Abstract
In holometabolous insects, developmentally controlled programmed cell death (PCD) is a conserved process that destroys a subset of larval tissues for the eventual creation of new adult structures. This process of histolysis is relatively well studied in salivary gland and midgut tissues, while knowledge concerning larval muscle destruction is limited. Here, we have examined the histolysis of a group of Drosophila larval abdominal muscles called the dorsal external oblique muscles (DEOMs). Previous studies have defined apoptosis as the primary mediator of DEOM breakdown, whose timing is controlled by ecdysone signaling. However, very little is known about other factors that contribute to DEOM destruction. In this paper, we examine the role of thin (tn), which encodes for the Drosophila homolog of mammalian TRIM32, in the regulation of DEOM histolysis. We find that loss of Tn blocks DEOM degradation independent of ecdysone signaling. Instead, tn genetically functions in a pathway with the death-associated inhibitor of apoptosis (DIAP1), Dronc, and death-associated APAF1-related killer (Dark) to regulate apoptosis. Importantly, blocking Tn results in the absence of active Caspase-3 immunostaining, upregulation of DIAP1 protein levels, and inhibition of Dronc activation. DIAP1 and Dronc mRNA levels are not altered in tn mutants, showing that Tn acts post-transcriptionally on DIAP1 to regulate apoptosis. Herein, we also find that the RING domain of Tn is required for DEOM histolysis as loss of this domain results in higher DIAP1 levels. Together, our results suggest that the direct control of DIAP1 levels, likely through the E3 ubiquitin ligase activity of Tn, provides a mechanism to regulate caspase activity and to facilitate muscle cell death.
Introduction
Programmed cell death (PCD) governs the development and homeostasis of multicellular organisms by controlling the patterning of adult body structures and the removal of obsolete or damaged tissues1–4. Mechanisms by which cells die can be divided into three types based upon morphological criteria5. Type I PCD, or apoptosis, is characterized by the upregulation of caspases accompanied by DNA fragmentation, membrane blebbing, and cell rounding6. Autophagy is referred to as Type II PCD and is distinguished by the presence of double-membraned autophagosomes7. Necrosis, the third type of PCD, is an inflammatory response characterized by cell swelling and rupture of the cell membrane8.
PCD is particularly evident during the metamorphosis of holometabolous insects, including Drosophila3, 9. The first signs of apoptosis are observed in embryogenesis and persist into the pupal stages where many larval tissues are remodeled in response to pulses of the steroid hormone 20-hydroxy-ecdysone (ecdysone)9–11. A pulse of ecdysone in late third instar larvae (L3) promotes the larval to pupal transition, whereas a second pulse triggers PCD around 12 h after puparium formation (APF)12. After old tissues are histolysed, newly formed tissues grow in the remaining 3.5 days before adult eclosion.
Most knowledge about tissue histolysis stems from the analysis of salivary gland or midgut tissue in pupal metamorphosis9. Salivary gland histolysis is regulated by the late ecdysone pulse13–15, whereby ecdysone binding to its receptors promotes the expression of early genes, including the transcription factors Broad complex (Br-C), E74 A, and E9316–19. Subsequent activation of cell death genes, including hid, reaper, Dronc, Drice, and the autophagy-related gene 1 (Atg1), result in the elimination of this tissue9, 14, 15, 17, 20, 21. While salivary gland histolysis is mediated by both apoptosis and autophagy22, midgut histolysis is triggered by the early ecdysone pulse and is primarily regulated by autophagy23. Less is understood about muscle remodeling during pupation, including the signaling pathways that control whether muscle cells are fated to live or die.
Drosophila makes two sets of muscles during its life cycle, one in embryogenesis for larval movement and the other during pupation for adult life. During metamorphosis, most of the larval muscles are histolyzed and this pupal remodeling assures muscles are functional for adult-specific functions like flight and mating24, 25. Two sets of muscles that undergo remodeling during the pupal transition are the dorsal internal oblique muscles (DIOMs) and the dorsal external oblique muscles (DEOMs)26, 27. Both of these muscle groups are present in abdominal segments A1 to A5. The muscles closest to the midline are designated as DIOM1 or DEOM1, whereas more lateral muscles are classified as DIOM2 or DEOM2 (Fig. 1a, b). DIOMs fail to undergo histolysis and persist until adult stages, whereas the DEOMs are removed by PCD26, 28. DEOM1 histolysis is initiated by 8 h APF and the muscles are lost by 12 h APF. DEOM2 histolysis is delayed and is typically completed by 24 h APF28–30.
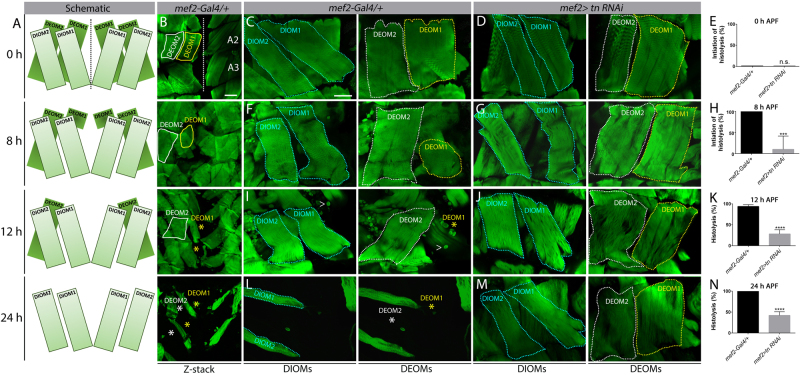
Fig. 1. Tn is required for DEOM histolysis.
a Schematic diagrams of the DEOMs during pupal development at 0, 8, 12, and 24 h APF. Dotted line denotes the midline. b Merged Z-stack images of DEOM histolysis that correspond to the same time points (a) in mef2-Gal4/+ control muscles stained to visualize F-actin (green). DEOM1 (yellow solid line) and DEOM2 (white solid line) are both present at 0 h APF. In WT muscles, DEOM1 starts to disintegrate at 8 h APF and is gone by 12 h APF (yellow asterisk). DEOM2 disappears by 24 h APF (white asterisk). A2 and A3 denotes abdominal segments 2 and 3, respectively. c–n Representative images and quantification of DEOM muscle histolysis at 0, 8, 12, and 24 h APF in mef2-Gal4/+ control or mef2>tn RNAi muscles stained with phalloidin (green). Substacks of single confocal planes separate out the DIOMs (cyan dotted lines) from the DEOMs. c, f, i, l DEOM1 (yellow dotted lines) and DEOM2 (white dotted lines) muscles degenerate (asterisks) by 24 h in control muscles. d, g, j, m However, reduction of tn by RNAi mostly blocks DEOM histolysis. e, h, k, n DEOM1 histolysis is not initiated at 0 h in mef2-Gal4/+ control or mef2>tn RNAi muscles (e). By 8 h, all control DEOM1s have started to breakdown, while most of these muscles are still present in tn RNAi pupae (h). By 12 h (k) or 24 h (n), most DEOM1s are still intact in tn RNAi muscles. White carets indicate remnants of fat body tissue that stain positive for F-actin. Mean ± SEM (n.s., not significant, ****p < 0.001, ***p < 0.005). Scale bars, 100 µm (b); 50 µm (c, d, f, g, i, j, l, m)
Destruction of the DEOMs is mediated solely by apoptosis28. Transmission electron microscopy studies show myonuclei containing condensed chromatin in dying DEOMs and ectopic expression of the anti-apoptotic protein p35 in DEOMs is sufficient to prevent muscle breakdown. In contrast, muscle-specific reduction of the autophagy gene Atg1 has no effect28. Like salivary gland and midgut histolysis, ecdysone signaling mediates DEOM breakdown, which is evidenced by a reduction in caspase staining and the absence of muscle histolysis upon loss of the ecdysone receptor28.
