Abstract
Allostery tweaks innumerable biological processes and plays a fundamental role in human disease and drug discovery. Exploration of allostery has thus been regarded as a crucial requirement for research on biological mechanisms and the development of novel therapeutics. Here, based on our previously developed allosteric data and methods, we present an interactive platform called AlloFinder that identifies potential endogenous or exogenous allosteric modulators and their involvement in human allosterome. AlloFinder automatically amalgamates allosteric site identification, allosteric screening and allosteric scoring evaluation of modulator–protein complexes to identify allosteric modulators, followed by allosterome mapping analyses of predicted allosteric sites and modulators in human proteome. This web server exhibits prominent performance in the reemergence of allosteric metabolites and exogenous allosteric modulators in known allosteric proteins. Specifically, AlloFinder enables identification of allosteric metabolites for metabolic enzymes and screening of potential allosteric compounds for disease-related targets. Significantly, the feasibility of AlloFinder to discover allosteric modulators was tested in a real case of signal transduction and activation of transcription 3 (STAT3) and validated by mutagenesis and functional experiments. Collectively, AlloFinder is expected to contribute to exploration of the mechanisms of allosteric regulation between metabolites and metabolic enzymes, and to accelerate allosteric drug discovery. The AlloFinder web server is freely available to all users at http://mdl.shsmu.edu.cn/ALF/.
INTRODUCTION
Allostery, or allosteric regulation, underlies a plethora of biological processes, encompassing cellular metabolism, enzyme catalysis, gene expression and transcription (1,2). Because of the omnipresence of allosteric regulation in cellular signaling and disease, it has been referred to as the ‘second secret of life’ (3). Allostery is mainly rooted in the population shift of the conformational ensemble of a biomolecule, as perturbation at allosteric sites in the structure that are topologically and spatially distinct from orthosteric sites causes a shift in the redistribution of the conformational states across the entire population (4–6). The quintessential event is involved in the binding of effectors, also named allosteric modulators, to allosteric sites, which triggers structural and/or dynamical alterations in orthosteric sites, allowing for exquisite control of biomolecule functional activity.
Recent studies have elucidated that the majority of proteins bind specific allosteric metabolites (also endogenous allosteric modulators) (7) and that allosteric metabolite–protein interactions control enzyme activity in vivo (7–9). Characterizing the detailed allosteric metabolite–protein interactions will thus deepen our understanding of the molecular underpinnings for metabolites in feedback modulation of pathways and has profound importance in human health as a consequence of the strong interplay between metabolites and the pathogenesis of various diseases (10). Meanwhile, exogenous allosteric modulators (also allosteric compounds produced from outside organism) (11), by targeting the structural diversity of allosteric sites, have clear therapeutic advantages such as high selectivity and low toxicity compared to traditional orthosteric ligands by attaching to the conserved orthosteric sites (11–14). Accumulating evidence has indicated that allosteric modulator discovery could produce huge demands in the coming years, witnessed by exponential growth in the number of disclosed allosteric proteins, particularly for G protein-coupled receptors, protein kinases, ion channels, phosphatases and peptidases (15).
Although allosteric modulators play important roles in metabolic mechanisms and novel therapeutics, current discovery of allosteric modulators presents several key challenges. By systematic analysis of known allosteric modulators from our constructed Allosteric Database (ASD) (16–18), it is recognized that the majority of reported allosteric modulators are confined to the allosteric proteins whose allosteric sites have been validated by the three-dimensional (3D) structures of allosteric modulator–protein complexes (19). This evidence highlights a conundrum in the identification of allosteric sites by crystallographic methods, and once the allosteric site of a target protein is disclosed, the discovery of allosteric modulators directed at the allosteric site becomes accessible. An alternative approach is the development of computational methods to identify allosteric sites (20,21). To this end, we previously developed Allosite (22,23) for the detection of allosteric sites on proteins and successfully found novel allosteric sites on several targets using the method (23,24). Furthermore, it is well established that the physicochemical characteristics of allosteric modulators are divergent from orthosteric ligands (25,26). This indicates that the current scoring functions are unable to efficiently assess the allosteric modulator–protein interactions for ranking of docked poses. To resolve these obstacles, we have recently developed Alloscore (27), which predicts the binding affinities of allosteric modulator–protein interactions. In addition to identification of allosteric sites, allosteric screening and evaluation of allosteric interactions, we have also recently collected allosterome (18,28) data, which present the relationship of allostery in a protein family, such as kinase and GPCR families.
