Abstract
Loss of monoubiquitination of histone H2B (H2Bub1) was found to be associated with poor-differentiation and enhanced malignancy of lung adenocarcinoma. This study, investigated the association and impact of the ubiquitin specific peptidase 22 (USP22), an H2Bub1 deubiquitinase, on stem cell-like characteristics and cisplatin resistance in cancer-initiating cells (CICs) from primary lung adenocarcinoma. CICs were isolated, enriched, and characterized from patient-derived cancer tissues using both in vitro tumorsphere formation and in vivo xenograft assays. USP22 was determined to be predominantly expressed in CICs, a subpopulation of cells with high expression of the stem cell biomarkers, CD133 and CD44. The expression of USP22 in CICs is markedly reduced upon FBS/retinoic acid-induced differentiation. Moreover, knockdown of USP22 significantly suppressed tumorsphere formation and xenograft growth in NOD-SCID gamma (NSG) mice. Notably, USP22 and aldehyde dehydrogenase (ALDH) activity were elevated in tumorsphere cells that survived cisplatin treatment, while knockdown of USP22 significantly sensitizes tumorsphere cells to cisplatin. Interestingly, ALDH1A3, a predominant ALDH isozyme implicated in enhancing cisplatin resistance in lung adenocarcinoma, is significantly down-regulated upon knockdown of USP22 in tumorsphere cells. Furthermore, knockdown of ALDH1A3 significantly sensitizes tumorsphere cells to cisplatin. Combined, these data demonstrate that USP22, predominantly expressed in CD133+ CICs, plays a critical role in tumorigenicity and cisplatin resistance in lung adenocarcinoma.
Implications
Targeting USP22 represents a potential therapeutic approach to suppress CICs in lung adenocarcinoma partially through down-regulation of ALDH1A3 expression.
Keywords: Lung adenocarcinoma, USP22, cancer-initiating cells (CICs), ALDH1A3, cisplatin resistance
Introduction
Lung cancer is the most common cause of cancer death worldwide. Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancers and about 50% of NSCLC are lung adenocarcinoma. Despite the advances in targeted therapies and most recently in immunotherapy, the 5-year survival rates for NSCLC remains only 15% (1). Clearly, new approaches are required to develop more effective therapies for this devastating disease. Platinum chemotherapy remains the first-line of treatment for most patients diagnosed with advanced NSCLC (2). Although most lung cancers are initially sensitive to chemotherapy, drug resistance is inevitable. Understanding the mechanism by which chemotherapy resistance occurs and developing drugs that decrease chemotherapy resistance has the potential to have a major impact on improving survival and quality of life in lung cancer patients.
H2B monoubiquitination (H2Bub1) levels are regulated by the reverse activities of deubiquitylases including USP22 and the E3 ubiquitin ligase RNF20/40 (3–6). USP22 is the most important and intensively studied deubiquitinase for H2Bub1 (7). H2Bub1 signaling pathway in particular has been implicated in regulating the differentiation of embryonic stem cells (8). Our previous studies showed that H2Bub1 levels are depleted in cancer cells and that manipulation of H2Bub1 affects sensitivity to chemotherapy in lung cancer cell lines (9). In cancer, H2Bub1 appears to act as a tumor suppressor (7). H2Bub1 is low or undetectable in many cancers, including breast, colon and lung cancers, and markedly higher in differentiated, normal tissue (10–13). A study by Glinsky et al identified an 11-gene signature that distinguishes tumors with aggressive growth, metastasis, and therapy resistance from less malignant tumors for diverse human cancers, and this signature included USP22 (14). Accordingly, knockdown of USP22 suppresses cell proliferation in several cancer cell lines (15). Conversely, overexpression of USP22 is detected in numerous cancers, including lung cancer (16), and is related to poor survival in patients (12, 17). Together, these studies suggest that USP22 may play a role in H2Bub1-mediated regulation of lung adenocarcinoma proliferation, differentiation and therapy resistance, and might be a candidate therapeutic target in cancer (15).
Cancer-initiating cells (CICs) are small subpopulations of cells found in cancers, including lung cancers, possessing stem cell-like properties, i.e., the ability to self-renew and to differentiate into heterogeneous cell populations (18, 19). Experimentally, CICs are defined by their ability to initiate tumors after transplantation into immunodefficient mice with a diabetes-susceptible Non-Obese Diabetic (NOD-SCID mice) (20, 21). A number of additional assays have been described to characterize CICs, including aldehyde dehydrogenase (ALDH) activity and sphere colony formation (tumorspheres) (22, 23). Several surface markers have also been described to aid in the isolation of CIC with flow cytometry, including CD133, CD44 and ALDH (24). However, no cell surface marker specifically isolates a phenotypically pure CIC population, though CD133 is most commonly used. As a result, there is a lack of consensus on the most specific manner to define CICs. Nevertheless, CICs are thought to play a critical role in the development of chemotherapy/radiotherapy resistance (18–20, 25, 26), however how these are regulated remains poorly understood.
ALDH1A3 is a member of ALDH superfamily of enzymes with the major function of detoxification with the participation of NAD(P)+ (27–31). Studies show that the expression of ALDH1A3 has intimate correlation with tumorigenesis, metastatic abilities and angiogenic activity of tumor (32–34), and ALDH activity was shown to promote therapy resistance through several mechanisms (35, 36). Although the important role of H2Bub1 in regulating growth and differentiation in embryonic stem cells is well described (17), little is known about the roles of H2Bub1 and USP22 in the growth, response to therapy in CICs derived from lung adenocarcinoma. In the study, we for the first time demonstrated that knockdown of USP22 suppresses ALDH activity, and targeting USP22 may potentially eliminate these CICs in lung adenocarcinoma through down-regulation of ALDH1A3 expression.
