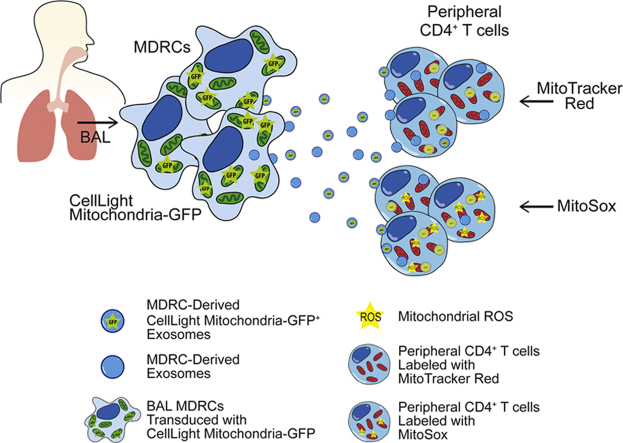
Abstract
Chronic inflammation involving both innate and adaptive immune cells is implicated in the pathogenesis of asthma. Intercellular communication is essential for driving and resolving inflammatory responses in asthma. Emerging studies suggest that extracellular vesicles (EVs) including exosomes facilitate this process. In this report, we have used a range of approaches to show that EVs contain markers of mitochondria derived from donor cells which are capable of sustaining a membrane potential. Further, we propose that these participate in intercellular communication within the airways of human subjects with asthma. Bronchoalveolar lavage fluid of both healthy volunteers and asthmatics contain EVs with encapsulated mitochondria; however, the % HLA-DR+ EVs containing mitochondria and the levels of mitochondrial DNA within EVs were significantly higher in asthmatics. Furthermore, mitochondria are present in exosomes derived from the pro-inflammatory HLA-DR+ subsets of airway myeloid-derived regulatory cells (MDRCs), which are known regulators of T cell responses in asthma. Exosomes tagged with MitoTracker Green, or derived from MDRCs transduced with CellLight Mitochondrial GFP were found in recipient peripheral T cells using a co-culture system, supporting direct exosome-mediated cell-cell transfer. Importantly, exosomally transferred mitochondria co-localize with the mitochondrial network and generate reactive oxygen species within recipient T cells. These findings support a potential novel mechanism of cell-cell communication involving exosomal transfer of mitochondria and the bioenergetic and/or redox regulation of target cells.
Abbreviations: BAL, Bronchoalveolar lavage; DCs, Dendritic cells; EVs, Extracellular vesicles; HLA-DR, Human Leukocyte Antigen – antigen D Related, MHC class II cell surface receptor encoded by the human leukocyte antigen complex; MDRCs, Myeloid-derived regulatory cells; MHC-II, Major histocompatibility complex-II; Mito-GFP, CellLight Mitochondria GFP; MitoT-Green, MitoTracker Green; MitoT-Red, MitoTracker Red; MSCs, Mesenchymal stem cells; NTA, Nanoparticle tracking analysis; ROS, Reactive oxygen species
Keywords: Exosomes, Mitochondria, Asthma, Myeloid-derived, Derived Regulatory Cells
Graphical abstract
Highlights
-
•
BAL and MDRC-derived exosomes contain mitochondria.
-
•
Airway exosomes contain mtDNA.
-
•
MDRC-derived exosomes transfer mitochondria to CD4+ T cells.
-
•
Exosomal mitochondria co-localize with host cell mitochondria.
1. Introduction
Asthma is a chronic disease characterized by persistent inflammation and partially reversible expiratory airflow obstruction [1], [2]. A diverse immune cell repertoire is involved in orchestrating inflammatory responses in asthma [3], [4], [5]. Thus, investigating novel mechanisms of immune cell regulation that underlie the pathophysiology of asthma may lead to the development of improved asthma therapies that target inflammatory responses.
Recent studies have suggested an immunoregulatory role for extracellular vesicles (EVs) in chronic inflammatory diseases, including acute lung injury [6], [7], [8], [9], [10], cancer [11], [12], [13], [14], [15], [16], [17] and asthma [18], [19], [20]. Exosomes are endosomally-derived EVs that facilitate intercellular transfer of nucleic acids [21], [22], proteins [23], lipids [24], and metabolites [25]. Although their biological functions beyond intercellular communication are still being elucidated, the transfer of bioactive cargo by exosomes has potential implications for disease pathogenesis [18], [24], [26], [27], [28], [29].
The content and activities of airway exosomes from the bronchoalveolar lavage (BAL) fluid of asthmatic subjects have been reported to be distinct from healthy controls [22], [30], and thus, exosomes may mediate important immunomodulatory effects in asthma [18], [19], [20]. Peripheral antigen presenting dendritic cells (DCs) have been shown to release exosomes expressing peptide-MHC II that activate T cells to produce cytokines [31], [32], [33]. We and others have shown that free radical generating myeloid-derived regulatory cells (MDRCs) regulate T cell proliferation and airway responses in asthma [34], [35], [36], [37], [38]. Our group has demonstrated that reactive oxygen species (ROS)-producing HLA-DR+ subsets of human MDRCs promote CD4+ T cell proliferation [34]. Although oxidant pathways and soluble mediators are the primary mechanisms by which MDRCs modulate T cell responses, exosome-mediated intercellular transfer of bioactive cargo to T cells has not yet been explored as a potential mechanism of immune regulation by MDRCs in asthma.