Despite the improvement in some specific allosteric applications, there is still a lack of an efficient and convenient platform for the rational discovery of allosteric modulators for therapeutic targets. In addition, exploration of the mechanisms underlying the allosteric regulation of metabolic enzymes by metabolites via computational tools also remains scarce. Based on our previous allosteric data and methods, here we present a platform called AlloFinder that discovers allosteric modulators and assesses biological functions by mapping allosterome data. AlloFinder utilizes feature and dynamic perturbations to uncover putative allosteric sites for a query protein, followed by in silico screening of potential allosteric modulators at the predicted allosteric site according to accurate allosteric scoring function. Importantly, the identified allosteric sites and modulators can be submitted to allosterome mapping analysis, which unmasks the specificity and evolution of the identified allosteric site in the human proteome as well as the similarity between the identified allosteric modulators and known allosteric modulators. Testing 60 allosteric proteins, AlloFinder successfully reemerged at least one known allosteric modulator at its own allosteric site within the top 1% of the ranked library in 53 cases. Significantly, we employed AlloFinder to discover allosteric modulators of signal transduction and activation of transcription 3 (STAT3), supporting the feasibility of AlloFinder in the discovery of allosteric modulators.
MATERIALS AND METHODS
Workflow of AlloFinder
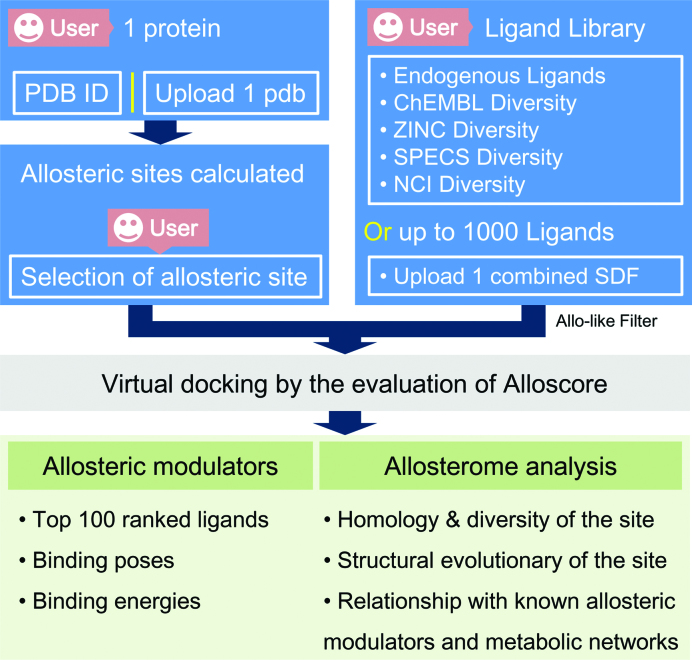
AlloFinder is deployed as a built-in computational workflow that is devoted to two allosteric functions: allosteric modulator screening and allosterome mapping. The function of allosteric modulator screening consists of a set of methods developed mainly in our laboratory, including an Allosite algorithm (23) for predicting allosteric sites on a query protein, an Allolike filter (29) for filtering ‘allosteric-like’ compounds in a ligand library, a pocket-generated pharmacophore method (30) for ruling out unbound compounds and an Alloscore algorithm (27) for scoring allosteric modulator–protein complexes. Based on the known allosteric data, an allosterome mapping analysis was designed here for structural and functional analyses of identified allosteric sites and modulators (Figure 1). The website is free and open to all, and there is no login requirement.
Figure 1.
The workflow of the AlloFinder. The user input is shown in red.
First, the user uploads a query protein to AlloFinder, and the Allosite algorithm, a feature-based regression combined with Normal Mode Analysis (NMA)-based perturbation, is used to predict all putative allosteric sites on the protein. The user can cherry-pick one allosteric site for virtual screening of a predefined ligand library. Then, the pocket-generated pharmacophore model for the selected allosteric site is generated for quickly ruling out unbound compounds in the library. Afterward, conformational sampling of an ensemble of docked conformations for each compound from the pharmacophore-filtered subset is executed using the genetic algorithm from AutoDock Vina (31,32). The Alloscore algorithm is subsequently utilized to evaluate the best binding energy from the conformational ensemble of each compound. All compounds of the library in screening were ranked by the energies and the top 100 compounds were outputted in the AlloFinder server. At last, the predicted allosteric sites and modulators are harnessed to perform allosterome mapping analyses in the human proteome. The runtime of the submitted jobs depends on which library is selected in AlloFinder and can vary between 30 min and several hours. More detailed procedure of the workflow is described in the ‘Materials and Methods’ section of Supplementary file.
AlloFinder input
The users can specify the protein of interest either with a PDB ID or by uploading PDB files in PDB format under ‘Query Protein’. Considering the running time, the server does not presently accept proteins with more than 2000 residues. A ‘Job Name’ is compulsory, which allows the users to find their queries in the ‘Job Queue’. For virtual database screening, the users can select one of the following databases, including Endogenous Ligands, ChEMBL Diversity (33), ZINC Diversity (34), SPECS Diversity (http://www.specs.net) and NCI Diversity (http://dtp.cancer.gov) under ‘Ligand Library’. The properties of the libraries and their differences are provided for user selection in the Help of the AlloFinder server. In addition, users are encouraged to provide their own ligand library with numbers up to 1000 compounds in SDF format. Details of the ligand library construction are provided in the Supplementary Materials and Methods.
