Abstract
Importin 13 is a member of the importin β family of transport receptors. Unlike most family members, importin 13 mediates both, nuclear protein import and export. To search for novel importin 13 cargoes, we used stable isotope labeling of amino acids in cell culture (SILAC) and mass spectrometry. Using stringent criteria, we identified 255 importin 13 substrates, including the known cargoes Ubc9, Mago and eIF1A, and validate many of them as transport cargoes by extensive biochemical and cell biological characterization. Several novel cargoes can also be transported by the export receptor CRM1, demonstrating a clear redundancy in receptor choice. Using importin 13 mutants, we show that many of the novel substrates contact regions on the transport receptor that are not used by Ubc9, Mago or eIF1A. Together, this study significantly expands the repertoire of importin 13 cargoes and sets the basis for a more detailed characterization of this extremely versatile transport receptor.
Keywords: Nuclear Translocation, SILAC, Mass Spectrometry, Cell biology*, Affinity proteomics, eIF1A, importin 13, nucleus, Ubc9
Selective transport of macromolecules between the cytoplasm and the nucleus occurs through nuclear pore complexes (NPCs) 1, which are built of ∼30 different nucleoporins and serve as the only gateway across the nuclear envelope (1). Only small molecules can rapidly move through the NPC by passive diffusion, whereas larger macromolecules typically require nuclear transport receptors (NTRs), also referred to as importins or exportins, for efficient translocation. Transport selectivity is achieved by a permeability barrier constituted of phenylalanine-glycine (FG)-repeat domains, which are present in ∼1/3 of all nucleoporins and which interact with NTRs. Directionality of transport is mediated by the small GTPase Ran, which is required for the assembly and disassembly of transport complexes. Importins bind their cargoes in the cytoplasm via nuclear localization signals and are disassembled in the nucleus by binding of RanGTP. In contrast, exportins bind their cargoes via nuclear export signals in the nucleus together with RanGTP to form a trimeric export complex and are disassembled in the cytoplasm upon GTP hydrolysis on Ran. The intrinsic GTPase activity of Ran is enhanced by the Ran GTPase activating protein (RanGAP) and the Ran binding proteins RanBP1 and Nup358, also known as RanBP2. For recent reviews on nucleocytoplasmic transport see (1–4).
In humans, about 20 different NTRs have been identified that function in import, export or bidirectional transport (5). The best-studied transport receptors are the importin α/β heterodimer and the export receptor CRM1. CRM1 typically recognizes its cargoes through a short leucine-rich or hydrophobic nuclear export signal (NES; (6)), whereas the importin α/β heterodimer recognizes a so called classical nuclear localization signal (cNLS) rich in lysine or arginines (5). Three nuclear transport receptors have been reported to mediate both nuclear import and export of proteins, namely exportin 5 (Msn5 in yeast) (7, 8), exportin 4 (9, 10) and importin 13. Importin 13 was first identified as a member of the NTR-family in the year 2000 (termed LGL2; (11)). Shortly after, a few importin 13 cargoes were identified that bound to immobilized importin 13 from a HeLa cell extract in a RanGTP-dependent manner (12). RanGTP abolished binding of the import cargoes Ubc9, an E2 SUMO-conjugating enzyme, the heterodimer Mago-RBM8A, a core component of the exon junction complex, the transcription factor subunit NF-YB and the ribosomal protein RL5. Conversely, RanGTP promoted binding of the translation initiation factor eIF1A to importin 13, identifying it as a bidirectional transport receptor. Later, a few additional import cargoes of importin 13 were characterized, e.g. components of the chromatin accessibility complex (13), subunits of the transcription factor NF-Y (14) and of the negative cofactor 2 (NC2; (15)).
Importin 13 adopts a highly flexible, superhelical structure ranging from an open to a tight ring-like conformation, which allows for the accommodation of a range of cargoes. Crystal structures of importin 13 transport complexes suggest that importin 13 recognizes its cargoes through different interaction modes (16–18). Cargo binding by importin 13 relies on folded domains rather than short linear nuclear localization signals as known for importin α/β and CRM1. Furthermore, different regions in importin 13 are involved in binding to different cargoes. Mago/RBM8A, for example, interacts primarily with the C-terminal, whereas Ubc9 binds to the N-terminal region of importin 13 (17), largely overlapping the RanGTP binding site of the transport receptor. The binding mode of Ubc9 is rather unusual, as most importins bind their cargoes primarily through their C-terminal arch. An exception is importin β that binds the parathyroid hormone-related protein (PTHrP) primarily through its N terminus (19). In contrast to CRM1, importin 13 interacts with its export cargo eIF1A also in the absence of RanGTP (12) and cooperative binding as described for CRM1 is not observed in vitro. The general cargo release mechanisms of importin 13 compared with other transport receptors are clearly distinct. The import receptor importin α/β, for example, releases its cargoes in the nucleus upon RanGTP binding, which induces a conformational change in importin β. CRM1, on the other hand, employs GTP-hydrolysis on Ran to initiate dissociation of the cargo in the cytoplasm. For importin 13, by contrast, RanGTP binding seems to facilitate the formation of export complexes in the nucleus by displacing bound import cargo. In the cytoplasm, on the other hand, import cargoes may displace eIF1A or other export cargoes, leading to the concomitant formation of a novel import complex.
Comprehensive lists of potential transportin- (20), importin α/β- (21–24), and CRM1- cargoes (25–27) are available. By contrast, only few importin 13 import cargoes were reported, as described above. For a long time, eIF1A remained the only identified export cargo. Only recently, eukaryotic translation initiation factor 4γ2 (eIF4G2) and high mobility group protein 20A (HMG20A) were described as export cargoes (28), underlining the bidirectional transport capacity of importin 13. Several potential importin 13 cargoes were identified in three systematic screens very recently (28–30). To further expand the cargo spectrum of importin 13, we applied stable isotope labeling of amino acids in cell culture (SILAC), to identify proteins that interact with immobilized importin 13 from a HeLa cell lysate in a Ubc9- or RanGTP-dependent manner. In total, 255 proteins were identified as importin 13 cargo candidates according to our most stringent criteria. Of these, 117 were enriched in the presence of RanGTP, suggesting them as potential export cargoes. Several candidates were validated using importin 13 overexpression experiments and/or protein-protein interaction studies. Ultimately, the large cargo spectrum should lead to a better understanding of importin 13-dependent nuclear transport.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA3.1(+)-HA and pcDNA3.1(+)-FLAG were generated by insertion of C-terminal tag-sequences into the EcoRI- and XhoI-sites of pcDNA3.1(+) (Invitrogen, Carlsbad, CA). pMal-His-MCS-MBP was generated by inserting a C-terminal MBP-tag via SalI and HindIII and by replacing the N-terminal MBP from the pMal-c2 vector (New England Biolabs, Ipswitch, MA) with an N-terminal His-tag via EheI and EcoRI. pGEX-6P-1-MCS2 was generated by insertion of the multiple cloning site of pEGFP-GST into pGEX-6P-1 (Amersham Biosciences, Buckinghamshire, United Kingdom) via EcoNI and BamHI.
pET23a-Ubc9 (obtained from Frauke Melchior, Heidelberg (31)), pQE80-His-importin 13 and pQE80-RanQ69L(1–180) (obtained from Dirk Görlich, Göttingen (12, 32), pCS2-FLAG-importin 13 (obtained from Detlef Doenecke, Göttingen (15)), pEGFP-N3-eIF1A (obtained from Flavia Bono, Tübingen (18)) were described previously. pdEGFP-GST-cNLS and pEGFP-GST were obtained from Detlef Doenecke, Göttingen.
For generation of pEF-HA-snurportin 1, the coding sequence of human snurportin 1 was inserted into a pEF-HA-vector (33) via NcoI/EcoRI. The coding sequence of human importin 13 was amplified by PCR from pQE80-His-importin 13 and inserted into a modified pET-vector containing ten N-terminal His- and two Z- tags (the IgG binding domain of Protein A), followed by a TEV-protease cleavage site (34), via AgeI and XhoI. FLAG-importin 13-E436R/D481R was generated by cutting the mutated region of importin 13 from HA-importin 13-E436R/D481R (18) via SmaI and EcoRV and insertion into pCS2-FLAG-importin 13. FLAG-importin 13-D426R and FLAG-importin 13-K802E/R803E were obtained by site-directed mutagenesis on the wild type plasmid pCS2-FLAG-importin 13. The coding sequence of Ubc9 was amplified by PCR from pET23a-Ubc9 and inserted into pEGFP-GST via EcoRI and SalI. The coding sequence of Mago was amplified by PCR from HeLa cDNA and inserted into pEGFP-GST via HindIII and EcoRI.
Coding sequences of importin 13 cargo candidates were amplified by PCR from HeLa cDNA and cloned with a C-terminal HA-tag into pcDNA3.1(+)-HA, an N-terminal GFP-GST-tag into pEGFP-GST, an N-terminal GST-tag into pGEX-6P-1-MCS2 and/or a C-terminal MBP-tag into pMal-His-MCS-MBP.
N-terminal GST and C-terminal MBP expression constructs of eIF1A and Ubc9 were generated by PCR amplification of the coding sequences and cloning into pGEX-6P-1 (Amersham Biosciences) and pMal-His-MCS-MBP, respectively. The His-tagged expression construct of eIF1A was generated by PCR amplification of the coding sequence and cloning into pET28a (Merck Biosciences, Darmstadt, Germany) via EcoRI and SalI.
Protein Expression and Purification
Expression and purification of RanQ69L (35), RanQ69L(aa1–180) (36), importin β (37) and Ubc9 (31) was adapted from published protocols. GTP-loading of Ran was described previously (38).
His-importin 13 was transformed in E. coli JM109 and grown in 2× YT-medium supplemented with 2% glycerol and 30 mm K2HPO4 to an OD600 of 0.6. The culture was diluted 1:2.5 with cold medium, grown to an OD600 of 0.75 and protein expression was induced with 0.1 mm IPTG overnight at 16 °C. HZZ-importin 13 was expressed in BL21 (DE3) codon+ cells in LB medium with 1 mm IPTG overnight at 16 °C. His- and HZZ-tagged importin 13 were purified in buffer A (50 mm Tris pH 7.5, 500 mm NaCl, 10 mm Mg(OAc)2, 5% glycerol, 10 mm β-mercaptoethanol, 0.1 mm PMSF, and 1 μg/ml each of leupeptin, pepstatin, and aprotinin) over Ni-NTA Sepharose (Qiagen, Hilden, Germany), followed by separation over a HiLoad 26/60 Superdex 200 prep grade column connected to a ÄKTApurifier system (GE Healthcare, Chicago, IL) using buffer B (50 mm Tris pH 7.4, 200 mm NaCl, 5% glycerol, 2 mm DTT).
MBP-tagged proteins were expressed in E. coli BL21 (DE3) codon+ cells in MBP medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl and 0.2% glucose). Expression was induced with 0.3 mm IPTG and cells were grown overnight at 18 °C. MBP-tagged proteins were purified in buffer C (50 mm Tris, pH 7.4 (NELFCD, PFKFB2, NSUN2, XRCC5) or pH8.8 (NOSIP, TBPL1), 200 mm NaCl, 1 mm MgCl2, 10% glycerol, 10 mm β-mercaptoethanol, 0.1 mm PMSF, and 1 μg/ml each of leupeptin, pepstatin and aprotinin) over Ni-NTA Sepharose (Qiagen) followed by purification over amylose resin (New England Biolabs).