The core machinery required for apoptosis is conserved among flies, worms, and mammals2, 31. The caspase family of proteins are the principal mediators of cell death and are present in most cells in an inactive form32, 33. Under normal conditions, caspase activity is blocked by the inhibitor of apoptosis (IAP) family of proteins to prevent cell death34, 35. In Drosophila, Dronc (Caspase-9) acts as the principal initiator caspase and is inhibited by DIAP136–38. Upon receiving a cell death stimulus, the IAP antagonists Reaper, Hid, and Grim (RGH) promote the degradation of DIAP1, resulting in Dronc release39–43. A Dronc–Dark complex form the apoptosome to activate the executioner caspases Drice and Dcp-1, which cleave cellular substrates to promote cell death44, 45.
Herein, we have discovered that Tn acts in a pathway with DIAP1 and Dronc to regulate abdominal muscle breakdown. tn, also called another B-box affiliate (abba), is homologous to mammalian TRIM32 and is characterized by an N-terminal RING domain followed by six NHL repeats at the C terminus46, 47. The RING domain provides E3 ubiquitin ligase activity, whereas the NHL repeats are predicted to facilitate protein–protein interactions. Pleiotropic roles exist for the ubiquitously expressed TRIM32 protein in regulating muscle physiology, muscle regeneration, and tumor suppression46, 47. Our results here demonstrate a role for Tn in controlling the fate of muscle cells, acting as a switch to control whether muscle cells live or die.
Results
Tn is required for muscle histolysis and is not an ecdysone target
We previously showed that Tn is required for myofibril stability and costamere integrity in Drosophila larval muscles48. However, the late pupal lethality of tn mutants suggested that Tn may be required during pupal metamorphosis. Indeed, targeting tn RNAi in the musculature using the Gal4/UAS system produced defects in abdominal muscle histolysis.
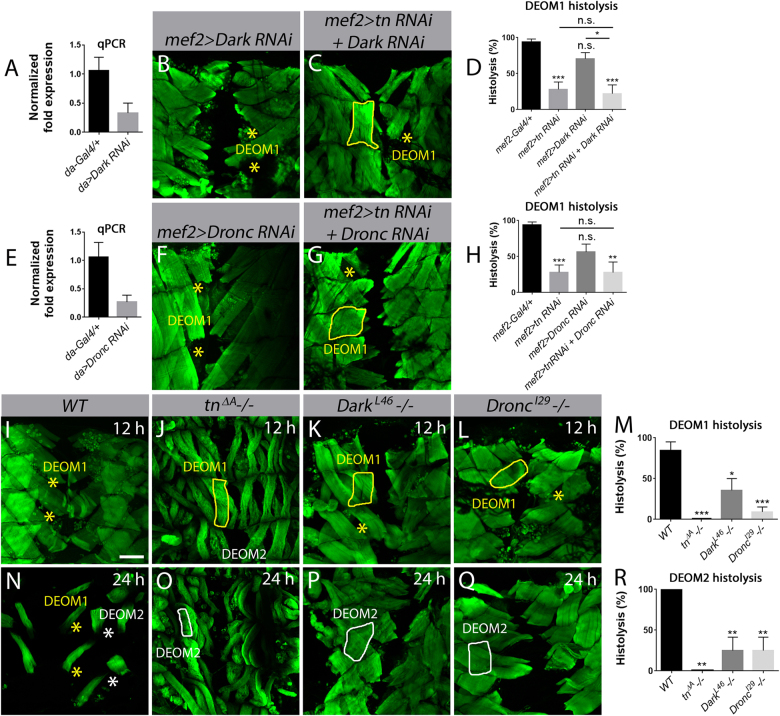
At 0 h APF, all DEOMs were present in both control (mef2-Gal4/+) and experimental (mef2>tn RNAi) genotypes (Fig. 1c–e). Around 8 h APF, smaller, rounded muscles indicated the onset of DEOM1 histolysis in controls (Fig. 1f, h), while DEOM1 was still fully intact upon a decrease in Tn (Fig. 1g, h). By 12 h APF, DEOM1 was absent in nearly all mef2-Gal4/+ individuals (Fig. 1i, k), while loss of DEOM1 was observed only in approximately 28% of tn RNAi abdominal segments (Fig. 1j, k). Complete histolysis of DEOM1 (and DEOM2) were apparent in controls at 24 h APF (Fig. 1l, n). In contrast, a partial block in degradation was observed in DEOMs with disrupted Tn function at 24 h APF (Fig. 1m, n). This block in DEOM histolysis was verified in tnΔA−/− mutants (tn−/−) and an additional tn RNAi line (Fig. S1). Thus, we conclude that the abrogation of Tn results in impaired muscle histolysis.
As ecdysone signaling directs tissue histolysis during metamorphosis9, we sought to examine if Tn is an ecdysone target. tn mRNA levels were measured in pupae with blocked ecdysone signaling prior to (0 h APF), during (12 h APF), or after (24 h APF) completion of histolysis using quantitative PCR (qPCR). There was no significant difference in tn transcript levels between mef2-Gal4/+ or mef2>DN-EcRB1 samples at any stage of development (Fig. 2a). The reduction in tn transcript levels in mef2>tn RNAi muscles is consistent with previous results49, further demonstrating the specificity of inducing tn RNAi in pupal muscles and the sensitivity of our qPCR approach. Our data demonstrate that ecdysone does not globally regulate tn expression during metamorphosis.
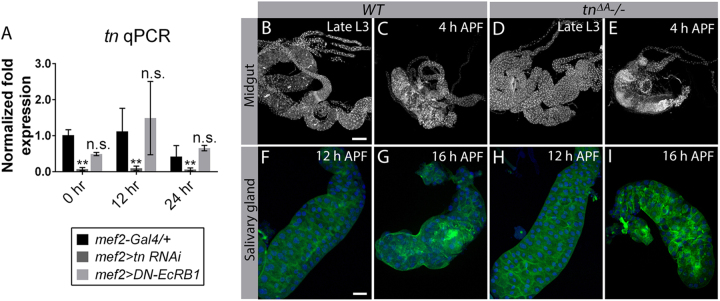
Fig. 2. Tn is not an ecdysone target and is not required for general tissue histolysis.
a tn mRNA levels measured by qPCR are not significantly different from controls (mef2-Gal4/+) when a dominant-negative version of isoform EcRB1 is expressed in muscle tissue during pupal morphogenesis. Note that tn transcript levels are decreased as expected upon muscle-specific tn RNAi knockdown. N = 3 biological replicates and 3 technical replicates for each genotype. b–i Z-stack confocal images of midguts (b–e) or salivary glands (f–i). Midgut histolysis proceeds normally by 4 h APF in WT (b, c) or tn mutants (d, e) as assayed by DAPI staining. PCD still occurs in salivary glands (F-actin, green and DAPI, blue) by 16 h APF upon loss of Tn (h, i) compared to WT controls (f, g). Mean ± SEM (n.s., not significant, **p < 0.01). Scale bars, 200 µm (b–e) and 100 µm (f–i)
To test if Tn plays a broader role in general tissue histolysis, we examined whether Tn regulates midgut or salivary gland breakdown. The midgut undergoes a drastic reduction in size between the late L3 and early pupal stages, whereas salivary gland histolysis takes place between 12 h APF and 16 h APF50. WT and tn−/− midguts appeared similar at 4 h APF (Fig. 2b–e) and there was no obvious delay or impairment in salivary gland histolysis in tn mutants compared to WT at 16 h APF (Fig. 2f–i). Therefore, unlike abdominal muscle, Tn does not play a broader role in general tissue breakdown during metamorphosis.