AlloFinder output
AlloFinder outputs interactive charts that show the top 100 predicted allosteric modulators ranked by the ‘Alloscore Score’ and the 2D structure and molecular weight for each modulator. Clicking the ‘Show Ligand’ button, a 3D representation of the modulator–protein complex can be viewed. Importantly, AlloFinder provides interactive pages for displaying site and ligand allosterome analyses.
In the site allosteric analyses, a 3D representation of the predicted allosteric site is shown in the left panel, together with a table showing the site properties in the right panel, including ‘Protein Name’, ‘PDB ID’, ‘Allosteric Site Score’, ‘Drug-like Score’, ‘Perturbation score’ and ‘Volume’. Clicking the ‘Site Allosterome Mapping’ button links to the site allosteric analyses. Further clicking the ‘Homology’ tab gives an interactive chart showing the similarity of the predicted allosteric site with the top 20 known allosteric sites and a table summarizing general information for each known allosteric site, such as allosteric site ID, gene name and site similarity score. Clicking the ‘View Pocket’ in the table shows detailed information of the allosteric site, such as protein name, site residues, site view and reference. Alternatively, clicking the ‘Structural Evolutionary’ tab provides an interactive chart showing the evolutionary information of the predicted allosteric site with the known allosteric sites.
In the ligand allosteric analyses, clicking the ‘Endogenous’ (‘Exogenous’) button offers an interactive chart showing the distribution of the relationship between the predicted allosteric modulator with the top 100 known endogenous (exogenous) allosteric modulators and a table summarizing general information for each endogenous (exogenous) allosteric modulator, such as allosteric modulator ID, 2D structure and ligand similarity score. Further clicking the ‘View Properties’ in the table shows the comprehensive properties of each endogenous (exogenous) allosteric modulator, such as formula, IUPAC name, and detailed physiochemical properties. Concomitantly, the metabolic network for the metabolite most similar to the predicted allosteric modulator is shown using the KEGG pathway (35). The user can download each result for offline analysis by clicking the ‘Download predicted allosteric site(s)’, ‘Download predicted allo-ligand(s)’ and ‘Download allosterome’ buttons, respectively.
There are several quantifiable items generated by AlloFinder, which can be harnessed by users in the page of Job Result for further analysis. First, the druggable score of the predicted allosteric site, referred to ‘Drug-like Score’, determines whether this allosteric site is suitable for design of allosteric modulators. Second, the predicted binding affinities of allosteric modulator–protein complexes, referred to ‘AlloScore Score’, are useful, which can guide the choice of potential allosteric modulators for experimental testing. Third, in the allosterome mapping of allosteric site, the calculation of the similarity of the predicted allosteric site with known human allosteric sites, referred to ‘Site Similarity Score’, determines whether this allosteric site is specific, which is helpful for the design of selective allosteric modulators for a highly homologous protein subtype. Fourth, for the allosterome mapping of allosteric modulators, the calculated similarity of the predicted allosteric modulator with known allosteric modulators, referred to ‘Ligand Similarity Score’, provides the clues whether the predicted allosteric modulator has the possibility of polypharmacology, which facilitates the design of more specific allosteric modulators. Expediently, the AlloFinder website offers a step-by-step Tutorial in the ‘Help’ page. The user can consult the ‘Help’ for detailed information.
Testing datasets
For testing the performance of AlloFinder, we selected 60 diverse allosteric proteins with the structure of an annotated allosteric site from the ASD, which are widely distributed across multiple classes of proteins (see Supplementary Table S1). Among the 60 proteins, 9 are regulated by allosteric metabolites and 51 are regulated by exogenous allosteric modulators according to the previous literature. For each protein system, a ligand dataset for benchmark was constructed to contain up to 1000 compounds that are first prepared by non-redundant allosteric modulators of the protein from the ASD and unique substrate-competitive ligands (non-allosteric modulators) of the protein from the ZINC if available. The remaining compounds of the ligand dataset were randomly taken from ZINC Diversity (see Supplementary Materials and Methods). As a result, there are 10–50 known allosteric modulators in the ligand datasets of 22 proteins, 2–9 known allosteric modulators in 13 proteins and only 1 known allosteric modulator in 25 proteins.
PERFORMANCE OF AlloFinder
To validate our implementation of AlloFinder, we assessed the performance of AlloFinder to identify endogenous/exogenous allosteric modulators from a ligand-dataset screening on the benchmark of 60 diverse allosteric proteins including kinases, proteases, transferases and GPCRs etc. The enrichment factor (EF) is the most common metrics for screening, which is the concentration of the known ligands among the top-scoring hits compared to their concentration throughout the entire library (36). The results showed that AlloFinder is capable of reemerging at least 1 known allosteric modulator at its own allosteric site in the top 1% of the ranked benchmark dataset (EF1) in 53 of 60 proteins (Supplementary Table S1), including 4 allosteric metabolites and 49 exogenous allosteric modulators. Meanwhile, more than 50% of the known allosteric modulators can be found in the top 20% of the ranked benchmark dataset (EF20) in all 60 proteins (Supplementary Table S1), and the result was further supported by a good performance (≥0.5) of Hit Rate, which evaluates the normalized EF20 of the screened library (37), in the proteins (Supplementary Table S2). The enrichment performance indicates that known allosteric modulators can be generally well-enriched against its target protein by AlloFinder.