GST-Ubc9 and GST-eIF1A were expressed in E. coli BL21 (DE3) in LB medium and GST-tagged importin 13 cargo candidates in BL21 (DE3) codon+ cells in LB medium overnight at 16 °C with 1 mm IPTG. GST-tagged proteins were purified in buffer D (50 mm Tris, pH 6.8, 300 mm NaCl, 1 mm MgCl2, 2 mm DTT, 0.1 mm PMSF, and 1 μg/ml each of leupeptin, pepstatin and aprotinin) over glutathione-Sepharose (GE Healthcare).
Antibodies
The polyclonal rabbit anti-importin 13 antibody was raised in rabbits by injection of His-importin 13 and affinity purified using GST-importin 13 coupled to CNBr beads. The rabbit polyclonal anti-importin β antibody was raised in rabbits by injection of GST-importin β and affinity purified using His-importin β coupled to CNBr beads. For the detection of tagged proteins by indirect immunofluorescence, a polyclonal rabbit anti-HA antibody (Sigma, St. Louis, MI, H6908, 1:500) or a monoclonal mouse anti-FLAG antibody (Sigma, F3165, 1:3,000) were used. Endogenous eIF1A was detected with a monoclonal rabbit anti-eIF1A antibody (Abcam, Cambridge, UK, ab177939, 1:500). For immunoblotting, mouse monoclonal anti-transportin- (BD Biosciences, Franklin Lakes, NJ, 558660), anti-Ran- (BD Biosciences, 610340, 1:20,000), anti-Ubc9- (Santa Cruz, Dallas, TX, sc-271057, 1:5,000) and rabbit monoclonal anti-eIF1A- antibodies (Abcam, ab177939, 1:200,000) were used. Horseradish peroxidase-coupled secondary antibodies for Western blotting were obtained from Jackson ImmunoResearch (West Grove, PA). Alexa Fluor secondary antibodies for immunofluorescence were obtained from Thermo Fisher Scientific (Waltham, MA).
Experimental Design and Statistical Rationale
The goal of the quantitative triple SILAC approach was to compare binding of cargo proteins from a labeled HeLa cell extract to importin 13 under different conditions. The rationale for the assay workflow is described in the Results section (Figs. 1 and 2). For SILAC 1, a single experiment was performed. SILAC 2–4 correspond to three biological replicates, including label-swap experiments. p values were calculated for SILAC 2–4 with a right-sided one-sample t test. See below (“Data processing for identification of importin 13 cargoes”) for details on our significance criteria.
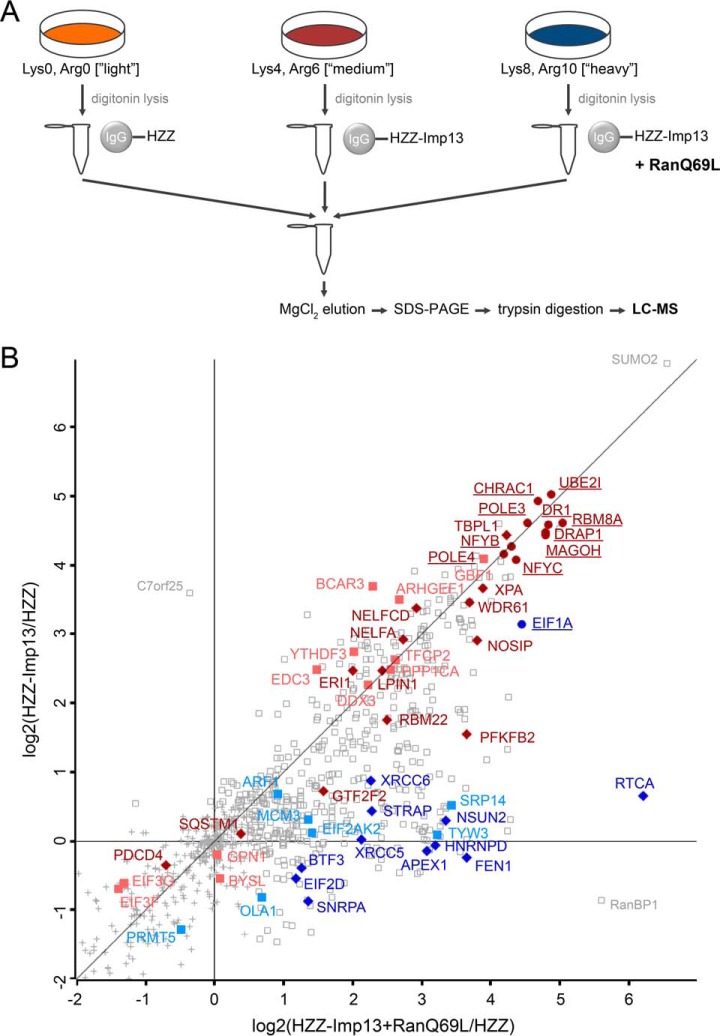
Fig. 1.
SILAC 1: identification of importin 13 cargoes using mass spectrometry. A, Experimental workflow. HZZ-tagged importin 13 or the HZZ-tag alone were immobilized on IgG-Sepharose and incubated with HeLa cell extracts containing either “light” (Lys0, Arg0), “medium” (Lys4, Arg6) or “heavy” (Lys8, Arg10) isotopes of lysine and arginine, with or without exogenous RanQ69L, as indicated. B, Scatter plot showing log2-ratios of importin 13 binding proteins enriched against the HZZ-affinity matrix in the presence (x axis) or absence (y axis) of RanQ69L. Colored proteins are either known importin 13 cargoes (underlined) or were further analyzed in this study. Gray squares correspond to proteins enriched with a log2 ratio ≥ 0.5. Gray crosses mark proteins that were considered insignificant. Red, proteins reduced by Ubc9 in SILAC 2–4. Blue, proteins enriched by RanQ69L in SILAC 2–4 (compare Fig. 2B, 2C). Proteins in dark colors were affected by overexpression of importin 13.
Fig. 2.
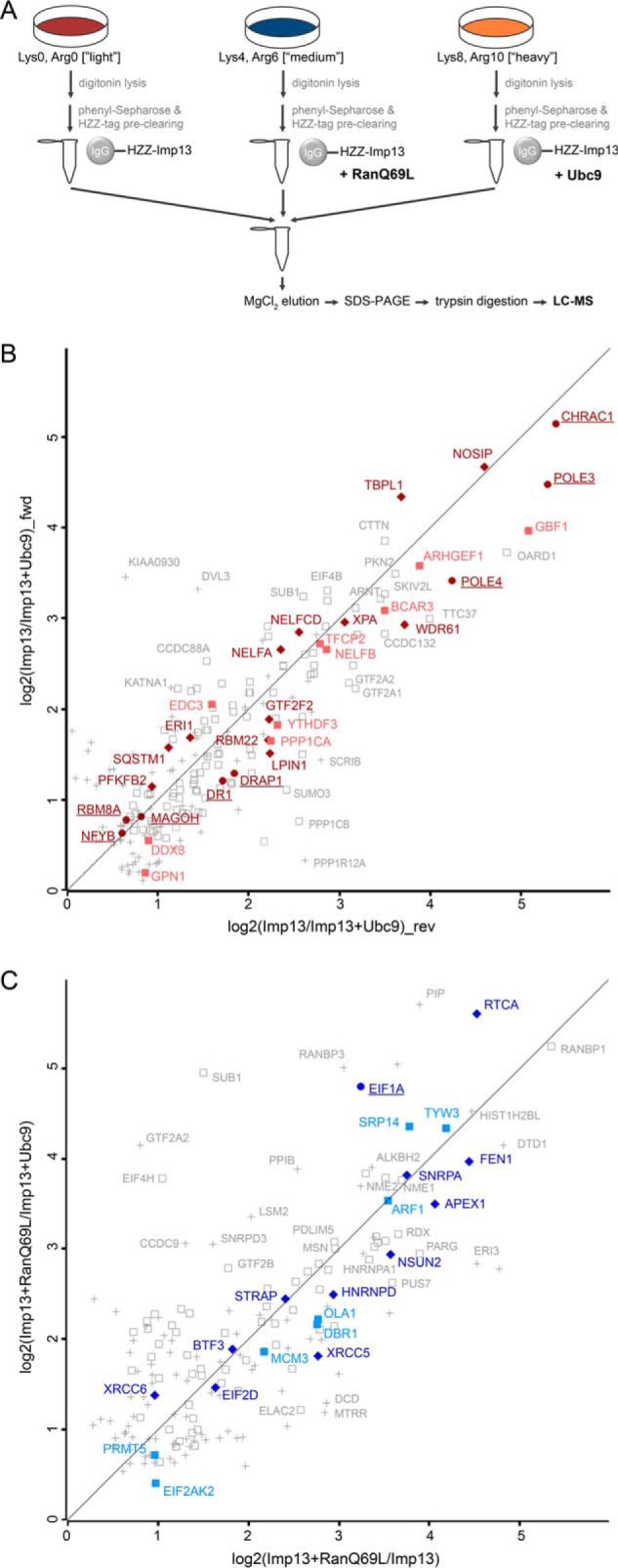
SILAC 2–4: importin 13 binding proteins from HeLa cell extracts. A, Experimental workflow. HZZ-tagged importin 13 was immobilized on IgG-Sepharose and incubated with labeled HeLa cell extracts (“light”, “medium”, “heavy”) supplemented with either RanQ69L, Ubc9 or buffer. B, Scatter plot showing log2-ratios of proteins binding to the importin 13 matrix in the absence or presence of Ubc9 in forward (fwd, y axis) and reverse (rev, x axis) reactions. C, Scatter plot showing log2-ratios of proteins binding to the importin 13 matrix in the presence or absence of RanGTP, with (y axis) or without (x axis) the addition of Ubc9. (B, C) Data are from three independent biological replicates with two experiments under identical labeling conditions (reverse) and one with a label switch (forward). Underlined proteins are known importin 13 cargoes. Proteins highlighted in red (B) and blue (C) but not underlined are cargo candidates that were further analyzed, and were (dark color, filled diamonds) or were not (light color, filled squares) affected by overexpression of importin 13. Gray open squares and gray crosses correspond to importin 13 cargo candidates identified in all three or in two out of three SILAC replicates with a log2 ratio ≥ 0.5, respectively.
SILAC Labeling of HeLa Cells and Preparation of Cell Extracts
Hela P4 cells (39) were grown in Dulbecco's modified Eagle medium (DMEM), high glucose, no glutamine, no lysine, no arginine (Thermo Fisher Scientific) supplemented with 10% dialyzed fetal bovine serum (FBS), 6 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 4,4,5,5-D4-l-Lysine 2HCl (Lys4) and 13C6-l-arginine HCl (Arg6) (“medium condition,” Silantes, München, Germany) or 13C615N2-l-lysine HCl (Lys8) and 13C615N4-l-arginine HCl (Arg10) (“heavy condition,” Silantes) at final concentrations of 73 mg/l and 42 mg/l, respectively. For “light conditions,” DMEM, high glucose, no glutamine, no lysine, no arginine was mixed in a 1:1 ratio with DMEM, high glucose, containing light l-lysine (Lys0) and l-arginine (Arg0) at final concentrations of 146 mg/l and 84 mg/l, respectively. FBS, glutamine and antibiotics were added as above. To achieve high labeling efficiencies (>95%), cells were subcultivated for at least five passages.