Material and Methods
CICs isolation, sphere formation cell culture and in vitro differentiation
We isolated putative CICs from three cases of lung adenocarcinoma who underwent surgical resection with curative intent without preoperative chemotherapy or radiation therapy at City of Hope. The clinical characteristics of the three cases are as follow: the sample-1 was from a patient with moderately differentiated adenocarcinoma with mutated p53 and KRAS at stage Ib; the sample-2 was from a patient with moderately differentiated squamous cell lung cancer with no molecular testing at stage Ib; the sample-3 was from a patient with poorly differentiated squamous cell lung cancer with no molecular testing at stage Ia. The tissue was cut into pieces and dissociated with 400 U/ml collagenase IV (Gibco) (19, 37) in DMEM/F12 medium (Gibco) in 37°C for 2 hours. After that we lysed red blood cells with ACK lysis buffer (Gibco) in room temperature for 2 min and spin down the remaining cell suspension. The cells (referred to in this study as Sample 1, Sample 2 and Sample 3) were transferred into ultra-low-attachment 6-well plates (Corning) and cultured in complete growth medium at 37°C in a humidified incubator. The complete growth medium consisted of DMEM/F12 medium (Gibco) containing 2% B-27 supplement (Gibco), 25 ng/ml FGF (Peprotech), 25 ng/ml EGF (Peprotech), 20 μg/ml insulin (Calbiochem), 2 μg/ml heparin (EDQM), 100 U/mL penicillin (Gibco), and 100 mg/mL streptomycin (Gibco), which has previously been reported to promote the growth of non-adherent cell spheres (18, 19, 38). The cells were then transferred to 10 cm ultra-low-attachment dish (Corning) for further expansion and experimentation.
For induced differentiation, cells were cultured in DMEM/F12 medium containing 10% FBS and 50 nM retinoic acid (RA) for 5 days in regular cell culture dishes. This study was reviewed and approved by the Institutional Review Board (IRB) 17196 of City of Hope National Medical Center, and informed consent for the collection of tumor tissues for the study were obtained from all patients.
Fluorescence activated cell sorting (FACS), apoptosis detection and ALDH assay
Lung CICs were dissociated into single-cell suspensions by Accumaxtm cell counting solution (EMD Millipore) and stained with mouse anti-CD133-PE antibody (clone AC133, Miltenyi Biotec) and mouse anti-CD44-FITC antibody (clone DB105, Miltenyi Biotec). The cells were sorted by flow cytometry. Apoptosis was measured by flow cytometry analysis of PE-labeled Annexin-V (BD Biosciences) and 7-AAD (BD Biosciences) co-staining, according to the manufacturer’s protocol. ALDH assay was performed by flow cytometry analysis with ALDEFLUOR™ Kit (STEMCELL), following the recommendation of the manufacturer.
Lentivirus infection, siRNA transfection and cisplatin treatment
New generation lentivirus vectors own the ability to transduce a broad range of cell types including cancer stem cells (39). A 3rd Generation Packaging Mix and USP22-set siRNA/shRNA/RNAi Lentivector (Human) Cat No. i026814 (target sequences: A: 244 - TTCGGCTGTTTCACAAAGAAGCATATTCA B: 830 - ACTGCAAAGGTGATGACAATGGGAAGAAG were purchased from Applied Biological Materials Inc. (Richmond, BC, Canada). Both shRNA were used to knock down USP22. The USP22-shRNA-lentivirus and GFP-control-lentivirus were transfected into 293T cells with Lipofectamine™ 3000 Reagent (Invitrogen) for packaging, and CICs were infected with an MOI of 50 using polybrene (8 μg/ml). The infected cells were selected with 5 μg/ml puromycin.
For knocking down ALDH1A3, ALDH1A3 siRNA (Cat No. sc-43611) was purchased from Santa Cruz. It is a mixture of three sense/antisense products (A Sense: 5′-CAGAGAGCUUAGUCAAAGAtt-3′, Antisense: 5′-UCUUUGACUAAGCUCUCUGtt-3′; B Sense: 5′-GGAAAGUAGUGCUUAAGUUtt-3′, Antisense: 5′-AACUUAAGCACUACUUUCCtt-3′; C Sense: 5′-CCUUGAUAGUGAUACGUUAtt-3′; Antisense: 5′-UAACGUAUCACUAUCAAGGtt-3′) applied to silence ALDH1A3 expression using the protocol of Lipofectamine® RNAiMAX reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). USP22 knockdown/GFP-CTRL (control GFP-specific shRNA) cells and ALDH1A3 knockdown/control (control-siRNA) cells were treated for 72 h with 1–8 μM cisplatin, when cell numbers were counted. Cisplatin-induced apoptosis was measured as mentioned above.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
Total RNAs extraction and cDNA were generated as we previously described (9). The primer for USP22 used were (5′-CTGGACGTGCTCCACCGACA-3′ (forward primer) and 5′-GTTGGCCTTCTTCCCATTGTCAT-3′ (reverse primer), and for ALDH1A3 were 5′-GAATGGCACGAATCCAAGAG-3′ (forward primer) and 5′-CCTCTGGAAGGCAACCTGT-3′ (reverse primer). Other primer sequences are available upon request. GAPDH gene was used as internal control for mRNA expression. Data were presented as the relative quantity of targets, normalized with respect to internal control, or relative to a calibrator control sample.
Western blot analysis
Mouse monoclonal anti-H2Bub1 antibody clone 7B4 (MABE453,) was from EMD Millipore (Merck, KGaA, Darmstadt, Germany). Rabbit anti-RNF20 antibody (#9425) was purchased from Cell Signaling Technology (Beverly, CA, USA). Rabbit anti-USP22 antibody (ab195289) and anti-ALDH1A3 antibody (ab129815) were purchased from Abcam (Cambridge, MA, USA). Rabbit anti-CD133 antibody (18470-1-AP) was from Proteintech (Rosemont, IL, USA). Mouse anti-CD133-pure antibody (clone AC133) was from Miltenyi Biotec (Auburn, CA, USA).
For Western blot analysis, total cellular protein was extracted by SDS sample buffer 2X and heated for 5 min at 95°C. The proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (4%–12% SDS PAGE), transferred onto polyvinylidenefluoride (PVDF) membranes, blocked with 5% nonfat milk in PBST, and blotted with the appropriate primary and secondary antibodies.
