Abstract
Antibody repertoire analysis by high throughput sequencing is now widely used, but a persisting challenge is enabling immunologists to explore their data to discover discriminating repertoire features for their own particular investigations. Computational methods are necessary for large-scale evaluation of antibody properties. We have developed BRepertoire, a suite of user-friendly web-based software tools for large-scale statistical analyses of repertoire data. The software is able to use data preprocessed by IMGT, and performs statistical and comparative analyses with versatile plotting options. BRepertoire has been designed to operate in various modes, for example analysing sequence-specific V(D)J gene usage, discerning physico-chemical properties of the CDR regions and clustering of clonotypes. Those analyses are performed on the fly by a number of R packages and are deployed by a shiny web platform. The user can download the analysed data in different table formats and save the generated plots as image files ready for publication. We believe BRepertoire to be a versatile analytical tool that complements experimental studies of immune repertoires. To illustrate the server’s functionality, we show use cases including differential gene usage in a vaccination dataset and analysis of CDR3H properties in old and young individuals. The server is accessible under http://mabra.biomed.kcl.ac.uk/BRepertoire.
INTRODUCTION
In recent years, the advent of new experimental techniques in the field of immune receptor sequencing has enabled researchers to obtain and analyse large collections (so-called repertoires) of immunoglobulin (Ig) genes. These datasets are representative of an individual’s antibody arsenal and enable comparisons between individuals, e.g. to estimate differences in response intensities to a given event, or between time points. Studies of repertoires have provided novel information on normal human immune development (1), responses to vaccines (2) or infection (3,4), changes observed in autoimmune diseases (5,6) or allergy (7) and age-related differences in the immune system (8). The complementary-determining regions (CDRs) of both the Ig light and heavy chains, are particularly important to study because they are (as their name suggests) the pre-eminent factors which determine binding specificity. Ig sequence repertoire collections are minimally in the range of tens of thousands of sequences, thus requiring the application of computational tools and statistical models for analysis. A number of solutions have been developed, mainly to investigate V(D)J gene usage (9–16), clonotype clustering (10–12,14–16), diversity (9,10,13,14) and CDR length distributions of antibody repertoires. While some software is distributed in a stand-alone manner (12,14,15,17,18), or as R packages (10,11,13), other solutions are available as web servers (9,15,16) which offer the key advantage of direct utilization with little or no necessary preparatory steps. To the best of our knowledge, however, no other web server offers: (i) BRepertoire’s flexibility in data handling, (ii) Its wide-ranging support of physico-chemical properties and (iii) Power in terms of statistical analyses. The server makes minimal assumptions on the nature of the data provided. In particular and despite the name BRepertoire, it can be used for T cell and non-human sequencing data as well. The calculation and analysis of physico-chemical properties provides a representation of amino acid sequences (e.g. from CDRs) on a fundamental level by which chemical commonalities and differences between sub-partitions of the data can be easily observed. To assess these features, BRepertoire also supports the calculation of a set of statistical significance and effect size measures. Moreover, the use of clonotype clustering groups the observations into families of common lineage in order to access the variety in a repertoire and identify sequences subject to clonal expansion, affinity maturation and class switching.
MATERIALS AND METHODS
Server implementation and architecture
The server has been implemented in R using shiny (http://shiny.rstudio.com) and various other R packages (21–23) (further packages are stated in the caption of Supplementary Table S1) to extend its functionality, most notably Peptides (24) and effsize (https://cran.r-project.org/package=effsize). The open source edition of the shiny server software has been installed on a Linux machine, using an Intel CPU (2.8 GHz, four cores) and 16 GB RAM. Currently, there is no explicit limit to the number of simultaneous users, except for the boundary imposed by finite computational resources (which will typically allow for four parallel sessions to run smoothly, depending on their respective demands).
