Abstract
Exosomal microRNAs modulate cancer cell metabolism and the immune response. Specific exosomal microRNAs have been reported to be reliable biomarkers of several solid and hematologic malignancies. We examined the possible diagnostic and prognostic values of exosomal microRNAs in two human bone marrow failure diseases: aplastic anemia and myelodysplastic syndromes. After screening 372 microRNAs in a discovery set (n=42) of plasma exosome samples, we constructed a customized PCR plate, including 42 microRNAs, for validation in a larger cohort (n=99). We identified 25 differentially expressed exosomal microRNAs uniquely or frequently present in aplastic anemia and/or myelodysplastic syndromes. These microRNAs could be related to intracellular functions, such as metabolism, cell survival, and proliferation. Clinical parameters and progression-free survival were correlated to microRNA expression levels in aplastic anemia and myelodysplastic syndrome patients before and after six months of immunosuppressive therapy. One microRNA, mir-126-5p, was negatively correlated with a response to therapy in aplastic anemia: patients with higher relative expression of miR-126-5p at diagnosis had the shortest progression-free survival compared to those with lower or normal levels. Our findings suggest utility of exosomal microRNAs in the differential diagnosis of bone marrow failure syndromes. (Registered at clinicaltrials.gov identifiers: 00260689, 00604201, 00378534, 01623167, 00001620, 00001397, 00217594).
Introduction
The bone marrow (BM) failure syndromes are a heterogeneous group of hematologic diseases characterized by peripheral blood (PB) cytopenia due to hematopoietic stem and progenitor cell (HSPC) destruction and dysfunction, and/or constitutional syndromes due to genetic lesions.1–3 Blood count improvement after immunosuppressive therapies (IST) implicates autologous immune-mediated HSPC destruction in aplastic anemia (AA).4 Myelodysplastic syndromes (MDS) are a heterogeneous group of clonal pre-malignant diseases characterized by ineffective hematopoiesis, progressive cytopenias, increased risk of developing acute myeloid leukemia (AML), and poor overall survival.2,5 However, approximately 10-15% of MDS cases share clinical and pathological features with AA, in a subtype known as hypocellular MDS; conversely, 10-15% of patients with BM failure ultimately develop secondary MDS after IST, suggesting that AA and MDS have common mechanisms in pathophysiology and disease progression.6,7 Despite the discovery of new molecular biomarkers, the differential diagnosis between BM failure syndromes and their prognosis is still challenging.8
In an effort to improve the definition of these hematologic disorders, circulating microRNAs (miRNAs, small non-coding RNA molecules of about 22 nucleotides), have been described as potential biomarkers of AA and MDS.9,10 There is evidence that miRNAs are crucial in controlling and modulating immunity, promoting survival and growth of malignant cells, and in cancer metastasis.11,12 Some studies have demonstrated the presence of cancer-specific miRNAs in plasma or serum, suggesting utility of these molecules as potential biomarkers of diseases.13,14
Exosomes are small extracellular vesicles directly released into the extracellular space by all cell types;15 RNAs (protected from degradation16) and proteins are contained within these vesicles, allowing the transfer of genetic material and signaling proteins to recipient cells.17,18 Indeed, exosomal miRNAs influence biological functions, and they have been proposed as specific and reliable biomarkers of many malignant disorders, including colon cancer, melanoma, and AML, and to track minimal residual diseases in hematologic malignancies.19–27
In the current work, we investigated plasma exosomal miRNAs as potential minimally-invasive biomarkers of AA and MDS. Detection of specific exosomal miRNAs in these diseases might aid in understanding the pathophysiology of BM failure as well as diagnosis and prognosis.
Methods
Human plasma samples
Whole PB was collected in ethylenediaminetetraacetic acid (EDTA) tubes from patients and healthy subjects after informed consent was obtained in accordance with the Declaration of Helsinki28 and protocols approved by the National Heart, Lung, and Blood Institute Institutional Review Board [National Institutes of Health (NIH), Bethesda, MD, USA]. All patients had a diagnosis of severe AA (SAA) or MDS according to standard criteria.29,30 Healthy controls were recruited from donors of the NIH Clinical Center Department of Transfusion Medicine.
A discovery set (n=42), which was used for an initial screening of an exosomal miRNA signature, included 16 healthy controls, 16 SAA patients, and 10 MDS patients. For a validation set (n=99), 36 healthy controls, 54 SAA patients, and 20 MDS patients were recruited. Additionally, 10 patients from the discovery set and 30 SAA patients from the validation set were screened for exosomal miRNA expression before and after six months of IST. Clinical characteristics are summarized in Online Supplementary Table S1. Specimens were collected at the time of diagnosis and after six months of IST. After centrifugation at 2000 RPM for 10 min, plasma was collected and stored at −80°C until use.
Exosome extraction
Isolation of exosomes from 800 μL of plasma was performed using the PureExo Exosome Isolation kit (101Bio, Palo Alto, CA, USA) according to the manufacturer’s instructions. Flow-through was collected and stored at −80°C until further use. To confirm the presence of exosomes in the flow-through, protein content, particle size, CD63 expression, and transmission electron microscopy were assessed (Online Supplementary Appendix and Online Supplementary Figure S1).31
RNA extraction and cDNA synthesis
To obtain high-quality RNA, a bead-based RNA purification protocol was performed using the Direct-zol™ −96 MagBead RNA kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions with minor modifications. Purified RNA was subjected to cDNA synthesis with the miScript® II RT kit (Qiagen, Hilden, Germany), and undiluted cDNA stored at −20°C until further use. To confirm that RNA was confined within exosomes, exosomes extracted from several samples were treated with RNase A, as described in the Online Supplementary Appendix.
miRNA profiling
Initial screening of 372 miRNAs and 12 miRNA controls was performed in the discovery set using the miScript® miRNA PCR Array Human Serum & Plasma 384HC array (MIHS-106ZE, Qiagen) and the miScript® SYBR® Green PCR kit (Qiagen). Data were analyzed using the real-time thermal cycler 7900HT Fast Real-Time PCRs (Applied Biosystems, Thermo Fisher Scientific). A custom miScript miRNA PCR array (CMIHS02531E, Qiagen) including 42 candidate miRNAs and 6 controls was designed for validation.