Tumorigenicity assay
In vivo tumorigenic potential was assessed by transplanting cells into NOD-SCID mice. To assess the tumorigenic potential of different cell populations, 103 – 104 cells were suspended in Matrigel (BD Biosciences) and injected subcutaneously into the right flank of four NOD-SCID mice respectively. After 2 weeks of the injection, the tumor size was measured every 3 days. Mice were euthanized 45 days after cell injection. All mice in this study were in a pathogen-free environment and fed ad lib. The use of animals was approved by the IRB-17196 of City of Hope National Medical Center and all applicable institutional and governmental regulations concerning the ethical use of animals were followed.
Statistical analysis
In the study, 3 CICs were derived from 3 independent patients (Samples 1, 2 and 3). All experiments were performed in duplicates or triplicates and repeated at least two times in each experiment. Two group comparisons were analyzed for variation and significance using a Student’s t test or Pearson χ2 test. The half maximal inhibitory concentration (IC50) was calculated by Probit Analysis. The value of P < 0.05 was considered significant.
Results
USP22 is highly expressed in CD133+ and CD44+ cancer cells in tumorspheres from primary lung adenocarcinoma tissues
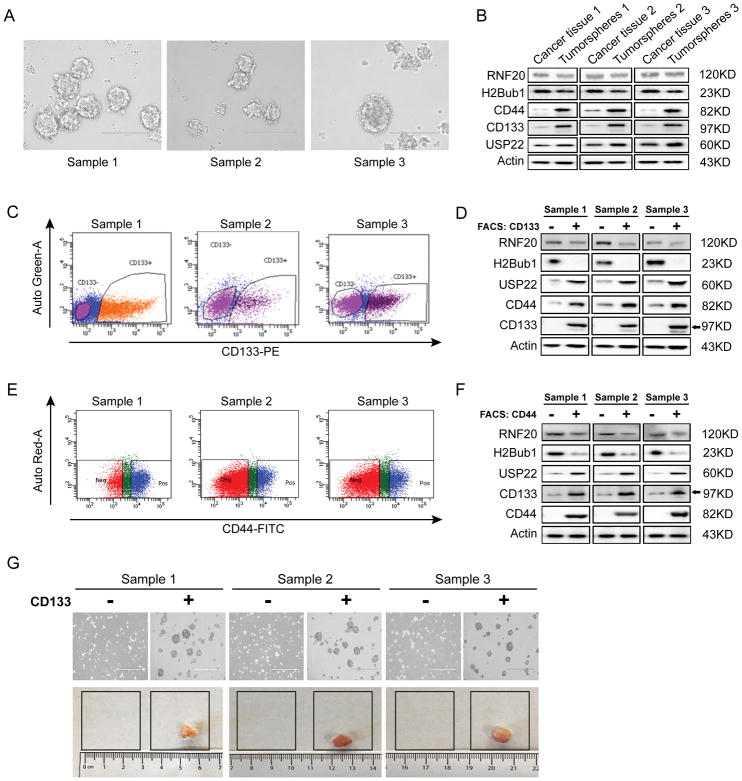
We recently reported that decreased H2Bub1 was associated with poor differentiation of lung adenocarcinoma (9). Herein, we further investigated the involvement of H2Bub1 deubiquitinase USP22 in CICs maintenance and drug resistance of lung adenocarcinoma. To do so, we first isolated and enriched a cancer cell subpopulation with stem cell properties from 3 primary lung adenocarcinoma tissues using serum-free tumorsphere culture approach. Typical tumorspheres formed by cells isolated from these primary tissues are shown in Figure 1A. To confirm that CICs with stem cell properties were enriched, we analyzed the protein expression levels of CD133 (a widely accepted surface marker of CICs), USP22, H2Bub1 and RNF20 in both tumorspheres and original lung cancer tissues by Western blot analysis. We observed that both CD133 and CD44 two putative lung CICs marker were markedly elevated in tumorspheres compared to the original tissues (Figure 1B). We observed that the protein level of USP22 was also moderately elevated in tumorsphere cells, while the levels of H2Bub1 and RNF20 proteins were significantly or moderated decreased in tumorspheres compared with the original tissues (Figure 1B). Together, the results suggest that H2Bub1 signaling pathway may be involved in regulation of CICs proliferation and maintenance.
Figure 1. Elevated USP22 protein in CD133+ cancer cells tumorspheres derived from primary lung adenocarcinoma tissues.
A. Images of tumorspheres from 3 lung adenocarcinoma tissues (Samples 1, 2, and 3). B. Western Blot analysis of CD133 (as a surface biomarker of CICs), CD44 (as another surface biomarker of CICs), USP22, RNF20, and H2Bub1, shows enriched USP22 protein in tumorspheres compared to original cancer tissues. C. Flow cytometry analysis of CD133 expression demonstrates a strong CD133 expression in about 30% of tumorsphere cells. D. Western Blot analysis demonstrates USP22 is markedly elevated, while H2Bub1 were significantly deceased in CD133+ cancer cells compared to CD133− cells sorted from tumorspheres. E. Flow cytometry analysis of CD44 expression demonstrates a strong CD44 expression in about 12% to 44% of tumorsphere cells. F. Western Blot analysis demonstrates USP22 and CD133 are markedly elevated, while H2Bub1 and RNF20 were markedly or moderately deceased in CD44+ cancer cells compared to CD44− cells sorted from tumorspheres. G. Mouse xenografts generated by both CD133− and CD133+ cancer cells isolated from tumorspheres, indicating CD133+ cells may represent CICs with stem cell characteristics in tumorspheres.