For the importin 13 binding assays, cells from two 15-cm plates per labeling condition were collected, adjusted to 1 × 108 cells/ml in transport buffer (TPB, 20 mm HEPES, 110 mm KOAc, 2 mm Mg(OAc)2, 1 mm EGTA, pH 7.3, 2 mm DTT, 1 μg/ml of each aprotinin, leupeptin and pepstatin, and 1× complete protease inhibitor mixture (Roche, Basel, Switzerland)) and permeabilized with 0.07% digitonin on ice for 5 min. Cell lysates were cleared by centrifugation at 1500 × g for 15 min at 4 °C, followed by ultracentrifugation at 100,000 × g for 30 min at 4 °C. Supernatants were incubated with 60 μg HZZ immobilized to 20 μl IgG-Sepharose 6 Fast Flow (GE Healthcare), to remove unspecific interaction partners, and 80 μl phenyl-Sepharose (GE Healthcare) to deplete the cell extract of nuclear transport receptors (40) for 2 h at 4 °C. Flow-throughs were collected and immediately used for binding experiments.
Binding Assays for the Identification of Importin 13 Cargoes
For SILAC experiments 2–4, HZZ-importin 13 (0.5 nmol) was immobilized on 20 μl IgG-Sepharose equilibrated in TPB containing 10 mg/ml BSA for one hour at 4 °C. The HZZ-importin 13 affinity matrix, preincubated for one hour with either 10 μm RanQ69L-GTP (RanQ69L for short), 5 μm Ubc9 or TPB alone in a final volume of 500 μl, was incubated with 350 μl labeled HeLa cell extract from two 15-cm plates (precleared with IgG-HZZ and phenyl-Sepharose) and either 10 μm RanQ69L-GTP, 5 μm Ubc9 or TPB alone in a final volume of 500 μl for two hours at 4 °C, respectively. The beads were washed three times with TPB and beads from all three binding conditions were combined for the last washing step. Bound proteins were eluted with 150 μl 1.5 m MgCl2 in 50 mm Tris, pH 7.4 and precipitated with isopropanol. The protein pellet was dissolved in 4× SDS sample buffer. Eluates were analyzed by SDS-PAGE, followed by mass spectrometry.
For SILAC 1, a condition with an HZZ-affinity matrix (0.5 nmol HZZ immobilized on 20 μl IgG-Sepharose) was included to detect proteins binding unspecifically to the HZZ-tag. Furthermore, the affinity matrices were not preincubated with RanQ69L-GTP nor Ubc9 and incubated with labeled HeLa cell extract that had not been precleared with IgG-HZZ and phenyl-Sepharose.
Sample Preparation for Mass Spectrometry
One-third of the magnesium chloride eluate from the importin 13 binding reaction was separated by SDS-PAGE. Each sample lane was split into 6 parts and in-gel digestion was performed as published (41). In brief, proteins were reduced with 10 mm DTT, 100 mm NH4HCO3 for 1 h at 56 °C, followed by alkylation with 55 mm iodoacetamide, 100 mm NH4HCO3 for 45 min at room temperature in the dark and digestion with 20 μg/ml trypsin (Serva Electrophoresis GmbH, Heidelberg, Germany) overnight at 37 °C. Peptides were extracted from the gel pieces (41) and desalted with C18 cartridges (3M, Maplewood, MN) as published (42, 43). Peptide eluates were dried by vacuum evaporation in a SpeedVac and dissolved in 20 μl of 2% acetonitrile, 0.1% formic acid for LC-MS analysis.
LC-MS Analysis
Peptides of 1 to 2 μl sample solution were loaded and washed on an Acclaim® PepMap 100 pre-column (100 μm × 2 cm, C18, 5 μm, 100 Å, Thermo Fisher Scientific) at a flow rate of 25 μl/min for 6 min in 100% solvent A (98% water, 2% acetonitrile, 0.07% TFA). Analytical peptide separation by reverse phase chromatography using the RSLCnano Ultimate 3000 system (Thermo Fisher Scientific) was performed on an Acclaim® PepMap RSLC column (75 μm × 50 cm, C18, 2 μm, 100 Å, Thermo Fisher Scientific) running a gradient from 98% solvent A (water, 0.1% formic acid) and 2% solvent B (80% acetonitrile, 20% water, 0.1% formic acid) to 32% solvent B within 110 min and to 65% solvent B within the next 16 min at a flow rate of 300 nl/min (solvents and acids from Fisher Chemicals). Peptides eluting from the chromatography were on-line ionized by nano-electrospray (nESI) using the Nanospray Flex Ion Source (Thermo Fisher Scientific) at 2.4 kV and continuously transferred into the Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific). Full scans in the mass range of 300–1850 m/z were acquired with the Orbitrap-FT analyzer and a resolution of 60,000. For parallel data-dependent top 10 fragmentation, the LTQ Velos Pro linear ion trap (CID) was used. LC-MS method programming and data acquisition was performed with the software XCalibur 2.2 (Thermo Fisher Scientific).
MS Data Analysis
Raw mass spectrometry files were analyzed with the MaxQuant software (version 1.5.1.0) (44) and searched against a reviewed human protein database derived from UniProt (Proteome ID: UP000005640, 20,243 entries, download August 2017). Search parameters were set as follows: trypsin/P digestion mode (tryptic specificity with no proline restriction) with maximum of two missed cleavages, oxidation of methionine and N-terminal protein acetylation were set as variable modifications and carbamidomethylation of cysteine was set as a fixed modification. Arg6 and Lys4 were defined as medium peptide labels and Arg10 and Lys8 as heavy peptide labels. The mass tolerance for peptide precursors was 4.5 ppm and for fragment ions 0.5 Da. Fourier transform-based mass spectrometer (FTMS) requantification and FTMS recalibration were enabled. Protein quantification was performed with a minimum ratio count of two and unique plus razor peptides were considered. False discovery rates were calculated by MaxQuant, using the revert-decoy mode and the filter for valid peptide sequence matches was set to 0.01. MaxQuant output data was further processed using Perseus software version 1.5.0.15 (45, 46).
Data Processing for Identification of Importin 13 Cargoes
Three biological replicates were performed including label-swap experiments (SILAC 2–4) to increase the specificity of the identified proteins and to exclude experimental bias. Note that for SILAC 1 only one experiment was performed. Proteins identified with “only identified by site,” “reverse,” and “potential contaminants” (Perseus version 1.5.0.15) were excluded from further analysis. An overview of the proteome data evaluation with Perseus can be found in supplemental Tables S3 (SILAC 1) and S4 (SILAC 2–4). In total, 1390 proteins were quantified, and for each protein, the normalized log2-ratios of reactions containing or lacking RanQ69L-GTP or Ubc9 were calculated (SILAC 2–4). In the case of SILAC 1, normalized log2-ratios for each protein were calculated for reactions with HZZ-importin 13 in the presence or absence of RanQ69L or the HZZ-tag alone. For easier visualization, the log2-ratios were plotted against each other to generate the scatter plots depicted in Fig. 1B (SILAC 1) and 2B and 2C (SILAC 2–4). Proteins were significantly enriched if they had normalized log2 SILAC ratios ≥ 0.5 in at least two out of three SILAC replicate experiments (SILAC 2–4), which corresponds to an enrichment of more than 1.4-fold. A right-sided one-sample t test was done for SILAC 2–4 to determine if the log 2 ratios were significantly different to the value zero. Proteins that were statistically significant with a threshold p value ≤ 0.01 or a threshold p value ≤ 0.05 are highlighted in supplemental Table S1. Peptides that were identified in SILAC 1–4 are listed in supplemental Table S2.
Transfection, Immunofluorescence, and Confocal Microscopy
HeLa P4 cells (39) were grown on coverslips in 24-well plates and cotransfected with plasmids coding for tagged importin 13 cargo candidates and either FLAG-tagged importin 13 or an empty vector using the calcium phosphate method (47). For indirect immunofluorescence, cells were fixed with 3.7% formaldehyde in PBS, permeabilized with 0.5% Triton X-100 in PBS for 5 min on ice, blocked with 1% BSA in PBS for 10 min and incubated with primary antibodies diluted in 1% BSA/PBS for one hour. After three washing steps with PBS, cells were incubated with Alexa Fluor secondary antibodies (Thermo Fisher Scientific) diluted in 1% BSA/PBS for one hour. Samples were mounted with Mowiol-DAPI and analyzed using a LSM 510-META confocal laser scanning microscope (Zeiss, Oberkochen, Germany) with a 63× Plan-Neofluar 1.3 NA water-corrected objective. Images were processed using Fiji (48).
For leptomycin B (LMB) treatment, cells were incubated with 10 nm LMB (Enzo Life Sciences, Farmingdale, NY) for 2 h, 24 h post transfection, followed by fixation and immunofluorescence staining as described above.
Image Analysis
Confocal microscopy images were analyzed using CellProfiler (49). Cell nuclei were identified using the adaptive Otsu thresholding method on DAPI-stained images. The identified nuclei were expanded by 20 pixels to yield the cell area followed by subtraction of the nuclear area to obtain the cytoplasmic area. Cells touching the border of an image were excluded from the analysis. Fluorescence intensities were measured in each cell and cells were filtered to have a minimum mean intensity of 0.05 for each overexpressed protein. Filtered cells were used to measure the fluorescence intensity in both the nucleus and cytoplasm and generate the ratio of the mean intensities in the nucleus and cytoplasm. For statistical analysis, a one-way analysis of variance (ANOVA) followed by Bonferroni post-test was performed.
Binding Assays with HeLa Cell Lysate
HZZ-tagged importin 13 (20 μg) immobilized on 20 μl IgG-Sepharose equilibrated with 5 mg/ml BSA in TPB (supplemental Fig. S1B) or PBS (supplemental Fig. S1C) was incubated with a HeLa cell extract prepared from two 15-cm plates in a total volume of 500 μl for one hour. Cell extracts were generated either by treatment with 0.07% digitonin on ice for 3–5 min (supplemental Fig. S1B) or three freeze/thaw cycles in liquid nitrogen (supplemental Fig. S1C). The digitonin lysate was cleared with phenyl-Sepharose and the IgG-HZZ affinity matrix as described above. Lysates were cleared by centrifugation at 1500 × g for 15 min to remove nuclei and cell debris, followed by ultracentrifugation at 100,000 × g for 30 min. Beads were washed three times with PBS or TPB and bound proteins were eluted with either magnesium chloride followed by isopropanol precipitation (supplemental Fig. S1B) as described above or with 0.1 m glycine, pH 3 (supplemental Fig. S1C). Glycine eluates were neutralized with 5 μl 1 m Tris, pH 10.4 and mixed 1:1 with 4x SDS sample buffer. Eluates were analyzed by SDS-PAGE, followed by Western blotting.
Pulldowns
GST- or MBP- fusion proteins (100 pmol) were immobilized on 10 μl glutathione-Sepharose (GE Healthcare) or amylose resin (New England Biolabs), respectively, equilibrated in TPB supplemented with 20 mg/ml BSA. The beads were incubated with 100 pmol of His-importin 13 in the presence or absence of 300 pmol Ubc9 or RanQ69L(aa1–180)-GTP in a total volume of 500 μl for 2 h at 4 °C and washed 3 times with 500 μl TPB lacking BSA. Bound proteins were eluted in 4× SDS sample buffer and analyzed by SDS-PAGE followed by Coomassie staining or Western blotting.
