Abstract
In Huntington's disease (HD), accurate estimates of expected future motor impairments are key for clinical trials. Individual prognosis is only partially explained by genetics. However, studies so far have focused on predicting the time to clinical diagnosis based on fixed impairment levels, as opposed to predicting impairment in time windows comparable to the duration of a clinical trial. Here we evaluate an approach to both detect atrophy patterns associated with early degeneration and provide a prognosis of motor impairment within 3 years, using data from the TRACK-HD study on 80 premanifest HD (pre-HD) individuals and 85 age- and sex-matched healthy controls. We integrate anatomical MRI information from gray matter concentrations (estimated via voxel-based morphometry) together with baseline data from demographic, genetic and motor domains to distinguish individuals at high risk of developing pronounced future motor impairment from those at low risk. We evaluate the ability of models to distinguish between these two groups solely using baseline imaging data, as well as in combination with longitudinal imaging or non-imaging data. Our models show improved performance for motor prognosis through the incorporation of imaging features to non-imaging data, reaching 88% cross-validated accuracy when using baseline non-longitudinal information, and detect informative correlates in the caudate nucleus and the thalamus both for motor prognosis and early atrophy detection. These results show the plausibility of using baseline imaging and basic demographic/genetic measures for early detection of individuals at high risk of severe future motor impairment in relatively short timeframes.
Keywords: Future motor impairment prediction, Premanifest Huntington's disease, Classification, Structural MRI, TRACK-HD
Highlights
-
•
Detection of pre-HD subjects at high risk of impairment is key for clinical trials.
-
•
Prognostic models of motor impairment can aid the detection of this population.
-
•
Genetics only partially explains disease progression (need for other correlates).
-
•
We achieve improved prognosis with baseline imaging, demographics and motor data.
1. Introduction
Huntington's disease (HD) is a neurodegenerative disorder that manifests as a triad of motor, cognitive and behavioral impairments that typically develop in adulthood. Current treatments for HD target symptom management. However, evidence of neurodegeneration beginning in HD many years before clinical diagnosis (Sheinerman and Umansky, 2013; Stout et al., 2011; Tabrizi et al., 2013) calls for interventions that will delay its onset or slow its progression prior to symptom manifestation (Lang, 2010; Paulsen et al., 2006). In that context, it is very important to consider alternative estimates of HD progression at presymptomatic stages of the disease (pre-HD) (Long et al., 2017) or assessments of future impairment for reliable pre-HD prognosis.
HD is caused by a single genetic mutation, a CAG repeat expansion in the huntingtin gene that allows its detection before symptom manifestation. Despite existing evidence that the CAG repeat length and age of onset are associated with disease progression (Penney et al., 1997), the heterogeneous clinical nature of this disease can only be partially explained by these factors (Ross et al., 2014). Therefore, it is important to find potential biomarkers of disease progression in addition to the number of CAG repeats.
In an effort to find biomarkers from multiple domains, observational studies such as TRACK-HD (Tabrizi et al., 2009) and PREDICT-HD (Paulsen et al., 2008) have followed hundreds of subjects across several years. Several publications based on these studies have focused on addressing two research problems: the characterization of early degeneration in HD and the estimation of motor impairment through HD progression. In the context of the first research question, localized atrophy patterns were detected through descriptive statistical observations using gray matter concentration (GMC) estimates through voxel-based morphometry (VBM) in pre-HD (Ciarochi et al., 2016), early HD (Hobbs et al., 2010; Kassubek et al., 2004) and throughout multiple stages of HD (Tabrizi et al., 2009, Tabrizi et al., 2013). VBM-based predictive multivariate approaches were also tested to infer the condition of unseen subjects using cross-validation strategies (Klöppel et al., 2009; Kostro et al., 2014). In longitudinal analyses, changes in brain volumes (Aylward et al., 2011) and white matter microstructure (Domínguez et al., 2016; Shaffer et al., 2017) were analyzed to detect variations specific to pre-HD (in juxtaposition to healthy controls) in an effort to detect clinically relevant biomarkers to measure disease progression. The other problem of interest, the characterization of motor impairment in HD, has been extensively studied due to the nature of HD diagnosis (Biglan et al., 2009; Long et al., 2014). In fact, most approaches aimed at the detection of potential structural neuroimaging biomarkers have been designed to find correlates of time-to-motor onset. These studies include descriptive statistical approaches based on volumetric analyses of pre-defined regions of interest (ROI) (Biglan et al., 2009; Paulsen et al., 2010), as well as predictive univariate (Paulsen et al., 2014) and multivariate strategies (Long and Paulsen, 2015) for motor onset estimation. While the inference of motor conversion is informative given the nature of HD diagnosis, prognostic models of future motor impairment that transcend the conversion phenotype are essential to detect pre-HD individuals at higher risk of deterioration.
This study addresses the aforementioned research problems. The first aim of this work is to identify regions with aberrant GMC covariation that provide an enriched characterization of pre-HD, giving complementary information to that obtained with descriptive statistics. The discrimination of pre-HD subjects from healthy controls allows us to detect signatures of atrophy at early disease stages and assess the information carried by GMC for future motor impairment prediction. Our classification approach, which makes use of VBM processing, achieves results comparable to previous work (Klöppel et al., 2009; Kostro et al., 2014). For the second research question, the characterization of motor impairment in HD, we propose a prognostic model within a relatively short time frame. To this end, we explore methods for motor impairment prognosis in pre-HD on a follow-up visit 3 years later in two tasks. First, we evaluate the prognostic prediction power achieved with imaging data only, with future motor impairment being quantified via the total motor score (TMS) of the unified Huntington's disease rating scale (UHDRS) (Huntington Study Group, 1996). By analyzing baseline GM integrity and longitudinal atrophy progression we can detect the information carried by accumulated atrophy effects and changes over time, as well as interactions between them. For the next task, we analyzed the effects of incorporating baseline GMC from each subject along with baseline data from other modalities (age, CAG repeats and TMS). Our results show that GMC from a single visit, along with basic demographic and genetic measurements, has prognostic power and can identify brain regions with aberrant baseline structure that could be useful for target-based clinical trials.
2. Methods
2.1. Study
TRACK-HD is a multinational longitudinal observational study whose goal is to identify changes that occur from health to early-stage HD through measurements within the genetic, clinical, motor and brain-imaging domains, among others. The cohort is evenly divided into pre-HD individuals, early HD individuals and age- and sex-matched controls.