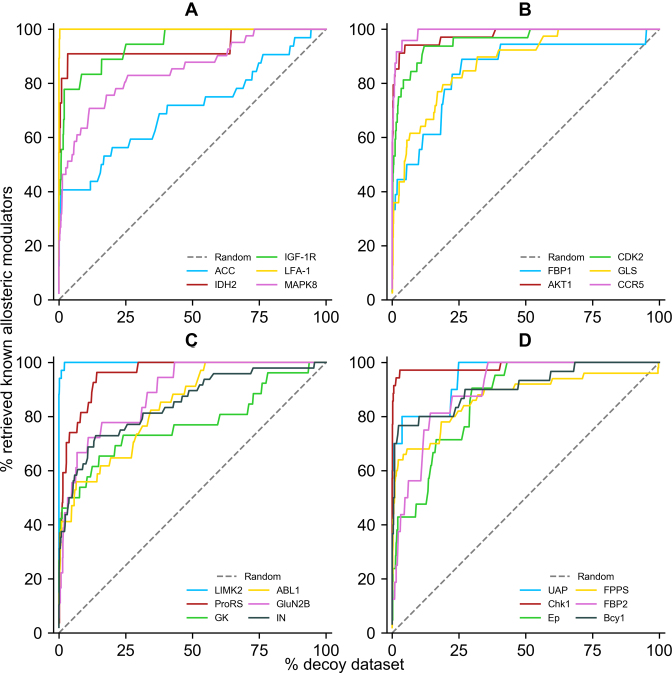
Furthermore, analyses of the receiver operating characteristic (ROC) curves of virtual screening performed with AlloFinder on proteins with ≥10 known allosteric modulators in the ligand dataset (see testing datasets above) were evaluated. This curve describes the tradeoff between sensitivity and specificity of a screening. As another supporting measure, the area under an ROC curve, i.e., the AUC value, was also calculated to reveal the quality of enrichment (38). AlloFinder exhibited prominent performance at any given percentage in the ROC curve (Figure 2) and >0.7 of AUC value (Supplementary Table S3) for each protein. Meanwhile, more than 40% of known allosteric modulators were retrieved in the top 5% ranked benchmark dataset in the proteins. During the analyses, we also found that for proteins with a common size of allosteric site, AlloFinder can efficiently retrieve low-MW known allosteric modulators bound to their allosteric site at the top of ranked ligands in a library. Exceptionally, for proteins with a large allosteric site such as Acetyl-CoA carboxylase (ACC) and glucokinase (GK), a few low-MW known allosteric modulators (e.g. ASD00431001 in ACC and ASD01208645 in GK) were not enriched at the top of ranked ligands perhaps because of the deficiency of allosteric modulator–protein interactions at a large site. Therefore, AlloFinder is able to discover diverse active allosteric modulators even at a large site, and users could still pay attention to some false-negative cases from low-MW ligands screened at such large allosteric sites of proteins.
Figure 2.
ROC curves of screening for proteins with ≥10 known allosteric modulators in the ligand dataset. Decoy dataset represent the ligands except known allosteric modulators in the dataset for the benchmark of a protein. Retrieved known allosteric modulators are the known allosteric modulators that are successfully found in the benchmark of a protein. Proteins are represented by different colored lines and labels. (A) ACC, Acetyl-CoA carboxylase; IDH2, Isocitrate dehydrogenase [NADP], mitochondrial; IGF-1R, Insulin-like growth factor 1 receptor; LFA-1, Integrin alpha-L; MAPK8, Mitogen-activated protein kinase 8; (B) FBP1, Fructose-1,6-bisphosphatase 1; AKT1, RAC-alpha serine/threonine-protein kinase; CDK2, Cyclin-dependent kinase 2; GLS, Glutaminase kidney isoform, mitochondrial; CCR5, C-C chemokine receptor type 5; (C) LIMK2, LIM domain kinase 2; ProRS, Proline–tRNA ligase; GK, Glucokinase; ABL1, Tyrosine-protein kinase ABL1; GluN2B, Glutamate receptor ionotropic, NMDA 2B; IN, HIV-1 integrase; (D) UAP, UDP-N-acetylglucosamine pyrophosphorylase; Chk1, Serine/threonine-protein kinase Chk1; Ep, Glucose-1-phosphate thymidylyltransferase; FPPS, Farnesyl pyrophosphate synthase; FBP2, Fructose-1,6-bisphosphatase isozyme 2; Bcy1, cAMP-dependent protein kinase regulatory subunit.
