Abstract
WEGO (Web Gene Ontology Annotation Plot), created in 2006, is a simple but useful tool for visualizing, comparing and plotting GO (Gene Ontology) annotation results. Owing largely to the rapid development of high-throughput sequencing and the increasing acceptance of GO, WEGO has benefitted from outstanding performance regarding the number of users and citations in recent years, which motivated us to update to version 2.0. WEGO uses the GO annotation results as input. Based on GO’s standardized DAG (Directed Acyclic Graph) structured vocabulary system, the number of genes corresponding to each GO ID is calculated and shown in a graphical format. WEGO 2.0 updates have targeted four aspects, aiming to provide a more efficient and up-to-date approach for comparative genomic analyses. First, the number of input files, previously limited to three, is now unlimited, allowing WEGO to analyze multiple datasets. Also added in this version are the reference datasets of nine model species that can be adopted as baselines in genomic comparative analyses. Furthermore, in the analyzing processes each Chi-square test is carried out for multiple datasets instead of every two samples. At last, WEGO 2.0 provides an additional output graph along with the traditional WEGO histogram, displaying the sorted P-values of GO terms and indicating their significant differences. At the same time, WEGO 2.0 features an entirely new user interface. WEGO is available for free at http://wego.genomics.org.cn.
INTRODUCTION
Gene Ontology (GO) was started by the GO Consortium in 1998 to focus on studies of the genome of three model organisms: Drosophila Melanogaster (fruit fly), Mus musculus (mouse) and Saccharomyces cerevisiae (brewer’s or baker’s yeast) (1–9). As a result of its unified and well-structured vocabulary, GO was quickly adopted across an array of genome projects (10), transcriptome projects (11–14), proteome projects (15) and more. GO consists of three sub-ontologies: biological process, cellular component and molecular function. These sub-ontologies and the terms therein were designed as a Directed Acyclic Graph (DAG). In order to calculate and present gene enrichment statistics and gene expression levels, the calculation of gene numbers of each GO ID requires a significant understanding of DAG structures. Some tools were created to carry out these analyses, such as agriGO 2.0 (16), BiNGO (17), g:Profiler (18), Gorilla (19), etc. (20,21).
WEGO (Web Gene Ontology Annotation Plot) (22) is a tool that focuses on analyzing GO annotations in a comparative manner. It was created in 2006 and was quickly accepted and put to use by a large number of researchers. In the past 12 years the website has been visited more than 12 636 545 times by users in more than 186 countries and regions (as of the end of 2017). WEGO was cited over 1536 times by publications covering research topics focusing on various types of species from Bryum argenteum (Bryophytes) (23) to Polynoidae (scale worms) (24) and from Gossypium (cotton) (25) to Bombus terrestris (bumblebee) (26). We have also benefitted from a great deal of positive feedback and some very constructive suggestions from users worldwide.
With the rapid development of high-throughput sequencing, the use of genome (10), transcriptome (11–14) and proteome (15) big data has become a major factor in downstream annotation and data analyses. In following this trend and answering user feedback, WEGO was updated to version 2.0 in 2018. It is now more applicable for big data, while its original characteristics of user friendliness and graphical presentation have been enhanced. Some major changes include the ability to upload several input files for analysis, the addition of a reference dataset of nine species for WEGO analyses and an additional output plot showing inconsistent terms.
WEGO INTERFACE
WEGO 2.0 uses the Tomcat 7.0 application server and the MariaDB 5.5 backend database, which is a branch version of the popular open source MySQL database system. The entire WEGO 2.0 service was developed using Java and JavaScript, specifically the NodeJS, Bootstrap 3, JQuery, eCharts 3 and DropzoneJS libraries and frameworks.
External2GO Query and GO Archive Query remain unchanged from the original version of WEGO and can be found in the ‘Tools’ tab. These features aid in translating GO terms between different biological databases and selecting the corresponding GO archives. If the GO vocabulary version adopted in WEGO analysis is different from the annotation process, some outdated GO numbers will appear. You can find such GO numbers under the ‘View error’ option. Using demo data as an example, GO:0004785 is an unmatched GO number listed in the view error. We looked up this number in the GO Archive Query and found that it only existed before the March 2008 version of GO vocabulary. This helps to find the correct GO file version.
A WEGO analysis workflow consists of the submission of input files, selection of GO terms and editing of output results. In addition to the improvements in user interface and user friendliness made in WEGO 2.0, some substantial updates were also included and are explained in further detail in the next sessions.
INPUT
Input files for WEGO are uploaded by way of a drag-and-drop action (Figure 1A)—an unlimited number of files can be uploaded for any one analysis. WEGO supports WEGO native, InterproScan result (XML, TXT, RAW) (27) and GAF (GO Annotation file format) (1–9) formats. There are three optional parameters before submitting:
The file format: The GAF format is the GO consortium’s standard format for GO annotation data, so we set GAF as the default WEGO input format. WEGO provides a demo analysis in the submission area for new users to familiarize themselves with the operation of WEGO. Input samples could be found in documentation.