To further characterize the expression of USP22 in lung adenocarcinoma CICs, we used CD133 as a CICs surface marker to define and sort CD133+ cells from tumorspheres by flow cytometry. Cells from tumorspheres were stained for CD133, and flow cytometric analysis showed two distinct phenotypic subpopulations of CD133+ and CD133− in these tumorspheres (Figure 1C). The percentage of CD133+ cells ranged from 26.10% to 29.60% for the total population of the three tumorspheres (Samples 1, 2, and 3). To determine the association of USP22, H2Bub1 and RNF20 protein expression with CICs, we measured the expression level of these proteins in both CD133+ and CD133− cells. As shown in Figure 1D, compared with CD133− cells of three samples, USP22 and CD44 were markedly higher, while H2Bub1 and RNF20 proteins were significantly decreased, or slightly lower, in the CD133+ cells. CD44+ cells in tumorspheres were analyzed and enriched by flow cytometric analysis, and the results showed that the percentage of CD44+ cells ranged from 17.9% to 44.6% for the total population of the three samples (Figure 1E). Western Blot analysis showed the expression of USP22 and CD133 were significantly higher, while H2Bub1 and RNF20 were markedly or moderately lower in the CD44+ cells (Figure 1F). To assess the cancer stemness of the CD133+ cells sorted by FACS, 103 of CD133+ and CD133− cells were transplanted into four NOD-SCID mice respectively. The results of in vivo xenograft transplantation experiments showed that CD133+ cells of all three samples could form tumors, but CD133− cells could not (Figure 1G). The average weight of xenografts formed by 103 CD133+ cells isolated from these three tumorsphere cells were 217.0 ± 10.0 mg; 326.5 ± 22.5 mg; and 346.0 ± 19.0 mg, respectively; while none of CD133− cells isolated from these cells formed detectable xenografts. The above results indicated that the CD133+ cells possessed cancer stem cell properties, as well as higher levels of USP22 protein. Consistently, using immunofluorescent microscope analysis, we found that expression of USP22 was dramatically higher in cells with high expression of CD133, while the expression of H2Bub1 displayed an opposite trend of low expression (Figure S1). Therefore, the data suggests that USP22 may be positively correlated with high expression of CD133 cells and potentially represent a cancer stem cell marker in lung adenocarcinoma.
USP22 down-regulation impairs the tumorigenecity of lung CICs
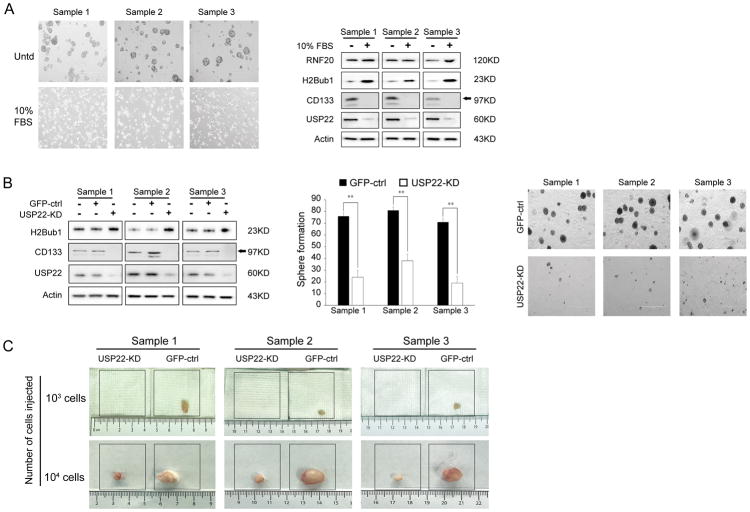
A previous study showed that the USP22-regulated H2Bub1 signaling pathway is required for differentiation of normal multipotent stem cells (17). To test the involvement of USP22 in the differentiation of lung CICs, the tumorspheres were further cultured in complete growth medium with 10% FBS and 50 nM RA to induce CICs differentiation. After culturing for 5–7 days, we found that tumorspheres attached to the dish and their morphology resembled adherent cells (Figure 2A, left panel). We analyzed the expression of USP22, CD133, H2Bub1 and RNF20 in lung CICs and differentiated cells. Western Blot analysis demonstrated that USP22 and CD133 expression were markedly decreased, after cells were differentiated; consistently, H2Bub1 expression was significantly elevated, while RNF20 expression was only mildly increased in differentiated cells compared with lung CICs (Figure 2A, right panel).
Figure 2. Impact of USP22 knockdown on tumorsphere formation.
A. Morphology changes of tumorspheres in differentiating culture medium (left panel); Western Blot analysis of USP22, CD133, RNF20 and H2Bub1 in tumorspheres in differentiating conditions (right panel). B. Western Blot analysis of USP22, CD133 and H2Bub1 in GFP-CTRL and USP22-KD cells (left panel); Quantitative analysis of tumorsphere formation upon USP22 knockdown, GFP-CTRL for cells transfected with control shRNA, USP22-KD for specific shRNA targeting USP22 (**P < 0.01, compared with Control, middle panel) and Tumorspheres (right panel). C. In vivo tumorigenesis of GFP-CTRL and USP22-KD tumorsphere cells, demonstrating USP22 knockdown suppressed both tumorsphere formation and tumorigenesis of lung adenocarcinoma CICs.
To investigate the role of USP22 in stemness of lung CICs, we first generated lung CICs with stable knock-down of USP22 using lentivirus expressing USP22-specific shRNA (USP22-KD). We first assessed the impact of knockdown of USP22 on in vitro tumorsphere formation. Tumorsphere formation assays showed a reduction in number and size of the spheres that were formed by USP22-KD cells, which were seeded at the same initial number and in the same duration of CICs infected by lentivirus expressing a control GFP-specific shRNA (GFP-CTRL) (Figure 2B, middle and right panels). As shown in Figure 2B left panel, compared to GFP-CTRL CICs, USP22 protein was markedly decreased in USP22-KD cells, and concurrently, CD133 was highly downregulated and H2Bub1 was moderately increased in all three cells. The knockdown of USP22 in CICs by USP22-shRNA B lentivirus also upregulated ALDH1A3 and sensitized CISs to cisplatin. The results were showed in Figure S2A.
Moreover, the impact of USP22 knockdown on the in vivo tumorigenic potential of CICs was further evaluated by injecting 103 and 104 USP22-KD cells or GFP-CTRL cells into four NOD-SCID mice respectively. Consistent with the tumorsphere results, the results of xenograft growth demonstrated that USP22-KD cells only formed visible xenografts when seeded at 104 cells, while GFP-CTRL lung CICs could form tumors at 103 cells (Figure 2C). The average weight of three samples were showed as follows:
103 cells: Sample 1GFP-CTRL: 135.0±14.0 mg; Sample 1USP22-KD: 0 mg; Sample 2GFP-CTRL: 93.5±6.5 mg; Sample 2USP22-KD: 0 mg; Sample 3GFP-CTRL: 99.5±13.5 mg; while none of 103 USP22-KD cells formed detectable xenografts.