Loss of Tn affects DIAP1 protein levels and Caspase-3 activity
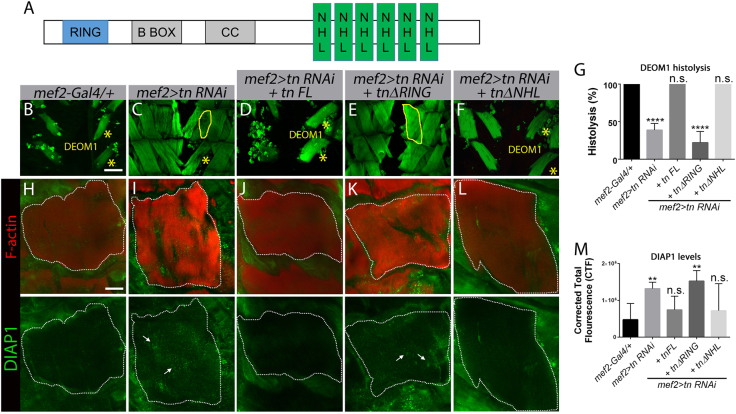
Zirin et al.28 previously demonstrated that disintegration of the DEOMs relies on apoptosis. To understand if Tn facilitates DEOM cell death, we assayed protein levels of the initiator caspase Dronc since antibodies against cleaved-Caspase-3 are a read-out of Dronc activity51. Elevated Caspase-3 was present in mef2-Gal4/+ muscles undergoing histolysis at 12 h APF (Fig. 3a, c). In contrast, there was an overall reduction in puncta corresponding to active Caspase-3 in muscles with disrupted Tn function (Fig. 3b, c). Next we examined DIAP1 protein levels. At 12 h APF, DIAP1 was present in DEOM controls at a basal level (Fig. 3d, f), whereas significantly higher DIAP1 levels were observed in tn RNAi DEOMs (Fig. 3e, f). This observed reduction in Dronc activity and the elevation of DIAP1 in tn mutants was not due to altered mRNA expression of DIAP1, Dronc, Drice, or Dark (Fig. S2). These data suggest that Tn post-transcriptionally regulates some of the cell death genes to direct muscle histolysis.
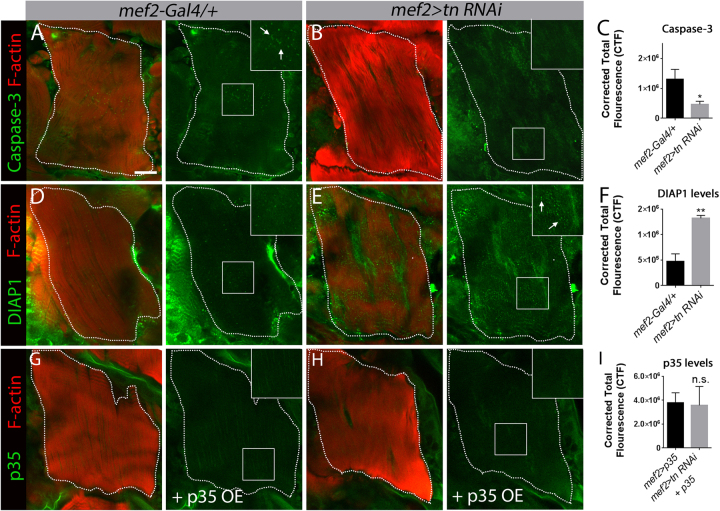
Fig. 3. Tn affects DIAP1 and Caspase-3 activity during DEOM histolysis at 12 h APF.
a–c In vivo labeling of Caspase-3 (green) in mef2-Gal4/+ control or mef2>tn RNAi DEOMs co-labeled with phalloidin (red). a Puncta corresponding to Caspase-3 are present in degenerating DEOM controls (white arrows in inset). b No Caspase-3(+) puncta are present in tn RNAi muscles. c Quantification of the relative fluorescence intensities of Caspase-3 in DEOMs reflects the general decrease observed upon a reduction in Tn. d–f Immunostaining for DIAP1 (green) and F-actin (red) is higher upon disruption of Tn function (e, white arrows in inset) than in mef2-Gal4/+ control muscles (d). f Bar graph showing that the relative DIAP1 fluorescence levels are increased upon reduced Tn function. g–i Overall p35 (green) levels in DEOMs labeled with F-actin (red) are not changed upon a reduction in Tn. mef2>p35 appears similar when co-expressed in a WT (g) or tn RNAi (h) background. Quantification of fluorescence intensity reveals no significant difference between WT or experimental DEOMs (i). Mean ± SEM (n.s., not significant, **p < 0.01, *p < 0.05). Scale bar, 50 µm
Since overexpression (OE) of the cell death inhibitor p35 blocks DEOM breakdown28, we next examined p35 to assess if Tn generally influences protein levels during histolysis or specifically functions in DIAP1-mediated apoptosis. The baculovirus p35 protein is not endogenously expressed in Drosophila, but inhibits effector caspases52. Thus, we used the Gal4/UAS system to target p35 in muscle alone (mef2>p35) or in a tn RNAi background (mef2>tn RNAi+ p35 OE). There was no significant difference in p35 levels in either genotype at 12 h APF (Fig. 3g–i). Moreover, there was no increase in the fluorescence intensity of DIAP1 upon p35 OE in the DEOMs (Fig. S3), proving that the observed accumulation of DIAP1 protein in tn−/− is not a secondary consequence of a block in muscle histolysis. Collectively, these findings show that Tn regulates DEOM cell death by influencing DIAP1 protein levels and Dronc activity.
tn functions in a pathway with cell death genes
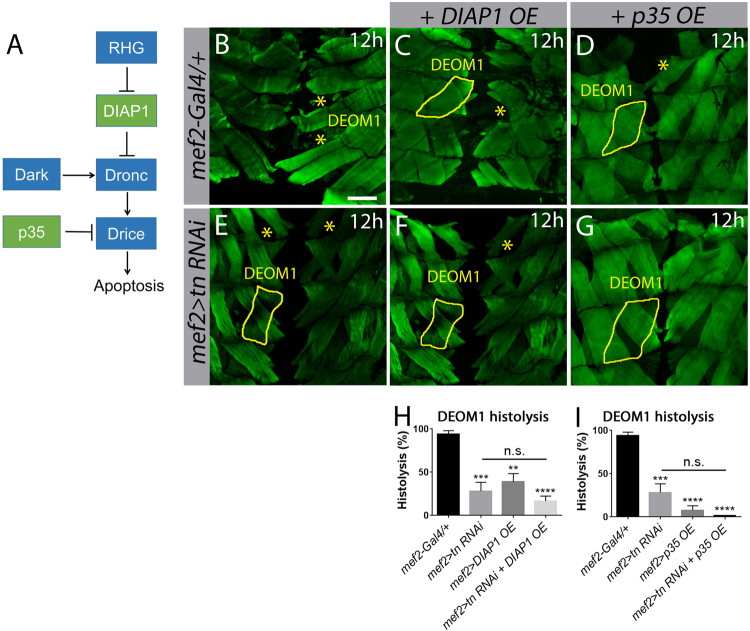
In the presence of cell death signals, the pro-apoptotic RGH proteins block DIAP1 inhibition, thereby enabling Dronc activation2, 35. Stable Dronc protein is activated by Dark and this Dronc–Dark apoptosome complex promotes the activity of effector caspases such as Drice for the execution of cell death (Fig. 4a)44, 45. Since our results show that tn RNAi blocks DEOM histolysis by altering DIAP1 and Caspase-3, we next sought to genetically manipulate cell death pathway components in a tn RNAi background to determine if Tn directly functions in apoptosis. Cell death was first blocked by expressing the cell death inhibitors DIAP1 (mef2>DIAP1 OE) or p35 (mef2>p35 OE). By 12 h APF, all DEOM1s underwent histolysis in mef2-Gal4/+ controls (Figs. 1i, k and 4b). Muscle-targeted DIAP1 OE inhibited degradation in approximately 50% of DEOM1s (Fig. 4c, h). In a tn RNAi background, DIAP1 OE did not enhance the extent of DEOM1 histolysis (Fig. 4f, h), suggesting that tn and DIAP1 may be acting in concert to regulate muscle degradation. In contrast, exogenous expression of p35 was more effective in the prevention of DEOM1 degeneration alone (Fig. 4d, i) and in combination with tn RNAi (Fig. 4g, i), presumably due to the ability of p35 to block three different effector caspases (Drice, Dcp-1, and Decay)3. The addition of exogenous UAS elements (UAS-GFP OE or UAS-GFP RNAi) in the same tn RNAi background did not alter DEOM1 histolysis, indicating that sufficient Gal4 protein is present to drive all UAS-based constructs in the presence of UAS-tn RNAi (Fig. S1).
