Abstract
BACKGROUND: Widespread infiltration of tumor cells into surrounding brain parenchyma is a hallmark of malignant gliomas, but little data exist on the overall invasion pattern of tumor cells throughout the brain. METHODS: We have studied the invasive phenotype of malignant gliomas in two invasive mouse models and patients. Tumor invasion patterns were characterized in a patient-derived xenograft mouse model using brain-wide histological analysis and magnetic resonance (MR) imaging. Findings were histologically validated in a cdkn2a−/− PDGF-β lentivirus-induced mouse glioblastoma model. Clinical verification of the results was obtained by analysis of MR images of malignant gliomas. RESULTS: Histological analysis using human-specific cellular markers revealed invasive tumors with a non-radial invasion pattern. Tumors cells accumulated in structures located far from the transplant site, such as the optic white matter and pons, whereas certain adjacent regions were spared. As such, the hippocampus was remarkably free of infiltrating tumor cells despite the extensive invasion of surrounding regions. Similarly, MR images of xenografted mouse brains displayed tumors with bihemispheric pathology, while the hippocampi appeared relatively normal. In patients, most malignant temporal lobe gliomas were located lateral to the collateral sulcus. Despite widespread pathological fluid-attenuated inversion recovery signal in the temporal lobe, 74% of the “lateral tumors” did not show signs of involvement of the amygdalo-hippocampal complex. CONCLUSIONS: Our data provide clear evidence for a compartmental pattern of invasive growth in malignant gliomas. The observed invasion patterns suggest the presence of preferred migratory paths, as well as intra-parenchymal boundaries that may be difficult for glioma cells to traverse supporting the notion of compartmental growth. In both mice and human patients, the hippocampus appears to be a brain region that is less prone to tumor invasion.
Introduction
Malignant gliomas are the most common intra-axial primary brain tumors and, despite multimodal treatment, survival rates remain poor [1]. Surgical resection is often the primary treatment for these tumors; however, it is not curative due to the widespread infiltration of glioma cells. Such invasive cells are also relatively resistant to radio- and chemotherapy [2], further complicating the management of these tumors. To better understand the mechanisms underlying the invasive behavior of tumor cells and to tailor future therapies targeting invasive glioma cells, more knowledge is needed about tumor cell migratory trajectories and their preferred sites of accumulation in the brain.
Early histopathological studies of brains from glioma patients showed that tumor invasion does not occur in a random manner; glioma cells follow distinct anatomical structures with a propensity to migrate along white matter tracts (WMTs), in perivascular spaces and the subependymal layers while avoiding certain gray matter regions [3], [4]. Despite extensive invasion into the brain parenchyma and the perivascular spaces, tumor seeding along cerebrospinal fluid (CSF)-routes is seen in only 2% of the cases [5] and metastasis outside the neuraxis have rarely been reported [6], [7]. This supports the notion that glioma cell invasion occurs within certain tissue compartments. The spread of glioma cells within the brain appears to respect some anatomical boarders, giving rise to defined subtypes. As such, optic pathway glioma is a well-known tumor entity residing in the centrobasal midline region, whereas the limbic gliomas are predominantly confined to gray matter structures of the mediobasal temporal lobe, illustrating the diversity of glial tumors [8], [9].
Considering that the extracellular space is much smaller than an invasive glioma cell and the astrocytic end-feet cover approximately 99% of the vasculature [10], it is remarkable that a tumor cell is able to move through the brain at all. To this end, studies have shown that glioma cells undergo several geno- and phenotypic changes that enable them to switch to an invasive phenotype. These changes are facilitated by chemo-attractive and repulsive cues that act in a stringent interplay between tumor cells and their microenvironment [11], [12], [13].
Orthotopic xenograft studies where human malignant glioma cells are transplanted into rodents have given some indications as to which brain regions are preferred (e.g., corpus callosum and internal capsule) and avoided (e.g., thalamus) by invasive tumor cells [14], [15], [16]. These rather old studies were, however, restricted by the methods available for visualization of tumor cells at the time. Many xenograft studies are based on chemically induced glioblastoma (GBM) models and serum-cultured commercial cell lines which either form circumscribed tumors (e.g., U87) or show limited peri-tumoral infiltration of the brain parenchyma (e.g., GL261) without recapitulating the invasive phenotype of gliomas [17]. Although a few studies have used genetically engineered mouse models or xenografts of patient-derived serum-free primary cell cultures that do display invasion of the brain parenchyma [17], a systematic brain-wide characterization of glioma migration patterns has not been performed.
To address this biological and clinically relevant subject, we have investigated the invasive growth pattern of malignant gliomas using two invasive GBM animal models and clinical patient imaging. Analyses were based on histological sections of xenograft tumors formed by transplanting patient-derived GBM-initiating cells (GICs) into severe combined immunocompromised (SCID-) mice and structural magnetic resonance (MR) images. GICs have been shown to reiterate the geno- and phenotype of the parental tumor and to form highly invasive tumors upon orthotopic transplantation [18], [19], [20]. Our key findings were validated in a cdkn2a−/− PDGF-β-GFP lentivirus-induced GBM mouse model. In both models, we observed systematic patterns of invasive growth and regional differences in the propensity to harbor or avoid invasive tumor cells.
Material and Methods
Study Populations and Ethical Approvals
Male C.B.-17 SCID mice (Taconic, Ejby, Denmark) were used for intracranial tumor transplantation (n = 22) and serial MR imaging studies (n = 40). Male cdkn2a−/− mice (n = 6) were used to generate PDGF-β-GFP lentivirus models of GBM. All animal experimental procedures were approved by the respective Animal Research Authorities.
Clinical data used for this study included prospectively registered data and MR images from patients who underwent operations for malignant temporal lobe gliomas (TLGs) at Oslo University Hospital, Norway, in a three-year period (July 2013–June 2016; n = 90). GIC cultures were established from GBM biopsies harvested from consenting patients (n = 5). Data collection and the use of human tissue was approved by institutional data protection officials and the Norwegian National Committee for Medical Research Ethics (#07321b).
Tissue Specimens and Cell Cultures
GIC cultures were derived from histologically confirmed GBMs and grown as spheres in serum-free neurosphere medium [21]. We have previously shown that GICs maintain their phenotype and tumorigenicity after in vitro culturing in these conditions [20]. The two GIC cultures used the most in this study (T08 and T65) have been characterized in detail with regards to growth, sphere forming ability, and expression of stem cell associated markers [20], [22], [23]. To identify transplanted human cells and verify human-specific antibody staining, we labeled two GIC cultures with green fluorescent protein (GFP) [23]. GL261, a murine GBM cell line, was obtained from the National Cancer Institute (NCI, Bethesda, MD, USA) and cultured in serum-containing adherent conditions [24].
Intracranial Transplantation, Brain Tissue Processing and Immunolabeling
GICs were stereotactically transplanted into 8–9-week-old C.B.-17 male SCID mice (Taconic, Ejby, Denmark) [20]. Two microliters of a cell suspension containing 100,000 cells/μl was inoculated into the right striatum at a position 0 mm anterior, 2 mm ventral and 2 mm right of the bregma; 2 μl of a 5000 cells/μl solution was transplanted for GL261 grafting. The right and left hemispheres are hereafter referred to as “transplanted” and “non-transplanted” hemispheres, respectively. Mice were sacrificed when humane endpoints were reached (defined as >10% weight loss and/or manifestation of neurological symptoms). Preparation of brain sections (12 μm) and immunolabeling were performed as previously described [21], see Suppl. File 1.
Transgenic GBM Model
A transgenic GBM model of cdkn2a−/− mice injected with PDGF-B-GFP lentivirus was established as previously described [25]. In the current study, a retroviral vector system containing mouse PDGF-B was kindly provided by Prof. Paolo Malatesta [26]. Vector was injected into cdkn2a-deficient mice as previously described [27]. After 12 weeks, mice were sacrificed by paraformaldehyde perfusion, post-fixed in formaldehyde, dehydrated and paraffin embedded. Brain sections (4 μm) from tumor-bearing brains (n = 6) were counterstained using hematoxylin and eosin (HE) or a GFP antibody.