2.2. Participants
We used data from 179 subjects (HD gene carriers and healthy controls) enrolled in the TRACK-HD study, each of them with imaging data available for at least one visit other than baseline and having reported TMS in visits 1 and 4, as shown in Fig. 1a. The average number of visits per subject with available MRI data was 3.9. Our predictive models were only trained with healthy controls and pre-HD individuals with absence of HD diagnosis throughout all the visits. Data from those subjects that converted to HD were used only for post-hoc validation of the trained models. The set of healthy controls was selected to be closely matched to the pre-HD sample in terms of group size, age and sex. The resulting training dataset was composed of 80 pre-HD individuals and 85 controls. The number of individuals who were not part of the training set because they converted to manifest HD during the study was 14. Basic demographics and CAG-based information for pre-HD individuals are reported in Table 1.
Fig. 1.
Available multimodal data features and schematics of classification tasks. (a) Multimodal features extracted from subjects with MRI scans for at least two given visits (baseline visit inclusive) were used. Baseline information of interest for each subject was composed of structural brain measures, number of CAG repeats, age, CAG-Age Product (CAP) score and TMS. TMS at visit 4 was used to infer future motor impairment. (b) Visual representation of GMC imaging features. (c) Schematics of cross-validation approach applied to classify subjects based on genetics (healthy controls and pre-HD individuals) or future TMS (pronounced and mild impairment).
Table 1.
Basic demographics and genetics-based information of study participants. Information is itemized for healthy controls, pre-HD individuals with no HD diagnosis througout the study and pre-HD individuals who converted to manifest HD. Age, CAG repeats and CAP scores are reported in terms of mean ± SD.
| Age | Sex (M:F) | CAG | CAP | |
|---|---|---|---|---|
| Controls | 44.1 ± 8.9 | 38:47 | ||
| Pre-HD (no diagnosis) | 41.0 ± 8.3 | 37:43 | 42.9 ± 2.1 | 45.9 ± 7.1 |
| Pre-HD (converted) | 40.6 ± 10.5 | 8:6 | 43.8 ± 3.1 | 51.0 ± 8.3 |
TRACK-HD exclusion criteria included age below 18 or above 65, major psychiatric, neurological or medical disorder or a history of severe head injury, as described elsewhere (Klöppel et al., 2015). The study was approved by the local ethics committees and all participants gave written informed consent according to the Declaration of Helsinki.
2.3. TMS adjustment and discretization
The focus of this work is to detect pre-HD individuals at high risk of motor deterioration. For this reason, we trained models that could discriminate subjects with pronounced future motor impairment (high risk) from those with mild future motor impairment (low risk), rather than devising models to estimate their actual motor scores. To account for the age dependent increase of TMS in healthy controls (Paulsen et al., 2014), we made a linear fit between the TMS of healthy controls and their age and removed the “normal aging” effects from the TMS estimated for pre-HD subjects (i.e., used residualized scores). High (low) risk groups were composed of subjects with TMS values above (below) the third (first) quartiles of the TMS distribution at visit 4 in pre-HD. Each group is composed of 20 subjects, the data from the remaining 40 subjects (intermediate risk) being used only for post-hoc validation of the trained models.
2.4. MRI data acquisition
3 T MRI data from 2 different scanner systems (Siemens Tim Trio and Philips Achieva) and 4 sites were acquired as described elsewhere (Klöppel et al., 2015; Tabrizi et al., 2009).
T1-weighted image volumes were acquired using a 3D MPRAGE acquisition sequence with the following imaging parameters: TR = 2200 ms (Siemens)/7.7 ms (Philips), TE = 2.2 ms(S)/3.5 ms(P), FA = 10°(S)/8°(P), FOV = 28 cm(S)/24 cm(P), matrix size 256 × 256(S)/224 × 224(P), 208(S)/164(P) sagittal slices to cover the entire brain with a slice thickness of 1.0 mm with no gap. T2-weighted image volumes were acquired with identical field of view, acquisition matrix, and slice thickness as T1-weighted images, and were used to provide complementary information for improved brain extraction from raw images. All volumes passed rigorous quality control by IXICO.
2.5. MRI data processing and feature extraction
A longitudinal extension of the “optimized” VBM protocol (Good et al., 2001) of the FMRIB Software Library (FSL) (Jenkinson et al., 2012) was used to evaluate interactions of baseline GM patterns and their changes across visits to discriminate groups of interest. We followed a procedure similar to the one proposed by Douaud and colleagues (Douaud et al., 2009) with some modifications. First, the T1-weighted anatomical volumes were adjusted for subject orientation changes across visits through a robust, inverse consistent rigid body registration between time points (Reuter et al., 2010), generating a template space and an associated image for each subject. These images were used along with masks estimated from their T2-weighted volumes to do brain extraction of raw T1-weighted volumes (see Fig. 2). The resulting images were then segmented into 3 tissue partial volumes (GM, white matter and cerebrospinal fluid concentrations) that represent the probability of each voxel belonging to a given tissue. GMC volumes were averaged across visits and the resulting mean volumes from each subject were incorporated on the aforementioned optimized VBM protocol to generate a study-specific GM template. Then, GMC volumes of each subject/visit were nonlinearly normalized into this template and smoothed with an isotropic Gaussian kernel with sigma of 1.7 mm (~4 mm full width at half maximum). From these images, we retrieved estimates of baseline GMC and its longitudinal decline (GMC slopes) across time.
Fig. 2.
Longitudinal processing to generate baseline GMC images and to estimate their decline across visits.
All preprocessing steps were implemented via FSL and FreeSurfer (Fischl, 2012) utilities, parallelized for large-scale computation using Nipype (Gorgolewski et al., 2011) in Python.
To correct for natural GM shrinkage associated to healthy aging in our pre-HD-specific analysis, we made a voxel-wise linear estimation of the baseline GMC changes as a function of age for healthy controls and regressed out this effect from GMC images from pre-HD subjects. This estimation is based on evidence of linear degradation of GM across time during adulthood (Giorgio et al., 2010; Walhovd et al., 2005).
In addition to imaging-based features, we used basic baseline information from demographic, genetic and motor domains to detect individuals at high risk of deterioration in pre-HD. This information is represented by the individuals' number of CAG repeats together with their baseline age and TMS. Another non-imaging feature of interest is the CAG-age product (CAP) score, which is the product of the number of CAG repeats of an individual and its age (Zhang et al., 2011). Here we use a normalized CAP score such that CAP = 100 × AGE × [(CAG − 35.5) ÷ 627], where the score at disease onset is approximately 100.