Statistical analysis
Data were analyzed using Prism (v.7.02; GraphPad software, La Jolla, CA, USA). miScript miRNA PCR Array Data Analysis software was utilized to analyze data from miRNA PCR arrays. Ingenuity Pathway Analysis (v.33559992, Qiagen Bioinformatics)32 and miRWalk 2.033 software were used for pathway and/or prediction analyses.
Results
Exosomal miRNA content profiling shows a distinct signature in SAA and MDS
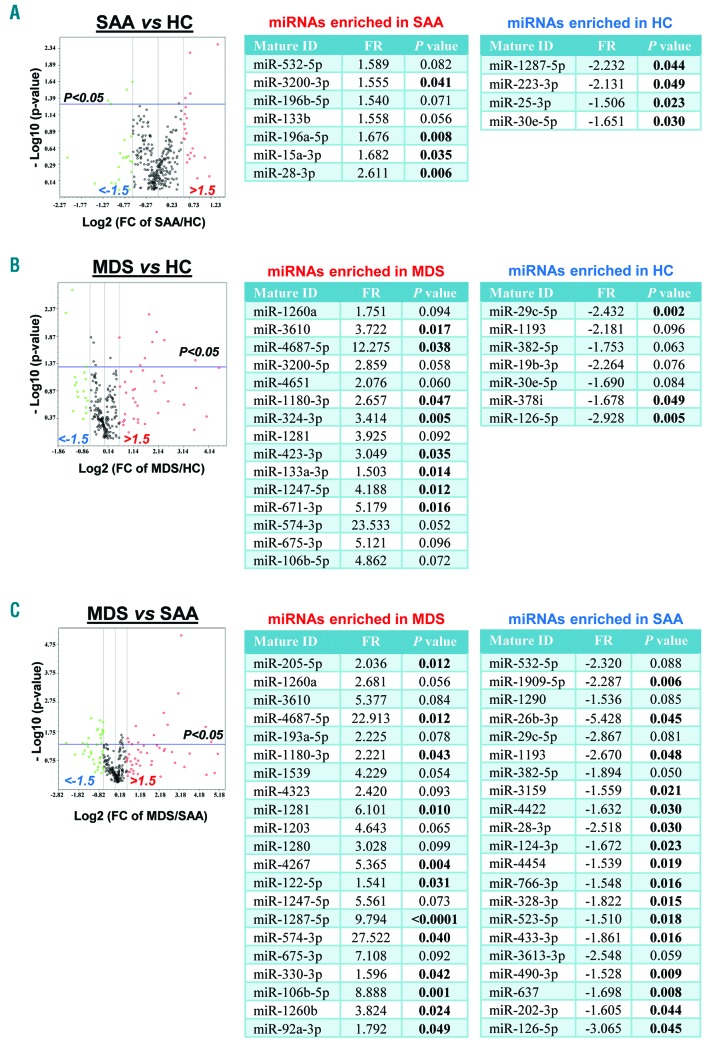
Previous studies investigated circulating miRNA expression levels in the plasma of AA and MDS patients,9,10 but no characteristic exosomal miRNAs have been described to date in these diseases. We first screened a large number of miRNAs (372 miRNAs associated with serum and plasma and 12 control miRNAs; miScript® miRNA PCR Array MIHS-3106Z; http://www.sabiosciences.com/genetable.php?pcatn=MIHS-3106Z) in plasma exosome samples in the discovery set by principal component analysis (PCA). We found different miRNAs in SAA (7 up- and 4 down-regulated) and MDS (15 up- and 7 down-regulated) (Figure 1A and B). Exosomal miRNA expression profiles were compared between SAA and MDS, resulting in 42 miRNAs (21 and 21 up-regulated in MDS and SAA, respectively) (Figure 1C).
Figure 1.
miRNAs in severe aplastic anemia (SAA) and myelodysplastic syndromes (MDS) patients compared to healthy controls (HC) in the discovery set. Principal component analysis was performed to compare 384 exosomal miRNA expression levels: SAA versus HC (A), MDS versus HC (B), and MDS versus AA (C). Results are shown using volcano plots and tables. In volcano plots, x- and y-axes show estimated expression difference measured in Log2(FC) and the significance of the expression difference measured in −Log10(P-value), respectively. In the plots, horizontal and vertical lines indicate cut-off of significance (P<0.05) and expression levels greater than ±1.5-fold regulation (FR), respectively. For each comparison, miRNAs with ±1.5 FR and P<0.1 are displayed in correspondent tables in which P<0.05 is highlighted in bold.