Since mitochondria are capable of generating ROS and participating in pro-inflammatory signaling, we hypothesized that they may be packaged and transferred within exosomes to influence T cell function. In this study, we identify and characterize mitochondria-containing exosomes isolated from the airways of control subjects and asthmatics. We demonstrate, for the first time, that airway MDRCs transfer mitochondria to T cells via exosomes. This study provides proof-of-concept that intercellular mitochondrial transfer by MDRCs represents a novel mode of immune regulation in asthma.
2. Materials and methods
2.1. Human subjects
Asthmatic and healthy volunteers were recruited from the University of Alabama at Birmingham Lung Health Center. All study subjects were non-smokers and were screened for IgE titer, blood eosinophil frequencies, past medical history, and FEV1. Healthy controls did not have any prior history of asthma, pulmonary infection or other known lung diseases. All asthmatic subjects enrolled in the study were previously diagnosed with asthma with 12% or more change in FEV1 30 min post-administration of 400 μg of albuterol, as described in the GINA guidelines (Global Strategy for Asthma Management and Prevention; http://www.ginasthma.org/) [39]. Healthy controls with high IgE were excluded from the study. Patients with serum cotinine levels greater than10 ng/ml or who had received treatment with inhaled or systemic corticosteroids within 6 weeks of enrollment were excluded from participation.
Samples collected from 22 study subjects (11 healthy controls and 11 asthmatics) were included in this study. Bronchoscopies on recruited participants were performed as described previously [34], [40]. 200 ml of sterile saline solution was administered with an average return of 113.6 ± 17.2 ml among healthy controls, and 91.2 ± 41.4 ml for asthmatics. Extracellular vesicles were purified from 80 ml of BAL fluid collected from each subject. This study was approved by the University of Alabama at Birmingham Institutional Review Board (IRB-151209005), and all procedures were conducted in accordance with relevant guidelines and regulations. Written informed consent was obtained from all participants.
2.2. Isolation of extracellular vesicles from BAL fluid
Extracellular vesicles (EVs) were purified from the BAL fluid using an established differential centrifugation method, as previously described [22], [30], [32], [41] with minor modifications. Briefly, 80 ml of BAL fluid was centrifuged at 300 ×g for 10 min at 4 °C to remove cells. Supernatant was further centrifuged at 2000 ×g for 10 min at 4 °C to remove any apoptotic bodies. The supernatant was centrifuged again at 10,000 ×g for 30 min at 4 °C and filtered through a 0.2 µm cellulose acetate filter (Corning, New York). The filtrate was then centrifuged at 100,000 ×g for 70 min at 4 °C. The resulting pellet was washed with fresh PBS to remove large protein aggregates. The washed pellet was centrifuged at 100,000 ×g for 70 min at 4 °C and resuspended in 200 µl of fresh sterile PBS. The purified EVs were stored at −80 °C.
2.3. Preparation of exosome-depleted media
RPMI media was supplemented with 20% Corning Human AB Serum (Corning, NY) and centrifuged on an ultracentrifuge overnight at 100,000 ×g at 4 °C. Supernatant was filtered through a 0.2 µm cellulose acetate filter (Corning, New York). The exosome depleted media was then diluted 1:1 with RPMI to make a final concentration of 10% Human AB Serum and additional supplements added (1% Penicillin-Streptomycin, 1% L-Glutamine). Exosome-depleted media was then stored at 4 °C until use.
2.4. Isolation of exosomes from isolated human airway HLA-DR+ MDRCs
Human HLA-DR+ myeloid-derived regulatory cells (MDRCs) were sorted by FACS from the BAL of healthy volunteers and asthmatics as described before [34]. BAL cells were first incubated with RPMI media containing 10% Corning Human AB Serum (Corning, NY) for 30 min on ice and then stained with the following antibodies: CD11b APC Cy7 (ICRF44, BD Biosciences, Franklin Lakes, NY), HLA-DR APC (LN3) CD163 PE (eBioGHI/61), CD33 PE-Cy7 (WM53), CD14 Percpcy5.5 (61D3) and CD11c PECy5 (3.9) (all other antibodies from eBioscience, Waltham, MA). HLA-DR+ MDRCs positive for these markers were sorted using BD FACS Aria. Sorted cells were cultured in exosome-depleted media for 48-h. Conditioned media was collected for exosome isolation. The conditioned media was centrifuged twice at 2000 ×g to remove any cell pellets and apoptotic bodies. The supernatant was then incubated with the Total Exosome Isolation Reagent for Cell Culture Media kit (ThermoFisher, Waltham, MA) per the manufacturer's protocol. Purified exosomes were stored in 50 µl of PBS at −80 °C.