2.6. Group analyses
In this work we applied statistical tests and classification approaches to estimate the information embedded in imaging data (baseline GMC or GMC slopes) and clinic-demographic variables (baseline age, TMS, CAG and CAP) to detect pre-HD individuals and to infer future motor impairment within pre-HD. Specifically, we focused in 3 aims: 1) detection of atrophy patterns and classification of pre-HD with imaging data, 2) prognosis of motor impairment in pre-HD individuals with imaging data, and 3) prognosis of motor impairment in pre-HD individuals with multimodal data. For the first aim, we evaluated the information carried by GMC differences between pre-HD individuals and healthy controls by using baseline GMC maps only. For the second aim, we analyzed whether baseline GMC and longitudinal changes of GM integrity throughout visits could be used to infer future TMS within the population of pre-HD individuals. Finally, after assessing the information carried by imaging data, we evaluated the predictive power of non-imaging features alone and in combination with baseline structural imaging to infer future motor impairment in pre-HD.
2.6.1. Classification and feature selection
Discrimination between 2 groups (either pre-HD vs controls or mild vs pronounced motor impairment) was performed using logistic regression. For our models to make correct predictions both in training and new data we included a regularization term (Tikhonov, 1963). Regularization controls for model complexity which typically leads to overfitting the training data, yielding poor results on test data. In order to detect a subset of features relevant for classification we applied elastic-net regularization (Zou and Hastie, 2005). Fig. 1c shows this approach being applied to imaging data, though it can be generalized to data from other modalities. Whole-brain voxel-level imaging data was used as input to the classifier, unless otherwise stated. The optimal value of regularization parameters was determined within each round of cross-validation. We used a nested cross-validation scheme composed of an outer leave-one-out cross-validation with an internal 5-fold cross-validation for performance estimation and parameter selection, respectively. For the case in which whole-brain data was not used, top-ranked voxels that conveyed optimal performance were also selected via univariate selection in the internal cross-validation procedure. To detect the location of the strongest effects for classification, we computed maps of the mean voxel weights assigned by the classifier.
2.6.2. Additional post-hoc assessments of classification
For aim 3, the contribution of multimodal features to classification was assessed by also computing the weights of non-imaging features to the estimation of future motor impairment. In addition, we evaluated the estimated prediction of our model for data that were not included in the training set, such as samples from pre-HD individuals with moderate future motor impairment and converted subjects.
2.6.3. Statistical analyses
Between-group univariate effects were estimated for comparison with multivariate effects. In this way we can detect additional patterns identified by multivariate techniques. Univariate tests were used under certain conditions as a first-stage feature selection approach prior to elastic-net regularization (see Classification and feature selection above). All univariate statistical tests for group comparisons as well as for feature selection (where applicable) were Wilcoxon rank-sum tests. For all brain maps, we controlled for multiple comparisons by estimating a false discovery rate (FDR) through the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995). For the estimation of confidence intervals for classification performance metrics, we used bootstrap resampling (105 resamples) of test predictions.
2.6.4. Classification performance metrics
The following measures were used to evaluate the performance of the trained models for both classification of pre-HD individuals and healthy controls and detection of subjects at high and low risk of deterioration within pre-HD.
Accuracy: Fraction of correct classifications among the total number of analyzed samples.
Sensitivity: Fraction of correct classifications among positive samples (pre-HD or high-risk).
Specificity: Fraction of correct classifications among negative samples (controls or low-risk).
Positive predictive value (PPV, or hits): Fraction of correct classifications among samples given a positive prediction (pre-HD or high-risk).
Negative predictive value (NPV, or correct rejections): Fraction of correct classifications among samples given a negative prediction (controls or low-risk).
Brier score: Average square deviation of probability of predictions and actual outcomes.
3. Results
3.1. Atrophy patterns in pre-HD
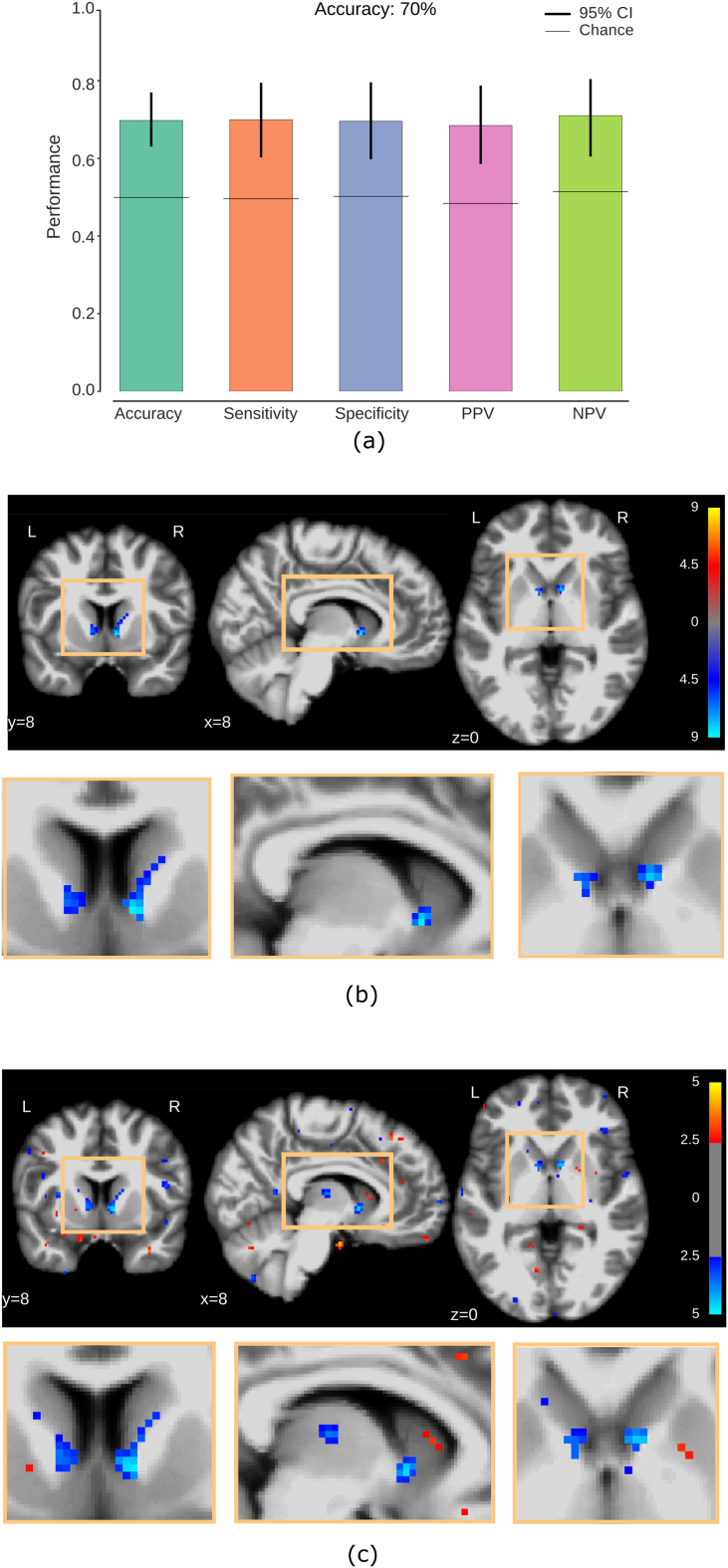
First we evaluated the information carried by GMC differences between pre-HD individuals and healthy controls by using full-brain GMC maps from these two groups. The results of the logistic regression classifier with elastic-net regularization are shown in Fig. 3a, the obtained classification accuracy being 70% between pre-HD and controls. After performing a voxel-wise univariate test of GMC between pre-HD individuals and healthy controls (Fig. 3b), we found significant group differences (Wilcoxon rank-sum test, FDR corrected, q < 0.05) localized bilaterally in the inferior part of the head of the caudate nucleus, with stronger effects showing up on the right hemisphere. In addition, we found that the mean voxel weight maps assigned by the classifier (Fig. 3c) show spatial patterns that are consistent with those of the univariate maps. However, our multivariate maps have a larger spatial extent, also showing GMC deficits in pre-HD in voxels that exhibit insignificant individual effects such as those located in the thalamus. Notably, these maps do not rely on potential site effects (see Supplementary Material). In summary, we find a consistent pattern of GM deterioration bilaterally in the inferior part of the caudate nucleus and we show that this spatial pattern plays an important role in the detection of pre-HD.
