Abstract
In multiple myeloma (MM), abnormal plasma cells accumulate and proliferate in the bone marrow. Recently, we observed that Runx2, a bone-specific transcription factor, is highly expressed in MM cells and is a major driver of MM progression in bone. The primary goal of the present study was to identify Runx2-targeting miRNAs that can reduce tumor growth. Expression analysis of a panel of miRNAs in MM patient specimens, compared with healthy control specimens, revealed that metastatic MM cells express low levels of miR-342 and miR-363 but high levels of Runx2. Reconstituting MM cells (CAG) with miR-342 and miR-363 reduced the abundance of Runx2 and the expression of metastasis-promoting Runx2 target genes RANKL and DKK1, and suppressed Runx2-downstream signaling pathways Akt/β-catenin/survivin, which are required for MM tumor progression. Intravenous injection of MM cells (5TGM1), stably overexpressing miR-342 and miR-363 alone or together, into syngenic C57Bl/KaLwRij mice resulted in a significant suppression of 5TGM1 cell growth, decreased osteoclasts and increased osteoblasts, and increased anti-tumor immunity in the bone marrow, compared to mice injected with 5TGM1 cells expressing a miR-Scramble control. In summary, these results demonstrate that enhanced expression of miR-342 and miR-363 in MM cells inhibits Runx2 expression and MM growth, decreases osteolysis, and enhances anti-tumor immunity. Thus, restoring the function of Runx2-targeting by miR-342 and miR-363 in MM cells may afford a therapeutic benefit by preventing MM progression.
Implications
miR-342 and miR-363-mediated downregulation of Runx2 expression in MM cells prevents MM progression.
Introduction
Multiple myeloma (MM) is a hematological malignancy characterized by high infiltration and accumulation of malignant plasma cells in the bone marrow (1-3). Bone disease occurs in approximately 90% of patients with MM and is the main cause of patient mortality (4). The consequences of MM progression in bone can be devastating for patients and include osteolytic bone lesions, hypercalcemia, renal insufficiency, and spinal cord and nerve-compression syndromes (5). However, the aggressive mechanisms driving MM progression in bone remain unclear. Runx2 is a bone-specific transcription factor that promotes osteoblastogenesis and bone formation (6, 7). Runx2 is also expressed in many cancer cells, including breast and prostate cancers, and has been shown to promote bone metastasis (8, 9). Recently, our studies have demonstrated that Runx2 expression is significantly higher in primary MM cells than in the plasma cells of bone marrow from patients with monoclonal gammopathy of undetermined significance or in normal plasma cells from healthy bone marrow donors (10). Inhibition of Runx2 expression in MM cell lines reduces tumor growth and prevents MM dissemination to bone, thus providing a basis for Runx2 as a potential therapeutic target against MM progression and dissemination (10). Unfortunately, transcription factors are often considered to be non-druggable, as targeting these intracellular proteins lacking enzymatic activity remains challenging (11, 12). Nevertheless, a potential novel avenue to target nuclear factors might be provided by miRNA technology.
MiRNAs are a class of small, non-coding RNAs that function by binding to the 3′ untranslated region (3′ UTR) of target mRNAs and repressing mRNA expression by interfering with the mRNA stability and/or by blocking protein translation (13). MiRNAs have critical roles in processes such as cell proliferation, differentiation, and survival and also during normal development, homeostasis, and disease (14, 15). Aberrant miRNA expression is frequently observed in various human tumors, including MM, indicative of critical roles in tumorigenesis (16, 17). Recently, it was demonstrated that miRNAs targeting osteoclast function can inhibit bone metastatic disease (18, 19). In addition, in vivo delivery of miRNAs or miRNA antagonists can prevent cancer-induced bone diseases (20). Some reports indicate that miRNAs can inhibit the growth of various tumors by directly suppressing Runx2 function (21). However, miRNA-mediated regulation of Runx2 in the context of MM progression has never been studied. Thus, miRNAs could be used as therapeutic agents, and identifying specific miRNAs that suppress Runx2 would provide a novel approach for the treatment of MM.
In this study, we show that reduced expression of miR-342 and miR-363 is responsible for the upregulation of Runx2 in MM progression. We demonstrate the effects of miR-342 and miR-363 on MM inhibition through direct downregulation of Runx2 using in vitro and in vivo approaches. Importantly, our data provides strong evidence that targeting Runx2 by a miRNA-based approach can be used to suppress MM progression.
Materials and Methods
Cell Culture
The mouse MM 5TGM1 cell line expressing luciferase (Luc) was a kind gift from Dr. Fenghuang Zhan (University of Iowa Health Care, Iowa), the human MM cell line CAG was from Dr. Sanderson's laboratory, and the human MM cell line U266 was purchased from ATCC (Manassas, VA). We have stably transfected Runx2 shRNA into 5TGM1 cells and the methods were published previously (10). Cells were cultured in RPMI 1640 with 10% fetal bovine serum, 2 mmol/L L-glutamine, 1 U/mL penicillin, and 10 mg/mL streptomycin at 37°C and 5% CO2. Cells were established as free of mycoplasma and bacteria following instructions of ATCC. Cells were thawed from liquid nitrogen and media changed after 24 h, followed by culturing for 48-96 h before the cells were used for actual experiments. Cells were not passaged more than 2 times after initial thawing. Human MM cells CAG and U266 and mouse MM cell line 5TGM1 were authenticated routinely and before the in vivo experiments by measuring the levels of kappa, lambda and IgG2bκ (soluble markers of CAG, U266 and 5TGM1 cells respectively) in the conditioned medium of cell culture by ELISA and the levels of CD138 expression (surface marker of MM cell) by flow cytometry.
Transfection and transduction
Double-stranded RNA oligos representing mature sequences that mimic endogenous miR-342 and miR-363 and a non-specific (NS) miRNA negative control (obtained from IDT, Coralville, Iowa) were electrotransfected at concentrations of 50 or 100 nm into CAG human myeloma cells using program EO-117 on the 4D-Nucleofector system and the Amaxa SF cell line 4D-Nucleofector X Kit (Lonza). Cells were harvested 48 h after transfection for protein and mRNA analysis. For miRNA lentiviral clone transfection, miRNA Lenti-miR vectors (System Biosciences) were used to produce vectors encoding miR-Scramble control or mature miR-342, miR-363, or miR-342+363 under control of the CMV promoter with a GFP reporter to allow monitoring of miRNA-expressing cells. The viruses were packaged in HEK293T cells (ATCC) (22). 5TGM1 cells were infected at 70% confluency for 48 h with lentiviral particles and polybrene (8 μg/mL). A second infection was repeated for another 48 h. Cells were selected by culturing for 14 days in puromycin (8 μg/mL). Protein and mRNA levels were determined in control and miRNA-overexpressing cells by Western blotting and reverse transcriptase quantitative PCR (RT-qPCR).
