Summary
An increasing number of studies have established hydrogen sulfide (H2S) gas as a major cytoprotectant and redox modulator. Following its discovery, H2S has been found to have pleiotropic effects on physiology and human health. H2S acts as a gasotransmitter and exerts its influence on gastrointestinal, neuronal, cardiovascular, respiratory, renal, and hepatic systems. Recent discoveries have clearly indicated the importance of H2S in regulating vasorelaxation, angiogenesis, apoptosis, ageing, and metabolism. Contrary to studies in higher organisms, the role of H2S in the pathophysiology of infectious agents such as bacteria and viruses has been less studied. Bacterial and viral infections are often accompanied by changes in the redox physiology of both the host and the pathogen. Emerging studies indicate that bacterial-derived H2S constitutes a defense system against antibiotics and oxidative stress. The H2S signaling pathway also seems to interfere with redox-based events affected on infection with viruses. This review aims to summarize recent advances on the emerging role of H2S gas in the bacterial physiology and viral infections. Such studies have opened up new research avenues exploiting H2S as a potential therapeutic intervention.
Keywords: hydrogen sulfide, cytoprotectant, antioxidant, metabolism, infection, Mycobacterium tuberculosis, HIV
Introduction
Early life forms first appeared on an anoxic earth in the Archean eon, approximately 3.8 billion years ago (1,2). Among them, were the dissimilatory sulfate-reducing bacteria which constitute one of the oldest forms of bacterial life on earth. These bacteria utilized inorganic sulfur substrates and produced hydrogen sulfide (H2S) as the end product of anaerobic respiration (3). Before the “great oxidation event” which occurred 2.5 billion years ago leading to an increase in atmospheric oxygen, H2S remained the most abundant and versatile chemical on the primitive earth (4,5). In fact, H2S is widely believed to be the primordial sustainable energy source (6). Primitive photolithotrophs used sulfide as the terminal electron acceptor, similar to today’s green and purple sulfur bacteria. Therefore, sulfide-based metabolism may have preceded the present, oxygen-based life on the planet by billions of years (7–9).
In sharp contrast to its pivotal role early in the evolutionary timeline, H2S is known mostly for being a foul smelling poisonous gas, associated with sewers, septic tanks, and as a weapon of chemical warfare during the First World War. Consequently, majority of the research pertaining to this gas has been conducted from a toxicology point of view (10,11). With studies published as long back as 1803 highlighting the detrimental effect of H2S on animals, along with the recent gene expression data, H2S was condemned as a respiratory and metabolic poison (12). It was not until the 1940s that the “transsulfuration” pathway involving the production of H2S by interconversion between cysteine, homocysteine via cystathionine was described for the first time in liver homogenates (13,14). Further studies led to detailed biochemical characterization of the enzymes cystathionine beta synthase (CBS) and cystathionine gamma lyase (CSE) involved in the transsulfuration reaction. Later, another enzyme, 3-mercaptopyruvate sulfurtansferase (3-MST) was identified as a part of H2S biogenesis pathways (15–18). However, the functional implication of the H2S biogenesis remained elusive for a long time. First glimpse of H2S involvement in cellular physiology emerged from the studies demonstrating measurable levels of endogenous H2S within brain tissues of healthy individuals (0.65–0.73 mg/g) and animals (1.57 ± 0.04 μg/g) (19–21). Along these lines, higher levels of neuronal H2S was found to be due to greater expression of CBS in the brain tissues. Additionally, H2S production in the brain tissue was efficiently reduced using pharmacological CBS inhibitors (hydroxylamine and amino-oxyacetic acid). Further studies proposed that H2S facilitates the induction of hippocampal long-term potentiation (LTP) by enhancing the activity of N-methyl d-aspartate (NMDA) receptors (22). Later, H2S was found to relax vascular smooth muscle by activating ATP-sensitive K+, intermediate conductance Ca2+ sensitive K+, and small conductance Ca2+-sensitive K+ channels (23–25). Importantly, H2S was identified to protect from oxidative stress and ischemia-reperfusion injury by multiple mechanisms such as restoring the levels of GSH and direct scavenging of mitochondrial ROS (Fig. 1) (26,27). These discoveries further led to the disclosure of mechanisms by which H2S protects various organs, including the heart and kidney from oxidative stress and ischemia-reperfusion injury (28). Based on these studies, H2S was inducted as the newest member of the family of small molecule gaseous transmitters or “gasotransmitters” alongside nitric oxide (NO) and carbon monoxide (CO) (29). Along this line hydrogen gas (H2) has also emerged as a potential gaseous signaling molecule with therapeutic antioxidant function (30,31). Recently, H2S was found to have a protective role in airway epithelial cells infected with respiratory syncytial virus (RSV) demonstrating for the first time that this molecule might be used as a therapeutic agent. Needless to say, the role of H2S has permeated to areas of metabolism, redox physiology, neurophysiology, apoptosis, angiogenesis, ageing, inflammation, atherosclerosis, pulmonary diseases among others with a whole spectrum of physiological implications (32–34).
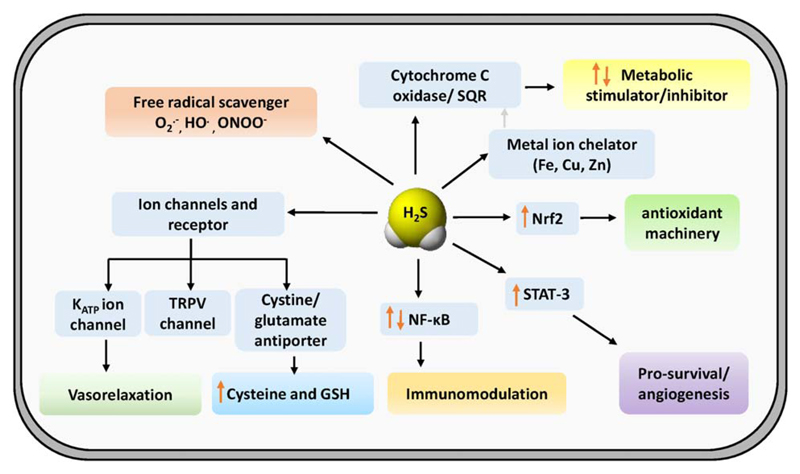
Fig 1.
Molecular targets of H2S in mammalian system. Top left: H2S has the ability to directly scavenge reactive oxygen and nitrogen species (ROS/RNS). Top right: H2S targets metal cofactor of cytochrome c oxidase and leads to inhibition of cellular respiration. Oxidation of H2S by sulfide quinone oxidoreductase (SQR) couples catabolism of H2S with mitochondrial electron transport chain (ETC) and thus modulates cellular metabolism. Bottom left: ion channels which are involved in systemic responses to H2S in blood vessels, heart, and neurons. Bottom right: H2S targets cysteine thiols by S-persulfidation of intracellular signaling proteins and transcription factors which likely accounts for the downstream effects on inflammation, antioxidant response, cellular proliferation, and survival.
H2S producing bacteria were discovered way back in 1877. Many investigators demonstrated bacterial production of H2S by its rotten egg smell and its ability to react with lead acetate resulting in the blackening of paper strips impregnated with lead acetate (12,35). In fact, lead acetate test was successfully exploited to distinguish between the paratyphoid and enteritidis groups and still remains an indispensable diagnostic tool (36). In the area of marine microbiology, H2S emitted from deep sea vents is often referred to as the “sunlight of the deep ocean” (37,38). H2S forms an important source of metabolic energy for the microorganisms inhabiting such niches, reminiscent of the primordial earth (6). Many bacterial species were demonstrated to possess orthologs of the transsulfuration pathway enzymes (CBS and CSE) and of 3-MST (36,39–41). However, the significance of H2S biogenesis in bacteria remained poorly characterized. In 1960s, co-culture experiments with Desulfovibrio desulfuricans/Pseudomonas aeruginosa and Escherichia coli/Staphylococcus aureus provided the first evidence of a possible “protective” role of H2S in bacteria. H2S produced by D. desulfuricans was demonstrated to be the “diffusible” factor responsible for imparting pseudomonads the ability to resist heavy metal (e.g., mercury) toxicity (42). Similarly, H2S produced by E. coli protected S. aureus from merbromin and mercuric chloride (43). Surprisingly, it took more than four decades to discover additional roles of H2S in protecting diverse bacterial species from oxidative stress and antibiotics (44). Similar reports of protective influence of H2S have emerged in plants and nematodes, however, they are beyond the scope of this review. Altogether, it appears that H2S is an important biological effector molecule with diverse roles in organisms ranging from bacteria to mammals.
While there has been a steady rise in the number of studies pertaining to the physiological role of H2S in mammalian systems, there is a significant lack in our understanding of the same when it comes to infectious agents like bacteria and viruses. A survey of PubMed also shows that the studies on H2S and bacterial/viral infections are relatively fewer as compared to mammalian systems (Fig. 2). Keeping this in mind, our effort is to compile a summary of the existing studies providing insights on how H2S exerts its biological influence on infectious diseases caused by bacterial and viral agents. First, we will provide a brief description of H2S biogenesis pathways and chemical properties of H2S. This will be followed by a description of studies on the contribution of H2S in influencing the physiology of bacteria and virus infected cells.
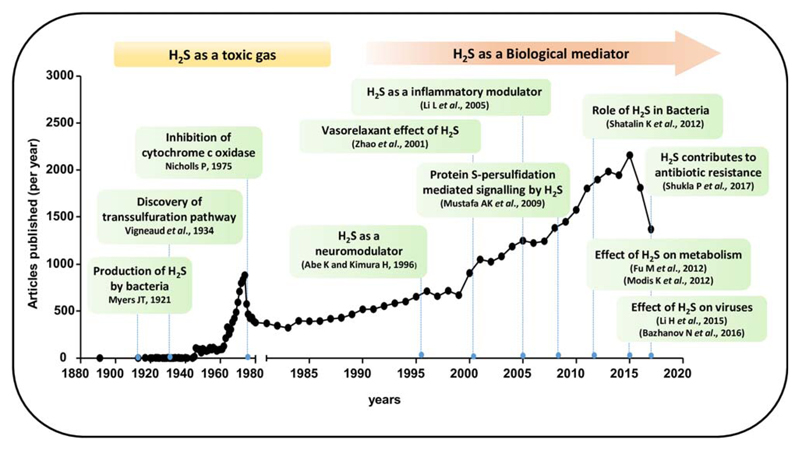
Fig 2.
Survey of literature on H2S. The figure shows number of published articles per year till 2017 (source: PubMed). Some of the landmarks discoveries are highlighted in the figure inset.
Biogenesis of H2S
The biogenesis of H2S has been mainly attributed to the transsulfuration pathway (45). This pathway has been known for many years and is evolutionarily conserved and present in many lower species as well as in mammals (46). Two enzymes constitute this pathway namely cystathionine beta synthase or CBS (EC 4.2.1.22) and cystathionine gamma lyase or CSE (EC 4.4.1.1). Both the enzymes uses pyridoxal l- phosphate (PLP) as cofactor and are hence sensitive to common PLP dependent enzyme inhibitors like hydroxylamine (47,48). Apart from transsulfuration, an additional pathway also exists which leads to the biogenesis of H2S. Cysteine aminotransferase (CAT, EC 2.6.1.3) catalyses the reaction of cysteine with keto acids (e.g., α-ketoglutarate) to form 3-mercaptopyruvate, which is subsequently desulfurated by 3-mercaptopyruvate sulfurtransferase (3-MST, EC 2.8.1.2) to form H2S (Fig. 3) (45). More recently, a new pathway to generate H2S using d-cysteine has been identified. The enzymes d-amino acid oxidase (DAO) along with 3-MST carry out biogenesis of H2S from d-cysteine (49). Nonenzymatic reactions also lead to the generation of H2S inside cells, however, their contribution is minor and remain poorly understood and characterized.
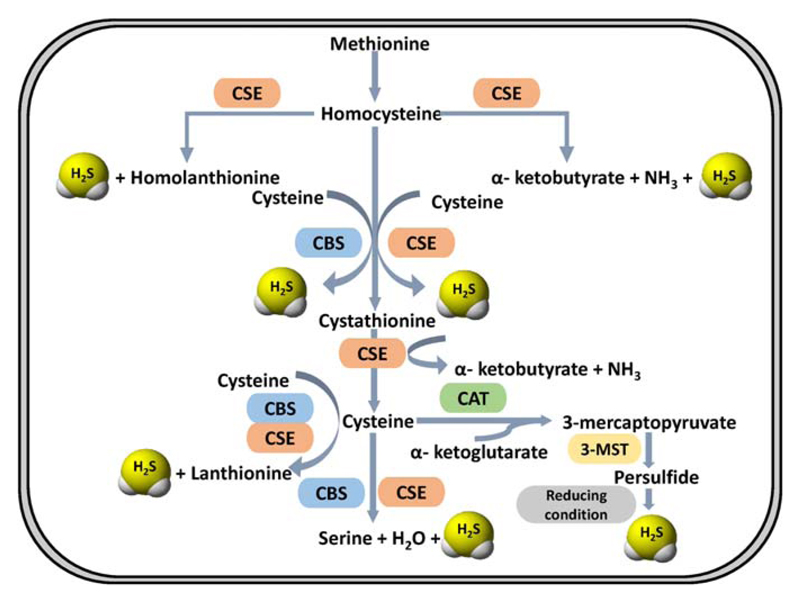
Fig 3.
Pathways involved in enzymatic biogenesis of H2S. The transsulfuration pathway consisting of the enzymes cystathionine β-synthase (CBS) and Cystathionine γ-lyase (CSE) is the major pathway for biological H2S production. In addition to this 3-mercaptopyruvate sulfurtransferase/cysteine aminotransferase (3-MST/CAT) pathway also contribute significantly to the production of H2S.