Description of functionalities
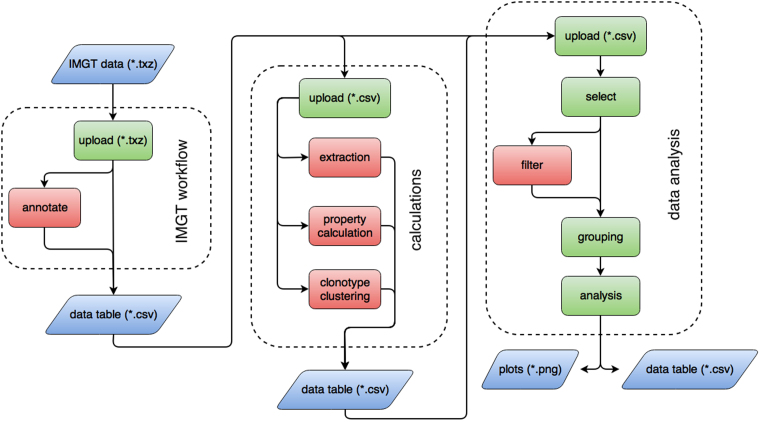
A full list of the currently supported calculations and plotting capabilities in BRepertoire is available in Supplementary Tables S1 and S2. The server’s functions are grouped into three branches, please see Figure 1. In the ‘IMGT’ branch output data from IMGT/V-Quest (19) can be loaded and transcribed into a data table including the columns specified by the user. Moreover, existing tables can be merged using the annotation tab. The ‘Calculation’ branch offers the extraction of data from columns, the calculation of 23 physico-chemical properties and a clonotype clustering interface. Finally, the ‘Analysis’ branch implements data selection, filtering and grouping and seven analysis tabs, which can be selected from a drop-down menu. The following section gives insight into some of the more complex functionalities. For the two most resource-intense functions, the clonotype clustering and the t-SNE analysis, we provide runtime and memory benchmarks (see Supplementary Figure S1).
Figure 1.
BRepertoire’s workflow. The server consists of three branches, which can be used sequentially or independently from one another. Mandatory and optional steps and downloadable artefacts such as data tables and figure files are highlighted in green, red and blue, respectively. The ‘IMGT’ branch accepts IMGT/V-Quest input (19,20) and allows to combine selected columns into a data table. Calculations performed in the second branch (physico-chemical properties and clonotype clustering) can be of use in the ‘Analysis’ branch.
Clonotype clustering
When B cells are activated (e.g. through infection or vaccination), some clones undergo clonal expansion making them much more prevalent in the dataset (see Supplementary Table S3). When analysing an individual’s repertoire, this poses a difficulty. For example, if one is interested in a certain gene distribution in a repertoire, a predominant clone would strongly distort the result. To overcome this problem, clonotype clustering can be applied to group the observations into families which are derived from the same ancestor by inferring relations at the nucleotide sequence level. Subsequently, one observation per clonotype can be computed which will represent the clone in further analysis (the modal sequence). This approach prevents the skewing of data by the over-representation of some clones. A variety of methods have been proposed to cluster the clones in repertoire data, based on protein or DNA sequences and the respective gene families (10,11,17). A widely used distance metric for the comparisons of the individual sequences is the Hamming distance (25). In BRepertoire’s implementation, however, we suggest the use of the Levenshtein distance (26) as it allows varying sequence lengths due to indels that might occur in the sequencing process. For further details, see Supplementary Figure S2.
Physico-chemical properties
Currently, the following properties are supported: the frequencies of tiny (A, C, G, S, T), small (A, C, G, S, T, D, N, P, V), aliphatic (A, I, L, V), aromatic (F, H, W, Y), non-polar (A, C, F, G, I, L, M, P, V, W, Y), polar (D, E, H, K, N, Q, R, S, T), charged (D, E, H, K, R), basic (H, K, R) and acidic (D, E) amino acids in a given sequence of amino acids, the aliphatic index (27), the Boman (potential protein interaction) index (28), the pI (isoelectric point) according to EMBOSS (29), a hydrophobicity measure according to the Kyte–Doolittle scale (30), the instability scale index proposed by Guruprasad et al. (31) and the Kidera factors, a ten-dimensional framework combining various characteristics (32). These properties, which allow for the description of a given amino acid sequence on a fundamental chemical level, have repeatedly proven to be useful (1,33) and can be calculated for any column in the dataset holding amino acid sequences in single-letter code.
