanding cell mechanics, they are generally complex, costly and labour-intensive, and they present throughput challenges. Micro/nano tools have been rapidly growing and spreading in the studies of cell mechanics due to their low-cost, easy adaptation and operation, portability, and high-throughput. In this context, MEMS devices for biological studies, which are also known as BioMEMS, provide a great opportunity to study the mechanical aspects of cells (Figure 2).
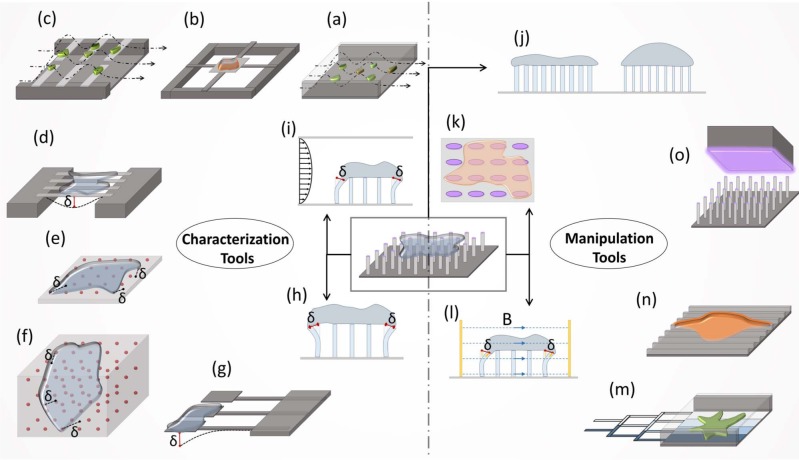
Figure 2.
BioMEMS devices in cell mechanics. The tools can be divided into two main categories: characterization tools, for the measurement of the different physical properties of cells, and manipulation tools, for the exertion of an extrinsic effect. (a) The adhesion strength characterization of cells in microfluidic channels is performed by simply counting the cells remaining after shear flow application. (b-c) Measurement of cell mass (b) in microfluidic chip and (c) on pedestals. Both tools are based on the resonance frequency change of the cantilevers or pad after cell attachment. (d) Cellular deformation measurement is performed by using piezoelectric nanoribbons. (e-i) The characterization of traction forces; (e-f) on 2D or in 3D bead embedded gels from the relative displacement of beads on (g) cantilever pads and (h) vertical micropillars is performed by measuring the deflection of cantilevers or micropillars, and (i) on micropillars under shear flow from micropillar displacement. (j-k) The manipulation of the cells by substrate alterations with micropillar configurations of (j) variable stiffness or (k) anisotropic pillar geometry. (l) Deformation application is performed using magnetic nanowires embedded in micropillars in a magnetic field. (m) The generation of substrate gradients is performed via microfluidics. (n) The manipulation of cell shape and phenotype is performed using nanoridge topography. (o) The generation of substrate patterns is performed using microcontact printing. Micropillar and microfluidic based approaches were found to have a variety of applications as both characterization and manipulation tools.
4.1 Measurement of cellular mechanical properties
As discussed in Section 2, cells maintain a biophysical equilibrium with their microenvironment by probing their surroundings in a sensitive and continuous manner. This equilibrium is interrupted by cells in case of any transformational change such as growth, migration, adhesion and differentiation. A biophysical imbalance between a cell and its environment emerges as traction forces, cell deformation and changes in cell mass, which are discussed in the following sections.
4.1.1 Cellular traction
Researchers promoted various methods for measuring traction forces, such as ultrathin silicone films [80,81], and polyacrylamide (PAA) gels cross-linked at different levels [82,83]. The ultrathin film approach measures the amount of traction force by examining the wrinkling of the film by the cells. Even though this method provided an important insight in earlier studies in the 1980s and 90s, measuring forces from wrinkles is complicated [84]. On the other hand, fluorescent microbead embedded PAA gels provide a more accurate quantification of the traction forces (Figure 2e). For example, Dembo et al. [82] studied the traction forces at focal adhesions during the locomotion of single 3T3 fibroblast cells using collagen conjugated polyacrylamide gels with embedded fluorescent marker beads. Moreover, gel stiffness can be tuned by changing the cross-linking level. However, changing the cross-linking level not only alters the mechanical properties of the substrate, but also has an effect on the porosity, surface chemistry and binding properties of the ligands [85]. Thus, this process makes it hard to isolate the effects of one substrate property change from others.
In the last decade, other methods based on micro/nano cantilevers or pillars have been used extensively to study traction forces (Figure 2g-l) [86,87]. When the cells are cultured on top of functionalized (adding ECM proteins) pillar arrays, they form focal adhesions with the pillar top surface and apply traction forces through these adhesion points. Under these traction forces, pillars behave like simple springs, which translate into forces that are linearly correlated with the deflections of the pillars. Thus, by measuring these deflections, the traction forces of the cells can be calculated.
Pioneers of this approach measured the traction forces of fibroblast cells during migration by utilizing flexible horizontal cantilevers [88]. The BioMEMS device used in the study incorporated mounted horizontal cantilevers and pads, whereby cell-surface interactions occur at the tip of these cantilevers. By imaging the deflection of these cantilevers, they were able to calculate traction forces (Figure 2g). Even though this method overcomes the computational and material complexities of the bead embedded gel approach, it is limited to forces generated at only one direction and location.
Tan et al. [86] investigated the interaction between cells and their substrates by seeding cells on micro pillars of 3 μm in diameter and 11 μm in height (Figure 2h). From the deflection of the pillars, they determined the traction forces applied by the cells and correlated these traction forces with the distribution of the focal adhesion on each post. They found that there were two groups of adhesions causing the traction forces. Forces generated by the first group increased with an adhesion size greater than 1 μm2, whereas there were no such correlations for adhesion sizes less than 1 μm2.
Rabodzey et al. [89] investigated the shear forces induced at cell-cell junctions during the neutrophil transmigration of vascular endothelium by growing endothelium on micro pillars (2 μm in diameter and 3.3 μm to 4.7 μm in height) in an in vitro laminar flow chamber (Figure 2i). They showed an increase in traction force during the intercellular penetration of neutrophils and gap formation. There was also an increase in traction forces applied by endothelial cells in response to the penetration and destruction of cell junctions. Based on these results, they suggested that a successful transmigration of a neutrophil through the endothelial monolayer depends on the competition between the cell-cell junctions and the cell-substrate.
Even though micropillars have great advantages due to their inherently simple structure, there are some limitations associated with them. For example, the nontrivial topology of the micropillars might affect cell adhesion for certain geometrical configurations. Moreover, some cell types require extra soft substrate stiffness which can be hard to achieve using micropillars due to manufacturing requirements [84]. Furthermore, both micropillar and gel approaches employ two dimensional (2D) substrates, which can cause cells to behave in a different way than in their native three dimensional (3D) environment.
In a recent study, Legant et al. [90] investigated the traction forces of EGFP-expressing 3T3 fibroblast cells in 3D elastic hydrogel matrices by exploiting the relative deformation of embedded fluorescent beads (Figure 2f). Even though this study employed a 3D environment for the cells, this approach is still susceptible to computationally intensive data processing. In a different approach, Marelli et al. [91] fabricated flexible curved cantilevers that suspend cells in 3D to measure traction forces. A limitation of this method is the restriction of cells in a confined configuration, which prevents cell migration and cell-cell interactions.
4.1.2 Cellular deformation
Cell deformation can be a significant indicator of the various vital functions of cells such as growth, locomotion and depolarization. Nguyen et al. [92] developed a new method to measure the mechanical responses of cells to electrical stimulations. The authors fabricated piezoelectric PbZrxTi1-xO3 (PZT) nanoribbons to measure the deformation of neuronal cells undergoing electrical excitations. The results showed a 1 nm cell deformation in response to a 120 mV stimulus. This result was in agreement with a theoretical model of a depolarized cell membrane experiencing tension (Figure 2d).
4.1.3 Cell mass
Cell mass can be used as an indicator of protein synthesis, DNA replication and other large molecule accumulation inside the cell during growth and differentiation [93]. Researchers have developed several approaches to measure cell mass incorporating both micro structures and microfluidics (Figure 2c).
Park et al. [93] developed a BioMEMS device that involves a microfluidic channel and horizontal cantilever arrays mounted on the sidewalls of the channel (Figure 2c). In this study, HeLa cells were injected into the channel and captured on the cantilevers by using positive dielectrophoresis. After culturing adhered cells on cantilevers for a period of time, the resonance frequency of cantilevers was measured with a Laser Doppler Vibrometer. Since the resonance frequency is correlated with the spring constant and the mass of the system, cell mass was calculated from the resonance frequency shift of the cantilevers.
Grover et al. [94] also used microfluidics, cantilevers and resonance frequency to measure cell mass using a different approach. The authors developed a microfluidic chip with two different fluid flows at the lateral sides and incorporated a cantilever at the location where the two fluids mixed. When a cell was flowing with the first fluid and passed through the cantilever, the resonance frequency was measured. Next, the fluid flow was reversed and the cell passed through the cantilever a second time, and the resonance frequency was measured again. The resonance frequency of the cantilever was proportional to the cell's buoyant mass in the fluid when the cell passed by the cantilever. From these measurements, absolute density, mass and the volume of a cell can be calculated according to Archimedes' law.
4.1.4 Cellular adhesion
The adhesion strength of cells can be quantified and used for practical purpose. Singh et al. [95] developed a microfluidic chip to measure cell adhesion strength and, then, isolate stem cells based on their specific adhesion strengths. In the microfluidic chip (Figure 2a), cells were subjected constantly to a fluid flow that could detach the cells from the surface. The authors showed significant difference in the adhesion strength between somatic, pluripotent, partially programmed and differentiated progeny cells. They used this variation in adhesion strength to isolate specific cell populations with 95%–99% purity and >80% survival.
4.2 Mechanical manipulation of cells
Initial studies on cell mechanics focused on the mechanical characterization of cells. Later, it was realized that the field required micro and nano instruments that can manipulate and simulate the mechanical environment of cells to further investigate cell mechanobiology. The properties and characteristics of some of these methods are reviewed in this section.
4.2.1 Magnetic pillars
Using a micropillar array (Figure 2l), Sniadecki et al. [96] applied an external magnetic field force to adhere cells to micropillars, some of which had embedded magnetic nanowires. It was reported that applying a magnetic force, which deflects the magnetic pillars, increases the focal adhesion size only at these pillars, but not at the nearby nonmagnetic pillars. The results showed that applying such a force caused a loss in contractility at discrete locations of the cell's periphery.
4.2.2 Shear flow
Ting et al. [97] investigated the effects of fluid shear stress on the cytoskeleton and cell-cell contacts of endothelial cells. In this study, endothelial monolayers were grown on two different micropillar arrays in a flow chamber. One of the micropillar arrays was placed close to the inlet and the other one was positioned around the middle of the chamber, to achieve both disturbed and laminar flow conditions, respectively. It was reported that while laminar flow conditions increased cytoskeletal tension, disturbed flow had a reverse effect.
4.2.3 Protein micropatterning
Protein micropatterning has been used to control and manipulate cell geometry, traction, migration and adhesion. To pattern proteins on a substrate, microfluidic and microcontact printing methods have been used (Figure 2m-o). In microfluidic patterning, proteins are immobilized on a substrate placed inside a microfluidic channel in which gradients are obtained by using a series of serpentine channels mixing different solutions at different ratios (Figure 2m). Dertinger et al. [98] generated laminin gradients to study axonal specification in neuronal cells. In another study, Rhoads et al. [99] investigated fibroblast haptotaxis using fibronectin gradients produced via microfluidics. A challenge of microfluidic patterning is that the patterned substrate remains in a closed system after the pattering is completed.
Another method of protein microcontact printing works by absorbing the protein of interest on a stamp and putting this stamp in conformal contact with a microengineered substrate (Figure 2o). Tan et al. [86] functionalized the top surface of micropillar arrays with ECM proteins by using microcontact printing. Protein functionalization was applied either to all of the pillar arrays or to a constrained smaller area. This approach allowed cell behaviour and traction forces to be analyzed in a restricted area.
4.2.4 Topographic modification
Topographical cues on a surface are crucial for cell morphology, migration and differentiation. Therefore, Teixeira et al. [100] investigated the morphological and biophysical behaviour of human corneal epithelial cells on substrates with nano grooves and ridges (Figure 2n). The study utilized topographically patterned substrates with feature dimensions ranging from as small as 70 nm to 2.1 μm, feature pitch between 400 nm to 4 μm, and groove depths of 150 nm and 600 nm. It was reported that epithelial cells elongated and aligned on substrates with ridge and groove features, whereas smooth surfaces caused the cells to assume round morphologies.
Bucaro et al. [101] investigated the relationship between the geometry of nanopillars (spacing and aspect ratio) and stem cell morphology. They used nanopillar arrays ranging from 0.2 to 0.5 μm in diameter, 5 μm to 10 μm in height, and 0.8 μm to 5μm in spacing. Based on the findings, they proposed a critical spacing distance at which the extensions of cells could only grow in the direction where the inter-pillar distances were the shortest. On the other hand, sub-critical spacing led cells to spread radially as focal adhesions could be established in every direction. However, cells showed no bridging over the nano pillars when over the critical spacing distance. Instead they spread at the base of the nano pillars with increased branching of the extensions. Moreover, a dramatic increase in cell polarization and alignment were observed with the increase in pillar aspect ratio (thus a reduction in the bending stiffness of the pillars).