RNA isolation and real-time RT-qPCR
Total RNA was isolated using RNeasy Mini kits (Qiagen Inc) according to the manufacturer's specifications. cDNA was synthesized using MMLV reverse transcriptase (Clontech). For miRNA detection, poly(A) tailing was performed using a poly(A) polymerase (Pol) and reverse transcription was carried out with reverse transcriptase (System Biosciences, Mountain View, CA) according to the manufacturer's instructions. Gene expression was determined by real-time RT-qPCR using Fast SYBR Green Master Mix (Applied Biosystems, Inc.) and gene-specific primers in an ABI StepOnePlus fast thermocycler. For each gene, expression levels were normalized to 28S ribosomal RNA (rRNA) or U6 RNA expression. Experiments were performed in triplicate, and results are given as mean values ± SEM. Nucleotide sequences of primers are provided in Supplementary Table 1.
Western blot analysis
Equal amounts of protein (50 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4% to 12% gradient gels (Bio-Rad) and transferred to nitrocellulose membranes (Schleicher and Schuell). Transferred proteins were probed with appropriate antibodies (Supplementary Table 2) and detected using enhanced chemiluminescence reagents (Amersham Biosciences). Band intensity was quantified by NIH Image J software version 1.45 (rsb.info.nih.gov/ij).
Gene expression profiles
Gene expression profile (ID: GSE17498) (23) was downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo). GSE17498, based on the platform of GPL8227 Agilent-019118 Human miRNA Microarray 2.0 G4470B (miRNA ID version), included a collection of 43 (normal: 3 and MM patients: 40) samples of purified plasma cells from bone marrow specimens for which miRNA profiles were compared.
Cell proliferation assay
The number of viable cells after miRNA overexpression was evaluated by a colorimetric MTT assay. CAG and 5TGM1 cells (2×104 cells per well) were seeded separately in 96-well plates in triplicate. After 72 h, cells were treated with 0.5 mg/mL MTT for 2 h. The optical density (OD) was then measured at 540 nm. The mean value and standard deviation (SD) were calculated from triplicate experiments.
Cell migration assay
CAG cells were transfected with control miRNA or miR-342 or miR-363 mimics as described above; 24 h later, the cells were counted and resuspended in RPMI 1640 serum-freemedium. Cell migration assays were conducted in 24-well, 15-mm internal-diameter multiwell notched tissue culture plates (Corning Inc, New York). Cells (2×105/500 μL) in RPMI 1640 serum-free medium were added into inserts (8-μm pore size), in triplicate for each group, and allowed to migrate toward complete medium (RPMI 1640 with 10% fetal bovine serum) in the bottom wells at 37°C and 5% CO2 for 24 h. Cells migrated into bottom wells were enumerated after 24 h, in triplicate, using a Z1 Dual Threshold Coulter Counter (Beckman Coulter).
Mouse models
C57BL/KalwRij mice were purchased from Harlan Laboratories, Inc. The 5TGM1 model is a syngenic model of murine myeloma that faithfully replicates the human myeloma disease (24-26). 5TGM1-luc cells overexpressing miR-Scramble control, miR-342-, miR-363-, or miR-342+363 were intravenously (IV) injected into 6-week-old C57BL/KalwRij mice (males and females) via the tail vein. Serum was collected biweekly for measurement of IgG2bκ levels (a soluble marker of 5TGM1 cells), and bioluminescent imaging was performed weekly to monitor tumor homing and growth. At the end of the experiment, femurs and tibias were harvested, fixed, decalcified, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E) or with a tartrate-resistant acid phosphatase (TRAP) staining kit. All animal studies were performed in accordance with UAB and National Institutes of Health (NIH) guidelines after institutional review and approval.
TRAP staining
TRAP staining was used to detect osteoclasts in bone sections from mice IV injected with tumor cells. Paraffin-embedded murine bone sections were deparaffinized in xylene and then rehydrated in a descending alcohol series (100%, 90%, 70%, and 50% alcohol). Sections were then washed in distilled water. Bone sections were stained using a TRAP staining kit (1). Osteoclast number was determined by counting TRAP-positive osteoclasts adjacent to the bone surface as described previously (27).
ELISA
The levels of serum IgG2bκ (a soluble marker of 5TGM1 cells) in mice were measured using a mouse IgG2bκ ELISA Quantitation kit (Bethyl Laboratories, Montgomery, TX) according to the manufacturer's instructions. All steps were performed at room temperature and the reaction product was quantified using a spectrophotometer at 450 nm, with each sample measured in duplicate.
Flow cytometry
Bone marrow was flushed out from tibia and femurs of each mouse in 1 mL of PBS, followed by centrifugation of PBS to pellet bone marrow cells. Red blood cells from the harvested bone marrow cells were lysed using ACK lysis buffer (Lonza), and bone marrow cells without red blood cells were then stained for 30 min at 4°C in the dark with anti-mouse B220-PECy7, IgM-FITC, and CD23-PE antibodies to detect regulatory B cells (Bregs); anti-mouse CD3-FITC, CD4-PerCP Cy5.5, CD127-BV510, and CD25-BV650 antibodies to detect regulatory T cells (Tregs); anti-mouse CD11c-AF700, CD11b-FITC, and B220-AF647 antibodies to detect plasmacytoid dendritic cells (pDCs); anti-mouse CD3-FITC, CD8-BV605, and PD-1-BV421 to detect CD8+ specific PD-1; anti-mouse CD3-FITC, CD8-BV605, and Tim-3-PE to detect CD8+ specific Tim-3; and anti-mouse CD3-FITC and IFN-γ-APC to detect CD3+ IFN-γ cells. Cells were then washed twice with FACS buffer (PBS with 1% bovine serum albumin and 0.1% sodium azide), resuspended in 1× PBS, and analyzed by LSRII (BD Biosciences) and FlowJo software.