104 cells: Sample 1GFP-CTRL: 427.0±18.0 mg; Sample 1USP22-KD: 188.0±13.0 mg; Sample 2GFP-CTRL: 512.5±33.5 mg; Sample 2USP22-KD: 214.0±10.0 mg; Sample 3GFP-CTRL: 443.5±20.5 mg; Sample 3USP22-KD: 173.0±16.0 mg (P=0.0040<0.01).
The above data suggest that knockdown of USP22 significantly suppressed the stemness of lung CICs.
USP22 down-regulation sensitizes tumorsphere cells to cisplatin
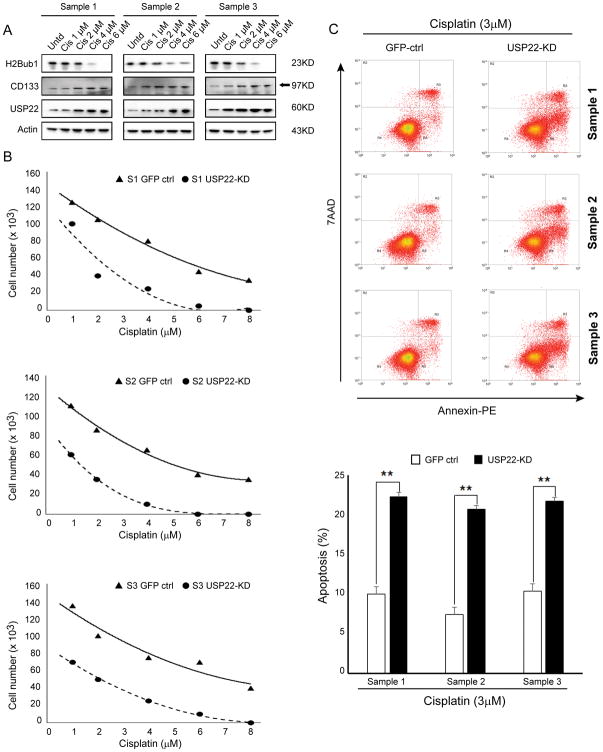
CICs are well-known for high resistance to treatment (17). Interestingly, we found that USP22 and CD133 were dramatically upregulated, while H2Bub1 was highly downregulated in tumorsphere cells that survived 1, 2, 4 and 6 μM cisplatin treatment (Figure 3A); suggesting USP22 may contribute to resistance to cisplatin in lung CICs. We hypothesized that knockdown of USP22 would sensitize CICs to cisplatin. We treated the GFP-CTRL and USP22-KD tumorsphere cells in undifferentiating culture medium with 1.0–8.0 μM cisplatin for 72 hours and measured cell viability using cell counting by trypan blue exclusion. The results showed that IC50 of cisplatin for all three USP22-KD cancer cells were significantly decreased compared with GFP-CTRL control cancer cells (Figure 3B, 2.2 μM vs 6.1 μM for sample 1, P = 0.0078; 1.3 μM vs 5.3 μM for sample 2, P = 0.0005; 1.8 μM vs 7.1 μM for sample 3, P = 0.0004). The same results were also found in CICs transfected with shRNA B lentivirus to knock down USP22 (Figure S2B). Consistently, apoptosis induced by 3.0 μM cisplatin for 72 h was also significantly increased in all USP22-KD cancer cells (Figure 3C). The above data strongly suggest that USP22 plays an important role in sensitivity of lung adenocarcinoma CICs to cisplatin.
Figure 3. Impact of USP22 knockdown on sensitivity of tumorspheres to cisplatin treatment.
A. Western Blot analysis of USP22, CD133 and H2Bub1 in the parental tumorsphere cells and tumorsphere cells that survived a 72h of 1, 2, 4 and 6 μM cisplatin treatment, indicating survived tumorsphere cells have elevated USP22 protein with the increase of cisplatin concentration. B. Cytotoxic effects of cisplatin in GFP-CTRL and USP22-KD cells. C. Flow cytometry analysis of apoptosis (upper panel) and quantitative analysis of induced apoptosis (lower panel) in GFP-CTRL and USP22-KD cells cisplatin treatment (**P < 0.01, compared with control), demonstrating USP22 knockdown significantly sensitized CICs to cisplatin.
ALDH1A3 is a downstream molecule of USP22 in lung adenocarcinoma tumorsphere cells
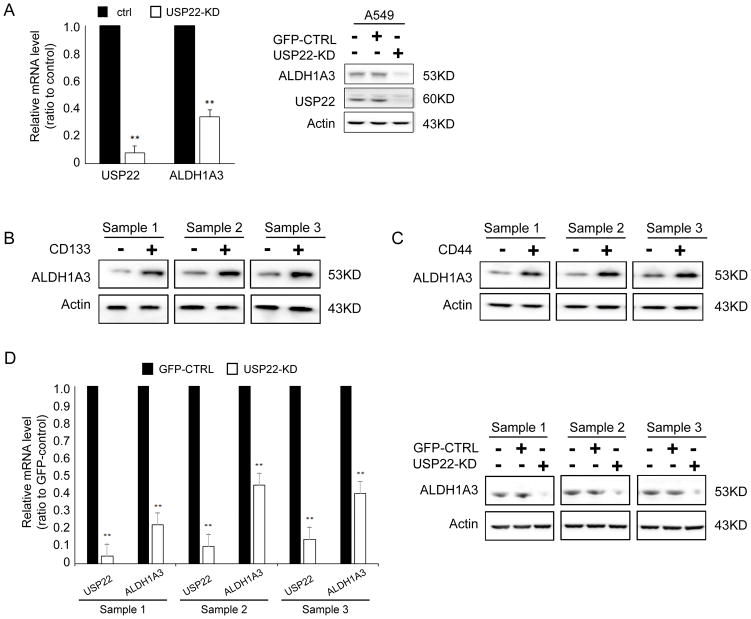
By using RNAseq analysis and quantitative PCR, we identified that knockdown of USP22 in A549, H1299, and H838 lung cancer cells significantly modified these genes involved in cell cycle, angiogenesis, epithelial mesenchymal transition, Myc, and K-Ras signaling pathways (40). Representative genes were shown in Figure S3; Interestingly, we found that USP22 knockdown ubiquitously down-regulated ALDH1A3 mRNA expression in these cells, indicating ALDH1A3 may be modified by USP22 at transcription level in an unidentified way. To further explore the correlation between USP22 with ALDH1A3 mRNA expression levels, we downloaded global gene expression RNAseq data from the cancer genome atlas (TCGA) gene expression data portal. Interestingly, the correlation analysis showed that USP22 mRNA level slightly trends to be positively correlated with ALDH1A3 mRNA level in a total of 592 lung adenocarcinoma samples, (Figure S4, P = 5.9e-09, R2= 0.155), while no correlation between CD133 and ALDH1A3 mRNA levels was observed in these samples (Figure S5, P = 3.5e-04, R2 = 0.02). Consideration of the high degree of heterogeneity of samples and cells in tissues, as well as RNAseq data quality, we consider the correlation is still clinically meaningful. More importantly, herein, we found that USP22 knockdown markedly downregulated mRNA (Figure 4A, left panel) and protein levels of ALDH1A3 (Figure 4A, right panel) in the lung cancer cell line A549. Consistently, in the study, we also found that ALDH1A3 was observably elevated in CD133+ cells (Figure 4B), while its mRNA level (Figure 4D, left panel) and protein level (Figure 4C, right panel) significantly decreased in USP22-KD lung tumorsphere cells.