4.2.5 Substrate stiffness
Saez et al. [102] fabricated elliptical micropillars to obtain anisotropic stiffness characteristics in each pillar to study the directional epithelial growth and the migration of epithelial cells. It was observed that cells migrated in the direction of the greatest stiffness (Fig 2k). Fu et al. [85] investigated the effect of substrate rigidity on cell morphology, focal adhesion, cytoskeletal contractility and stem cell differentiation by using micropillar arrays with different stiffness values (Fig 2j). In this study, stiffness was controlled by varying the pillar height. Results showed that cells displayed spherical morphology at lower stiffness arrays, whereas they displayed spreading on more rigid arrays. MSCs cultured on rigid substrates tended to an osteogenic fate, whereas on soft micropillar arrays, they favoured an adipogenic fate.
5. Emerging areas of application in biology and medicine
Micro and nano technologies are used in a broad range of applications, including single cell analysis, cancer cell mechanics, organ-on-a-chip systems, pathogen detection, implantable devices, and neurobiology. These emerging applications are reviewed in this section.
5.1 Single cell analysis
Single cell isolation is a micro-scale mechanical technique crucial for understanding processes at a cellular level. With single cell analysis, important variations in a cellular population can be detected and analyzed, which is not possible in a bulk analysis. It opens new dimensions for the study of rare cells, such as stem cells, circulating tumour cells, and biological samples collected from patients. The mechanical trapping of single cells in microfluidic channels include lateral and planar trapping using side structures or U shaped micro-apertures or microwells located under the cells [103-110]. Another way to trap single cells is to employ pneumatic valves integrated in microfluidic systems [111]. Nanolitre volumes of reagents can be applied to the isolated cells by using integrated valves and pumps. Several assays based on this system were developed including cell viability. These include assay and ionophore-mediated intracellular Ca2+ flux measurements, and multistep receptor-mediated Ca2+ measurements.
A third method to trap single cells is droplet encapsulation, in which individual cells can be isolated in extremely small volumes [112]. After capturing the cells inside droplets, they can be easily used in various assays, including, cytotoxicity screening and viability [113], isolation and protein/DNA purification [114-116], therapeutic applications [117] and subcellular organelle studies [118].
The biochemical analysis of single cells has been considered one of the holy grails in cell biology and has become possible with microfluidic technology. There are different protocols for dissolving the cellular membrane to access the intracellular contents. Chemical lysis, using detergents, using alkaline conditions, electrical lysis, laser lyses, mechanical lysis and thermal lysis can be used for lysing the cellular membrane [119-125].
Important developments have been made in microfluidic systems and these have been integrated with major analytical methods currently used for genomics and proteomics for single cell analysis. The gene expression analysis of single cells has been demonstrated in various studies such as the quantification of mRNA from two distinct populations [126] and the combination of microfluidic systems with qRT-PCR [127]. Microfluidic systems can be used to study single cell proteomics. A challenge in single cell proteomics is the amount of protein that can be isolated from an individual cell. The concentration of some proteins in a cell may be extremely low, and they cannot be amplified as in DNA and RNA isolation. Therefore, sensitivity is essential in single cell proteomics. In such a system, a microfluidic approach was used to manipulate, lyse, label, separate, and quantify the b2 adrenergic receptor contents of a single cell using single-molecule fluorescence counting [128]. As another approach, a microfluidic system incorporated with a mass spectroscopy was used to analyze proteins in a single cell [129].
5.2 Cancer cell mechanics
Cancer cell mechanics provides a promising opportunity to understand how cancer cells malignantly grow, transform, aggressively spread and invade normal tissues. Cancer cells are known as malfunctioning biological cells in the human body, which can uncontrollably proliferate and disrupt the organization of tissue [26]. Cancer cells tend to adapt themselves to squeeze and spread in normal tissues or blood vessels. Therefore, they deform more easily than normal cells, which was observed in many studies using breast cancer cells [12,67,130], hepatoma cells [131,132], and HeLa cells [133,134]. Such differences in mechanical properties may be regarded as an inherent marker for cancer diagnosis and treatment. On the other hand, cancer cells may respond and behave differently to external stimulations (extracellular matrices and induced forces). For example, cancer cells were found to exhibit a larger traction force than normal cells by approximately 20% for HeLa cells and 50% for L929 cells on micropatterned substrates [134]. Cancer cell behaviours such as adhesion, migration and division were studied under different mechanically and magnetically induced stimulations [135-137], which help us understand metastatic mechanisms.
Currently, the integration of MEMS and imaging techniques [133,136-140] are inspiring more interest in exploring how cancer cells respond to ECM and external forces. The conversion of these mechanical signals into chemical signals results in adaptive changes in cellular behaviours, such as cell adhesion, migration and division.
BioMEMS devices, to date, have played a dominant role in the studies of cancer cell mechanics due to the following reasons: (1) BioMEMS devices provide a platform which can better mimic the in vivo environment. For example, micropatterned matrices [133] for studying cell exerted traction forces and migration can better mimic the microenvironment of cancer cells. (2) BioMEMS devices exhibit higher precisely-controlled and spatially-resolved forces. Micropost arrays [86,134,141] and microfluidic assays [142] can precisely control forces by changing geometry or fluid velocity. (3) BioMEMS devices enable the analysis of cancer cells with higher accuracy and throughput compared to conventional tools.
Techniques to study cancer cell mechanics can be categorized in terms of their applications in cancer cell mechanics and cell-ECM interactions (Figure 3). Principles, advantages, limitations and applications are summarized in Table 2. AFM-based methods (Figure 3a) [67,68], magnetic twisting cytometry (Figure 3b) [131,143] and cytoindentation (Figure 3c) [12,144] usually exert local forces on a cell. Therefore, they suffer from low throughput and direct contact with the cell surface may cause active cellular responses. Single-cell-based techniques such as microplate stretcher (Figure 3d) [145,146] and micropipette aspiration (Figure 3e) [132,147,148] have modest throughput, but they also cannot avoid cell-tool interactions either. Optical techniques such as optical tweezers (Figure 3f) [149-152] and optical stretchers (Figure 3g) [153-155] minimize active cell responses when deforming cancer cells without contact. The optical stretcher takes advantage of a microfluidic channel to mimic the in vivo environment and uses two counter-propagating divergent laser beams to suspend and deform cancer cells. The laser induced forces can be precisely controlled by output power. For example, using this technique, Guck et al. found that the deformability of SV-T2 cells was significantly higher compared to BALB/3T3 cells [155].
Figure 3.
Major techniques for cancer cell mechanics study. (a) Atomic force spectroscopy; (b) magnetic twisting cytometry; (c) cytoindentation; (d) microplate stretcher; (e) micropipette aspiration; (f) laser/optical tweezers; (g) optical stretcher; (h) shear flow; (i) microfluidic assay; (j) microfabricated post array; (k) particle tracking microrheology; (l) magnetic nanoparticle-based stimuli. This figure [26] is reused with permission from Elsevier.
Table 2.
Methods for studying cancer cells mechanics and mechanical properties of cancer cells reported in the literature
| Methods | Induced force | Advantages | Disadvantages | Applications | References | ||||
|---|---|---|---|---|---|---|---|---|---|
| Case study | Major observations | Typical values | |||||||
| Cancer cells | Normal cells | ||||||||
| Atomic force microscopy (AFM) | Partial of cell | 10−7–10−11N | 1) High resolution; 2) Low forces with minimal disruption; 3) Three dimensional surface profile. |
1) Can be used only for cells that adhered to a substrate; 2) Relatively slow scanning rate; 3)Mechanical contact may lead to cellular response. |
Stiffness of metastatic cancer cells from lung, breast and pancreas [67] | Stiffness of metastatic cancer cells is more than 70% softer than the benign cells | Lung: 0.56±0.09kPa; Breast: 0.50±0.08kPa; Pancreas: 0.54±0.08kPa; | Lung: 2.10±0.79kPa; Breast: 1.93±0.50kPa; Pancreas: 0.54±0.12kPa; | [67,68,257] |
| Stiffness and adhesion forces of metastatic cancer cells and benign mesothelial cells [68] | Metastatic tumour cells are more than 80% softer than benign cells and surface adhesion is ∼33% less than normal cells. | Stiffness: 0.38±0.20kPa; Adhesion force: 34.2±5.3pN. | Stiffness: 2.53±1.30kPa; Adhesion force: 51.1±15.2pN. | ||||||
| Magnetic twisting cytometry (MTC) | Partial of cell | 10−10–10−12N | Probe the single cell with very small deformations and over wide ranges of time scale and amplitude. | 1) The bead localization on the cell is random; 2) The induced bead rotation and displacement are strongly dependent on the bead attachment angle. |
The effects of tubeimoside I (TBMS I) on human hepatoma (HepG2) cells[131] | The stiffness of HepG2 cells decreased consistently with the increased concentration of TBMS I exposure. In addition, the HepG2 cells responded to TBMS I much faster than the normal liver (L-02) cells. | Stiffness (HepG2): 0.44±0.01 Pa/nm; Respond time: 73s. | Stiffness(L-02): 0.88±0.04 Pa/nm, Respond time: 109s. | [131,143,257] |
| Cytoindentation | Partial of cell | 10−7–10−9N | 1) Simple modelling of the viscoelastic behaviour of the cells; 2) Detection of differences in cell mechanics; 3) The probe is large to measure bulk cellular properties. |
The response may depend significantly on the precise probing location. | The elasticity of benign (MCF-10A) and cancerous (MCF-7) human breast epithelial cells [12] | Apparent Young's modulus of malignant (MCF-7) cells significantly decreased, (1.4–1.8 times) than that of non-malignant (MCF-10A) cells at physiological temperature (37°C), and their apparent Young's modulus increased with loading rate. | – | – | [12,144,257] |
| Microplate stretcher | Single cell | 10−7–10−9N | Sufficient to induce significant deformation of an entire cell. | 1) Time consuming; 2) Low throughput; 3) Can only be applied to extremely well adhering cells. |
Elastic response and energy dissipation under repeated tensile loading of epithelial pancreatic cancer cells [146] | The elastic modulus of Panc-1 pancreatic cancer cells decreased after treatment with SPC. | Before SPC: 28.8 ± 2.6 mN m−1. After SPC: 16.3 ± 1.1 mN m−1. | – | [145,146,257] |
| Micropipette aspiration(MA) | Single cell | 10−7N- 10−10N | 1) Allows for real time correlation of pressure and whole cell deformation; 2) High accuracy; 3) Aspiration pressure can be maintained over a specified duration. |
Analytical or computational models are often necessary to derive material properties and the underlying assumptions may at times be difficult to validate. | Viscoelastic properties of human hepatocytes and hepatocellular carcinoma (HCC) cells[132] | HCC cells have higherelastic coefficients but not viscous coefficients compared to than hepatocytes. | Hepatocellular carcinoma (HCC): K1=103.6±12.6N.m−2; K2=42.5±10.4N.m−2; μ=4.5±1.9Pa.s. | Hepatocytes: Kl=87.5±12.1N.m−2; K2=33.3±10.3N.m−2; μ=5.9±3.0Pa.s. | [132,147,148, 156,257] |
| Laser/optical tweezers(OT) | Single cell | 10−11–10−14N | A focused laser beam allows precise bead manipulation in all directions. | 1) Force level is limited to induce larger deformation; 2) Larger force would require higher laser power that could excessively heat the cell. |
Elasticity of myeloblasts (62–71 CD33+CD34+cells and 57–63 CD33+CD34-cells) from AML patients[150] | The induced deformation of CD33+CD+cells is greater than CD33+CD34-cells under the same stretching force. | The elastic area compressibility modulus, kα= 1.40±0.71 N/m (CD33+CD34-). | The elastic area compressibility modulus, kα= 0.25±0.15 N/m (CD33+CD34+). | [168–171, 267] |
| Optical stretcher | Single cell | 10−9–10−11N | 1) Cells can be suspended to eliminate mechanical contact; 2) Very small numbers of cells are required for distinction; 3) Relatively high throughput using a microfluidic channel. |
1) Laser power should be controlled without damaging the cells; 2) Limitations may exist when probing stiffer cells. |
Optical deformation (OD) of mouse fibroblasts and human breast epithelial cells [155] | Optical deformability of the SV-T2 cells is significantly increased compared to the BALB/3T3 cells; the cancerous MCF-7 cells are deformed more than the normal MCF-10 cells, and the metastatic modMCF-7 are deformed even more than the nonmetastatic MCF-7. | ODSV-T2 = 11.7±1.1 ODMCF-7 = 21.4±1.1; ODmodMCF-7 = 30.4±1.8. | ODBALB/3T3 = 8.4±1.0; ODMCF-10 = 10.5±0.8. | [153-156,257] |
| Shear flow | Cell populations | 1–100Pa | Cone and plate rheometers allow precise control over the applied shear stress. | 1) Difficult to visualize induced cellular deformations; 2) Small variations in the cell height and topology can cause local variations of shear stress. |
Influence of shear flow on the adhesion of nonmetastatic (MCF-7) and highly metastatic (MDA-MB-435) cells[135] | Detachment of the nonmetastatic MCF-7 cell line decreased significantly while detachment of the highly metastatic MDA-MB-435 significantly increased after 15 hour exposure of a 15 dyn/cm2 shear stress. | Detachment (MCF-7) decreased from 44.0±4.6% to 12.1±3.7%; Detachment (MDA-MB-435) ncreased from 37.2±6.3% to 36.2±2.1%. | – | [135,257,258] |
| Microfluidic assay | Cell populations | – | 1) High throughput; 2) Can mimic in vivo environment; 3) High accuracy; 4) Easy to fabricate and low cost. |
Microfluidic channels need to be properly designed. | Deformability of benign breast epithelial cells (MCF-10A) and nonmetastatic tumour breast cells (MCF-7) [130]. | Transit velocity is not significantly affected by cell type. MCF-10A cells were found to have longer entry time than MCF-7 cells of similar sizes, MCF-10A is stiffer than MCF-7 cells. | MCF-10A intry time: 1.698±0.201s; ilongation index: 1.231±0.01191; Transit velocity: 187.0±7.920μm/s. | MCF-7 Entry time: 0.433±0.045s; Elongation index 1.281±0.01505; Transit velocity: 177.3±9.836 μm/s. | [130,142,259] |
| Microfabricated post array | Single cell/Cell populations | 10−7–10−9N | 1) Force application is localized and can be measured with high resolution; 2) The geometry and stiffness of micropillars can be independently adjusted; 3)Can be applied for cell adhesion force, traction force and migration studies. |
1) Substrate has a nontrivial topology that might affect cell adhesion and bias the measurements; 2) Substrate may cause active cell-substrate response. |
Traction forces exerted by cancer cells [134] | Cancer cell exhibits a larger traction force than the normal cell by ∼20% for a HeLa cell and ∼50% for a L929 cell. | Traction forces of Hela cell: 2.84±0.49 μN and L929 cell: 3.48±0.46 μN. | Traction forces: 2.3210.16μN. | [86,134,141,257] |
| Nanoparticle-based techniques | Cell populations | – | It can provide insight into intracellular dynamics and structure, as well as into active transport processes. | 1) Hard transferring of experimental observations to theoretical and phenomenological models; 2) Nonlinear effects. |
ECM stiffness on the intracellular rheology of cancer cells [33]. | In 3D matrices, the intracellular effective creep compliance of prostate cancer cells is shown to increase with increasing ECM stiffness, whereas modulating ECM stiffness does not significantly affect the intracellular mechanical state when cells are attached to 2D matrices. | – | – | [133,136-140,257] |
| Partial of cell/Cell population | 10−7 −10−9N | 1) High spatially and resolved force with long duration and repeats. 2) Localized stimuli and cell population measurement are both achieved, which increases the throughput. |
1) System is complex, which limits its real application; 2) Both cells and magnetic elements are needed to be patterned, and the alignment between them should be precisely controlled. |
The influence of mechanical forces on single-cell behaviour [137]. | Nanoparticle-induced tension generates filopodia asymmetry and bias metaphase-plate orientation of Hela cells. | – | – | ||
Microfluidic techniques provide versatility in cancer cell mechanics under precisely controlled fluid flow. For example, using the shear flow technique (Figure 3h), Moss et al. [135] found that the detachment of the nonmetastatic MCF-7 cell line decreased significantly while the detachment of the highly metastatic MDA-MB-435 significantly increased after a 15 hour exposure of a 15 dyn/cm2 shear stress. In another example, Tan et al. [86] used microfluidic assays (Figure 3i) as a “deformation passage” for the cells to pass through, and MCF-10A cells were found to have a longer entry time than MCF-7 cells of similar sizes indicating that MCF-10A was stiffer than MCF-7 cells.