Acquisition and Analysis of Mouse Brain Slide Images
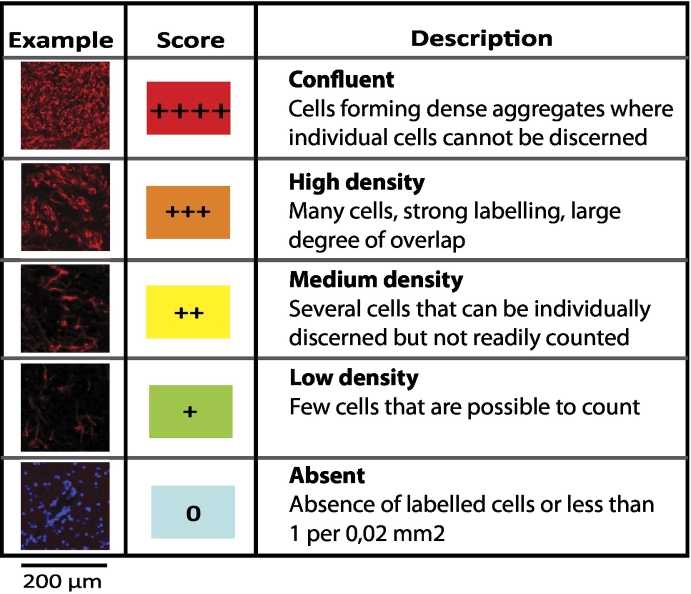
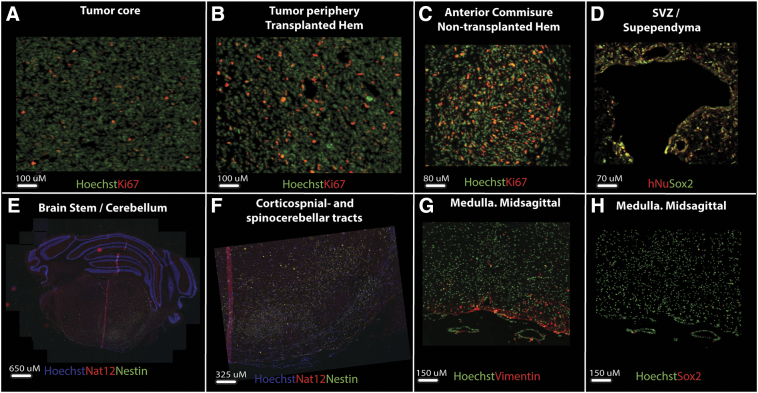
High-resolution brightfield or fluorescence images of histological sections were acquired through a 20× Plan-Apochromat/0.8 objective using an automated slide scanner system (Axio Scan Z1, Carl Zeiss Microscopy, Munich, Germany). Images were inspected using the Zen Lite Blue software from Carl Zeiss Microscopy. The density of labeled cells in different brain regions was assessed using a 0–4 point grading system adopted from Yetman et al. [28] (Table 1A). The anatomical location of the observed labeling was determined by identification of multiple prominent anatomical landmarks with reference to the mouse brain atlas of Franklin and Paxinos [29]. For additional confirmation of these observations in the two mouse models, a quantitative image analysis workflow [30] was used to quantify human-specific mitochondrial (hMit) or GFP labeling in selected brain regions. In each of these brain regions, three square regions-of-interest (ROIs) with size 200 × 200 μm or 50 × 50 μm (depending on the area of the region investigated) were randomly placed well within the external boundaries of each region. For each of these ROIs, the fluorescent signal was separated from background using the machine learning ilastik software [31], which segments images into classes-of-interest based on supervised random forest learning algorithms and classifies labeling into objects of interest. Segmented images were processed in ImageJ to extract measurements of labeled areas in each ROI.
Table 1A.
Semi-Quantitative Scale Used to Grade Cell Labeling Density (Vimentin)
Five-point grading scale used to grade density of immunolabeled cells shown in Table 1B
Mouse Brain MR Imaging
Mice were scanned at regular intervals in a 9.4 T small-animal MR scanner (Agilent, Palo Alto, CA, US) with a 35-mm quadrature driven birdcage radiofrequency coil (Rapid Biomedical, Rimpar, Germany) under inhalation anesthesia (1.5% isoflurane and atmospheric oxygen). End-stage structural MR images were routinely acquired <24 hours prior to sacrifice.
The MR protocols included coronal multislice T1-weighted fast spin echo sequence (TR/TEeff = 1500/8.3 ms, 6 averages, slice thickness 0.75 mm with 62.5 x 62.5 μm in-plane resolution) and T2-weighted fast spin echo (TR/TEeff = 4000/46 ms, 4 averages, slice thickness 0.75 mm with 62.5 x 62.5 μm in-plane resolution). Magnevist™ Gd-DTPA (Bayer, Whippany, NJ, USA) was injected intraperitoneally at 0.5 mmol/kg. T1-weighted images were acquired before and 15 min after contrast injection. MR images were evaluated for areas containing contrast-enhancing tumor on T1 or T2-hyperintensity using anatomical landmarks as described in a standard mouse brain reference atlas [29].
Evaluation of Clinical MR Images of Malignant TLGs
Immediate preoperative MR images were available for 70 treatment-naive cases. Tumor location was evaluated using contrast-enhanced T1-weighted images and tumor extension was assessed by pathological hyperintense signal on fluid attenuated inversion recovery (FLAIR)-weighted images since it has previously been suggested that T2 and FLAIR hyperintense non-enhancing regions could contain histopathological features corresponding to biologically active tumor [32]. The TLGs were categorized into four subgroups based on the location of the dominant contrast-enhancing lesion by using a modified version of Yasargil's classification [8]. Furthermore, Visually AcceSsible Rembrandt Images (VASARI) were used as a guideline for scoring the following MR features: site of tumor epicenter, presence of satellites, multifocality or -centricity and ependymal enhancement [33]. Details are provided in Suppl. File 2.
Results
Brain-wide Invasion Patterns in GIC-Derived Tumors
Coronal tissue sections sampled across six mouse brains carrying xenograft tumors from five GIC cultures were examined for the presence of grafted tumor cells. In addition to using GFP-labeled cells, we immunofluorescently stained for a panel of cellular markers (Suppl. File 1). All samples were immunolabeled with antibodies against human mitochondria and the intermediate filament protein vimentin (expressed in invasive cells).
Transplanted Hemisphere (Table 1B)
Table 1B.
Brain-Wide Distribution and Density of Immunolabeled Tumor Cells
The Five-Point Grading Scale is Defined in Table 1A
| Transplanted hemisphere |
Non-transplanted hemisphere |
|||
|---|---|---|---|---|
| Brain regions (BRs) | hMit | Vim | hMit | Vim |
| Tumor Core | Non-adjacent BRs | |||
| Caudate Putamen | ++++ | +++ | ++ | ++ |
| Globus Pallidus | ++++ | ++++ | ++ | + |
| Adjacent BRs | ||||
| Ventral striatum | +++ | ++ | + | + |
| Thamalus | +++ | ++ | + | + |
| Hypothalamus | + | + | − | − |
| Amygdala | + | − | − | − |
| Septal nuclei (medial and lateral) | +++ | +++ | ++ | ++ |
| Subependymal space of the Lateral ventricle | ++++ | ++++ | +++ | +++ |
| Cortical areas | ||||
| Limbic cx | ++ | ++ | ++ | + |
| Cingulate cx | ++ | + | + | + |
| Motor cx | +++ | ++ | ++ | + |
| Somatosensory cx | +++ | ++ | ++ | + |
| Insular cx | +++ | +++ | + | + |
| Auditory cx | ++ | ++ | + | − |
| Visual cx | ++ | ++ | + | − |
| Piriform cx | +++ | ++ | − | − |
| Parahippocampal white matter | ||||
| Fornix | +++ | +++ | +++ | +++ |
| Fimbriae | +++ | +++ | +++ | ++ |
| Hippocampal Commisure | +++ | +++ | +++ | +++ |
| Alveus | +++ | +++ | ++ | ++ |
| Hippocampal formation | ||||
| CA1–3⁎ | + | − | + | − |
| Dentate Gyrus | + | − | + | − |
| Subiculum | + | + | + | − |
| Non-adjacent BRs | ||||
| Subpial space over | ||||
| the convexities | +++ | +++ | + | + |
| Pons | ++ | ++ | ++ | ++ |
| Cerebellar cortex | (+) | (+) | (+) | (+) |
| White matter tracts | ||||
| Lateral olfactory tract | +++ | ++ | − | − |
| Anterior Commisure (A&P) | +++ | +++ | ++ | ++ |
| Corpus callosum | ||||
| Truncus | ++++ | ++++ | ++++ | ++++ |
| External capsule | ++++ | ++++ | +++ | +++ |
| Internal capsule | +++ | +++ | ++ | ++ |
| Optic pathway | ||||
| Optic nerve | +++ | +++ | +++ | ++ |
| Optic chiasm | ++++ | +++ | ++++ | +++ |
| Optic tract | ++++ | ++++ | ++ | ++ |
| Optic nerve layer of superior colliculus | ++ | + | − | − |
| Cerebral peduncle | ++++ | ++++ | ++ | ++ |
| Spinocerebellar tracts | +++ | +++ | + | + |
| Pyramidal tracts | +++ | +++ | ++ | ++ |
| Cerebellar peduncles | ++ | ++ | + | + |
Stratum oriens of CA: hMit ++/Vim+/hMit +/Vim+.
In most cases, the tumor implantation site in the dorsal striatum featured a compact nuclear-dense tumor core that included the caudate putamen complex and the globus pallidus (Figure 1, A–D). The striatum was enlarged due to the densely packed tumor cells. This mass effect resulted in distortion of the surrounding structures (Figure 1, A–C), including elevation of the corpus callosum and external capsule and displacement of the lateral ventricles, thalamus, septal nuclei and hippocampus.
