Abstract
Correlated mutations between residue pairs in evolutionarily related proteins arise from constraints needed to maintain a functional and stable protein. Identifying these inter-related positions narrows down the search for structurally or functionally important sites. MISTIC is a server designed to assist users to calculate covariation in protein families and provide them with an interactive tool to visualize the results. Here, we present MISTIC2, an update to the previous server, that allows to calculate four covariation methods (MIp, mfDCA, plmDCA and gaussianDCA). The results visualization framework has been reworked for improved performance, compatibility and user experience. It includes a circos representation of the information contained in the alignment, an interactive covariation network, a 3D structure viewer and a sequence logo. Others components provide additional information such as residue annotations, a roc curve for assessing contact prediction, data tables and different ways of filtering the data and exporting figures. Comparison of different methods is easily done and scores combination is also possible. A newly implemented web service allows users to access MISTIC2 programmatically using an API to calculate covariation and retrieve results. MISTIC2 is available at: https://mistic2.leloir.org.ar.
INTRODUCTION
Evolutionary pressure to maintain a functional and stable protein structure gives rise to correlated mutations between residue pairs. For example, mutations of essential residues in a protein sequence may occur, only if a compensatory mutation takes place elsewhere within the protein to preserve its structure and/or activity. Those inter relationships can guide the identification of structurally or functionally important positions in a given protein fold or family (1–3).
Correlated mutations can be observed in multiple sequence alignments of homologous proteins (MSAs). Several methods have been developed to capture the sequence variability in these MSAs and have been successfully applied to contact and structure prediction (these are some examples, the list is not exhaustive (4–13)). Early covariation methods were based on mutual information (MI) while modern approaches make use of global statistical methods (e.g. inverse potts models, sparse inverse covariance estimation). Some applications include contact-guided ab initio protein structure prediction, model ranking, evaluation and improvement (14–18), and detection of allosteric communication (3,19,20). Covariation signal is made up of phylogeny, structure, function, interactions and stochastic components (21). High covariation scores are not proof of coevolution but suggestive of it. Statistical dependency between amino acid positions may arise either from direct or indirect correlated residues. This fact can be used to classify covariation methods in two categories: traditional methods, that consider all covarying interactions as independent between each other, and direct coupling methods that deconvolute the covariation signal in order to infer only direct interactions.
Several servers are available to calculate covariation between positions (11,22–24), most of them are only focused in predicting residue contacts. Scripting tools are available for most methods as standalone binaries or other scripting interfaces, some of them providing functions to retrieve and preprocess sequence and structural data (25,26).
MISTIC2 offers support for four covariation methods: (i) Corrected Mutual Information (MIp) (25,27), (ii) mean field Direct Coupling Analysis (mfDCA) (6,28), (iii) pseudo-likelihood maximization DCA (plmDCA) (5,8) and (iv) multivariate Gaussian modeling DCA (gaussianDCA) (29). It makes the comparison between them possible in a simple way, and at the same time it provides a framework to visualize and explore the results interactively.
MATERIALS AND METHODS
Input
For covariation analysis a Multiple Sequence Alignment is the primary requirement. Typically, the alignment should contain a large number of diverse sequences for accurate contact prediction (6,7,14,27). Pfam domain families are frequently used since they contain extensive alignments of homologous sequences. The server accepts as inputs a Pfam accession, an Uniprot ID and custom alignments in FASTA, Stockholm, Clustal, Nexus, PIR and Phylip format. If an Uniprot ID is provided, the server will list the Pfam models for the query protein and also recommend PDB structures. In each case, a reference sequence must be selected for mapping the MSA couplings, conservation and other scores onto the PDB structure.
Output
Completed jobs can be accessed through the submission page using its job id. Expected running times for each algorithm are included in the supplementary file. A brief summary of the input data will appear along with the results for calculated covariation methods.