Determination of Endogenous Protein Concentrations
HeLa cells were lysed by boiling in 4× SDS sample buffer and defined amounts of HeLa cell lysate and His-tagged recombinant proteins were analyzed by SDS-PAGE followed by Western blotting and densitometry. Endogenous protein concentrations were determined assuming a HeLa cell volume of ∼2500 μm3.
RESULTS AND DISCUSSION
A Proteomic Screen for the Identification of Importin 13 Cargoes
Compared with our detailed understanding of importin 13-cargo interactions at a structural level, our knowledge on importin 13-mediated transport in general and the importin 13 cargo spectrum in particular is rather limited. To identify novel binding partners of the transport receptor, we set up proteomic screens using immobilized importin 13 fused to an HZZ-tag (i.e. a His-tag linked to two IgG-binding ZZ-domains of Protein A from Staphylococcus aureus) as bait and HeLa cell extracts as a source of potential substrates. Functionality of HZZ-importin 13 was confirmed with in vitro nuclear import assays (data not shown). In a first version of the screen (“SILAC 1”), which served as a feasibility test toward the more sophisticated screens (“SILAC 2–4”), we either immobilized the HZZ-importin 13 fusion protein or the HZZ-tag alone and compared binding of cytosolic proteins to the two different matrices and to importin 13 in the presence or absence of RanGTP.
To this end, we devised triple-SILAC experiments, where immobilized proteins were incubated with extracts from HeLa cells that had been grown under three different conditions: in “light” medium, containing the “normal” amino acids lysine and arginine (Lys0, Arg0); in “medium” medium containing 4,4,5,5-D4-lysine and 13C6-arginine (Lys4, Arg6); in “heavy” medium, containing 13C615N2-lysine and 13C615N4-arginine (Lys8, Arg10). The “heavy” extract was supplemented with RanQ69L-GTP (RanQ69L for short), a Ran mutant that is resistant to the GTPase activating protein RanGAP (50), and the mixture was added to immobilized HZZ-importin 13. Note that endogenous RanGTP is expected to hydrolyze its bound nucleotide in the presence of RanGAP from the cell extract. The “medium” extract was added to HZZ-importin 13 beads, whereas the “light” extract was added to HZZ-beads, both without exogenous Ran. After the binding reactions, the beads were washed and combined, and bound proteins were eluted and analyzed by LC-MS. A comparison of ratios of tryptic peptides derived from “light,” “medium,” and “heavy” extracts should allow conclusions about specific binding of individual proteins to importin 13 in the absence or presence of RanQ69L.
A flow chart of the experimental setup is depicted in Fig. 1A and the result of this experiment (“SILAC 1”) in Fig. 1B. For SILAC 1 only one replicate was performed and the data was not corrected for intrinsic noise. Although many proteins showed unspecific binding (log2-ratio ∼0) to the affinity matrix or were even enriched (log2-ratio < 0) on the HZZ-tag, the majority of proteins (506 out of 931 that were quantified) interacted specifically with importin 13 (log2-ratio > 0.5). Clearly, many of the known importin 13 substrates were strongly enriched on the importin 13 matrix as compared with the HZZ-matrix (Fig. 1B, proteins underlined). The major importin 13-import cargoes, Ubc9 (depicted with its gene name UBE2I), CHRAC1 (also known as CHRAC15), a component of the chromatin accessibility complex, and POLE3 (CHRAC17), a subunit of DNA-polymerase epsilon, for example, were enriched ∼30-fold (i.e. with a log2-ratio ∼5). Likewise, the major export cargo of importin 13, eIF1A, bound strongly to importin 13, even in the absence of added RanQ69L (y axis; log2-ratio ∼3), but only weakly to the HZZ-tag. Ran-independent binding of purified eIF1A to importin 13 has been described previously (12). Note that in the presence of RanQ69L (x axis; log2-ratio ∼5), binding of eIF1A was further promoted. Another protein showing a strong enrichment on the importin 13 matrix in the presence of RanQ69L was RanBP1 with an enrichment factor higher than 32 (log2 > 5). RanBP1 is a well-described RanGTP-binding protein that seems to interact indirectly with immobilized importin 13 under our experimental conditions.
Besides the known binding partners of importin 13, a large number (∼500) of proteins were identified in SILAC 1 that interacted with HZZ-importin 13 but not with the HZZ-tag alone (Table I and supplemental Table S1). Somewhat unexpected, the effect of RanQ69L on known import cargoes was rather low under our experimental conditions. Ubc9, CHRAC1 and POLE3, for example, showed very similar enrichment levels in the absence or presence of RanQ69L. Nevertheless, several proteins (e.g. APEX1 and BTF3) were found to be enriched on the importin 13 matrix only in the presence of RanQ69L, implicating them as potential export cargoes. Possibly, RanGTP-binding proteins in the cell extract, such as other nuclear transport receptors, compete with importin 13 for RanGTP binding. Together, RanQ69L-dependence should not be considered as a simple criterion for identification of and differentiation between importin 13 import and export cargoes. However, cargoes that bind to importin 13 only in the presence of RanQ69L are considered potential export cargoes.
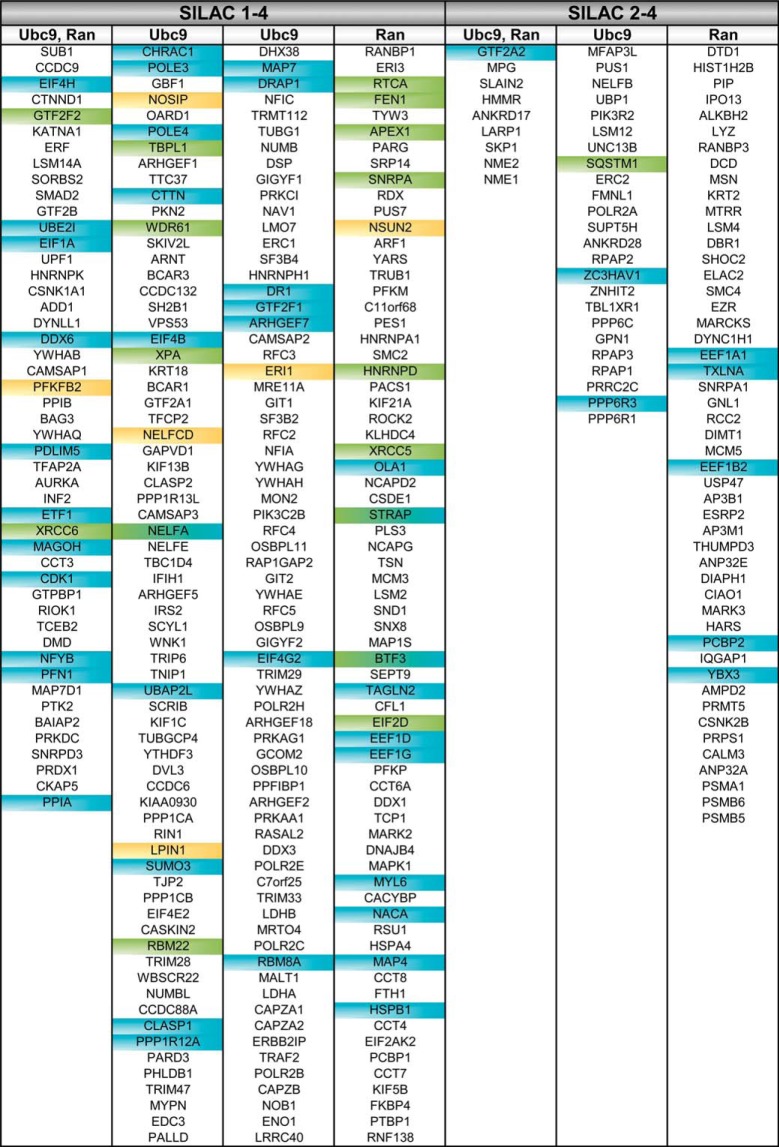
Table I. Importin 13 cargo candidates. Proteins (shown with their gene names) that were significant in SILAC 1–4 or only in SILAC 2–4 were extracted from supplemental Table S1 and sorted from higher to lower confidence cargoes. Ubc9: proteins reduced by Ubc9 in SILAC 2–4. Ran: proteins enriched by RanQ69L in SILAC 2–4. Green: proteins validated in importin13 overexpression experiments. Yellow: proteins validated in importin 13 overexpression and binding experiments. Blue: previously described importin 13 cargoes. See supplemental Fig. S2A and supplemental Table S1 for a complete list of cargo candidates and additional information.
Ubc9 as a Competitor for Cargo Binding to Importin 13
To get a better idea about specific importin 13 binding partners and, ideally, to discriminate between import and export cargoes, we first treated the cell lysates with phenyl-Sepharose to deplete endogenous transport receptors, which are known to interact with RanGTP and might affect binding of potential substrates to importin 13 (40). Furthermore, several nuclear transport receptors were identified in SILAC 1 that could interact with importin 13 indirectly via certain nucleoporins. Second, we preincubated the lysates with an HZZ-matrix to preabsorb proteins that might unspecifically interact with the tag. These treatments resulted in quantitative depletion of the transport receptors importin 13, transportin and importin β from the individual lysates, whereas endogenous Ran and eIF1A were hardly affected (supplemental Fig. S1A). Third, we included a condition, where Ubc9, a major binding partner of importin 13, was added in excess to the cell extract. Ubc9 has previously been shown to compete with other cargoes for importin 13 binding. Importantly, also cargoes with different binding sites on importin 13 than Ubc9 like Mago-RBM8A should be affected (17). Together with the RanQ69L-criterion, this approach should allow discrimination between specific and nonspecific interactors.
Before the SILAC-experiments, we validated our experimental approach and analyzed the binding of the export cargo eIF1A from a HeLa cytosolic extract to immobilized HZZ-tagged importin 13. As shown in supplemental Fig. S1B, eIF1A interacted with importin 13 to some extent without the addition of RanQ69L. RanQ69L clearly enhanced binding of eIF1A to importin 13, as expected. Ubc9, when added as a recombinant protein in large excess to the reaction, reduced binding of eIF1A to its export receptor, as described previously (12). Next, we analyzed binding of cytosolic Ubc9 to immobilized importin 13. Here, RanQ69L largely abolished binding of the cargo to the transport receptor, as expected for an import substrate (supplemental Fig. S1C). Like the observation made for the export cargo eIF1A, exogenous Ubc9 is expected to compete with other potential import cargoes for importin 13 binding. Together, a careful comparison of proteins binding to importin 13 in the absence or presence of RanQ69L or Ubc9 should allow the identification of specific import or export cargoes.