EXAMPLES
Example 1: TR:RXR heterodimers
Thyroid hormone receptors (TRs) form heterodimers with retinoic acid receptor (RXR) to regulate the transcriptional activity of TRs. Binding of 9-cis retinoic acid (9c) to RXR allosterically inhibits TR-T3:RXR transactivation by the effect of T3 (3,3′,5 triiodo-L-thyronine) binding on the TR (39). Using a TR-T3:RXR heterodimer complex (PDB ID: 3UVV) (39) and the Endogenous Ligands library as inputs, AlloFinder perfectly predicted only one allosteric site on the RXR that was identical to the crystal allosteric site of 9-cis retinoic acid, followed by the discovery of the 9-cis retinoic acid at the ranked second of the output hits (Supplementary Figure S1 and Table S4). The binding information of 9-cis retinoic acid at the site is shown in Supplementary Figure S2. Site allosterome analyses unraveled that the allosteric site of 9-cis retinoic acid had the highest similarity score of 0.40 with other known allosteric sites, reflecting the specificity of the RXR allosteric site. Ligand allosterome analyses showed that the endogenous 9-cis retinoic acid had the highest similarity score of 0.83 with the endogenous 9-cis retinal. The 9-cis retinoic acid is the oxidation product of 9-cis retinal, catalyzed by retinal dehydrogenase (40,41). Indeed, 9-cis retinal was ranked 15th of the hits, indicating that both 9-cis retinoic acid and 9-cis retinal could have a similar effect on the regulation of retinol metabolism by binding to the predicted allosteric site of RXR.
Example 2: GluN1b:GluN2B NMDA receptor
NMDA (N-methyl-D-aspartate) receptors are heteromeric ion channels consisting of GluN1 and GluN2 subunits. Ifenprodil allosterically inhibits the GluN1b:GluN2B NMDA receptor by binding to the interface between the amino-terminal domains (ATDs) of GluN1b and GluN2B (42). The crystal structure of GluN1b:GluN2B ATD heterodimer (PDB ID: 3QEL) (42) was uploaded to AlloFinder, which generated two putative allosteric sites. The crystal allosteric site of ifenprodil was ranked first and chosen as a target for virtual screening of the ZINC Diversity Library. Ifenprodil was recaptured and ranked third in the top 100 hits (Supplementary Figure S3 and Table S5). The binding feature of Ifenprodil at the site is provided in Supplementary Figure S4. Subsequent site and ligand allosterome analyses exhibited that the allosteric site and ifenprodil are structurally distinct from other known allosteric sites and modulators. Detailed information of example 1 and 2 is further provided in the online Tutorial under the Help of the AlloFinder srever.
Discovery of STAT3 allosteric inhibitor
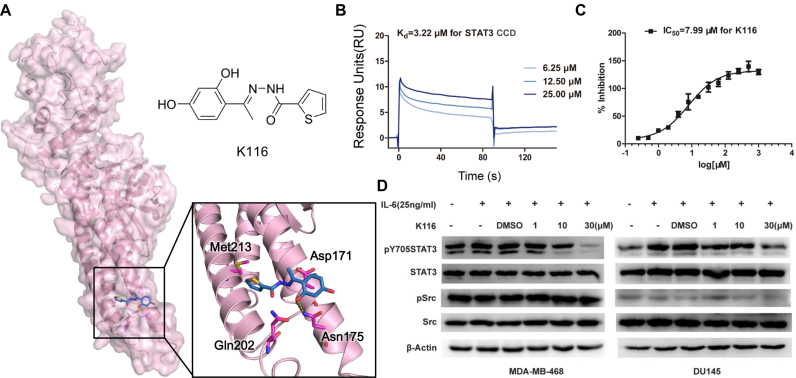
STAT3, a multi-domain protein, is activated by tyrosine phosphorylation in response to cytokine-receptor IL-6 binding and plays an important role in cell growth and apoptosis (43). Figure 3 shows the results of using AlloFinder to identify allosteric sites and modulators of STAT3. To discover novel allosteric inhibitors of STAT3, the crystal structure of STAT3 (PDB ID: 3CWG) (44) was uploaded to AlloFinder and five putative allosteric sites (Sites 1–5) on STAT3 were predicted. Site 5 situated in the coiled-coil domain (CCD) of STAT3 was selected as a viable target for in silico screening of the SPECS Diversity Library because previous biochemical experiments provided clues that the CCD distant from the STAT3-DNA interface indirectly regulates STAT3 activation and function (45,46). With about 2 h of the running time, 100 hits ranked by the Alloscore Score were outputted in the page of the AlloFinder server (Supplementary Table S6). The top 15 hits predicted by AlloFinder were purchased and tested with bioassays. Among them, K116 (AH-034/11963955) (Figure 3A) enabled binding to STAT3 CCD (Kd = 3.22 μM) (Figure 3B) and inhibited STAT3 binding to the phosphopeptide ligand (IC50 = 7.99 μM) (Figure 3C). In the binding model at Site 5, K116 showed several hydrophobic effects with Asp171, Gln202 and Met213 as well as a hydrogen bond with Asn175 in STAT3 (Figure 3A). Site-directed mutagenesis indicated Site 5 surrounded by residues Asp171, Asn175, Gln202 and Met213 as a bona fide site for K116 (Supplementary Figure S5). Furthermore, cellular and functional experiments confirmed that K116 both inhibited STAT3 phosphorylation at Tyr705 (Figure 3D) and promoted STAT3-mediated apoptosis in a dose-dependent manner (Supplementary Figure S5). Taken together, these data support the tractability and feasibility of AlloFinder in the discovery of allosteric sites and modulators for STAT3 as well as other therapeutic targets.