Gene Ontology Files: Since GO vocabulary is frequently updated, WEGO offers users the ability to select the correct version that exactly matches what has been adopted in their GO annotations. The default GO file is the latest version
Reference data: WEGO provides the reference data of nine model species including: baker’s yeast, Caenorhabditis elegans, Escherichia coli, house mouse, human, fruit fly, brown rat, rice and zebrafish (http://www.geneontology.org/page/download-go-annotations). The backend data for these nine species is obtained from the GO Consortium website (1–9). By default, no reference data is selected.
Figure 1.
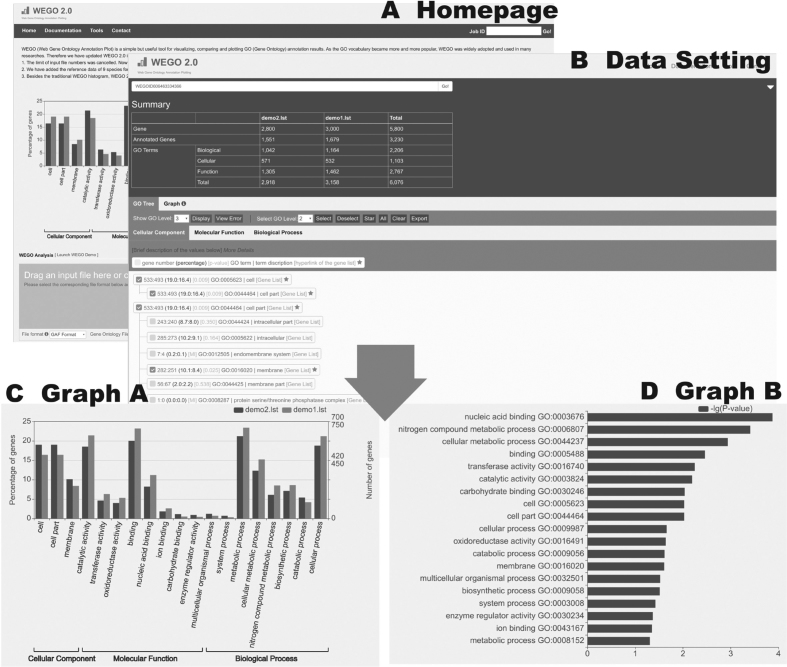
The WEGO interface. This combination chart is just a demonstration of the use of WEGO. Panel (A) is the homepage, uploading the input file. Panel (B) shows the settings of the data. Panels (C) and (D) are two different output plots. (A) Homepage with an example of submission; (B) Data Setting—GO tree tab, showing the statistical summary and GO term selections; (C) Graph A: traditional WEGO histogram: comparisons in gene numbers and percentages of selected GO terms. More datasets uploaded, there will be more column serials in the histogram. (D) Graph B: log (10) of P-values obtained from all datasets of selected GO terms in descending order, indicating the data differences, especially significant differences.
ANALYSIS OUTPUT
A serial number is generated for every job submitted, which is called a job ID. It could be entered on the top right corner of the homepage to re-access the editing page. The job ID is valid for 3 months therefore users can use the serial ID to retrieve the results, instead of re-analyzing big datasets.
A foldable summary of basic statistics of the input file is shown in a tabular form, as in Figure 1B, where the numbers of genes of the three sub-ontologies for each sample, are listed correspondingly.
GO terms are listed as a hierarchical GO tree as shown in Figure 1B. Each GO term is presented in a row, shown in the following order: gene number, percentage, P-value, GO term, term description and hyperlink of the gene list. Three sub-ontologies of GO annotations are listed in separate tabs, which is easier for the users to switch between them. There are two sets of buttons for displaying and selecting GO terms. These buttons work globally which means all three sub-ontologies are effected. The ‘View Error’ page lists GO terms that are not contained in the GO tree due to the mismatching of GO archive versions used in annotation and WEGO. Chi-square tests are carried out for all datasets of particular GO terms. P-values are obtained to indicate the sample differences. A sample difference is considered significant when the P-value < 0.05, thus a star is added for this GO term. By clicking on the ‘Star’ button all the starred GO terms are automatically selected. This function makes it easier for users to identify significantly different GO terms in all the input datasets.
Two output graphs automatically synchronize with GO tree editing. In the ‘Graphs’ tab the properties of the graphs could be edited, including the sizes, colors and legends of graphs, as shown in Figure 1. The users are welcome to export the graphs in SVG, PNG or JPEG formats using the ‘Export’ button.