Fig. 4. tn genetically interacts with DIAP1 during DEOM histolysis.
a Schematic of Drosophila core cell death machinery. b–g Merged Z-stack confocal images of the abdominal muscles stained with phalloidin (green) at 12 h APF. DEOM1 is outlined with a solid yellow line and histolysed DEOM1s are marked by yellow asterisks. b Histolysis proceeds normally in mef2-Gal4/+ muscles. Overexpression of DIAPI (c) or p35 (d) in muscles partially blocks DEOM1 histolysis. e–g There is no significant difference in DEOM1 histolysis upon RNAi knockdown of tn alone (e), or with overexpression of DIAP1 (f) or p35 (g) in a tn RNAi background. h A bar graph showing similar levels of muscle histolysis in mef2>tn RNAi+DIAP1 OE compared to mef2>tn RNAi muscles. i Quantification showing no significant difference in DEOM1 breakdown between mef2>tn RNAi and mef2>tn RNAi+p35 OE pupae. Mean ± SEM (n.s., not significant, ****p < 0.001, ***p < 0.005, **p < 0.01). Scale bar, 100 µm (b–g)
Next, we utilized muscle-specific RNAi to silence Dronc (mef2>Dronc RNAi) or Dark (mef2>Dark RNAi) to mimic a reduction in apoptotic signaling at 12 APF. In contrast to blocking apoptosis through OE of DIAP1 or p35, RNAi knockdown of Dark (Fig. 5b, d) or Dronc (Fig. 5f, h) alone was less effective at blocking DEOM1 histolysis. qPCR analysis revealed that Dark (Fig. 5a) and Dronc (Fig. 5e) transcript levels were decreased by ~60% under control of the ubiquitous daughterless (da)-Gal4 promoter, likely accounting for the weaker block in histolysis than DIAP1 OE. The extent of muscle histolysis was not enhanced in a tn RNAi background upon a further reduction in Dark (Fig. 5c, d) or Dronc (Fig. 5g, h), further supporting the model that Tn acts with cell death genes to regulate DEOM histolysis.
Fig. 5. Dronc and Dark are required for DEOM histolysis.
Merged Z-stack confocal images of the abdominal muscles stained with phalloidin (green) at 12 h APF (b, c, f, g, i–l) or 24 h APF (n–q). DEOM1s are outlined with a solid yellow line, DEOM2s with a solid white line. Histolyzed DEOM1 and DEOM2 muscles are marked by yellow and white asterisks, respectively. a Dark transcript levels are reduced by over 50% using the ubiquitous da-Gal4 driver. N = 3 biological replicates and 3 technical replicates for each genotype. b, c DEOM1 histolysis is partially blocked upon RNAi knockdown of Dark alone (b) or if Dark is reduced in a tn RNAi background at 12 h APF. d Quantification of DEOM1 histolysis reveals a slight enhancement in muscle degeneration in mef2>tn RNAi+Dark RNAi compared to mef2>Dark RNAi alone. e Quantification of Dronc transcript levels show over a 60% decrease upon expression of UAS-Dark RNAi under control of da-Gal4. N = 3 biological replicates and 3 technical replicates for each genotype. f–h There is no significant difference in the histolysis of DEOM muscles in mef2>tn RNAi+Dronc RNAi pupae compared to Dronc RNAi. i–l WT DEOM1s have histolyzed by 12 h APF (i). There is a strong block in muscle breakdown in tnΔA (j), DarkL46 (k), and DroncI29 (l) homozygous mutants at this same time point. m A bar graph showing the quantification of DEOM1 histolysis in (i–l). n–q All DEOM muscles are absent in WT pupae at 24 h APF (n). tnΔA (o), DarkL46−/− (p), and DroncI29−/− (q) show significantly reduced DEOM2 breakdown at 24 h APF. r Quantification of DEOM2 histolysis in tnΔA, DarkL46, and DroncI29 mutants. Mean ± SEM (n.s., not significant, ***p < 0.005, **p < 0.01, *p < 0.05). Scale bar, 100 µm (b, c, f, g, i–l, n–q)
Due to this partial block of Dark or Dronc caspase activity using RNAi, we next examined loss-of-function alleles. Both homozygous DarkL46 and DroncI29 individuals are partially pupal lethal53, 54 but survive through muscle remodeling. At 12 APF, DEOM1 histolysis was blocked in both Dark (Fig. 5k, m) and Dronc (Fig. 5l, m) mutants, although to a lesser extent than tn−/−, where muscle degradation was completely abolished (Fig. 5j, m). At 24 h APF when DEOM1 and DEOM2 were normally absent in WT pupae (Fig. 5n, r), the majority of DEOM2s failed to undergo histolysis upon loss of Tn (Fig. 5o, r), Dark (Fig. 5p, r), or Dronc (Fig. 5q, r). Together, these results show that Tn mediates muscle breakdown by acting through the DIAP1-Dronc pathway.
The RING domain of Tn is required for DEOM histolysis
Drosophila Tn contains a conserved N-terminal RING domain and six NHL repeats in the C terminus (Fig. 6a). The B-Box and coiled-coiled (CC) regions are poorly conserved48. The requirement for the RING and NHL regions was investigated using genetic rescue experiments. We examined the effects of expressing full-length Tn (Tn FL) or versions of Tn lacking the RING (tnΔRING) or NHL domains (tnΔNHL) in DEOMs subjected to tn RNAi at 24 h APF. Transgene expression was confirmed by Tn immunostaining in the DEOMs and qPCR for tn transcript quantitation (Fig. S4). As expected, restoration of DEOM histolysis was observed upon the introduction of Tn FL in a tn RNAi background (Fig. 6d, g) compared to tn RNAi alone (Fig. 6c, g). DEOM1 was still intact at 24 h APF upon loss of the RING domain in Tn (Fig. 6e, g), while the presence of the RING domain (mef>tn RNAi+tnΔNHL) restored DEOM histolysis (Fig. 6f, g). These results indicate that the RING domain of Tn is required to prevent muscle breakdown.