Statistical analysis
Typically, data is grouped into several samples (e.g. by patient, time points, tissues) to identify differences between them. This is often done by comparing the V(D)J gene usage but BRepertoire also provides the calculation of physico-chemical features (see above). The ‘Distribution analysis’ tab assists users in finding sample-specific features by providing a variety of statistical hypothesis tests and effect size measures. The user is able to specify subsets for comparison. Further details are reported in Supplementary Figure S3. For the analysis of the V(D)J gene usage distributions, we provide Kullback-Leibler divergence (34) and cosine similarity calculations.
Input and output formats
The server accepts IMGT/V-Quest (19,35) archives, tables in the comma-separated values (CSV) or tab-delimited formats as input (see Figure 1). The former may be obtained by uploading a set of DNA sequences to the IMGT/V-Quest server which performs a series of data annotations, including the assignment of V(D)J-genes, while for the latter a range of format options, such as the usage of quotation marks, can be specified. In order to load data pre-processed by other tools than IMGT, such as IgBlast (36) and MiXCR (17), the user should take care that these options are set appropriately (e.g. MiXCR produces tab-delimited output files). For all types of input, the current upload size limit is 256 MB. For our own datasets, this relates to approximately 200 000 reads (including columns for annotation and calculated properties), a size that has been successfully processed by BRepertoire. Data calculated by the server can be downloaded as CSV files and plots may be retrieved as image files in the PNG format. The ‘Calculation’ branch attaches new features (such as the physico-chemical properties) in new columns at the rear of the input table. Results obtained from the ‘Analysis’ branch (such as output from statistical tests) can be downloaded as separate files if required.
Tutorials and tooltips
In order to assist the user with their first steps, we provide three different kinds of tutorials, explaining the branches and tabs in detail: (i) A text-based tutorial, including screen-shots of the interfaces, (ii) Video tutorials, which allow users to follow the configuration of the interfaces step-by-step and (iii) A live tutorial. The latter represents a special mode of the server, that displays an additional interface on the left hand side of every tab. Once engaged, the user is able to navigate through the tutorial in a step-wise manner by clicking a series of buttons, which trigger actions described below these buttons. As the original interface remains fully functional, one might, in the course of the tutorial, try to alter some of the parameters to test their effect, e.g. by changing the clustering threshold. In addition, we provide tooltips next to most input elements, which show help-text messages once the cursor is hovered above them.
Use cases
This section describes a realistic selection of analysis steps applied to two genuine datasets abbreviated ‘vaccination’ (use case 1) and ‘PBMC’ (use case 2). A detailed description of these datasets is given in Supplementary Table S3. For both, sequences were submitted to the IMGT/V-Quest server (19) to retrieve information on gene usage and CDR3 regions which was then subjected to the ‘Analysis’ branch of BRepertoire. Note, that column names in the following section are encapsulated by single quotation marks (‘’), while data levels / values in these columns are shown in italic to enhance readability.
Preparation
To obtain the physico-chemical properties used in these examples, the ‘Calculate’ branch of the server has been used. The ‘Select’ and ‘Filter’ tabs (Supplementary Figures S4 and S5) can be applied to reduce the dataset to certain columns and values (here, the following columns have been used: ‘Sample.ID’ (Day 0, Day 7 or Day 28), ‘Age.Group’ (Young or Old), ‘Vfamily’ (IGHV1 to IGHV7, V gene family), ‘Jfamily’ (IGHJ1 to IGHJ6, J gene family), ‘Pepstats_length’ (length of CDR3H in numbers of amino acids), ‘Isotype’ (A, M and G) and ‘Kidera1’ to ‘Kidera10’ (32)). Finally, in order to compare different subgroups of the data to one another, at least one grouping column has to be set (Supplementary Figure S6).