Validation of miRNA content profiles
For validation of results obtained in the discovery set, 42 candidate miRNAs were selected and examined in a larger cohort of SAA, MDS, and healthy controls (n=99) (see Online Supplementary Table S1 for clinical characteristics and Online Supplementary Table S2 for selected miRNAs). To assess reaction quality and for data normalization, 6 control miRNAs (miRTC, PPC, SNORD61, miR-339-3p, miR-211-5p, and miR-30c-5p) were included in the custom PCR array plate, and analysis was performed as described in the Online Supplementary Appendix.
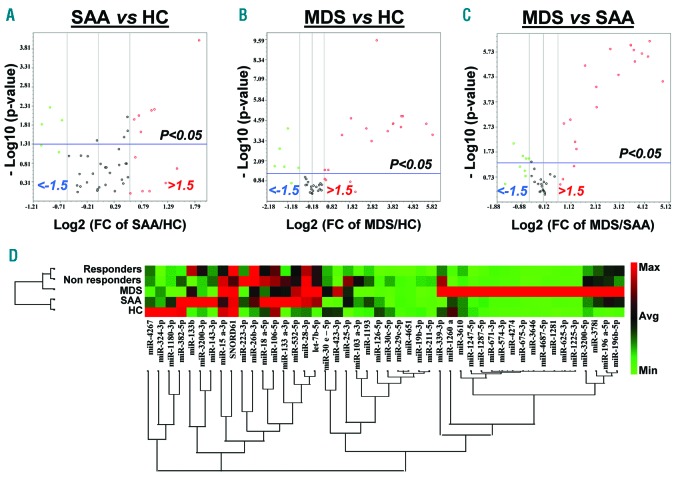
PCA of the validation set revealed 9 miRNAs present in SAA (6 up- and 3 down-regulated) (Figure 2A). When MDS patients were compared to healthy controls, 15 and 5 miRNAs were significantly up-regulated and down-regulated, respectively (Figure 2B). In addition, MDS patients were compared to the SAA group, and 20 miRNAs were found to be differentially expressed (Figure 2C). Hierarchical clustering displayed differences in exosomal miRNA signatures between SAA and MDS (Figure 2D).
Figure 2.
Validation of miRNA signatures in severe aplastic anemia (SAA) and myelodysplastic syndromes (MDS) patients. Principal component analysis was employed to compare the 48 miRNA expression levels in the validation set: SAA versus healthy controls (HC) (A), MDS versus HC (B), and MDS versus SAA (C). These results are shown with volcano plots in a similar manner as described in Figure 1. (D) Hierarchical clustering visualizes the 48 exosomal miRNAs in SAA, MDS, SAA-responders, SAA-non-responders, and HC. A red-green color scale indicates normalized miRNA expression levels (red: maximum; green: minimum).
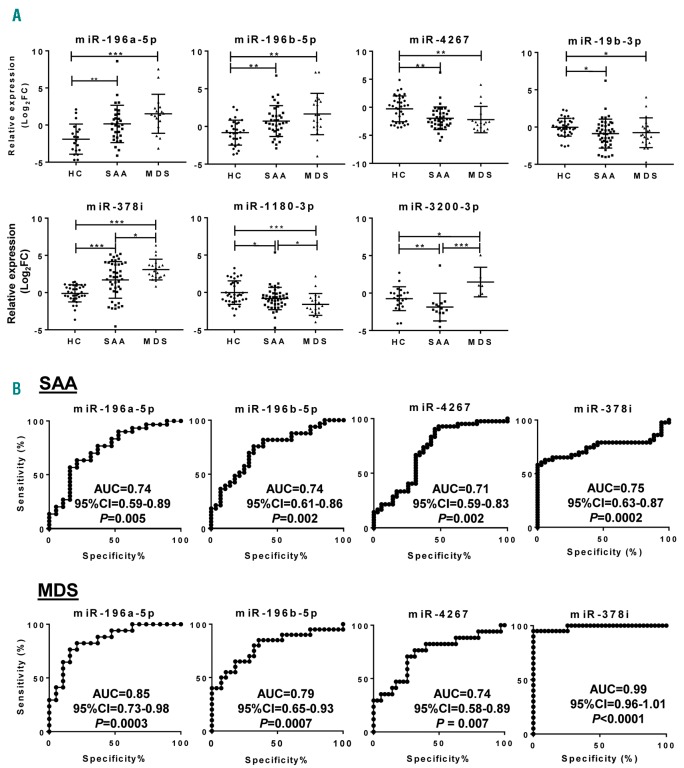
Relative expression (Log2FC) values of 42 selected miRNAs were calculated for each sample; data were compared among SAA, MDS, and control groups by one-way ANOVA using the Kruskal-Wallis test and multiple comparisons by the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli (Figure 3 and Online Supplementary Figure S2). miR-196a-5p and miR-196b-5p levels were higher in SAA (P=0.009 and P=0.002, respectively) and MDS (P<0.0001 and P=0.0003, respectively) compared to controls, but there were no differences between SAA and MDS groups (details of the analysis in Online Supplementary Table S3). miR-4267, miR-19b-3p, and miR-1180-3p levels were lower in both SAA and MDS patients compared to controls. miR-378i was increased in both SAA and MDS, with higher levels in MDS. miR-532-5p was increased in SAA (P=0.026) but not in MDS (P=0.098) compared to controls. Relative levels of 13 miRNAs were higher in MDS compared to controls and SAA patients, while miR-126-5p and miR-382-5p were lowest in MDS. miR-3200-3p was decreased in SAA and increased in MDS patients compared to controls. miR-423-3p, miR-1193, and miR-143-3p were higher in MDS patients relative to SAA and controls. Other miRNAs did not show variations in either SAA or MDS.
Figure 3.