Fig. 6. The RING domain is required for DEOM histolysis and to maintain DIAP1 protein levels.
a Schematic diagram showing the conserved RING and NHL domains in Tn. The B-box and coiled-coil domains are predicted, but poorly conserved. b–f Confocal Z-stack micrographs of abdominal muscles at 24 h APF stained for F-actin (green). b DEOM1 underwent histolysis (*) in the mef2-Gal4/+ control genotype. c DEOM1 histolysis is incomplete (yellow solid line) in mef2>tn RNAi animals. d–f Expression of Tn FL (d) or TnΔNHL (f) restores histolysis, while removal of the RING domain completely blocks DEOM degeneration, indicating this region is essential for normal histolysis (e). g Bar graph showing a significant decrease in DEOM histolysis in mef2>tn RNAi; tnΔRING flies. h–l Confocal Z-stack merged images of DEOM1 (white dotted line) at 12 h APF co-stained for F-actin (red) and DIAP1 (green). Low DIAP1 levels are observed in genotypes in which DEOM histolysis proceeds normally, including mef2-Gal4 (h), mef2>tn RNAi+tn FL (j), and mef2>tn RNAi+tnΔNHL (l). Increased DIAP1 levels are observed in mef2>tn RNAi (i) or mef2>tn RNAi+tnΔRING (k) DEOMs. m Quantification of relative fluorescence intensity levels reveals significantly higher DIAP1 levels in mef2>tn RNAi+tnΔRING muscles. Mean ± SEM (n.s., not significant, ****p < 0.001, **p < 0.01). Scale bar, 100 µm (b–f) and 50 µm (h–l)
The RING domain in E3 ubiquitin ligases such as Tn is required to transfer ubiquitin moieties from an E2 enzyme to a target substrate for proteasomal degradation55. Thus, to test if the RING domain of Tn may regulate DIAP1 protein levels through a similar mechanism, we examined DIAP1 immunostaining in DEOMs at 12 h APF again using truncated Tn constructs. Consistent with previous results in Fig. 3e, DIAP1 protein was elevated upon a reduction in Tn (Fig. 6i, m). Expression of Tn FL (Fig. 6j, m) or Tn lacking the NHL region (Fig. 6l, m) reduced DIAP1 levels similar to control muscles (Fig. 6h, m). In contrast, significantly higher DIAP1 was present in DEOMs expressing TnΔRING in a tn RNAi background (Fig. 6k, m). These results substantiate the importance of the RING domain in DEOM histolysis, specifically suggesting that DIAP1 is a substrate for Tn-mediated E3 activity.
Tn acts via DIAP1 to regulate Dronc activity
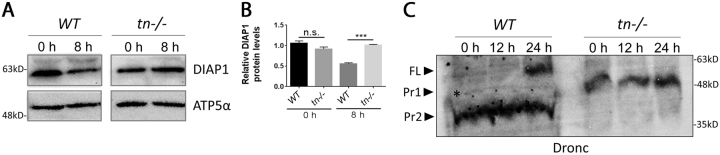
To further verify that loss of Tn alters DIAP1 levels, we performed western blots to quantify DIAP1 protein in WT or tn−/− pupae. Before the initiation of DEOM destruction (0 h APF), there was no significant difference in DIAP1 levels between WT or tn mutants (Fig. 7a, b). However, at 8 h APF, DIAP1 levels were approximately 2-fold higher upon loss of Tn. Since a primary role for DIAP1 is to bind and inhibit Dronc activity, we next examined Dronc processing. During apoptosis, FL Dronc is cleaved to produce Pr1 and Pr2 forms56–58. DIAP1 physically interacts with the FL and Pr1 forms of Dronc, thus preventing further cleavage to the active Pr2 protein. In WT pupae at 24 h APF, both the FL and processed Pr1 and Pr2 Dronc proteins were present (Fig. 7c). However, only the active form of Dronc was present from 0 h to 12 h APF, consistent with the normal timing of DEOM muscle histolysis. Interestingly, loss of Tn resulted in solely the Pr1 form (Fig. 7c). These data support a model whereby the upregulation of DIAP1 protein upon Tn deficiency prevents full Dronc activation, thus preventing apoptosis.
Fig. 7. Loss of Tn affects DIAPI protein levels and Dronc activation.
a Total protein extracted from pupae at 0 or 8 h APF was subjected to western blotting to assess DIAP1 or ATP5α protein levels. DIAP1 levels at 8 h APF are increased in tn mutants. b Densitometry quantification of the relative DIAP1 protein levels using ATP5α as a loading control reveals a significant increase in DIAP1 protein at 8 h upon loss of Tn (n = 3). c Dronc processing in WT or tn−/− pupae. Dronc FL (~55 kDa) is present at 24 h APF in WT samples. The cleaved Pr1 form (~40 kDa) is weakly observed during WT metamorphosis (black asterisk), while the active Pr2 cleavage product of Dronc (~37 kDa) predominates from 0 to 24 h APF. Only the Pr1 form is present upon loss of Tn throughout DEOM histolysis. Mean ± SEM (n.s., not significant, ***p < 0.005)
Discussion
PCD is required for the destruction of certain larval tissues during metamorphosis4, 9, 59. Zirin et al.28 established that histolysis of the abdominal muscles is regulated by apoptosis, while blocking autophagy does not affect muscle breakdown28. In addition to the ecdysone receptor, only a handful of nuclear proteins are known to function in DEOM histolysis. Loss of East results in a partial block in DEOM degeneration, whereas premature muscle destruction is observed in muscles that lack Chromator27. Moreover, the two nuclear receptors, FTZ-F1 and HR39, antagonistically function to regulate the timing of DEOM histolysis28. Here we have further identified Tn as a novel protein in pupal muscle remodeling. However, loss of Tn does not affect salivary gland and midgut histolysis, highlighting an exclusive muscle role for Tn during Drosophila metamorphosis.
Our genetic assays demonstrate that tn functions with core components of the cell death machinery to regulate DEOM destruction. It was surprising that inhibition of apoptotic activity in Dark or Dronc mutants was not sufficient to completely block histolysis by 24 h APF. One explanation is the existence of additional cell death mechanisms other than apoptosis. While histolysing DEOMs contained autophagic vesicles, a reduction in autophagy components did not block or delay muscle degradation at 8 h APF28. We tested if a decrease in Tn-mediated apoptosis could sensitize muscle cells to initiate autophagy as a compensatory mechanism to assure cell death. However, this does not seem to be the case as RNAi knockdown of Atg1, Atg5, or Atg18 does not further block DEOM histolysis in a tn RNAi background (Fig. S5). A second explanation is that the hypomorphic nature of these alleles may not completely abrogate Dark and Dronc function. Alternatively, additional effector caspases, including Dcp-1, Decay, and/or Damm, may be operating in the latter stages of DEOM histolysis since these caspases may function redundantly or act independent of the DIAP1–Drice axis3.