Fig. 3.
GMC detects localized patterns of brain atrophy in pre-HD individuals and discriminates them from healthy controls. (a) Full-brain cross-validated classification performance of controls and pre-HD individuals. Error bars indicate 95% confidence intervals (bootstrap resampling of classifier predictions). Horizontal lines show chance performance of the classifier (randomization of test labels). (b) Univariate GMC differences between pre-HD and controls. Two bilateral clusters are located in the inferior part of the head of the caudate nucleus (Wilcoxon rank-sum test, FDR corrected at q < 0.05). Color bar: red: pre-HD > controls, blue: pre-HD < controls. Units: common logarithm of uncorrected p-value. (c) Maps of the mean voxel weights assigned by the classifier. Multivariate weights are compatible with univariate maps but extend to other regions such as the thalamus. Weights shown at an arbitrary threshold of 2.5, in units of the standard deviation across all voxels, for illustration purposes.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Prognosis of pre-HD population with imaging data
We then extended our analysis of imaging data by incorporating voxel-level longitudinal estimates of GMC change for future motor impairment detection in pre-HD. Fig. 4a shows the classification performance to discriminate groups of individuals with mild and pronounced future motor impairment using baseline GMC, achieving a classification accuracy of 70%. By using longitudinal changes via GMC slopes, the model's accuracy is reduced to 67% (Fig. 4b). In addition, Fig. 4c shows the performance of the classifier when both baseline GMC and GMC slopes are used in combination, yielding a classification accuracy of 70%.
Fig. 4.
GM integrity at baseline visit along with GM atrophy rates discriminate pre-HD individuals with mild and pronounced future motor impairment. Classification performance to estimate future motor impairment (a) using GMC at baseline visit only, (b) using GMC slopes and (c) by combining them. All conventions as in Fig. 3a. Maps of the mean voxel weights assigned by the classifier for (d) GM integrity and (e) GM atrophy rates. GMC at baseline and GM rate of change show spatial complementarity of multivariate effects in the inferior/superior part of the head of the caudate nucleus, respectively.
In terms of the localization of correlated effects of future motor deterioration, it is evident that these have a spatial complementary nature for baseline and longitudinal imaging. The location of strong effects for GM integrity at baseline are mostly found bilaterally in the inferior part of the head of the caudate nucleus, showing a negative correlation between GMC in these regions and the severity of future motor impairment (Fig. 4d). On the other hand, patterns of negative correlation between longitudinal GM change and future motor deterioration are located in the superior part of the head of the caudate nucleus, meaning that GM atrophy in these regions is positively correlated with motor impairment.
In summary, by incrementing the number of imaging features to be twice as much as that of a single data source (baseline GMC) the accuracy of the classifier is not degraded. Furthermore, interesting patterns of spatial complementarity are detected for GMC at baseline and GMC slopes. These findings suggest that these two data sources provide complementary information. Nonetheless, it seems like the usage of GMC at baseline visit suffices to obtain good prediction of future motor decline.
3.3. Prognosis of pre-HD population with multimodal data
The set of non-imaging features composed of baseline metrics such as age, CAG repeats, CAP score and TMS achieved a classification accuracy of 75% (Fig. 5a). In order to evaluate whether imaging data could complement non-imaging features for future motor impairment detection, we integrated non-imaging features on top of baseline GMC. A group of GMC voxels chosen via feature selection by an internal cross-validation procedure (see Methods section) was incorporated to the non-imaging features for future motor impairment inference, and improved the accuracy rate to 88% (with information from 5 voxels, total set of 9 features as shown in Fig. 5b and c). It can be seen that informative patterns of GMC are highly localized, with the classifier performance gradually decreasing as more voxels are combined with non-imaging features.
Fig. 5.
The incorporation of baseline GMC to multimodal data significantly increases the estimation of future motor impairment. Classification performance to estimate future motor impairment (a) using baseline non-imaging features (CAG repeats, age, CAP score and TMS) and (b) by adding baseline GMC information. All conventions as in Fig. 3a. (c) Classification accuracy rate decreases as more voxels are combined with non-imaging features. As less informative voxels for future motor impairment estimation (according to internal cross-validation) are incorporated to non-imaging features, the classification accuracy gradually decreases, with the optimal value being obtained for the 5 most informative voxels (9 features total). (d) Most informative voxels and their spatial extent as additional voxels are incorporated in the classification. The red cluster conveys the location of the 5 voxels that provide improved classification accuracy of future motor impairment, while the voxels in the blue cluster show the spatial extent of the informative voxels as they grow up to 1000. The voxels in the blue cluster clearly span the caudate nucleus bilaterally, including the thalamic region. (e) Unitary-scaled mean weights of non-imaging features for the estimation of future motor impairment. Error bars indicate 95% confidence intervals.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The location of atrophy patterns that are relevant for classification of motor impairment is shown in Fig. 5d, the most informative patterns of GMC being located in the right head of the caudate nucleus and part of its boundary with the nucleus accumbens. While the best classification is achieved by using a small set of voxels, an improved interpretation of the location of informative patterns of GMC would be achieved by relaxing the spatial constraint for voxel selection. For that purpose, we arbitrarily allowed our model to select the 1000 most informative voxels. These voxels span the head of the caudate nucleus bilaterally and expand into the thalamus. Importantly, the extent of these areas is not influenced by site variability (see Supplementary material). By analyzing the relative contributions of non-imaging features to the estimation of future motor impairment, it can be seen that baseline TMS, CAG and CAP score are positively correlated with future motor impairment (weights significantly different from 0 as shown in Fig. 5e), the highest contribution being provided by baseline TMS, which is to be expected.