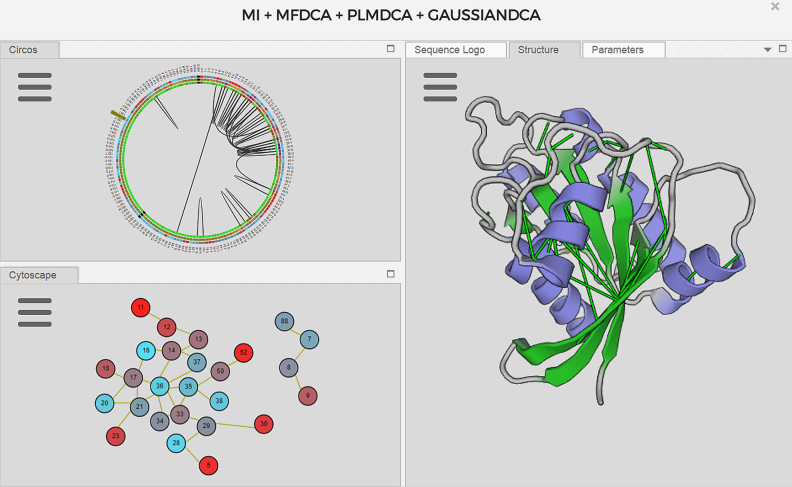
Results are explored through a window layout made of several tabs, each of them containing one ‘component’. Available components include: Circos, Structure Viewer, Covariation Network, Reference Sequence Viewer, Sequence Logo, ROC Curve, Filters and Data Tables. The user can accommodate the tabs sharing the workspace for practical purposes (Figure 1).
Figure 1.
MISTIC2 workspace: upper left: circo visualization of the MSA information. Amino acid names and positions are in the outer ring. Conservation (second ring) from light blue (lower) to red (higher); cScore (third ring) from yellow (lower) to violet (higher); pScore (inner ring) from green (lower) to red (higher). Inner lines are the top 5% covariation scores. Bottom left: covariation network colored by conservation. Right: covariation network's selected edges mapped onto the 3D structure (ribbon representation, pdb code: 4LPK_B).
Covariation network
Covariation data is presented as an interactive network (30) with many information sources embedded in it, such as conservation, other evolutionary-based scores, secondary structure, markov clustering of the network, and it is interactively linked to a 3D structure viewer. Nodes represent columns of the input MSA numbered according to the reference sequence, and edges (links between nodes) illustrate covariation between these positions.
A Markov cluster algorithm (MCL) (31,32) applied on the covariation network topology allows to identify clusters of positions. The MCL algorithm has been extensively used for biological network clustering (33). It has been shown that clusters of correlated amino acids define ‘sectors’ within the protein that have functional roles (34,35). Although many clustering methods could be applied instead, the tool is intended to give a preliminary overview for hypothesis generation.
Structure viewer
From the network viewer, nodes and edges can be selected and mapped onto a reference PDB structure, including the coloring scheme of its attributes like conservation or cluster membership. Residue selections can be saved with different standard representations. This is useful for applying different filtering criteria and visualizing them at the same time.
Settings and filter tools
Filtering tools are offered for selecting specific subsets of nodes from the network for visualization. For example the user might want to filter the covariation network made up by the highest conserved residues, the subnetwork with the strongest covarying pairs, or contacting residue pairs, among others.
In the Filters tab, thresholds can be changed for any score and new results will be updated across all visualizations. Users can adjust filters for conservation and covariation scores, residue distance cutoff, and choose cScore and pScore thresholds. Raw data and tables can be downloaded in JSON and CSV format, respectively.
In the original MISTIC, derived Mutual Information scores were calculated for each position; these were named cumulative and proximity Mutual Information (cMI and pMI, respectively). These scores have been proved to be predictive of functional positions (1). MISTIC2 extends these derived scores to all available methods renamed as cumulative Score and proximity Score (cScore and pScore, respectively). Thus, the cScore for each position is the sum of its first neighbours covariation scores, being suggestive of how much information a residue shares. The pScore for a given position is calculated as the average of the cScore of every residue within 7.5 Å distance (by default). It gives an idea of the information accumulated in the proximity of a residue in the 3D structure. This score can only be calculated if a 3D structure is available. Both scores have an intrinsic threshold: for cScore the user has to choose the covariation limit to be considered (either a value or a top X% of the covariation scores), while for pScore a distance threshold has to be selected. An important remark is that cScore and pScore thresholds have been optimized only for catalytic residue prediction with MIp (1), whereas for DCA-like methods these thresholds are not optimized. MISTIC2 allows the user to adjust these cutoffs manually and all scores are updated dynamically.
Covariation circos
The covariation Circos is a circular representation of the information contained in an MSA. It summarizes the conservation of each MSA position, the cumulative score (cScore), the proximity score (pScore) and the covariation between positions in a concise and intuitive way. Lines in the center of the circle connect pairs of positions within the top 5% covariation score (by default) though this value can be changed by the user in the parameters tab (see Figure 1).