Cell traction force is important for many biological processes, such as mechanical signal transmission and cell migration. Therefore, measuring cells' exerted traction forces may provide a better understanding of cancer cell metastasis [154,156]. The micropost array technique (Figure 3j) has been previously used to measure the traction forces of cancer cells [86,134,141]. Similar to the micropost array, Li et al. [134] presented a silicon-nanowire-array-based technique for quantifying the traction forces of three distinct groups of cells: normal mammalian cells, benign cells (L929) and malignant cells (HeLa). The results indicated that cancer cells exhibited a larger traction force than normal cells [134].
To study the interactions between the mechanical properties of ECM and those of the cancer cells, Baker et al. [133,136,138-140] developed an innovative fluorescent nanoparticle-based tracking microrheology method (Figure 3k) [136]. They showed that the intracellular effective creep compliance of prostate cancer cells increased with increasing ECM stiffness in 3D matrices, whereas modulating ECM stiffness did not significantly affect the intracellular mechanical state when the cells are adhered to 2D matrices [136].
Magnetic nanoparticle induced stimuli (Figure 3l) [137] is an innovative approach whereby magnetic fields are used to exert highly localized and spatially resolved forces on the cell membrane. Using this technique, Tseng et al. [137] observed that magnetic nanoparticle induced tension could generate asymmetrical filopodia. Furthermore, as particle-applied forces increased, filopodia protrusions appeared more frequently, emanating from the region to which the forces were applied.
5.3 Organ-on-a-chip systems
Cell culture technologies have a broad range of applications in cell and molecular biology research, tissue engineering and drug screening assays. The traditional, yet-still-prevalent 2D cell culture typically seeds cells on the surface of plastic flasks, petri dishes or well plates, where a cell monolayer is formed within the bulk culturing medium. Despite the 2D nature of these systems, numerous biological studies have been performed based on this platform [157-160]. 2D cell cultures have certain limitations: the cell culture conditions poorly mimic the cellular environment in vivo, soluble growth factors can be present at abnormally high concentrations, 3D cues are largely absent, oxygen tension can be too high, and cell–cell interactions are rarely seen [161].
The pursuit of gaining a better understanding of the effect of living tissue environment on cells, cell-cell interactions and the response of cells in a natural environment has led to the wave of research efforts aiming to build more natural conditions, termed 3D cell cultures. Leveraging microfabrication techniques such as soft lithography, microfluidics and micropatterning, superior 3D cell culture systems have been developed [162-166]. These techniques can create more natural environments by mimicking 3D ECM structures, applying microfluidic networks capable of transporting nutrients and oxygen, and exerting mechanical loads on cells. The study of the seeding, stimulating and proliferating of cells under these conditions has provided a more accurate understanding of cell mechanics and the effect of environmental cues [167-170].
Microfabrication has enabled co-cultures of various cell types in a single platform with physiological environmental conditions. These platforms are called organ-on-a-chip devices when these sytems are specialized to mimic the function of a specific organ. The development of these microengineering approaches has opened up new possibilities for creating in vitro models that reconstitute more complex 3D organ-level structures and for integrating crucial dynamic mechanical cues as well as chemical signals [166,170,171].
There have been encouraging breakthroughs in devising and constructing devices that resemble the structure of the human lungs [172-174], liver [175-178], intestines [164,179,180], kidneys [181,182], cancer tissue [183] and artificial cells in regenerative medicine [117,166,170]. These devices are able to perform either specific healthy physiology functions or pathological conditions of human organs. As an example, the human lung-on-a-chip system was used to show that lung tissue responds differently to bacteria and inflammatory cytokines in the presence of a cyclic mechanical strain [172]. Mechanical cues were shown to accentuate the toxic and inflammatory responses of the lungs to silica nanoparticles, as similar effects were observed in a whole mouse model, proving the system's potential for drug screening and toxicology applications [172].
Recently, a multi-organ-chip with co-cultures of 3D human artificial liver and skin tissues has been designed and tested [184]. The system was shown to support two different culture modes, with tissue exposed to fluid flow or tissue shielded by standard cultures from the underlying fluid flow. This system has provided long-term cultures over 28 days, while supporting tissue crosstalk.
The development of microfabrication technologies to maintain cells in their 3D natural environments in vitro and construct higher organ-level microsystems is still in its primitive stages. Researchers are still exploring how to optimize the properties of culturing systems by exploiting more biocompatible materials that better mimic ECMs, to improve cell viability, to form organized cellular structures and to integrate more functions into a single platform. Micro and nano technologies hold enormous possibilities, and they provide powerful and promising approaches for the next generation of organon-chip platforms.
5.4 Pathogen detection based on physical properties
Pathogens, including bacteria, viruses, microbes and other microorganisms, are associated with many different fields of research, including diagnostics, pathology, drug discovery, clinical research, biological warfare, disease outbreaks and food safety [185]. Conventional pathogen detection methods involve cell culture, polymerase chain reaction (PCR)-based methods and enzyme-linked immunosorbent assays (ELISA) [186]. Despite the high accuracy and sensitivity, conventional pathogen detection methods are time-consuming due to the complex procedures involved. Recent advances in BioMEMS techniques provide new opportunities to develop biosensors for pathogen detection with simpler processes and smaller dimensions [187-190]. A biosensor is an analytical device that combines a sensitive bioreceptor element with a biological signal transducer to detect an analyte [191]. A bioreceptor (e.g., cell, microorganism, enzyme, antibody, nucleic acid) is used for interactions with the analyte, while a biological signal transducer can convert a signal introduced by the interaction of the analyte with a biological element into another signal that can be more easily measured and quantified. Generally, transduction principles can be classified into three primary categories: optical, electrochemical, and mechanical.
Optical-based biosensors share the advantages of high sensitivity, flexibility and resistance to electrical noise [192]. The most popular optical-based biosensors can be divided into three types: surface plasmon resonance (SPR), chemiluminescence and fluorescence. Due to the advantages of label-free, high sensitivity and real-time measurements, SPR biosensors have attracted much attention recently, and many commercial biosensors based on SPR have been employed in a range of applications, from fundamental studies to clinical diagnosis [193].
Chemiluminescence is a kind of light generated during a chemical reaction, containing electrochemiluminescence (where the luminescence comes from the electrochemical reaction) and bioluminescence (where the emission is produced by living organisms). Chemiluminescence-based methods are superior to other optical methods, because the absence of an external light source not only simplifies the method, but also reduces the detection noise [194]. Paper-based chemiluminescence ELISA is capable of achieving a good sensitivity and linear range for different antigens, which is needed in clinical applications [195]. Based on the chemiluminescence technique, Wolter et al. [196] developed the first flow-through chemiluminescence microarray. This semi-automated readout system provided rapid and simultaneous detections of Escherichia coli, Salmonella typhimurium and Legionella pneumophila within 13 mins, with detection limits of 105, 3×103 and 3×106 cells/mL. A chemiluminescence (CL) flow-through DNA microarray assay was later reported by the same research group. By introducing the stop-PCR method, this system achieved a lower detection limit of 10–100 cells/mL compared to an antibody microarray [197].
Fluorescence is the dominant optical detection method for pathogen detection, which is based on the relaxation process whereby the excited electron returns to its ground state [194]. In addition to its general merits, such as high sensitivity and easy incorporation into microfluidic devices, fluorescence detection displays a distinct advantage in terms of its detection limit for low signal cross-talk from other species, because only structurally rigid compounds with unsaturated or aromatic functional groups can emit fluorescence [194].
Mechanical-based biosensors are generally composed of microcantilever systems based on different sensing principles: stress detection or mass detection. For the stress detection mode, biochemical interactions between pathogens and the sensitized surface of a cantilever result in the alteration of surface-free energy and surface stress on both sides. Consequently, the mechanical deflection of the cantilever can be measured along with the label-free detection of the bioanalyte [204]. For the mass detection mode, a biosensor equipped with a piezoelectric surface immobilized with antibodies is placed in a solution containing pathogens. Then an increase of the crystal mass is generated because of the attachment of the agent to the antibody coated surface, resulting in a measurable corresponding resonance frequency shift [197]. This detection method generally includes three types of applications: quartz crystal microbalance (QCM), surface acoustic wave (SAW) and magnetoelastic detection [210-212].
Implantable medical devices, some of which are BioMEMS-based systems, have been used to restore lost or damaged organ functions in the body, benefiting many people and improving their quality of life. The most important considerations concerning the implantation are safety and the reliable performance of the devices in their designed lifetime. Furthermore, the implanted devices have to be compatible with the mechanical properties of the organ, tissue and the microenvironment of the cells within [171,198]. In some cases, the systems stay inside the body for years and some even stay for a life time. Therefore, biocompatibility is always at the top of the list of critical requirements. Neural prostheses are an important application of BioMEMS devices [199,200]. Using electrical pulses to simulate intercellular communication can help restore the lost neural activity. For example, retinal implants are designed to restore sight [201,202] and cochlear implants are designed to restore hearing [203,204]. There are two types of retinal implants, epiretinal implants and subretinal implants. The selection is based on the condition of the patient. If the patient's photoreceptors are not functional, then a subretinal implant is selected, otherwise, an epiretinal implant can be used [201,202]. If the optic nerve is damaged or degenerated, retinal implants are no longer an option and, in this case, brain implants are utilized through the occipital lobe, which is responsible for processing visual signals. Compared to retinal implants, this approach is more challenging due to the complex organization of the nerve cells and the mechanical properties of the soft brain tissue. There have been significant research efforts to design and develop the next generation of implantable BioMEMs devices, which embody complex functionalities and better adapt to the physical environment of the host tissue or organ [205-207].