Due to the high reported contribution (relative to other features) of baseline TMS and localized patterns of GMC to the correct estimation of future motor impairment, we considered it suitable to do a post-hoc analysis of the performance of our classification approach when it is trained with baseline TMS only, a set of base attributes (CAG, age, CAP score) and through the incremental inclusion of TMS and imaging features to the model. Table 2 shows that a consistent classification improvement is achieved through the aggregation of these features. In addition to the already reported accuracy rate of up to 88%, the incorporation of baseline TMS and imaging features improves the Brier score from 0.23 to 0.16.
Table 2.
Aggregation of baseline TMS and imaging to genetic/demographic features provides improved estimation of future motor impairment. Standard features used for estimation of motor onset (CAG, age and CAP score) are enriched with baseline TMS and GMC features. A consistent improvement of the classifier performance is obtained when these features are sequentially incorporated for classification.
| Features | Accuracy | Sensitivity | Specificity | PPV | NPV | Brier |
|---|---|---|---|---|---|---|
| CAG, CAP & age | 0.66 | 0.65 | 0.66 | 0.62 | 0.65 | 0.23 |
| TMS | 0.73 | 0.70 | 0.75 | 0.74 | 0.70 | 0.21 |
| CAG, CAP, age & TMS | 0.75 | 0.75 | 0.75 | 0.75 | 0.75 | 0.21 |
| Imaging + CAG, CAP, age & TMS | 0.88 | 0.85 | 0.90 | 0.89 | 0.85 | 0.16 |
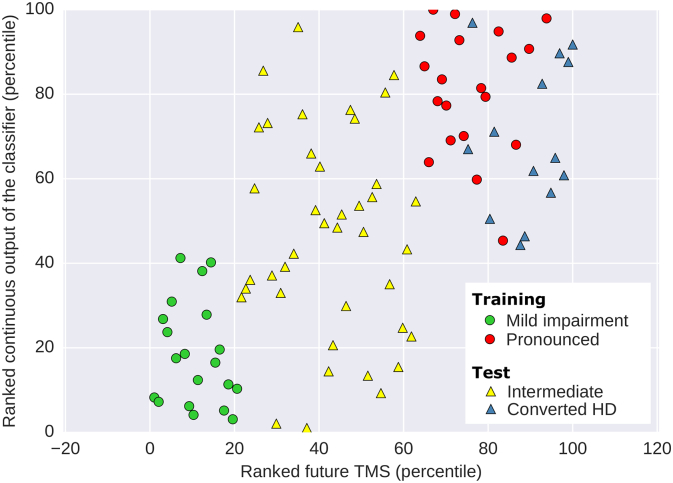
It should be stressed that while the presented model has been trained with a subset of pre-HD subjects with mild and pronounced motor impairment, it is still capable of providing information about subjects that do not belong to either of these two groups, such as prospective patients that will develop moderate impairment. Furthermore, the model can also infer the amount of motor deterioration of pre-HD individuals who became manifest throughout the study. This is possible as the raw output provided by a linear classifier is continuous and represents the (signed) distance of an observation to the decision hyperplane. To illustrate this point, Fig. 6 shows all the subjects (including those who will develop moderate impairment and converted subjects) arranged according to the continuous output of the classifier. Despite not being provided with the future outcome from moderate and converted subjects, the majority of individuals that develop moderate impairment are located between those with mild and pronounced impairment. In addition, converted subjects are ranked similarly to those with future pronounced impairment. This shows that even if the classifier is trained with individuals from two clearly different groups to improve its performance, it is still capable of providing information about unseen subjects with various degrees of future motor impairment.
Fig. 6.
Model learned from groups with mild and pronounced motor impairment within pre-HD can be applied to intermediate and converted HD cases. Scatter plot of the ranked continuous output of the classifier and the ranked future TMS using 5 most informative voxels for classification of future motor impairment (according to internal cross-validation) and non-imaging features (baseline age, TMS, CAG repeats and CAP score). Individuals with intermediate future motor impairment are assigned a continuous output by the classifier that lies between groups with future mild and pronounced impairment. Furthermore, converted HD individuals are assigned consistently along with pronounced HD subjects.
4. Discussion
In this work we evaluate the capacity of GMC to detect aberrant structural patterns in pre-HD and to estimate future motor impairment at early stages of the disease using a cross-validated classification approach. We also demonstrate an improved detection of individuals at high risk of developing pronounced motor deterioration through the integration of multimodal data (baseline demographic, genetic and motor information combined with GMC volumes). The generated prognostic models estimate future motor impairment of individuals at preclinical stages in HD several years before clinical onset. Our study focused on the characterization of future motor impairment of pre-HD individuals who did not manifest symptoms throughout the study. However, post-hoc analyses show that reasonable predictions can be made for pre-HD subjects that converted to manifest HD in this time period. Our approach also shows the potential of GMC information in identifying structural correlates of motor impairment, indicating potential regions with abnormal baseline structure that could be used for target-based clinical trials that aim to slow disease progression in pre-HD.
We first evaluated our method for the detection of atrophy patterns in pre-HD compared to control subjects. For this task we obtained a whole-brain classification accuracy rate of 70%. This result is comparable with those obtained for inter-site classification performance in pre-HD within TRACK-HD, which used information from voxels in the striatum only (Kostro et al., 2014), while it surpasses the results presented for the PREDICT-HD study (Klöppel et al., 2009). Patterns of reduced GM in pre-HD detected in the caudate nucleus confirm findings in earlier studies and the aforementioned classification approaches. While GM loss in the thalamus is evident in early HD (Douaud et al., 2006; Kassubek et al., 2004), findings in pre-HD are not in entire agreement (Thieben et al., 2002; Van Den Bogaard et al., 2011). This may be explained to some extent by the fact that non-negligible GM multivariate effects were detected in the thalamus in spite of showing insignificant individual effects for pre-HD detection. In fact, this finding is consistent with results from another multivariate analysis, which detected GMC reduction covariation in both the thalamus and the caudate nucleus in pre-HD (Ciarochi et al., 2016). These findings reassured that our features extracted with VBM were informative enough to extend our work from pre-HD detection to motor deterioration prediction within pre-HD.