Figure 4. Decreased expression of ALDH1A3 in USP22 knockdown tumorsphere cells.
A. Quantitative reverse transcription PCR (qRT-PCR) and Western Blot analysis show both mRNA and protein of ALDH1A3 were downregulated in lung cancer cell line A549 upon USP22 knockdown. B. Western Blot analysis of co-expression of ALDH1A3 and USP22 in CD133+/CD133− cancer cells. C. Western Blot analysis of co-expression of ALDH1A3 and USP22 in CD44+/CD44− cancer cells. D. qRT-PCR and Western Blot analysis show both mRNA and protein of ALDH1A3 were decreased in USP22-KD tumorspheres cells, indicating USP22 might also modulate ALDH1A3 in tumorsphere cells.
ALDH1A3 is associated with resistance to cisplatin in lung adenocarcinoma tumorsphere cells
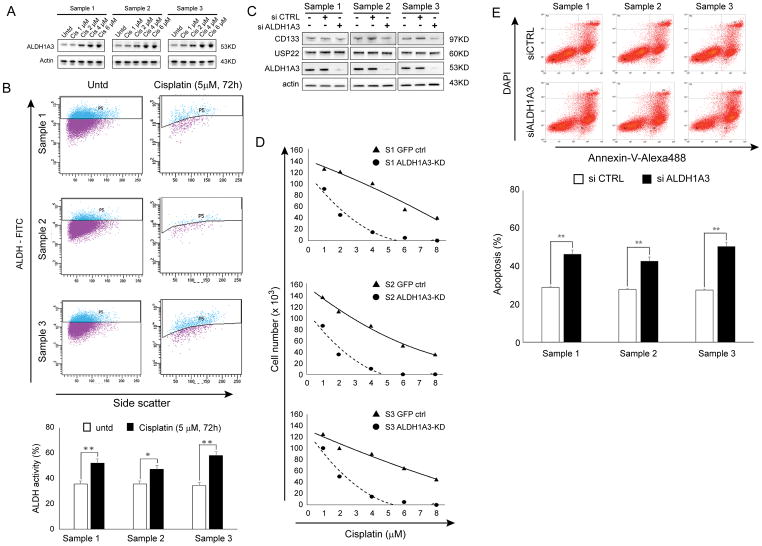
We found that the expression of ALDH1A3 was significantly increased in tumorsphere cells survived 1, 2, 4 and 6 μM cisplatin treatment (CICs surviving a 72h cisplatin course) compared with untreated cancer cells (Figure 5A). ALDH activity analyzed by flow cytometric ALDH assay (Figure 5B) was also markedly increased in tumorsphere cells survived a 72h 5 μM cisplatin treatment compared with untreated cancer cells. It reported that ALDH positive cells isolated from lung cancer cells lines demonstrated a high resistance to cisplatin and gemcitabine, when compared to ALDH− cells (41). Therefore, we further determined the impact of ALDH1A3 knockdown on the three lung CICs sensitivity to cisplatin. ALDH1A3 protein was depleted in CICs by transfecting them with ALDH1A3-specific siRNA (ALDH1A3-KD). As shown in Figure 5C, Western blot analysis revealed that ALDH1A3 was significantly decreased; CD133 was slightly reduced, while USP22 was unchanged in ALDH1A3-KD compared to GFP-CTRL cells (Figure 5C). We treated the control siRNA/ALDH1A3-KD lung CICs with 1.0–8.0 μM cisplatin for 72 hours. The results demonstrated that the IC50 was significantly decreased in ALDH1A3-KD cells compared with control cells (Figure 5D, 1.9 μM vs 6.9 μM for sample 1, P = 0.0094; 1.6 μM vs 6.3 μM for sample 2, P = 0.0046; 2.2 μM vs 7.3 μM, P = 0.0113 for sample 3). Consistently, apoptosis induced by 6.0 μM cisplatin for 72 h was significantly increased upon knockdown of ALDH1A3 in these CICs (Figure 5E). Therefore, ALDH1A3 may be a downstream target of USP22, and be associated with USP22-mediated resistance to cisplatin in lung adenocarcinoma CICs.
Figure 5. Contribution of ALDH1A3 to sensitivity of tumorspheres to cisplatin.
A. Western Blot analysis of ALDH1A3 protein in survival tumorsphere cells after 1, 2, 4 and 6 μM cisplatin treatment and B. Flow cytometric analysis of ALDH activity in survival tumorsphere cells after 5 μM cisplatin treatment (**P < 0.01, compared with untreated), suggest ALDH1A3 is associated with resistance to cisplatin in lung adenocarcinoma tumorspheres. C. Western Blot analysis of USP22, CD133, and ALDH1A3 in three paired control siRNA and ALDH1A3-KD cells. D. Cytotoxic effects of cisplatin in control and ALDH1A3-KD sphere cells. E. Cisplatin induced apoptosis and quantitative analysis of induced apoptosis in control and ALDH1A3-KD cells (*P < 0.05, **P < 0.01, compared with control), indicating ALDH1A3 knockdown also sensitized tumorspheres to cisplatin.