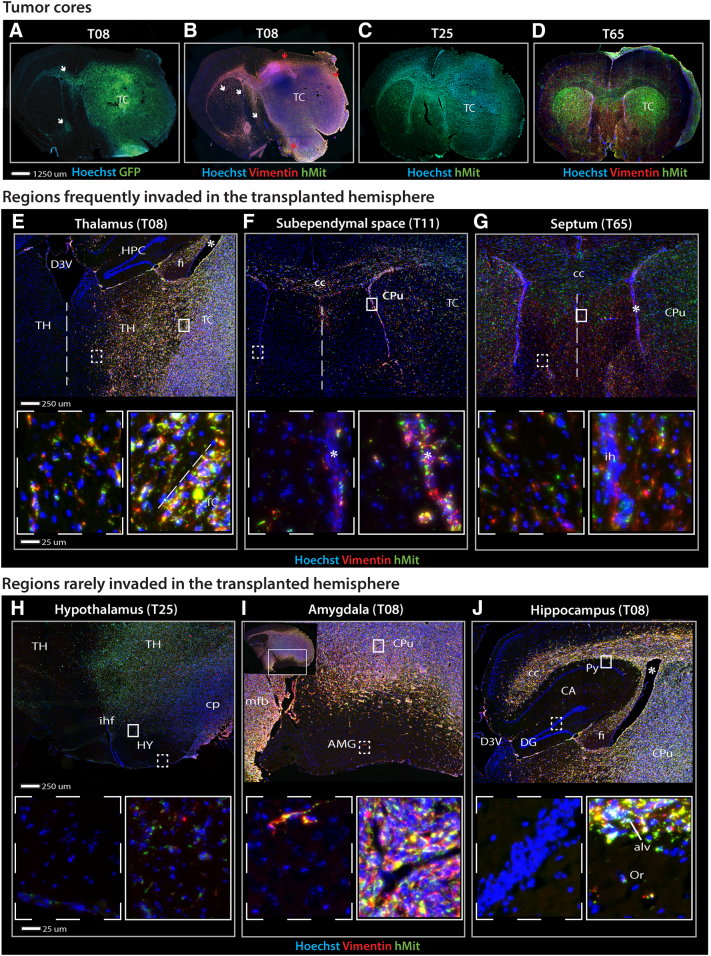
Figure 1.
Tumor cell distribution in the transplanted hemisphere of GIC-derived tumors.
Fluorescence microscopy images of immunolabeled coronal sections from GIC-derived tumors derived from T08, T11, T25 or T65 cultures. (A) T08 formed tumors with a large tumor core and moderate tumor cell migration mainly along white matter tracts (corpus callosum and anterior commissure) (white arrows). (B) Vimentin-labeling (red) of invasive tumor cells showed an accumulation of tumor cells in the septal area and the subpial lining (interhemispheric fissure) – not contiguous with the tumor core- covering the medial surfaces of both hemispheres. Tumor-infiltrated regions were divided in areas adjacent (red arrows) and non-adjacent (white arrows) to the tumor core. Counter-staining with a human-specific mitochondrial antibody (hMit) (green). (C) T25 xenograft tumors with a large tumor core and bihemispheric tumor invasion. (D) T65 displayed a much smaller tumor core with extensive intraparenchymal invasion and distribution of tumor cells resembling a butterfly glioma. (E-G) The thalamus and septal areas were frequently invaded by tumor cells. The subependymal lining was infiltrated (A-C), and even when tumor cores were not contiguous with the subventricular zone, tumor cells were detected on the contralateral side. (H-J) Certain brain regions adjacent to the tumor core, such as the hypothalamus, amygdala and hippocampus, were rarely invaded. (J) The white matter surrounding the hippocampus was packed with tumor cells, whereas the cornu ammonis and the dentate gyrus were spared.
Abbreviations: aca, anterior part of anterior commissure; AMG, amygdala; alv, alveus; CA, cornu ammoni; cc, corpus callosum; cp, cerebral peduncle; CPu, caudate putamen; D3V, dorsal part of the third ventricle; DG, dentate gyrus; fi, fimbria; HPC, hippocampus; HY, hypothalamus; ihf, interhemispheric fissure; Or, oriens layer of the hippocampus; Py, pyramidal layer of the hippocampus; py, pyramidal tracts; TC, tumor core; TH, thalamus; * lateral ventricle.
Outside the striatal tumor core, we found bihemispheric dispersal of tumor cells (Figure 1, A–D). Tumor cell infiltration was consistently seen in regions adjacent to the tumor core, such as the thalamus and ventral striatum (Figure 1, E). Particularly high densities of tumor cells were seen in the subependymal layers of the lateral ventricles. This prominent feature (Figure 1, B–D) was also present in cases with small tumor cores (Figure 1F). Outside the tumor core and its adjacent regions, a differential pattern of invasion was seen. Tumor cell accumulation was identified in the septal nuclei (Figure 1G), while the hypothalamus, amygdala and hippocampus, located at approximately the same distance from the tumor core, were remarkably free of tumor cells (Figure 1, H–J). These tumor-free islands were surrounded by white matter with extensive tumor infiltration (Figure 1, H–J).
In the cerebral cortex, various degrees of tumor cell invasion were seen in the deep layers (IV-VI) overlying the corpus callosum, while only a few cells were found in the superficial layers (I-III) (Figure 1, A–D). In regions where the tumor extended to the convexity surface, tumor cells also accumulated in the space between the cortex and the meningeal coverings, resulting in cortical impingement (Figure 1, B–C).
Tumor cell infiltration was found along WMTs, resulting in dispersion to more distant parts of the brain (Figure 2, A–E). In cases with large tumor cores, tumor cells infiltrated extensively into the white matter enveloping the striatal core, causing thickening of the corpus callosum and partial loss of the structural integrity of the ipsilateral internal capsule and the anterior commissure. Interestingly, the optic white matter stood out as one of the brain regions with the highest tumor cell density. A large number of tumor cells was found in the optic tract, chiasm and even in the distal part of the optic nerves (Figure 2, A and C). Similarly, the projection fibers of the cerebral peduncle were packed with tumor cells en route to the brain stem (Figure 2A). The brain stem was moderately infiltrated with tumor cells accumulating primarily in white matter regions, following the fiber bundles into the cerebellar cortex and medulla (Figure 2, D and E).
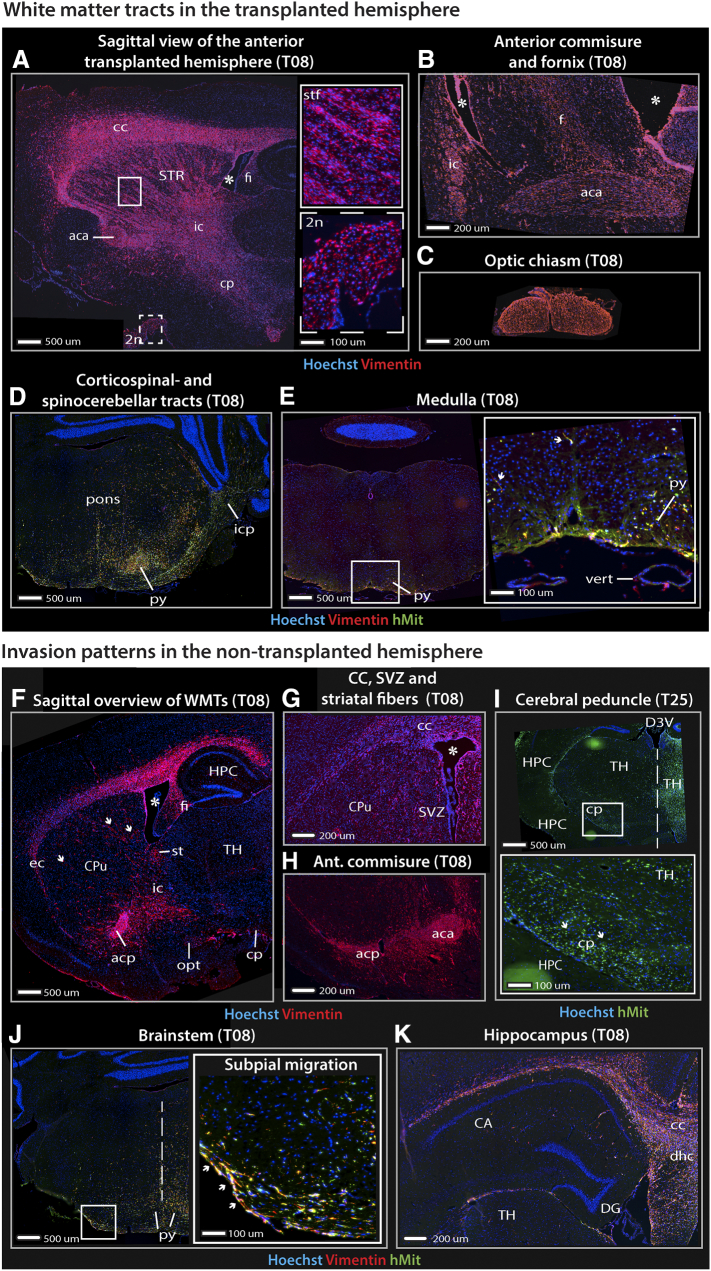
Figure 2.