SILAC-experiments with Ubc9 as a Competitor
We included the above criteria for specific cargo binding in our subsequent experiments. Again, triple-SILAC experiments were performed, where immobilized importin 13 was incubated with extracts from HeLa cells that had been grown under three different conditions as described above. The extracts were supplemented with RanQ69L (e.g. the “medium” extract), Ubc9 (e.g. the “heavy” extract) or buffer (e.g. the “light” extract). We performed three independent biological replicates with two SILAC experiments under identical labeling conditions (SILAC 2 and 3) and a third one (SILAC 4) with a switch in the labeling scheme (“reverse” and “forward” reactions). After the binding reactions, the beads were washed and combined, and bound proteins were eluted and analyzed by LC-MS. A flow chart of the experimental setup is depicted in Fig. 2A. Fig. 2B shows the result for proteins that compete with Ubc9 for binding to immobilized importin 13 in forward (fwd) and reverse (rev) reactions. As described above, binding of import and export cargoes to the importin 13 matrix is expected to be reduced by Ubc9 (supplemental Fig. S1B). Indeed, many of the previously described importin 13 import cargoes were re-identified with high scores. CHRAC1, for example, which forms a heterodimer with POLE3 (13), was enriched 39-fold (log 2 = 5.3) on the HZZ-importin 13 matrix in reactions in the absence of Ubc9 as compared with reactions in its presence (mean value of SILAC 2–4). Other known importin 13 cargoes that were found in this screen (underlined) are MAGOH (Mago (12)), NFYB (14), DR1 (NC2β (15)) and DRAP1 (NC2α (15), some of them with a rather low enrichment factor. Potentially, these cargoes have a high affinity for importin 13, resulting in efficient competition with exogenous Ubc9, and, consequently, a low enrichment (Imp13/Imp13+Ubc9). Indeed, in SILAC 1 all these proteins showed a strong enrichment for importin 13 compared with the novel cargo candidates. Note that Ubc9 itself was identified with a high log2-ratio in the reverse reaction (see supplemental Table S1), because the protein expressed in bacteria that was added in excess (i.e. Ubc9 with “normal,” light amino acids) could compete with the isotopically labeled forms of Ubc9, as present in the “medium” and the “heavy” extracts. The forward reaction, however, would not result in meaningful ratios, because high concentrations of “light” recombinant Ubc9 were present in the “light” extract. With our significance criteria, 217 proteins, with reduced importin 13 binding in the presence of Ubc9 were identified, 186 of which were also identified in SILAC 1. One protein with very high log2-ratios in forward and reverse reactions was the nitric oxide synthase interacting protein (NOSIP).
RanGTP Dependent Cargo Binding to Importin 13
To specifically address potential importin 13 export substrates, we next analyzed proteins that were enriched on the importin 13 matrix by the addition of RanQ69L, either in the absence or presence of Ubc9. Fig. 2C shows a plot of log2-ratios for individual proteins obtained from reactions containing or lacking RanQ69L (x axis) and reactions containing either RanQ69L or Ubc9 (y axis). As in SILAC 1, RanBP1 was strongly enriched on the importin 13 matrix in the presence of RanQ69L. Another protein with very high ratios is eIF1A, the classic importin 13 export cargo, confirming the robustness of our experimental approach. In total, we identified 175 proteins that were enriched by RanQ69L in SILAC 2–4, of which 117 were also significant in SILAC 1, e.g. APEX1, NSUN2 and RTCA (see Tables I and supplemental Table S1). Therefore, we included the results of SILAC 1 in our analysis, where proteins were identified that interact with HZZ-importin 13 but not with the tag alone. The combined results of SILAC 1 and SILAC 2–4 are assembled in Tables I and supplemental Table S1. In total, 255 proteins were identified as potential importin 13 cargoes in all four SILAC experiments. In addition, 82 proteins were significant in the three experiments SILAC 2–4, but not in SILAC 1. A Venn-diagram depicting the results of the two sets of experiments is shown in supplemental Fig. S2A. We also compared our importin 13 binding partners as identified in SILAC 2–4 with proteins that had been described in two previous proteomic studies (29, 30). As shown in supplemental Fig. S2B, a significant overlap (87 proteins) was observed. In addition, we found 250 proteins that had not been identified before. Several reasons for only partially overlapping sets of importin 13 cargoes are obvious: Imamoto and coworkers (29) used a very different experimental setup, searching for proteins that are imported into the nucleus in the presence of importin 13. Beck and coworkers (30), on the other hand, used a proximity ligation approach (BioID).
Importin 13 is a Rate-limiting Factor for Nuclear Import and Export
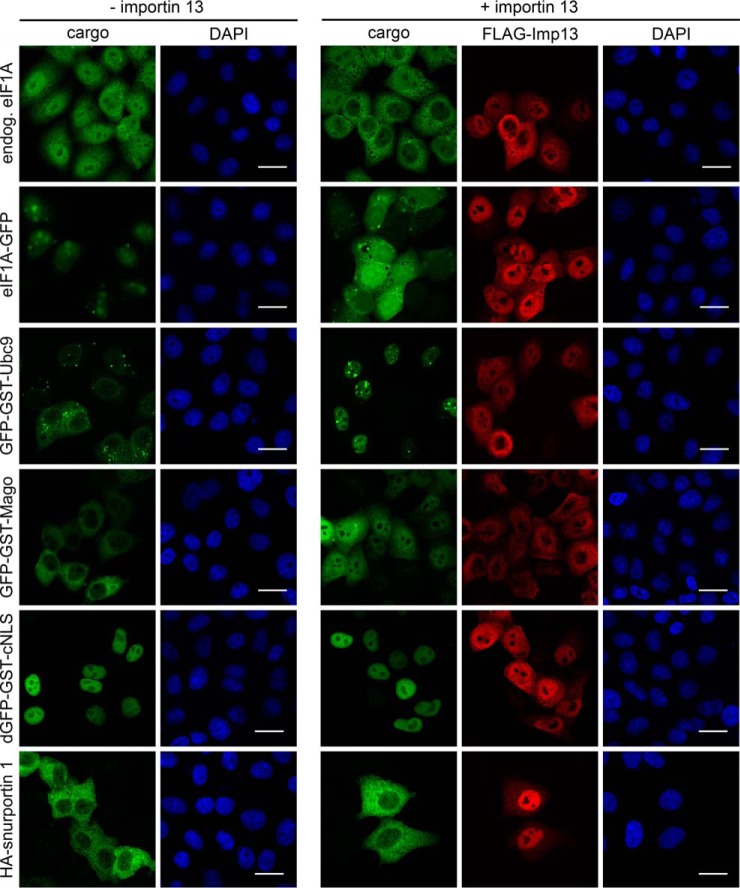
The CRM1-specific export inhibitor leptomycin B (LMB) (51) has been an extremely useful tool for the analysis of CRM1-dependent nuclear export. No such inhibitor is available for importin 13 or, as a matter of fact, for any other transport receptor. Therefore, other means are required to analyze the role of importin 13 in nucleocytoplasmic transport of candidate cargoes. The cellular concentration of many nuclear transport factors is in the micromolar range (52) and may not be rate-limiting for transport, even of overexpressed cargoes. In our HeLa cells, we measured a total concentration of importin 13 of ∼80 nm, i.e. about one order of magnitude lower than that of other NTRs (supplemental Fig. S3). For importin β, for example, we measured a concentration of ∼1.7 μm. Hence, importin 13 might be rate-limiting for transport. Indeed, endogenous eIF1A, the classic importin 13 export cargo, was found to localize largely to the nucleus in control cells (Fig. 3). Upon overexpression of importin 13, however, the protein was found in the cytoplasm. Essentially the same observation was made for overexpressed, GFP-tagged eIF1A, as reported previously (18). Likewise, overexpressed GFP-GST-Ubc9 and GFP-GST-Mago were cytoplasmic in control cells, but nuclear in importin 13-overexpressing cells. Our control proteins (dGFP-GST-cNLS, a cargo of the classic importin α/β pathway, and HA-snurportin 1, an established cargo of the export receptor CRM1 (53)) were not affected by exogenous importin 13. Note that the distribution of FLAG-importin 13 was heterogeneous. This may result from different expression levels, overexpressed cargo proteins and/or different stages of the cell cycle. Together, these results show that importin 13 is rate-limiting for nucleocytoplasmic transport of endogenous and exogenous cargoes, because of low total or active importin 13 concentrations. The activity of importin 13 could be affected by a cofactor and/or post-translational modifications. Hence, overexpression of importin 13 could be used to analyze the effect of the transport receptor on candidate importin 13 cargoes.
Fig. 3.
Importin 13 is rate-limiting for transport of cargo proteins. HeLa cells were co-transfected with plasmids coding for eIF1A-GFP, GFP-GST-Ubc9, GFP-GST-Mago, dGFP-GST-cNLS or HA-snurportin 1 and FLAG-importin 13 or an empty vector. For endogenous eIF1A, HeLa cells were transfected with FLAG-importin 13 or an empty vector and stained with an antibody against eIF1A. FLAG-importin 13 and HA-snurportin 1 were visualized by indirect immunofluorescence using anti-FLAG and anti-HA antibodies, respectively. DNA was stained with DAPI. Scale bars, 20 μm. Note that eIF1A-GFP is enriched in nucleoli whereas endogenous eIF1A appears to be excluded from these sites. This, however, can be ascribed to the eIF1A antibody, which does not detect nucleolar eIF1A (data not shown).
Functional Validation of Importin 13 Cargoes
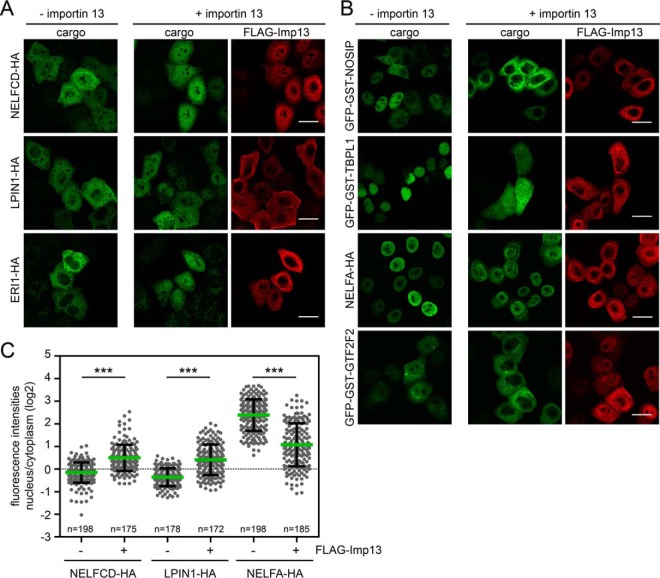
To validate our novel cargo candidates, we performed importin 13 overexpression experiments. The coding sequences of 45 candidate proteins from different categories of significance were amplified from HeLa cDNA and cloned into expression vectors containing C-terminal HA- or N-terminal GFP-GST-tags. The latter tag was chosen to significantly increase the molecular weight of the protein of interest, restricting its passive diffusion across the NPC, whereas the small HA-tag was chosen to modify the protein as little as possible. Tags were fused to either the N- or C terminus to ascertain that the tag position does not interfere with importin 13 interaction. We then analyzed the subcellular localization of the individual proteins in the absence or presence of overexpressed FLAG-tagged importin 13. First, we analyzed putative cargoes, which showed reduced binding to importin 13 in the presence of Ubc9 (see Fig. 2B and Table I and supplemental Table S1). Of 25 proteins tested, only few showed a clear shift toward the nucleus in importin 13 overexpressing cells, namely the 3–5′ exoribonuclease 1 (ERI1-HA), the phosphatidic acid phosphohydrolase lipin-1 (LPIN1-HA), and the negative elongation factor C/D (NELFCD-HA; Figs. 4A and 4C). These proteins had been identified as candidates with high log2-values for the importin 13/importin 13 + Ubc9 ratio. Interestingly, importin 13 affected only the localization of HA- but not of GFP-GST-tagged substrates (data not shown). This observation suggests that the transport receptor may bind to N- or C-terminal regions of the respective proteins, an interaction that could be compromised by artificial tags.
Fig. 4.