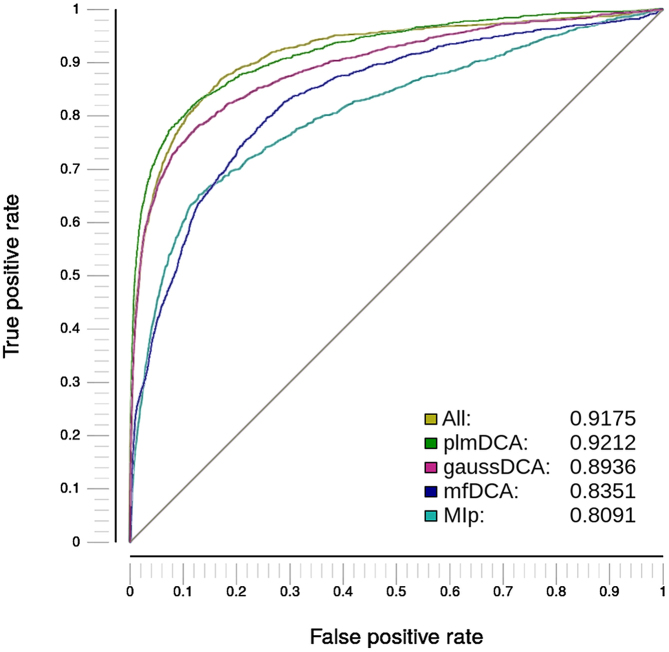
Area under the ROC curve
This tab lets the user evaluate each method's performance for residue contact prediction at a distance threshold set by the user (6.05 Å as default) (an AUC equal to 1 means a perfect predictor, AUC equal to 0.5 means random predictor) (Figure 2).
Figure 2.
AUC for residue contacts prediction. It can be observed that in this case (Pfam family: PF00071, pdb: 4LPK_B), there are methods that outperformed others. plmDCA method has the best predictive performance: plmDCA > all (plmDCA + Gaussian DCA + mfDCA + MI) > Gaussian DCA > mfDCA > MI.
Application programming interface
MISTIC2 web server now supports a web service by implementing an Application Programming Interface (API). It provides a programmatic access to MISTIC services without the need to go through the web interface to submit new jobs or retrieve results. Specific operations have been implemented to upload and validate the input data, as well as selecting which algorithms to run and change default parameters. An example python script is available in the supplementary material (Supplementary file ‘Programmatic access to MISTIC2’ section). The API endpoints are listed in Supplementary Table S1.
Case study KRas protein
Human Kras protein (Uniprot accession RASK_HUMAN) is a member of the RAS family of small monomeric GTPases. They function as molecular binary switches, with their biological activities being determined by their nucleotide-binding state. When bound to GTP, RAS proteins engage in a variety of downstream ‘effector’ pathways involved in cell growth, differentiation and survival. Mutations in Ras genes can lead to the production of permanently activated Ras proteins, resulting in an overactive downstream signaling inside the cell, even in the absence of incoming signals. Ras genes are key players in tumor pathogenesis being the three genes (HRas, KRas and NRas) the most common oncogenes in human cancer (36).
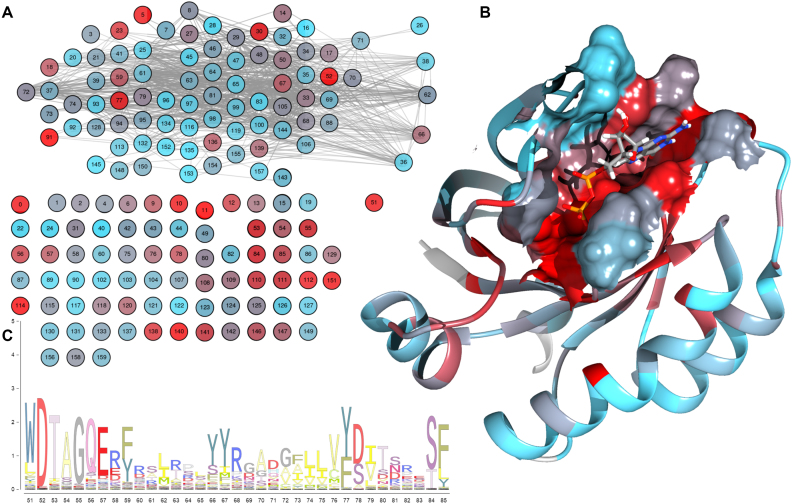
Different aspects of the Kras family of proteins such as conservation, coevolution and functionally important residues among others, can be studied with MISTIC2. Figure 3 shows that Kras-GTP/GTP interaction site is enriched in conserved residues, highlighting the functional relevance of the interaction site.