5.5 Neuroscience
Neurons are widely regarded as one of the most complex cell types within the body. Their unique anatomy strongly influences their electrochemical function of signal propagation and transmission.
The shape of an axon greatly affects signal conductivity spatially or temporally. As such, the physical geometry of a chip on which neurons are interfaced is of critical importance. Another consideration in basic design is the electrode; it is an essential tool that can both measure neuronal performance and apply stimuli. Since both platform geometry and electrodes are generally the most critical aspects in neuroscience research, the primary driving factor in the choice of topography and electrode type of BioMEMS is the type of research being performed.
The actual physical geometry of the micro-scale platform remains the most crucial feature for the development and research of BioMEMS neurons. Neurons are commonly segregated from each other and isolated into individual chambers. The design of these small compartments allows for the direct confinement of neuronal somas, axons and dendritic branches in separate chambers connected by a series of micro tunnels [208-210]. By using this basic confinement technique, a wide variety of research options becomes possible, as specific structures can be isolated and investigated.
Although electrodes have diversified into many different shapes and materials in order to fulfil different roles, their primary functions of stimulation and recording remain the same. A common two-dimensional multi-electrode array consists of a configuration of flat contacts on which neurons are cultivated or patterned [211]. An example of this is demonstrated with neural progenitor cells that are patterned directly onto a multielectrode array (MEA) in order to monitor elicited bursting activity [212]. (Other types of MEAs, such as needle arrays, will be mentioned later).
Developments in optogenetics have allowed for the creation of genetically modified neurons that experience fluoresce when activated or that can be stimulated using light. Recent developments in single-cell fluorescent manipulation have resulted in the creation of a probe with a microscopic tip that is resolute enough for individual cells [213]. The optical fibre can detect individual fluorescing neurons with high reliability in vivo; it can be used to optically stimulate an individual cell without affecting the neighbouring population [213].
Guiding axons in a specific direction using compartmentalization techniques can yield interesting research possibilities. A common technique in BioMEMS research is the culturing of neurons such that they produce rows of axons in parallel microchannels. Other methods for guiding axon propagation involve utilizing techniques such as soft lithography [214]; this process can be used for “inking” proteins or growth factors attracting or repelling the neurites, thus effecting their growth direction. Under certain conditions, axon growth can be polarized into specific directions, allowing for construction of neural circuits [215]. Compartmentalized culturing plays a key role in neuronal co-cultures. Using micro-scale platforms to create small cultures of neurons alongside other bodily cells allows for research into interactions between neurons and other cells. Investigations into behaviour can involve, for instance, circuitry originating from a central nervous system neuron to that of the peripheral nervous system, and ending on a myocyte, replicating a miniature signal pathway from the brain to a muscle [208].
Co-cultures of different types of cells are useful when attempting to mimic the environment in vivo. Takeuchi et al. [216] co-cultured rodent superior cervical ganglion neurons and ventricular myocytes, and then seeded them together; neural connections were made and the electrical activity monitored.
Areas of research that focus on axon manipulation frequently involve disease models where a neuron has sustained some sort of physical damage. Axotomy procedures are used to simulate the severing of the axon, something which could otherwise occur to victims of traumatic accidents. Methodologies developed for inflicting damage to the axon may involve processes such as laser transection, where a precision laser beam severs the axon [217]. Another method for axotomy is using vacuum aspiration, when a pinpoint vacuum is applied to an extended region of the axon in order to destroy that section [218]. Yet another methodology that has been demonstrated involved a miniature device that pinched the axon at variable forces to simulate compressive damage [219]. Once the axon has been damaged, the dynamics of the neuron can be studied in order to see how the cell responds to the trauma.
One attribute commonly associated with the development of Alzheimer's disease is the buildup of insoluble beta amyloids that occur as a result of amyloid protein cleaving. These beta amyloids plaque themselves between synaptic connections in the brain and inhibit signals between neurons. BioMEMS platforms were used to demonstrate how the beta amyloid proteins interfere with neurotrophin growth factors, a protein integral to neuron function [220]. Typical cultures in petri dishes produce unorganized tangles of neural fibres that normally hinder the results of these kinds of experiments, but with a BioMEMS neuron culture with a propagated parallel axon, beta amyloids can easily be introduced to the isolated synaptic branches.
5.6 Neurophysiology
Some of the most fundamental questions in the field of neuroscience concern how physiological and behavioural functions are controlled by neuronal circuits, and how these circuits affect each other. To answer these questions, several types of electrophysiological techniques have been developed over the past few decades, such as intracellular recordings with sharp or patch electrodes, or extracellular recordings with single electrodes or stereotrodes. These techniques can provide us with information regarding neural cell function in vivo and in vitro, and are capable of recording the activities of a single cell or several neurons at a time. Unfortunately, these existing methods are not capable of monitoring the complete repertoire of biophysical properties of each cell in a circuit, because of physical and electrical limitations, and the devices' relatively low throughput. It is becoming increasingly evident that in order to map the brain's synaptic characteristics at the micro- and nanoscale, the recording device also needs to be scaled accordingly. In the past decade there have been many attempts to miniaturize and further improve the existing recording tools. As part of this process, many laboratories introduced techniques that attempt to combine the advantages of extracellular electrode arrays with the benefits of intracellular electrodes creating the new era of micro-scale MEA.
In a network of cells, the neuron sending the action potential is called the presynaptic cell and the one receiving it is called the postsynaptic cell. Relationships between neurons can be inhibitory and excitatory depending on the type of neurotransmitter released by the cells [3]. Action potentials can be recorded with both extra- and intracellular techniques (Figure 4a-c), however, the readouts look different depending on the applied technique. When designing a MEA, it is critical that the device has the capability to detect supra- and subthreshold membrane potentials as well. Intracellular recording techniques, such as the ones illustrated by Figure 4b&c, can monitor changes in voltage in the membrane potentials, as well as the current changes caused by the ion flow in the cells, which is one of the main reasons why MEAs need to be able to penetrate the cytosol of the neurons.
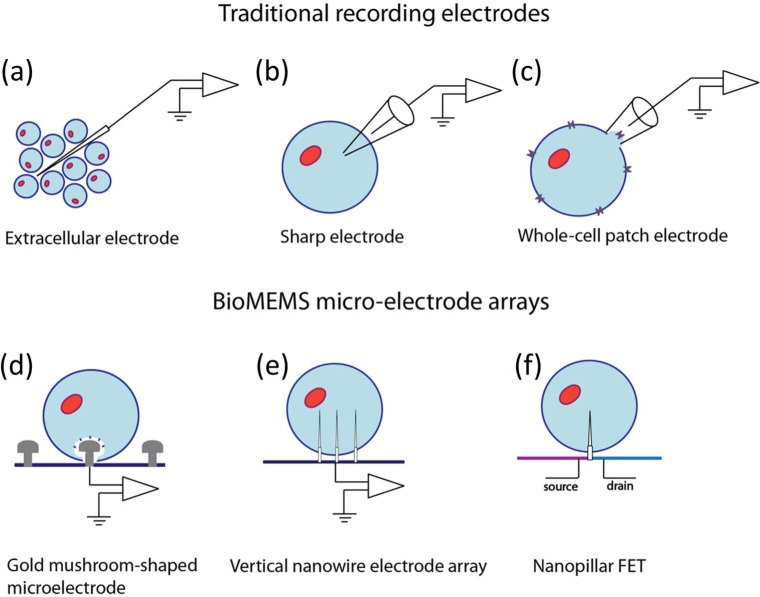
Figure 4.
Different forms of electrophysiological recording techniques. Shown above are schematic illustrations of traditional electrodeneuron interface configurations and BioMEMS microelectrode arrays. In the schematics, neurons are depicted in light blue (somas are marked with orange) (a) Extracellular recording electrode. The electrode does not penetrate any of the cells, thus it can record the activity of multiple neurons. (b) Intracellular recording with a sharp glass microelectrode. (c) Whole-cell patch clamp technique. This technique allows us to study single or multiple ion channels (marked with purple) located on a membrane patch of a single cell. (d) Gold mushroom-shaped microelectrodes are actively engulfed by neurons because of their dendritic spine-like shapes. The mushroom-shaped protrusion is 1.42μm high. (e) A vertical nanowire electrode array (VNEA) that penetrates the cell membrane providing direct contact with the cell. (f) A pillar-shaped protruding nanowire is the sensing gate electrode of the FET.
By utilizing extracellular techniques (Figure 4a) investigators can also monitor the synchronized activity of large ensembles of neurons. This type of additive signal is called local field potential (LFP). In this case, voltage is generated as the sum of the current flow in the population of local neurons [221]. Based on the LFPs recorded, investigators can analyze the network level changes in activity in the brain more generally. These oscillations combined with the intracellular properties and potentials of single neurons can carry substantial information regarding the network level and single cell modulations that can be highly meaningful when characterizing a brain region's connectivity.
As mentioned previously, an ideal MEA needs to utilize the advantages of intracellular and extracellular recording systems, and do so in a high throughput manner in order to maximize the amount of information gained from a recording. These arrays need to have great electrical coupling with each single cell they are recording from, like glass sharp and patch electrodes do, but without having to worry about the short duration of the recording due to mechanical or biophysical instabilities. Therefore, they need to be flexible enough that small distortions will not cause them to break, but stiff enough that they can keep a recording stable for longer periods of time (days or even months). An ideal device needs to be able to record the relevant transmembrane potentials, action potentials, EPSPs and IPSPs, as well as LFPs [222].
Since 2007, Spira et al. have been working on creating a new type of MEA that has an improved cell adhesion property compared to conventional recording techniques [223-227]. As a result of this project, they were able to create an electrode design that is capable of establishing not only chemical, but also biological adhesion with the cell. As illustrated by Figure 4d, the electrodes on the MEA are specifically shaped to a micrometre sized gold mushroom protrusion that is covered with a chemical attractant to create a tight connection with the neuron [227]. This camouflaged appearance contributes to a higher electrical coupling because the cells tend to actively engulf the electrode by endocytosis. The MEA when tested in Aplysia neuron culture showed no significant changes in input resistance before and after the recording, and stimulation session, thus the array can be used to stimulate the cells to fire action potentials without causing any damage to them. The array was capable of recording action potentials of up to 25 mV intracellularly from multiple cells over two days due to the stable cell-electrode coupling [227], which has great potential for application to in vivo chronic recordings.
A different approach was taken when Robinson et al. fabricated electrodes utilizing nanopillars for intracellular recordings [228-230]. In this study they created vertical nanowire electrode arrays (VNEAs) that allowed parallel electrical connections with ensembles of rat cortical neurons. The schematic illustration of the VNEAs is shown in Figure 4e. The VNEA's planar geometry makes it a great candidate for high throughput in vitro recordings, as well as recordings performed in slice preparations; however, it is not ideal for in vivo preparations. When tested in rat cortical neurons, approximately half of the electrode tips penetrated the cells spontaneously. The other 50% successfully penetrated after a short electroporating current was applied to the cells, which is one of the major design flaws. It has been shown that recordings following electroporation are only transient, because the current activates cell repair mechanisms that close up the membrane and consequently make the electrode extrude[225]. Despite this problem, the array provides a promising new method that is capable of short-term, multi-site, single cell, high-throughput, intracellular recordings [222,230].
6. Future directions
Cell mechanics as a research area is a crucial discipline that bridges cell biology and biochemistry with the help of micro and nano-engineered technologies. Experimental and computational mechanics provide a detailed understanding about essential connections among structures, mechanical properties and functions of cells. Although many efforts in cell mechanics have already been aimed towards understanding how cells move, sense, deform and interact with their microenvironment, we must continue to study how the mechanical properties of cells change during a state of disease and how these changes impact signaling processes. Despite remarkable progress to date, there are still many opportunities as yet unexplored to study cell mechanics in reproduction, tissue repair as well as in a long list of diseases. BioMEMS devices open new venues in studying mechanical aspects of cells due to their cost-effective, relatively easy fabrication, and user-friendly nature. Even though BioMEMS devices have given biomedical researchers unprecedented capabilities in cell mechanics, there is still room for advancement and improvement, in characterizing and manipulating cells mechanically. An important direction is nano and micro-scale sensing technologies that can adapt to the 3D environment of cells.
We are still confronted with the challenges of exploring and understanding the mechanotransduction scheme, and metastatic mechanism of cancers. The solutions to these problems lie in quantifying the interplay between cancer cell mechanics and the underlying chemistry. Cancer cell mechanics is still a fledgling field, which requires a better understanding of the mechanotransduction scheme and the metastatic mechanism, and is inspiring more innovative techniques to translate those experimental results into clinical applications.
Some areas of neuroscience have also benefited from BioMEMS. These miniature systems provide us with an improved ability to physically manipulate neurons, and initiate precision signal stimulation and recording. As we attempt to more closely mimic in vivo human body environments, it is likely that we will see more complex neural patterning, accurate three-dimensional cultures, better manipulation of neuronal behaviour, more efficient neural-electrode interfaces, and an increase in the number of implantable devices.
From a more general perspective, today's BioMEMS technologies are basically building small channels or simple solid structures, and most of these tools rely on the simplest mechanical laws. To put this in an analogy, the tools that we have so far are similar to primitive mechanical tools that people were using during the middle ages. Since the renaissance, with advancements in power generation, electrical and information breakthroughs, we have had versatile, automated, durable mechanisms in almost all of the tools we use every day. For the future of BioMEMS devices in biomedical areas, the incorporation of mechanical, electrical, optical and information technologies has great potential for developing efficient, versatile and comprehensive analysis tools. A single microengineered device that could measure multiple quantities in a short amount of time would be the ultimate quest of BioMEMS devices.