Use case 1: gene usage
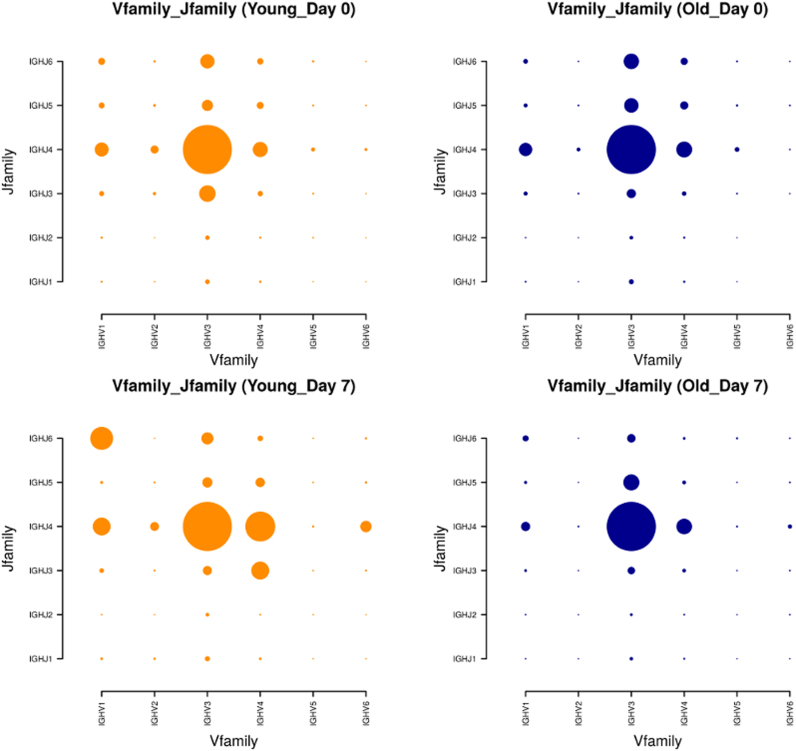
A common analysis focusses on the combination of V(D)J genes in repertoires. Using the ‘Gene frequency’ tab the data can be searched for differences in gene usage. Using the vaccination dataset, we applied this functionality to depict the gene usage of the age groups (Young and Old) at the time-points before and one week after vaccination (Day 0 and Day 7). The (combined) ‘Vfamily’ and ‘Jfamily’ frequencies have been calculated and are shown in Figure 2, resulting in four 2D frequency plots (see Supplementary Figure S7 for the results on Day 28). At Day 0, the gene usage between Young and Old seems to be comparable. However, at Day 7 there is a shift in the Young group towards IGHV1-IGHJ6, IGHV1-IGHJ4, IGHV4-IGHJ3 and IGHV4-IGHJ4 usage, respectively. In contrast, the gene usage for the Old group changes only slightly. This weaker change might reflect the known impairment of elderly people to effectively produce antibodies in response to a given stimulus (37). This tab can also be used to generate one- and three-dimensional plots, see Supplementary Figures S8 and S9.
Figure 2.
Gene usage plot (2D) showing the frequencies of gene families present in the repertoires studied at Day 0 and Day 7. The Young and Old groups are shown in orange and blue, respectively. The reported values are normalized, i.e. the values of each plot sum up to 100%. It is evident, that seven days after vaccination, a significant redistribution has taken place in the Young group, which is not matched in the older comparison group. Note, that gene family IGHV7 has been excluded from the analysis by using the filtering function because it holds only very few observations. A quantitative estimate of the difference is obtained by calculating the Kullback–Leibler divergence (34), the values are reported in Supplementary Table S4.