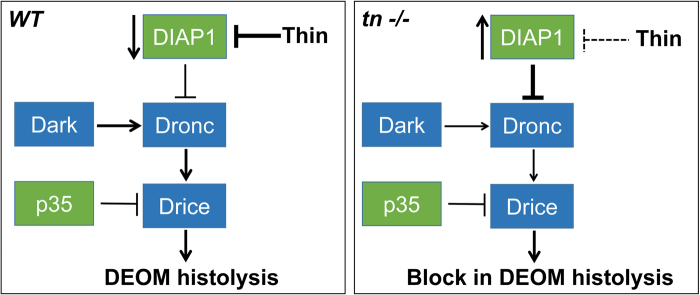
We expected more than a partial block in DEOM histolysis upon manipulation of DIAP1 (i.e., DIAP1 OE alone or tn RNAi+DIAP1 OE) at 12 h APF. It is possible that normal or overexpressed DIAP1 levels in the DEOMs are not high enough to block apoptosis, especially using RNAi approaches to reduce Tn levels. However, the use of tn-null alleles clearly shows a complete block in muscle degradation and a corresponding inhibition of active Dronc. Seemingly a delicate balance exists to regulate mRNA and protein expression, as well as protein turnover and proteolytic processing of active caspases. Cells must normally prevent cell death and only activate the apoptotic cascade upon a commitment to die. Thus, threshold levels of caspase activity must be reached for this terminal fate. There is evidence for stage or tissue-specific differential sensitivity to pro-apoptotic factors. Early L3 individuals are resistant to apoptosis, while wandering L3 larvae have elevated levels of Dark, Dronc, and Drice that are sufficient to trigger cell death under the appropriate stimuli28. We propose a model whereby Tn, through its RING domain, normally ubiquitinates DIAP1 for delivery to the proteasome during DEOM histolysis. This degradation of DIAP1 relieves Dronc inhibition, thereby initiating the caspase cascade for the execution of cell death (Fig. 8a). A general reduction in Tn, or loss of RING domain activity, prevents the addition of ubiquitin moieties and causes an increase in DIAP1 levels, effectively blocking cell death by limiting caspase activity (Fig. 8b).
Fig. 8. Model for Tn-mediated regulation of DEOM histolysis.
a In WT DEOMs, Tn targets DIAPI for degradation. Downregulation of DIAP1 results in DEOM histolysis via caspase-dependent apoptosis. b In the absence of Tn, DIAPI levels increase and block caspase activity. Lack of caspase activation blocks cell death and preserves DEOMs
Numerous roles have been identified for mammalian TRIM32 in normal and cancerous cells. In muscle, mutations in the NHL repeats result in limb-girdle muscular dystrophy type 2H or sarcotubular myopathy60–66. Several structural muscle proteins are targets of TRIM32 activity, including tropomyosin, desmin, α-actinin, and dysbindin67–69. However, it is not yet clear if regulation of these muscle substrates contributes to normal muscle physiology, is required to prevent atrophy, or plays a critical role in disease pathology46. The TRIM32-mediated degradation of additional protein substrates, including p53, Abi2, Piasy, and the X-linked IAP (XIAP), contribute to oncogenic or tumor suppressor activities that either confer resistance or susceptibility to apoptosis70–73. Tumor necrosis factor-α can trigger death receptor-mediated apoptosis through the regulation of XIAP activity73. TRIM32 colocalizes and directly interacts with XIAP in human kidney epithelial cells (HEK293). Moreover, TRIM32 induces apoptosis through the direct ubiquitination and subsequent protein turnover of XIAP degradation73. This control of apoptotic cell death mirrors our genetic results, strongly suggesting that this TRIM32-mediated regulation of IAP family members may be a conserved mechanism to regulate apoptosis. It would be interesting to further investigate if Tn and mammalian TRIM32 regulates apoptotic decisions in other contexts of muscle development and/or disease.
Herein, we have provided the first evidence for Tn in the regulation of muscle histolysis. Importantly, our genetic assays suggest that DIAP1 is a target of Tn and that regulation of DIAP1 and/or Caspase activity are crucial for a cell’s decision to execute cell death. These findings will further increase our general understanding about PCD during tissue destruction in Drosophila development and will provide a conserved framework to identify novel targets of Tn.
Materials and methods
Fly genetics
Drosophila melanogaster stocks were raised on standard cornmeal medium at 25 °C, unless otherwise indicated. The following fly stocks were used in this study: w1118 strain as WT; mef2-Gal4 (Bloomington Drosophila Stock Center (BDSC), BL27390); tnΔA 48, two different UAS-tn RNAi lines (Vienna Drosophila Resource Center (VDRC), v19290 and v19291); UAS-EcR.B1 (BL6469); UAS-DIAP1.H (BL6657); UAS-Dronc RNAi (BL32963)74; UAS-Dark RNAi (BL 33924); UAS-p35.BH2 (BL5073); UAS-tn FL48. The DarkL46 and DroncI29 alleles were generously provided by Bergmann and co-workers53, 54. The mef2-Gal4; UAS-tn RNAi (mef2>tn RNAi) line was created using standard recombination techniques and is maintained at 18 °C to maintain viability as 25 °C results in partial pupal lethality.
Immunostaining and microscopy
To examine the effect of Tn in DEOM histolysis, white prepupae at 0 h were collected and aged until 8 h APF, 12 h APF, or 24 APF. Muscle preparations were dissected, fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 30 min and immunostained as indicated75. To analyze Tn function in salivary gland and midgut histolysis, wandering L3 larvae were dissected, fixed with 4% formaldehyde in PBS, and stained with DAPI (4′,6-diamidino-2-phenylindole) and/or phalloidin. The following primary antibodies were used: rabbit anti-Caspase-3 (1:100, Cell Signaling Technology, Danvers, MA,USA), mouse anti-DIAP1 (1:200, B. Hay)76, guinea pig anti-p35 (1:10, P. Meier)40, and guinea pig anti-Tn48. Secondary antibodies used for fluorescent immunolabeling were Alexa Fluor anti-mouse 488, Alexa Fluor anti-rabbit 488, Alexa Fluor anti-mouse 594, and Alexa Fluor anti-guinea pig 488 (1:400, Molecular Probes, Eugene, OR, USA). Phalloidin 488 and 594 were used for F-actin labeling (1:400, Molecular Probes, Eugene, OR, USA). Immunostained preparations were imaged on a Zeiss LSM 700, processed using the Zeiss Zen software and assembled into figures in Photoshop Elements.
Western blotting
Five to ten pupae of the appropriate genotype were homogenized in 3× Laemmli buffer (150 mM Tris-HCl (pH 6.8), 300 mM dithiothreitol, 6% sodium dodecyl sulfate (SDS), 0.3% bromophenol blue, and 30% glycerol), boiled at 100 °C for 10 min, and centrifuged at 13,000 × g to remove cellular debris. The resulting proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinyl difluoride membrane, and probed with mouse anti-DIAP1 (1:500, B. Hays) or guinea pig anti-Dronc (1:400, P. Meier). Horse radish peroxidase-conjugated secondary antibodies (1:5000, GE Healthcare, UK) were used to detect the primary antibodies. Protein bands were visualized using the ECL Plus Western Blotting Detection Kit (ThermoFisher, Waltham, MA, USA) and analyzed with the FluorChem M system (Protein Simple). The blots were stripped (6.25 ml of 1 M Tris-HCl, pH 6.8, 10 ml of 20% SDS, and 700 μl β-mercaptoethanol) and reprobed with mouse anti-ATP5α (1:10,000, Abcam, UK). Densitometry analysis was performed by calculating the band intensities of DIAP1 relative to the ATP5α loading control using Image J.
Quantitative PCR
RNA was isolated from a pool of five pupae using the RNAeasy Mini Kit (Qiagen, Valencia, CA, USA) for each genotype. Three separate pools were used for each time point. Either mef2-Gal4/+ (Fig. 2) or w1118 (Fig. 6) were used as a control. After elution, RNA concentrations were determined and single strand complementary DNA (cDNA) was generated from 100 ng of RNA using the SuperScript VILO cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA). For the qPCR reactions, each cDNA sample was diluted to 1:50 and mixed with Power UP SYBR Green Master mix and the appropriate primers (Applied Biosystems, Foster City, CA, USA). rp49 was used as the reference gene. Primers were synthesized by Integrated DNA Technologies (IDT): rp49: 5′F-GCCCAAGGGTATCGACAACA-3′, 5R′-GCGCTTGTTCGATCCGTAAC-3′; Dark: 5′F-AAGTACAATGTGAGCCGCCT-3′, 5′R-CCCAAGTCTTTCCCGATCCC-3′; Dronc: 5′F-AGTCGGCCGATATTGTGGAC-3′, 5′R-ACATAAGGGGTGAGTGCTCC-3′; Drice: 5′F-GACTGCCGCTACAAGGACAT-3′, 5′R-TGATTGGCCGTGAAGAAGCT-3′; Diap1: 5′F-GCGTGGAAATCGGTTGCTG-3′, 5′R-GATGCGATCTAATGCTTCGGC-3′; tn: 5′F-GAGCTGCATATCGAAATCACCG-3′; 5′R-AGATAGGCTTTTTCCGAGCAAAC-3′.