7. Acknowledgement
The authors wish to thank Daniela Solomon from Case Western Reserve University Kelvin Smith Library for her help in performing the literature search.
8. Conflict of Interest
The authors declare no conflicts of interest. No part of this study was performed on any human or animal subjects.
References
- [1].Gurkan UA, Akkus O. (2008) The Mechanical Environment of Bone Marrow: A Review. Ann Biomed Eng 36: 1978–1991. [DOI] [PubMed] [Google Scholar]
- [2].Bershadsky A, Kozlov M, Geiger B. (2006) Adhesion-mediated mechanosensitivity: A time to experiment, and a time to theorize. Current opinion in cell biology 18: 472–481. [DOI] [PubMed] [Google Scholar]
- [3].Krishnan R, Park CY, Lin Y-C, Mead J, Jaspers RT, et al. (2009) Reinforcement versus fluidization in cytoskeletal mechanoresponsiveness. PLoS One 4: e5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vogel V, Sheetz MP. (2009) Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Current opinion in cell biology 21: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bukoreshtliev NV, Haase K, Pelling AE. (2012) Mechanical cues in cellular signalling and communication. Cell and tissue research: 1–18. [DOI] [PubMed]
- [6].Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, et al. (2011) Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 469: 226–230. [DOI] [PubMed] [Google Scholar]
- [7].Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, et al. (2005) Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell motility and the cytoskeleton 60: 24–34. [DOI] [PubMed] [Google Scholar]
- [8].Brock A, Chang E, Ho C-C, LeDuc P, Jiang X, et al. (2003) Geometric determinants of directional cell motility revealed using microcontact printing. Langmuir 19: 1611–1617. [DOI] [PubMed] [Google Scholar]
- [9].Thiery JP, Acloque H, Huang RY, Nieto MA. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890. [DOI] [PubMed] [Google Scholar]
- [10].Zhu C, Bao G, Wang N. (2000) Cell mechanics: Mechanical response, cell adhesion, and molecular deformation. Annu Rev Biomed Eng 2: 189–226. [DOI] [PubMed] [Google Scholar]
- [11].Cross SE, Jin Y-S, Rao J, Gimzewski JK. (2007) Nanomechanical analysis of cells from cancer patients. Nature nanotechnology 2: 780–783. [DOI] [PubMed] [Google Scholar]
- [12].Li QS, Lee GY, Ong CN, Lim CT. (2008) AFM indentation study of breast cancer cells. Biochem Biophys Res Commun 374: 609–613. [DOI] [PubMed] [Google Scholar]
- [13].Ingber DE, Prusty D, Sun Z, Betensky H, Wang N. (1995) Cell shape, cytoskeletal mechanics, and cell cycle control in angiogenesis. J Biomech 28: 1471–1484. [DOI] [PubMed] [Google Scholar]
- [14].Ingber DE. (1993) Cellular tensegrity: Defining new rules of biological design that govern the cytoskeleton. Journal of Cell Science 104: 613–613. [DOI] [PubMed] [Google Scholar]
- [15].Bao G, Suresh S. (2003) Cell and molecular mechanics of biological materials. Nature materials 2: 715–725. [DOI] [PubMed] [Google Scholar]
- [16].Chen CS, Tan J, Tien J. (2004) Mechanotransduction at cell-matrix and cell-cell contacts. Annu Rev Biomed Eng 6: 275–302. [DOI] [PubMed] [Google Scholar]
- [17].Huang S, Ingber DE. (1999) The structural and mechanical complexity of cell-growth control. Nature cell biology 1: E131–E138. [DOI] [PubMed] [Google Scholar]
- [18].Bausch A, Kroy K. (2006) A bottom-up approach to cell mechanics. Nature Physics 2: 231–238. [Google Scholar]
- [19].Gardel M, Nakamura F, Hartwig J, Crocker JC, Stossel T, et al. (2006) Stress-dependent elasticity of composite actin networks as a model for cell behavior. Physical Review Letters 96: 088102. [DOI] [PubMed] [Google Scholar]
- [20].Kasza KE, Rowat AC, Liu J, Angelini TE, Brangwynne CP, et al. (2007) The cell as a material. Current opinion in cell biology 19: 101–107. [DOI] [PubMed] [Google Scholar]
- [21].Kishore V, Bullock W, Sun X, Van Dyke WS, Akkus O. (2012) Tenogenic differentiation of human MSCs induced by the topography of electrochemically aligned collagen threads. Biomaterials 33: 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chien S. (2007) Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. American Journal of Physiology-Heart and Circulatory Physiology 292: H1209–H1224. [DOI] [PubMed] [Google Scholar]
- [23].Geiger B, Bershadsky A. (2002) Exploring the neighborhood: Adhesion-coupled cell mechanosensors. Cell 110: 139–142. [DOI] [PubMed] [Google Scholar]
- [24].Orr AW, Helmke BP, Blackman BR, Schwartz MA. (2006) Mechanisms of mechanotransduction. Developmental cell 10: 11–20. [DOI] [PubMed] [Google Scholar]
- [25].Discher DE, Janmey P, Wang Y-L. (2005) Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 310: 1139–1143. [DOI] [PubMed] [Google Scholar]
- [26].Suresh S. (2007) Biomechanics and biophysics of cancer cells. Acta Materialia 55: 3989–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Makale M. (2007) Cellular mechanobiology and cancer metastasis. Birth Defects Research Part C: Embryo Today: Reviews 81: 329–343. [DOI] [PubMed] [Google Scholar]
- [28].Klein-Nulend J, Bacabac R, Veldhuijzen J, Van Loon J. (2003) Microgravity and bone cell mechanosensitivity. Advances in Space Research 32: 1551–1559. [DOI] [PubMed] [Google Scholar]
- [29].Judex S, Gross TS, Bray RC, Zernicke RF. (1997) Adaptation of bone to physiological stimuli. J Biomech 30: 421–429. [DOI] [PubMed] [Google Scholar]
- [30].Affonce DA, Lutchen KR. (2006) New perspectives on the mechanical basis for airway hyperreactivity and airway hypersensitivity in asthma. Journal of applied physiology 101: 1710–1719. [DOI] [PubMed] [Google Scholar]
- [31].Tan J, Kalapesi F, Coroneo M. (2006) Mechanosensitivity and the eye: Cells coping with the pressure. British journal of ophthalmology 90: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Park Y, Diez-Silva M, Popescu G, Lykotrafitis G, Choi W, et al. (2008) Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum. Proceedings of the National Academy of Sciences 105: 13730–13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gm A, Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. (2000) Endothelial dysfunction, hemodynamic forces, and atherogenesisa. Annals of the New York Academy of Sciences 902: 230–240. [DOI] [PubMed] [Google Scholar]
- [34].Lee GY, Lim CT. (2007) Biomechanics approaches to studying human diseases. Trends in biotechnology 25: 111–118. [DOI] [PubMed] [Google Scholar]
- [35].Stossel TP. (1993) On the crawling of animal cells. Science 260: 1086–1094. [DOI] [PubMed] [Google Scholar]
- [36].Janmey PA. (1998) The cytoskeleton and cell signaling: Component localization and mechanical coupling. Physiological reviews 78: 763–781. [DOI] [PubMed] [Google Scholar]
- [37].Dahl KN, Ribeiro AJ, Lammerding J. (2008) Nuclear shape, mechanics, and mechanotransduction. Circulation research 102: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guilak F, Tedrow JR, Burgkart R. (2000) Viscoelastic properties of the cell nucleus. Biochemical and biophysical research communications 269: 781–786. [DOI] [PubMed] [Google Scholar]
- [39].Lecuit T, Lenne P-F. (2007) Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nature Reviews Molecular Cell Biology 8: 633–644. [DOI] [PubMed] [Google Scholar]
- [40].Costa KD, Sim AJ, Yin F. (2006) Non-Hertzian approach to analyzing mechanical properties of endothelial cells probed by atomic force microscopy. J Biomech Eng 128: 176–184. [DOI] [PubMed] [Google Scholar]
- [41].Takai E, Landesberg R, Katz R, Hung C, Guo X. (2006) Substrate modulation of osteoblast adhesion strength, focal adhesion kinase activation, and responsiveness to mechanical stimuli. Molecular and Cellular Biomechanics 3: 1. [PubMed] [Google Scholar]
- [42].Wirtz D, Konstantopoulos K, Searson PC. (2011) The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nature Reviews Cancer 11: 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Maciaszek JL, Lykotrafitis G. (2011) Sickle cell trait human erythrocytes are significantly stiffer than normal. Journal of Biomechanics 44: 657–661. [DOI] [PubMed] [Google Scholar]
- [44].Kollmannsberger P, Fabry B. (2011) Linear and nonlinear rheology of living cells. Annual Review of Materials Research 41: 75–97. [Google Scholar]
- [45].Petersen NO, McConnaughey WB, Elson EL. (1982) Dependence of locally measured cellular deformability on position on the cell, temperature, and cytochalasin B. Proceedings of the National Academy of Sciences 79: 5327–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ingber DE. (1997) Tensegrity: The architectural basis of cellular mechanotransduction. Annual Review of Physiology 59: 575–599. [DOI] [PubMed] [Google Scholar]
- [47].Forgacs G. (1995) Biological specificity and measurable physical properties of cell surface receptors and their possible role in signal transduction through the cytoskeleton. Biochemistry and cell biology 73: 317–326. [DOI] [PubMed] [Google Scholar]
- [48].Schwarz US, Gardel ML. (2012) United we stand–integrating the actin cytoskeleton and cell–matrix adhesions in cellular mechanotransduction. Journal of Cell Science 125: 3051–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vogel V, Sheetz M. (2006) Local force and geometry sensing regulate cell functions. Nature Reviews Molecular Cell Biology 7: 265–275. [DOI] [PubMed] [Google Scholar]
- [50].Théry M, Racine V, Piel M, Pépin A, Dimitrov A, et al. (2006) Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proceedings of the National Academy of Sciences 103: 19771–19776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sukharev S, Sachs F. (2012) Molecular force transduction by ion channels–diversity and unifying principles. Journal of Cell Science 125: 3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Galbraith CG, Yamada KM, Sheetz MP. (2002) The relationship between force and focal complex development. The Journal of cell biology 159: 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rose DM, Alon R, Ginsberg MH. (2007) Integrin modulation and signaling in leukocyte adhesion and migration. Immunological reviews 218: 126–134. [DOI] [PubMed] [Google Scholar]
- [54].Wang JH-C, Thampatty BP. (2006) An introductory review of cell mechanobiology. Biomechanics and modeling in mechanobiology 5: 1–16. [DOI] [PubMed] [Google Scholar]
- [55].Harris AK, Wild P, Stopak D. (1980) Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science 208: 177–179. [DOI] [PubMed] [Google Scholar]
- [56].Harris AK, Stopak D, Wild P. (1981) Fibroblast traction as a mechanism for collagen morphogenesis. Nature 290: 249–251. [DOI] [PubMed] [Google Scholar]
- [57].Couchman JR, Rees DA. (1979) The behaviour of fibroblasts migrating from chick heart explants: Changes in adhesion, locomotion and growth, and in the distribution of actomyosin and fibronectin. Journal of Cell Science 39: 149–165. [DOI] [PubMed] [Google Scholar]
- [58].Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. (1994) Traction forces generated by locomoting keratocytes. The Journal of cell biology 127: 1957–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Davies PF. (1995) Flow-mediated endothelial mechanotransduction. Physiological reviews 75: 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dewey C, Jr, Bussolari S, Gimbrone M, Jr, Davies PF. (1981) The dynamic response of vascular endothelial cells to fluid shear stress. J Biomech Eng 103: 177–185. [DOI] [PubMed] [Google Scholar]
- [61].Diamond S, Eskin S, McIntire L. (1989) Fluid flow stimulates tissue plasminogen activator secretion by cultured human endothelial cells. Science 243: 1483–1485. [DOI] [PubMed] [Google Scholar]
- [62].Nerem RM, Harrison DG, Taylor RW, Alexander WR. (1993) Hemodynamics and vascular endothelial biology. Journal of cardiovascular pharmacology 21: S6–S10. [DOI] [PubMed] [Google Scholar]
- [63].Levesque MJ, Liepsch D, Moravec S, Nerem RM. (1986) Correlation of endothelial cell shape and wall shear stress in a stenosed dog aorta. Arteriosclerosis, Thrombosis, and Vascular Biology 6: 220–229. [DOI] [PubMed] [Google Scholar]
- [64].Davies PF, Barbee KA, Volin MV, Robotewskyj A, Chen J, et al. (1997) Spatial relationships in early signaling events of flow-mediated endothelial mechanotransduction. Annual Review of Physiology 59: 527–549. [DOI] [PubMed] [Google Scholar]
- [65].White GE, Fujiwara K. (1986) Expression and intracellular distribution of stress fibers in aortic endothelium. The Journal of cell biology 103: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Toborek M, Kaiser S. (1999) Endothelial cell functions. ¶ Relationship to atherogenesis. Basic research in cardiology 94: 295–314. [DOI] [PubMed] [Google Scholar]
- [67].Cross SE, Jin YS, Rao J, Gimzewski JK. (2007) Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol 2: 780–783. [DOI] [PubMed] [Google Scholar]
- [68].Cross SE, Jin YS, Tondre J, Wong R, Rao J, et al. (2008) AFM-based analysis of human metastatic cancer cells. Nanotechnology 19: 384003. [DOI] [PubMed] [Google Scholar]
- [69].Wu H, Kuhn T, Moy V. (1998) Mechanical properties of L929 cells measured by atomic force microscopy: Effects of anticytoskeletal drugs and membrane crosslinking. Scanning 20: 389–397. [DOI] [PubMed] [Google Scholar]