Runx2 3′UTR-reporter constructs and transfection
The Runx2 3′UTR (80-100 bp) fragment containing sites for miR-342 and miR-363 binding (seed sequences) was amplified using the In-fusion HD Cloning Kit (Clontech, Mountain View, CA) with overhanging 5′ and 3′ primers homologous to the target, pMIR REPORT. DNA fragments were inserted into the SpeI site of the pMIR REPORT-Luc mRNA expression plasmid (Cat # AM5795, Thermo Fisher Scientific, MA) using the In-fusion HD Cloning Kit manufacturer's protocol to generate the Runx2 3′UTR-Luc reporter plasmid. Also, mutations were introduced into miR-342 and miR-363 binding sites in the Runx2 3′UTR and sub-cloned as described above into pMIR-REPORT-Luc plasmids to generate Runx2 3′UTR-Luc reporter plasmids with mutated miR-342 and miR-363 binding sites. Wild-type (WT) and mutant (MT) sequences of each 3′UTR are shown in Supplementary Table 3. Luc assays were performed with HEK293 cells co-transfected with the specified miRNA mimics (100 nM each) or NS control miRNA (100 nM) and the specified Runx2 3′UTR-Luc reporter (200 ng) plasmid using 6 μL of Fugene transfection reagent (Promega, Madison, WI) in triplicate in six independent experiments. Transfection with Renilla Luc plasmid (Promega, Madison, WI) was used to normalize the relative Luc values. Relative Luc activity (firefly Luc activity/Renilla Luc activity) was expressed in relative luminescence units and plotted.
Results
miR-342 and miR-363 expression negatively correlates with Runx2 expression in MM cells
To seek the miRNAs that inhibit Runx2 expression in MM cells, a panel of miRNAs that were differentially expressed between plasma cells from bone marrow specimens of patients with MM and those of normal donors was examined by downloading the miRNA microarray dataset GSE17498 from the Gene Expression Omnibus database. Two miRNAs, miR-342 and miR-363, were found significantly downregulated in MM patient samples compared with normal donor samples (Fig. 1A). Targetscan analysis revealed binding sites for miR-342 and miR-363 at the 3′UTR region of Runx2 (Fig. 1B), suggesting Runx2 regulatory roles for miR-342 and miR-363. This inverse correlation between miR-342/miR-363 and Runx2 expression was also confirmed in CAG and U266 human MM cell lines (Figs. 1C and D).
Figure 1. Runx2 expression in patients with MM and MM cell lines inversely correlates with Runx2-targeting miRNAs.
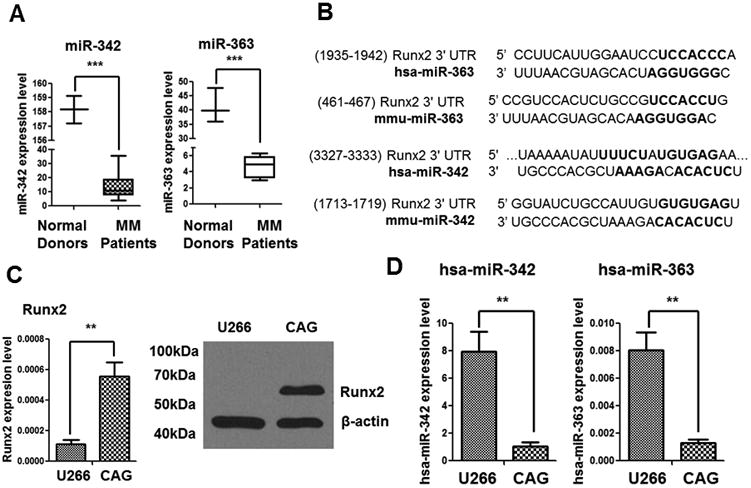
(A) Expression of miR-342 and miR-363 is significantly downregulated in plasma cells of bone marrow specimens from patients with MM. The data were derived from three normal donors and 40 MM patients at diagnosis [data provided by Lionetti and co-workers (ref. accession number GSE17498 to GEO datasets)]. The bars and box represent the Tukey distribution from each group. (B) TargetScan analysis revealed the binding sites of miR-342 and miR-363 on the Runx2 3′UTR. Complementary miRNA seed sequence binding sites are shown in bold black (hsa: human; mmu: mouse). (C)Relative expression of Runx2 in human MM cell lines U266 and CAG was measured using RT-qPCR (left) and Western blotting (right). (D) Relative expression of miR-342 and miR-363 in U266 and CAG cells as measured by RT-qPCR. The P values were obtained with two-tailed Student t test (**P<0.005; ***P<0.0001).
miR-342 and miR-363 mimics markedly suppress Runx2, along with cell proliferation and migration, in CAG MM cells
Based on the relatively high abundance of miR-342 and miR-363 in the plasma cells of normal donors, and the strong downregulation of these miRNAs in the plasma cells of patients with MM and in MM cell lines (Figs. 1A, 1D), we hypothesized that reconstitution of miR-342 and miR-363 in MM cells could reverse Runx2-enhanced MM progression. To test this hypothesis, we transiently transfected miR-342 and miR-363 mimics into CAG MM cells, both individually and in combination, and examined the levels of Runx2 expression by RT-qPCR and Western blot. The results showed a decrease in Runx2 expression upon miRNA mimic transfection (Figs. 2A and B). Interestingly, the miR-342+363 mimic was more effective than either individual mimic was in suppressing Runx2 expression. However, despite the strikingly decreased expression of Runx2 protein in cells co-expressing miR-342 and miR-363 mimics, Runx2 mRNA expression decreased by less than two-fold compared with that in controls, suggesting that Runx2 protein is regulated through both transcription-dependent and alternate mechanisms, such as control of protein translation.
Figure 2. miR-342 and miR-363 suppress Runx2 expression and the proliferation and migration of MM cells in vitro.

(A, B) Relative expression of Runx2 in CAG cells transfected with NS-miR-control or miR-342, miR-363, or miR-342+363 mimics was assessed by Western blotting (A) and RT-qPCR (B). (C, D) Non-specific (NS) miR-control or miR-342 (C) or miR-363 mimics (D) were co-transfected with either WT or mutant Runx2 3′UTR-Luc reporter plasmids into HEK293 cells (MT: mutant). Firefly Luc activity was normalized to co-transfected Renilla Luc activity and presented in relative luminescence units. (E) Cell viability was measured by MTT assay in CAG cells transfected with miR-Scramble control or miR-342, miR-363, or miR-342+363 mimics. (F) Transwell migration assays were conducted to assess cell migration in CAG cells transfected with miR-Scramble control or miR-342, miR-363, or miR-342+363 mimics. The P values were obtained by one-way ANOVA followed by Tukey-Kramer post hoc test (ns: non-significant; *P<0.05; **P<0.005).