Cystathionine Beta Synthase
Human cystathionine beta synthase (CBS) is a tetramer and is allosterically stimulated by S-adenosyl methionine (SAM) which binds to a conserved “CBS pair domain” in the C-terminal end of the protein (50,51). CBS catalyses the first and committed step of the transsulfuration pathway which canonically, leads to the production of cystathionine from serine and homocysteine (52). However, when serine is replaced by cysteine, H2S is produced (Fig. 3). CBS can generate H2S by additional reactions including β replacement of cysteine by water to form serine and β replacement of cysteine by a second molecule of cysteine to form lanthionine. In kinetic terms, the β replacement of cysteine with homocysteine remains the most favorable (52). As mentioned earlier, CBS utilizes PLP as a cofactor and is a type-II PLP binding protein (53). The cofactor remains covalently linked to the active site lysine via Schiff base formation and is pertinent to the enzymatic activity of the protein. In addition to PLP, human CBS also contains heme which acts as a redox dependent gas sensor (54). Apart from this the heme moiety also functions as a “metabolic switch” committing the pathway toward H2S production (55). Under ER stress, heme oxygenase is induced, which catabolises heme in presence of molecular oxygen to produce biliverdin and CO, the later one binds to heme cofactor of CBS and inhibits its activity. This inhibition leads to low levels of cystathionine and increased levels of homocysteine. These metabolites cue the second enzyme, CSE, to increase the production of H2S from cysteine and homocysteine further highlighting the metabolic flexibility of this pathway (55).
Cystathionine Gamma Lyase
As the name suggests, the second enzyme of the pathway CSE, primarily catalyses the cleavage of cystathionine to form cysteine, ammonia and α-ketobutyrate (52). Human CSE is a homotetramer and a PLP binding protein. It can catalyse the production of H2S from cysteine and homocysteine alone or in combination (Fig. 3). The substrate promiscuity of CSE permits the accommodation of cysteine, homocysteine, and cystathionine in the same binding pocket. Regulation of CSE is not very well known but as mentioned earlier, on ER stress it upregulates the production of H2S by utilizing cysteine and homocysteine over cystathionine as substrates (55).
Mercaptopyruvate Sulfurtransferase
3-mercaptopyruvate is generated by a transamination reaction between cysteine and α-ketoglutarate catalysed by aspartate/cysteine aminotransferase (56). This 3 mercaptopyruvate (3-MP) is subsequently used as a substrate by 3-MST to form H2S (Fig. 3). 3-MST transfers the sulfur to a nucleophilic cysteine in the active site. This leads to the formation of a bound persulfide which acts as a source of H2S under reducing conditions or in presence of acceptors like thioredoxin. 3-MST is localized to the mitochondria unlike CBS and CSE which are cytosolic, where it is believed to contribute bioenergetically via sulfide oxidation. Also, unlike the transsulfuration enzymes, 3-MST is inhibited under oxidizing conditions due to a labile active site cysteine which gets converted to cysteine sulfonate leading to enzyme inactivation (57).
MST/DAO Pathway
In addition to l-cysteine, H2S production was observed in brain homogenates when d-cysteine was used as a substrate. This led to the discovery of new pathway involving peroxisomal enzyme d-amino oxidase (DAO) in H2S biogenesis. d-cysteine is metabolized by DAO to 3-mercaptopyruvate (3MP), which then translocates to mitochondria where it is converted to H2S and pyruvate. It has been reported that the production of H2S from d-cysteine is ~ 60 times greater in comparison to l-cysteine. Since DAO is only localized to the brain and the kidney, the functionality of the 3MST/DAO pathway for the production of H2S is only relevant to these tissues (49,58).
Chemical Biology of H2S
H2S gas was discovered in 1777, by Carl Wilhelm Scheele, and was largely considered as a toxic gas for over hundred years (59). Based on toxicological studies, the permissible exposure limit of H2S is 10 ppm and 800 ppm exposure for 5 min is the lethal concentration for 50% of humans (LC50) (60,61). Much of its toxicity is owed to the fact that H2S is known to inhibit respiration thereby acting as a metabolic poison. When present at higher concentrations it causes reversible inhibition of cytochrome c oxidase (complex IV), thereby perturbing mitochondrial respiration and oxidative phosphorylation (46). This is further exemplified by the observations that H2S induces a state of “suspended animation” with consequent lowering of metabolic rate and body temperature in mice (61).
The standard two electron redox potential of H2S/S0 couple, −0.23 V (noted +0.140 value in acidic condition) at pH 7 (versus the standard hydrogen electrode) is comparable to that of major cellular antioxidant buffers, glutathione disulfide/glutathione (E°′= −0.262 V) and cystine/cysteine (E°′= −0.245 V) redox potentials (62–65). While the physiological concentration of H2S is a matter of ongoing controversy, it seems that low nM concentrations are most likely (66). Only in case of aorta, the reported concentration of free H2S is ~ 20–100 fold higher than that of other tissues (67). Interestingly, the flux of sulfur into H2S is comparable to that of GSH, indicating that the low nM levels are maintained as a consequence of higher sulfide clearance rate (59,68). The low steady-state concentration of H2S than GSH (~10 mM) precludes its involvement in counteracting oxidative stress by acting as an antioxidant buffer (46). Alternatively, it is proposed that H2S can modulate intracellular redox signaling by modifying cysteine thiols of the various cellular proteins (S-persulfidation) coordinating redox homeostasis. However, since a direct reaction of H2S with thiols is unlikely, the mechanism by which persulfides are formed intracellularly is poorly understood (69,70).
H2S is lipophilic and is known to permeate freely through biological membranes without any assistance from membrane channels (lipid bilayer permeability PM ≥ 0.5 ± 0.4 cm/s) (71). Being a weak acid, it dissociates immediately and equilibrates with its anion HS– and S2– in aqueous solution as shown in Eq. (1).
| (1) |
The pKa1 for H2S dissociation ranges from 6.97 to 7.06 at 25 °C, while pKa2 is estimated to be between 12.20 to 15.00 at 25 °C (72). Based on these values it is calculated that the ratio of HS–:H2S is 3:1 at physiological pH of 7.4 (72). Nevertheless, total intracellular H2S levels is referred to as total free sulfide pool (i.e., H2S + HS– + S2–). Based on its chemical features, H2S can influence cellular redox physiology via four mechanisms: (1) scavenging of ROS and RNS, (2) reaction with metal centres, (3) modulation of cellular respiration, and (4) reaction with protein cysteine thiols to generate persulfides (S-persulfidation- an oxidative posttranslational modification [oxPTM]) (69,73). These mechanisms are described in the following section.
H2S as a Free Radical Scavenger
H2S acts as a cytoprotective molecule and has the ability to directly scavenge free radical species (74). Owing to its nucleophilic properties, H2S has been shown to react with oxygen (O2), ROS, peroxynitrite (ONOOH/ONOO–), and hypochlorite (HOCL/–OCL) (65,75). The apparent second-order rate constants of H2S with various oxidants have been summarized in Table 1. While these studies indicate a direct scavenging of oxidants by H2S in vitro, the low concentrations of H2S (10 nM to 3 μM) compared to other antioxidants (GSH; 1–10 mM) in vivo raised substantial concerns about its role in remediating ROS/RNS under biologically relevant conditions (66,76–79). Alternatively, H2S has been shown to increase GSH production by enhancing the inward transport of cystine and inducing the expression of GSH-biosynthetic enzyme, GCL (γ-GCS) (80,81). This increase in intracellular GSH could be another mechanism by which H2S indirectly participates in protection from oxidative stress.
Table 1. Second-order rate constants of H2S with oxidants (Refs. 65,75).
| Reaction with oxidants | Rate constants (M−1 s−1) at 37 °C |
|---|---|
| + H2S/HS− → H2O2 + S− | 1.5 × 106 |
| HO. + H2S/HS− → S− + H2O | 1.5 × 1010 |
| H2O2 + HS− → HSOH + OH− | 0.73 |
| ONOOH + HS− → HSOH + N | 4.8 × 103 |
| HSOH + HS− → HSSH + OH− | 1.0 × 105 |
| HOCl + HS− → HSCl + OH− | 8.0 × 107 |
| S− + S− + 2H+ → HSSH | 6.5 × 109 |
Reaction with Metal Centers
The interaction of H2S with metals falls into two categories: (i) electron-transfer reaction and (ii) coordinate complex formation (65). In the first category, complete electron transfer occurs between the sulfide species and the metal, whereas coordinate complex formation involves binding of the sulfur species to the metal ligand (65). These reactions are predicted on the basis of chemical properties of H2S to act as a nucleophile. Interestingly, a wine-like model was used to study the reaction mechanism of metals with H2S. Sulfidic off-odors encountered during wine production are due to the presence of H2S and low-molecular-weight thiols (82). These off-odors are usually removed in a process called Cu fining, wherein Cu (II) is added to selectively and rapidly form ~1.4:1 H2S/Cu and ~2:1 thiol/Cu complexes, resulting in oxidation of H2S and reduction of Cu (II) to Cu (I) (82). The CuS precipitate formed is than subsequently removed from the wine by racking and/or filtration (82).
In a biological setup, interaction of H2S with the mitochondrial heme protein-cytochrome C oxidase (CcO) is extensively studied. It has been demonstrated that high concentrations of H2S competitively binds to CcO, resulting in inhibition of O2 binding (83–85). H2S interacts with CcO through the O2-binding copper (CuB)/heme (a3) iron binuclear site in oxidized state (Cu2+/Fe3+) and reduces the enzyme (86). The Ki for this reaction is 0.2 μM with purified CcO (86). Most of the studies demonstrating inhibitory effect of H2S on respiration via interaction with CcO were done using very high/nonphysiological concentrations of H2S. However, it was shown that the liver mitochondria of H2S treated rats show biphasic respiration profile (87). Low concentrations (0.1–3.0 μM) of H2S induces respiration whereas higher concentrations (30–100 μM) inhibits (87). At lower concentrations, H2S acts as a mitochondrial electron donor and stimulates electron transport chain (87,88).
Other than CcO, H2S is known to covalently modify ferryl/peroxo heme within hemoglobin and myoglobin resulting in the formation of green colored sulfhemoglobin and sulfmyoglobin species, both of which are indicators of H2S poisoning (89). Additionally, H2S can react with nonheme iron present in iron-sulfur cluster containing proteins to generate insoluble precipitates (65). Lastly, H2S is reported to react with a copper-containing protein (Cu—Zn—SOD) (90). This reaction involves copper-catalysed reduction of to H2O2 Oxidation of H2C to S0 (90).
H2S and Cellular Bioenergetics
The effects of H2S on cellular bioenergetics are largely derived from examining mitochondrial function. The effect of H2S on mitochondria is complex, exhibiting two opposing effects; inhibition and stimulation of mitochondrial bioenergetics (88). Oxidation of H2S by mitochondrial inner membrane localized Sulfide-Quinone oxidoreductase (SQR) leads to transfer of electron from H2S to ubiquinone and increases the flux of electron transport to mitochondrial respiratory complex III and IV, thereby leading to enhanced oxygen consumption and cellular respiration (91,92).
Recently, it was demonstrated that H2S has a biphasic effect on cellular oxygen consumption/mitochondrial electron transport (87,88). These investigators creatively adapted Seahorse XF technology to precisely measure dynamic changes in mitochondrial bioenergetics in real-time in response to a gradient of H2S. Interestingly, treatment of isolated mitochondria with low H2S concentrations, NaHS (<1 μM) enhances mitochondrial oxygen consumption rate (OCR), ATP turnover rate and leads to increased maximal respiratory capacity (87). In contrast, treatment with high concentrations of NaHS (30–300 μM) causes reduced mitochondrial OCR and ATP generation, which is consistent with previous studies showing the inhibitory effect of H2S on mitochondrial respiration when present at high concentrations (84,93). The low endogenous concentrations of H2S in mitochondria is primarily maintained by mitochondrial localized 3-Mercaptopyruvate sulfurtransferase (3-MST) (87). Supplementation of isolated mitochondria with 3-mercaptopyruvate (3-MP) leads to enhanced H2S production by 3-MST pathway and induces mitochondrial bioenergetic parameters (87). Subsequently, genetic silencing of 3-MST leads to reduced basal OCR and due to absence of 3-MST there is no stimulatory effect of 3-MP on mitochondrial bioenergetic parameters (87).
Furthermore, it was found that the stimulatory effect of 3-MP on mitochondrial bioenergetic parameters was absent in mitochondria isolated from aged mice as compared to young mice (88). Moreover, exposure of H2S was shown to cause physiological alterations which enhanced thermotolerance and life span of Caenorhabditis elegans (94). The other two PLP-dependent cytosolic enzymes CBS and CSE also maintain endogenous H2S levels. Under stress condition, CSE is known to translocate into mitochondria and stimulate mitochondrial H2S generation. This subsequently results in an increased ATP generation and resistance to hypoxia (95). Furthermore, elevated endogenous H2S in colon cancer cells have been shown to regulate cell migration and invasion (96). Additionally, pharmacological inhibition of H2S production diminished the growth of cancer cells by suppressing basal respiration, ATP production, spare respiratory capacity, and glycolysis (96,97).
Protein Persulfidation as a Mechanism of H2S Mediated Signaling
H2S exerts its signaling property via oxidative posttranslational modification (oxPTM) of cysteine residues, called S-persulfidation (98–100). Persulfidation (R-SSH) is the oxidation of thiol (—SH) from −2 to −1 oxidation state (101). Being a reductant, H2S/HS– can carry out the persulfidation reaction only when one of the reagents (—SH group of proteins or H2S) is in the oxidized form (101). The proposed reactions for the formation of S-persulfide bond by the nucleophilic attack of HS– anion on reversibly oxidized protein —SH group is shown in (Fig. 4).