Figure 3.
Discovery and experimental validation of the STAT3 allosteric modulator. (A) Predicted allosteric site and modulator K116 of STAT3. (B) Binding affinity (Kd) of K116 with STAT3 CCD detected by Biacore T200. (C) IC50 of K116 measured with the fluorescence polarization assay. The K116 concentrations ranged from 1 to 0.18 mM continually during the indicated time for 24 h. (D) Western blot of STAT3, phospho-STAT3 (pY705STAT3), Src, phospho-Src in MADMB-468 and DU145 cells treated with or without K116. Data are representative of three independent determinations.
DISCUSSION AND CONCLUSION
Allostery is currently regarded as a unifying mechanism for metabolic function and regulation (14), and it is also a novel tactic for drug discovery (47). Because of the immense significance of allostery, it has been the subject of intense investigation over the past few years (15). However, a comprehensive insight into allostery by experimental methods is unsatisfactory owing to the complex nature of allostery in biological process. Computation-based strategies that previously focus on the prediction of allosteric sites and communication pathways, and on the elucidation of allosteric mechanism have emerged as alternatives (48). For example, a number of web servers or algorithms based on the normal model analysis methods (e.g. AlloPred (49), DynOmics ENM (50), PARS (51), SPACER (52) and STRESS (53)), machine learning methods (e.g. Allosite (22) and a random forest model (54)), molecular dynamics (MD) methods (e.g. MD simulations (55) and AllosMod (56)), and a distance geometry method (e.g. ExProSE) (57) have become available for the prediction of allosteric sites. In addition, web servers such as AlloSigMA (58) and MCPath (59) have been developed to investigate allosteric communication pathways between allosteric and orthosteric sites.
Despite the current availability of the prediction of allosteric sites and communication pathways via various methods, they are not practically achievable for allosteric screening of putative modulators for proteins. To this end, the AlloFiner platform aims to provide the biologists and medicinal chemists with a free and user-friendly web server to automatically identify allosteric modulators for a particular protein by the user, followed by subsequent allosterome mapping analyses of predicted allosteric sites and modulators. Specially, it is applicable to unearth allosteric mechanism for metabolites, which is helpful for a large-scale investigation of the allosteric protein–metabolite interactome and a better understanding of biological pathways regulated by metabolite interactions.
AlloFinder’s predictability was extensively validated by retrospective evaluations and a real case of STAT3. However, there is still the existence of limitations for this server in the identification of allosteric modulators. The most challenge is derived from the use of rigid conformation of a protein for the conformational sampling of modulators, which may underestimate the search space of some modulators. Considering the progress of computing power, we will update the AlloFinder to utilize the flexible docking to improve the search space in the future. Collectively, the AlloFinder web server, to the best of our knowledge, is the first of its kind that will not only be exploited to probe allosteric mechanisms of metabolic enzymes by allosteric metabolites but will also provide valuable guidance to allosteric drug discovery.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Yingli Wu at the Shanghai Jiaotong University School of Medicine for fruitful discussions on the allosteric modulators.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (973 Program) [2015CB910403 to J.Z.]; National Natural Science Foundation of China [81322046 to J.Z., 81473137 to J.Z., 91753117 to J.Z., 81721004 to G.C., J.Z.]. Funding for open access charge: National Basic Research Program of China (2015CB910403); National Natural Science Foundation of China (81322046).
Conflict of interest statement. None declared.