Tumor cell distribution in white matter tracts and non-transplanted hemisphere of GIC-derived tumors.
Fluorescence microscopy images of immunolabeled sagittal (A, F) and coronal (B-E, G-K) sections from GIC-derived tumors derived from T08 or T25 cultures. (A-B) Tumor cells followed white matter tracts and high tumor cell densities were seen in the internal capsule, anterior commissure and the cerebral peduncle. (C) The optic chiasm and distal parts of the optic nerves (inlet with dashed line in A) were among the most tumor-infiltrated regions. (D) Tumor cells accumulated in the ventral regions of the pons close to the pyramidal tracts and projected laterally towards the spinocerebellar tracts following the cerebellar peduncles into the cerebellum. (E) Island of tumor cells in the medullary pyramidal tracts. Migration of single tumor cells can even be seen here (white arrows). (F) A large number of tumor cells were also detected in the white matter contralateral to the side of transplantation. Notably, tumor cells were detected in the optic tract, while the surrounding cortical areas harbored only a few cells. (F-H) The commissural fibers were extensively infiltrated by tumor cells and comprised the primary path of cell migration into the non-transplanted hemisphere. Crossing striatal fibers were visualized by the presence of tumor cells (white arrows). In most cases, the subventricular zone of the lateral ventricles and their subependymal lining were infiltrated by tumor cells. (I) Projection fibers of the cerebral peduncle and medial lemniscus harbored invasive tumor cells (inlet). (J) Tumor cells were detected in the ventral part of the pons. The inlet shows a clear tendency of tumor cells to accumulate in the subpial space (white arrows). (K) Despite the accumulation of tumor cells surrounding the hippocampus, the cornu ammonis and the dentate gyrus were remarkably free of tumor cells.
Abbreviations: 2n, optic nerve; aca, anterior part of anterior commissure; acp, posterior part of the anterior commissure; CA, cornu ammoni; cc, corpus callosum; cp, cerebral peduncle; CPu, caudate putamen; D3V, dorsal part of the third ventricle; DG, dentate gyrus; dhc, dorsal hippocampal commissure; f, fornix; fi, fimbria; HPC, hippocampus; HY, hypothalamus; ic, internal capsule; icp, inferior cerebellar peduncle; ihf, interhemispheric fissure; ml, medial lemniscus; opt, optic tract; Py, pyramidal layer of the hippocampus; py, pyramidal tracts; st, stria terminalis; stf, striatal fibers; STR, striatum; SVZ, subventricular zone; TC, tumor core; TH, thalamus; vert, vertebral artery; * lateral ventricle.
Non-Transplanted Hemisphere (Table 1B)
The primary path of cell dispersal to the non-transplanted hemisphere was via the commissural fibers, dominated by the corpus callosum (Figure 2, F–H). From the corpus callosum and the external capsule, the tumor cells infiltrated the dorsal parts of the cortical regions such as the motor- and somatosensory cortex (Figure 2F). Similarly, the striatum was infiltrated by direct invasion from the overlying white matter. However, islands of tumor cells representing crossed myelinated fiber tracts were the most dominant trait of cellular distribution in the striatum (Figure 2G). The anterior commissure also facilitated migration of tumor cells into the non-transplanted hemisphere (Figure 2H). The brain region comprising the cerebral peduncle and medial lemniscus harbored tumor cells, whereas the neighboring structures (e.g., hippocampus) were remarkably tumor-free (Figure 2I).
The subependymal lining of the lateral ventricle was one of the most tumor-infiltrated regions of the non-transplanted hemisphere (Figure 2G). As in the transplanted hemisphere, these observations confirmed the propensity of tumor cells to migrate towards the subventricular zone. Similar to the contralateral side, structures belonging to the optic pathway contained the highest density of invasive tumor cells (Figure 2F) and tumor cells were readily detected in the ventral part of the brain stem (Figure 2J).
Evaluating the white matter regions surrounding the hippocampus, we observed the same high density of malignant cells as in the transplanted hemisphere (Figure 2, F and K). Therefore, it was remarkable that the invasive tumor cells appeared to avoid the hippocampal formation (Figure 2K). These findings were verified in a separate group of xenograft tumors (Figure 3, A and B). Analysis of the staining patterns of other cellular markers is shown in Suppl. Figure 1.
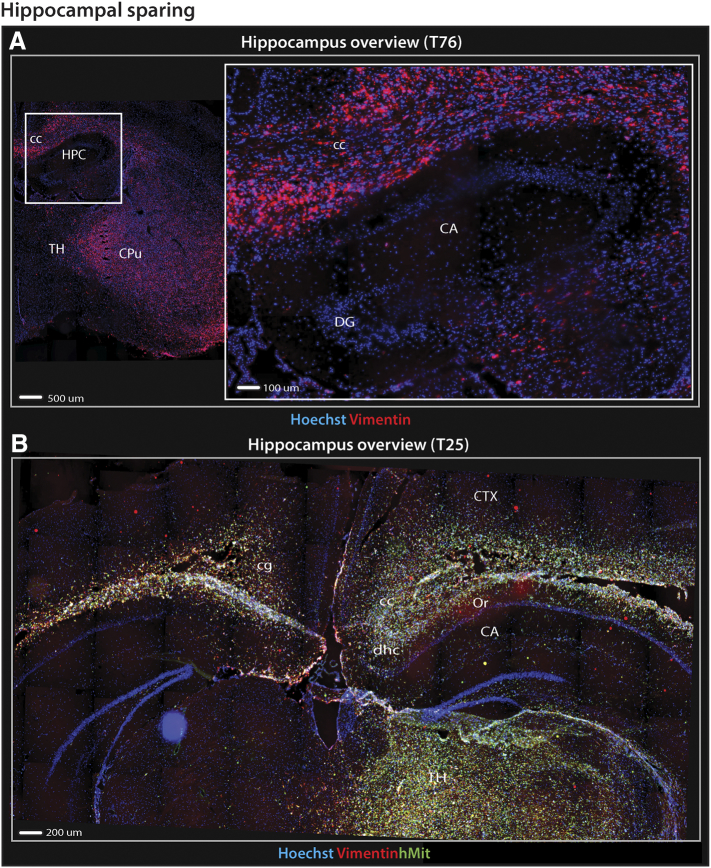
Figure 3.
Hippocampal sparing in GIC-derived tumors.
Fluorescence microscopy images of immunolabeled coronal sections from GIC-derived tumors derived from T25 or T76 cultures revealed that hippocampal sparing was a prominent feature present in several xenograft tumors. (A) Overview of the transplanted hemisphere shows a large bulky tumor in the transplanted hemisphere while the hippocampus (inlet) is remarkably free of tumor invasion. (B) Overview of both hemispheres at the level of the hippocampi. T25 displayed intense tumor cell infiltration of both grey (e.g. cortical layers of the convexity and thalamus) and white matter structures (e.g. corpus callosum, hippocampal commissure and alveus) surrounding the hippocampi compared to the cornu ammonis and dentate gyri which harbored relatively few tumor cells.
Abbreviations: CA, cornu ammoni; cc, corpus callosum; cg, cingulum; cp, cerebral peduncle; CPu, caudate putamen; CTX, cortex; DG, dentate gyrus; dhc, dorsal hippocampal commissure; HPC, hippocampus; Or, oriens layer of the hippocampus; TH, thalamus.
Supplementary Figure 1.
Immunolabeling of xenografted tumors
Cellular Distribution is Not Correlated With Migratory Distance
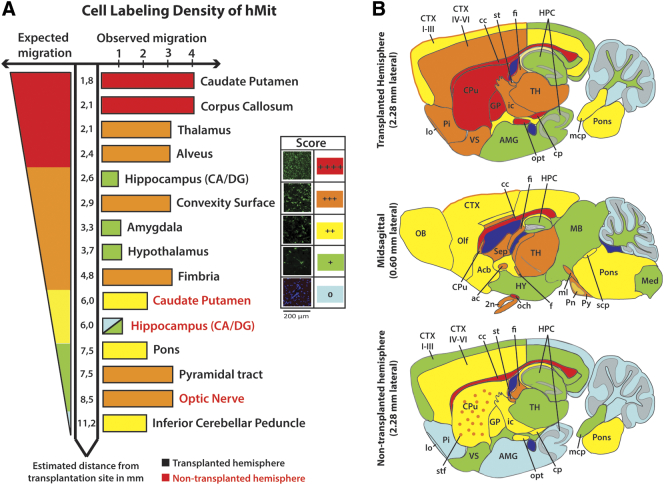
Our data indicated that, despite the widespread dispersion of tumor cells, glioma cell invasion followed specific patterns respecting certain intra-parenchymal barriers. Thus, we explored the correlation between cellular distribution by assessing hMit staining and migratory distance.