Further, to examine the specificity and efficacy of miR-342 and miR-363 on Runx2 activity, we cloned an 80- to 100-bp fragment of the Runx2 3′UTR containing binding sites for miR-342 and miR-363 into the 3′UTR of a Luc reporter gene to create the Runx2 3′UTR-Luc plasmid. Also, we mutated the miR-342 and miR-363 binding sites in the Runx2 3′UTR and cloned the sequence into the 3′UTR of the Luc reporter gene to create a Runx2 3′UTR-Luc plasmid with mutated binding sites for miR-342 and miR-363. In HEK293 cells, ectopic expression of miR-342 or miR-363 mimics substantially repressed Luc activity when co-transfected with the WT Runx2 3′UTR-Luc plasmid but not when co-transfected with the mutant Runx2 3′UTR-Luc plasmid (Fig. 2C and D), suggesting that miR-342 and miR-363 bind to and inhibit the activity of the Runx2.
We then assessed the MM cell proliferation and migration abilities after transfection of miR-342 and miR-363 mimics. MTT and transwell migration assays showed miR-342, miR-363, and miR-342+363 mimics markedly suppress both proliferation and migration of CAG cells (Fig. 2E and F). We then assessed the expression of DKK1 and RANKL, which promote cancer cell migration and cause osteolytic bone lesions (28, 29), and found decreased expression of DKK1 and RANKL in CAG cells treated with miR-342, miR-363, or miR-342+363 mimics (Supplementary Figure. S1). Together, these data reveal that miR-342 and miR-363 suppress Runx2 expression and function, the proliferation of CAG cells, and the expression of molecules that promote cancer cell migration and thereby inhibit migration of CAG cells. Our results consistently demonstrate that co-transfection of miR-342 and miR-363 mimics had a more robust effect than the individual miRNA mimics had.
miR-342 and miR-363 downregulate Akt/β-catenin/survivin signaling in MM cells
We have previously demonstrated that MM cell-derived Runx2 promotes tumor progression through upregulation of Akt/β-catenin/survivin signaling (10). Therefore, we examined whether miR-342 and miR-363 downregulate these Runx2 downstream signaling pathways in CAG cells. Western blot results showed suppressed expression of phosphorylated Akt (p-Akt) and survivin upon miR-342 and miR-363 mimic transfection (Fig. 3A). Real-time PCR results showed β-catenin and GSK-3β to be downregulated upon miR-342+363 mimic transfection (Fig. 3B). These results demonstrate the effectiveness of targeting the abnormally and highly expressed transcription factor Runx2 by miR-342 and miR-363 mimics. In combination, miR-342 and miR-363 had a synergistic effect on downregulating Runx2 and its target genes and signaling, which was likely the cause of reduced MM proliferation and migration.
Figure 3. Akt/β-catenin/survivin signaling pathways downregulated by miR-342 and miR-363.
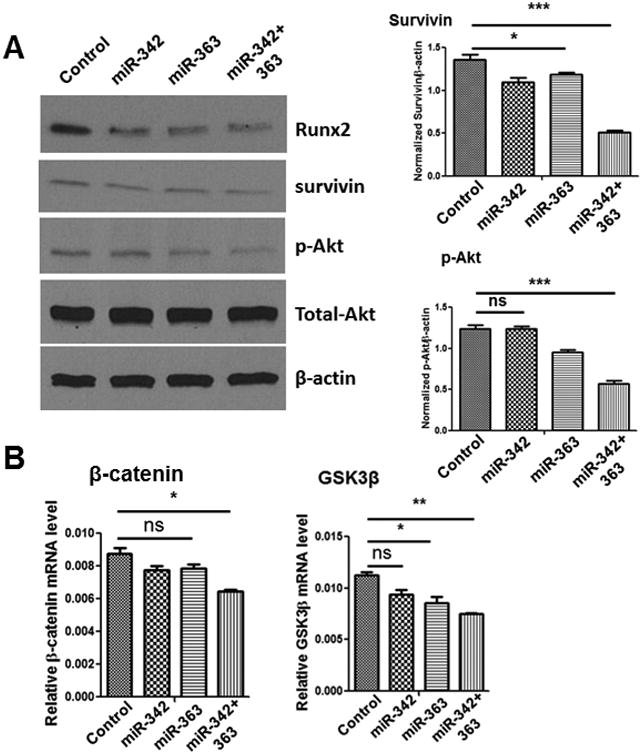
(A) Representative Western blot showing the relative expression of Runx2, survivin, and p-Akt in CAG cells transfected with miR-Scramble control, miR-342, miR-363, or miR-342+363 (left). Survivin and p-Akt expression levels were quantified and normalized to β-actin levels using ImageJ software (right). (B)Relative expression of Wnt-signaling pathway components β-catenin and GSK3β in CAG cells transfected with miR-Scramble control or miR-342, miR-363, or miR-342+363 mimics, measured by RT-qPCR. The P values were obtained by one-way ANOVA followed by Tukey-Kramer post hoc test (ns: non-significant; *P<0.05; **P<0.005; ***P<0.0001).
Overexpression of miR-342 and miR-363 in 5TGM1 MM cells inhibits MM cell growth in vitro and in vivo
To investigate the effects of miR-342 and miR-363 in vitro and in vivo, mouse MM 5TGM1 cells were stably transduced with miR-Scramble control, miR-342, miR-363, or miR-342+363 (Supplementary Figure. S2A). RT-qPCR showed miR-342, miR-363, and miR-342+363 overexpression in 5TGM1 cells, and Western blotting and RT-qPCR confirmed that Runx2 expression in 5TGM1 cells was significantly reduced after miR-342, miR-363, or miR-342+miR-363 transduction, compared to miR-Scramble control cells (Supplementary Figure. S2B-C). We also assessed 5TGM1 cell proliferation after stable transduction. MTT assays showed that overexpression of miR-342, miR-363, or miR-342+363 markedly suppresses proliferation of 5TGM1 cells in vitro (Supplementary Figure. S2D). Stably transduced cells were IV injected into C57BL/KaLwRij mice via the tail vein (106 cells per mouse, n=10 mice per group). Bioluminescent imaging showed that mice injected with 5TGM1-miR-342 or 5TGM1-miR-363 cells had much smaller tumors in bone than mice injected with 5TGM1-miR-Scramble control cells had, and mice injected with 5TGM1-miR-342+363 cells had the least tumor growth (Fig. 4A). Serum IgG2bk levels (a reliable marker for total tumor burden) confirmed the bioluminescent imaging results (Fig. 4B). Bioluminescent imaging and ELISAs were performed 4 weeks after MM cell injection. In a separate 5TGM1-IV injection experiment (n=4 mice per group), the survival rates of mice bearing 5TGM1-miR-342, -miR-363, or -miR-342+363 tumors were significantly higher than those of mice bearing 5TGM1-Scramble control tumors (Fig. 4C). Furthermore, H&E staining demonstrated that all mice in the 5TGM1-miR-Scramble group had tumors in tibia/femur, whereas less than 20% of the mice in the 5TGM1-miR-342, -miR-363, and -miR-342+363 groups had detectable tumors in bone (Fig. 4D). These results demonstrate that miRNA-mediated Runx2 knockdown suppresses MM cell growth in vitro and in vivo.