Analysis of importin 13 cargo candidates. HeLa cells were transfected with plasmids coding for C-terminal HA-tagged or N-terminal GFP-GST-tagged importin 13 cargo candidates (i.e. proteins with reduced binding to importin 13 in the presence of Ubc9; compare Fig. 2B) and FLAG-importin 13 or an empty vector. Expressed proteins were detected by indirect immunofluorescence using anti-HA or anti-FLAG antibodies, or directly via the GFP-tag. NELFCD, LPIN1, and ERI1 showed a more nuclear localization upon importin 13 overexpression (A), and NOSIP, TBPL1, NELFA and GTF2F2 a more cytoplasmic localization (B). C, Dot plot of the log2-ratios of nuclear and cytoplasmic intensities of importin 13 cargo candidates with average values (green bars) and standard deviation (error bars). Data were scored from a minimum of three independent experiments. n, number of cells analyzed. ***, p value < 0.001. See Fig. 2B and Table I and S1 for additional proteins tested. Scale bars, 20 μm.
Several other proteins including NOSIP (nitric oxide synthase interacting protein), TBPL1 (TATA box-binding protein-like protein 1), NELFA (negative elongation factor A) and GTF2F2 (general transcription factor IIF subunit 2; Figs. 4B and 4C) as well as WDR61, XPA, SQSTM1, RBM22 (data not shown) were rather shifted toward the cytoplasm in importin 13-overexpressing cells or were not affected (e.g. GBF1; compare color code in Fig. 2B and supplemental Table S1). Of these, only GTF2F2 and TBPL1 were slightly enriched on importin 13 beads in SILAC 2–4 in the presence of RanQ69L, implicating them as bona fide export cargoes, albeit with rather low scores.
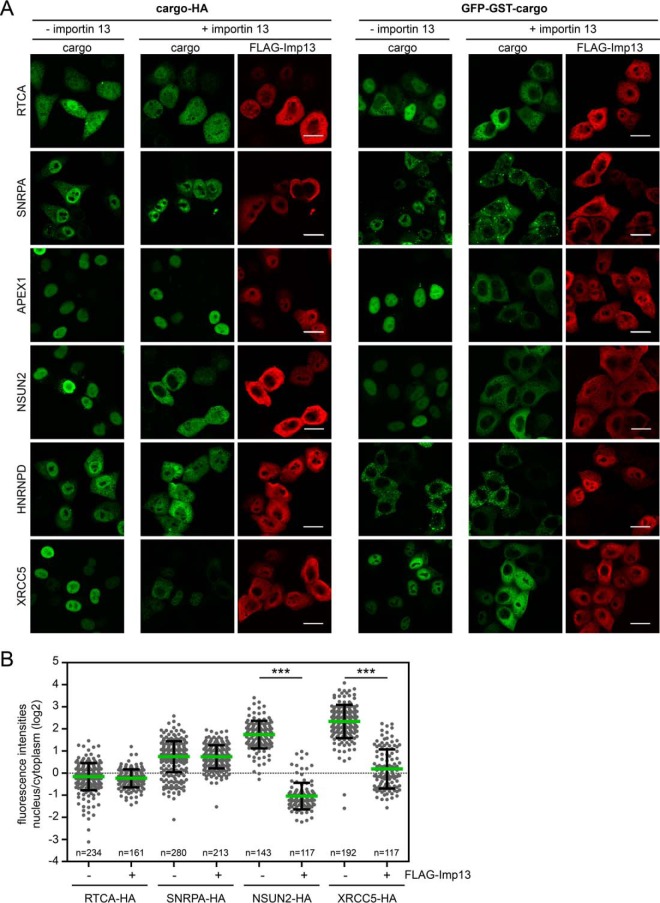
A somewhat clearer picture emerged when we analyzed putative importin 13 export cargoes, i.e. proteins enriched in the presence of RanQ69L (see Fig. 2C and Tables I and supplemental Table S1). As shown in Figs. 5A and 5B, many of the candidate proteins with different significance levels were affected by overexpression of importin 13, with all proteins showing a shift from the nucleus to the cytoplasm. Again, some proteins only changed their subcellular localization when expressed as either N- or C-terminal tagged proteins. RTCA-HA, SNRPA-HA and APEX1-HA, for example, showed a similar distribution in the absence or presence of overexpressed importin 13, whereas the GFP-GST-tagged proteins were clearly shifted toward the cytoplasm by exogenous importin 13. For the heterogeneous nuclear ribonucleoprotein HNRNPD, a shift toward the cytoplasm was observed for the HA-tagged version, whereas GFP-GST-HNRNPD was always cytoplasmic. Likewise, the serine-threonine kinase receptor-associated protein STRAP was only affected if fused to a C-terminal HA-tag (data not shown). For NSUN2 and XRCC5, a shift toward the cytoplasm was observed with both tags. Curiously, importin 13 had opposing effects on the negative elongation complex subunits NELFA and NELFCD. Whereas NELFA shifted toward the cytoplasm, NELFCD showed a more nuclear localization in cells coexpressing exogenous importin 13. Possibly, importin 13 can mediate transport of a single NELF subunit in one direction and of the entire NELF complex in the opposite direction.
Fig. 5.
Analysis of importin 13 cargo candidates enriched with RanQ69L. A, HeLa cells were transfected with plasmids coding for C-terminal HA- or N-terminal GFP-GST-tagged importin 13 cargo candidates (compare Fig. 2C) and FLAG-importin 13 or an empty vector. Expressed proteins were detected by indirect immunofluorescence using anti-HA or anti-FLAG antibodies or directly via the GFP-tag. Scale bars, 20 μm. B, Dot plot of the log2-ratios of nuclear and cytoplasmic intensities of importin 13 cargo candidates with average values (green bars) and standard deviation (error bars). Data were scored from a minimum of three independent experiments. n, number of cells analyzed. ***, p value < 0.001. See Fig. 2C and Table I and S1 for additional proteins tested.
Of 45 proteins tested, 21 were not affected by importin 13 overexpression. Several explanations appear plausible: First, some of these proteins exhibited rather unspecific binding to the HZZ-matrix in SILAC 1 (i.e. PRMT5, EIF3F, EIF3G, GPN1, BYSL; compare Fig. 1B). Second, posttranslational modifications like phosphorylation or SUMOylation, or additional cofactors might become rate-limiting for transport under our experimental conditions. Finally, some of the interaction partners could reflect transport-independent functions of importin 13. In this regard, additional cellular roles have been described for importin β (54, 55) and CRM1 (56) that are not related to nucleocytoplasmic transport.
Direct Interaction of Cargo Proteins with Importin 13
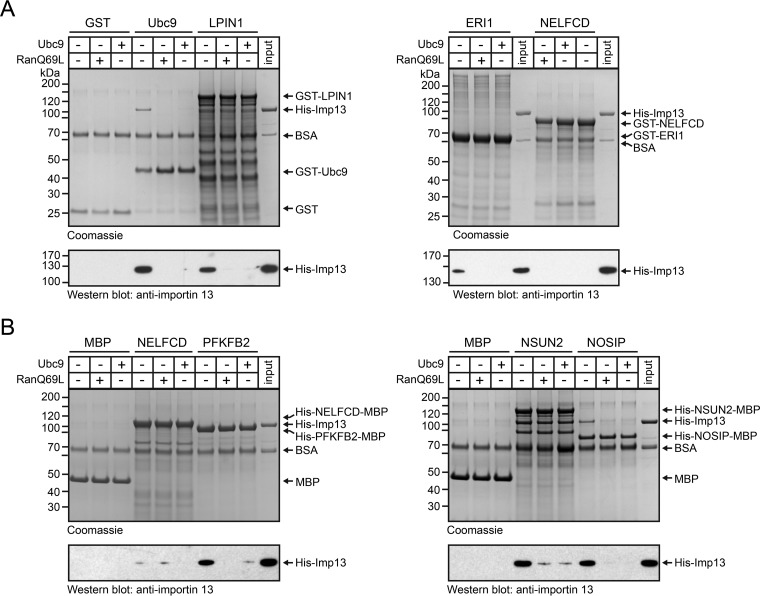
To corroborate our findings, we performed binding experiments with purified proteins. Several candidate substrates were expressed as GST- (Ubc9 as a control and LPN1, ERI1 and NELFCD) or MBP- (NELFCD, PFKFB2, NSUN2, NOSIP, TBPL1, XRCC5) tagged fusion proteins, immobilized on beads and tested for interaction with importin 13 in the absence or presence of Ubc9 or RanQ69L, respectively. For these experiments, we used a C-terminally truncated version of RanQ69L (RanQ69L(aa1–180)), which is frequently used in binding studies and crystallization trials (36, 57). No binding of importin 13 to immobilized GST or MBP was observed (Fig. 6A and 6B). Likewise, we could not detect significant binding of the transport receptor to GST-NELFCD (Fig. 6A), NELFCD-MBP (Fig. 6B) or XRCC5-MBP (data not shown). These proteins might only interact with importin 13 in the context of a larger complex containing additional components. In this regard, several known importin 13 cargoes have been reported to bind only as heterodimers (e.g. CHRAC1/POLE3) but not as individual subunits to importin 13 (13). This could also apply to XRCC5, which forms a heterodimer with XRCC6 (58), and NELFCD, a subunit of the negative elongation complex (59). Note that NELFA and NELFCD show a similar enrichment on the affinity matrix (compare Fig. 2B), as do many of the known importin 13 cargo heterodimers. Ubc9, ERI1, LPIN1 (Fig. 6A) and PFKFB2, NSUN2 and NOSIP (Fig. 6B) and TBPL1 (data not shown), by contrast, did form a complex with the transport receptor in the absence of RanQ69L and Ubc9, but not in the presence of either protein, demonstrating the specificity of the interaction. Note that some interactions could only be detected by Western-blotting. Perhaps, stable binding to importin 13 requires a posttranslational modification of the cargo protein and/or the transport receptor that is not present in our bacterially expressed proteins. Furthermore, binding of physiological cargoes to importin 13 might require the presence of certain co-factors (“co-cargoes”) that are present in the cellular extracts but not in our binding reactions with purified proteins.
Fig. 6.
Importin 13 cargo candidates interact directly with importin 13. GST- (A) or MBP- (B) tagged importin 13 cargo candidates or the tags alone were immobilized on glutathione or amylose beads, respectively, and incubated with His-importin 13 in the absence or presence of RanQ69L(aa1–180)-GTP or Ubc9, as indicated. Bound proteins were analyzed by SDS-PAGE, followed by Coomassie staining (top) and immunoblotting with anti-importin 13 antibodies (bottom). GST-Ubc9 served as a positive control for importin 13 binding. The input corresponds to 10% or 1% of His-importin 13 used in the binding reactions for the Coomassie gels or the Western blots, respectively.
Together, several candidate proteins were verified as novel, bona fide transport cargoes of importin 13. Our results, however, suggest that the RanQ69L-criterion does not always allow a clear distinction between import and export substrates under our experimental conditions, although in the pilot experiment (supplemental Fig. S1C), the classic import cargo Ubc9 was affected as expected. Binding of NOSIP to importin 13, for example, was not enhanced by RanQ69L, although the protein showed a more cytoplasmic localization in cells overexpressing importin 13, indicative of nuclear export. Apart from direct export, other mechanisms could account for a more cytoplasmic localization of NOSIP and other cargo candidates in importin 13 overexpression experiments, including indirect stimulation of export or inhibited nuclear import, as has been suggested for the testis specific form of importin 13 (28). Together, functional experiments in cells are required to understand the effects of importin 13 on the subcellular localization of such proteins.