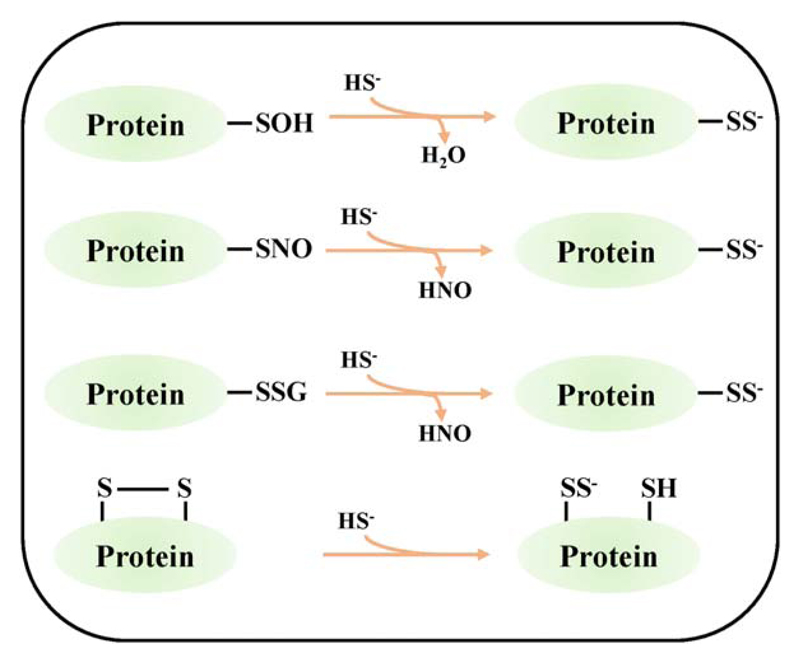
Fig 4.
Proposed reaction mechanisms for H2S mediated S-persulfidation of proteins. From Top: H2S reacts with sulfenylated (—SOH), S-nitrosylated (—SNO), S-glutathionylated (—SSG) and disulfide form of cysteine in proteins by S-persulfidation.
Recently, it was reported that persulfidation protects proteins from oxidative stress-induced damage and the over oxidized persulfidated cysteine sulfonic acid can be reversed to thiol (—SH) by the depersulfidase activity of thioredoxin (102). Thus, persulfidation can act as a protective mechanism against oxidative stress-induced protein damage (102). Using LC–MS-based techniques, S-persulfidation of proteins involved in fatty acids and carbohydrate metabolism, cellular response to stress, cell redox homeostasis, translation, and cell cycle were identified (73). Similarly, H2S was shown to modify cysteines in about 10–25% of liver proteins including actin, tubulin, and glyceraldehyde-3-phosphate (GAPDH) by persulfidation under physiological conditions and modulate their activities (103). Several key host transcription factors, Nrf2 and Nf-κB, are directly targeted by H2S via S-persulfidation (98). Apart from eukaryotic systems, a recent study, for the first time, assessed widespread S-persulfidation of proteins in Staphylococcus aureus (S. aureus) (104). More importantly, H2S exposure increased the level of S-persulfidation, whereas mutations in transsulfuration pathways (cysM and metB) had an opposite effect (104). Lastly, this study revealed extensive persulfidation of transcription factors involved in virulence regulation (SarA family) and interaction with host immune response (superantigen-like proteins [SSLs]) (104).
Detection of H2S and Protein S-Persulfidation
Tools for Detection and Quantitation of H2S
To achieve a better understanding of the physiological effects of H2S, it is imperative to determine the levels of this gasotransmitter in its free and other biological forms. This becomes especially important as the role of H2S as a protectant versus poison has been contended throughout the course of its study as a biological effector molecule. The suitability of any detection method relies heavily on three pertinent aspects namely sensitivity, reproducibility and experimental ease. A multitude of detection methodologies are available for qualitative and quantitative measurement of H2S levels. While this may seem advantageous at first glance, it has led to a huge variability in the reported levels of “bioavailable” H2S throughout literature. Furthermore, additional factors influencing detection arise due to experimental aspects like biological sample (tissue, sera, and cells), pH, and oxygen. The latter have been demonstrated to exert a direct effect on the stability of H2S (105).
The detection methods range from simple colorimetric assays to techniques like gas chromatography (GC), High pressure liquid chromatography (HPLC), electrochemical, polarographic, fluorescent, and recently developed nanotechnology-based systems. Some of these techniques are summarized in Table 2. Although these methods have been widely used for biological H2S detection, they are not devoid of pitfalls. The issues of sensitivity, invasiveness, artefactual readout, half-life, stability, permeability, lack of spatio-temporal insight, and cumbersome experimental setup cannot be overlooked. Therefore, the reporting and interpretation of experimental data becomes immensely influenced by the method that a researcher chooses to adopt. Many authors have argued that for a dynamic gaseous effector molecule like H2S, the absolute numbers in terms of concentration may actually not matter as much as the determination of the qualitative trend of its rise and fall under different physiological conditions. This may, however, not hold true for pathophysiological conditions wherein accurate determination of H2S levels may become indispensable for diagnosis of certain diseases.
Table 2. Tools for detection and quantification of H2S.
| Detection method | Description | References |
|---|---|---|
| Colorimetric | H2S reacts with metal salts like lead acetate, bismuth chloride, silver nitrate to form the lead sulfide which can be detected and quantified using UV-VIS spectroscopy. | (106–108) |
| “Zinc trap” method in which zinc acetate reacts with H2S forming zinc sulfide with subsequent acidification with N, N-dimethyl phenylenediamine. The product can be detected and quantified using UV-VIS spectroscopy. | (109,110) | |
| Chromatographic | Gas chromatography has been combined with flame photometric detectors, ion chromatography, silver particle trapping, and chemiluminescent detectors. | (20,67,111–114) |
| HPLC of sulfide derivatized with monobromobimane, dibromobimane, p-phenylenediamine, and Fe3+. Reverse Phase HPLC of methylene blue formed by the zinc trap assay. |
(115–118) | |
| Electrochemical | Sulfide ion specific electrode measures S2– form of sulfide which requires alkaline environment using Ag/Ag2S electrodes. | (119,120) |
| Polarographic real time measurement of H2S using a polarographic oxygen sensor as anode and platinum wire as cathode and alkaline K3Fe(CN)6 as electrolyte. An H2S permeable membrane allows diffusion of H2S into the electrolyte solution reducing it. The electrolyte subsequently gets re-oxidized on the surface of the platinum electrode to produce a current proportional to H2S concentration. | (121,122) | |
| Fluorescent sensors | All fluorescent probes consist of a fluorescence signal transducer and the fluorescence modulator. The transducer is a suitable fluorescent moiety while the modulator is chosen based on the chemical nature of H2S and physiologically permissible reaction kinetics. Fluorescent moieties like rhodamine, BODIPY, dansyl, 7-hydroxy-4-methylcoumarin, naphthilamide, cyanine, etc, have been used as transducers. The modulators have been designed based on the selective reduction of nitro groups to amines and azide groups to amines by H2S, thiolysis, addition and cyclization reactions based on the nucleophilic nature of H2S and copper sulfide precipitation resulting from the affinity of H2S for copper. Others include selenium-based probes for reversible detection of H2S. Probes have been designed based on the selenide-selenoxide redox reaction of many selenoenzymes. These probes can monitor redox cycling between H2S and ROS. | (123) |
| Nanotechnology-based sensors | Single walled carbon nanotube networks, gold nanoclusters, nanorods, nanocomposites of FAM DNA etched on the surface of silver nanoparticles have been employed for the detection of H2S. | (124–127) |
Apart from the aforementioned techniques, recent studies using H2S sensitive fluorescent proteins as genetically encoded biosensors have garnered immense interest. GFP molecule has been reengineered to contain the unnatural amino acid p-azidophenylalanine (pAzF). This azido group can be reduced specifically by H2S imparting selectivity to the GFP molecule, now termed as hsGFP. An orthogonal tRNA-tRNA synthetase system from E. coli was used for the selective incorporation of pAzF into hsGFP molecule in response to the Amber (TAG) codon. In presence of both H2S and pAzF, the chromophore p-azidoben-zylideneimidazolidone of hsGFP is converted to p-aminobenzy-lideneimidazolidone with fluorescence excitation and emission maxima at 454 nm and 500 nm, respectively. This genetically encoded probe has been used in mammalian cell line HEK293T for the detection of intracellular H2S. Such a genetically encoded biosensor is noninvasive and can reflect the dynamic nature of the turnover of H2S inside cells (128). Furthermore, such sensors can be targeted to specific cellular locations. Taken together, a genetically encoded biosensor appears to be a promising tool to study the levels of H2S under physiological conditions.
Tools for Detection of Protein S-Persulfidation
In addition to H2S, the detection of the post translational modification caused by it is also of the utmost importance to fully appreciate the functional relevance of this gasotransmitter. Protein persulfidation was identified to be the mechanism by which H2S exerts its signaling function (103). Detection of this modification, however, poses a significant challenge as the persulfide group exhibits similar reactivity to free thiols (129). Table 3 summarizes some of the widely used methods for the detection of intracellular protein persulfidation levels.
Table 3. Tools for detection of protein S-persulfidation.
| Detection method | Description | References |
|---|---|---|
| Biotin switch assay | This was the first assay developed for the detection of persulfidation. It was based on the premise that persulfides would not react with electrophilic thiolblocking reagent S-methyl methanethiosulfonate (MMTS). Persulfides were subsequently labeled with N-[6-(biotinamido)hexyl]- 30 -(20 -pyridyldithio)propionamide (biotin-HPDP). The biotin labeled species were pulled down by streptavidin beads and analysed by MS. It was demonstrated that upto 25% of proteins in the liver were persulfidated under basal conditions. The chemistry was later on proved to be incorrect as persulfides were shown to react with both electrophilic and nucleophilic species. | (103) |
| IAA assay | According to this assay, Iodoacetic acid (IAA) a thiol blocking agent would react with both free thiols and protein persulfides. Subsequently, DTT would cleave the alkylated persulfide. This was followed by labeling of that particular cysteine with iodoacetamide-linked biotin (IAP). However, how this method distinguishes the persulfides from intramolecular and intermolecular disulfides and S-nitrosothiols, which would also be reduced by DTT, remains poorly understood. | (130) |
| Maleimide assay | This assay was based on the fact that N-ethyl maleimide, a thiol-blocking agent, would block both free thiol and persulfide. Maleimide was conjugated to Cy5. DTT was used to cleave the alkylated moieties. Fluorescence signal would decrease if the sample contains persulfides. Ratiometric decrease in fluorescence can be used for quantification. | (131) |
| Biotin thiol assay | Maleimide-PEG2-Biotin was used to alkylate both thiols and persulfides followed by binding of the proteins on an avidin column. Elution was done by DTT which cleaved the disulfide bond leaving the biotin tag bound to the column. The eluate containing the persulfidated proteins was analysed by Western blotting and LC-MS/MS. | (132) |
| Pro-Per-DP –Protein persulfide detection protocol | Iodoacteyl PEG2 Biotin was used to alkylate both free thiols and persulfides. These species were pulled down by streptavidin coated magnetic beads and subsequently reduced with DTT. The supernatant, containing the persulfidated proteins were analysed by MS. | (133) |
| Tag switch assay | This assay exploits the difference in chemical and physical properties of the thiol and the persulfide group. A thiol-blocking reagent methylsulfonyl benzothiazole, reacts with free thiol and persulfide. When compared to disulfides in proteins, the disulfide bond in persulfide adducts react strongly to nucleophiles. Subsequently, a new tag-switch reagent, biotinylated cyanoacetic acid was used to label persulfidated proteins specifically. This method has been further improved by the use of cyanoacetic acid derivatives with BODIPY moiety (CN-BOT) for labeling cells and Cy3-dye (CN-Cy3) for labeling cell lysates, respectively. | (102,134) |
Role of H2S in Bacterial Physiology
While the initial co-culture experiments described earlier provided a valuable clue with regard to potential of H2S in protecting bacteria from toxic compounds, in depth examination of these findings was never attempted (42,43). It was only in 2011 that a study highlighted the importance of H2S in protecting bacteria from antibiotics and oxidative stress (44). In this context, H2S has been termed as a “double edged sword” mitigating not only the effects of antibiotics but also the resulting oxidative stress caused by them. To ascertain the role of H2S in E coli, the authors compared wild type and 3-MST deficient E. coli by a phenotypic microarray. While these strains showed no difference with respect to growth defects in vitro, the 3-MST deficient strain became highly susceptible to structurally and functionally different classes of antibiotics. Similar results were obtained for CBS/CSE deficient strains of P. aeruginosa, S. aureus, and B. anthracis, establishing the protective role of H2S across gram negative and gram-positive bacteria. Overexpression of 3-MST led to enhanced protection against spectinomycin whereas chemical inhibition on 3-MST, CBS, and CSE rendered them highly susceptible to a variety of antibiotics. NaHS, an H2S donor chemically complemented these mutant strains establishing the role of endogenously generated H2S as a protective mechanism against antibiotics.
Several studies have shown that a wide range of antibiotics exert killing by triggering ROS generation in addition to inhibiting the function of their primary targets (135,136). Antibiotics have been shown to stimulate respiration, which increases generation of toxic hydroxyl radicals via Fe2+-catalysed Fenton reaction (137). Consistent with these observations the authors have shown that pretreatment with Fe-chelator (dipyridyl) or ROS scavenger (thiourea) induces gentamycin resistance to both the wild type and H2S deficient strains of E. coli. Interestingly, a similar degree of protection from gentamycin was observed when the cells were treated with NaHS. Additionally, all the H2S deficient mutant strains exhibited severe susceptibility to H2O2 which was mitigated when they were pretreated with NaHS. DNA damage is one of the direct consequences of oxidative stress generated by antibiotics (138,139). On treatment with sublethal levels of ampicillin, which is known to cause oxidative stress, 3-MST deficient strain of E. coli showed tell-tale signs of DNA damage. Overexpression of 3-MST and pretreatment with NaHS ameliorated this damage. Additionally, the antioxidant effect of H2S was also shown to be in part due to the stimulation of antioxidant enzymes such as catalase and superoxide dismutase (SOD). Consistent with this, the rate of degradation of H2O2 was significantly greater in the crude cell lysates of wild type as compared to 3-MST deficient E. coli. However, the precise biochemical mechanism by which H2S modulates the activity of antioxidants was left unaddressed. Recently, direct sequestration of Fe2+ ions by H2S has been shown to counteract antibiotics triggered oxidative stress (140). This mechanism became very apparent in E. coli strain deficient in an iron uptake regulator, Fur. The fur mutant cells constitutively take up iron resulting in iron accumulation and consequent DNA damage and killing by H2O2. Interestingly, susceptibility to ROS is further exacerbated in the fur/3-mst double mutant, whereas overexpression of 3-mst in Fur deficient cells completely reversed both DNA damage and killing. All of these findings indicate that H2S can mitigate oxidative stress by potentiating the activity of antioxidant enzymes and sequestering Fe2+ (Fig. 5).