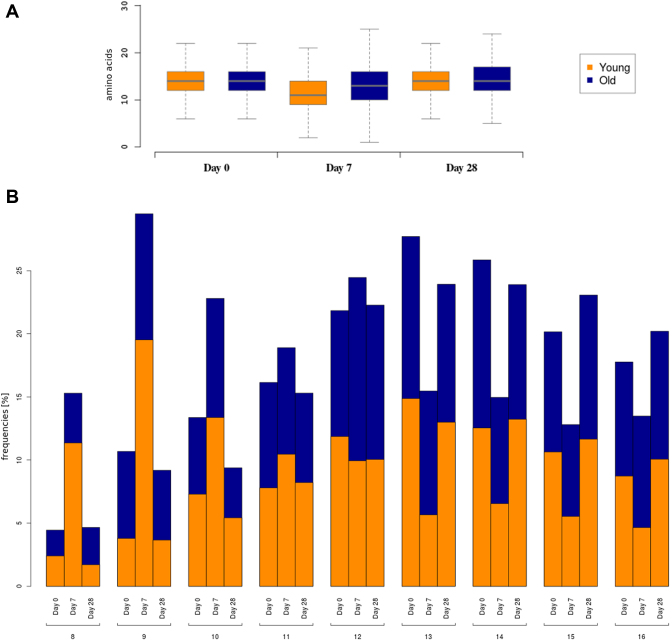
Use case 1: CDR3H sizes
To establish whether the repertoires of the Old and Young groups are affected in different ways by the vaccination, one might investigate the distributions of heavy chain CDR3 amino acid lengths (column ‘Pepstats_length’). As shown in Figure 3A (a boxplot), the median and the spreads are similar for both age groups at Day 0. At Day 7, however, there is a considerable change in the distribution for the Young, resulting from the clonal expansion of cells with comparably smaller CDR3H lengths in response to the vaccine. For the elderly participants’ repertoires, there is an increase in the spread in both directions with only a slight change in the median. Using the ‘Distribution tab’, this change between Day 0 and Day 7 can be quantified to represent effect sizes (Cliff’s Δ (38), see Supplementary Figure S3) of 0.38 and 0.14 for the Young and Old groups, respectively. After 28 days, the Young group has completely returned to the original state while the Old group shows a small deviation. In Figure 3B (a multi-dimensional barplot), it becomes clear that the shift of the box observed for the Young group on Day 7 results mainly from a strong relative increase of clones with CDR3H lengths of 8, 9 and 10. Moreover, it is clearly evident that this change is fully reversed at Day 28. The corresponding bars for the Old group indicate much less pronounced differences.
Figure 3.
The plots show the CDR3H lengths as frequencies for the Young and Old groups of the vaccination data set. The boxplot in (A) indicates the weaker response of the Old group at Day 7 and the longer abating time compared to the Young group (see main text for an interpretation). In (B), the same data are shown in a multi-dimensional barplot, with the length of the CDR3H and the respective day of sample collection on the outer and inner x-axis, respectively, and the relative frequency of the observations on the y-axis. The normalization has been performed using the ‘by all data in group’ setting, thus all bars of the Young and Old groups sum up to 100% for each day respectively, allowing a direct comparison. Only the lengths with a significant relative population are shown. For both plots, IGHV7 and CDR3H loops with a length over 35 have been excluded.