REFERENCES
- 1. Goodey N.M., Benkovic S.J.. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008; 4:474–482. [DOI] [PubMed] [Google Scholar]
- 2. Nussinov R., Tsai C., Liu J.. Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 2014; 136:17692–17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fenton A.W. Allostery: an illustrated definition for the ‘second secret of life’. Trends Biochem. Sci. 2008; 33:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Motlagh H.N., Wrabl J.O., Li J., Hilser V.J.. The ensemble nature of allostery. Nature. 2014; 508:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu S., Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R., Zhang J.. Ras conformational ensembles, allostery, and signaling. Chem. Rev. 2016; 116:6607–6665. [DOI] [PubMed] [Google Scholar]
- 6. Kornev A.P., Taylor S.S.. Dynamics-Driven allostery in protein kinases. Trends Biochem. Sci. 2015; 40:628–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Link H., Kochanowski K., Sauer U.. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat. Biotechnol. 2013; 31:357–361. [DOI] [PubMed] [Google Scholar]
- 8. Li X., Gianoulis T.A., Yip K.Y., Gerstein M., Snyder M.. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010; 143:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gallego O., Betts M.J., Gvozdenovic-Jeremic J., Maeda K., Matetzki C., Aguilar-Gurrieri C., Beltran-Alvarez P., Bonn S., Fernández-Tornero C., Jensen L.J. et al. . A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010; 6:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nicholson J.K., Holmes E., Elliott P.. The metabolome-wide association study: a new look at human disease risk factors. J. Proteome Res. 2008; 7:3637–3638. [DOI] [PubMed] [Google Scholar]
- 11. Wootten D., Christopoulos A., Sexton P.M.. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat. Rev. Drug Discov. 2013; 12:630–644. [DOI] [PubMed] [Google Scholar]
- 12. Guarnera E., Berezovsky I.N.. Allosteric sites: remote control in regulation of protein activity. Curr. Opin. Struct. Biol. 2016; 37:1–8. [DOI] [PubMed] [Google Scholar]
- 13. Jeffrey Conn P., Christopoulos A., Lindsley C.W.. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009; 8:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Changeux J.-P., Christopoulos A.. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell. 2016; 166:1084–1102. [DOI] [PubMed] [Google Scholar]
- 15. Lu S., Li S., Zhang J.. Harnessing allostery: a novel approach to drug discovery. Med. Res. Rev. 2014; 34:1242–1285. [DOI] [PubMed] [Google Scholar]
- 16. Huang Z., Zhu L., Cao Y., Wu G., Liu X., Chen Y., Wang Q., Shi T., Zhao Y., Wang Y. et al. . ASD: a comprehensive database of allosteric proteins and modulators. Nucleic Acids Res. 2011; 39:D663–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang Z., Mou L., Shen Q., Lu S., Li C., Liu X., Wang G., Li S., Geng L., Liu Y. et al. . ASD v2.0: Updated content and novel features focusing on allosteric regulation. Nucleic Acids Res. 2014; 42:D510–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen Q., Wang G., Li S., Liu X., Lu S., Chen Z., Song K., Yan J., Geng L., Huang Z. et al. . ASD v3.0: Unraveling Allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res. 2016; 44:D527–D535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang W., Wang G., Shen Q., Liu X., Lu S., Geng L., Huang Z., Zhang J.. ASBench: benchmarking sets for allosteric discovery. Bioinformatics. 2015; 31:2598–2600. [DOI] [PubMed] [Google Scholar]
- 20. Wagner J.R., Lee C.T., Durrant J.D., Malmstrom R.D., Feher V.A., Amaro R.E.. Emerging computational methods for the rational discovery of allosteric drugs. Chem. Rev. 2016; 116:6370–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu S., Huang W., Zhang J.. Recent computational advances in the identification of allosteric sites in proteins. Drug Discov. Today. 2014; 19:1595–1600. [DOI] [PubMed] [Google Scholar]
- 22. Huang W., Lu S., Huang Z., Liu X., Mou L., Luo Y., Zhao Y., Liu Y., Chen Z., Hou T. et al. . Allosite: a method for predicting allosteric sites. Bioinformatics. 2013; 29:2357–2359. [DOI] [PubMed] [Google Scholar]
- 23. Song K., Liu X., Huang W., Lu S., Shen Q., Zhang L., Zhang J.. Improved method for the identification and validation of allosteric sites. J. Chem. Inf. Model. 2017; 57:2358–2363. [DOI] [PubMed] [Google Scholar]
- 24. Jiang H., Dong J., Song K., Wang T., Huang W., Zhang J.-M., Yang X., Shen Y., Zhang J.. A novel allosteric site in casein kinase 2α discovered using combining bioinformatics and biochemistry methods. Acta Pharmacol. Sin. 2017; 38:1691–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Westen G.J.P., Gaulton A., Overington J.P.. Chemical, target, and bioactive properties of allosteric modulation. PLoS Comput. Biol. 2014; 10:e1003559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith R.D., Lu J., Carlson H.A.. Are there physicochemical differences between allosteric and competitive ligands. PLOS Comput. Biol. 2017; 13:e1005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li S., Shen Q., Su M., Liu X., Lu S., Chen Z., Wang R., Zhang J.. Alloscore: A method for predicting allosteric ligand-protein interactions. Bioinformatics. 2016; 32:1574–1576. [DOI] [PubMed] [Google Scholar]
- 28. Shen Q., Cheng F., Song H., Lu W., Zhao J., An X., Liu M., Chen G., Zhao Z., Zhang J.. Proteome-Scale investigation of protein allosteric regulation perturbed by somatic mutations in 7,000 cancer genomes. Am. J. Hum. Genet. 2017; 100:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Q., Zheng M., Huang Z., Liu X., Zhou H., Chen Y., Shi T., Zhang J.. Toward understanding the molecular basis for chemical allosteric modulator design. J. Mol. Graph. Model. 2012; 38:324–333. [DOI] [PubMed] [Google Scholar]