While motor impairment has been extensively analyzed in HD to find putative markers based on imaging and clinical assessments or to characterize its progression (Biglan et al., 2009; Long et al., 2014; Paulsen et al., 2014), these analyses rely on the knowledge of the time to disease manifestation of the evaluated individuals. Such an approach focuses on the estimation of the chances of motor conversion in a given time period in the future as opposed to assessing the degree of future motor impairment developed at early disease stages. Here we have presented a multivariate, cross-validated approach that incorporates baseline GMC information on top of non-imaging baseline metrics for an accurate detection of individuals at higher risk of developing pronounced motor deterioration in pre-HD, as measured by future TMS. Accuracy improves from 75% to up to 88% when imaging data is incorporated. The only precedent of the use of cross-validated classification approaches using imaging data for future motor impairment detection was applied for the prediction of HD motor diagnosis (Long and Paulsen, 2015), which reported a Brier score of 0.11 (the lower the score, the more accurate the model is) when incorporating imaging features to predict the probability of an individual becoming manifest in the next 5 years. The Brier score achieved by our best model (0.16, see Table 2) is very close to this value. It should be stressed though that our approach is more challenging for two reasons: we estimate future motor impairment within a shorter time frame (3 years from baseline visit) and we do so for motor impairment within pre-HD, where impairment signals are generally less pronounced (Biglan et al., 2009). We argue that the inclusion of baseline GMC makes our model more robust to the inherent inter- and intra-rater variability of TMS (Acton, 2012). Overall, we show that baseline GM integrity is informative enough to boost future motor impairment estimation in pre-HD within the TRACK-HD study population relative to genetics and baseline impairment.
Further analysis of our findings let us detect informative patterns for later motor impairment estimation in the imaging and non-imaging feature domains, as well as the interactions among them. Reduced baseline GM integrity in the head of the caudate nucleus and the thalamus on both hemispheres is a strong correlate of future motor impairment in pre-HD, in addition to being informative for pre-HD detection (see Fig. 3, Fig. 5). We also show that the nucleus accumbens is informative for future motor impairment estimation, consistent with previous findings for pre-HD and early HD detection (Douaud et al., 2006; Hobbs et al., 2010; Van Den Bogaard et al., 2011), suggesting that high GM atrophy in these regions within pre-HD can also be associated to high risk of pronounced future motor impairment. Evidence of atrophy in the thalamus and the nucleus accumbens being correlated to TMS in pre-HD (Van Den Bogaard et al., 2011) supports this claim. It should also be noticed that the effects on the thalamus would have been neglected if our model was based on single variable effects, as it can be seen by comparing Fig. 3b and c. In terms of the influence of non-imaging features, we observe that the highest contribution to motor impairment estimation is provided by baseline TMS. Furthermore, TMS, CAG and CAP score (along with loss of GMC in the caudate nucleus and thalamus) are positively correlated with future motor impairment. In contrast, age exhibits a consistently negative multivariate association ([−0.64, −0.36], CI = 95%) with the predicted variable. This observation may be related to the exclusion of manifest subjects, which imposes a negative correlation between CAG repeats and age. This shows that our multivariate approach is able to detect interactions between features, providing a fully integrated interpretation of data from multiple domains. To our knowledge this is the first VBM-based predictive approach that looks at interactions between multimodal features for future motor impairment prediction in pre-HD. Our multimodal analysis could be further enriched by incorporating additional measures of brain structure integrity such as white matter microstructure, it being highly relevant for pre-HD characterization as it seems to precede gray matter degeneration (Gregory et al., 2015; Shaffer et al., 2017). Despite being appealing, such an analysis is beyond the scope of our work.
We also analyzed the localization of correlates of future motor impairment when combining GM integrity at baseline and longitudinal atrophy progression. Our results show that reduced baseline GMC in the inferior part of the head of the caudate nucleus is predictive of future motor onset, whereas longitudinal atrophy patterns are localized in the superior part of the head of the caudate nucleus. This may suggest that loss in GM integrity in the ventral striatum is a first-stage putative marker of motor impairment and we speculate that this area is typically affected in early stages of the disease before symptom onset. On the other hand, longitudinal atrophy in the dorsal striatum later over the course of pre-HD would become a second-stage correlate of impairment. In any case, the found association between the caudate nucleus longitudinal GM reduction and future motor impairment is consistent with previous evidence of changes in caudate volume being associated with cognitive decline and disease severity (Domínguez et al., 2016). In terms of the attained classification results, the inclusion of atrophy rates to GM integrity did not boost the performance of our model. This may be explained by the limited amount of visits for which both imaging and TMS are available (at most 4). We hypothesize that a more robust estimation of structural atrophy would have been complementary to baseline GMC for future motor impairment detection provided that more time points were available. Unfortunately the extension of TRACK-HD, the TRACK-ON study, does not provide TMS information.
Our proposed methodology has shown the predictive power of localized patterns of gray matter integrity and non-imaging features to characterize pre-HD and, within this group, to predict future motor impairment. The influence of confounders on the reported imaging patterns has been reduced by closely matching healthy controls and pre-HD individuals in terms of number of samples, age and sex. Alternatively, we have removed effects of normal aging within pre-HD for future motor decline prediction. However, spurious associations could be related to other potential confounders. An important factor to control for are scanner differences or even differences in imaging locations (sites). We discovered that there are no associations between groups of interest and site, as it is shown in the Supplementary Material. Nevertheless, we rerun our classification analyses after accounting for site variability to verify that the reported brain locations associated to the presence of HD gene mutation and increased risk of future motor impairment remained unaltered, which is indeed the case (please refer to the Supplementary material for those results). Similarly, we evaluated the influence of HD comorbidities in our results. These show that it is unlikely that comorbidities influenced the spatial patterns of gray matter integrity associated to the groups of interest in this work. We should also keep in mind that the more confound factors are considered in a study setup, the more complicated its design becomes. In other words, there is a trade-off between the number of considered confounders to control for on a study and its actual viability.
It is important to point out that while this work uses data from various sites from the TRACK-HD study, our results should be tested on data from an independent study to validate the replicability of these findings. It is also worth mentioning that in order to investigate motor impairment at early stages of the progression of HD we focused on its pre-symptomatic stage, which could potentially introduce sample selection bias. An additional caveat concerns the small number of subjects (165) relative to other studies, which is further reduced to discern between pre-HD individuals with later mild or pronounced motor impairment. The number of samples is reduced at the cost of achieving an improved signal-to-noise ratio for the detection of motor impairment. We would also like to raise awareness about the interpretation of GMC; it does not represent volumetric differences in GM, but differences in voxel concentration in the acquisition space (not on the generated study-specific template) (Henley et al., 2010). Even if they represented volumetric differences, there is some evidence that suggests that neuron density does not correlate to GMC (Eriksson et al., 2009).