The transplantation site was considered the reference point for cell migration. Euclidian distances to various brain structures were calculated using a mouse brain atlas [29] based on observed migration paths (Suppl. File 3). Cellular distribution was not defined by migratory distances, as some regions relatively close to the tumor core (hypothalamus, hippocampus) were avoided by tumor cells, while more remote brain regions comprising both gray and white matter in the brain stem, were highly infiltrated (Figure 4A). All tumor cell migration observations are summarized in Figure 4B. Formal quantification of tumor cell burden was performed in selected regions and revealed significant enrichment of tumor cells in structures within the same distance range as the hippocampus in both hemispheres (Suppl. Figure 2).
Figure 4.
Tumor cell distribution in different brain regions.
(A) Schematic illustration comparing expected and observed labeling density, showing that the distribution of tumor cells was not related to the distance from the site of cell transplantation. Distribution of tumor cells was not related to the distance from the site of cell transplantation. (B) Schematic maps showing the brain-wide distribution of tumor cells derived from Table 1B.
Abbreviations: 2n, optic nerve; ac, anterior commissure; Acb, accumbens nuclei; AMG, amygdala; CA, cornu ammoni; cc, corpus callosum; CTX I-III, cortical layers 1–3; CTX IV-VI, cortical layers 4–6; DG, dentate gyrus; f, fornix; fi, fimbria; GP, globus pallidus; HY, hypothalamus; ic, internal capsule; lo, lateral olfactory tract; MB, midbrain; mcp, middle cerebellar peduncle; Med, medulla; ml, medial lemniscus; OB, olfactory bulb; och, optic chiasm; Olf, olfactory cortex; Pi, piriform cortex; Pn, pontine nuclei; scp, superior cerebellar peduncle; Sep, septum; st, stria terminalis; stf, striatal fibers; TH, thalamus; VS, ventral striatum.
The Hippocampal Formation is an Invasion-Privileged Brain Region in a Transgenic GBM Model
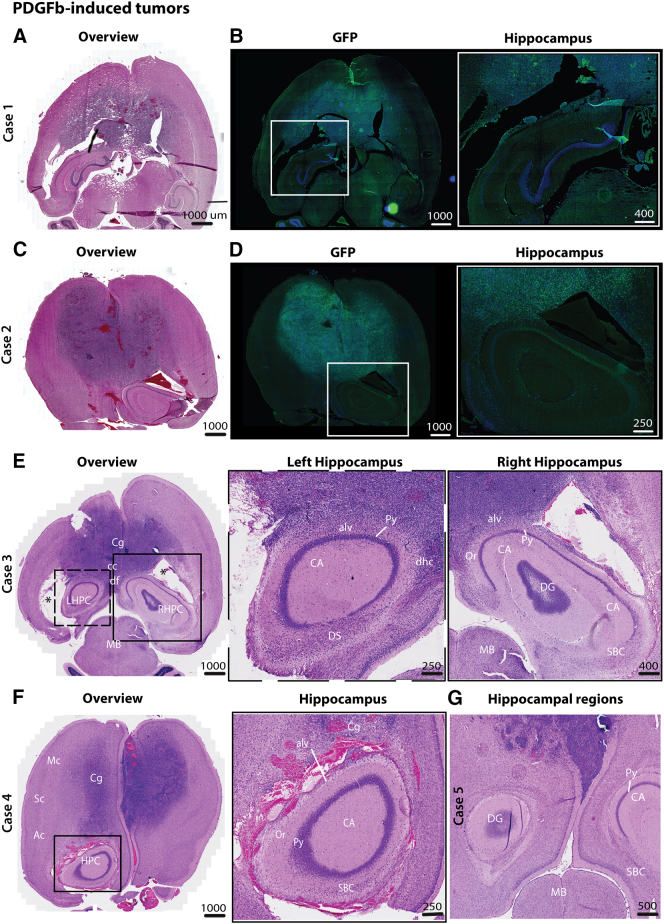
We also studied invasion patterns in the invasive GBM model using transgenic cdkn2a−/− mice injected with PDGF-β-GFP lentivirus. Despite extensive tumor cell infiltration in both hemispheres and the abundant presence of tumor cells in the para-hippocampal regions, very few tumor cells were observed within the hippocampus proper (Figure 5, Suppl. Figure 3).
Figure 5.
Hippocampal sparing was also present in an invasive transgenic mouse GBM-model.
(A-B, C-D) HE-stained and corresponding GFP-stained axial sections of PDGF-induced tumors in cdkn2a−/− mice showed highly invasive tumors that affected both hemispheres. Tumors demonstrated widespread dissemination of tumor cells in several neo- and mesocortical regions as well as the corpus callosum. Enlarged images of the hippocampal regions showed that tumor cells extensively infiltrated the para-hippocampal regions, including cortical regions and white matter, whereas the cornu ammonis and dentate gyri appeared to be spared (inlet). A high density of tumor cells was also observed in the subependymal lining as well as in the mesencephalon. (E-G) Similar invasion patterns were observed in the other cases, and in one case, intense neovascularization was seen outside the hippocampus.
Abbreviations: Ac, auditory cortex; alv, alveus; CA, cornu ammoni; cc, corpus callosum; Cg, cingulate cortex; df, dorsal fornix; DS, dorsal subiculum; LHPC, left hippocampus; MB, midbrain; Mc, motor cortex; Oc, olfactory cortex; Or, oriens layer of the hippocampus; Py, pyramidal layer of the hippocampus; RSG, retrosplenial granular cortex; RHPC, right hippocampus; SBC, subiculum; Sc, somatosensory cortex.
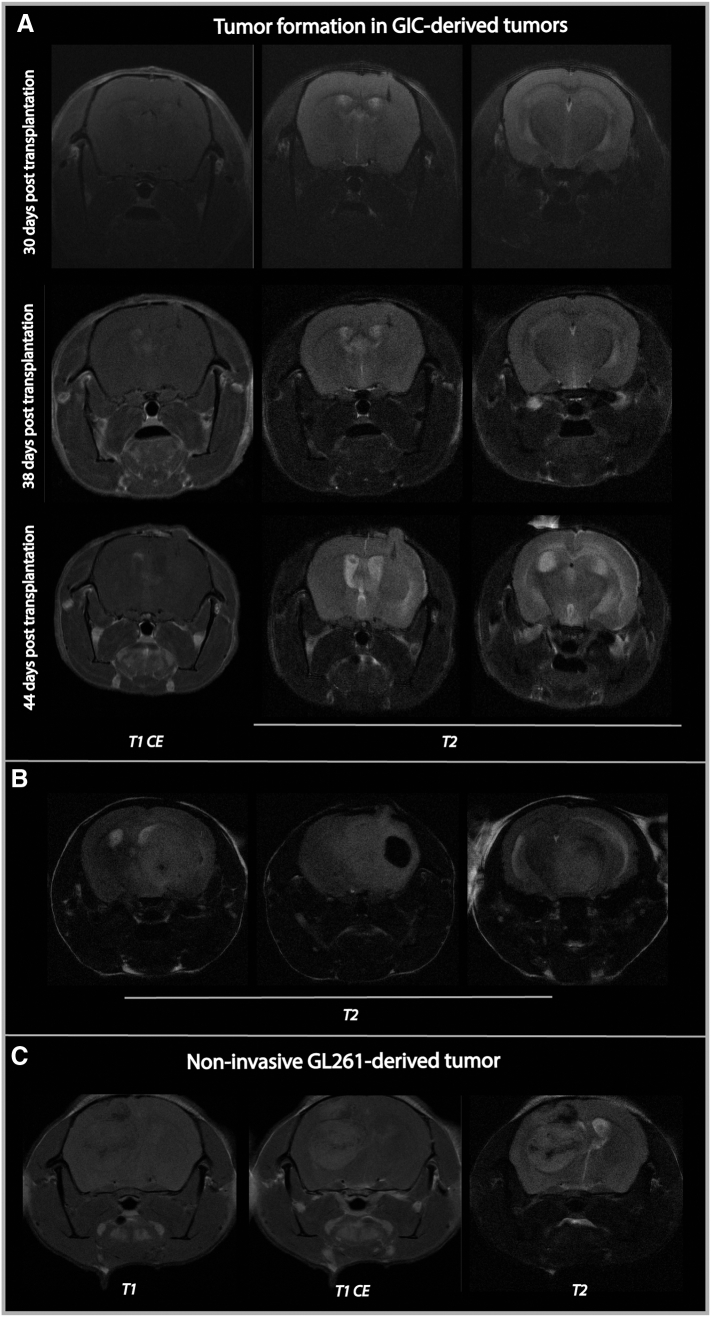
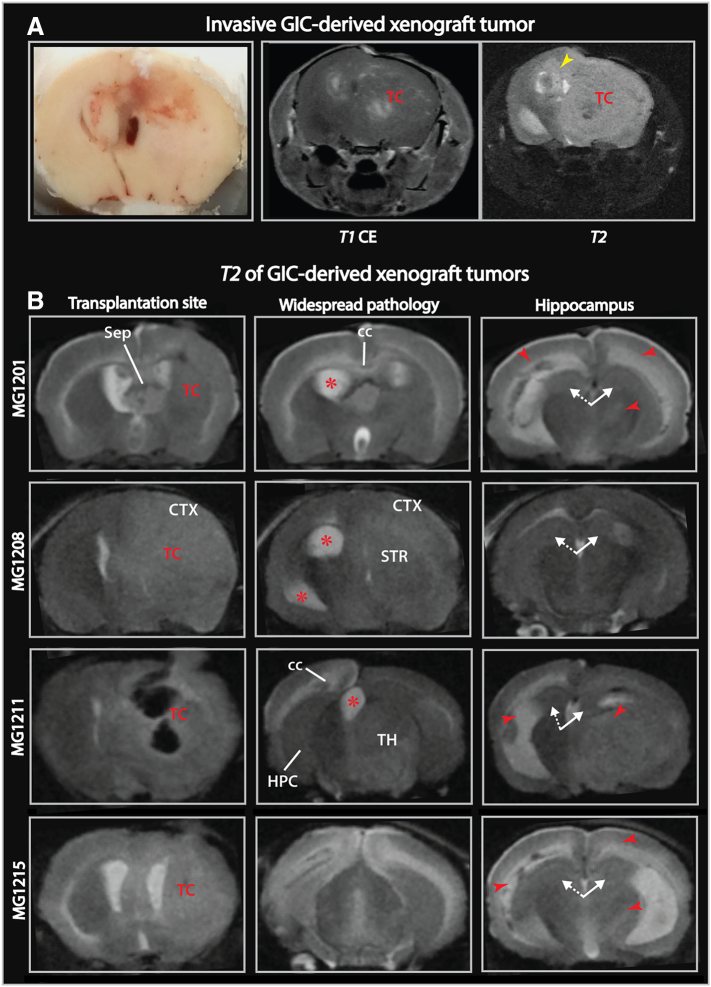
MR Imaging of GIC-Derived Xenograft Tumors Show Sparing of the Hippocampus
The first detectable signal abnormality in GIC-derived xenograft tumors (T08), was hyperintensity on T2-weighted images (Suppl. Figure 4A), whereas end-stage MR images typically displayed edema/tumor infiltration, midline shift and hydrocephalus (Figure 6). Scattered contrast-enhancement and spontaneous intratumoral hemorrhage were also observed in the transplanted hemisphere (Figure 6, Suppl. Figure 4B). These features are also commonly seen on clinical MR images of GBM patients [34]. Similar to our clinical findings, the hippocampus was relatively spared from T2 hyperintensity (Figure 6B). Such T2-signal abnormalities were otherwise consistently observed in mouse brains harboring these xenograft tumors. In contrast, mice transplanted with the GL261 glioma cell line showed well-demarcated lesions with limited T2-signal abnormality (Suppl. Figure 4C).
Supplementary Figure 4.
Additional MR images of xenograft tumors
Figure 6.
MR imaging of GIC-derived xenograft tumors.