Three independent biological replicates were processed for each genotype and reactions were run in triplicate using the Quant Studio 3 Applied Biosystem with Quant studio design and analysis software. The average of the triplicates was used to calculate the 2−ΔΔCt values (normalized fold expression). Quantification of mRNA levels between different genotypes at the same time point was performed using multiple t tests. Two-way analysis of variance (ANOVA) was used to compare transcript levels of the same genotype at different developmental time points.
Transgenic fly lines
PCR amplification from the tn cDNA GH06739 (Drosophila Genomic Resource Center) was used to generate UAS-tn FL, UAS-tnΔRING, and UAS-tnΔNHL using the following primers: UAS-Tn FL—5′F-TAACGCGTCGACAATGGAGCAATTCGAGCAGCTGTTGAC-3′, 5′R-CTAGTCTAGATCAGAAGACTTGGACGCGGTGATTC 3′; UAS-Tn ΔRING—5′F-AATAAGAATAGCGGCCGCATGAATCTGCGACGAGACATCACG-3′, 5′R- CTAGTCTAGATCAGAAGACTTGGACGCGGTGATTC 3′; UAS-TnΔNHL—5′F-TAACGCGTCGACAATGGAGCAATTCGAGCAGCTGTTGAC 3′. 5′R-CTAGTCTACATGCTGGCGCTTGCGCAGGTACACCTG 3′.
Each of the amplified regions was digested with SalI/XbaI or NotI/XbaI (restriction sites underlined), subcloned into the pUAST Drosophila transformation vector, and verified by sequencing. Transgenic fly lines were generated by Genetic Services, Inc.
Quantitative image analysis
Initiation of muscle histolysis (%): The number of DEOM1 muscles that were morphologically smaller or fragmented (indicative of histolysis initiation) in abdominal segments A2 and A3 in control or experimental genotypes at 0 or 8 h APF was quantitated. The percent of muscle histolysis initiation is illustrated as a percent of smaller or fragmented DEOM1 muscles/total number of DEOM1 muscles analyzed.
Histolysis (%): The extent of muscle histolysis was determined by counting the number of DEOM1 muscles present in abdominal segments A2 and A3 in control and experimental samples at the indicated time points (12 or 24 h APF). All values are portrayed in percentages as DEOM1 muscles absent/total number of DEOM1 muscles analyzed.
Corrected total fluorescence (CTF): To measure the fluorescence intensities of Caspase-3, DIAP1, and p35 protein levels in dying DEOM1 or DEOM2 muscles, the CTF method was used77, 78. The net average fluorescence intensity in a region of interest was measured in single section planes (1 µm slices) inside the DEOMs and in an area without fluorescence for background subtraction. All measurements were performed in Image J.
Statistical analysis: All raw data were imported into GraphPad Prism 6.0 for statistical analysis and graph production. All error bars represent mean ± standard error of the mean (SEM). Statistical significances were determined using either a Student's t test, Mann–Whitney tests, or one-way ANOVA. Differences were considered significant if p < 0.05 and are indicated in each figure legend. All “n” values are listed in Supplementary Tables.
Electronic supplementary material
Acknowledgements
We thank Bruce Hay and Pascal Meier for sharing antibodies. We also thank Andreas Bergmann, the VDRC, and the BDSC for fly lines used in this study. Monoclonal antibodies from the Developmental Studies Hybridoma Bank were created by the National Institute of Child Health and Human Development of the National Institutes of Health and are maintained at The University of Iowa for monoclonal antibodies. This work was supported by a K-INBRE postdoctoral grant through the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (NIGMS) to K.V. (P20GM103418) and a grant through the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) to E.R.G (R01AR060788). Both are sponsored through the National Institutes of Health (NIH).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Baehrecke
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/21/2022
Editor's Note: Concerns have been raised about the integrity of the data reported in this article. This is currently being investigated. Further editorial action may be taken as appropriate once the investigation into the concerns is complete and all parties have been given an opportunity to respond in full.
Change history
09/30/2022
Editor's Note: Following investigation into concerns that were previously raised the editors have determined that no further editorial action is needed.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-0756-x).
References
- 1.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell. Biol. 2015;16:329–344. doi: 10.1038/nrm3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denton D, Aung-Htut MT, Kumar S. Developmentally programmed cell death in Drosophila. Biochim. Biophys. Acta. 2013;1833:3499–3506. doi: 10.1016/j.bbamcr.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Baehrecke EH. How death shapes life during development. Nat. Rev. Mol. Cell. Biol. 2002;3:779–787. doi: 10.1038/nrm931. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/S0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 6.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker NI, Harmon BV, Gobé GC, Kerr JF. Patterns of cell death. Methods Achiev. Exp. Pathol. 1988;13:18–54. [PubMed] [Google Scholar]
- 9.Nicolson S, Denton D, Kumar S. Ecdysone-mediated programmed cell death in Drosophila. Int. J. Dev. Biol. 2015;59:23–32. doi: 10.1387/ijdb.150055sk. [DOI] [PubMed] [Google Scholar]
- 10.Baehrecke EH. Ecdysone signaling cascade and regulation of Drosophila metamorphosis. Arch. Insect Biochem. Physiol. 1996;33:231–244. doi: 10.1002/(SICI)1520-6327(1996)33:3/4<231::AID-ARCH5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 11.Baehrecke EH. Autophagic programmed cell death in Drosophila. Cell Death Differ. 2003;10:940–945. doi: 10.1038/sj.cdd.4401280. [DOI] [PubMed] [Google Scholar]
- 12.Thummel CS. Flies on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- 13.Jiang C, Baehrecke EH, Thummel CS. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 1997;124:4673–4683. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- 14.Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell. 2000;5:445–455. doi: 10.1016/S1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- 15.Lee CY, Simon CR, Woodard CT, Baehrecke EH. Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 2002;252:138–148. doi: 10.1006/dbio.2002.0838. [DOI] [PubMed] [Google Scholar]
- 16.DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila broad-complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cakouros D, Daish T, Martin D, Baehrecke EH, Kumar S. Ecdysone-induced expression of the caspase DRONC during hormone-dependent programmed cell death in Drosophila is regulated by Broad-Complex. J. Cell Biol. 2002;157:985–995. doi: 10.1083/jcb.200201034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- 19.Baehrecke EH, Thummel CS. The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev. Biol. 1995;171:85–97. doi: 10.1006/dbio.1995.1262. [DOI] [PubMed] [Google Scholar]
- 20.Daish TJ, Cakouros D, Kumar S. Distinct promoter regions regulate spatial and temporal expression of the Drosophila caspase dronc. Cell Death Differ. 2003;10:1348–1356. doi: 10.1038/sj.cdd.4401312. [DOI] [PubMed] [Google Scholar]
- 21.Kilpatrick ZE, Cakouros D, Kumar S. Ecdysone-mediated up-regulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J. Biol. Chem. 2005;280:11981–11986. doi: 10.1074/jbc.M413971200. [DOI] [PubMed] [Google Scholar]
- 22.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denton D, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 25.Currie DA, Bate M. The development of adult abdominal muscles in Drosophila: myoblasts express twist and are associated with nerves. Development. 1991;113:91–102. doi: 10.1242/dev.113.1.91. [DOI] [PubMed] [Google Scholar]