Although we do not use subjects with future motor impairment between mild and pronounced levels to train our model, this does not prevent us from including them in the evaluation of our classifier as it gives a continuous output before applying a cutoff value to assign categorical classes. By evaluating this continuous output for individuals with later intermediate motor impairment, we see that this group is approximately located between clusters with mild or severe impairment (see Fig. 6) in spite of the model not having access to their future motor scores. We also analyzed the response of the classifier on those pre-HD individuals that transition to HD manifestation during the study and show that they are assigned to a cluster of subjects that is closely linked to individuals with pronounced future motor impairment. This suggests that our model is able to detect a continuum of motor impairment in pre-HD that may potentially be used to assess future impairment of pre-HD individuals that transition to HD between baseline and follow up visits, as outlined in Fig. 7. Such estimates could provide a baseline against which both active and placebo treatment could be compared.
Fig. 7.
Schematics of the hypothetical application of imaging data to detect pre-HD individuals at high risk of future motor decline. At the moment a prospective trial participant is recruited, a structural MRI volume and other clinical measurements (TMS, age, number of CAG repeats) are collected. Then the GMC map estimated from the MRI volume is combined along with the other measurements and provided to the pre-trained classifier. Patients for which the classifier estimated severe motor impairment would become candidates for treatment on a clinical trial. The motor impairment achieved after active or placebo treatment could be compared with that estimated by the classifier, allowing an evaluation of the tentatively improved outcome with respect to the estimated impairment.
Overall, this work provides strong evidence that GMC used on top of standard demographic, genetic and motor features can be used to accurately detect future motor impairment within pre-HD. In fact, information from a single visit is sufficient for this purpose and the model is also capable of detecting brain regions associated with impairment within pre-HD. In that sense the presented approach could potentially be used in clinical trials for early detection of pre-HD individuals at high risk of developing pronounced future motor impairment. Targeted therapies could be applied to at-risk individuals on brain impairment correlates detected by our model to potentially make future motor symptoms milder or even slow disease progression.
Acknowledgements
Data used in this work was generously provided by the participants in the TRACK-HD study and made available by Dr. Sarah Tabrizi, Principal Investigator, University College London. The authors would also like to thank CHDI Foundation for their financial support. Special thanks to Sara Berger for her valued input in the elaboration of this manuscript and Pouya Bashivan for sharing computer code with us.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.05.008.
Appendix A. Supplementary data
Supplementary material
References
- Acton Q.A. 2012. Advances in Chorea Research and Treatment: 2012 Edition: Scholarly Brief. (ScholarlyEditions) [Google Scholar]
- Aylward E.H., Nopoulos P.C., Ross C.A., Langbehn D.R., Pierson R.K., Mills J.A.…Andrew R. Longitudinal change in regional brain volumes in prodromal Huntington disease. J. Neurol. Neurosurg. Psychiatry. 2011;82(4):405–410. doi: 10.1136/jnnp.2010.208264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995 [Google Scholar]
- Biglan K.M., Ross C.A., Langbehn D.R., Aylward E.H., Stout J.C., Queller S.…Nance M. Motor abnormalities in premanifest persons with Huntington's disease: the PREDICT-HD study. Mov. Disord. 2009;24(12):1763–1772. doi: 10.1002/mds.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarochi J.A., Calhoun V.D., Lourens S., Long J.D., Johnson H.J., Bockholt H.J.…Turner J.A. Patterns of co-occurring gray matter concentration loss across the Huntington disease prodrome. Front. Neurol. 2016 doi: 10.3389/fneur.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez J.F., Stout J.C., Poudel G., Churchyard A., Chua P., Egan G.F., Georgiou-Karistianis N. Multimodal imaging biomarkers in premanifest and early Huntington's disease: 30-month IMAGE-HD data. Br. J. Psychiatry. 2016 doi: 10.1192/bjp.bp.114.156588. [DOI] [PubMed] [Google Scholar]
- Douaud G., Gaura V., Ribeiro M.J., Lethimonnier F., Maroy R., Verny C.…Remy P. Distribution of grey matter atrophy in Huntington's disease patients: a combined ROI-based and voxel-based morphometric study. NeuroImage. 2006;32(4):1562–1575. doi: 10.1016/j.neuroimage.2006.05.057. [DOI] [PubMed] [Google Scholar]
- Douaud G., MacKay C., Andersson J., James S., Quested D., Ray M.K.…James A. Schizophrenia delays and alters maturation of the brain in adolescence. Brain. 2009;132(9):2437–2448. doi: 10.1093/brain/awp126. [DOI] [PubMed] [Google Scholar]
- Eriksson S.H., Free S.L., Thom M., Symms M.R., Martinian L., Duncan J.S., Sisodiya S.M. Quantitative grey matter histological measures do not correlate with grey matter probability values from in vivo MRI in the temporal lobe. J. Neurosci. Methods. 2009;181(1–9):111–118. doi: 10.1016/j.jneumeth.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio A., Santelli L., Tomassini Valentina, Bosnell R., Smith S., De Stefano N., Johansen-Berga H. Age-related changes in grey and white matter structure throughout adulthood. NeuroImage. 2010;51(3–2):943–951. doi: 10.1016/j.neuroimage.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good C.D., Johnsrude I.S., Ashburner J., Henson R.N., Friston K.J., Frackowiak R.S. A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage. 2001 doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Gorgolewski K., Burns C.D., Madison C., Clark D., Halchenko Y.O., Waskom M.L., Ghosh S.S. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Front. Neuroinform. 2011;5(August) doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S., Scahill R.I., Seunarine K.K., Stopford C., Zhang H., Zhang J.…Campbell C. Neuropsychiatry and white matter microstructure in Huntington's disease. J. Huntington's Dis. 2015 doi: 10.3233/JHD-150160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley S.M.D., Ridgway G.R., Scahill R.I., Klöppel S., Tabrizi S.J., Fox N.C., Kassubek J. Pitfalls in the use of voxel-based morphometry as a biomarker: examples from Huntington disease. Am. J. Neuroradiol. 2010;31(4):711–719. doi: 10.3174/ajnr.A1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs N.Z., Henley S.M., Ridgway G.R., Wild E.J., Barker R.A., Scahill R.I.…Tabrizi S.J. The progression of regional atrophy in premanifest and early Huntington's disease: a longitudinal voxel-based morphometry study. J. Neurol. Neurosurg. Psychiatry. 2010;81(7):756–763. doi: 10.1136/jnnp.2009.190702. [DOI] [PubMed] [Google Scholar]