Exosomal miRNAs in severe aplastic anemia (SAA) and myelodysplastic syndromes (MDS) patients. Relative expression levels of the 48 miRNAs were calculated as Log2FC and shown for each group [SAA, MDS, and healthy controls (HC)]. (A) 7 exosomal miRNAs differentially expressed in both SAA and MDS compared to HC. (B) Receiver operating characteristic (ROC) curves for 4 miRNAs in AA and MDS. The ROC curve of the miRNA panel was generated based on the predicted probability for each patient and using the healthy group as a control. P<0.05 was considered statistically significant. AUC: area under the curve; CI: Confidence Interval.
Association of exosomal miRNAs with diseases
To assess specificity and sensitivity of exosomal miRNAs for the diagnosis of SAA and MDS, a receiver operating characteristic (ROC) curve analysis was employed using the validation set samples (Figure 3 and Online Supplementary Table S3). miR-196a-5b [area under the curve (AUC), 0.74], miR-196b-5p (AUC, 0.74), miR-4267 (AUC, 0.71), miR-378i (AUC, 0.75), miR-19b-3p (AUC, 0.68), miR-1180-3p (AUC, 0.64), miR-423-3p (AUC, 0.63), miR-532-5p (AUC, 0.65), miR-574-3p (AUC, 0.64), and miR-3200-3p (AUC, 0.77) showed strong associations with SAA. Other miRNAs were not statistically significantly associated with the disease.
In MDS patients, 21 exosomal miRNAs displayed strong association with the disease (Figure 3B). Strong association was observed in all 7 miRNAs present in both SAA and MDS patients with the highest AUC value (0.99) of miR-378i, followed by miR-574-3p (AUC, 0.87), miR-196a-5p (AUC, 0.85), miR-3200-3p (AUC, 0.83), miR-196b-5p (AUC, 0.79), miR-1180-3p (AUC, 0.79), miR-4267 (AUC, 0.74), and miR-19b-3p (AUC, 0.67).
Correlation with clinical parameters
Pearson correlation analysis was used to investigate correlation of exosomal miRNAs with clinical parameters of hemoglobin (Hb), white blood cell (WBC) count, platelet (Plt) count, absolute neutrophil count (ANC), absolute lymphocyte (ALC) and absolute reticulocyte counts (ARC), and lactate dehydrogenase (LDH) level in MDS patients and in SAA patients (before and after IST) from the validation set (Online Supplementary Table S4). Correlation with LDH was included in the analysis because higher LDH has been related to hemolysis and to disease progression in many malignant hematologic disorders.34 No association was found between any exosomal miRNA and Hb at diagnosis and after treatment. miR-574-3p and miR-4274 were positively associated with WBC (r=−0.332, P=0.021) and Plt (r=0.330, P=0.021) counts at diagnosis, respectively, but not after IST. miR-15a-3p (r=0.312, P=0.026), miR-532-5p (r=0.368, P=0.010), and miR-26b-3p (r=0.364, P=0.017) positively correlated with an LDH level before therapy. miR-103a-3p and miR-29c-3p positively correlated with ANC before IST, while miR-126-5p negatively correlated after treatment (r=−0.326, P=0.040). miR-4651 was positively related to ARC before therapy, but no miRNAs were found after IST. However, SAA patients after treatment displayed negative correlations between ALC and several miRNAs: miR-3200-3p, miR-196a-5p, miR-28-3p, miR-133b, miR-26b-3p, let-7b-5p, miR-133a-3p, and miR-106b-5p (Online Supplementary Table S4). In MDS, miR-1180-3p was positively correlated with a Hb level (r=0.483, P=0.036) and WBC count (r=0.561, P=0.013), while miR-3200-3p (r=0.963, P=0.002), miR-196b-5p (r=0.485, P=0.035), miR-378i (r=0.498, P=0.030), and miR-1260a (r=0.495, P=0.037) only to WBC count. No miRNAs showed a correlation with Plt count or ARC. miR-223-3p and miR-19b-3p were positively associated with an LDH level. miR-3200-3p was also positively associated with ANC (r=0.911, P=0.012), and miR-196a-5p, miR-15a-3p, miR-133b, miR-106b-5p, and miR-4267 with ALC (Online Supplementary Table S4).