- [70].Neuman KC, Nagy A. (2008) Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods 5: 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lenormand G, Hénon S, Richert A, Siméon J, Gallet F. (2001) Direct measurement of the area expansion and shear moduli of the human red blood cell membrane skeleton. Biophys J 81: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kee Y-S, Robinson DN. (2013) Micropipette Aspiration for Studying Cellular Mechanosensory Responses and Mechanics. Dictyostelium discoideum Protocols: Springer. pp. 367–382. [DOI] [PubMed] [Google Scholar]
- [73].Thoumine O, Cardoso O, Meister J-J. (1999) Changes in the mechanical properties of fibroblasts during spreading: A micromanipulation study. European Biophysics Journal 28: 222–234. [DOI] [PubMed] [Google Scholar]
- [74].Bacabac RG, Smit TH, Cowin SC, Van Loon JJ, Nieuwstadt F, et al. (2005) Dynamic shear stress in parallel-plate flow chambers. Journal of biomechanics 38: 159–167. [DOI] [PubMed] [Google Scholar]
- [75].Coisne C, Lyck R, Engelhardt B. (2013) Live cell imaging techniques to study T cell trafficking across the blood-brain barrier in vitro and in vivo. Fluids and Barriers of the CNS 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Barabino GA, McIntire L, Eskin SG, Sears DA, Udden M. (1987) Endothelial cell interactions with sickle cell, sickle trait, mechanically injured, and normal erythrocytes under controlled flow. Blood 70: 152–157. [PubMed] [Google Scholar]
- [77].Montes RA, Eckman JR, Hsu LL, Wick TM. (2002) Sickle erythrocyte adherence to endothelium at low shear: Role of shear stress in propagation of vaso-occlusion. American journal of hematology 70: 216–227. [DOI] [PubMed] [Google Scholar]
- [78].Han S-K, Colarusso P, Herzog W. (2009) Confocal microscopy indentation system for studying in situ chondrocyte mechanics. Medical engineering & physics 31: 1038–1042. [DOI] [PubMed] [Google Scholar]
- [79].Ragan T, Huang H, So P, Gratton E. (2006) 3D particle tracking on a two-photon microscope. Journal of Fluorescence 16: 325–336. [DOI] [PubMed] [Google Scholar]
- [80].Harris AK, Wild P, Stopak D. (1980) Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science 208: 177–179. [DOI] [PubMed] [Google Scholar]
- [81].Helfman DM, Levy ET, Berthier C, Shtutman M, Riveline D, et al. (1999) Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol Biol Cell 10: 3097–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Dembo M, Wang YL. (1999) Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J 76: 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, et al. (2001) Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat Cell Biol 3: 466–472. [DOI] [PubMed] [Google Scholar]
- [84].Yang MT, Fu J, Wang YK, Desai RA, Chen CS. (2011) Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat Protoc 6: 187–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fu J, Wang YK, Yang MT, Desai RA, Yu X, et al. (2010) Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods 7: 733–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, et al. (2003) Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci U S A 100: 1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lemmon CA, Sniadecki NJ, Ruiz SA, Tan JL, Romer LH, et al. (2005) Shear force at the cell-matrix interface: Enhanced analysis for microfabricated post array detectors. Mech Chem Biosyst 2: 1–16. [PMC free article] [PubMed] [Google Scholar]
- [88].Galbraith CG, Sheetz MP. (1997) A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci U S A 94: 9114–9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Rabodzey A, Alcaide P, Luscinskas FW, Ladoux B. (2008) Mechanical forces induced by the transendothelial migration of human neutrophils. Biophys J 95: 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, et al. (2010) Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods 7: 969–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Marelli M, Gadhari N, Boero G, Chiquet M, Brugger J. (2013) Cell force measurements in 3D microfabricated environments based on compliant cantilevers. Lab Chip. [DOI] [PubMed]
- [92].Nguyen TD, Deshmukh N, Nagarah JM, Kramer T, Purohit PK, et al. (2012) Piezoelectric nanoribbons for monitoring cellular deformations. Nat Nanotechnol 7: 587–593. [DOI] [PubMed] [Google Scholar]
- [93].Park K, Jang J, Irimia D, Sturgis J, Lee J, et al. (2008) ‘Living cantilever arrays' for characterization of mass of single live cells in fluids. Lab Chip 8: 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Grover WH, Bryan AK, Diez-Silva M, Suresh S, Higgins JM, et al. (2011) Measuring single-cell density. Proc Natl Acad Sci U S A 108: 10992–10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Singh A, Suri S, Lee T, Chilton JM, Cooke MT, et al. (2013) Adhesion strength-based, label-free isolation of human pluripotent stem cells. Nat Methods 10: 438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sniadecki NJ, Anguelouch A, Yang MT, Lamb CM, Liu Z, et al. (2007) Magnetic microposts as an approach to apply forces to living cells. Proc Natl Acad Sci U S A 104: 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ting LH, Jahn JR, Jung JI, Shuman BR, Feghhi S, et al. (2012) Flow mechanotransduction regulates traction forces, intercellular forces, and adherens junctions. Am J Physiol Heart Circ Physiol 302: H2220–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Dertinger SK, Jiang X, Li Z, Murthy VN, Whitesides GM. (2002) Gradients of substrate-bound laminin orient axonal specification of neurons. Proc Natl Acad Sci U S A 99: 12542–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Rhoads DS, Guan JL. (2007) Analysis of directional cell migration on defined FN gradients: Role of intracellular signaling molecules. Exp Cell Res 313: 3859–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. (2003) Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci 116: 1881–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Bucaro MA, Vasquez Y, Hatton BD, Aizenberg J. (2012) Fine-tuning the degree of stem cell polarization and alignment on ordered arrays of high-aspect-ratio nanopillars. ACS Nano 6: 6222–6230. [DOI] [PubMed] [Google Scholar]
- [102].Saez A, Ghibaudo M, Buguin A, Silberzan P, Ladoux B. (2007) Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc Natl Acad Sci U S A 104: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Di Carlo D, Wu LY, Lee LP. (2006) Dynamic single cell culture array. Lab on a Chip 6: 1445–1449. [DOI] [PubMed] [Google Scholar]
- [104].Kim MC, Isenberg BC, Sutin J, Meller A, Wong JY, et al. (2011) Programmed trapping of individual bacteria using micrometre-size sieves. Lab on a Chip 11: 1089–1095. [DOI] [PubMed] [Google Scholar]
- [105].Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, et al. (2009) Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab on a Chip 9: 2659–2664. [DOI] [PubMed] [Google Scholar]
- [106].Park MC, Hur JY, Cho HS, Park SH, Suh KY. (2011) High-throughput single-cell quantification using simple microwell-based cell docking and programmable time-course live-cell imaging. Lab on a Chip 11: 79–86. [DOI] [PubMed] [Google Scholar]
- [107].Hosokawa M, Hayashi T, Mori T, Yoshino T, Nakasono S, et al. (2011) Microfluidic Device with Chemical Gradient for Single-Cell Cytotoxicity Assays. Analytical Chemistry 83: 3648–3654. [DOI] [PubMed] [Google Scholar]
- [108].Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. (2009) Microfluidic control of cell pairing and fusion. Nature Methods 6: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kurosawa O, Oana H, Matsuoka S, Noma A, Kotera H, et al. (2006) Electroporation through a microfabricated orifice and its application to the measurement of cell response to external stimuli. Measurement Science & Technology 17: 3127–3133. [Google Scholar]
- [110].Kovac JR, Voldman J. (2007) Intuitive, image-based cell sorting using optofluidic cell sorting. Analytical Chemistry 79: 9321–9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, et al. (2003) Microfluidic device for single-cell analysis. Analytical Chemistry 75: 3581–3586. [DOI] [PubMed] [Google Scholar]
- [112].Chang TM. (1964) Semipermeable microcapsules. Science 146: 524–525. [DOI] [PubMed] [Google Scholar]
- [113].Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, et al. (2009) Droplet microfluidic technology for single-cell high-throughput screening. Proceedings of the National Academy of Sciences of the United States of America 106: 14195–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Koster S, Angile FE, Duan H, Agresti JJ, Wintner A, et al. (2008) Drop-based microfluidic devices for encapsulation of single cells. Lab on a Chip 8: 1110–1115. [DOI] [PubMed] [Google Scholar]
- [115].Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, et al. (2008) Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms (vol 15, pg 427, 2008). Chemistry & Biology 15: 875–875. [DOI] [PubMed] [Google Scholar]
- [116].Gu SQ, Zhang YX, Zhu Y, Du WB, Yao B, et al. (2011) Multifunctional Picoliter Droplet Manipulation Platform and Its Application in Single Cell Analysis. Analytical Chemistry 83: 7570–7576. [DOI] [PubMed] [Google Scholar]
- [117].Chang TMS. (2005) Therapeutic applications of polymeric artificial cells. Nature Reviews Drug Discovery 4: 221–235. [DOI] [PubMed] [Google Scholar]
- [118].Gadd JC, Kuyper CL, Fujimoto BS, Allen RW, Chiu DT. (2008) Sizing subcellular organelles and nanoparticles confined within aqueous droplete. Analytical Chemistry 80: 3450–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].El-Ali J, Gaudet S, Gunther A, Sorger PK, Jensen KF. (2005) Cell stimulus and lysis in a microfluidic device with segmented gas-liquid flow. Analytical Chemistry 77: 3629–3636. [DOI] [PubMed] [Google Scholar]
- [120].Di Carlo D, Ionescu-Zanetti C, Zhang Y, Hung P, Lee LP. (2005) On-chip cell lysis by local hydroxide generation. Lab on a Chip 5: 171–178. [DOI] [PubMed] [Google Scholar]
- [121].Lu H, Schmidt MA, Jensen KF. (2005) A microfluidic electroporation device for cell lysis. Lab on a Chip 5: 23–29. [DOI] [PubMed] [Google Scholar]
- [122].Wang HY, Bhunia AK, Lu C. (2006) A microfluidic flow-through device for high throughput electrical lysis of bacterial cells based on continuous dc voltage. Biosensors & Bioelectronics 22: 582–588. [DOI] [PubMed] [Google Scholar]
- [123].Le Gac S, Zwaan E, van den Berg A, Ohl CD. (2007) Sonoporation of suspension cells with a single cavitation bubble in a microfluidic confinement. Lab on a Chip 7: 1666–1672. [DOI] [PubMed] [Google Scholar]
- [124].Quinto-Su PA, Lai HH, Yoon HH, Sims CE, Allbritton NL, et al. (2008) Examination of laser microbeam cell lysis in a PDMS microfluidic channel using time-resolved imaging. Lab on a Chip 8: 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Di Carlo D, Jeong KH, Lee LP. (2003) Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab on a Chip 3: 287–291. [DOI] [PubMed] [Google Scholar]
- [126].Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, et al. (2008) Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proceedings of the National Academy of Sciences of the United States of America 105: 20173–20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Petriv OI, Kuchenbauer F, Delaney AD, Lecault V, White A, et al. (2010) Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proceedings of the National Academy of Sciences of the United States of America 107: 15443–15448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Huang B, Wu HK, Bhaya D, Grossman A, Granier S, et al. (2007) Counting low-copy number proteins in a single cell. Science 315: 81–84. [DOI] [PubMed] [Google Scholar]
- [129].Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. (2006) Profiling signaling peptides in single mammalian cells using mass spectrometry. Analytical Chemistry 78: 7267–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Hou HW, Li QS, Lee GY, Kumar AP, Ong CN, et al. (2009) Deformability study of breast cancer cells using microfluidics. Biomed Microdevices 11: 557–564. [DOI] [PubMed] [Google Scholar]
- [131].Zhao H, Wang Y, Jiang X, Shi X, Zhong H, et al. (2013) In vitro assay of cytoskeleton nanomechanics as a tool for screening potential anticancer effects of natural plant extract, tubeimoside I on human hepatoma (HepG2) cells. Chinese Science Bulletin 58: 2576–2583. [Google Scholar]
- [132].Zhang G, Long M, Wu ZZ, Yu WQ. (2002) Mechanical properties of hepatocellular carcinoma cells. World J Gastroenterol 8: 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Baker EL, Srivastava J, Yu D, Bonnecaze RT, Zaman MH. (2011) Cancer cell migration: Integrated roles of matrix mechanics and transforming potential. PLoS One 6: e20355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Li Z, Song J, Mantini G, Lu MY, Fang H, et al. (2009) Quantifying the traction force of a single cell by aligned silicon nanowire array. Nano Lett 9: 3575–3580. [DOI] [PubMed] [Google Scholar]
- [135].Moss MS, Sisken B, Zimmer S, Anderson KW. (1999) Adhesion of nonmetastatic and highly metastatic breast cancer cells to endothelial cells exposed to shear stress. Biorheology 36: 359–371. [PubMed] [Google Scholar]