- Huntington Study Group Unified Huntington's disease rating scale: reliability and consistency. Mov. Disord. 1996;I(2):136–142. doi: 10.1002/mds.870110204. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C., Behrens T., Woolrich M., Smith S. FSL. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Kassubek J., Juengling F.D., Kioschies T., Henkel K., Karitzky J., Kramer B.…Landwehrmeyer G.B. Topography of cerebral atrophy in early Huntington's disease: a voxel based morphometric MRI study. J. Neurol. Neurosurg. Psychiatry. 2004;75(2):213–220. [PMC free article] [PubMed] [Google Scholar]
- Klöppel S., Chu C., Tan G.C., Draganski B., Johnson H., Paulsen J.S.…Frackowiak R.S.J. Automatic detection of preclinical neurodegeneration: presymptomatic Huntington disease. Neurology. 2009;72(5):426–431. doi: 10.1212/01.wnl.0000341768.28646.b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S., Gregory S., Scheller E., Minkova L., Razi A., Durr A.…Orth M. Compensation in preclinical Huntington's disease: evidence from the track-on HD study. EBioMedicine. 2015;2(10):1420–1429. doi: 10.1016/j.ebiom.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostro D., Abdulkadir A., Durr A., Roos R., Leavitt B.R., Johnson H.…Klöppel S. Correction of inter-scanner and within-subject variance in structural MRI based automated diagnosing. NeuroImage. 2014;98:405–415. doi: 10.1016/j.neuroimage.2014.04.057. [DOI] [PubMed] [Google Scholar]
- Lang A.E. Clinical trials of disease-modifying therapies for neurodegenerative diseases: the challenges and the future. Nat. Med. 2010;16(11):1223–1226. doi: 10.1038/nm.2220. Retrieved from. [DOI] [PubMed] [Google Scholar]
- Long J.D., Mills J.A., Leavitt B.R., Durr A., Roos R.A., Stout J.C.…Track-HD and Track-On Investigators Survival end points for huntington disease trials prior to a motor diagnosis. JAMA Neurol. 2017;74(11):1352–1360. doi: 10.1001/jamaneurol.2017.2107. (Retrieved from) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.D., Paulsen J.S. Multivariate prediction of motor diagnosis in Huntington's disease: 12 years of PREDICT-HD. Mov. Disord. 2015;30(12):1664–1672. doi: 10.1002/mds.26364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.D., Paulsen J.S., Marder K., Zhang Y., Kim J.I., Mills J.A., Pallai J. Tracking motor impairments in the progression of Huntington's disease. Mov. Disord. 2014;29(3):311–319. doi: 10.1002/mds.25657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J.S., Hayden M., Stout J.C., Langbehn D.R., Aylward E., Ross C.A., Group, P.-H. I. of the H. S Preparing for preventive clinical trials. Arch. Neurol. 2006;63(13):883–890. doi: 10.1001/archneur.63.6.883. [DOI] [PubMed] [Google Scholar]
- Paulsen J.S., Langbehn D.R., Stout J.C., Aylward E., Ross C.a, Nance M.…Hayden M. Detection of Huntington's disease decades before diagnosis: the predict-HD study. J. Neurol. Neurosurg. Psychiatry. 2008;79(8):874–880. doi: 10.1136/jnnp.2007.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J.S., Long J.D., Ross C.A., Harrington D.L., Erwin C.J., Williams J.K.…Barker R.A. Prediction of manifest huntington's disease with clinical and imaging measures: a prospective observational study. Lancet Neurol. 2014;13(12):1193–1201. doi: 10.1016/S1474-4422(14)70238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J.S., Nopoulos P.C., Aylward E., Ross C.A., Johnson H., Magnotta V.A.…Nance M. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res. Bull. 2010;82(3–4):201–207. doi: 10.1016/j.brainresbull.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney J.B., Vonsattel J.P., MacDonald M.E., Gusella J.F., Myers R.H. CAG repeat number governs the development rate of pathology in Huntington's disease. Ann. Neurol. 1997;41(5):689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- Reuter M., Rosas H.D., Fischl B. Highly accurate inverse consistent registration : a robust approach. NeuroImage. 2010;53(4):1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. a, Aylward E.H., Wild E.J., Langbehn D.R., Long J.D., Warner J.H., Tabrizi …., J S. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 2014;10(4):204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- Shaffer J.J., Ghayoor A., Long J.D., Kim R.E.Y., Lourens S., O'Donnell L.J.…Johnson H.J. Longitudinal diffusion changes in prodromal and early HD: evidence of white-matter tract deterioration. Hum. Brain Mapp. 2017 doi: 10.1002/hbm.23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman K.S., Umansky S.R. Early detection of neurodegenerative diseases: circulating brain-enriched microRNA. Cell Cycle. 2013;12(1):1–2. doi: 10.4161/cc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J.C., Paulsen J.S., Queller S., Solomon A.C., Whitlock K.B., Campbell J.C.…Aylward E.H. Neurocognitive signs in prodromal Huntington disease. Neuropsychology. 2011;25(1):1–14. doi: 10.1037/a0020937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi S.J., Langbehn D.R., Leavitt B.R., Roos R.A., Durr A., Craufurd D.…Stout J.C. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi S.J., Scahill R.I., Owen G., Durr A., Leavitt B.R., Roos R.A.…Langbehn D.R. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12(7):637–649. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- Thieben M.J., Duggins A.J., Good C.D., Gomes L., Mahant N., Richards F.…Frackowiak R.S.J. The distribution of structural neuropathology in pre-clinical Huntington's disease. Brain J. Neurol. 2002;125(Pt 8):1815–1828. doi: 10.1093/brain/awf179. [DOI] [PubMed] [Google Scholar]
- Tikhonov A. Solution of incorrectly formulated problems and the regularization method. Soviet Math. Dokl. 1963;5:1035/1038. http://ci.nii.ac.jp/naid/10004315593/en/ Retrieved from. [Google Scholar]
- Van Den Bogaard S.J.A., Dumas E.M., Acharya T.P., Johnson H., Langbehn D.R., Scahill R.I.…Roos R.A.C. Early atrophy of pallidum and accumbens nucleus in Huntington's disease. J. Neurol. 2011;258(3):412–420. doi: 10.1007/s00415-010-5768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd K., Fjell A., Reinvang I., Lundervold A., Dale A., Eilertsen D.…Fischl B. Effects of age on volumes of cortex, white matter and subcortical structures. Neurobiol. Aging. 2005;26(9):1275–1278. doi: 10.1016/j.neurobiolaging.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Long J.D., Mills J.A., Warner J.H., Lu W., Paulsen J.S., Group, the P.-H. I. and C. of the H. S Indexing disease progression at study entry with individuals at-risk for Huntington disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156(7):751–763. doi: 10.1002/ajmg.b.31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005;67(2):301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material