miR-126-5p as a candidate biomarker of response to IST in SAA
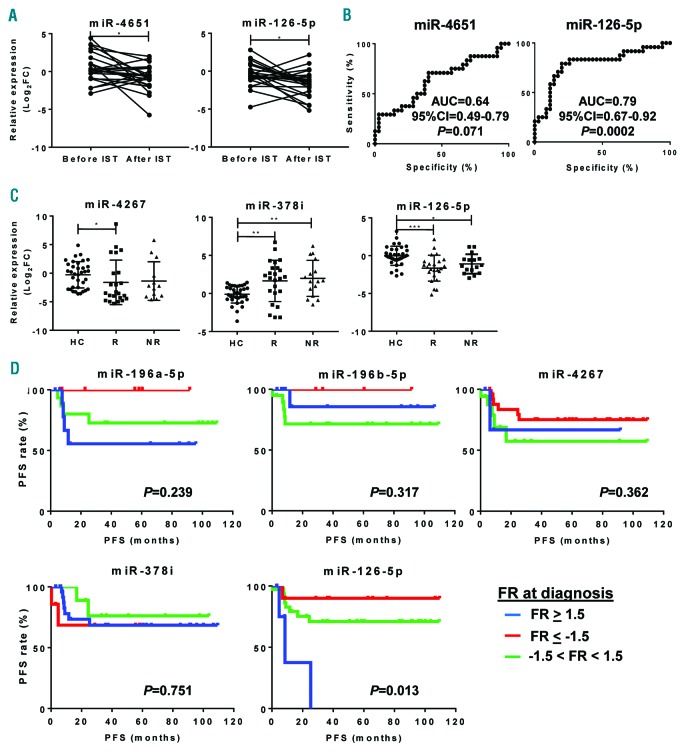
We next sought to investigate effects of IST on exosomal miRNA profiles after six months of treatment in 40 SAA patients. PCA was performed to detect miRNAs in SAA patients before and after IST (Online Supplementary Figure S3). miR-143-3p (P=0.033), miR-324-3p (P=0.001), miR-1180-3p (P=0.009), miR-126-5p (P=0.008), and miR-382-5p (P=0.009) were significantly decreased after treatment (Online Supplementary Figure S4). Subsequently, SAA patients were classified into either SAA-responders (complete or partial response) or SAA-non-responders after six months of therapy based on standard clinical parameters, and miRNA expression profiles were then compared before and after IST. No variations were observed after IST for any miRNAs in SAA-non-responders. In SAA-responders, miR-4651 (P=0.015) and miR-126-5p (P=0.025) were significantly reduced after treatment (Figure 4A). However, only miR-126-5p (AUC, 0.79) displayed a strong association with the response to IST in SAA patients by ROC analysis (Figure 4B). When exosomal miRNA levels of SAA patients were compared at post treatment to those of healthy controls, miR-126-5p was significantly decreased in responders compared to healthy controls (Figure 4C). However, no differences were observed before therapy.
Figure 4.
Exosomal miRNAs and their correlation with prognosis in severe aplastic anemia (SAA) responders. Relative expression levels were calculated as Log2FC for all 48 miRNAs in SAA patients and compared before and after immunosuppressive therapies (IST). (A) Only miR-4651 and miR-126-5p were significantly decreased after treatment in SAA-responders. (B) For these 2 exosomal miRNAs, receiver operating characteristic (ROC) curves were generated as described in Figure 3 using a healthy group as a control. (C) Relative expression of differentially expressed exosomal miRNAs at diagnosis and after treatment in responders (R) and non-responders (NR) of SAA patients were compared to healthy controls (HC) using one-way ANOVA with Kruskal-Wallis and the two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli tests. Data are shown as mean+Standard Deviation (SD). (D) After calculation of progression-free survival (PFS), patients were divided into three groups according to miRNA relative expression shown as Log2FC at diagnosis of selected miRNAs. P<0.05 was considered statistically significant. AUC: area under the curve; FR: fold regulation.
SAA patients were classified based on fold regulation (FR) values before treatment, and progression-free survival (PFS) of each group was determined as shown in Figure 4D. When miRNA levels were undetermined, patients were removed from the analysis. There were no significant variations in PFS based on relative expression of miR-196a-5p (P=0.239), miR-196b-5p (P=0.317), miR-4267 (P=0.362), miR-378i (P=0.751), miR-19b-3p (P=0.093), miR-1180-3p (P=0.069), and miR-3200-3p (P=0.744). However, SAA patients with higher miR-126-5p at diagnosis experienced decreased PFS (8.5 months; n=6; median follow up 6.9 months; range 3.2-25.3 months) compared to those with normal or lower levels (n=36 and n=11; median follow up 22.7 and 37.4 months; range 0.3-109.3 and 3-109.5 months, respectively; P=0.013) (Figure 4D).
Pathway analysis of exosomal miRNAs
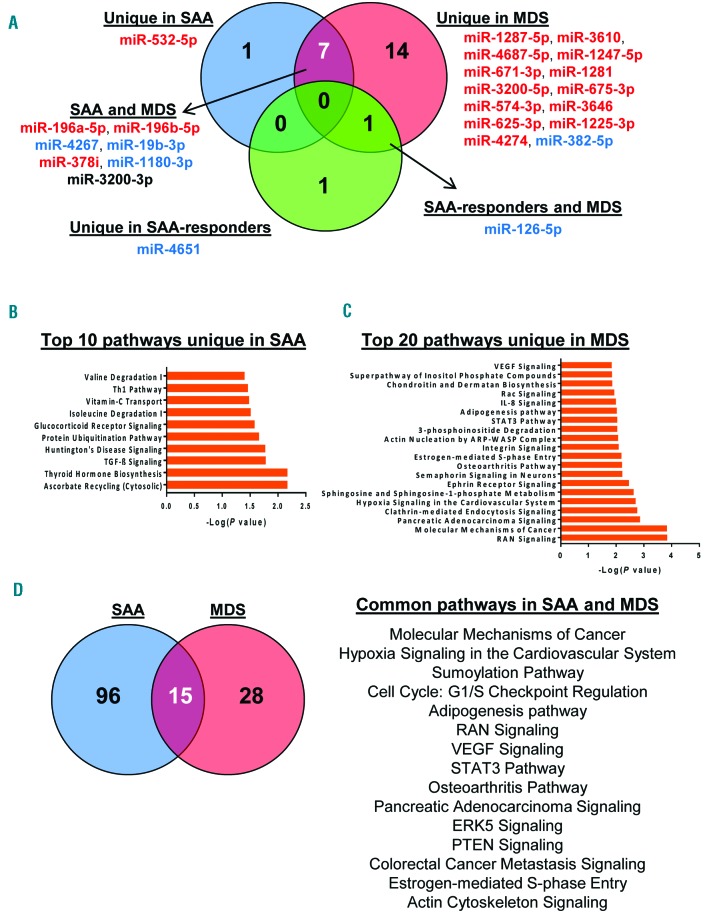
Using available software,35 increased or decreased miRNAs in each group (SAA, MDS, and SAA-responder patients) were interpolated (Figure 5). Venn diagrams displayed that miR-532-5p was unique in SAA, miR-4651 unique in SAA-responders, 14 miRNAs were unique in MDS, 7 miRNAs common in SAA and MDS, and miR-126-5p was common in MDS and SAA-responders to IST (Figure 5A). Next, predicted target genes for each miRNA were identified using miRWalk 2.0 database, followed by filtering to extract genes that were exclusively present in SAA, MDS, or SAA-responders, or present in both MDS and SAA or SAA-responders (Figure 5B and C, Online Supplementary Table S5 and Online Supplementary Figure S5). Common genes targeted by the 7 shared miRNAs but not present in MDS were selected for analysis, and the top 20 pathways were reported (Figure 5D). miR-4651 and miR-126-5p were used for pathway analysis in SAA-responders, after removing the common targeted genes from MDS and SAA at diagnosis (Online Supplementary Figure S5C). The analysis revealed involvement of several intracellular functions, related to cell cycle, DNA damage response, intracellular signaling, and metabolic pathways.