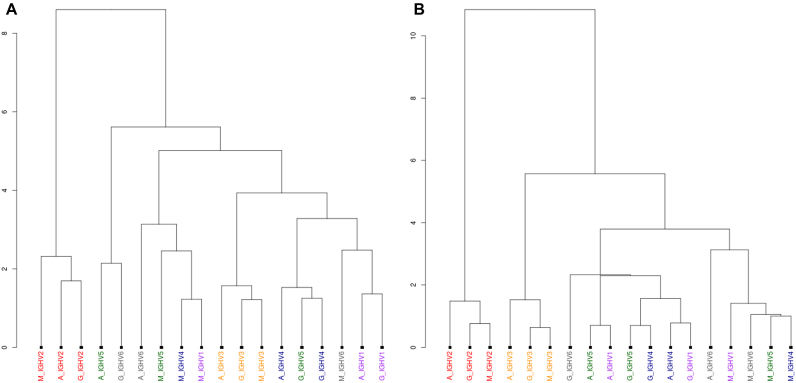
Use case 2: IGHV2 separation
The server also supports the calculation and analysis of physico-chemical properties. In this example, the PBMC dataset has been used. Hierarchical clustering of the ten Kidera factors (Figure 4A) shows a separation of IGHV2 from the other V gene families—a result stable for all three isotypes. The ‘Distribution analysis’ tab can be used (see Supplementary Table S5 and Figure S9) to compare sequences incorporating IGHV2 with all other sequences (pooled together). From this analysis, Kidera factors 2, 4, 5, 6, 7 and 9 can be identified to be the features which contribute most to the observed separation (estimated by their associated effect sizes). If the analysis is repeated, this time using only these six Kidera factors, the separation becomes even more distinct (see Figure 4B and Supplementary Figure S11). A PCA plot (Supplementary Figure S12) shows the same separation.
Figure 4.
Dendrogram plots, showing the distances between the sequences grouped by their V gene family and the isotype, calculated by hierarchical clustering of the Kidera factors for the PBMC dataset. In (A), all 10 Kidera factors have been used in a first attempt, showing a separation of CDR3H sequences encoded by IGHV2 from the rest. Sub-plot (B) uses only Kidera factors 2, 4, 5, 7 and 9, as these have been identified to contribute most to the separation (see main text). By selecting these features only, the separation becomes even more pronounced. For both plots, IGHV7 and CDR3H loops with a length over 35 have been excluded.
CONCLUSION
The analysis of high-throughput sequencing has paved the way for quantitative studies in immunology, where adaptive immune repertoires can contain millions of different variants of the receptor genes. Analysis of repertoires from individuals is now routinely possible and affordable, resulting in many useful applications in biology and medical research. The BRepertoire web server presented has been used to manipulate, analyse and visualize antibody repertoire sequence data from two different case studies that represent typical scenarios of interest in the study of immune responses, thereby proving its usefulness in the analysis of this kind of data. We demonstrate here that BRepertoire can be used to process and analyse data coming directly from IMGT/V-Quest, which is the international standard web software for parsing antibody sequence data. We also demonstrate the user-friendliness and versatility of the developed suite of tools, particularly in the statistical analyses of large-scale data and their visualization and comparison. The server is not limited to the analysis of specific features of repertoire-derived data, such as the V(D)J-gene usage frequencies, but can be applied to any type of data (numerical, nominal, character strings) if grouped into subsets. BRepertoire offers a number of statistical tests, effect size measures (including Monte-Carlo permutation and Kolmogorov–Smirnoff) and a variety of adaptable plots and analysis functions (including PCA, t-SNE, dendrograms, histograms, boxplot and barplots). The flexibility of these tools enables users to explore their data interactively rather than rely on predetermined outputs. Because almost all calculations can be performed in real-time and the results are shown immediately on the screen, the user can analyse complex data very efficiently and test a range of different input parameters quickly. To assist the user with the handling of the more comprehensive interfaces, we provide video-, text-based and live tutorials. The live tutorial familiarises the user with the server and the available workflows in a step-by-step manner using the sample input provided.