VISUALIZATION OF OUTPUT
An example of the two types of graphs as WEGO output is shown in Figure 1C and D. Graph A is the traditional WEGO histogram remaining from the previous version. The x-axis displays the GO terms selected from the GO trees. The right y-axis shows the gene numbers of selected GO terms, while the left y-axis shows the percentages. The y-axis could be either linear or log scaled. The log scaled y-axis is recommended when the gene numbers differ too much. Graph B is the newly added graph in 2018 update. The y-axis shows the user selected GO terms and the x-axis shows the log of the P-values from Chi-square tests of all samples. The Chi-square test of independence is applied to determine whether there is a significant difference between the expected frequencies of genes with GO terms and their observed frequencies. When the P-value < 0.05, it is concluded that there is a sample difference in proportions of GO-enriched genes.
The graphs are easily exported using the ‘Export’ button at the top right corners of both graphs. WEGO supports SVG format as output since it is a vector figure format that does not lose its clarity in data transmission. PNG and JPEG formats are also supported. The graphs only show data that is selected in the ‘GO Tree’ tab; if the selection is changed both graphs automatically update. The GO term selecting settings are used in both figures.
UPDATES IN 2018
In order to improve the user-friendliness of WEGO, as well as to keep up with the big data era, WEGO has updated to its 2.0 version. The following three updates greatly improve the functions and usability of WEGO:
WEGO now supports unlimited number of input files, where in contrast the previous version had the restriction of three files. As high-throughput sequencing becomes cheaper and easier, it is common now that 8–10 files have to be analyzed at the same time (28–33). Therefore, this optimization is considered to be very applicable. Moreover, the Chi-square tests can now be applied to multiple datasets (instead of applying to every two datasets), which means only one P-value is calculated for each GO term.
In WEGO 2.0 the genomic annotations of nine species are provided as reference data, which are used as the baseline in genomic comparative analyses. The data are obtained from GO annotations in the Gene Ontology Consortium website, providing comprehensive and non-redundant annotation files for each organism (1–9).
Another important update of WEGO 2.0 is the additional bar graph (Figure 1D) that shows the GO terms (of user’s interests) with the most significant differences in descending order. The horizontal axis is designed to show the log10 of P-values. These P-values are calculated from Chi-square tests of the gene numbers of a particular GO term in all datasets. Therefore, graph B aids in identifying and visualizing the GO terms with most significant differences in all datasets.
Besides these three points, some other slight improvements in WEGO 2.0 include a tabular brief summary of the statistics of input datasets, and a totally new interface. To improve the efficiency and user experience, the analysis workflow is reduced from four to two steps. WEGO now supports some modern user interface technologies, such as web-based drag-and-drop style of file uploading and interactive chart editing and faster switching of GO trees.
PROSPECTIVE DISCUSSION
The large number of WEGO visitors and citations in the past 12 years was beyond the authors’ expectation. It is fully acknowledged that the wide acceptance of WEGO did not stand on its own. It was greatly related to the extensive use of GO vocabulary (1–9). It was concluded that the most important features for a tool, especially a web-based tool, a clear and user-friendly interface, the ease to use and the constant maintenance and improvement rather than complex backend statistical methods and development techniques.
In order to keep up with the development trend of the field of genomics, WEGO now allows for any number of input files and provides enhanced visual presentations, including the visualization of significantly different GO terms. More extended functions such as supporting other well-structured annotation results (e.g. KEGG) (34,35) are likely to be developed in the future. In the future, the maintenance of WEGO will be constantly considered as the most important task, therefore it is greatly appreciated to receive feedback from users. The authors sincerely welcome any feedback through the contact page (http://wego.genomics.org.cn/contact).
DATA AVAILABILITY
WEGO 2.0 is available at http://wego.genomics.org.cn. This website is free and open to all users and there is no login requirement.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the BGI platform and our colleagues for their encouragement and suggestions, as well as the help and support from colleagues in our co-operating organizations and departments. We would also like to sincerely thank Yuxin Chen for the technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Collaborative Innovation Center of High Performance Computing; Critical Patented Project of the Science and Technology Bureau of Fujian Province, China [2013YZ0002–2]; Zhejiang Province for Public Welfare [2016C33122]; Joint Project of the Natural Science and Health Foundation of Fujian Province, China [2015J01397]. Funding for open access charge: Collaborative Innovation Center of High Performance Computing.
Conflict of interest statement. None declared.