Figure 5.
Pathway analysis using differentially expressed exosomal miRNAs. (A) VENNY (an interactive tool for comparing lists with Venn Diagrams) was used to find common or unique miRNAs among severe aplastic anemia (SAA), myelodysplastic syndromes (MDS), and SAA-responder patients. miRNAs classified into individual groups are listed accordingly. Red: increased exosomal miRNAs; blue: decreased exosomal miRNAs; miR-3200-3p is shown in black because of different expression profiles between SAA (down-regulated) and MDS (up-regulated). Predicted targeted genes of miRNAs exclusively expressed in SAA or MDS were used for pathway analysis by IPA software. Top 10 pathways in SAA (B) and the top 20 in MDS (C) are shown. (D) Venn diagram shows the number of unique or common pathways in SAA and MDS and a list of the 15 common signaling pathways.
Discussion
We investigated exosomal miRNA profiles in SAA and MDS in order to find potential biomarkers for diagnosis and disease progression in BM failure syndromes. Based on screening of 372 miRNAs in the discovery set of plasma exosome samples, a custom miRNA PCR plate was designed, including 42 miRNAs for validation in a larger cohort. The analysis revealed 25 exosomal miRNAs that were uniquely or commonly present in SAA and/or MDS patients; they were involved in several biological functions, such as HSC differentiation.
Recently published work from our laboratory describes circulating miRNAs using whole plasma samples of AA and MDS patients;9 however, no distinctive signatures have been reported for exosomal miRNAs to date. In our current study, we identified exosomal miRNAs exclusively present in SAA (miR-532-5p), MDS (14 miRNAs), and SAA responders to IST (miR-4651) patients, or common to SAA and MDS (7 miRNAs), and to SAA responders to IST and MDS (miR-126-5p). The miRNAs we identified have not been reported to be different in AA and/or MDS plasma samples. However, circulating miRNAs are composed of passively released nucleic acids from different cell types,15,36 and actively secreted in exosomes. Exosomal miRNAs are not randomly loaded into vesicles; rather, mature miRNAs are specifically sorted by diverse mechanisms and based on sequence, reflecting the tissue of origin.17,20,24 Because they are tissue-specific and stable under different conditions, exosomal miRNAs have been proposed as better biomarkers. Their significance in AA and MDS has not yet been examined. Plasma circulating miRNAs have been related to extracellular pro-inflammatory signaling pathways, such as Toll-like receptor or tumor necrosis factor α.9 Dysregulated exosomal miRNAs in our cohort were associated with numerous intracellular functions.
Circulating miR-532-5p (unique in SAA) is related to response to chemotherapy in AML, and associated with decreased expression of Runt-related (RUNX) 3 protein in melanoma.37,38 miR-196a and miR-196b (present in our SAA and MDS cases) are differentially expressed in the long-term HSC compartment compared to more mature populations, and miR-196 induces the promotion of HSC differentiation through the inhibition of Homeobox (HOX) family members.39 miR-19b (decreased in our SAA and MDS cases) together with other miRNAs promotes the differentiation of progenitor cells into lymphoid precursors through inhibition of PTEN.39 Additionally, cytokines modulate miRNA expression, such as miR-196 by interferons during hepatitis C infection.40 Based on this, increased exosomal miR-532-5p and miR-196, and decreased miR-19b in our cohort of SAA and MDS patients may indicate attempted expansion of the stem and progenitor cell compartments. miR-1180-3p has an anti-apoptotic function through inhibition of the BCL protein family;41 this miRNA was decreased in our SAA and MDS, patients more apoptotic in marrow with active disease. Other exosomal miRNAs present in our cohort do not have known functions in hematopoiesis or the immune response.