BRepertoire is freely available to all users, without any registration or login requirement. Any uploaded or generated data are only stored temporarily, as required by the server’s functions. New features and bug fixes will be bundled to new versions of the software as indicated by the version number (details are reported on the server’s ‘Releases’ page).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Prof. David Kipling, Prof. Ton Coolen and Dr Alexander Mozeika for fruitful discussions and advice, Emma Sinclair for feedback, Joseph C.F. Ng for generating the graphical abstract and Sophie Krecht for her help in deriving the video tutorials.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MRC/BBSRC Systems Immunology of the Lifecourse programme grant: MABRA ‘Multiscale analysis of B cell responses in ageing’ [MR/L01257X/1]. Funding for open access charge: MRC/BBSRC Systems Immunology of the Lifecourse programme grant [MABRA MR/L01257X/1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Martin V.G., Wu Y.-C.B., Townsend C.L., Lu G.H.C., O'Hare J.S., Mozeika A., Coolen A.C.C., Kipling D., Fraternali F., Dunn-Walters D.K.. Transitional B cells in early human B cell development time to revisit the paradigm. Front. Immunol. 2016; 7:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu Y.-C.B., Kipling D., Dunn-Walters D.K.. Age-related changes in human peripheral blood IGH repertoire following vaccination. Fron. Immunol. 2012; 3:doi:10.3389/fimmu.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breden F., Watson C.T.. Using High-Throughput sequencing to characterize the development of the antibody repertoire during Infections: A case study of HIV-1. Adv. Exp. Med. Biol. 2017; 1053:245–263. [DOI] [PubMed] [Google Scholar]
- 4. Wendel B.S., He C., Qu M., Wu D., Hernandez S.M., Ma K.-Y., Liu E.W., Xiao J., Crompton P.D., Pierce S.K. et al. Accurate immune repertoire sequencing reveals malaria infection driven antibody lineage diversification in young children. Nat. Commun. 2017; 8:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vander Heiden J.A., Stathopoulos P., Zhou J.Q., Chen L., Gilbert T.J., Bolen C.R., Barohn R.J., Dimachkie M.M., Ciafaloni E., Broering T.J. et al. Dysregulation of B cell repertoire formation in myasthenia gravis patients revealed through deep sequencing. J. Immunol. 2017; 198:1460–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourcy C.F.A.d., Dekker C.L., Davis M.M., Nicolls M.R., Quake S.R.. Dynamics of the human antibody repertoire after B cell depletion in systemic sclerosis. Sci. Immunol. 2017; 2:eaan8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He J.-S., Subramaniam S., Narang V., Srinivasan K., Saunders S.P., Carbajo D., Wen-Shan T., Hidayah Hamadee N., Lum J., Lee A. et al. IgG1 memory B cells keep the memory of IgE responses. Nat. Commun. 2017; 8:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin V., Wu Y.-C.B., Kipling D., Dunn-Walters D.. Ageing of the B-cell repertoire. Phil. Trans. R. Soc. B. 2015; 370:20140237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. IJspeert H., Schouwenburg P.A.v., Zessen D.v., Pico-Knijnenburg I., Stubbs A.P., Burg M.v.d.. Antigen receptor galaxy: A user-friendly, web-based tool for analysis and visualization of T and B cell receptor repertoire data. J. Immunol. 2017; 198:4156–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bischof J., Ibrahim S.M.. bcRep: R package for comprehensive analysis of B cell receptor repertoire data. PLoS One. 2016; 11:e0161569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta N.T., Heiden V.A.J., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H.. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015; 31:3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schaller S., Weinberger J., Jimenez-Heredia R., Danzer M., Oberbauer R., Gabriel C., Winkler S.M.. ImmunExplorer (IMEX): a software framework for diversity and clonality analyses of immunoglobulins and T cell receptors on the basis of IMGT/HighVQUEST preprocessed NGS data. BMC Bioinform. 2015; 16:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nazarov V.I., Pogorelyy M.V., Komech E.A., Zvyagin I.V., Bolotin D.A., Shugay M., Chudakov D.M., Lebedev Y.B., Mamedov I.Z.. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinform. 