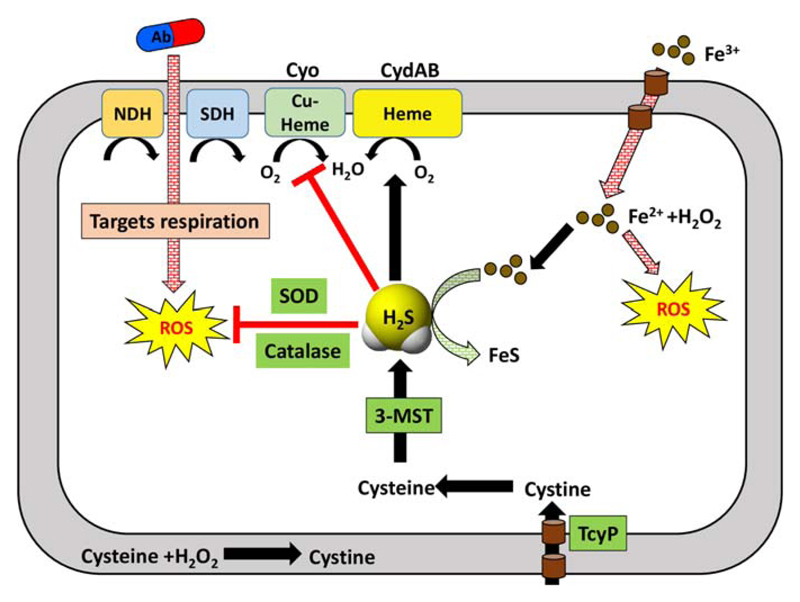
Fig 5.
Hydrogen sulfide and its cytoprotective effect in E. coli. H2S produced by 3-MST can directly sequester Fe2+ ions thereby preventing the detrimental Fenton reaction which lead to the production of ROS. H2S promotes tolerance to antibiotics by upregulation of cytochrome bd oxidase (CydAB) and downregulating cytochrome bo oxidase (Cyo) to maintain respiratory flux and redox poise. Additionally, H2S augments catalase and super oxide dismutase (SOD) activities. Abbreviations: NDH, NADH dehydrogenase; SDH, succinate dehydrogenase; Ab, antibiotics; TcyP, Transporter of l-cystine.
Apart from the above studies, another mechanism was put forward to explain the protective role of H2S. This involved the master regulator CysB which is involved in regulation of a number of sulfur metabolism genes in E. coli. CysB regulates the expression of TcyP, a cystine importer. During oxidative stress, H2O2 interacts with cysteine leading to its depletion and induction of CysB regulon including TcyP. As a consequence, TcyP leads to an increased influx of cystine/cysteine inside the cytoplasm resulting in the enhanced production of H2S via 3-MST. More recently, using chemical-biology approaches, we developed a series of bacteria specific H2S donors to explain the mechanism of H2S mediated protection (141). On pretreatment of E. coli with one such H2S donor (1c), we showed increased resistance to bactericidal antibiotics (ampicillin and amikacin) and H2O2. Furthermore, using a noninvasive biosensor of cytoplasmic redox potential (roGFP2) (142,143), we for the first time, precisely measured real-time changes in the redox physiology of E.coli in response to antibiotics in the presence or absence of H2S. Importantly, we showed that while elevation in endogenous H2S levels does not influence redox physiology of E.coli, it efficiently reversed antibiotics induced oxidative shift in the cytoplasmic redox potential of bacteria. To further probe mechanistic aspects of these findings, we discovered a functional association between H2S-directed cytoprotection and alternate mode of cellular respiration catalysed by cytochrome bd oxidase (CydB). H2S, due to its strong affinity for metals such as copper, is known to inhibit copper-heme containing cytochrome bo oxidase (CyoA). Under these conditions, the respiration proceeds via a less energy efficient CydB. In agreement with this, treatment of the cells with 1c led to a downregulation of cyoA transcript, whereas the transcripts of alternate respiratory oxidases such as cydB and appY were either maintained or enhanced, respectively. This realignment of respiratory oxidases mimics the expression profile of E. coli grown under respiratory arrest conditions (e.g., hypoxia), implicating H2S in respiration inhibition and metabolic slow down. In contrast, ampicillin treatment enhanced the expression of the energy efficient cyoA and repressed cydB, consistent with the reported hyperactivation of electron transport chain by bactericidal antibiotics (144). However, pretreatment of the cells with 1c reversed the influence of ampicillin on cyoA and cydB transcripts. In agreement with these findings, the cyoA mutant pretreated with 1c remained protected from ampicillin toxicity, whereas 1c-derived H2S remained completely ineffective in protecting cydB mutant. The cydB from E. coli has also been shown to reduce H2O2 by acting as catalase and quinol peroxidase (145). Therefore, sustenance of cydB expression by H2S can potentiate antibiotic tolerance by bolstering the bacterial antioxidant capacity. Altogether, these observations put forth a central role of alternate respiration and oxidant mitigating mechanisms in resisting the activity of antibiotics by H2S.
The association of H2S and drug resistance is not new. Nearly 40 years ago, several studies have reported the presence of plasmid-borne genetic elements enhancing both basal H2S production and antibiotic resistance in multidrug-resistant strains of E. coli isolated from patients suffering from urinary tract infection (146,147). Recently, we confirmed these findings and demonstrated that clinical strains of multidrug resistant uropathogenic E. coli possesses greater levels of endogenous H2S as compared to wild-type. Intriguingly, pretreating these strains with aspartate, a 3-MST inhibitor significantly reduced endogenous H2S levels and markedly reversed (50%) resistance toward ampicillin. More importantly, exposure to H2S donor neutralized the adverse effect of aspartate on drug resistance. Our work provided a strong pharmacological foundation for design of inhibitors of H2S biogenesis as a possible adjuvant to antibiotics. Further, it appears that targeting antioxidant enzymes and alternate respiratory complexes (Cyd/App) is likely to potentiate the killing efficiency of antibiotics.
The effect of H2S on intracellular human pathogens such as Mycobacterium tuberculosis (Mtb) is largely unknown. However, a recent study aimed at identifying genetic components involved in protection from oxidative stress showed that H2S donor (NaHS) can complement the defects in recycling of the major mycobacterial antioxidant, mycothiol (MSH) (148). Using a specific biosensor of mycothiol redox potential (EMSH; Mrx1-roGFP2) (143), authors have shown that NaHS restored steady-state MSH/MSSM ratio (basal EMSH) in the mutants showing oxidative EMSH. It is likely that NaHS increased mycothiol biosynthesis similar to H2S-mediated increased biogenesis of GSH in eukaryotes. This supplementation with NaHS also enhanced the survival of the Mtb mycothiol recycling mutants in both activated macrophages and animals. Thus, the contribution of endogenous H2S biogenesis pathways in regulating redox balance, stress response, and virulence is eagerly awaited in several human pathogens including Mtb.
Oxidative Stress is an Integral Part of Infection with Diverse Class of Viruses
Oxidative stress has been linked to vast group of etiological agents that cause acute and chronic diseases such as infection with viruses, bacteria, and parasites (149). Viral and bacterial infections in particular have been linked to induce ROS/RNS production, alteration in metabolic pathways, and leading to several disease associated complications (149,150). However, this field still lacks mechanistic insight. The impact of these infectious agents on host redox physiology and how this could be targeted for therapeutic benefits remains a challenging area of research.
In case of viral infections, induction of oxidative stress inside host is a prerequisite for successful infection and long term viral replication. RNA viruses such as influenza and paramyxovirus infection generates ROI via activation of monocytes and polymorphonuclear leukocytes (149). A study indicates that the oxidative state of the host cells provides an environment permissive for viral replication (151). Using mice models, it has been shown that influenza A (RNA virus) infection creates redox imbalance by decreasing the levels of GSH, vitamins C, and vitamin E (152). Similarly, a transgenic mouse model of hepatitis B virus (DNA virus) was exploited to show that this virus induces ROS and leads to hepatocarcinogeneis and oxidative DNA damage during chronic necroinflammatory conditions (153). Based on these studies, it has been proposed that antioxidant strategies can be utilized to target viral replication and to decrease viral induced oxidative stress to control pathological manifestations (154).
The role of oxidative stress has been extensively studied in retroviruses such as HIV-1 (Human Immunodeficiency Virus). Studies have shown that HIV-1 replication induces ROS generation and decreases cellular antioxidants such as GSH and Trx, and modulates immunopathogenesis during AIDS progression (155,156). Plasma and peripheral blood mononuclear cells (PBMCs) of AIDS patients show reduction in concentration of other major antioxidants like cysteine, methionine, vitamins C and E, along with elevated levels of lipid peroxidation products (157). At molecular level, ROS has been shown to induce the activity of redox sensitive transcription factors Nf-κB, AP-1, and Sp1, which regulates HIV-1 gene transcription by binding to 5′-LTR promoter (158,159). Despite these studies, for a very long time, the relation between HIV-1 and oxidative stress remained circumstantial. This was largely owing to the lack of sophisticated and sensitive tools to measure intracellular redox potential of HIV-1 infected cells during various stages of infection. To fill this knowledge gap, we exploited a noninvasive biosensor of GSH redox potential (Grx1-roGFP2; EGSH) and accurately measured oxidative stress in the cytoplasm and mitochondria of HIV-1 infected monocytes (160). We demonstrated that monocytes latently infected with HIV-1 are intrinsically resistant to oxidative stress and displayed reductive EGSH (160). More importantly, we showed that a marginal oxidative shift in EGSH (25 mV) triggers reactivation of HIV-1 without adversely affecting cellular physiology. Furthermore, supplementation with antioxidants such as N-acetylcysteine kept HIV-1 in a silent state by preventing an oxidative shift in EGSH required for viral activation (160). Lastly, global expression analysis revealed that pathways associated with redox metabolism are significantly affected during HIV-1 latency and reactivation (Fig. 6) (160). Taken together, these results highlight the central role of host redox physiology in modulating HIV-1 life cycle.
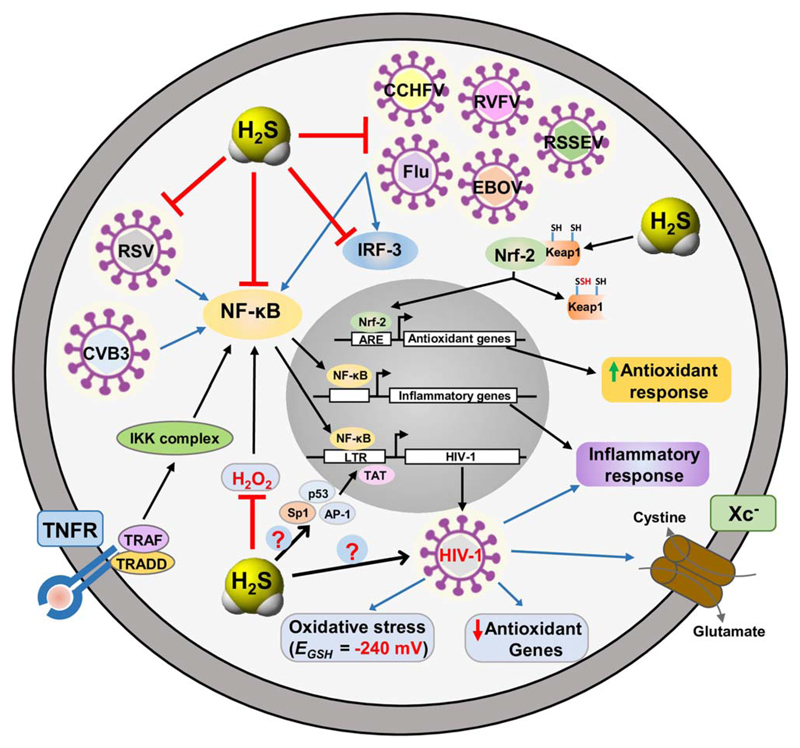
Fig 6.
H2S targets key host factors and viral steps to perturb replication of enveloped RNA viruses. H2S acts as an antioxidant gaseous signaling molecule by directly scavenging ROS and upregulating cellular antioxidant capacity via Nrf-2/ARE pathway. Moreover, H2S has been shown to exert broad range antiviral and anti-inflammatory activity. Redox sensitive transcription factors Nf-κB, Ap-1, p53, and Sp1 mediates HIV-1 reactivation from latency in conjunction with viral Trans-activator of transcription (Tat). As pathways related to redox metabolism are significantly affected on HIV-1 replication it would be of great interest to characterize the role H2S in HIV-1 latency and reactivation. Abbreviations: RSV, Respiratory Syncytial Virus; CVB3, Coxsackie virus B3; Flu, Influenza virus (H1N1 (A strain), H3N2 (A strain) and Brisbane (B strain)); EBOV, Ebola virus; RVFV, Rift valley fever virus; CCHFV, Crimean-Congo hemorrhagic fever virus; RSSEV, Far-eastern subtype tick-borne flavivirus; Nf-κB, Nuclear factor kappa-light-chain-enhancer of activated B cells; IRF-3, Interferon regulatory factor 3; TNFR, tumor necrosis factor receptor; Xc-, cystine/glutamate transporter; TRADD, Tumor necrosis factor receptor type 1-associated DEATH domain protein; TRAF, TNF receptor associated factor.