Discussion
Our results show that CD133+ cells have higher expression of USP22 than CD133− cells. USP22 levels are increased with cisplatin treatment, however when cells are grown under differentiating conditions, there is a significant decrease in USP22 protein levels. Accompanied by downregulation of USP22, the stem cells surface markers, CD133 and ALDH1A3, are also downregulated. Importantly, downregulation of USP22 significantly impaired tumorigenicity and cisplatin resistance of lung CICs. These results suggest that USP22 may be a key regulator of stemness of lung CICs.
CICs are quiescent cells that are resistant to chemotherapy compared with more differentiated cancer cells(25, 26, 42). CICs are thought to be responsible for disease recurrence and progression. The importance of CICs in cancer progression, metastasis and chemoresistance is being gradually understood. CICs are a highly attractive target for the development of new lung cancer treatment strategies. Currently, there is a lack of consensus on the most specific manner to define CICs, and no cell surface marker specifically isolates a phenotypically pure CIC population. Although several surface markers have been described to aid in the isolation of CIC with flow cytometry, CD133 being the most commonly used, CICs still need to be defined by their ability to initiate tumors after transplantation into NOD-SCID mice (43, 44). In our study, CD133 was used as stem cell-like cell biomarker, and using tumorspheres formation and in vivo transplantation assays, we confirmed that CD133+ or CD133High cancer cells own stem cell-like characteristics. Furthermore, we found that USP22 is predominantly highly expressed in CD133+ cancer cells, while H2Bub1 is depleted in CD133+ cancer cells. Consistently, we previously showed that H2Bub1 levels are depleted in cancer cells and that manipulation of H2Bub1 affects sensitivity to chemotherapy in lung cancer cell lines (9). When H2Bub1 is lost, cancer cells proliferate more readily, and de-differentiation of normal epithelial cells may occur (9). A number of studies suggest that elevation of USP22 was involved in tumor progression and highly malignant clinico-biological features, correlates with therapy resistance and poor prognosis (14, 17, 36). In view of the correlation between USP22 and stem cell-like features in a variety of cancers, it is conceivable that high expression of USP22 could decrease H2Bub1 levels, negatively regulating the differentiation of human lung CICs and maintaining stemness, thereby promoting tumorigenesis and drug resistance.
ALDH1A3 activity is important for the stem-cell characteristics of CICs, such as sphere formation, tumorigenesis abilities and chemotherapy resistance (32, 34, 45). In the current study, by using RNAseq and gene set enrichment analysis, we surprisingly found silencing of USP22 had no significant effect on c-Myc pathway and global c-Myc levels, a well-known downstream molecule of USP22(10, 46), while ubiquitously down-regulated ALDH1A3 mRNA expression in several lung cancer cell lines, indicating ALDH1A3 may be transcriptionally regulated by USP22. A previous study showed that ALDH1A3 is the predominant ALDH isozyme responsible for ALDH1 activity and tumorigenicity in most NSCLCs; and that inhibiting ALDH1A3 may eliminate the ALDH+ subpopulation in NSCLCs (41). Interestingly, in current study, we observed that ALDH activity and ALDH1A3 was markedly elevated CD133+ cells and tumorsphere cells that survived cisplatin treatment. This may indicate that USP22 regulates ALDH activity through transcriptionally regulates ALDH1A3 levels. Consistently, a recent study also showed that targeting ALDH1 significantly re-sensitized resistant lung cancer cells to the cytotoxic effects of cisplatin (47). It is possible that CICs may escape cisplatin-mediated cytotoxicity by increasing ALDH activity. Alternatively, it is possible that only the CICs with high ALDH1A3 expression and consequently high ALDH activity can survive in the presence of cisplatin. Questions about how USP22 transcriptionally regulates ALDH1A3 and how much ALDH1A3 contributes to USP22-mediated drug resistance are remained unclear. We plan to further investigate the apparent interaction between USP22 and ALDH1A3 expression. We consider knockdown of USP22 may increase global H2Bub1, alter H3K4 and H3K79 methylation to affect chromatin structure change, and then to modulate gene expression in a context specific manner. It is meaningful to develop approaches to suppress USP22 to treat. Nevertheless, we demonstrated that lung CICs were significantly sensitized to cisplatin by knockdown of USP22; indicating USP22 may represent a potential therapeutic target to eliminate CICs in lung adenocarcinoma through down-regulation of ALDH1A3 expression.
Supplementary Material
Acknowledgments
Financial support: This study was supported by the V Foundation for Cancer Research (D.J.Raz); the Stop Cancer Foundation (D.J.Raz); NIH grants 5K12CA001727-20 (D.J.Raz); and P30CA33572 that supports Analytical Cytometry Core, Animal Tumor Models Program, and Intergrative Genomics Core.
The authors acknowledge the generous support of the Baum Family Foundation for this lab. The authors thank Lucy Brown at the flow cytometry core for cytometric analysis, Loren Quintanar at Light Microscope core of City of Hope for imaging analysis, Qi Cui Ph.D for tumorsphere formation assay, Yahong Shi Ph.D at Division of Neurosciences, City of Hope for critical discussion, and Indra M. Newman, PhD, from the Department of Surgery at City of Hope for discussion and manuscript editing.