(A) Photograph of a coronally sectioned brain with xenograft tumor and corresponding MR images displayed scattered contrast enhancement and widespread pathological T2-hyperintensity in the transplanted hemisphere. The T2-hyperintensity was found along the corpus callosum extending into the non-transplanted hemisphere (yellow arrowhead). (B) Coronal MR slices showing T2-signal abnormality (red arrowheads) involved several regions outside the striatal tumor core: thalamus, cortical areas of the convexity, parts of occipital lobes and white matter structures, whereas the hippocampi in the transplanted (white arrow with solid line) and non-transplanted hemispheres (white arrow with dashed line) were spared. Red asterisks mark dilated ventricles.
Abbreviations: cc, corpus callosum; HPC, hippocampus; Sep, septum; STR, striatum; TC, tumor core; TH, thalamus.
Growth Patterns of Malignant TLGs and the Relative Sparing of the Hippocampus
In the human brain, the temporal lobes are the most common sites for gliomas when adjustments are made for tissue volume. In fact, TLGs account for 30% of all glial tumors [35]. To evaluate whether these tumors are involved with the amygdala-hippocampal complex, we analyzed MR images of 70 cases of malignant TLGs. A detailed characterization of the clinical cohort is provided in Suppl. File 4.
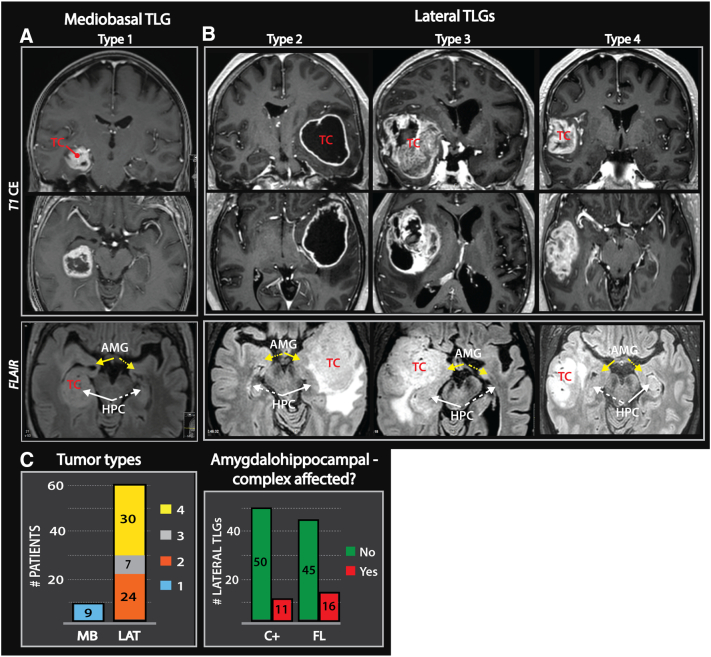
The first group, comprising nine of seventy cases, included tumors with contrast-enhancing lesions occupying the medio-basal temporal regions (type 1), such as the uncus, amygdalo-hippocampal complex and the parahippocampal- and/or fusiform gyrus (Figure7A). The second group, comprising the remaining sixty-one patients, included tumors primarily located lateral to the aforementioned structures. These tumors were considered “lateral” and further subdivided into insular-temporo-opercular (type 2), fronto-orbital-insular-temporopolar (type 3) and non-limbic lateral tumors (type 4) (Figure 7B).
Figure 7.
MR imaging of malignant temporal lobe gliomas.
(A) Contrast-enhanced T1 (T1CE)- weighted images of a mediobasal TLG (type 1). Pathological FLAIR- hyperintensity typically involved the amygdalo-hippocampal complex. (B) T1CE- weighted images of more lateral temporal tumors with the three subtypes: insular-temporo-opercular (type 2), orbitofrontal-insular-temporal pole (type 3) and non-limbic lateral (type 4) tumors. Pathologic FLAIR- hyperintensity occupied large parts of the temporal lobe in these tumors. (B-C) 74% of the examined TLGs were “lateral” and FLAIR-images revealed that the amygdalo-hippocampal complex (white and yellow arrow with solid lines) was frequently spared. Arrows with dashed lines point toward the amygdalo-hippocampal complex contralateral to the tumor core.
Abbreviations: AMG, amygdala; HPC, hippocampus; TC, tumor core.
Tumor extension beyond the contrast-enhancing lesion was based on FLAIR-weighted images. The analysis showed that the FLAIR signal abnormalities in most “lateral” tumors occupied large parts of the temporal lobe. Furthermore, the tumors frequently extended into the adjacent lobes. Indeed, ipsilateral ependymal enhancement was detected in 40% of the cases. Despite the growth of these tumors into various brain compartments, the amygdalo-hippocampal complex appeared to be spared of pathological FLAIR-signal in 74% of these cases (Figure 7, B and C and Suppl. File 5). In the medio-basal temporal tumors the pathological FLAIR-hyperintensity was either confined to the medial structures or spread within the limbic system to the insula (Figure 7A).
Discussion
In this study, we have demonstrated that xenografted patient-derived GICs from tumor-sphere cultures form highly invasive tumors that recapitulate the phenotype of malignant gliomas.
A key finding is the presence of a non-radial migratory pattern of tumor cells within the brain, contrasting with the notion of isotropic tumor propagation. Invasive tumor cells were found to migrate along several of the classical secondary structures of Scherer [10] and tended to accumulate extensively along WMTs located ipsi- and contralaterally to the primary tumor. Of particular interest is the enrichment of tumor cells in the optic and pontine white matter structures, despite their distance from the transplantation site. In pediatric patients, these two regions are well known for their preponderance to harbor diffuse gliomas [36]. We also found tumor cell accumulation in the periventricular areas, an observation that fits well with neurosurgical [37] and oncological [38] clinical data. In contrast, few tumor cells invaded the hippocampus and certain limbic structures. These findings were consistent in patient material as well as in two mouse models. The propensity of tumor cells to inhabit and avoid certain brain regions suggests the presence of molecular cues that guide tumor cell migration towards or away from certain brain regions. The clear differences in accumulation of tumor cells, not related to migratory distances from the tumor bulk, highlight this phenomenon. Knowledge about the tumor load in various brain regions may also have a potential implication for radiotherapy; regions/structures enriched with tumor cells could potentially receive higher dosage coverings, whereas the areas free of invasive tumor cells could be relatively spared [38].