- 30. Chen J., Lai L.. Pocket v.2: Further developments on receptor-based pharmacophore modeling. J. Chem. Inf. Model. 2006; 46:2684–2691. [DOI] [PubMed] [Google Scholar]
- 31. Trott O., Olson A.J.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang R., Lu Y., Wang S.. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003; 46:2287–2303. [DOI] [PubMed] [Google Scholar]
- 33. Gaulton A., Hersey A., Patr A., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibri E., Davies M. et al. . The ChEMBL database in 2017. Nucleic Acids Res. 2018; 45:945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Irwin J.J., Shoichet B.K.. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005; 45:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang N., Shoichet B.K., Irwin J.J.. Benchmarking sets for molecular docking. J. Med. Chem. 2006; 49:6789–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roy A., Srinivasan B., Skolnick J.. PoLi: a virtual screening pipeline based on template pocket and ligand similarity. J. Chem. Inf. Model. 2015; 55:1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Triballeau N., Acher F., Brabet I., Pin J.P., Bertrand H.. Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J. Med. Chem. 2005; 48:2534–2547. [DOI] [PubMed] [Google Scholar]
- 39. Putcha B.-D.K., Wright E., Brunzelle J.S., Fernandez E.J.. Structural basis for negative cooperativity within agonist-bound TR:RXR heterodimers. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6084–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Napoli J.L. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim. Biophys. Acta. 2012; 1821:152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang P., Chandra V., Rastinejad F.. Retinoic acid actions through mammalian nuclear receptors. Chem. Rev. 2014; 114:233–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karakas E., Simorowski N., Furukawa H.. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011; 475:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chai E.Z.P., Shanmugam M.K., Arfuso F., Dharmarajan A., Wang C., Kumar A.P., Samy R.P., Lim L.H.K., Wang L., Goh B.C. et al. . Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol. Ther. 2016; 162:86–97. [DOI] [PubMed] [Google Scholar]
- 44. Ren Z., Mao X., Mertens C., Krishnaraj R., Qin J., Mandal P.K., Romanowski M.J., McMurray J.S., Chen X.. Crystal structure of unphosphorylated STAT3 core fragment. Biochem. Biophys. Res. Commun. 2008; 374:1–5. [DOI] [PubMed] [Google Scholar]
- 45. Zhang T., Kee W.H., Seow K.T., Fung W., Cao X.. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol. Cell. Biol. 2000; 20:7132–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma J., Zhang T., Novotny-Diermayr V., Tan A.L.C., Cao X.. A novel sequence in the coiled-coil domain of Stat3 essential for its nuclear translocation. J. Biol. Chem. 2003; 278:29252–29260. [DOI] [PubMed] [Google Scholar]
- 47. Nussinov R., Tsai C.-J.. Allostery in disease and in drug discovery. Cell. 2013; 153:293–305. [DOI] [PubMed] [Google Scholar]
- 48. Greener J.G., Sternberg M.J.. Structure-based prediction of protein allostery. Curr. Opin. Struct. Biol. 2018; 50:1–8. [DOI] [PubMed] [Google Scholar]
- 49. Greener J.G., Sternberg M.J.. AlloPred: prediction of allosteric pockets on proteins using normal mode perturbation analysis. BMC Bioinformatics. 2015; 16:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H., Chang Y.Y., Lee J.Y., Bahar I., Yang L.W.. DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res. 2017; 45:W374–W380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Panjkovich A., Daura X.. PARS: a web server for the prediction of protein allosteric and regulatory sites. Bioinformatics. 2014; 30:1314–1315. [DOI] [PubMed] [Google Scholar]
- 52. Goncearenco A., Mitternacht S., Yong T., Eisenhaber B., Eisenhaber F., Berezovsky I.N.. SPACER: Server for predicting allosteric communication and effects of regulation. Nucleic Acids Res. 2013; 41:W266–W272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Clarke D., Sethi A., Li S., Kumar S., Chang R.W.F., Chen J., Gerstein M.. Identifying allosteric hotspots with dynamics: application to inter- and intra-species conservation. Structure. 2016; 24:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen A.S.Y., Westwood N.J., Brear P., Rogers G.W., Mavridis L., Mitchell J.B.O.. A random forest model for predicting allosteric and functional sites on proteins. Mol. Inform. 2016; 35:125–135. [DOI] [PubMed] [Google Scholar]
- 55. Dror R.O., Green H.F., Valant C., Borhani D.W., Valcourt J.R., Pan A.C., Arlow D.H., Canals M., Lane J.R., Rahmani R. et al. . Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013; 503:295–299. [DOI] [PubMed] [Google Scholar]
- 56. Weinkam P., Pons J., Sali A.. Structure-based model of allostery predicts coupling between distant sites. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:4875–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Greener J.G., Filippis I., Sternberg M.J.E.. Predicting protein dynamics and allostery using multi-protein atomic distance constraints. Structure. 2017; 25:546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guarnera E., Tan Z.W., Zheng Z., Berezovsky I.N.. AlloSigMA: allosteric signaling and mutation analysis server. Bioinformatics. 2017; 33:3996–3998. [DOI] [PubMed] [Google Scholar]
- 59. Kaya C., Armutlulu A., Ekesan S., Haliloglu T.. MCPath: Monte Carlo path generation approach to predict likely allosteric pathways and functional residues. Nucleic Acids Res. 2013; 41:W249–W255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.