Figure 4. miRNA-mediated Runx2 knockdown impairs MM progression in vivo.
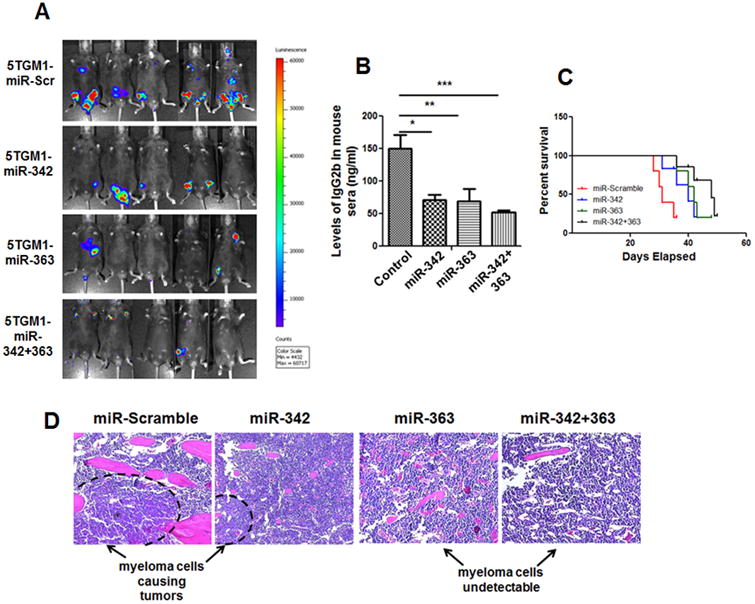
(A) Representative bioluminescent images obtained 4 weeks after IV injection of 5TGM1 cells stably transfected with miR-Scramble control or miR-342, miR-363, or miR-342+363 mimics (n=10 mice per group). (B) Serum IgG2bκ concentration measured by ELISA 4 weeks after IV injection of 5TGM1-miR-Scramble control, -miR-342, -miR-363-, or -miR-342+363 cells (n=10 mice per group). Significant differences between groups are indicated by P value (*P<0.05; **P<0.005; ***P<0.0001). (C) Kaplan-Meier survival curves in mice injected IV with 5TGM1-miR-Scramble control, -miR-342, -miR-363, or -miR-342+363 cells. Survival curves were significantly different (**P<0.005), according to the Log-rank (Mantel-Cox) test. (D) Representative micrographs of H&E-stained bone sections from mice injected IV with 5TGM1-miR-Scramble control, -miR-342, -miR-363, or -miR-342+363 cells. Tumors were present, with abundant myeloma cells, in mice injected with 5TGM1-miR-Scramble control cells, whereas very few tumors and myeloma cells were found in mice injected with 5TGM1-miR-342, -miR-363, or -miR-342+363 cells.
Overexpression of miR-342 and miR-363 in 5TGM1 cells decreases the number of MM cell-induced osteoclasts and increases the number of osteoblasts in vivo
MM is associated with osteolytic bone disease mediated by increased osteoclastic bone resorption and impaired osteoblastic bone formation (30, 31). To understand the effects of miR-342 and miR-363 expression in MM-related bone disease, the number of osteoblasts and osteoclasts was assessed in animals injected with 5TGM1-miR-342 or 5TGM1-miR-363 cells versus 5TGM1-miR-Scramble control cells. The number of osteoblasts was evaluated by immunostaining for osteocalcin (a marker for osteoblasts) and osteoclasts were visualized by TRAP staining as previously described (27). Enumeration of osteoblasts showed a trend toward increased osteoblast numbers in mice injected with 5TGM1-miR-342 and 5TGM1-miR-363 cells compared with mice injected with 5TGM1-Scramble control cells, but the increase was statistically significant only in those injected with the 5TGM1-miR-342+363 cells. Interestingly, osteoclast number was significantly decreased in mice injected with 5TGM1 cells overexpressing any of the miRNAs compared with 5TGM1-miR-Scramble control cells (Fig. 5). Together, these data indicate that miR-342 and miR-363 expression in MM cells can impact the BM microenvironment and bone destruction, possibly with a greater effect on osteoclasts than on osteoblasts.
Figure 5. Overexpression of miR-342 or miR-363 in 5TGM1 cells suppresses osteoclast formation and enhances osteoblastogenesis in a C57BL/KalwRij mouse model system.
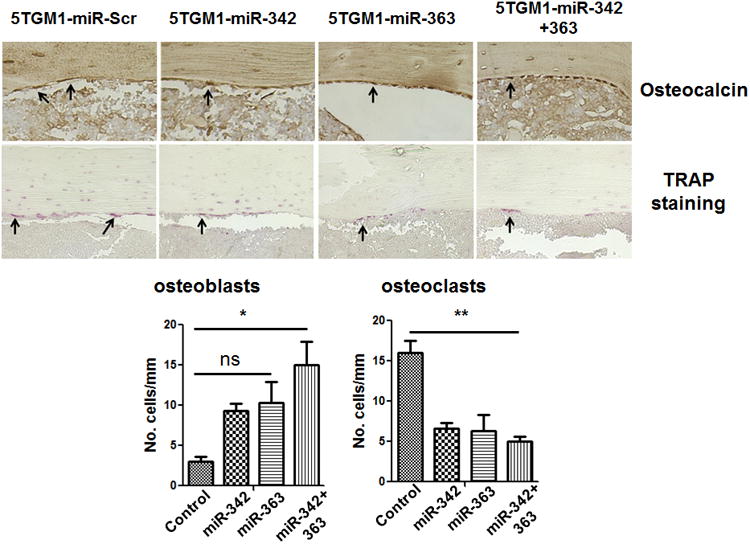
Immunostaining for osteocalcin (osteoblasts) and TRAP staining for osteoclasts were performed on bones harvested from C57BL/KalwRij mice injected with 5TGM1-miR-Scramble control, -miR-342, -miR-363, or -miR-342+363 cells. Representative micrographs are shown. Arrows indicate osteoblasts and osteoclasts in the respective panels. Osteocalcin-positive and TRAP-positive cells were photographed and counted (lower panel). The P values were obtained by one-way ANOVA followed by Tukey-Kramer post hoc test (Scr: Scramble; ns: non-significant; *P<0.05; **P<0.005).