As previously described, the migratory patterns of gliomas, at least in early stages, respect anatomical borders such as the cortex and pia [39]. Since the pial lining represents a barrier for further spread of tumor cells, surgical dissection techniques have been developed to leave the pia intact during resection of gliomas. A glioma subtype that appears to spread within the limbic system [8] was noted by Yasargil et al. and later described in detail by Schramm et al. [9]. Similar efforts were performed by Capizzano et al. [40] for diffuse gliomas; however, GBMs were not included in their tumor series. The mediobasal tumors were confined to the medial limbic structures, whereas the more heterogeneous “lateral” group comprised both para- and non-limbic structures. In the “lateral” tumor types, 75–80% did not show radiological signs of growth into the amygdalo-hippocampal complex and in the remaining cases this may represent a late event in tumor growth. Histological confirmation could not be done, as it was not amenable to perform a biopsy of a brain region without radiological signs of tumor involvement.
Among the hypotheses to explain such a specific invasive pattern, Engelhorn et al. suggested intracerebral segregated areas that could represent relative hindrances for invasive distribution or edema development [41]. Such borders could be a result of regional organization of blood vessels or white matter orientation secondary to brain development but are more likely defined through certain molecular cues such as netrins and bone morphogenic proteins [42]. Such morphogenetic fields are suggested to influence the loco-regional tumor spread in other solid cancers [43] and could explain the success of newer surgical approaches in resecting such anatomical regions instead of more localized tumor resection [44].
The spread of glioma cells through the brain has been suggested to follow the same embryonically defined regions [45] but supportive data has been scant. Interestingly, we have previously described a similar interaction between GICs and the microenvironment. In chicken embryos, GICs induced tumor formation if they were transplanted into the brain but failed to do so in the developing spinal cord [46]. Exploring this hypothesis in the human brain is, however, very demanding. Imaging modalities have limited ability to demonstrate invasive tumor cells and whole-brain preparations from patients cannot be examined at early stages of the disease. Previous autopsy studies have been limited by lack of methods for visualization of invasive tumor cells [3], [47].
A strength of our study is the use of two different mouse models including both patient-derived xenografts into SCID mice and cdkn2a−/− mice injected with PDGF-β-GFP lentivirus. In both models, we observed widespread invasion of tumor cells, as well as a relative paucity of tumor cell migration into the hippocampus. Other groups have also presented a general notion of a widespread invasive pattern and the described pattern of migration along WMTs. Though not described, sparing of the hippocampus can even be seen in these papers [19], [41]. A systematic description of the brain-wide invasion patterns has, however, not yet been done. The main weakness of the GIC-derived GBM model is the lack of immune cells, which could play a role in glial cell migration and tumor formation [48]. Additionally, it is possible that the target chosen for transplantation of cells could affect tumor cell migratory patterns. However, our finding of similar migratory patterns in the transgenic mouse model suggests that these factors, if anything, are of subordinate importance. The Cancer Genome Atlas (TCGA) research network classified GBMs into four molecular subtypes mainly based on the tumor's gene expression patterns [49]. As proneural expression patterns tend to be over-represented in GIC cultures and is also found in the PDGF induced tumors [22], [50], the presented data could be more relevant for this tumor subtype. The presented patient MRI findings, however, suggests a broader validity, as the clinical cohort most likely includes all glioblastoma subtypes.
Migratory patterns within the brain have been hypothesized to be dominated by invasion along vessels in the perivascular spaces [10]. This suggests a primary spread of tumor cells along a path of minimal physical resistance, supporting the importance of physical factors such as tumor pressure and mechanical strain in the invasive pattern of tumor cells. The data presented in this paper do not support these previous observations. The migratory patterns we observed were dominated by spread through the brain parenchyma with a predilection for WMTs. This fits well with our earlier finding that adult human neural stem cells migrate along WMTs rather than accumulating in the perivascular spaces [51]. There was, however, one striking difference. In this model, with ischemic injury to the hippocampus, the normal stem cells exhibited selective migration into the hippocampus [51]. This suggests that migration into the hippocampus is not primarily hampered by mechanical constraints but is rather regulated through molecular signaling. The extra cellular matrix composition is brain region-specific [52] and may explain the observed migration patterns. Further studies are needed to highlight this molecular machinery, and we are now actively pursuing this in ex vivo as well as in vivo models.
The following are the supplementary data related to this article.
The following are the supplementary data related to this article.
List of antibodies
MR classification criteria of malignant temporal lobe gliomas
Migration paths and distance calculations
Detailed characterization of the clinical cohort
Summary of the histological, molecular and MR imaging features of the clinical cohort
Quantification of tumor cells in GIC-derived xenografts
Quantification of tumor cells in PDGF- β induced tumors
Funding
This study was funded by Oslo University Hospital and the Norwegian Research Council through CAST-SFI. Histological images were acquired at the Norbrain Slide Scanning Facility at the Institute of Basic Medical Sciences, University of Oslo, a resource funded by the Research Council of Norway. Additional support was provided by funding through the Anni-Hofmann Stiftung (contribution by RG) and the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement No. 720270, HBP SGA1 (contributions by TBL).
Acknowledgments
Acknowledgements
We thank Grazyna Babinska, Emily Telmo, Hong Qu and Sharon C. Yates for expert technical assistance, Biljana Stangeland and Mrinal Joel for the GFP-transfection procedures and Jinan Behnan for the culturing of GL261 cells. We also thank Ivar Sjaastad and the preclinical MR imaging facility at the Institute for Experimental Medical Research; Sissel Reinlie and the Department of Neurosurgery at Oslo University Hospital; and Håvard Attramadal at the Institute for Surgical Research at Oslo University Hospital for support and collaboration. Donation of a PDGF-β expression plasmid by Professor Paolo Malatesta (Department of Experimental Medicine, University of Genoa, Italy) is gratefully acknowledged.
Conflicts of Interest
None.