2015; 16:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shugay M., Bagaev D.V., Turchaninova M.A., Bolotin D.A., Britanova O.V., Putintseva E.V., Pogorelyy M.V., Nazarov V.I., Zvyagin I.V., Kirgizova V.I. et al. VDJtools: unifying post-analysis of T cell receptor repertoires. PLOS Comput. Biol. 2015; 11:e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duez M., Giraud M., Herbert R., Rocher T., Salson M., Thonier F.. Vidjil: A web platform for analysis of high-throughput repertoire sequencing. PLoS ONE. 2016; 11:e0166126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bagaev D.V., Zvyagin I.V., Putintseva E.V., Izraelson M., Britanova O.V., Chudakov D.M., Shugay M.. VDJviz: a versatile browser for immunogenomics data. BMC Genom. 2016; 17:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M.. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015; 12:380–381. [DOI] [PubMed] [Google Scholar]
- 18. Bystry V., Reigl T., Krejci A., Demko M., Hanakova B., Grioni A., Knecht H., Schlitt M., Dreger P., Sellner L. et al. ARResT/Interrogate: an interactive immunoprofiler for IG/TR NGS data. Bioinformatics. 2017; 33:435–437. [DOI] [PubMed] [Google Scholar]
- 19. Brochet X., Lefranc M.-P., Giudicelli V.. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008; 36:W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alamyar E., Duroux P., Lefranc M.-P., Giudicelli V.. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012; 882:569–604. [DOI] [PubMed] [Google Scholar]
- 21. Loo M.P.J.v.d. The stringdist package for approximate string matching. R J. 2014; 6:111–122. [Google Scholar]
- 22. Müllner D. fastcluster: Fast hierarchical, agglomerative clustering routines for R and python. J. Stat. Softw. 2013; 53:1–18. [Google Scholar]
- 23. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2009; NY: Springer-Verlag. [Google Scholar]
- 24. Osorio D., Rondón-Villarreal P., Torres R.. Peptides: A package for data mining of antimicrobial peptides. R J. 2015; 7:4–14. [Google Scholar]
- 25. Hamming R. Error detecting and error correcting codes. Bell Syst. Tech. J. 1950; 29:147–160. [Google Scholar]
- 26. Levenshtein V.I. Binary codes capable of correcting deletions, insertions, and reversals. Dokl. Phys. 1966; 10:707–710. [Google Scholar]
- 27. Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980; 88:1895–1898. [PubMed] [Google Scholar]
- 28. Boman H.G. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003; 254:197–215. [DOI] [PubMed] [Google Scholar]
- 29. Rice P., Longden I., Bleasby A.. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000; 16:276–277. [DOI] [PubMed] [Google Scholar]
- 30. Kyte J., Doolittle R.F.. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982; 157:105–132. [DOI] [PubMed] [Google Scholar]
- 31. Guruprasad K., Reddy B.V., Pandit M.W.. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990; 4:155–161. [DOI] [PubMed] [Google Scholar]
- 32. Kidera A., Konishi Y., Oka M., Ooi T., Scheraga H.A.. Statistical analysis of the physical properties of the 20 naturally occurring amino acids. J. Protein Chem. 1985; 4:23–55. [Google Scholar]
- 33. Laffy J.M.J., Dodev T., Macpherson J.A., Townsend C., Lu H.C., Dunn-Walters D., Fraternali F.. Promiscuous antibodies characterised by their physico-chemical properties: From sequence to structure and back. Prog. Biophys. Mol. Biol. 2017; 128:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kullback S., Leibler R.A.. On information and sufficiency. Ann. Math. Stat. 1951; 22:79–86. [Google Scholar]
- 35. Ruiz M., Giudicelli V., Ginestoux C., Stoehr P., Robinson J., Bodmer J., Marsh S.G.E., Bontrop R., Lemaitre M., Lefranc G. et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2000; 28:219–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ye J., Ma N., Madden T.L., Ostell J.M.. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013; 41:W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ademokun A., Wu Y.-C., Martin V., Mitra R., Sack U., Baxendale H., Kipling D., Dunn-Walters D.K.. Vaccinationinduced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging Cell. 2011; 10:922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cliff N. Dominance statistics: ordinal analyses to answer ordinal questions. Psychol. Bull. 1993; 114:494–509. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.