There is increasing evidence of essential roles of circulating and exosomal miRNAs in modulating cancer cell metabolism, the immune response, and altering the local and distant tumor environments.12,17,42 miR-126-5p and its 3’ UTR counterpart are regulators of angiogenesis, cell metabolism, and glucose homeostasis, and they are down-regulated in many tumors. By targeting the PI3K/AKT/mTOR pathway, overexpression of miR-126 results in reduction of proliferation, driving malignant and normal HSCs into quiescence.43,44 Decreased exosomal miR-126 is not only related to increased proliferation of leukemic cells and differentiation44 but also decreases autophagy in cancer cells, leading to accumulation of damaged mitochondria and reactive oxygen species.44,45 In SAA, increased amino acid biosynthesis has been reported.46 It is plausible that miR-126-5p modulates metabolic programming during BM failure and recovery, especially as miR-126-5p was decreased in MDS and SAA responders to IST. Pathway analysis of predicted target genes using miRWalk 2.0 and IPA software revealed that downstream signaling could be regulated by exosomal miRNAs, as for MAPK, p38, JAK/STAT, and ERK. These pathways can be activated by many different extracellular stimuli, such as cytokines and growth factors, and exosomal miRNAs by interfering with this signaling could impair intracellular responses.12,17 However, miRNAs from circulating exosomes may be derived from different tissues, such as BM or T cells. Therefore, additional functional studies and exosomal miRNA profiling in BM samples will resolve the origin and biological functions of these miRNAs during active disease.
Utilization of circulating and exosomal miRNAs in clinical practice is uncertain, mainly due to technical issues such as data normalization, exosome preparation, and RNA extraction.9,47–49 For circulating miRNAs, normalization is performed using small nucleolar RNAs (snoRNAs) and other methods such as GeNormPlus, NormFinder, and global mean of miRNA expression. For exosomal miRNAs, there are no clear guidelines for data normalization. RNU6 used for fresh samples for miRNA normalization is not a reliable endogenous control for frozen samples.47 The mean expression value is often applied for nor-malization47 but employing only one method could result in missing significant differences.48 In our study, we normalized our data using the geometric mean of 2 control miRNAs within each group: one snoRNA (SNORD61) and one miRNA (miRNA-211-5p) homogeneously expressed in the discovery set. This approach achieved high concordance across plates and for replicates. Ultracentrifugation remains the standard assay for extracellular vesicles isolation and enrichment, as no commercial kit achieves a high-yield exosome with more than 99% purity and therefore allows exclusion of lipoproteins. Treating extracted exosomes with RNase or proteinase K can increase the purity of samples, but there are no generally accepted guidelines for such manipulations.49 In consideration of these limitations, we utilized the PureExo Exosome Isolation kit for vesicle extraction because the ultra-centrifugation step can be eliminated and exosomes can be obtained from small volume biological samples.
Biomarkers allow better evaluation of disease in general and improve prognosis, thus helping clinicians in the decision-making process. In SAA, age, sex, and pre-treatment PB counts are established as prognostically valuable markers for response to IST.50 In the current study, we observed positive correlation of some exosomal miRNAs with WBC and Plt counts, and LDH levels at diagnosis in SAA, but not with pre-treatment PB counts. These results suggest that their expression was not affected by transfusion history or disease severity, and that miRNAs could be used as independent diagnostic or prognostic markers. Similarly, positive correlation of miRNAs with clinical parameters was also seen in MDS.
Exosomal miRNAs are available from PB, and they are considered reliable biomarkers because they are protected from degradation and specifically loaded into vesicles from proliferating or apoptotic cells. We propose further study of measurement of exosomal miRNAs in marrow failure syndromes in general. We have identified different miRNA signatures in SAA and MDS, and a candidate biomarker of responsiveness to IST. However, a larger cohort of patients combined with an ultra-pure exosome isolation technique are needed to validate the clinical utility of exosomal miRNAs.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the support of the Electron Microscopy Core of the National Heart, Lung, and Blood Institute (NHLBI) at National Institutes of Health (NIH, Bethesda, MD, USA). The authors would like to thank Xingmin Feng and Sawa Ito (Hematology Branch, NHLBI, NIH), Nastaran Rezaie and Anisha Kharkia (Qiagen, Germantown, MD, USA), and Leonid Margolis (National Institute of Child Health and Human Development, NIH, Bethesda, MD, USA) for technical assistance; and Kinneret Broder and Therese Intrater (Hematology Branch, NHLBI, NIH) for assistance in obtaining samples from healthy volunteers.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/7/1150
Funding
This research was supported by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
References
- 1.Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108(8):2509–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenaux P, Adès L. How we treat lower-risk myelodysplastic syndromes. Blood. 2013;121(21):4280–4286 [DOI] [PubMed] [Google Scholar]
- 3.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 4.Young NS. Hematopoietic cell destruction by immune mechanisms in aquired aplastic anemia. Semin Hematol. 2000;37(1):3–14. [DOI] [PubMed] [Google Scholar]
- 5.Zeidan AM, Al Ali N, Barnard J, et al. Comparison of clinical outcomes and prognostic utility of risk stratification tools in patients with therapy-related vs. de novo myelodysplastic syndromes: A report on behalf of the MDS clinical research consortium. Leukemia. 2017;31(6):1391–1397. [DOI] [PubMed] [Google Scholar]
- 6.Maciejewski JP, Selleri C. Evolution of clonal cytogenetic abnormalities in aplastic anemia. Leuk Lymphoma. 2004;45(3):433–440. [DOI] [PubMed] [Google Scholar]
- 7.Serio B, Risitano A, Giudice V, Montuori N, Selleri C. Immunological derangement in hypocellular myelodysplastic syndromes. Transl Med UniSa. 2014;8:31–42. [PMC free article] [PubMed] [Google Scholar]