miR-342 and miR-363 overexpression enhances antitumor immunity in vivo
Cancer cells, including MM cells, actively suppress the immune system and recruit immune-suppressive cells to evade immune detection and killing (32, 33). To determine whether miR-342 and miR-363 overexpression in MM cells can enhance antitumor immunity, bone marrow cells were harvested from hind limbs of mice injected with 5TGM1-miR-342, -miR-363, or -miR-342+363 cells or 5TGM1-miR-Scramble control cells, stained with specific antibodies against immune cells, and analyzed by flow cytometry (n=6 mice in each of the four groups). Flow cytometry analysis revealed a significant decrease in immunosuppressive Tregs and Bregs, and an increase in antigen-presenting pDCs in the bone marrow of mice injected with 5TGM1-miR-342+363 cells compared with the bone marrow of mice injected with 5TGM1-miR-Scramble control cells (Fig. 6A-C). Previous studies have demonstrated that blockade of programmed cell death-1 (PD-1) and T-cell immunoglobulin domain and mucin domain-containing molecule-3 (Tim-3) signaling in CD8+ T cells enhances the cytotoxic response of CD8+ T cells by production of effector cytokines such as IFN-γ and TNF-α (34, 35). Our flow cytometry analysis revealed that, although the number of cytotoxic CD3+ CD8+ T cells did not significantly differ, there was a significant decrease in CD8+ T cells expressing the exhaustion markers PD-1 and Tim-3 in the bone marrow of mice injected with 5TGM1-miR-342+363 cells compared with mice injected with 5TGM1-miR-Scramble control cells (Supplementary Figure. S3A-C). Also, there was a significant increase in expression of IFN-γ from CD3+ T cells in the bone marrow of mice injected with 5TGM1 miR-342+363 cells compared with mice injected with 5TGM1-miR-Scramble control cells (Supplementary Figure. S3D). Finally, we assessed the expression of PD-L1 in MM cells after Runx2 knockdown by shRNA as well as in 5TGM1-miR-342, -miR-363, or –miR-342+363 cells. PD-L1 expression was suppressed upon Runx2 knockdown by shRNA or miRNA (Supplementary Figure. S4A-C). These results are consistent with our hypothesis that MM cells overexpressing Runx2 also upregulate PD-L1 and are responsible for CD8+ T cell exhaustion by ligand-receptor interactions between PD-L1 and PD-1. Together, these results suggest that overexpressing miR-342 and miR-363 in 5TGM1 cells can enhance antitumor immunity in the bone marrow microenvironment.
Figure 6. Immune cell profile in bone marrow of C57BL/KalwRij mice injected with 5TGM1-miR-Scramble control, -miR-342, -miR-363, or -miR-342+363 cells.
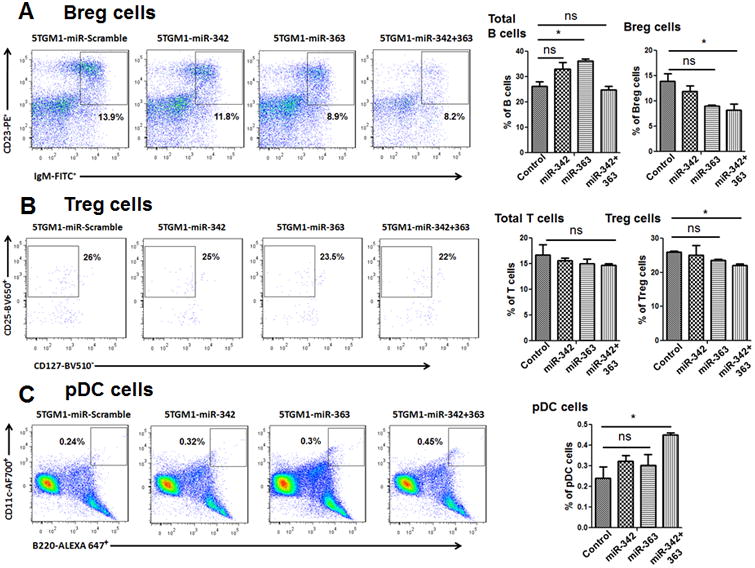
Total bone marrow cells were analyzed by flow cytometry. Numbers shown in each plot represent relative percentages of cells within the indicated gate. Total B cells, total T cells, and pDCs represent the respective population in all live cells. Bregs are shown as a population in total B cells, and Tregs as a population of CD4+ cells. (A-left) Breg cells (IgM+ CD23+) displayed in the gated area for each mouse group; (A-right) quantification of the percentage of total B-lymphocytes (B220+) and Breg cells in each group. (B-left) Treg cells (CD4+ CD25hi CD127lo) displayed in the gated area for each group; (B-right) quantification of the percentage of total T-lymphocytes (CD3+) and Treg cells in each group. (C-left) pDCs (B220+ Cd11c+) displayed in the gated area; (C-right) quantification of the percentage of pDCs in each group. Percentages are reported as the mean ± SEM. The P values were obtained by one-way ANOVA followed by Tukey-Kramer post hoc test (Scr: Scramble; ns: non-significant; *P<0.05).
Discussion
Runx2, as a transcription factor, is critical for regulating genes that support bone formation (36, 37). However, recent studies have demonstrated that Runx2 is aberrantly expressed in several cancer types, including MM (38, 39). Our earlier in vivo data demonstrated that enhanced expression of Runx2 in MM cells is associated with a more aggressive phenotype of MM, inhibiting MM-Runx2 significantly decreases the progression of MM, and high Runx2 expression in MM cells is associated with a high-risk population of patients with MM (10). These novel findings point to Runx2 as a new target for MM therapy.