Figure 3.
(A) MIp covariation network with nodes colored by conservation, from light blue (lower) to red (higher). (B) Ribbon representation of the overall structure (PDB 4LPK_B) with surface representation of the GDP/GTP interaction site, colored as in the network. It is worth noting the high conservation in the binding site. (C) A section of the sequence logo which translates to the network conservation coloring schema.
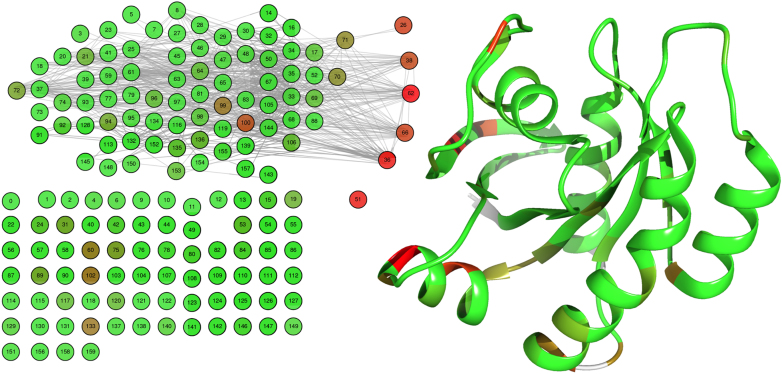
Coloring the covariation network by the pScore (pMIp in this case) reveals that the highest scoring set of residues is different than the set of highly conserved ones. The pScores point out positions structurally close to hub nodes (high cScore). The residues located at the right side of the network have low conservation (blue color in the network depicted in Figure 3) and high pScore (red in Figure 4), suggesting that they might be functionally important as they have a high information content in their structural proximity (1), yet this would never be noticed looking at their conservation values. We do not intend to prove their functionality, but to show the potential of the server.
Figure 4.
Left panel:network; right panel: structure. Colored by pMI from green to red (lower to higher scores).
DISCUSSION AND CONCLUDING REMARKS
We present MISTIC2 web server, an important update over the previous version that includes additional methods to calculate covariation, the possibility of running them all at once and compare them, a reworked results visualization and a web service API for programmatic access. The new interface can display sequence and structural aspects of protein families through a number of interactive representations, and includes tools for filtering and exporting figures and tables.
MISTIC2 is a simple and unique server that provides a powerful tool for non-bioinformaticians end-users to calculate and analyze the covariation signal contained within protein families. It also provides a programmatic access without the need to manually interact with the web interface. MISTIC2 presents itself as an essential tool for exploring protein family evolution, discovery of important residues, biological hypothesis generation and a tool to help guide rational experiment design.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agencia Nacional de promocion cientifica y tecnologica. Mincyt [PICT 2014-1087]. Funding for open access charge: Agencia National de Promocion cientifica y tecnologica. Mincyt.
Conflict of interest statement. None declared.
REFERENCES
- 1. Marino Buslje C., Teppa E., Di Domenico T., Delfino J.M., Nielsen M.. Networks of high mutual information define the structural proximity of catalytic sites: implications for catalytic residue identification. PLoS Comput. Biol. 2010; 6:e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McMurrough T.A., Dickson R.J., Thibert S.M.F., Gloor G.B., Edgell D.R.. Control of catalytic efficiency by a coevolving network of catalytic and noncatalytic residues. PNAS. 2014; 111:E2376–E2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stetz G., Verkhivker G.M.. Computational analysis of residue interaction networks and coevolutionary relationships in the Hsp70 Chaperones: a Community-Hopping model of allosteric regulation and communication. PLoS Comput. Biol. 2017; 13:e1005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunn S.D., Wahl L.M., Gloor G.B.. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008; 24:333–340. [DOI] [PubMed] [Google Scholar]
- 5. Seemayer S., Gruber M., Soding J.. CCMpred–fast and precise prediction of protein residue-residue contacts from correlated mutations. Bioinformatics. 2014; 30:3128–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morcos F., Pagnani A., Lunt B., Bertolino A., Marks D.S., Sander C., Zecchina R., Onuchic J.N., Hwa T., Weigt M.. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. PNAS. 2011; 108:E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones D.T., Buchan D.W., Cozzetto D., Pontil M.. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012; 28:184–190. [DOI] [PubMed] [Google Scholar]
- 8. Ekeberg M., Lovkvist C., Lan Y., Weigt M., Aurell E.. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013; 87:012707. [DOI] [PubMed] [Google Scholar]