REFERENCES
- 1. The Gene Ontology Consortium The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006; 34:D322–D326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The Gene Ontology Consortium The Gene Ontology project in 2008. Nucleic Acids Res. 2008; 36:D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The Gene Ontology Consortium The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010; 38:D331–D335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The Gene Ontology Consortium The Gene Ontology: enhancements for 2011. Nucleic Acids Res. 2012; 40:D559–D564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015; 43:D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017; 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. . Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blake J.A., Dolan M., Drabkin H., Hill D.P., Li N., Sitnikov D., Bridges S., Burgess S., Buza T., McCarthy F. et al. . Gene Ontology annotations and resources. Nucleic Acids Res. 2013; 41:D530–D535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C. et al. . The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004; 32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krosch M.N., Bryant L.M., Vink S.. Differential gene expression of Australian Cricotopus draysoni (Diptera: Chironomidae) populations reveals seasonal association in detoxification gene regulation. Sci. Rep. 2017; 7:14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Govender N., Senan S., Mohamed-Hussein Z.A., Ratnam W.. Transcriptome analysis of reproductive tissue differentiation in Jatropha curcas Linn. Genomics Data. 2017; 13:11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoang N.V., Furtado A., Mason P.J., Marquardt A., Kasirajan L., Thirugnanasambandam P.P., Botha F.C., Henry R.J.. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genomics. 2017; 18:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee W.K., Namasivayam P., Ong Abdullah J., Ho C.L.. Transcriptome profiling of sulfate deprivation responses in two agarophytes Gracilaria changii and Gracilaria salicornia (Rhodophyta). Sci. Rep. 2017; 7:46563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rotllant G., Nguyen T.V., Sbragaglia V., Rahi L., Dudley K.J., Hurwood D., Ventura T., Company J.B., Chand V., Aguzzi J. et al. . Sex and tissue specific gene expression patterns identified following de novo transcriptomic analysis of the Norway lobster, Nephrops norvegicus. BMC Genomics. 2017; 18:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roy A., George S., Palli S.R.. Multiple functions of CREB-binding protein during postembryonic development: identification of target genes. BMC Genomics. 2017; 18:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z.. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017; 45:W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maere S., Heymans K., Kuiper M.. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005; 21:3448–3449. [DOI] [PubMed] [Google Scholar]
- 18. Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J.. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016; 44:W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M.. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005; 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- 21. Supek F., Bosnjak M., Skunca N., Smuc T.. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L. et al. . WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006; 34:W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao B., Li X., Zhang D., Liang Y., Yang H., Chen M., Zhang Y., Zhang J., Wood A.J.. Desiccation tolerance in bryophytes: the dehydration and rehydration transcriptomes in the desiccation-tolerant bryophyte Bryum argenteum. Sci. Rep. 2017; 7:7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y., Sun J., Chen C., Watanabe H.K., Feng D., Zhang Y., Chiu J.M., Qian P.Y., Qiu J.W.. Adaptation and evolution of deep-sea scale worms (Annelida: Polynoidae): insights from transcriptome comparison with a shallow-water species. Sci. Rep. 2017; 7:46205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cai C., Wu S., Niu E., Cheng C., Guo W.. Identification of genes related to salt stress tolerance using intron-length polymorphic markers, association mapping and virus-induced gene silencing in cotton. Sci. Rep. 2017; 7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li L., Su S., Perry C.J., Elphick M.R., Chittka L., Sovik E.. Large-scale transcriptome changes in the process of long-term visual memory formation in the bumblebee, Bombus terrestris. Sci. Rep. 2018; 8:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zdobnov E.M., Apweiler R.. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001; 17:847–848. [DOI] [PubMed] [Google Scholar]
- 28. Looi Q.H., Amin H., Aini I., Zuki M., Omar A.R.. De novo transcriptome analysis shows differential expression of genes in salivary glands of edible bird's nest producing swiftlets. BMC Genomics. 2017; 18:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manchon L., Chebli K., Papon L., Paul C., Garcel A., Campos N., Scherrer D., Ehrlich H., Hahne M., Tazi J.. RNA sequencing analysis of activated macrophages treated with the anti-HIV ABX464 in intestinal inflammation. Sci. Data. 2017; 4:170150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang D., Li L., Wu G., Vasseur L., Yang G., Huang P.. De novo transcriptome sequencing of Isaria cateniannulata and comparative analysis of gene expression in response to heat and cold stresses. PLoS One. 2017; 12:e0186040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang P., Xu L., Xu H., Tang Y., He G., Cao Y., Feng Y., Yuan S., Ming J.. Histological and transcriptomic analysis during bulbil formation in lilium lancifolium. Front. Plant Sci. 2017; 8:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Z., Li Y., Liu H., Zhai X., Deng M., Dong Y., Fan G.. Genome-wide expression analysis of salt-stressed diploid and autotetraploid Paulownia tomentosa. PLoS one. 2017; 12:e0185455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zuo C., Zhang W., Chen Z., Chen B., Huang Y.. RNA sequencing reveals that endoplasmic reticulum stress and disruption of membrane integrity underlie dimethyl trisulfide toxicity against fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 2017; 8:1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O’Donovan C., Phan I. et al. . The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003; 31:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WEGO 2.0 is available at http://wego.genomics.org.cn. This website is free and open to all users and there is no login requirement.