At molecular level HIV-1 infection or exposure to HIV-1 related proteins downregulates the Nuclear factor-erythroid-2 p45 related factor 2-Antioxidant Response Element (Nrf2/ARE) pathway, leading to reduced expression of antioxidant genes (161,162). Nrf2 is a constitutive transcription factor and master regulator of the antioxidant response (163,164). Nrf2 is inhibited in cytosol by Kelch-like ECH-associated protein-1 (Keap1), which is a redox-sensitive ubiquitin ligase substrate adaptor leading to ubiquitination and degradation of Nrf2 (165). Interestingly, Nrf2 inducer (Sulforaphane) has the ability to block HIV-1 infection in primary macrophages, which are the long-lived reservoirs of HIV-1 in infected individuals (166). In this direction, sulforaphane has also been shown to enhance phagocytic activity of HIV-1 infected monocytes-derived macrophages (MDMs) and alveolar macrophages (Ams) from HIV-1 transgenic rats, thereby reducing the severity of HIV-1 related pulmonary dysfunctions (161). Nrf2 activation has also been shown to assist Marburg virus ((-ss) RNA) (MARV), Kaposi’s sarcoma-associated herpesvirus (dsDNA) (KSHV), and Dengue virus ((+ss) RNA) replication. Activation of Nrf2 induced by VP24 and vFLIP proteins of MARV and KSHV, respectively, leads to dysregulation of host antiviral response and modulates viral gene expression, thereby ensuring a conducive environment for infection and also promotes the survival of infected cells (167,168). Dengue infection induced oxidative stress has been shown to activate Nrf2 thereby modulating the level of oxidative stress and affecting both antiviral and cell death response (169). While these studies clearly establish a connection between oxidative stress and infection with pathogenic viruses, the contribution of H2S in these process is poorly understood. Interestingly, Nrf2/Keap-1 pathway could provide the missing link between H2S, redox stress and virus infections. H2S has been shown to inhibit Keap1 by persulfidation of Cys 151, which leads to translocation of Nrf2 into the nucleus and its subsequent binding to Antioxidant Response Element (ARE). This results in the induction of genes encoding antioxidant and phase II detoxifying enzymes, such as heme oxygenase-1 (HO-1), NAD(P)H:quinone oxidoreductase-1 (NQO-1), glutamate cysteine ligase catalytic subunit (GCLC), and thioredoxin reductase-1 (TXNRD-1) (163,165). In relation to this H2S has been shown to protect against cellular senescence and oxidative stress via S-sulfhydration of Keap1 and resulting activation of Nrf2 (165). Further experimentation on H2S biogenesis and Nrf2/ARE pathway during viral infections will provide next stage of insight in this process. The following section provides recent knowledge in this direction.
H2S and Host–Pathogen Interaction
Host-derived H2S has a marked effect on the outcome of bacterial and viral infections. Blocking the host transsulfuration pathway in macrophages by propargylglycine increased the viability of Mycobacterium smegmatis. This impairment in bacterial clearance was shown to be due to defects in the phagolysosomal fusion during infection. Treatment with N-acetylcysteine (NAC), which is known to increase cysteine flux through H2S biogenesis pathway, significantly increased the phagolysosomal fusion resulting in vacuolar acidification and killing of mycobacteria (170). H2S was found to inhibit the induction of an inflammatory response on infection with Mycoplasma fermentans. Underlying mechanism revealed that H2S inhibited the activation and nuclear translocation of a redox sensitive transcription factor NF-κB, thereby diminishing the transcription of proinflammatory genes (171,172). One of the mechanism by which H2S affects the activity of a global transcriptional regulator, NF-κB, is by persulfidation of Cys-38 residue in the p65 subunit (131).
H2S production by gut microbiota presents itself as an interesting line of study to explore the effect of bacteria-derived effector molecules affecting host physiology and pathophysiology. Sulfide reducing bacteria (SRB) represent a major class of the normal gut microbiota (173). The predominant genera residing in gut are Desulfovibrio, Desulfobacter, Desulfolobus, and Desulfotomaculum. SRBs are the major contributors of nonenzymatic H2S produced in the human body (174). Some of the initial studies to explore the significance of gut microbiota-derived H2S came from experiments done on germ free animals. It was observed that fecal samples of germ free mice contained half the H2S as compared to control mice (175). In addition to this, free H2S levels in inferior vena cava, blood plasma and in gastrointestinal tissues, were shown to be diminished in germ free mice. Apart from this, sulfane sulfur levels of plasma, adipose, and lung tissues were also found to be lower in such mice. This implicated the gut microflora as a potential source of circulating H2S for the host (176). Recent studies have also shown that colonocytes are capable of using H2S as an energy source (177).
Gut bacteria-derived H2S has been shown to have both protective and detrimental effects on colonic health. Increased fecal sulfide levels have been found in patients with ulcerative colitis (UC) (178). Furthermore, it has been suggested that epithelial damage associated with UC is due to increased availability of dietary sulfate for SRBs (179). In contrast, using animal models of colitis, it was demonstrated that scavenging bacterial H2S by bismuth did not ameliorate the symptoms of colitis (180). In fact, the condition was shown to improve on exogenous H2S administration (181). The ability of luminal H2S to modify secreted defensive proteins like trefoil factor 3 (TFF3) by reduction of the disulfide bond is believed to be a potential mechanism of the anti-inflammatory role of H2S. TFF3 plays an important role in mucosal repair and regeneration (182).
Our survey of literature revealed the existence of very few reports on H2S and viral infections. Existing studies have highlighted the role of H2S in modulating Respiratory Syncytial Virus (RSV) ((-ss) RNA virus) infection (183). RSV causes lower and upper respiratory tract infection in infants for which there is no vaccine and only limited supportive measures for treatment exist with no real benefits (184). RSV mediates its influence on the host by upregulating the expression of various Nf-κB and IRF-3 dependent cytokines and chemokines (185). This leads to inflammation, cellular infiltration in lungs and other pulmonary dysfunctions. Importantly, RSV infection resulted in downregulation of expression and impaired activity of H2S biosynthesis enzymes. As a consequence, the endogenous levels of H2S were diminished in RSV infected cells (183). Pharmacological inhibition or genetic silencing of CSE (cystathionine gamma-lyase) enhances RSV multiplication and exacerbates disease condition, airway dysfunction, and pulmonary inflammation (186,187). Consistent with these findings, exogenous administration of H2S reduces the secretion of viral induced chemokines and cytokine through inhibition of NF-κB mediated activation of genes encoding proinflammatory cytokines (183). H2S treatment (using slow releasing H2S donor-GYY4137) significantly blocked RSV replication in vitro and in vivo by targeting viral assembly, release, viral spread, and replication (Fig. 6) (183,186). Moreover, H2S treatment significantly improved clinical disease parameters and pulmonary dysfunction on RSV infection (186). Similarly, H2S also exerts antiviral and anti-inflammatory effects on the viruses in the family of Paramyxoviridae; human metapneumovirus (hMPV) and Nipah virus (NiV) (183).
H2S has also been shown to affect replication of highly pathogenic enveloped RNA virus from Ortho-, Filo-, Flavi-, and Bunyavirus families (Fig. 6) (188). FDA approved antiviral treatment is available for influenza virus (Orthomyxoviridae), whereas there is no vaccine or therapeutic interventions to target Ebola virus (Filoviridae), Far-eastern subtype tick-borne flavivirus (Flaviviridae), Rift valley fever virus and Crimean-Congo hemorrhagic fever virus (Bunyaviridae) (189). H2S was shown to significantly reduce replication of all the above families of viruses (188). As explained earlier, studies confirmed that H2S targets the transcription factor Nf-κB and IRF-3 nuclear translocation to inhibit the release of viral induced proinflammatory mediators (188).
Lastly, H2S has been recently shown to modulate Coxsackie virus B3 (CVB3) infection induced inflammatory response, which is a predominant cause of human myocarditis and ultimately leads to heart failure (190). Treatment of CVB3 infected rats with H2S significantly resulted in downregulation of proinflammatory mediators, reduces myocardial injury, and alleviates damage of myocardial cells (191). H2S was shown to inhibit Nf-κB signaling by lowering IκBα degradation leading to reduced nuclear translocation and DNA binding ability of Nf-κB (191). CVB3 infection also induces MAPK signaling cascade by activating ERK1/2, p38 and JNK1/2 which are upstream signaling molecule involved in activation of Nf-κB (192). H2S treatment also showed reduced CVB3 induced activation of ERK1/2, p38, and JNK1/2 in rat myocardial cells, thereby supressing the expression of inflammatory mediators and alleviating myocardial damage (191).
Conclusion
A bourgeoning body of literature stands testament to the cytoprotective role of H2S in various organs and tissues, ameliorating a wide variety of stresses. In comparison, the importance of this gasotransmitter from the perspective of bacterial physiology and pathogenesis remains understudied. This aspect becomes increasingly important as H2S has been shown to confer protection from both antibiotic and oxidative stresses. In today’s scenario, where drug resistance is becoming one of the leading healthcare concerns, there is an immediate need to have a greater understanding of the role of H2S in bacterial antibiotic resistance. Additionally, it would be worthwhile to study the importance of this molecule in intracellular bacteria, which face a hostile oxidative host environment.
Viral infections are accompanied by host physiological perturbations like alteration in redox homeostasis, inflammation, metabolism, among others. Of particular interest to us is the observation that a number of viruses have been shown to adversely affect the host H2S biogenesis. Antiviral and anti-inflammatory effects of H2S highlight its potential as a therapeutic molecule. It can, therefore, bolster the efficacy of the regular drug regime used for viral infections. Despite all these observations there is a dearth of knowledge in terms of molecular mechanism of these effects which could form a promising line of research. Through our review, we have highlighted some of the important studies in this field pertaining to the role of H2S in physiology and pathogenesis of these infectious agents. Considering that the role of H2S as a cytoprotectant appears to be conserved across phyla, the significance of this ubiquitous biological effector molecule in bacteria and viruses should be pursued with renewed interest.
Acknowledgements
This work was supported by the Wellcome-DBT India Alliance grant, IA/S/16/2/502700, and in part by Department of Biotechnology (DBT) Grants BT/PR5020/MED/29/1454/2012, BT/PR11911/BRB/20/1327/2014, BT/PR13522/COE/34/27/2015, DBT-IISc program and Infosys Foundation. AS is a senior fellow of Wellcome DBT India Alliance. PB is grateful to the Council of Scientific and Industrial Research (CSIR), India for fellowship. VKP is supported by the Indian Institute of Science, India.
Abbreviations
- CBS
Cystathionine beta synthase
- CSE
Cystathionine gamma lyase
- 3-MST
3-mercaptopyruvate sulfurtransferase
- SQR
Sulfide quinone oxidoreductase
- LTP
Long term potentiation
- NMDA
N-methyl-D-aspartate
- GSH
Glutathione
- RSV
Respiratory syncytial virus
- HIV-1
Human immunodeficiency virus type 1
- PLP
Pyridoxal-5-phosphate
- SAM
S-adenosyl methionine
- DAO
D-amino oxidase
- 3-MP
3-mercaptopyruvate
- ox-PTM
Oxidative posttranslational modification
- ROS
Reactive oxygen species
- RNS
Reactive nitrogen species
- ROI
Reactive oxygen intermediates
- CcO
Cytochrome c oxidase
- SOD
Superoxide dismutase
- OCR
Oxygen consumption rate
- LC-MS
Liquid chromatography - Mass spectrometry
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- Nrf2
Nuclear factor-erythroid-2 p45 related factor 2
- ARE
Antioxidant response element
- Keap1
Kelch-like ECH-associated protein 1
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cell
- GC
Gas chromatography
- HPLC
High pressure liquid chromatography
- FAM
Fluorescein
- GFP
Green fluorescent protein
- pAzF
para-Azido phenylalanine
- MMTS
S-methyl methanethiosulfonate
- IAA
Iodoacetic acid
- IAM
Iodoacetamide
- DTT
Dithiothreitol
- CydB
Cytochrome bd oxidase
- CyoA
Cytochrome bo oxidase
- MSH
Mycothiol
- MSSM
Oxidized mycothiol
- RNA
Ribonucleic acid
- DNA
Deoxyribonucleic acid
- Trx
Thioredoxin
- PBMC
Peripheral blood mononuclear cells
- AP-1
Activator protein 1
- Sp1
Specificity protein 1
- LTR
Long terminal repeat
- roGFP
Redox-sensitive green fluorescent protein
- NAC
N-acetylcysteine
- SRB
Sulfur reducing bacteria
- UC
Ulcerative colitis
- TFF3
Trefoil factor 3
- IRF3
Interferon regulatory factor
- MAPK
Mitogen-activated protein kinase
- ERK
Extracellular signal regulated kinase
- JNK
c-Jun N-terminal kinase
Footnotes
Amit Singh: http://orcid.org/0000-0001-6761-1664
Conflict of Interest
The authors indicate no potential conflicts of interest.