- [136].Baker EL, Bonnecaze RT, Zaman MH. (2009) Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J 97: 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Tseng P, Judy JW, Di Carlo D. (2012) Magnetic nanoparticle-mediated massively parallel mechanical modulation of single-cell behavior. Nat Methods 9: 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Baker EL, Lu J, Yu D, Bonnecaze RT, Zaman MH. (2010) Cancer cell stiffness: Integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys J 99: 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Gal N, Weihs D. (2012) Intracellular mechanics and activity of breast cancer cells correlate with metastatic potential. Cell Biochem Biophys 63: 199–209. [DOI] [PubMed] [Google Scholar]
- [140].Yizraeli ML, Weihs D. (2011) Time-dependent micromechanical responses of breast cancer cells and adjacent fibroblasts to electric treatment. Cell Biochem Biophys 61: 605–618. [DOI] [PubMed] [Google Scholar]
- [141].Schoen I, Hu W, Klotzsch E, Vogel V. (2010) Probing cellular traction forces by micropillar arrays: Contribution of substrate warping to pillar deflection. Nano Lett 10: 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Huang Y, Agrawal B, Sun D, Kuo JS, Williams JC. (2011) Microfluidics-based devices: New tools for studying cancer and cancer stem cell migration. Biomicrofluidics 5: 13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Puig-De-Morales-Marinkovic M, Turner KT, Butler JP, Fredberg JJ, Suresh S. (2007) Viscoelasticity of the human red blood cell. Am J Physiol Cell Physiol 293: C597–605. [DOI] [PubMed] [Google Scholar]
- [144].Ahn BM, Kim J, Ian L, Rha KH, Kim HJ. (2010) Mechanical property characterization of prostate cancer using a minimally motorized indenter in an ex vivo indentation experiment. Urology 76: 1007–1011. [DOI] [PubMed] [Google Scholar]
- [145].Suresh S, Spatz J, Mills JP, Micoulet A, Dao M, et al. (2005) Connections between single-cell biomechanics and human disease states: Gastrointestinal cancer and malaria. Acta Biomater 1: 15–30. [DOI] [PubMed] [Google Scholar]
- [146].Beil M, Micoulet A, von Wichert G, Paschke S, Walther P, et al. (2003) Sphingosylphosphorylcholine regulates keratin network architecture and visco-elastic properties of human cancer cells. Nat Cell Biol 5: 803–811. [DOI] [PubMed] [Google Scholar]
- [147].Chen K, Li D, Jiang YH, Yao WJ, Wang XJ, et al. (2004) Influence of expressed TRAIL on biophysical properties of the human leukemic cell line Jurkat. Cell Res 14: 161–168. [DOI] [PubMed] [Google Scholar]
- [148].Xinyu L, Yifei W, Yu S. Real-Time High-Accuracy Micropipette Aspiration for Characterizing Mechanical Properties of Biological Cells; 2007 10–14 April 2007. pp. 1930–1935. [Google Scholar]
- [149].Tavano F, Bonin S, Pinato G, Stanta G, Cojoc D. (2011) Custom-Built Optical Tweezers for Locally Probing the Viscoelastic Properties of Cancer Cells. International Journal of Optomechatronics 5: 234–248. [Google Scholar]
- [150].Hong-Lian Guo LC-X, Jian-Fa Duan, Yu-Qiang Jiang, Xue-Hai Han, Li Zhao-Lin, Bing-Ying Cheng, Dao-Zhong Zhang. (2004) Mechanical Properties of Breast Cancer Cell Membrane Studied with Optical Tweezers. Chin Phys Lett 21: 2543–2546. [Google Scholar]
- [151].Youhua T, Leung AYH, Kaiqun W, Tsz-Kan F, Dong S. (2011) Optical Tweezer Technology. Nanotechnology Magazine, IEEE 5: 17–21. [Google Scholar]
- [152].LaFratta CN. (2013) Optical tweezers for medical diagnostics. Anal Bioanal Chem 405: 5671–5677. [DOI] [PubMed] [Google Scholar]
- [153].Bellini N, Vishnubhatla KC, Bragheri F, Ferrara L, Minzioni P, et al. (2010) Femtosecond laser fabricated monolithic chip for optical trapping and stretching of single cells. Opt Express 18: 4679–4688. [DOI] [PubMed] [Google Scholar]
- [154].Jonietz E. (2012) Mechanics: The forces of cancer. Nature 491: S56–S57. [DOI] [PubMed] [Google Scholar]
- [155].Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, et al. (2005) Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J 88: 3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Kim DH, Wong PK, Park J, Levchenko A, Sun Y. (2009) Microengineered platforms for cell mechanobiology. Annu Rev Biomed Eng 11: 203–233. [DOI] [PubMed] [Google Scholar]
- [157].Chen Christopher S., Mrksich Milan, Huang Sui, Whitesides George M., Ingber DE. (1997) Geometric control of cell life and death. Science 276: 1425–1428. [DOI] [PubMed] [Google Scholar]
- [158].Polte Thomas R., Eichler Gabriel S., Wang Ning, Ingber DE. (2004) Extracellular matrix controls myosin light chain phosphorylation and cell contractility through modulation of cell shape and cytoskeletal prestress. American Journal of Physiology Cell Physiology 286: C518–528. [DOI] [PubMed] [Google Scholar]
- [159].Feinberg Adam W., Feigel Alex, Shevkoplyas Sergey S., Sheehy Sean, Whitesides George M., et al. (2007) Muscular thin films for building actuators and powering devices. Science 317: 1366–1370. [DOI] [PubMed] [Google Scholar]
- [160].Ohgushi H, Dohi Y, Katuda T, Tamai S, Tabata S, et al. (1996) In vitro bone formation by rat marrow cell culture. Journal of Biomedical Materials Research 32: 333–340. [DOI] [PubMed] [Google Scholar]
- [161].El-Ali Jamil, Sorger PK, Jensen KF. (2006) Cells on chips. Nature 442: 403–411. [DOI] [PubMed] [Google Scholar]
- [162].Puleo CM, Ambrose WM, Takezawa T, Elisseeff J, Wang T-H. (2009) Integration and application of vitrified collagen in multilayered microfluidic devices for corneal microtissue culture. Lab Chip 9: 3221–3227. [DOI] [PubMed] [Google Scholar]
- [163].Torisawa Y-S, Spina CS, Mammoto T, Mammoto A, Weaver JC, et al. (2014) Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat Methods. [DOI] [PubMed]
- [164].Kim HJ, Huh D, Hamilton G, Ingber DE. (2012) Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a chip 12: 2165–2174. [DOI] [PubMed] [Google Scholar]
- [165].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, et al. (2010) Reconstituting organ-level lung functions on a chip. Science 328: 1662–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Chang TMS. (2007) Artificial cells: Biotechnology, nanomedicine, regenerative medicine, blood substitutes, bioencapsulation, cell/stem cell therapy: World Scientific. [DOI] [PMC free article] [PubMed]
- [167].Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, et al. (2012) A human disease model of drug toxicity–induced pulmonary edema in a lung-on-a-chip microdevice. Science translational medicine 4: 159ra147–159ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Huh D, Torisawa Y-S, Hamilton GA, Kim HJ, Ingber DE. (2012) Microengineered physiological biomimicry: Organs-on-chips. Lab Chip 12: 2156–2164. [DOI] [PubMed] [Google Scholar]
- [169].Wang G, McCain ML, Yang L, He A, Pasqualini FS, et al. (2014) Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nature medicine 20: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Chang TMS. (2013) Selected Topics in Nanomedicine: World Scientific.
- [171].Huh D, Hamilton GA, Ingber DE. (2011) From 3D cell culture to organs-on-chips. Trends Cell Biol 21: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Huh Dongeun, Matthews Benjamin D., Akiko Mammoto, Montoya-Zavala Martín, Hsin Hong Yuan, et al. (2010) Reconstituting organ-level lung functions on a chip. Science 328: 1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Huh D, Fujioka H, Tun Y-C, Futai N, Paine R, et al. (2007) Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proceedings of the National Academy of Sciences of the United States of America 104: 18886–18891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Douville Nicholas J., Zamankhan P, Tung Y-C, Li R, Vaughan Benjamin L., et al. (2011) Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab on a chip 11: 609–619. [DOI] [PubMed] [Google Scholar]
- [175].Carraro A, Hsu W-M, Kulig KM, Cheung WS, Miller ML, et al. (2008) In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomedical Microdevices 10: 795–806. [DOI] [PubMed] [Google Scholar]
- [176].Kane BJ, Zinner MJ, Yarmush ML, Toner M. (2006) Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Analytical Chemistry 78: 4291–4298. [DOI] [PubMed] [Google Scholar]
- [177].Lee Philip J., Hung Paul J., Lee LP. (2007) An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnology and Bioengineering 97: 1340–1346. [DOI] [PubMed] [Google Scholar]
- [178].Nakao Yosuke, Kimura Hiroshi, Sakai Yasuyuki, Fujii T. (2011) Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Imura Y, Asano Y, Sato K, Yoshimura E. (2009) A microfluidic system to evaluate intestinal absorption. Analytical Sciences 25: 1403–1407. [DOI] [PubMed] [Google Scholar]
- [180].Kimura H, Yamamoto T, Sakai H, Sakai Y, Fujii T. (2008) An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab on a chip 8: 741–746. [DOI] [PubMed] [Google Scholar]
- [181].Jang K-J, Suh K-Y. (2009) A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab on a chip 10: 36–42. [DOI] [PubMed] [Google Scholar]
- [182].Jang K-J, Cho HS, Kang DH, Bae Won Gyu, Kwon T-H, et al. (2011) Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integrative Biology 3: 134–141. [DOI] [PubMed] [Google Scholar]
- [183].Rizvi I, Gurkan UA, Tasoglu S, Alagic N, Celli JP, et al. (2013) Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proceedings of the National Academy of Sciences 110: E1974–E1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Wagner I, Materne E-M, Brincker S, Sußbier U, Fradrich C, et al. (2013) A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab on a chip 13: 3538–3547. [DOI] [PubMed] [Google Scholar]
- [185].Foudeh AM, Didar Fatanat T, Veres T, Tabrizian M. (2012) Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 12: 3249–3266. [DOI] [PubMed] [Google Scholar]
- [186].Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C. (2010) An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnology Advances 28: 232–254. [DOI] [PubMed] [Google Scholar]
- [187].Wang S, Tasoglu S, Chen PZ, Chen M, Akbas R, et al. (2014) Micro-a-fluidics ELISA for Rapid CD4 Cell Count at the Point-of-Care. Scientific reports 4. [DOI] [PMC free article] [PubMed]
- [188].Inci F, Tokel O, Wang S, Gurkan UA, Tasoglu S, et al. (2013) Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano 7: 4733–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Wang S, Inci F, Chaunzwa TL, Ramanujam A, Vasudevan A, et al. (2012) Portable microfluidic chip for detection of Escherichia coli in produce and blood. International journal of nanomedicine 7: 2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Gurkan UA, Moon S, Geckil H, Xu F, Wang S, et al. (2011) Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnology journal 6: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Turner APF, Karube I, Wilson GS, Worsfold PJ. (1987) Biosensors: Fundamentals and applications. Analytica Chimica Acta 201: 363–364. [Google Scholar]
- [192].Bhattacharya S, Jaesung J, Liju Y, Akin D, Bashir R. (2007) BioMEMS and Nanotechnology-Based Approaches for Rapid Detection of Biological Entities. Journal of Rapid Methods & Automation in Microbiology 15: 1–32. [Google Scholar]
- [193].Hoa XD, Kirk AG, Tabrizian M. (2007) Towards integrated and sensitive surface plasmon resonance biosensors: A review of recent progress. Biosensors and Bioelectronics 23: 151–160. [DOI] [PubMed] [Google Scholar]
- [194].Yi C, Zhang Q, Li CW, Yang J, Zhao J, et al. (2006) Optical and electrochemical detection techniques for cell-based microfluidic systems. Anal Bioanal Chem 384: 1259–1268. [DOI] [PubMed] [Google Scholar]
- [195].Wang S, Ge L, Song X, Yu J, Ge S, et al. (2012) Paper-based chemiluminescence ELISA: Lab-on-paper based on chitosan modified paper device and wax-screen-printing. Biosensors and Bioelectronics 31: 212–218. [DOI] [PubMed] [Google Scholar]
- [196].Wolter A, Niessner R, Seidel M. (2008) Detection of Escherichia coli O157:H7, Salmonella typhimurium, and Legionella pneumophila in water using a flow-through chemiluminescence microarray readout system. Analytical Chemistry 80: 5854–5863. [DOI] [PubMed] [Google Scholar]
- [197].Donhauser SC, Niessner R, Seidel M. (2011) Sensitive Quantification of Escherichia coil O157:H7, Salmonella enterica, and Campylobacter jejuni by Combining Stopped Polymerase Chain Reaction with Chemiluminescence Flow-Through DNA Microarray Analysis. Analytical Chemistry 83: 3153–3160. [DOI] [PubMed] [Google Scholar]
- [198].Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, et al. (2004) The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnology progress 20: 338–345. [DOI] [PubMed] [Google Scholar]
- [199].Hochberg LR, Serruya MD, Friehs GM, Mukand JA, Saleh M, et al. (2006) Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature 442: 164–171. [DOI] [PubMed] [Google Scholar]
- [200].Bosch JR, Groen J. (1995) Sacral (S3) segmental nerve stimulation as a treatment for urge incontinence in patients with detrusor instability: Results of chronic electrical stimulation using an implantable neural prosthesis. The Journal of urology 154: 504–507. [DOI] [PubMed] [Google Scholar]
- [201].Reddy BV, Swamy YSK, Usha N. (2011) Generate Vision in Blind People Using Suitable Neuroprosthesis Implant of BIOMEMS in Brain. In: Abraham A, Mauri J, Buford J, Suzuki J, Thampi S, editors. Advances in Computing and Communications: Springer; Berlin Heidelberg: pp. 309–317. [Google Scholar]
- [202].Meyer J-U. (2002) Retina implant—a bioMEMS challenge. Sensors and Actuators A: Physical 97–98: 1–9.