It is well known that miRNAs are deregulated in numerous diseases and cancer (16, 17). Herein, we identified miR-342 and miR-363 as suppressors of MM progression through direct targeting of Runx2 in MM cells. Expression analysis revealed decreased expression of miR-342 and miR-363 in MM cells that have high levels of Runx2, and TargetScan analysis revealed multiple binding sites of miR-342 and miR-363 at the 3′UTR region of Runx2, indicating that miR-342 and miR-363 are negative regulators of Runx2. Indeed, upon reconstitution of miR-342 and miR-363 in MM cells, Runx2 expression was suppressed and, more importantly, the growth of MM cells was impaired in vitro and in vivo. These data strongly suggest that miRNA delivery, namely delivery of miR-342 and miR-363, is a viable therapeutic strategy to inhibit progression and dissemination of MM.
MM is characterized by the development of progressive and destructive osteolytic bone disease, yet despite advances in treatment strategies, bone destruction is a major cause of morbidity in MM patients (4, 40). Osteolytic bone disease is driven by increased osteoclast activity and decreased osteoblast activity, which eventually destroys the bone. Current therapies to treat osteolytic bone disease are based on antiresorptives, which inhibit osteoclast activity via antibody-based blockade of RANKL (e.g., denosumab) or by inducing osteoclast apoptosis using bisphosphonates. Recent studies have demonstrated that miRNA-mediated targeting of osteoclasts can be used to treat osteolytic bone disease (18). RANKL is a key stimulator of osteoclast differentiation (41). Our prior work established that Runx2 in MM cells enhances the expression and secretion of RANKL by MM cells and promotes bone resorption (10). In the present study, we demonstrate that the expression of RANKL in CAG MM cells is dramatically decreased by the induction of miR-342 and miR-363. Furthermore, our studies in mice bearing miR-342-, miR-363-, or miR-Scramble-overexpressing MM tumors indicate that regulation of Runx2 by miR-342 and miR-363 ultimately affects the bone marrow microenvironment by increasing osteoblast and decreasing osteoclast numbers in vivo. In addition to decreased RANKL production from MM cells overexpressing miR-342 and miR-363, reduced tumor growth by miR-342 and miR-363 may contribute to the reduced bone resorption in these mice. Moreover, increases in the numbers of immunosuppressive Tregs and Bregs and a decrease in the number of pDCs in the tumor microenvironment predicts worse survival for patients with various types of cancer (42, 43). In our study, compared with bone marrow from mice injected with 5TGM1-miR-Scramble cells, bone marrow from mice injected with 5TGM1 cells overexpressing miR-342 and miR-363 had a higher percentage of Tregs, Bregs, and PD-1+/Tim-3+ CD8+ cells, along with a lower percentage of pDCs and CD3+ IFN-γ cells. This improved immune environment in bone marrow is another likely reason for the decreased tumor growth in mice bearing miR-342- and miR-363-overexpressing tumors.
Together, our studies open new avenues for the development of miRNA-based therapeutic strategies in patients with MM. For example, miRNA replacement therapy aimed at restoring the levels of repressed miRNA is currently being developed with miR-34 and has entered phase I clinical trials (44). Additionally, formulated miRNA mimics are distinct from molecularly targeted drugs because miRNAs target a broad range of genes, rather than an individual gene product. Thus, our study demonstrates an opportunity to block multiple signaling pathways that are crucial for MM progression and dissemination, in addition to inhibiting Runx2. As therapeutics, miR-342 and miR-363 are also clinically advantageous as they are less likely to induce adverse effects, since most normal cells already express these miRNAs. Improved delivery strategies that target MM cells directly, such as using MM-specific phage fusion proteins as targeting ligands for liposomal-mediated delivery of miRNAs (45), will greatly enhance targeted delivery and offer promise for MM therapy moving forward.
Supplementary Material
Acknowledgments
We thank Dr. Ralph D. Sanderson (UAB, AL) for CAG MM cells, Dr. Fenghuang Zhan (UI, IA) for 5TGM1-luc MM cells. We also thank the UAB Animal Imaging Core for assistance with mouse bioluminescence imaging, and the UAB Histomorphometry and Molecular Analysis Core for tissue processing. This work was supported by National Institutes of Health (NIH) grant R01CA151538 (Y. Yang), an International Myeloma Foundation Senior Award (Y. Yang), American Society of Hematology (ASH) Bridge Grant Award (Y. Yang), and NIH Cancer Center Support Grant P30 CA13148 (Y. Yang).