References
- [1].Sleep NH. The Hadean-Archaean environment. Cold Spring Harb Perspect Biol. 2010;2:a002527. doi: 10.1101/cshperspect.a002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chyba CF. Rethinking earth’s early atmosphere. Science. 2005;308:962–963. doi: 10.1126/science.1113157. [DOI] [PubMed] [Google Scholar]
- [3].Dworkin M. In: The prokaryotes. Ecophysiology and Biochemistry. 3rd edn. Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. Vol. 2 Springer; New York: 2006. [Google Scholar]
- [4].Crowe SA, Døssing LN, Beukes NJ, Bau M, Kruger S, et al. Atmospheric oxygenation three billion years ago. Nature. 2013;501:535–538. doi: 10.1038/nature12426. [DOI] [PubMed] [Google Scholar]
- [5].Schopf JW. Geological evidence of oxygenic photosynthesis and the biotic response to the 2400-2200 Ma “Great Oxidation Event”. Biochemistry (Moscow) 2014;79:165–177. doi: 10.1134/S0006297914030018. [DOI] [PubMed] [Google Scholar]
- [6].Olson KR, Straub KD. The role of hydrogen sulfide in evolution and the evolution of hydrogen sulfide in metabolism and signaling. Physiology. 2016;31:60–72. doi: 10.1152/physiol.00024.2015. [DOI] [PubMed] [Google Scholar]
- [7].Fenchel TM, Riedl RJ. The sulfide system: a new biotic community underneath the oxidized layer of marine sand bottom. Mar Biol. 1970;7:255–268. [Google Scholar]
- [8].Sylvan JB, Toner BM, Edwards KJ. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio. 2012;3:e00279–e00311. doi: 10.1128/mBio.00279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mulkidjanian AY, Bychkov AY, Dibrova DV, Galperin MY, Koonin EV. Open questions on the origin of life at anoxic geothermal fields. Orig Life Evol Biosph. 2012;42:507–516. doi: 10.1007/s11084-012-9315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mitchell CW, Davenport SJ. Hydrogen sulphide literature. Public Health Rep. 1924;39:1–13. [Google Scholar]
- [11].Beauchamp RO, Buss JS, Popp JA, Boreiko CJ, gelkovich DA. A critical review of the literature on hydrogen sulfide toxicity. CRC Crit Rev Toxicol. 1984;13:25–97. doi: 10.3109/10408448409029321. [DOI] [PubMed] [Google Scholar]
- [12].Szabo C. A timeline of hydrogen sulfide (H2S) research: from environmental toxin to biological mediator. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.09.010. pii: S0006–2952(17)30606-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vigneaud V, Loring HS, Craft H. The oxidation of the sulfur of homocystine, methionine, and S-methylcysteine in the animal body. J Biol Chem. 1934;105:481. [Google Scholar]
- [14].Binkley F, Vigneaud V. The formation of cysteine from homocysteine and serine by liver tissue of rats. J Biol Chem. 1942;144:507. [Google Scholar]
- [15].Braunstein AE, Goryachenkova EV, Lac ND. Reaction catalysed by serine sulfhydrase from chicken liver. Biochim Biophys Acta. 1969;171:366–368. doi: 10.1016/0005-2744(69)90173-9. [DOI] [PubMed] [Google Scholar]
- [16].Hylin JW, Wood JL. Enzymatic formation of polysulfides from mercaptopyruvate. J Biol Chem. 1959;234:2141–2144. [PubMed] [Google Scholar]
- [17].Akopyan TN, Braunstein AE, Goryachenkova EV. Beta-cyanoalanine synthase: purification and characterization. Proc Natl Acad Sci USA. 1975;72:1617–1621. doi: 10.1073/pnas.72.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stipanuk MH, Beck PW. Characterization of the enzymic capacity for cysteine desulphhydration inliver and kidney of the rat. Biochem J. 1982;206:267–277. doi: 10.1042/bj2060267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Warenycia MW, Goodwin LR, Benishin CG, Reiffenstein RJ, Francom DM, et al. Acute hydogen sulfide. Demonstration of selective uptake of sulfide by the brainstem by measurement of brain sulfide levels. Biochem Pharmacol. 1989;38:973–981. doi: 10.1016/0006-2952(89)90288-8. [DOI] [PubMed] [Google Scholar]
- [20].Goodwin LR, Francom D, Dieken FP, Taylor JD, Warenycia MW, et al. Determination of sulfide in brain tissue by gas dialysis/ion chromatography: postmortem studies and two case reports. J Anal Toxicol. 1989;13:105–109. doi: 10.1093/jat/13.2.105. [DOI] [PubMed] [Google Scholar]
- [21].Savage JC, Gould DH. Determination of sulfide in brain tissue and rumen fluid by ion-interaction reversed-phase high-performance liquid chromatography. J Chromatogr. 1990;526:540–545. doi: 10.1016/s0378-4347(00)82537-2. [DOI] [PubMed] [Google Scholar]
- [22].Abe K, Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhao W, Zhang J, Lu Y, Wang R. The vasorelaxant effect of H(2)S as a novel endogenous gaseous K(ATP) channel opener. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhao W, Wang R. H(2)S-induced vasorelaxation and underlying cellular and molecular mechanisms. Am J Physiol Heart Circ Physiol. 2002;283:H474–H480. doi: 10.1152/ajpheart.00013.2002. [DOI] [PubMed] [Google Scholar]
- [25].Tang G, Wu L, Liang W, Wang R. Direct stimulation of K(ATP) channels by exogenous and endogenous hydrogen sulfide in vascular smooth muscle cells. Mol Pharmacol. 2005;68:1757–1764. doi: 10.1124/mol.105.017467. [DOI] [PubMed] [Google Scholar]
- [26].Elrod JW, Calvert JW, Morrison J, Doeller JE, Kraus D, et al. Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc Natl Acad Sci USA. 2007;104:15560–15565. doi: 10.1073/pnas.0705891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jha S, Calvert JW, Duranski MR, Ramachandran A, Lefer DJ. Hydrogen sulfide attenuates hepatic ischemia-reperfusion injury: role of antioxidant and antiapoptotic signaling. Am J Physiol Heart Circ Physiol. 2008;295:H801–H806. doi: 10.1152/ajpheart.00377.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wu D, Wang J, Li H, Xue M, Ji A, et al. Role of hydrogen sulfide in ischemia-reperfusion injury. Oxid Med Cell Longev. 2015;2015:186908. doi: 10.1155/2015/186908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang R. Two’s company, three’s a crowd: can H2S be the third endogenous gaseous transmitter? FASEB J. 2002;16:1792–1798. doi: 10.1096/fj.02-0211hyp. [DOI] [PubMed] [Google Scholar]
- [30].Ohsawa I, Ishikawa M, Takahashi K, Watanabe M, Nishimaki K, et al. Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat Med. 2007;13:688–694. doi: 10.1038/nm1577. [DOI] [PubMed] [Google Scholar]
- [31].Iio A, Ito M, Itoh T, Terazawa R, Fujita Y, et al. Molecular hydrogen attenuates fatty acid uptake and lipid accumulation through downregulating CD36 expression in HepG2 cells. Med Gas Res. 2013;3:6. doi: 10.1186/2045-9912-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang R. Physiological implication of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- [33].Hongfang J, Bailin C, Bin Z, Chunyu Z, Xinmin L, et al. effect of hydrogen sulfide on hypoxic pulmonary vascular structural remodeling. Life Sci. 2006;78:1299–1309. doi: 10.1016/j.lfs.2005.07.009. [DOI] [PubMed] [Google Scholar]
- [34].Qiao W, Chaoshu T, Hongfang J, Junbao D. Endogenous hydrogen sulfide is involved in the pathogenesis of atherosclerosis. Biochem Biophys Res Commun. 2010;396:182–186. doi: 10.1016/j.bbrc.2010.04.061. [DOI] [PubMed] [Google Scholar]
- [35].Durham HE. A simple method for demonstrating the production of gas by bacteria. Br Med J. 1898;1:1387. doi: 10.1136/bmj.1.1952.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Clarke PH. Hydrogen sulphide production by bacteria. J Gen Microbiol. 1953;8:397–407. doi: 10.1099/00221287-8-3-397. [DOI] [PubMed] [Google Scholar]
- [37].Gaill F. Aspects of life development at deep sea hydrothermal vents. FASEB J. 1993;7:558–565. doi: 10.1096/fasebj.7.6.8472894. [DOI] [PubMed] [Google Scholar]
- [38].Sylvan JB, Toner BM, Edwards KJ. Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio. 2012;3:e00279–e00311. doi: 10.1128/mBio.00279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Myers JT. The production of hydrogen sulphide by bacteria. J Bacteriol. 1920;5:231–252. doi: 10.1128/jb.5.3.231-252.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Artman M. The production of hydrogen sulphide from thiosulphate by Escherichia coli. Microbiology. 1956;14:315–322. doi: 10.1099/00221287-14-2-315. [DOI] [PubMed] [Google Scholar]
- [41].Seiflein TA, Lawrence JG. Two transsulfurylation pathways in Klebsiella pneumoniae. J Bacteriol. 2006;188:5762–5774. doi: 10.1128/JB.00347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bachenheimer AG, Bennett EO. The sensitivity of mixed population of bacteria to inhibitors. I. The mechanism by which desulfovibrio desulfuricans protects Ps. aeruginosa from the toxicity of mercurials. Antonie Van Leeuwenhoek. 1961;27:180–188. doi: 10.1007/BF02538438. [DOI] [PubMed] [Google Scholar]
- [43].Stutzenberger FJ, Bennett EO. Sensitivity of mixed populations of Staphylococcus aureus and Escherichia coli to Mercurials. Appl Microbiol. 1965;13:570–574. doi: 10.1128/am.13.4.570-574.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- [45].Kabil O, Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxid Redox Signal. 2014;20:770–782. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kabil O, Banerjee R. Redox biochemistry of hydrogen sulfide. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Taoka S, West M, Banerjee R. Characterization of the heme and pyridoxal phosphate cofactors of human cystathionine beta-synthase reveals nonequivalent active sites. Biochemistry. 1999;38:2738–2744. doi: 10.1021/bi9826052. [DOI] [PubMed] [Google Scholar]
- [48].Kery V, Poneleit L, Meyer JD, Manning MC, Kraus JP. Binding of pyridoxal 5’-phosphate to the heme protein human cystathionine beta-synthase. Biochemistry. 1999;38:2716–2724. doi: 10.1021/bi981808n. [DOI] [PubMed] [Google Scholar]
- [49].Shibuya N, Koike S, Tanaka M, Ishigami-Yuasa M, Kimura Y, et al. A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat Commun. 2013;4:1366. doi: 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
- [50].Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- [51].Finkelstein JD, Kyle WE, Martin JL, Pick AM. Activation of cystathionine synthase by adenosylmethionine and adenosylethionine. Biochem Biophys Res Commun. 1975;66:81–87. doi: 10.1016/s0006-291x(75)80297-x. [DOI] [PubMed] [Google Scholar]
- [52].Singh S, Padovani D, Leslie RA, Chiku T, Banerjee R. Relative contributions of cystathionine beta-synthase and gamma-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J Biol Chem. 2009;284:22457–22466. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Meier M, Janosik M. Structure of human cystathionine β-synthase: a unique pyridoxal 5’-phosphate-dependent heme protein. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kabil O, Weeks CL, Carballal S, Gherasim C, Alvarez B, et al. Reversible heme-dependent regulation of human cystathionine beta-synthase by a flavoprotein oxidoreductase. Biochemistry. 2011;50:8261–8263. doi: 10.1021/bi201270q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kabil O, Yadav V, Banerjee R. Heme-dependent metabolite switching regulates H2S synthesis in response to endoplasmic reticulum (ER) stress. J Biol Chem. 2016;291:16418–16423. doi: 10.1074/jbc.C116.742213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shibuya N, Mikami Y, Kimura Y, Nagahara N, Kimura H. Vascular endothelium expresses 3-mercaptopyruvate sulfurtransferase and produces hydrogen sulfide. J Biochem. 2009;146:623–626. doi: 10.1093/jb/mvp111. [DOI] [PubMed] [Google Scholar]
- [57].Nagahara N, Katayama A. Post-translational regulation of mercaptopyruvate sulfurtransferase via a low redox potential cysteine-sulfenate in the maintenance of redox homeostasis. J Biol Chem. 2005;280:34569–34576. doi: 10.1074/jbc.M505643200. [DOI] [PubMed] [Google Scholar]
- [58].Shibuya N, Kimura H. Production of hydrogen sulfide from D-cysteine and its therapeutic potential. Front Endocrinol. 2013;4:87. doi: 10.3389/fendo.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xie ZZ, Liu Y, Bian JS. Hydrogen sulfide and cellular redox homeostasis. Oxid Med Cell Longev. 2016;2016:6043038. doi: 10.1155/2016/6043038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lindenmann J, Matzi V, Neuboeck N, Komenda BR, Maier A, et al. Severe hydrogen sulphide poisoning treated with 4-dimethylaminophenol and hyperbaric oxygen. Diving Hyperb Med. 2010;40:213–217. [PubMed] [Google Scholar]
- [61].Blackstone E, Morrison M, Roth MB. H2S induces a suspended animation-like state in mice. Science. 2005;308:518. doi: 10.1126/science.1108581. [DOI] [PubMed] [Google Scholar]
- [62].Mishanina TV, Libiad M, Banerjee R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat Chem Biol. 2015;11:457–464. doi: 10.1038/nchembio.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Millis KK, Weaver HK, Rabenstein LD. Oxidation/reduction potential of glutathione. J Org Chem. 1993;58:4144–4146. [Google Scholar]
- [64].Giles GI, Jacob C. Reactive sulfur species: an emerging concept in oxidative stress. Biol Chem. 2002;383:375–388. doi: 10.1515/BC.2002.042. [DOI] [PubMed] [Google Scholar]
- [65].Li Q, Jr, Lancaster JR. Chemical foundations of hydrogen sulfide biology. Nitric Oxide. 2013;35:21–34. doi: 10.1016/j.niox.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shen X, Peter EA, Bir S, Wang R, Kevil CG. Analytical measurement of discrete hydrogen sulfide pools in biological specimens. Free Radic Biol Med. 2012;52:2276–2283. doi: 10.1016/j.freeradbiomed.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Levitt MD, Abdel-Rehim MS, Furne J. Free and acid-labile hydrogen sulfide concentrations in mouse tissues: anomalously high free hydrogen sulfide in aortic tissue. Antioxid Redox Signal. 2011;15:373–378. doi: 10.1089/ars.2010.3525. [DOI] [PubMed] [Google Scholar]