- 8.DeZern AE, Sekeres MA. The challenging world of cytopenias: distinguishing myelodysplastic syndromes from other disorders of marrow failure. Oncologist. 2014;19(7):735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosokawa K, Kajigaya S, Feng X, et al. A plasma microRNA signature as a biomarker for acquired aplastic anemia. Haematologica. 2017;102(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler JT, Abdelhamed S, Kurre P. Extracellular vesicles in the hematopoietic microenvironment. Haematologica. 2018;103(3):382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang WT, Chen YQ. Circulating miRNAs in cancer: from detection to therapy. J Hematol Oncol. 2014;7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasetti M, Santarelli L, Neuzil J, Dong L. MicroRNA regulation of cancer metabolism: role in tumour suppression. Mitochondrion. 2014;19 Pt A:29–38. [DOI] [PubMed] [Google Scholar]
- 13.Grasedieck S, Sorrentino A, Langer C, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood. 2013;121(25):4977–4984. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chen J, Sen S. MicroRNA as Biomarkers and Diagnostics. J Cell Physiol. 2016;231(1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. [DOI] [PubMed] [Google Scholar]
- 16.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. miRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19(2):1568–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med. 2017;49(1):e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franquesa M, Hoogduijn MJ, Ripoll E, et al. Update on controls for isolation and quantification methodology of extracellular vesicles derived from adipose tissue mesenchymal stem cells. Front Immunol. 2014;5:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleem SN, Abdel-Mageed AB. Tumor-derived exosomes in oncogenic reprogramming and cancer progression. Cell Mol Life Sci. 2015;72(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyiadzis M, Whiteside TL. Information transfer by exosomes: A new frontier in hematologic malignancies. Blood Rev. 2015;29(5):281–290. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Wang S, Sun K, Chng WJ. The emerging roles of exosomes in leukemogeneis. Oncotarget. 2016;7(31):50698–50707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turpin D, Truchetet ME, Faustin B, et al. Role of extracellular vesicles in autoimmune diseases. Autoimmun Rev. 2016;15(2):174–183. [DOI] [PubMed] [Google Scholar]
- 23.Buzas EI, György B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10(6):356–364. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, De Veirman K, Faict S, et al. Multiple myeloma exosomes establish a favourable bone marrow microenvironment with enhanced angiogenesis and immunosuppression. J Pathol. 2016; 239(2):162–173. [DOI] [PubMed] [Google Scholar]
- 25.Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeffer SR, Grossmann KF, Cassidy PB, et al. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J Clin Med. 2015;4(12):2012–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornick NI, Huan J, Doron B, et al. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci Rep. 2015;5:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194. [DOI] [PubMed] [Google Scholar]
- 29.International Agranulocytosis and Aplastic Anaemia Study. Incidence of aplastic anemia: the relevance of diagnostic criteria. By the International Agranulocytosis and Aplastic Anemia Study. Blood. 1987;70(6):1718–1721. [PubMed] [Google Scholar]
- 30.Greenberg PL, Stone RM, Al-Kali A, et al. Myelodysplastic Syndromes, Version 2.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2017;15(1):60–87. [DOI] [PubMed] [Google Scholar]
- 31.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit3.22. [DOI] [PubMed] [Google Scholar]
- 32.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014; 30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. [DOI] [PubMed] [Google Scholar]
- 34.Dumontet C, Drai J, Bienvenu J, et al. Profiles and prognostic values of lactate dehydrogenase isoenzymes in patients with non-hodgkin’s lymphoma. Leukemia. 1999;13(5):811–817. [DOI] [PubMed] [Google Scholar]
- 35.Oliveros JC. Venny. An interactive tool for comparing lists with Venn’s diagrams. (2007-2015). Available from: http://bioin-fogp.cnb.csic.es/tools/venny/index.html
- 36.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosakhani N, Räty R, Tyybäkinoja A, Karjalainen-Lindsberg ML, Elonen E, Knuutila S. MicroRNA profiling in chemoresistant and chemosensitive acute myeloid leukemia. Cytogenet Genome Res. 2013;141(4):272–276. [DOI] [PubMed] [Google Scholar]
- 38.Kitago M, Martinez SR, Nakamura T, Sim MS, Hoon DS. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin Cancer Res. 2009;15(9):2988–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montagner S, Dehó L, Monticelli S. MicroRNAs in hematopoietic development. BMC Immunol. 2014;15:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan G, Wu L, Tan J, et al. MiR-1180 promotes apoptotic resistance to human hepatocellular carcinoma via activation of NF- B signaling pathway. Sci Rep. 2016;6:22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356(2 Pt A):301–308. [DOI] [PubMed] [Google Scholar]
- 43.Ebrahimi F, Gopalan V, Smith RA, Lam AK. miR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol. 2014;96(1):98–107. [DOI] [PubMed] [Google Scholar]
- 44.Lechman ER, Gentner B, Ng SW, et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell. 2016;29(2):214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomasetti M, Monaco F, Manzella N, et al. MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma. Oncotarget. 2016; 7(24):36338–36352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong P, Zhang J, Cui X. Abnormal metabolites related to bone marrow failure in aplastic anemia patients. Genet Mol Res. 2015;14(4):13709–13718. [DOI] [PubMed] [Google Scholar]
- 47.Manier S, Liu CJ, Avet-Loiseau H, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood. 2017;129(17):2429–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bockmeyer CL, Säuberlich K, Wittig J, et al. Comparison of different normalization strategies for the analysis of glomerular microRNAs in IgA nephropathy. Sci Rep. 2016;6:31992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narita A, Kojima S. Biomarkers for predicting clinical response to immunosuppressive therapy in aplastic anemia. Int J Hematol. 2016;104(2):153–158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.