References
- 1.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16:1575–1584. doi: 10.1093/neuonc/nou147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsukado Y, Maccarty CS, Kernohan JW. The growth of glioblastoma multiforme (astrocytomas, grades 3 and 4) in neurosurgical practice. J Neurosurg. 1961;18:636–644. doi: 10.3171/jns.1961.18.5.0636. [DOI] [PubMed] [Google Scholar]
- 4.Scherer H. The forms of growth in gliomas and their practical significance. Brain. 1940;63(1):1–34. [Google Scholar]
- 5.Giese A, Westphal M. Glioma invasion in the central nervous system. Neurosurgery. 1996;39:235–250. doi: 10.1097/00006123-199608000-00001. [discussion 250-232] [DOI] [PubMed] [Google Scholar]
- 6.Hamilton JD, Rapp M, Schneiderhan T, Sabel M, Hayman A, Scherer A, Kropil P, Budach W, Gerber P, Kretschmar U. Glioblastoma multiforme metastasis outside the CNS: three case reports and possible mechanisms of escape. J Clin Oncol. 2014;32:e80–e84. doi: 10.1200/JCO.2013.48.7546. [DOI] [PubMed] [Google Scholar]
- 7.Muller C, Holtschmidt J, Auer M, Heitzer E, Lamszus K, Schulte A, Matschke J, Langer-Freitag S, Gasch C, Stoupiec M. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med. 2014;6:247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 8.Yasargil MG, von Ammon K, Cavazos E, Doczi T, Reeves JD, Roth P. Tumours of the limbic and paralimbic systems. Acta Neurochir. 1992;118:40–52. doi: 10.1007/BF01400725. [DOI] [PubMed] [Google Scholar]
- 9.Schramm J, Aliashkevich AF. Surgery for temporal mediobasal tumors: experience based on a series of 235 patients. Neurosurgery. 2007;60:285–294. doi: 10.1227/01.NEU.0000249281.69384.D7. [discussion 294-285] [DOI] [PubMed] [Google Scholar]
- 10.Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15:455–465. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fayzullin A, Tuvnes FA, Skjellegrind HK, Behnan J, Mughal AA, Langmoen IA, Vik-Mo EO. Time-lapse phenotyping of invasive glioma cells ex vivo reveals subtype-specific movement patterns guided by tumor core signaling. Exp Cell Res. 2016;349(2):199–213. doi: 10.1016/j.yexcr.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Glass R, Synowitz M. CNS macrophages and peripheral myeloid cells in brain tumours. Acta Neuropathol. 2014;128:347–362. doi: 10.1007/s00401-014-1274-2. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein JJ, Goldberg WJ, Laws ER., Jr. Immunohistochemistry of human malignant astrocytoma cells xenografted to rat brain: apolipoprotein E. Neurosurgery. 1989;24:541–546. doi: 10.1227/00006123-198904000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Horten BC, Basler GA, Shapiro WR. Xenograft of human malignant glial tumors into brains of nude mice. A histopatholgical study. J Neuropathol Exp Neurol. 1981;40:493–511. doi: 10.1097/00005072-198109000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Laws ER, Jr., Goldberg WJ, Bernstein JJ. Migration of human malignant astrocytoma cells in the mammalian brain: Scherer revisited. Int J Dev Neurosci. 1993;11:691–697. doi: 10.1016/0736-5748(93)90056-j. [DOI] [PubMed] [Google Scholar]
- 17.Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PO, Miletic H, Thorsen F, Bjerkvig R. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol. 2012;14:979–993. doi: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Wakimoto H, Mohapatra G, Kanai R, Curry WT, Jr., Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 2012;14:132–144. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vik-Mo EO, Sandberg C, Olstorn H, Varghese M, Brandal P, Ramm-Pettersen J. Brain tumor stem cells maintain overall phenotype and tumorigenicity after in vitro culturing in serum-free conditions. Neuro Oncol. 2010;12 doi: 10.1093/neuonc/noq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mughal AA, Grieg Z, Skjellegrind H, Fayzullin A, Lamkhannat M, Joel M, Ahmed MS, Murrell W, Vik-Mo EO, Langmoen IA. Knockdown of NAT12/NAA30 reduces tumorigenic features of glioblastoma-initiating cells. Mol Cancer. 2015;14:160. doi: 10.1186/s12943-015-0432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stangeland B, Mughal AA, Grieg Z, Sandberg CJ, Joel M, Nygard S, Meling T, Murrell W, Vik Mo EO, Langmoen IA. Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget. 2015;6:26192–26215. doi: 10.18632/oncotarget.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joel M, Mughal AA, Grieg Z, Murrell W, Palmero S, Mikkelsen B. Targeting PBK/TOPK decreases growth and survival of glioma initiating cells in vitro and attenuates tumor growth in vivo. Mol Cancer. 2015;14 doi: 10.1186/s12943-015-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behnan J, Isakson P, Joel M, Cilio C, Langmoen IA, Vik-Mo EO, Badn W. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells. 2014;32:1110–1123. doi: 10.1002/stem.1614. [DOI] [PubMed] [Google Scholar]
- 25.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calzolari F, Appolloni I, Tutucci E, Caviglia S, Terrile M, Corte G, Malatesta P. Tumor progression and oncogene addiction in a PDGF-B-induced model of gliomagenesis. Neoplasia (New York, NY) 2008;10:1373–1382. doi: 10.1593/neo.08814. [following 1382] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walzlein JH, Synowitz M, Engels B, Markovic DS, Gabrusiewicz K, Nikolaev E, Yoshikawa K, Kaminska B, Kempermann G, Uckert W. The antitumorigenic response of neural precursors depends on subventricular proliferation and age. Stem Cells. 2008;26:2945–2954. doi: 10.1634/stemcells.2008-0307. [DOI] [PubMed] [Google Scholar]
- 28.Yetman MJ, Lillehaug S, Bjaalie JG, Leergaard TB, Jankowsky JL. Transgene expression in the Nop-tTA driver line is not inherently restricted to the entorhinal cortex. Brain Struct Funct. 2015;221(4):2231–2249. doi: 10.1007/s00429-015-1040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franklin KBJ, Paxinos G. Third Edition. Acedemic Press; 2007. The Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- 30.Yates SCPM, Coello C, Kreshuk A, Leergaard TB, Bjaalie JG. 2017. Workflow for automated quantification and spatial analysis of Alzheimer's plaque labeling in microscopic rodent brain sections Program no 4415 2017 Neuroscience Meeting Planner Washington, DC: Society for Neuroscience; 2017 Online. [Google Scholar]
- 31.Sommer C, Straehle C, Köthe U, Hamprecht FA. 2011. ilastik: Interactive Learning and Segmentation Toolkit Eighth IEEE International Symposium on Biomedical Imaging (ISBI) Proceedings; pp. 230–233. [Google Scholar]
- 32.Barajas RF, Jr., Phillips JJ, Parvataneni R, Molinaro A, Essock-Burns E, Bourne G, Parsa AT, Aghi MK, McDermott MW, Berger MS. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro Oncol. 2012;14:942–954. doi: 10.1093/neuonc/nos128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(TCIA) TCIA . 2014–2016. VASARI MR feature key. [Google Scholar]
- 34.Rees JH, Smirniotopoulos JG, Jones RV, Wong K. Glioblastoma multiforme: radiologic-pathologic correlation. Radiographics. 1996;16:1413–1438. doi: 10.1148/radiographics.16.6.8946545. [quiz 1462-1413] [DOI] [PubMed] [Google Scholar]
- 35.Larjavaara S, Mantyla R, Salminen T, Haapasalo H, Raitanen J, Jaaskelainen J, Auvinen A. Incidence of gliomas by anatomic location. Neuro Oncol. 2007;9:319–325. doi: 10.1215/15228517-2007-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 37.Moon JH, Kim SH, Shim JK, Roh TH, Sung KS, Lee JH, Park J, Choi J, Kim EH, Kim SH. Histopathological implications of ventricle wall 5-aminolevulinic acid-induced fluorescence in the absence of tumor involvement on magnetic resonance images. Oncol Rep. 2016;36:837–844. doi: 10.3892/or.2016.4881. [DOI] [PubMed] [Google Scholar]
- 38.Nourallah B, Digpal R, Jena R, Watts C. Irradiating the subventricular zone in glioblastoma patients: is there a case for a clinical trial? Clin Oncol (R Coll Radiol) 2017;29:26–33. doi: 10.1016/j.clon.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Scherer H. Structural development in gliomas. Am J Cancer. 1938;34:333–351. [Google Scholar]
- 40.Capizzano AA, Kirby P, Moritani T. Limbic tumors of the temporal lobe: radiologic-pathologic correlation. Clin Neuroradiol. 2015;25:127–135. doi: 10.1007/s00062-014-0287-5. [DOI] [PubMed] [Google Scholar]
- 41.Engelhorn T, Savaskan NE, Schwarz MA, Kreutzer J, Meyer EP, Hahnen E, Ganslandt O, Dorfler A, Nimsky C, Buchfelder M. Cellular characterization of the peritumoral edema zone in malignant brain tumors. Cancer Sci. 2009;100:1856–1862. doi: 10.1111/j.1349-7006.2009.01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwadate Y, Fukuda K, Matsutani T, Saeki N. Intrinsic protective mechanisms of the neuron-glia network against glioma invasion. J Clin Neurosci. 2016;26:19–25. doi: 10.1016/j.jocn.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Hockel M. Morphogenetic fields of embryonic development in locoregional cancer spread. Lancet Oncol. 2015;16:e148–151. doi: 10.1016/S1470-2045(14)71028-9. [DOI] [PubMed] [Google Scholar]
- 44.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 45.Kalani MY, Kalani MA, Gwinn R, Keogh B, Tse VC. Embryological development of the human insula and its implications for the spread and resection of insular gliomas. Neurosurg Focus. 2009;27:E2. doi: 10.3171/2009.5.FOCUS0997. [DOI] [PubMed] [Google Scholar]
- 46.Joel M, Sandberg CJ, Boulland JL, Vik-Mo EO, Langmoen IA, Glover JC. Inhibition of tumor formation and redirected differentiation of glioblastoma cells in a xenotypic embryonic environment. Dev Dyn. 2013;242:1078–1093. doi: 10.1002/dvdy.24001. [DOI] [PubMed] [Google Scholar]
- 47.Maccarthy CS. Surgical treatment of gliomas of the brain. J Int Coll Surg. 1955;23:290–297. [PubMed] [Google Scholar]
- 48.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19:20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP. An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17:98. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 51.Olstorn H, Varghese M, Murrell W, Moe MC, Langmoen IA. Predifferentiated brain-derived adult human progenitor cells migrate toward ischemia after transplantation to the adult rat brain. Neurosurgery. 2011;68:213–222. doi: 10.1227/NEU.0b013e3181fd2c11. [discussion 222] [DOI] [PubMed] [Google Scholar]
- 52.Dauth S, Grevesse T, Pantazopoulos H, Campbell PH, Maoz BM, Berretta S, Parker KK. Extracellular matrix protein expression is brain region dependent. J Comp Neurol. 2016;524:1309–1336. doi: 10.1002/cne.23965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of antibodies
MR classification criteria of malignant temporal lobe gliomas
Migration paths and distance calculations
Detailed characterization of the clinical cohort
Summary of the histological, molecular and MR imaging features of the clinical cohort
Quantification of tumor cells in GIC-derived xenografts
Quantification of tumor cells in PDGF- β induced tumors