- 9. Feinauer C., Skwark M.J., Pagnani A., Aurell E.. Improving contact prediction along three dimensions. PLoS Comput. Biol. 2014; 10:e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eickholt J., Cheng J.. Predicting protein residue-residue contacts using deep networks and boosting. Bioinformatics. 2012; 28:3066–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones D.T., Singh T., Kosciolek T., Tetchner S.. MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins. Bioinformatics. 2015; 31:999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weigt M., White R.A., Szurmant H., Hoch J.A., Hwa T.. Identification of direct residue contacts in protein-protein interaction by message passing. PNAS. 2009; 106:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adhikari B., Cheng J.. Protein residue contacts and prediction methods. Methods Mol. Biol. 2016; 1415:463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marks D.S., Colwell L.J., Sheridan R., Hopf T.A., Pagnani A., Zecchina R., Sander C.. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011; 6:e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sathyapriya R., Duarte J.M., Stehr H., Filippis I., Lappe M.. Defining an essence of structure determining residue contacts in proteins. PLoS Comput. Biol. 2009; 5:e1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adhikari B., Bhattacharya D., Cao R., Cheng J.. CONFOLD: residue-residue contact-guided ab initio protein folding. Proteins. 2015; 83:1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Michel M., Hayat S., Skwark M.J., Sander C., Marks D.S., Elofsson A.. PconsFold: improved contact predictions improve protein models. Bioinformatics. 2014; 30:i482–i488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller C.S., Eisenberg D.. Using inferred residue contacts to distinguish between correct and incorrect protein models. Bioinformatics. 2008; 24:1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suel G.M., Lockless S.W., Wall M.A., Ranganathan R.. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003; 10:59–69. [DOI] [PubMed] [Google Scholar]
- 20. Sung Y.M., Wilkins A.D., Rodriguez G.J., Wensel T.G., Lichtarge O.. Intramolecular allosteric communication in dopamine D2 receptor revealed by evolutionary amino acid covariation. PNAS. 2016; 113:3539–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Atchley W.R., Wollenberg K.R., Fitch W.M., Terhalle W., Dress A.W.. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol. Biol. Evol. 2000; 17:164–178. [DOI] [PubMed] [Google Scholar]
- 22. Ovchinnikov S., Kamisetty H., Baker D.. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife. 2014; 3:e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marks D.S., Hopf T.A., Sander C.. Protein structure prediction from sequence variation. Nat. Biotechnol. 2012; 30:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oteri F., Nadalin F., Champeimont R., Carbone A.. BIS2Analyzer: a server for co-evolution analysis of conserved protein families. Nucleic Acids Res. 2017; 45:W307–W314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zea D.J., Anfossi D., Nielsen M., Marino-Buslje C.. MIToS.jl: mutual information tools for protein sequence analysis in the Julia language. Bioinformatics. 2017; 33:564–565. [DOI] [PubMed] [Google Scholar]
- 26. Bakan A., Meireles L.M., Bahar I.. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011; 27:1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buslje C.M., Santos J., Delfino J.M., Nielsen M.. Correction for phylogeny, small number of observations and data redundancy improves the identification of coevolving amino acid pairs using mutual information. Bioinformatics. 2009; 25:1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kajan L., Hopf T.A., Kalas M., Marks D.S., Rost B.. FreeContact: fast and free software for protein contact prediction from residue co-evolution. BMC Bioinformatics. 2014; 15:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baldassi C., Zamparo M., Feinauer C., Procaccini A., Zecchina R., Weigt M., Pagnani A.. Fast and accurate multivariate Gaussian modeling of protein families: predicting residue contacts and protein-interaction partners. PLoS One. 2014; 9:e92721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016; 32:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Enright A.J., Van Dongen S., Ouzounis C.A.. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002; 30:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Dongen S.M. Graph clustering by flow simulation. 2000; PhD Thesis.
- 33. Brohee S., Faust K., Lima-Mendez G., Vanderstocken G., van Helden J.. Network analysis Tools: from biological networks to clusters and pathways. Nat. Protoc. 2008; 3:1616–1629. [DOI] [PubMed] [Google Scholar]
- 34. Aguilar D., Oliva B., Marino Buslje C.. Mapping the mutual information network of enzymatic families in the protein structure to unveil functional features. PLoS One. 2012; 7:e41430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halabi N., Rivoire O., Leibler S., Ranganathan R.. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009; 138:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karnoub A.E., Weinberg R.A.. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 2008; 9:517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.