- 26.Kimura KI, Truman JW. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J. Neurosci. 1990;10:403–401. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasser M, Bte Osman Z, Chia W. EAST and Chromator control the destruction and remodeling of muscles during Drosophila metamorphosis. Dev. Biol. 2007;307:380–393. doi: 10.1016/j.ydbio.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Zirin J, et al. Ecdysone signaling at metamorphosis triggers apoptosis of Drosophila abdominal muscles. Dev. Biol. 2013;383:275–284. doi: 10.1016/j.ydbio.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleesha Y, Puah WC, Lin F, Wasser M. FMAj: a tool for high content analysis of muscle dynamics in Drosophila metamorphosis. BMC Bioinforma. 2014;15(Suppl. 16):S6. doi: 10.1186/1471-2105-15-S16-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuleesha Y, Puah WC, Wasser M. Live imaging of muscle histolysis in Drosophila metamorphosis. BMC Dev. Biol. 2016;16:12. doi: 10.1186/s12861-016-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier P, Finch A, Evan G. Apoptosis in development. Nature. 2000;407:796–801. doi: 10.1038/35037734. [DOI] [PubMed] [Google Scholar]
- 32.Thornberry NA. Caspases: key mediators of apoptosis. Chem. Biol. 1998;5:R97–R103. doi: 10.1016/S1074-5521(98)90615-9. [DOI] [PubMed] [Google Scholar]
- 33.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat. Rev. Mol. Cell. Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 35.Kocab AJ, Duckett CS. Inhibitor of apoptosis proteins as intracellular signaling intermediates. FEBS J. 2016;283:221–231. doi: 10.1111/febs.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Waldhuber M, Emoto K, Petritsch C. The Drosophila caspase DRONC is required for metamorphosis and cell death in response to irradiation and developmental signals. Mech. Dev. 2005;122:914–927. doi: 10.1016/j.mod.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Wang SL, Hawkins CJ, Yoo SJ, Müller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/S0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 40.Yoo SJ, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 41.Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- 43.Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat. Cell Biol. 2002;4:425–431. doi: 10.1038/ncb794. [DOI] [PubMed] [Google Scholar]
- 44.Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 45.Yuan S, et al. Structure of the Drosophila apoptosome at 6.9 å resolution. Structure. 2011;19:128–140. doi: 10.1016/j.str.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazzari E, Meroni G. TRIM32 ubiquitin E3 ligase, one enzyme for several pathologies: from muscular dystrophy to tumours. Int. J. Biochem. Cell Biol. 2016;79:469–477. doi: 10.1016/j.biocel.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 47.Shieh PB, Kudryashova E, Spencer MJ. Limb-girdle muscular dystrophy 2H and the role of TRIM32. Handb. Clin. Neurol. 2011;101:125–133. doi: 10.1016/B978-0-08-045031-5.00009-8. [DOI] [PubMed] [Google Scholar]
- 48.LaBeau-DiMenna EM, et al. Thin, a Trim32 ortholog, is essential for myofibril stability and is required for the integrity of the costamere in Drosophila. Proc. Natl. Acad. Sci. USA. 2012;109:17983–17988. doi: 10.1073/pnas.1208408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooks DS, Vishal K, Kawakami J, Bouyain S, Geisbrecht ER. Optimization of wrMTrck to monitor Drosophila larval locomotor activity. J. Insect Physiol. 2016;93-94:11–17. doi: 10.1016/j.jinsphys.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin VP, Thummel CS. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell Dev. Biol. 2005;16:237–243. doi: 10.1016/j.semcdb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 53.Srivastava M, et al. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tocchini C, Ciosk R. TRIM-NHL proteins in development and disease. Semin. Cell Dev. Biol. 2015;47-48:52–59. doi: 10.1016/j.semcdb.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 56.Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 2002;277:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- 57.Muro I, Monser K, Clem RJ. Mechanism of Dronc activation in Drosophila cells. J. Cell Sci. 2004;117:5035–5041. doi: 10.1242/jcs.01376. [DOI] [PubMed] [Google Scholar]
- 58.Lee TV, et al. Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation. PLoS Genet. 2011;7:e1002261. doi: 10.1371/journal.pgen.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conradt B. Genetic control of programmed cell death during animal development. Annu. Rev. Genet. 2009;43:493–523. doi: 10.1146/annurev.genet.42.110807.091533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frosk P, Del Bigio MR, Wrogemann K, Greenberg CR. Hutterite brothers both affected with two forms of limb girdle muscular dystrophy: LGMD2H and LGMD2I. Eur. J. Hum. Genet. 2005;13:978–982. doi: 10.1038/sj.ejhg.5201436. [DOI] [PubMed] [Google Scholar]
- 61.Schoser BG, et al. Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann. Neurol. 2005;57:591–595. doi: 10.1002/ana.20441. [DOI] [PubMed] [Google Scholar]
- 62.Borg K, et al. Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum. Mutat. 2009;30:E831–E844. doi: 10.1002/humu.21063. [DOI] [PubMed] [Google Scholar]
- 63.Cossée M, et al. Use of SNP array analysis to identify a novel TRIM32 mutation in limb-girdle muscular dystrophy type 2H. Neuromuscul. Disord. 2009;19:255–260. doi: 10.1016/j.nmd.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Nectoux J, et al. Detection of TRIM32 deletions in LGMD patients analyzed by a combined strategy of CGH array and massively parallel sequencing. Eur. J. Hum. Genet. 2015;23:929–934. doi: 10.1038/ejhg.2014.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neri M, et al. A patient with limb girdle muscular dystrophy carries a TRIM32 deletion, detected by a novel CGH array, in compound heterozygosis with a nonsense mutation. Neuromuscul. Disord. 2013;23:478–482. doi: 10.1016/j.nmd.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Saccone V, et al. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum. Mutat. 2008;29:240–247. doi: 10.1002/humu.20633. [DOI] [PubMed] [Google Scholar]
- 67.Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell Biol. 2012;198:575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 69.Locke M, Tinsley CL, Benson MA, Blake DJ. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 2009;18:2344–2358. doi: 10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kudryashova E, Kramerova I, Spencer MJ. Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J. Clin. Invest. 2012;122:1764–1776. doi: 10.1172/JCI59581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J, et al. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 2014;21:1792–1804. doi: 10.1038/cdd.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin TY, Huang CH, Chou WG, Juang JL. Abi enhances Abl-mediated CDC2 phosphorylation and inactivation. J. Biomed. Sci. 2004;11:902–910. doi: 10.1007/BF02254375. [DOI] [PubMed] [Google Scholar]
- 73.Ryu YS, et al. TRIM32 protein sensitizes cells to tumor necrosis factor (TNFα)-induced apoptosis via its RING domain-dependent E3 ligase activity against X-linked inhibitor of apoptosis (XIAP) J. Biol. Chem. 2011;286:25729–25738. doi: 10.1074/jbc.M111.241893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi Y, Liu H, Daniels MP, Zhang G, Xu H. Loss of Drosophila i-AAA protease, dYME1L, causes abnormal mitochondria and apoptotic degeneration. Cell Death Differ. 2016;23:291–302. doi: 10.1038/cdd.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 2013;11:e1001708. doi: 10.1371/journal.pbio.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson R, et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- 77.Burgess A, et al. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA. 2010;107:12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCloy RA, et al. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.