Common Cargoes of Importin 13 and CRM1
Several of the chosen export candidates were cytoplasmic even in control cells, precluding an export-promoting effect of overexpressed importin 13. We therefore tested if these proteins could be substrates of the classic export receptor CRM1. Indeed, the CRM1-inhibitor LMB led to a nuclear localization of the transcription factor BTF3-HA, the eukaryotic translation factor eIF2D (EIF2D-HA) and the ubiquitin-binding protein sequestosome 1 (SQSTM1-HA; Fig. 7A). Strikingly, these proteins were shifted back to the cytoplasm in the presence of LMB in importin 13 overexpressing cells, suggesting that at least two factors, CRM1 and importin 13, can serve as alternative export receptors. As a control, we transfected cells with a plasmid coding for snurportin 1, a classical CRM1-export cargo (53). Like the novel importin 13 cargoes, snurportin 1 also accumulated in the nucleus in the presence of LMB. This block in nuclear export, however, could not be overcome by overexpression of importin 13. In light of these results, we compared our list of putative importin 13 cargoes (SILAC 2–4) with two comprehensive lists of CRM1 cargoes (25, 26). Our analysis revealed 94 proteins that might use both transport pathways for nuclear export (supplemental Fig. S2C and supplemental Table S1). This redundancy could reflect differential usage of transport receptors in different cell types and/or during different stages of the cell cycle.
Fig. 7.
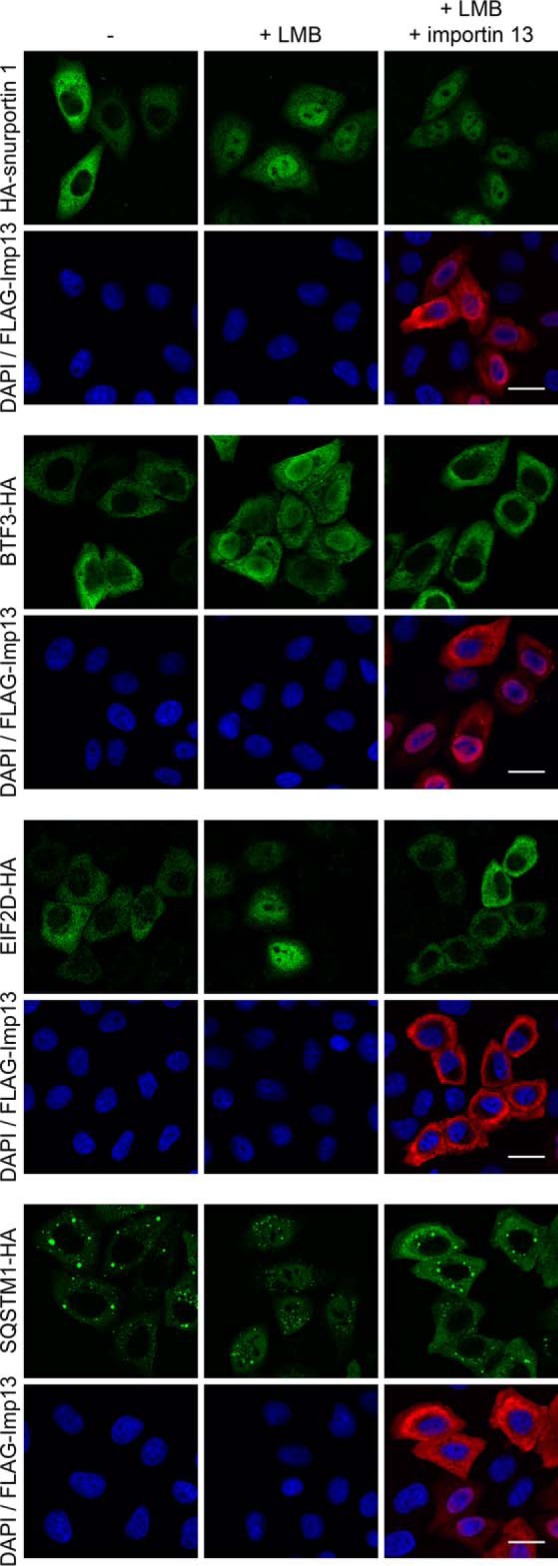
Redundancy in importin 13 and CRM1 mediated transport. HeLa cells were transfected with plasmids coding for HA-tagged importin 13 cargo candidates BTF3, EIF2D and SQSTM1 or HA-snurportin 1 and FLAG-importin 13 or an empty vector. Where indicated, cells were treated with 10 nm LMB for 2 h. Proteins were detected by indirect immunofluorescence using anti-HA or anti-FLAG antibodies. DNA was stained with DAPI. Scale bars, 20 μm.
Together, our results show that our experimental setup is a valid approach for the identification of importin 13-dependent nuclear export cargoes. With our criteria of statistical significance, 175 were identified as potential export substrates (SILAC2–4). Using functional assays, not only proteins with a high score, but also with intermediate (e.g. BTF3-HA) or rather low scores (e.g. GST-GFP-XRCC6) could be validated.
Different Importin 13 Interaction Modes of Individual Cargoes
As a bi-directional transporter, importin 13 uses different regions for interaction with import or export cargoes (17, 18). Curiously, however, also import cargoes bind differently to importin 13 (17). Importin 13 mutants deficient in importing either Ubc9 or Mago, for example, were still able to transport the other protein. We therefore tested different importin 13 mutants for their ability to import or export newly identified cargo proteins. Importin 13 E436R/D418R is deficient in exporting eIF1A, whereas the mutants E426R and K802E/R803E are deficient in importing Ubc9 and Mago, respectively (17, 18). As shown in Figs. 8A and 8B, our control substrates GFP-GST-Ubc9, GFP-GST-Mago and eIF1A-GFP were affected by the different importin 13 mutants as expected. Import of LPIN1-HA was promoted by wild type importin 13 and by the mutant K802E/R803E, but not by the mutants importin 13 E436R/D418R and E426R, suggesting that lipin-1 uses similar regions on importin 13 for interaction compared with two well-established transport cargoes. Nuclear import of NELFCD-HA, on the other hand, was equally efficient with wild type importin 13 or any of the three mutant proteins. Likewise, all forms of importin 13 promoted export of NSUN2-HA (Figs. 8A and 8B) and several other proteins that were tested (e.g. HNRNPD-HA and GFP-GST-RTCA; data not shown). Together, these results show that different regions in importin 13 can mediate the interaction with transport cargoes, demonstrating the versatility of this transport receptor. As already shown for Ubc9 and eIF1A by X-ray crystallography (17, 18), importin 13 may not recognize simple, linear transport signals, but rather complex structures as they occur only in correctly folded molecules or even in multimeric protein complexes.
Fig. 8.
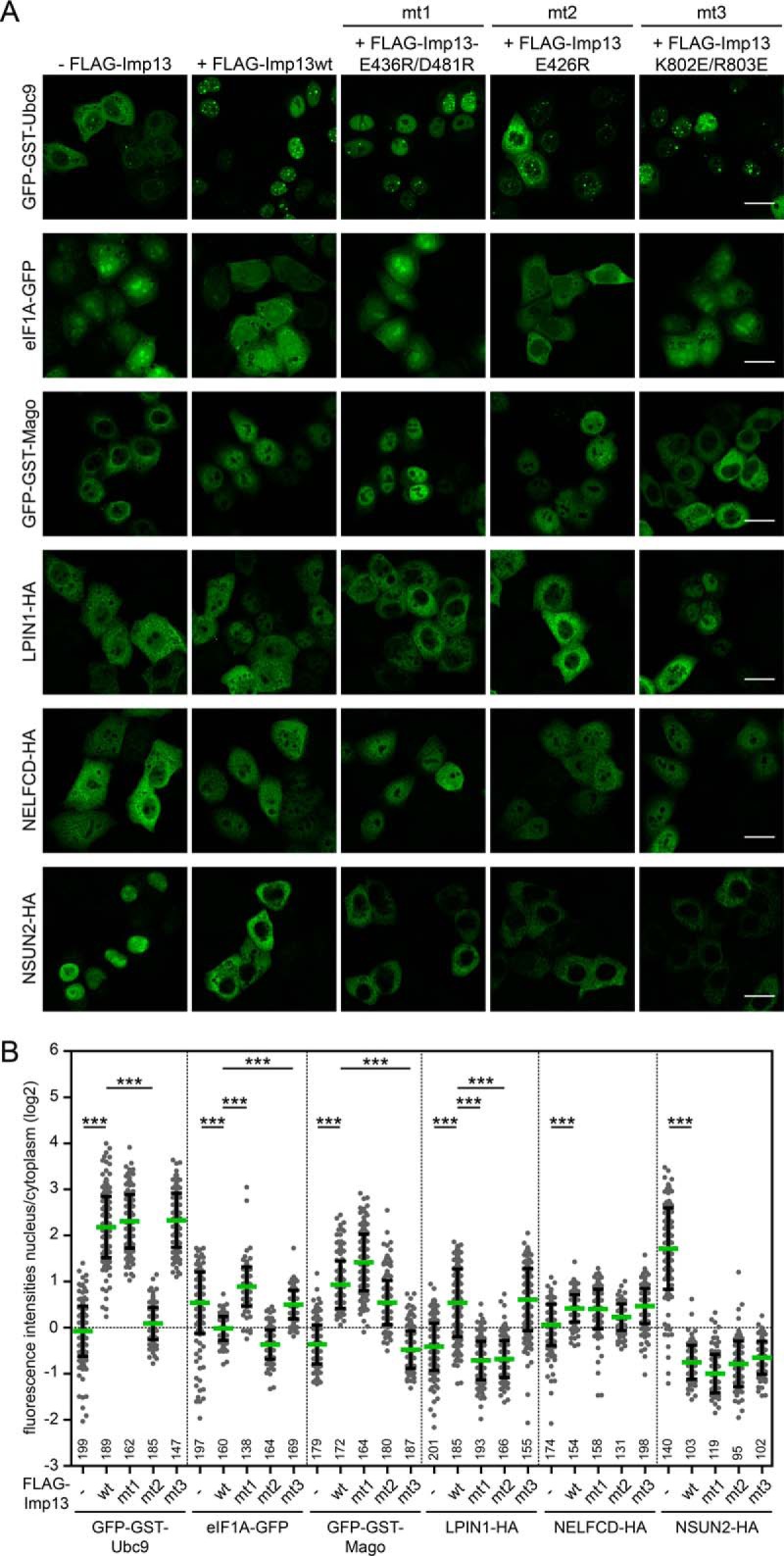
Importin 13 has different binding modes for cargo proteins. A, HeLa cells were transfected with plasmids coding for GFP-tagged known importin 13 cargoes (Ubc9, Mago and eIF1A) or HA-tagged importin 13 cargo candidates (LPIN1, NELFCD, NSUN2) and FLAG-tagged wildtype (wt) or mutant importin 13 (mt1: E436R/D481R, mt2: E426R, mt3: K802E/R803E) or an empty vector (-FLAG-Imp13). Expressed proteins were detected by indirect immunofluorescence using anti-HA antibodies or directly via the GFP-tag. Scale bars, 20 μm. See supplemental Fig. S4 for DAPI staining and FLAG-Imp13 co-transfection. B, Dot plot of the log2-ratios of nuclear and cytoplasmic intensities of importin 13 cargo candidates with average values (green bars) and standard deviation (error bars). The number below each dot plot corresponds to the number of cells analyzed. Data were scored from a minimum of three independent experiments. ***, p value < 0.001.