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest
References
- 1.Yang Y, Macleod V, Bendre M, Huang Y, Theus AM, Miao HQ, et al. Heparanase promotes the spontaneous metastasis of myeloma cells to bone. Blood. 2005;105:1303–9. doi: 10.1182/blood-2004-06-2141. [DOI] [PubMed] [Google Scholar]
- 2.Anderson KC, Carrasco RD. Pathogenesis of myeloma. Annual review of pathology. 2011;6:249–74. doi: 10.1146/annurev-pathol-011110-130249. [DOI] [PubMed] [Google Scholar]
- 3.Barlogie B, Mitchell A, van Rhee F, Epstein J, Morgan GJ, Crowley J. Curing myeloma at last: defining criteria and providing the evidence. Blood. 2014;124:3043–51. doi: 10.1182/blood-2014-07-552059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jagannath S. Pathophysiological underpinnings of multiple myeloma progression. J Manag Care Pharm. 2008;14:7–11. doi: 10.18553/jmcp.2008.14.S7-A.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350:1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 6.Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–13. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 7.Mori K, Kitazawa R, Kondo T, Maeda S, Yamaguchi A, Kitazawa S. Modulation of mouse RANKL gene expression by Runx2 and PKA pathway. J Cell Biochem. 2006;98:1629–44. doi: 10.1002/jcb.20891. [DOI] [PubMed] [Google Scholar]
- 8.van der Deen M, Akech J, Wang T, FitzGerald TJ, Altieri DC, Languino LR, et al. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFbeta in prostate cancer cells. J Cell Biochem. 2010;109:828–837. doi: 10.1002/jcb.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das K, Leong DT, Gupta A, Shen L, Putti T, Stein GS, et al. Positive association between nuclear Runx2 and oestrogen-progesterone receptor gene expression characterizes a biological subtype of breast cancer. Eur J Cancer. 2009;45:2239–2248. doi: 10.1016/j.ejca.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trotter TN, Li M, Pan Q, Peker D, Rowan PD, Li J, et al. Myeloma cell-derived Runx2 promotes myeloma progression in bone. Blood. 2015;125:3598–608. doi: 10.1182/blood-2014-12-613968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 12.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 14.Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR- 17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JH, Zheng WW, Cheng ST, Liu BX, Liu FR, Song JQ. Correlation between microRNA21 and sprout homolog 2 gene expression in multiple myeloma. Mol Med Rep. 2015;11:4220–4224. doi: 10.3892/mmr.2015.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, et al. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell. 2013;24:542–56. doi: 10.1016/j.ccr.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431–5. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181–7. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colden Melissa, Dar Altaf A, Saini Sharanjot, Dahiya Priya V, Shahryari Varahram, Yamamura Soichiro, et al. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death and Disease. 2017;8:e2572. doi: 10.1038/cddis.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards CM, Edwards JR, Lwin ST, Esparza J, Oyajobi BO, McCluskey B, et al. Increasing Wnt signaling in the bone marrow microenvironment inhibits the development of myeloma bone disease and reduces tumor burden in bone in vivo. Blood. 2008;111:2833–42. doi: 10.1182/blood-2007-03-077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–6. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 24.Bakkus MH, Asosingh K, Vanderkerken K, Thielemans K, Hagemeijer A, De Raeve H, et al. Myeloma isotype-switch variants in the murine 5T myeloma model: evidence that myeloma IgM and IgA expressing subclones can originate from the IgG expressing tumour. Leukemia. 2001;15:1127–32. doi: 10.1038/sj.leu.2402164. [DOI] [PubMed] [Google Scholar]
- 25.Zhu D, van Arkel C, King CA, Meirvenne SV, de Greef C, Thielemans K, et al. Immunoglobulin VH gene sequence analysis of spontaneous murine immunoglobulin-secreting B-cell tumours with clinical features of human disease. Immunology. 1998;93:162–70. doi: 10.1046/j.1365-2567.1998.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Akker TW, Radl J, Franken-Postma E, Hagemeijer A. Cytogenetic findings in mouse multiple myeloma and Waldenstrom's macroglobulinemia. Cancer Genet Cytogenet. 1996;86:156–61. doi: 10.1016/0165-4608(95)00169-7. [DOI] [PubMed] [Google Scholar]
- 27.Kelly T, Suva LJ, Huang Y, Macleod V, Miao HQ, Walker RC, et al. Expression of heparanase by primary breast tumors promotes bone resorption in the absence of detectable bone metastases. Cancer Res. 2005;65:5778–84. doi: 10.1158/0008-5472.CAN-05-0749. [DOI] [PubMed] [Google Scholar]
- 28.Schmiedel BJ, Scheible CA, Nuebling T, Kopp HG, Wirths S, Azuma M, et al. RANKL expression, function, and therapeutic targeting in multiple myeloma and chronic lymphocytic leukemia. Cancer Res. 2013;73:683–94. doi: 10.1158/0008-5472.CAN-12-2280. [DOI] [PubMed] [Google Scholar]
- 29.Yaccoby S, Ling W, Zhan F, Walker R, Barlogie B, Shaughnessy JD., Jr Antibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood. 2007;109:2106–11. doi: 10.1182/blood-2006-09-047712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roodman GD. Pathogenesis of myeloma bone disease. Leukemia. 2009;23:435–41. doi: 10.1038/leu.2008.336. [DOI] [PubMed] [Google Scholar]
- 31.Heider U, Hofbauer LC, Zavrski I, Kaiser M, Jakob C, Sezer O. Novel aspects of osteoclast activation and osteoblast inhibition in myeloma bone disease. Biochem Biophys Res Commun. 2005;338:687–93. doi: 10.1016/j.bbrc.2005.09.146. [DOI] [PubMed] [Google Scholar]
- 32.Pratt G, Goodyear O, Moss P. Immunodeficiency and immunotherapy in multiple myeloma. British Journal of Haematology. 2007;138:563–579. doi: 10.1111/j.1365-2141.2007.06705.x. [DOI] [PubMed] [Google Scholar]
- 33.Ferrone C, Dranoff G. Dual roles for immunity in gastrointestinal cancers. J Clin Oncol. 2010;28:4045–51. doi: 10.1200/JCO.2010.27.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. 2017;355:1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu W, Shi Y, Li S, Zhang Y, Liu Y, Wu Y, et al. Blockade of Tim-3 signaling restores the virus-specific CD8+ T-cell response in patients with chronic hepatitis B. Eur J Immunol. 2012;42:1180–91. doi: 10.1002/eji.201141852. [DOI] [PubMed] [Google Scholar]
- 36.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 37.Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 38.Pratap J, Imbalzano KM, Underwood JM, Cohet N, Gokul K, Akech J, et al. Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure: implications for breast cancer progression. Cancer Res. 2009;69:6807–6814. doi: 10.1158/0008-5472.CAN-09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onodera Y, Miki Y, Suzuki T, Takagi K, Akahira J, Sakyu T, et al. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Sci. 2010;101:2670–2675. doi: 10.1111/j.1349-7006.2010.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giuliani N, Colla S, Rizzoli V. New insight in the mechanism of osteoclast activation and formation in multiple myeloma: focus on the receptor activator of NF-kappaB ligand (RANKL) Exp Hematol. 2004;32:685–91. doi: 10.1016/j.exphem.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien CA. Control of RANKL gene expression. Bone. 2010;46:911–9. doi: 10.1016/j.bone.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Su YX, Lao XM, Liang YJ, Liao GQ. CD19(+)IL-10(+) regulatory B cells affect survival of tongue squamous cell carcinoma patients and induce resting CD4(+) T cells to CD4(+)Foxp3(+) regulatory T cells. Oral Oncol. 2016;53:27–35. doi: 10.1016/j.oraloncology.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka Atsushi, Sakaguchi Shimon. Regulatory T cells in cancer immunotherapy. Cell Research. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 45.Bedi D, Gillespie JW, Petrenko VA, Ebner A, Leitner M, Hinterdorfer P, et al. Targeted Delivery of siRNA into Breast Cancer Cells via Phage Fusion Proteins. Mol Pharm. 2013;10:551–559. doi: 10.1021/mp3006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