- [203].Wilson BS, Finley CC, Lawson DT, Wolford RD, Eddington DK, et al. (1991) Better speech recognition with cochlear implants. Nature 352: 236–238. [DOI] [PubMed] [Google Scholar]
- [204].Svirsky MA, Robbins AM, Kirk KI, Pisoni DB, Miyamoto RT. (2000) Language development in profoundly deaf children with cochlear implants. Psychological science 11: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Ravikumar M, Sunil S, Black J, Barkauskas DS, Haung AY, et al. (2014) The roles of blood-derived macrophages and resident microglia in the neuroinflammatory response to implanted Intracortical microelectrodes. Biomaterials. [DOI] [PMC free article] [PubMed]
- [206].Potter-Baker KA, Ravikumar M, Burke AA, Meador WD, Householder KT, et al. (2014) A comparison of neuroinflammation to implanted microelectrodes in rat and mouse models. Biomaterials 35: 5637–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Potter KA, Buck AC, Self WK, Callanan ME, Sunil S, et al. (2013) The effect of resveratrol on neurodegeneration and blood brain barrier stability surrounding intracortical microelectrodes. Biomaterials 34: 7001–7015. [DOI] [PubMed] [Google Scholar]
- [208].Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. (2006) Microfluidic culture platform for neuroscience research. Nat Protoc 1: 2128–2136. [DOI] [PubMed] [Google Scholar]
- [209].Taylor AM, Rhee SW, Tu CH, Cribbs DH, Cotman CW, et al. (2003) Microfluidic multicompartment device for neuroscience research. Langmuir 19: 1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Goyal G, Nam Y. (2011) Neuronal micro-culture engineering by microchannel devices of cellular scale dimensions. Biomedical Engineering Letters 1: 89–98. [Google Scholar]
- [211].Chadwick M H, Steve M P. (2010) How to culture, record and stimulate neuronal networks on micro-electrode arrays (MEAs). Journal of Visualized Experiments. [DOI] [PMC free article] [PubMed]
- [212].Stephens CL, Toda H, Palmer TD, DeMarse TB, Ormerod BK. (2012) Adult neural progenitor cells reactivate superbursting in mature neural networks. Experimental neurology 234: 20–30. [DOI] [PubMed] [Google Scholar]
- [213].LeChasseur Y, Dufour S, Lavertu G, Bories C, Deschênes M, et al. (2011) A microprobe for parallel optical and electrical recordings from single neurons in vivo. Nat Methods 8: 319–325. [DOI] [PubMed] [Google Scholar]
- [214].Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, et al. (2013) Simple precision creation of digitally specified, spatially heterogeneous, engineered tissue architectures. Advanced Materials 25: 1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Greene AC, Washburn CM, Bachand GD, James CD. (2011) Combined chemical and topographical guidance cues for directing cytoarchitectural polarization in primary neurons. Biomaterials 32: 8860–8869. [DOI] [PubMed] [Google Scholar]
- [216].Takeuchi A, Nakafutami S, Tani H, Mori M, Takayama Y, et al. (2011) Device for co-culture of sympathetic neurons and cardiomyocytes using microfabrication. Lab Chip 11: 2268–2275. [DOI] [PubMed] [Google Scholar]
- [217].Hellman AN, Vahidi B, Kim HJ, Mismar W, Steward O, et al. (2010) Examination of axonal injury and regeneration in micropatterned neuronal culture using pulsed laser microbeam dissection. Lab Chip 10: 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, et al. (2005) A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2: 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Hosmane S, Fournier A, Wright R, Rajbhandari L, Siddique R, et al. (2011) Valve-based microfluidic compression platform: Single axon injury and regrowth. Lab Chip 11: 3888–3895. [DOI] [PubMed] [Google Scholar]
- [220].Poon WW, Blurton-Jones M, Tu CH, Feinberg LM, Chabrier MA, et al. (2011) β-Amyloid impairs axonal BDNF retrograde trafficking. Neurobiology of aging 32: 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Wong R, Miles R, Traub R. (1984) Local circuit interactions in synchronization of cortical neurones. Journal of experimental biology 112: 169–178. [DOI] [PubMed] [Google Scholar]
- [222].Spira ME, Hai A. (2013) Multi-electrode array technologies for neuroscience and cardiology. Nat Nanotechnol 8: 83–94. [DOI] [PubMed] [Google Scholar]
- [223].Hai A, Dormann A, Shappir J, Yitzchaik S, Bartic C, et al. (2009) Spine-shaped gold protrusions improve the adherence and electrical coupling of neurons with the surface of micro-electronic devices. Journal of The Royal Society Interface 6: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Hai A, Kamber D, Malkinson G, Erez H, Mazurski N, et al. (2009) Changing gears from chemical adhesion of cells to flat substrata toward engulfment of micro-protrusions by active mechanisms. Journal of neural engineering 6: 066009. [DOI] [PubMed] [Google Scholar]
- [225].Hai A, Spira ME. (2012) On-chip electroporation, membrane repair dynamics and transient in-cell recordings by arrays of gold mushroom-shaped microelectrodes. Lab Chip 12: 2865–2873. [DOI] [PubMed] [Google Scholar]
- [226].Spira ME, Kamber D, Dormann A, Cohen A, Bartic C, et al. Improved neuronal adhesion to the surface of electronic device by engulfment of protruding micro-nails fabricated on the chip surface; 2007. IEEE. pp. 1247–1250. [Google Scholar]
- [227].Fendyur A, Spira ME. (2012) Toward on-chip, in-cell recordings from cultured cardiomyocytes by arrays of gold mushroom-shaped microelectrodes. Frontiers in neuroengineering 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn D-R, et al. (2010) Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proceedings of the National Academy of Sciences 107: 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Robinson JT, Jorgolli M, Shalek AK, Yoon M-H, Gertner RS, et al. (2012) Vertical nanowire electrode arrays as a scalable platform for intracellular interfacing to neuronal circuits. Nat Nanotechnol 7: 180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Robinson JT, Jorgolli M, Park H. (2013) Nanowire electrodes for high-density stimulation and measurement of neural circuits. Frontiers in neural circuits 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Hayashi K. (2006) Tensile properties and local stiffness of cells. Mechanics of biological tissue: Springer. pp. 137–152. [Google Scholar]
- [232].An SS, Laudadio RE, Lai J, Rogers RA, Fredberg JJ. (2002) Stiffness changes in cultured airway smooth muscle cells. American Journal of Physiology – Cell Physiology 283: C792–C801. [DOI] [PubMed] [Google Scholar]
- [233].Maxim Dokukin E, Nataliia Guz V, Sokolov I. (2013) Quantitative Study of the Elastic Modulus of Loosely Attached Cells in AFM Indentation Experiments. Biophysical journal 104: 2123–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Simon A, Cohen-Bouhacina T, Porte M, Aime J, Amedee J, et al. (2003) Characterization of dynamic cellular adhesion of osteoblasts using atomic force microscopy. Cytometry Part A 54: 36–47. [DOI] [PubMed] [Google Scholar]
- [235].Collinsworth AM, Zhang S, Kraus WE, Truskey GA. (2002) Apparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiation. American Journal of Physiology-Cell Physiology 283: C1219–C1227. [DOI] [PubMed] [Google Scholar]
- [236].Zhang JS, Kraus WE, Truskey GA. (2004) Stretch-induced nitric oxide modulates mechanical properties of skeletal muscle cells. American Journal of Physiology-Cell Physiology 287: C292–C299. [DOI] [PubMed] [Google Scholar]
- [237].Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. (2001) Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. Journal of biomechanics 34: 1545–1553. [DOI] [PubMed] [Google Scholar]
- [238].Mozhanova A, Nurgazizov N, Bukharaev A. (2003) Local elastic properties of biological materials studied by SFM. SPM-2003, Proceedings Nizhni Novgorod. [Google Scholar]
- [239].Dulinska I, Targosz M, Strojny W, Lekka M, Czuba P, et al. (2006) Stiffness of normal and pathological erythrocytes studied by means of atomic force microscopy. Journal of biochemical and biophysical methods 66: 1–11. [DOI] [PubMed] [Google Scholar]
- [240].Rosenbluth MJ, Lam WA, Fletcher DA. (2006) Force microscopy of nonadherent cells: A comparison of leukemia cell deformability. Biophysical journal 90: 2994–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Mahaffy R, Park S, Gerde E, Käs J, Shih C. (2004) Quantitative analysis of the viscoelastic properties of thin regions of fibroblasts using atomic force microscopy. Biophysical journal 86: 1777–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Rotsch C, Jacobson K, Radmacher M. (1999) Dimensional and mechanical dynamics of active and stable edges in motile fibroblasts investigated by using atomic force microscopy. Proceedings of the National Academy of Sciences 96: 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Pesen D, Hoh JH. (2005) Micromechanical architecture of the endothelial cell cortex. Biophysical journal 88: 670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Murakoshi M, Yoshida N, Iida K, Kumano S, Kobayashi T, et al. (2006) Local mechanical properties of mouse outer hair cells: Atomic force microscopic study. Auris Nasus Larynx 33: 149–157. [DOI] [PubMed] [Google Scholar]
- [245].Bausch AR, Möller W, Sackmann E. (1999) Measurement of Local Viscoelasticity and Forces in Living Cells by Magnetic Tweezers. Biophysical journal 76: 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Bausch AR, Ziemann F, Boulbitch AA, Jacobson K, Sackmann E. (1998) Local Measurements of Viscoelastic Parameters of Adherent Cell Surfaces by Magnetic Bead Microrheometry. Biophysical journal 75: 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Sagvolden G, Giaever I, Pettersen EO, Feder J. (1999) Cell adhesion force microscopy. Proceedings of the National Academy of Sciences 96: 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Thoumine O, Ott A, Louvard D. (1996) Critical centrifugal forces induce adhesion rupture or structural reorganization in cultured cells. Cell motility and the cytoskeleton 33: 276–287. [DOI] [PubMed] [Google Scholar]
- [249].Yamamoto A, Mishima S, Maruyama N, Sumita M. (1998) A new technique for direct measurement of the shear force necessary to detach a cell from a material. Biomaterials 19: 871–879. [DOI] [PubMed] [Google Scholar]
- [250].Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, et al. (2001) Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nature cell biology 3: 466–472. [DOI] [PubMed] [Google Scholar]
- [251].Galbraith CG, Sheetz MP. (1997) A micromachined device provides a new bend on fibroblast traction forces. Proceedings of the National Academy of Sciences 94: 9114–9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Davies PF, Dewey CF, Jr, Bussolari SR, Gordon EJ, Gimbrone MA., Jr (1984) Influence of hemodynamic forces on vascular endothelial function. In vitro studies of shear stress and pinocytosis in bovine aortic cells. Journal of Clinical Investigation 73: 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Morigi M, Zoja C, Figliuzzi M, Foppolo M, Micheletti G, et al. (1995) Fluid shear stress modulates surface expression of adhesion molecules by endothelial cells. Blood 85: 1696–1703. [PubMed] [Google Scholar]
- [254].Galbraith C, Skalak R, Chien S. (1998) Shear stress induces spatial reorganization of the endothelial cell cytoskeleton. Cell motility and the cytoskeleton 40: 317–330. [DOI] [PubMed] [Google Scholar]
- [255].Ji JY, Jing H, Diamond SL. (2003) Shear stress causes nuclear localization of endothelial glucocorticoid receptor and expression from the GRE promoter. Circulation research 92: 279–285. [DOI] [PubMed] [Google Scholar]
- [256].Chau L, Doran M, Cooper-White J. (2009) A novel multishear microdevice for studying cell mechanics. Lab on a Chip 9: 1897–1902. [DOI] [PubMed] [Google Scholar]
- [257].Bao G, Suresh S. (2003) Cell and molecular mechanics of biological materials. Nat Mater 2: 715–725. [DOI] [PubMed] [Google Scholar]
- [258].Avraham-Chakim L, Elad D, Zaretsky U, Kloog Y, Jaffa A, et al. (2013) Fluid-flow induced wall shear stress and epithelial ovarian cancer peritoneal spreading. PLoS One 8: e60965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Byun S, Son S, Amodei D, Cermak N, Shaw J, et al. (2013) Characterizing deformability and surface friction of cancer cells. Proc Natl Acad Sci U S A 110: 7580–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]