CONCLUSION
Three recent large-scale studies used different approaches for the identification of importin 13 cargoes: Imamoto and coworkers (29) used reconstituted nuclear import in permeabilized cells, Jans and coworkers (28) a yeast-two-hybrid approach and Beck and coworkers (30) proximity ligation coupled to mass spectrometry (BioID). Our study is based on specific interactions of importin 13 with cargo proteins. Hence, it nicely complements the three previous studies, yielding a significant overlap of cargoes but also many uniquely identified proteins (supplemental Fig. S2B). Using two different experimental setups in SILAC 1 (Fig. 1) and SILAC 2–4 (Fig. 2), we identified a total of 255 high-confidence importin 13 cargoes, which (i) bound to HZZ-importin 13 but not to the HZZ-tag, and whose binding was (ii) reduced by exogenous Ubc9 or (iii) affected by RanQ69L (see supplemental Table S1). The latter criterion was not a perfect predictor of a cargo protein being an import or an export substrate. For the very versatile transport receptor importin 13, detailed analyses are required to clarify its exact role in the regulation of the subcellular localization of cargo proteins. We initiated such a detailed analysis for a large number of proteins, using biochemical and cell biological assays.
DATA AVAILABILITY
The raw mass spectrometry data and search output files are deposited at ProteomeXchange via the PRIDE database with the accession number PXD008459.
Supplementary Material
Acknowledgments
We thank Drs. Frauke Melchior, Flavia Bono, Detlef Doenecke, Blanche Schwappach and Dirk Görlich for the generous gift of reagents and our former lab members Drs. Sarah Port and Sarah Wälde for the generation of expression plasmids. We also thank Annegret Nath and Mohamed Hamed for the purification of proteins and Claudia Nolte for cloning of LPIN1-plasmids.
Footnotes
* The project was funded by a grant from the Deutsche Forschungsgemeinschaft (DFG) to R.H.K. (KE 660/14-1) and also by Sonderforschungsbereich SFB 860 (project B06).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- NPC
- nuclear pore complex
- FBS
- fetal bovine serum
- NTR
- nuclear transport receptors
- FG
- phenylalanine-glycine
- RanGAP
- Ran GTPase activating protein
- SILAC
- stable isotope labeling in cell culture
- TPB
- transport buffer.
REFERENCES
- 1. Beck M., and Hurt E. (2017) The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18, 73–89 [DOI] [PubMed] [Google Scholar]
- 2. Dickmanns A., Kehlenbach R. H., and Fahrenkrog B. (2015) Nuclear pore complexes and nucleocytoplasmic transport: from structure to function to disease. Int. Rev. Cell Mol. Biol. 320, 171–233 [DOI] [PubMed] [Google Scholar]
- 3. Knockenhauer K. E., and Schwartz T. U. (2016) The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmidt H. B., and Görlich D. (2016) Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 41, 46–61 [DOI] [PubMed] [Google Scholar]
- 5. Fried H., and Kutay U. (2003) Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60, 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hutten S., and Kehlenbach R. H. (2007) CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 17, 193–201 [DOI] [PubMed] [Google Scholar]
- 7. Calado A., Treichel N., Muller E. C., Otto A., and Kutay U. (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21, 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshida K., and Blobel G. (2001) The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 152, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gontan C., Guttler T., Engelen E., Demmers J., Fornerod M., Grosveld F. G., Tibboel D., Görlich D., Poot R. A., and Rottier R. J. (2009) Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J. Cell Biol. 185, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lipowsky G., Bischoff F. R., Schwarzmaier P., Kraft R., Kostka S., Hartmann E., Kutay U., and Görlich D. (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19, 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang C., Sweezey N. B., Gagnon S., Muskat B., Koehler D., Post M., and Kaplan F. (2000) A novel karyopherin-beta homolog is developmentally and hormonally regulated in fetal lung. Am. J. Respir. Cell Mol. Biol. 22, 451–459 [DOI] [PubMed] [Google Scholar]
- 12. Mingot J. M., Kostka S., Kraft R., Hartmann E., and Görlich D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J. 20, 3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walker P., Doenecke D., and Kahle J. (2009) Importin 13 mediates nuclear import of histone fold-containing chromatin accessibility complex heterodimers. J. Biol. Chem. 284, 11652–11662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kahle J., Baake M., Doenecke D., and Albig W. (2005) Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol. Cell. Biol. 25, 5339–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kahle J., Piaia E., Neimanis S., Meisterernst M., and Doenecke D. (2009) Regulation of nuclear import and export of negative cofactor 2. J. Biol. Chem. 284, 9382–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bono F., Cook A. G., Grünwald M., Ebert J., and Conti E. (2010) Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of importin 13. Mol. Cell 37, 211–222 [DOI] [PubMed] [Google Scholar]
- 17. Grünwald M., and Bono F. (2011) Structure of Importin13-Ubc9 complex: nuclear import and release of a key regulator of sumoylation. EMBO J. 30, 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grünwald M., Lazzaretti D., and Bono F. (2013) Structural basis for the nuclear export activity of Importin13. EMBO J. 32, 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cingolani G., Bednenko J., Gillespie M. T., and Gerace L. (2002) Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell 10, 1345–1353 [DOI] [PubMed] [Google Scholar]
- 20. Kimura M., Kose S., Okumura N., Imai K., Furuta M., Sakiyama N., Tomii K., Horton P., Takao T., and Imamoto N. (2013) Identification of cargo proteins specific for the nucleocytoplasmic transport carrier transportin by combination of an in vitro transport system and stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics. Mol. Cell. Proteomics 12, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blazek E., and Meisterernst M. (2006) A functional proteomics approach for the detection of nuclear proteins based on derepressed importin alpha. Proteomics 6, 2070–2078 [DOI] [PubMed] [Google Scholar]
- 22. Fukumoto M., Sekimoto T., and Yoneda Y. (2011) Proteomic analysis of importin alpha-interacting proteins in adult mouse brain. Cell Structure Function 36, 57–67 [DOI] [PubMed] [Google Scholar]
- 23. Kimura M., Okumura N., Kose S., Takao T., and Imamoto N. (2013) Identification of cargo proteins specific for importin-beta with importin-alpha applying a stable isotope labeling by amino acids in cell culture (SILAC)-based in vitro transport system. J. Biol. Chem. 288, 24540–24549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyamoto Y., Baker M. A., Whiley P. A., Arjomand A., Ludeman J., Wong C., Jans D. A., and Loveland K. L. (2013) Towards delineation of a developmental alpha-importome in the mammalian male germline. Biochim. Biophys. Acta 1833, 731–742 [DOI] [PubMed] [Google Scholar]
- 25. Kirli K., Karaca S., Dehne H. J., Samwer M., Pan K. T., Lenz C., Urlaub H., and Görlich D. (2015) A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thakar K., Karaca S., Port S. A., Urlaub H., and Kehlenbach R. H. (2013) Identification of CRM1-dependent Nuclear Export Cargos Using Quantitative Mass Spectrometry. Mol. Cell. Proteomics 12, 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu D., Marquis K., Pei J., Fu S. C., Cagatay T., Grishin N. V., and Chook Y. M. (2015) LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 31, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fatima S., Wagstaff K. M., Lieu K. G., Davies R. G., Tanaka S. S., Yamaguchi Y. L., Loveland K. L., Tam P. P., and Jans D. A. (2017) Interactome of the inhibitory isoform of the nuclear transporter Importin 13. Biochim. Biophys. Acta 1864, 546–561 [DOI] [PubMed] [Google Scholar]
- 29. Kimura M., Morinaka Y., Imai K., Kose S., Horton P., and Imamoto N. (2017) Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. eLife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mackmull M. T., Klaus B., Heinze I., Chokkalingam M., Beyer A., Russell R. B., Ori A., and Beck M. (2017) Landscape of nuclear transport receptor cargo specificity. Mol. Sys. Biol. 13, 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pichler A., Gast A., Seeler J. S., Dejean A., and Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 32. Guttler T., Madl T., Neumann P., Deichsel D., Corsini L., Monecke T., Ficner R., Sattler M., and Gorlich D. (2010) NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 17, 1367–1376 [DOI] [PubMed] [Google Scholar]
- 33. Gasteier J. E., Madrid R., Krautkramer E., Schroder S., Muranyi W., Benichou S., and Fackler O. T. (2003) Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J. Biol. Chem. 278, 38902–38912 [DOI] [PubMed] [Google Scholar]
- 34. Favaloro V., Vilardi F., Schlecht R., Mayer M. P., and Dobberstein B. (2010) Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 123, 1522–1530 [DOI] [PubMed] [Google Scholar]
- 35. Melchior F., and Gerace L. (1995) Mechanisms of nuclear protein import. Curr. Opin. Cell Biol. 7, 310–318 [DOI] [PubMed] [Google Scholar]
- 36. Port S. A., Monecke T., Dickmanns A., Spillner C., Hofele R., Urlaub H., Ficner R., and Kehlenbach R. H. (2015) Structural and functional characterization of CRM1-Nup214 interactions reveals multiple FG-binding sites involved in nuclear export. Cell Rep. 13, 690–702 [DOI] [PubMed] [Google Scholar]
- 37. Hu T., Guan T., and Gerace L. (1996) Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 134, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kehlenbach R. H., Dickmanns A., Kehlenbach A., Guan T., and Gerace L. (1999) A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charneau P., Mirambeau G., Roux P., Paulous S., Buc H., and Clavel F. (1994) HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241, 651–662 [DOI] [PubMed] [Google Scholar]
- 40. Ribbeck K., and Görlich D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shevchenko A., Wilm M., Vorm O., and Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 42. Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 43. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 44. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 45. Cox J., and Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 47. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., and Struhl K. (1994) Current protocols in molecular biology. New York: Greene Publishing Associates and Wiley-Interscience [Google Scholar]
- 48. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., and Sabatini D. M. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klebe C., Bischoff F. R., Ponstingl H., and Wittinghofer A. (1995) Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34, 639–647 [DOI] [PubMed] [Google Scholar]
- 51. Wolff B., Sanglier J. J., and Wang Y. (1997) Leptomycin B is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4, 139–147 [DOI] [PubMed] [Google Scholar]
- 52. Riddick G., and Macara I. G. (2005) A systems analysis of importin-α-β mediated nuclear protein import. J. Cell Biol. 168, 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paraskeva E., Izaurralde E., Bischoff F. R., Huber J., Kutay U., Hartmann E., Lührmann R., and Görlich D. (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang C. C., Huang T. L., Shimamoto Y., Tsai S. Y., and Hsia K. C. (2017) Regulation of mitotic spindle assembly factor NuMA by Importin-beta. J. Cell Biol. 216, 3453–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., and Weis K. (2001) Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106 [DOI] [PubMed] [Google Scholar]
- 56. Wu Z., Jiang Q., Clarke P. R., and Zhang C. (2013) Phosphorylation of Crm1 by CDK1-cyclin-B promotes Ran-dependent mitotic spindle assembly. J. Cell Sci. 126, 3417–3428 [DOI] [PubMed] [Google Scholar]
- 57. Monecke T., Güttler T., Neumann P., Dickmanns A., Görlich D., and Ficner R. (2009) Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 324, 1087–1091 [DOI] [PubMed] [Google Scholar]
- 58. Walker J. R., Corpina R. A., and Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 59. Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D. K., Hasegawa J., Omori M., Inukai N., Endoh M., Yamada T., and Handa H. (2003) Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry data and search output files are deposited at ProteomeXchange via the PRIDE database with the accession number PXD008459.