- [68].Szabo C. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 2007;6:917–935. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- [69].Stein A, Bailey SM. Redox biology of hydrogen sulfide: implications for physiology, pathophysiology, and pharmacology. Redox Biol. 2013;1:32–39. doi: 10.1016/j.redox.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kimura H. Production and physiological effects of hydrogen sulfide. Antioxid Redox Signal. 2014;20:783–793. doi: 10.1089/ars.2013.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mathai JC, Missner A, Kugler P, Saparov SM, Zeidel M, et al. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci USA. 2009;106:16633–16638. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Vorobets VS, Kovach SK, Kolbasov GY. Distribution of ion species and formation of ion pairs in concentrated polysulfide solutions in photoelectrochemical transducers. Zhurnal Prikl Him. 2002;75:229–234. [Google Scholar]
- [73].Longen S, Richter F, Kohler Y, Wittig I, Beck KF, et al. Quantitative persulfide site identification (qPerS-SID) reveals protein targets of H2S releasing donors in mammalian cells. Sci Rep. 2016;6:29808. doi: 10.1038/srep29808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Predmore BL, Lefer DJ, Gojon G. Hydrogen sulfide in biochemistry and medicine. Antioxid Redox Signal. 2012;17:119–140. doi: 10.1089/ars.2012.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Carballal S, Trujillo M, Cuevasanta E, Bartesaghi S, Möller MN, et al. Reactivity of hydrogen sulfide with peroxynitrite and other oxidants of biological interest. Free Radic Biol Med. 2011;50:196–205. doi: 10.1016/j.freeradbiomed.2010.10.705. [DOI] [PubMed] [Google Scholar]
- [76].Shen X, Pattillo CB, Pardue S, Bir SC, Wang R, Kevil CG. Measurement of plasma hydrogen sulfide in vivo and in vitro. Free Radic Biol Med. 2011;50:1021–1031. doi: 10.1016/j.freeradbiomed.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Whitfield NL, Kreimier EL, Verdial FC, Skovgaard V, Olson KR. Reappraisal of H2S/sulfide concentration in vertebrate blood and its potential significance in ischemic preconditioning and vascular signaling. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1930–R1937. doi: 10.1152/ajpregu.00025.2008. [DOI] [PubMed] [Google Scholar]
- [78].Furne J, Saeed A, Michael DL. Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1479–R1485. doi: 10.1152/ajpregu.90566.2008. [DOI] [PubMed] [Google Scholar]
- [79].Vitvitsky V, Dayal S, Stabler S, Zhou Y, Wang H, et al. Perturbations in homocysteine-linked redox homeostasis in a murine model for hyperhomocysteinemia. Am J Physiol Regul Integr Comp Physiol. 2004;287:R39–R46. doi: 10.1152/ajpregu.00036.2004. [DOI] [PubMed] [Google Scholar]
- [80].Kimura Y, Kimura H. Hydrogen sulfide protects neurons from oxidative stress. FASEB J. 2004;18:1165–1167. doi: 10.1096/fj.04-1815fje. [DOI] [PubMed] [Google Scholar]
- [81].Kimura Y, Goto YI, Kimura H. Hydrogen sulfide increases glutathione production and suppresses oxidative stress in mitochondria. Antioxid Redox Signal. 2010;12:1–13. doi: 10.1089/ars.2008.2282. [DOI] [PubMed] [Google Scholar]
- [82].Kreitman GY, Danilewicz JC, Jeffery DW, Elias RJ. Reaction mechanisms of metals with hydrogen sulfide and thiols in model wine. Part 1: copper-catalyzed oxidation. J Agric Food Chem. 2016;64:4095–4104. doi: 10.1021/acs.jafc.6b00641. [DOI] [PubMed] [Google Scholar]
- [83].Hill BC, Woon TC, Nicholls P, Peterson J, Greenwood C, et al. Interactions of sulphide and other ligands with cytochrome c oxidase. An electron-paramagnetic-resonance study. Biochem J. 1984;224:591–600. doi: 10.1042/bj2240591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Petersen LC. The effect of inhibitors on the oxygen kinetics of cyto-chrome c oxidase. Biochim Biophys Acta. 1977;460:299–307. doi: 10.1016/0005-2728(77)90216-x. [DOI] [PubMed] [Google Scholar]
- [85].Nicholls P. Inhibition of cytochrome c oxidase by sulphide. Biochem Soc Trans. 1975;3:316–319. doi: 10.1042/bst0030316. [DOI] [PubMed] [Google Scholar]
- [86].Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- [87].Modis K, Coletta C, Erdelyi K, Papapetropoulos A, Szabo C. Intramitochondrial hydrogen sulfide production by 3-mercaptopyruvate sulfurtransferase maintains mitochondrial electron flow and supports cellular bioenergetics. FASEB J. 2012;27:601–611. doi: 10.1096/fj.12-216507. [DOI] [PubMed] [Google Scholar]
- [88].Szabo C, Ransy C, Módis K, Andriamihaja M, Murghes B, et al. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br J Pharmacol. 2014;171:2099–2122. doi: 10.1111/bph.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Román-Morales E, Pietri R, Ramos-Santana B, Vinogradov SN, Lewis-Ballester A, et al. Structural determinants for the formation of sulfhemeprotein complexes. Biochem Biophys Res Commun. 2010;400:489–492. doi: 10.1016/j.bbrc.2010.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Searcy DG, Whitehead JP, Maroney MJ. Interaction of Cu, Zn superoxide dismutase with hydrogen sulfide. Arch Biochem Biophys. 1995;318:251–263. doi: 10.1006/abbi.1995.1228. [DOI] [PubMed] [Google Scholar]
- [91].Goubern M, Andriamihaja M, Nubel T, Blachier F, Bouillaud F. Sulfide, the first inorganic substrate for human cells. FASEB J. 2007;21:1699–1706. doi: 10.1096/fj.06-7407com. [DOI] [PubMed] [Google Scholar]
- [92].Yong R, Searcy DG. Sulfide oxidation coupled to ATP synthesis in chicken liver mitochondria. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:129–137. doi: 10.1016/s1096-4959(01)00309-8. [DOI] [PubMed] [Google Scholar]
- [93].Bouillaud F, Blachier F. Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxid Redox Signal. 2011;15:379–391. doi: 10.1089/ars.2010.3678. [DOI] [PubMed] [Google Scholar]
- [94].Miller DL, Roth MB. Hydrogen sulfide increases thermotolerance and lifespan in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:20618–20622. doi: 10.1073/pnas.0710191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Fu M, Zhang W, Wu L, Yang G, Li H, et al. Hydrogen sulfide (H2S) metabolism in mitochondria and its regulatory role in energy production. Proc Natl Acad Sci USA. 2012;109:2943–2948. doi: 10.1073/pnas.1115634109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Szabo C, Coletta C, Chao C, Módis K, Szczesny B, et al. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci USA. 2013;110:12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hellmich MR, Coletta C, Chao C, Szabo C. The therapeutic potential of cystathionine beta-synthetase/hydrogen sulfide inhibition in cancer. Antioxid Redox Signal. 2015;22:424–448. doi: 10.1089/ars.2014.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li L, Rose P, Moore PK. Hydrogen sulfide and cell signaling. Annu Rev Pharmacol Toxicol. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]
- [99].Filipovic MR. Persulfidation (S-sulfhydration) and H2S. In: Moore P, Whiteman M, editors. Chemistry, Biochemistry and Pharmacology of Hydrogen Sulfide. Handb. Exp. Pharmacol. Vol. 230. Springer, Cham; 2015. pp. 29–59. [DOI] [PubMed] [Google Scholar]
- [100].Paul BD, Snyder SH. Gasotransmitter hydrogen sulfide signaling in neuronal health and disease. Biochem Pharmacol. 2017 doi: 10.1016/j.bcp.2017.11.019. S0006–2952(17)30700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Iciek M, Kowalczyk-Pachel D, Bilska-Wilkosz A, Kwiecień I, Górny M, et al. S-sulfhydration as a cellular redox regulation. Biosci Rep. 2016;36:e00304. doi: 10.1042/BSR20150147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wedmann R, Onderka C, Wei S, Szijártó IA, Miljkovic JL, et al. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci. 2016;7:3414–3426. doi: 10.1039/c5sc04818d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, et al. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72–ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Peng H, Zhang Y, Palmer LD, Kehl-Fie TE, Skaar EP, et al. Hydrogen sulfide and reactive sulfur species impact proteome S-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect Dis. 2017;3:744–755. doi: 10.1021/acsinfecdis.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kolluru GK, Shen X, Bir SC, Kevil CG. Hydrogen sulfide chemical biology: pathophysiological roles and detection. Nitric Oxide. 2013;35:5–20. doi: 10.1016/j.niox.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zobell CE, Feltham CB. A comparison of lead, bismuth, and iron as detectors of hydrogen sulphide produced by bacteria. J Bacteriol. 1934;28:169–176. doi: 10.1128/jb.28.2.169-176.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Basic A, Blomqvist S, Carlén A, Dahlén G. Estimation of bacterial hydrogen sulfide production in vitro. J Oral Microbiol. 2015;7:28166. doi: 10.3402/jom.v7.28166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ahn YJ, Lee YJ, Lee J, Lee D, Park HK, et al. Colorimetric detection of endogenous hydrogen sulfide production in living cells. Spectrochim Acta A Mol Biomol Spectrosc. 2017;177:118–124. doi: 10.1016/j.saa.2017.01.040. [DOI] [PubMed] [Google Scholar]
- [109].Fischer E. Bildung von Methylenblau als Reaktion auf Schwefelwasserstoff. Ber Dtsch Chem Ges. 1883;16:2234–2236. [Google Scholar]
- [110].Modis K, Asimakopoulou A, Coletta C, Papapetropoulos A, Szabo C. Oxidative stress suppresses the cellular bioenergetic effect of the 3-mercaptopyruvate sulfurtransferase/hydrogen sulfide pathway. Biochem Biophys Res Commun. 2013;433:401–407. doi: 10.1016/j.bbrc.2013.02.131. [DOI] [PubMed] [Google Scholar]
- [111].Tangerman A, Meuwese-Arends MT, Van Tongeren JHM. New methods for the release of volatile sulfur compounds from human serum: its determination by Tenax trapping and gas chromatography and its application in liver diseases. J Clin Lab Med. 1985;106:175–182. [PubMed] [Google Scholar]
- [112].Ubuka T. Assay methods and biological roles of labile sulfur in animal tissues. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;781:227–249. doi: 10.1016/s1570-0232(02)00623-2. [DOI] [PubMed] [Google Scholar]
- [113].Ishigami M, Hiraki K, Umemura K, Ogasawara Y, Ishii K, et al. A source of hydrogen sulfide and a mechanism of its release in the brain. Antioxid Redox Signal. 2009;11:205–214. doi: 10.1089/ars.2008.2132. [DOI] [PubMed] [Google Scholar]
- [114].Furne J, Saeed A, Levitt MD. Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. American Am J Physiol Regul Integr Comp Physiol. 2008;295:R1479–R1485. doi: 10.1152/ajpregu.90566.2008. [DOI] [PubMed] [Google Scholar]
- [115].Ogasawara Y, Ishii K, Togawa T, Tanabe S. Determination of trace amounts of sulphide in human red blood cells by high-performance liquid chromatography with fluorimetric detection after derivatization with p-phenylenediamine and iron(III) Analyst. 1991;116:1359–1363. doi: 10.1039/an9911601359. [DOI] [PubMed] [Google Scholar]
- [116].Newton GL, Fahey RC. Determination of biothiols by bromobimane labeling and high-performance liquid chromatography. Methods Enzymol. 1995;251:148–166. doi: 10.1016/0076-6879(95)51118-0. [DOI] [PubMed] [Google Scholar]
- [117].Togawa T, Ogawa M, Nawata M, Ogasawara Y, Kawanabe K, et al. High performance liquid chromatographic determination of bound sulfide and sulfite and thiosulfate at their low levels in human serum by pre-column fluorescence derivatization with monobromobimane. Chem Pharm Bull. 1992;40:3000–3004. doi: 10.1248/cpb.40.3000. [DOI] [PubMed] [Google Scholar]
- [118].Montoya LA, Shen X, McDermott JJ, Kevil CG, Pluth MD. Mechanistic investigations reveal that dibromobimane extrudes sulfur from biological sulfhydryl sources other than hydrogen sulfide. Chem Sci. 2015;6:294–300. doi: 10.1039/c4sc01875c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Mason J, Cardin CJ, Dennehy A. The role of sulphide and sulphide oxidation in the copper molybdenum antagonism in rats and guinea pigs. Res Vet Sci. 1978;24:104–108. [PubMed] [Google Scholar]
- [120].Khan SU, Morris GF, Hidiroglou M. Rapid estimation of sulfide in rumen and blood with a sulfide-specific ion electrode. Microchem J. 1980;25:388–395. [Google Scholar]
- [121].Jeroschewski P, Steuckart C, Kühl M. An Amperometric Microsensor for the Determination of H2S in Aquatic Environments. Anal Chem. 1996;68:4351–4357. [Google Scholar]
- [122].Kraus DW, Doeller JE. Sulfide consumption by mussel gill mitochondria is not strictly tied to oxygen reduction: measurements using a novel polarographic sulfide sensor. J Exp Biol. 2004;207:3667–3679. doi: 10.1242/jeb.01212. [DOI] [PubMed] [Google Scholar]
- [123].Yu F, Han X, Chen L. Fluorescent probes for hydrogen sulfide detection and bioimaging. Chem Commun. 2014;50:12234–12249. doi: 10.1039/c4cc03312d. [DOI] [PubMed] [Google Scholar]
- [124].Mubeen S, Zhang T, Chartuprayoon N, Rheem Y, Mulchandani A, et al. Sensitive detection of H2S using gold nanoparticle decorated single-walled carbon nanotubes. Anal Chem. 2010;82:250–257. doi: 10.1021/ac901871d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Dong Y, Wang L, Wang F, Li N, Jin Y, et al. An etching based fluorescent probe for sensitive detection of hydrogen sulfide in cells. Analyst. 2017;142:4703–4707. doi: 10.1039/c7an01394a. [DOI] [PubMed] [Google Scholar]
- [126].Zhang X, Zhou W, Yuan Z, Lu C. Colorimetric detection of biological hydrogen sulfide using fluorosurfactant functionalized gold nanorods. Analyst. 2015;140:7443–7450. doi: 10.1039/c5an01665g. [DOI] [PubMed] [Google Scholar]