Footnotes
Disclosure of Potential Conflicts of Interest: DR: Merck, grant funding; Cireca, LLC, consultant.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350(4):379–92. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. NAT CELL BIOL. 2008;10(4):483–8. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 4.Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, et al. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO REP. 2009;10(8):894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20(10):1343–52. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomic Proteomic. 2006;5(3):179–89. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- 7.Cole AJ, Clifton-Bligh R, Marsh DJ. Histone H2B monoubiquitination: roles to play in human malignancy. Endocr Relat Cancer. 2015;22(1):T19–33. doi: 10.1530/ERC-14-0185. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. MOL CELL. 2012;46(5):662–73. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, Wang J, Tong TR, Wu X, Nelson R, Yuan YC, et al. Loss of H2B monoubiquitination is associated with poor-differentiation and enhanced malignancy of lung adenocarcinoma. INT J CANCER. 2017;141(4):766–77. doi: 10.1002/ijc.30769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22(19):2664–76. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. CANCER RES. 2011;71(17):5739–53. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- 12.Liu YL, Yang YM, Xu H, Dong XS. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J SURG ONCOL. 2011;103(3):283–9. doi: 10.1002/jso.21802. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, et al. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J Cancer Res Clin Oncol. 2011;137(8):1245–53. doi: 10.1007/s00432-011-0998-9. [DOI] [PubMed] [Google Scholar]
- 14.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J CLIN INVEST. 2005;115(6):1503–21. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. MOL CELL. 2008;29(1):102–11. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ning J, Zhang J, Liu W, Lang Y, Xue Y, Xu S. Overexpression of ubiquitin-specific protease 22 predicts poor survival in patients with early-stage non-small cell lung cancer. EUR J HISTOCHEM. 2012;56(4):e46. doi: 10.4081/ejh.2012.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. MOL CELL. 2012;46(5):705–13. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Sarvi S, Mackinnon AC, Avlonitis N, Bradley M, Rintoul RC, Rassl DM, et al. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. CANCER RES. 2014;74(5):1554–65. doi: 10.1158/0008-5472.CAN-13-1541. [DOI] [PubMed] [Google Scholar]
- 19.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106(38):16281–6. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. NATURE. 1994;367(6464):645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. NAT REV CANCER. 2012;12(2):133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 22.Kim RJ, Park JR, Roh KJ, Choi AR, Kim SR, Kim PH, et al. High aldehyde dehydrogenase activity enhances stem cell features in breast cancer cells by activating hypoxia-inducible factor-2alpha. CANCER LETT. 2013;333(1):18–31. doi: 10.1016/j.canlet.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 23.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. CELL. 2012;148(1–2):259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 24.Fang L, Cai J, Chen B, Wu S, Li R, Xu X, et al. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/beta-catenin signalling. NAT COMMUN. 2015;6:8640. doi: 10.1038/ncomms9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison R, Schleicher SM, Sun Y, Niermann KJ, Kim S, Spratt DE, et al. Targeting the mechanisms of resistance to chemotherapy and radiotherapy with the cancer stem cell hypothesis. J Oncol. 2011;2011:941876. doi: 10.1155/2011/941876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leon G, MacDonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther. 2016;158:71–90. doi: 10.1016/j.pharmthera.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J CELL MOL MED. 2009;13(8B):2236–52. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh S, Arcaroli J, Thompson DC, Messersmith W, Vasiliou V. Acetaldehyde and retinaldehyde-metabolizing enzymes in colon and pancreatic cancers. ADV EXP MED BIOL. 2015;815:281–94. doi: 10.1007/978-3-319-09614-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. STEM CELLS. 2011;29(1):32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Torres M, Allan AL. Aldehyde dehydrogenase as a marker and functional mediator of metastasis in solid tumors. Clin Exp Metastasis. 2016;33(1):97–113. doi: 10.1007/s10585-015-9755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pors K, Moreb JS. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? DRUG DISCOV TODAY. 2014;19(12):1953–63. doi: 10.1016/j.drudis.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. CLIN CANCER RES. 2014;20(15):4154–66. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Chen Y, Chen Z. MiR-125a/b regulates the activation of cancer stem cells in paclitaxel-resistant colon cancer. CANCER INVEST. 2013;31(1):17–23. doi: 10.3109/07357907.2012.743557. [DOI] [PubMed] [Google Scholar]
- 34.Golubovskaya V, O’Brien S, Ho B, Heffler M, Conroy J, Hu Q, et al. Down-regulation of ALDH1A3, CD44 or MDR1 sensitizes resistant cancer cells to FAK autophosphorylation inhibitor Y15. J Cancer Res Clin Oncol. 2015;141(9):1613–31. doi: 10.1007/s00432-015-1924-3. [DOI] [PubMed] [Google Scholar]
- 35.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res Treat. 2012;133(1):75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- 36.Sladek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17(1):7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 37.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. CELL STEM CELL. 2007;1(4):389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Qiu X, Wang Z, Li Y, Miao Y, Ren Y, Luan Y. Characterization of sphere-forming cells with stem-like properties from the small cell lung cancer cell line H446. CANCER LETT. 2012;323(2):161–70. doi: 10.1016/j.canlet.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. NAT GENET. 2003;33(3):401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 40.Zhang K, Tong TR, Yun X, Nelson R, Isaac N, Salgia R, et al. Elevated USP22 is a potential therapeutic target for human non-small cell lung cancer [abstract] CANCER RES. 2017;13(Supplement 77):4451. [Google Scholar]
- 41.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. MOL CANCER RES. 2009;7(3):330–8. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garg M. Targeting microRNAs in epithelial-to-mesenchymal transition-induced cancer stem cells: therapeutic approaches in cancer. Expert Opin Ther Targets. 2015;19(2):285–97. doi: 10.1517/14728222.2014.975794. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. NATURE. 2007;445(7123):106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 44.Vermeulen L, Todaro M, de Sousa MF, Sprick MR, Kemper K, Perez AM, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105(36):13427–32. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong B, Wu W, Cheng T, Schlitter AM, Qian C, Bruns P, et al. A subset of metastatic pancreatic ductal adenocarcinomas depends quantitatively on oncogenic Kras/Mek/Erk-induced hyperactive mTOR signalling. GUT. 2016;65(4):647–57. doi: 10.1136/gutjnl-2014-307616. [DOI] [PubMed] [Google Scholar]
- 46.Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ, et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J CELL PHYSIOL. 2017;232(12):3664–76. doi: 10.1002/jcp.25841. [DOI] [PubMed] [Google Scholar]
- 47.MacDonagh L, Gallagher MF, Ffrench B, Gasch C, Breen E, Gray SG, et al. Targeting the cancer stem cell marker, aldehyde dehydrogenase 1, to circumvent cisplatin resistance in NSCLC. ONCOTARGET. 2017;8(42):72544–63. doi: 10.18632/oncotarget.19881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.