- [127].Yu Q, Gao P, Zhang KY, Tong X, Yang H, et al. Luminescent gold nanocluster-based sensing platform for accurate H2S detection in vitro and in vivo with improved anti-interference. Light Sci Appl. 2017;6:e17107. doi: 10.1038/lsa.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Chen Z-J, Ai H-W. A highly responsive and selective fluorescent probe for imaging physiological hydrogen sulfide. Biochemistry. 2014;53:5966–5974. doi: 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- [129].Pan J, Carroll KS. Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem Biol. 2013;8:1110–1116. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Krishnan N, Fu C, Pappin DJ, Tonks NK. H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, et al. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Gao X-H, Krokowski D, Guan B-J, Bederman I, Majumder M, et al. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. ELife. 2015;4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Doka E, Pader I, Biro A, Johansson K, Cheng Q, et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci Adv. 2016;2:e1500968. doi: 10.1126/sciadv.1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park CM, et al. Detection of protein S-sulfhydration by a tag-switch technique. Angew Chem Int Ed Engl. 2014;53:575–581. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- [137].Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Liochev SI, Fridovich I. Superoxide and iron: partners in crime. IUBMB Life. 1999;48:157–161. doi: 10.1080/713803492. [DOI] [PubMed] [Google Scholar]
- [139].Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Damage to the bases in DNA induced by hydrogen peroxide and ferric ion chelates. J Biol Chem. 1989;264:20509–20512. [PubMed] [Google Scholar]
- [140].Mironov A, Seregina T, Nagornykh M, Luhachack GL, Korolkova N, et al. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc Natl Acad Sci USA. 2017;114:6022–6027. doi: 10.1073/pnas.1703576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Shukla P, Khodade VS, SharathChandra M, Chauhan P, Mishra S, et al. “On demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem Sci. 2017;8:4967–4972. doi: 10.1039/c7sc00873b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- [143].Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10:e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Al-Attar S, Yu Y, Pinkse M, Hoeser J, Friedrich T, et al. Cyto-chrome bd displays significant quinol peroxidase activity. Sci Rep. 2016;6:27631. doi: 10.1038/srep27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Burkardt HJ, Mattes R, Schmid K, Schmitt R. Properties of two conjugative plasmids mediating tetracycline resistance, raffinose catabolism and hydrogen sulfide production in Escherichia coli. Mol Gen Genet. 1978;166:75–84. doi: 10.1007/BF00379731. [DOI] [PubMed] [Google Scholar]
- [147].Jones RT, Thai LP, Silver RP. Genetic and molecular characterization of an Escherichia coli plasmid coding for hydrogen sulfide production and drug resistance. Antimicrob Agents Chemother. 1978;14:765–770. doi: 10.1128/aac.14.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Nambi S, Long JE, Mishra BB, Baker R, Murphy KC, et al. The oxidative stress network of Mycobacterium tuberculosis reveals coordination between radical detoxification systems. Cell Host Microbe. 2015;17:829–837. doi: 10.1016/j.chom.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Ivanov AV, Bartosch B, Isaguliants MG. Oxidative stress in infection and consequent disease. Oxid Med Cell Longev. 2017;2017:3496043. doi: 10.1155/2017/3496043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Beck MA, Handy J, Levander OA. The role of oxidative stress in viral infections. Ann N Y Acad Sci. 2000;917:906–912. doi: 10.1111/j.1749-6632.2000.tb05456.x. [DOI] [PubMed] [Google Scholar]
- [151].Peterhans E, Grob M, Burge T, Zanoni R. Virus-induced formation of reactive oxygen intermediates in phagocytic cells. Free Radic Res Commun. 2009;3:39–46. doi: 10.3109/10715768709069768. [DOI] [PubMed] [Google Scholar]
- [152].Hennet T, Peterhans E, Stocker R. Alterations in antioxidant defences in lung and liver of mice infected with influenza A virus. J Gen Virol. 1992;73:39–46. doi: 10.1099/0022-1317-73-1-39. [DOI] [PubMed] [Google Scholar]
- [153].Hagen TM, Huang S, Curnutte J, Fowler P, Martinez V, et al. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci USA. 1994;91:12808–12812. doi: 10.1073/pnas.91.26.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Schwarz BK. Oxidative stress during viral infection: a review. Free Radic Biol Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- [155].Perl A, Banki K. Genetic and metabolic control of the mitochondrial transmembrane potential and reactive oxygen intermediate production in HIV disease. Antioxid Redox Signal. 2000;2:551–573. doi: 10.1089/15230860050192323. [DOI] [PubMed] [Google Scholar]
- [156].Muller F. Reactive oxygen intermediates and human immunodeficiency virus (HIV) infection. Free Radic Biol Med. 1992;13:651–657. doi: 10.1016/0891-5849(92)90039-j. [DOI] [PubMed] [Google Scholar]
- [157].Israel N, Gougerot-Pocidalo MA. Oxidative stress in human immunodeficiency virus infection. Cell Mol Life Sci. 1997;53:864–870. doi: 10.1007/s000180050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Flohe L, Brigelius-Flohe R, Saliou C, Traber MG, Packer L. Redox regulation of NF-κB activation. Free Radic Biol Med. 1997;22:1115–1126. doi: 10.1016/s0891-5849(96)00501-1. [DOI] [PubMed] [Google Scholar]
- [159].Trachootham D, Lu W, Ogasawara MA, Nilsa RD, Huang P. Redox regulation of cell survival. Antioxid Redox Signal. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Bhaskar A, Munshi M, Khan SZ, Fatima S, Arya R, et al. Measuring glutathione redox potential of HIV-1-infected macrophages. J Biol Chem. 2015;290:1020–1038. doi: 10.1074/jbc.M114.588913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Staitieh BS, Ding L, Neveu WA, Spearman P, Guidot DM, et al. HIV-1 decreases Nrf2/ARE activity and phagocytic function in alveolar macrophages. J Leukoc Biol. 2017;102:517–525. doi: 10.1189/jlb.4A0616-282RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Ivanov AV, Valuev-Elliston VT, Ivanova ON, Kochetkov SN, Starodubova ES, et al. Oxidative stress during HIV infection: mechanisms and consequences. Oxid Med Cell Longev. 2016;2016:8910396. doi: 10.1155/2016/8910396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Hourihan JM, Kenna JG, Hayes JD. The gasotransmitter hydrogen sulfide induces nrf2-target genes by inactivating the keap1 ubiquitin ligase substrate adaptor through formation of a disulfide bond between cys-226 and cys-613. Antioxid Redox Signal. 2013;19:465–481. doi: 10.1089/ars.2012.4944. [DOI] [PubMed] [Google Scholar]
- [164].Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Yang G, Zhao K, Ju Y, Mani S, Cao Q, et al. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid Redox Signal. 2013;18:1906–1919. doi: 10.1089/ars.2012.4645. [DOI] [PubMed] [Google Scholar]
- [166].Furuya AK, Sharifi HJ, Jellinger RM, Cristofano P, Shi B, et al. Sulforaphane Inhibits HIV Infection of Macrophages through Nrf2. PLoS Pathog. 2016;12:e1005581. doi: 10.1371/journal.ppat.1005581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Page A, Volchkova VA, Reid SP, Mateo M, Bagnaud-Baule A, et al. Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep. 2014;6:1026–1036. doi: 10.1016/j.celrep.2014.02.027. [DOI] [PubMed] [Google Scholar]
- [168].Gjyshi O, Bottero V, Veettil MV, Dutta S, Singh VV, et al. Kaposi’s sarcoma-associated herpesvirus induces Nrf2 during de novo infection of endothelial cells to create a microenvironment conducive to infection. PLoS Pathog. 2014;10:e1004460. doi: 10.1371/journal.ppat.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Olagnier D, Peri S, Steel C, van Montfoort N, Chiang C, et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog. 2014;10:e1004566. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Garg S, Vitvitsky V, Gendelman HE, Banerjee R. Monocyte differentiation, activation, and mycobacterial killing are linked to transsulfuration-dependent redox metabolism. J Biol Chem. 2006;281:38712–38720. doi: 10.1074/jbc.M606235200. [DOI] [PubMed] [Google Scholar]
- [171].Benedetti F, Curreli S, Krishnan S, Davinelli S, Cocchi F, et al. Anti-inflammatory effects of H(2)S during acute bacterial infection: a review. J Transl Med. 2017;15:1–11. doi: 10.1186/s12967-017-1206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Gemici B, Wallace JL. Anti-inflammatory and cytoprotective properties of hydrogen sulfide. Methods Enzymol. 2015;555:169–193. doi: 10.1016/bs.mie.2014.11.034. [DOI] [PubMed] [Google Scholar]
- [173].Wallace JL, Motta JP, Buret AG. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am J Physiol Gastrointest Liver Physiol. 2018;314:G143–G149. doi: 10.1152/ajpgi.00249.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Linden DR. Hydrogen sulfide signaling in the gastrointestinal tract. Antioxid Redox Signal. 2014;20:818–830. doi: 10.1089/ars.2013.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Flannigan KL, McCoy KD, Wallace JL. Eukaryotic and prokaryotic contributions to colonic hydrogen sulfide synthesis. Am J Physiol Gastrointest Liver Physiol. 2011;301:G188–G193. doi: 10.1152/ajpgi.00105.2011. [DOI] [PubMed] [Google Scholar]
- [176].Shen X, Carlstrom M, Borniquel S, Jadert C, Kevil CG, et al. Microbial regulation of host hydrogen sulfide bioavailability and metabolism. Free Radic Biol Med. 2013;60:195–200. doi: 10.1016/j.freeradbiomed.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lagoutte E, Mimoun S, Andriamihaja M, Chaumontet C, Blachier F, et al. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim Biophys Acta. 2010;1797:1500–1511. doi: 10.1016/j.bbabio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- [178].Levine J, Ellis CJ, Furne JK, Springfield J, Levitt MD. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol. 1998;93:83–87. doi: 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- [179].Blachier F, Davila AM, Mimoun S, Benetti PH, Atanasiu C, et al. Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids. 2010;39:335–347. doi: 10.1007/s00726-009-0445-2. [DOI] [PubMed] [Google Scholar]
- [180].Furne JK, Suarez FL, Ewing SL, Springfield J, Levitt MD. Binding of hydrogen sulfide by bismuth does not prevent dextran sulfate-induced colitis in rats. Dig Dis Sci. 2000;45:1439–1443. doi: 10.1023/a:1005580709390. [DOI] [PubMed] [Google Scholar]
- [181].Wallace JL, Vong L, McKnight W, Dicay M, Martin GR. Endogenous and exogenous hydrogen sulfide promotes resolution of colitis in rats. Gastroenterology. 2009;137:569–578. doi: 10.1053/j.gastro.2009.04.012. [DOI] [PubMed] [Google Scholar]
- [182].Albert TK, Laubinger W, Müller S, Hanisch F-G, Kalinski T, et al. Human intestinal TFF3 forms disulfide-linked heteromers with the mucus-associated FCGBP protein and is released by hydrogen sulfide. J Proteome Res. 2010;9:3108–3117. doi: 10.1021/pr100020c. [DOI] [PubMed] [Google Scholar]
- [183].Li H, Ma Y, Escaffre O, Ivanciuc T, Komaravelli N, et al. Role of hydrogen sulfide in paramyxovirus infections. J Virol. 2015;89:5557–5568. doi: 10.1128/JVI.00264-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Schmidt ME, Varga SM. Modulation of the host immune response by respiratory syncytial virus proteins. J Microbiol. 2017;55:161–171. doi: 10.1007/s12275-017-7045-8. [DOI] [PubMed] [Google Scholar]
- [185].Garofalo RP, Haeberle H. Epithelial regulation of innate immunity to respiratory syncytial virus. Am J Respir Cell Mol Biol. 2000;23:581–585. doi: 10.1165/ajrcmb.23.5.f204. [DOI] [PubMed] [Google Scholar]
- [186].Ivanciuc T, Sbrana E, Ansar M, Bazhanov N, Szabo C, et al. Hydrogen sulfide is an antiviral and antiinflammatory endogenous gasotransmitter in the airways. Role in respiratory syncytial virus infection. Am J Respir Cell Mol Biol. 2016;55:684–696. doi: 10.1165/rcmb.2015-0385OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Bazhanov N, Ansar M, Ivanciuc T, Garofalo RP, Casola A. Hydrogen sulfide: a novel player in airway development, pathophysiology of respiratory disease and antiviral defenses. Am J Respir Cell Mol Biol. 2017;57:403–410. doi: 10.1165/rcmb.2017-0114TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Bazhanov N, Escaffre O, Freiberg AN, Garofalo RP, Casola A. Broad-range antiviral activity of hydrogen sulfide against highly pathogenic RNA viruses. Sci Rep. 2017;7 doi: 10.1038/srep41029. 41029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Yen HL. Current and novel antiviral strategies for influenza infection. Curr Opin Virol. 2016;18:126–134. doi: 10.1016/j.coviro.2016.05.004. [DOI] [PubMed] [Google Scholar]
- [190].Yajima T, Knowlton KU. Viral myocarditis: from the perspective of the virus. Circulation. 2009;119:2615–2624. doi: 10.1161/CIRCULATIONAHA.108.766022. [DOI] [PubMed] [Google Scholar]
- [191].Wu Z, Peng H, Du Q, Lin W, Liu Y. GYY4137, a hydrogen sulfidereleasing molecule, inhibits the inflammatory response by suppressing the activation of nuclear factorkappa B and mitogenactivated protein kinases in Coxsackie virus B3infected rat cardiomyocytes. Mol Med Rep. 2015;11:1837–1844. doi: 10.3892/mmr.2014.2901. [DOI] [PubMed] [Google Scholar]
- [192].Yuen S, Smith J, Caruso L, Balan M, Opavsky MA. The coxsackie-adenovirus receptor induces an inflammatory cardiomyopathy independent of viral infection. J Mol Cell Cardiol. 2011;50:826–840. doi: 10.1016/j.yjmcc.2011.02.011. [DOI] [PubMed] [Google Scholar]