2.5. Nanoparticle tracking analysis (NTA) quantitation of EVs
The NanoSight NS300 (Cambridge, MA) was used to determine the concentration and size distributions of purified airway EVs [42]. Polystyrene latex microspheres (Malvern Instruments Ltd., Malvern, UK) of 100 nm diameter were used to calibrate the instrument. EVs were diluted with PBS 1:1000 to make a final volume of 1 ml and loaded in to a 1 ml syringe. The syringe was placed on a syringe pump and EVs infused at a flow rate of 25 µl/s at room temperature. The camera level was set to 7, gain to 1, and detection threshold to 5. A total of 5 videos were acquired with a duration of 1 min per video. A minimum of 2000–4000/video were processed.
2.6. ImageStream analysis of EVs
ImageStream combines flow cytometry with fluorescence imaging technology and can resolve much smaller particles. The tetraspanin marker, CD63 eFlour450 (clone H5C6; Affymetrix, Inc., Santa Clara) was used as a positive marker to distinguish exosomes from other types of EVs and calibration beads [32], [43]. The following antibodies were used to characterize EVs: HLA-DR APC (clone LN3; Affymetrix, Inc., Santa Clara), CD9 PE (clone M-L13; BD Biosciences, San Jose, CA), CD81 PE-Cy7 (clone 5A6; BioLegend, Inc., San Diego, CA), Grp94 DyLight 488 (clone 9G10; Enzo Life Sciences, Inc., Framingdale, NY), and ARF6 APC (AssayPro, LLC, St. Charles, MO) [43], [44], [45], [46]. The stained samples were imaged at 60 × magnification with extended depth of field (EDF), while acquiring data on channels Ch01, Ch03, Ch06, Ch07, Ch09, Ch11 and Ch12. For acquisition of MitoTracker Green, MitoTracker Red, MitoSox, or CellLight Mitochondrial GFP, channel Ch02 was used for green fluorescence and Ch03 for red. Appropriate controls, single color stains, and calibration beads were used to adjust spectral compensation. A total of 5000 events were acquired. Channels Ch01 and Ch09 were used as brightfields, and Ch12 was used for side-scatter. The acquired data was analyzed using IDEAS software version 6.2 (EMD Millipore, Bellerica, MA).
2.7. Flow cytometry analysis of EVs
Flow cytometry analysis was conducted by staining 1 × 107 particles of EVs with antibodies specific for CD63, HLA-DR, CD9, CD81, ARF6, Grp94, and TSG101 Alexa Fluor 647 (clone 4A10; Novus Biologicals, Littleton, CO). Data was acquired using a BD Becton Dickinson LSRII (Franklin Lakes, NJ). ApogeeBead Mix (Apogee Flow Systems Ltd., Hertfordshire, UK) was used to adjust the photomultiplier tube (PMT) voltage for forward and side scatters to 600 V and 286 V respectively. This allowed us to acquire particles between 50 and 100 nm on the cytometer. A total of 100,000 events were acquired for particles within the 50–100 nm gate. PBS with antibodies alone was used to rule out potential noise from free, unbound antibodies. Sheath fluid from the fluidics system was used to determine the threshold for removing background signal inherent to the cytometer. Thresholding on both forward and side scatters was set to 500 V. Single color stains of EVs were used to determine fluorescence compensation. Data was analyzed using FlowJo X.
2.8. Labeling of airway EVs with MitoTracker green
EVs were labeled with a final concentration of 100 nM MitoTracker Green (ThermoFisher, Waltham, MA) in PBS. Samples were stained for 15 min at 37 °C in the dark, followed by isolation using Total Exosome Isolation from Cell Culture Media kit (ThermoFisher, Waltham, MA) per the manufacturer's protocol. Final EV pellet was resuspended in 50 µl of sterile PBS.
2.9. Cryo-electron microscopy of EVs
For Cryo-EM, 3 µl of an EV sample was applied to glow-discharged 200 mesh Quantifoil R 2/2 nickel grids (Electron Microscopy Sciences, Hatfield, PA) and vitrified in liquid ethane using a FEI Vitrobot Mark IV (FEI; Eindhoven, Netherlands), as previously described [47]. The grids were transferred to a Gatan 622 cryo-holder and observed in a FEI Tecnai F20 electron microscope (FEI; Eindhoven, Netherlands) operated at 200 kV. Images were collected under low-dose conditions on a Gatan Ultrascan 4000 CCD camera at a magnification of 65,500 × and a typical defocus of 2.5 µm.
2.10. Labeling of MDRCs with MitoTracker green and transduction of MDRCs with CellLight Mitochondria GFP
Sorted MDRCs were labeled with MitoTracker Green (ThermoFisher, Waltham, MA) or transduced with CellLight Mitochondria GFP (a fusion construct of the leader sequence of the E1 alpha pyruvate dehydrogenase (a mitochondrial matrix protein) and emGFP (emerald Green Fluorescent protein), Thermofisher, Waltham, MA). For MitoTracker Green (MitoT-Green) staining, the stock dye (1 mM) was first diluted 1:1000 in PBS, and further diluted with 100 µl of cell suspension (1 ×105~1 ×106 cells) and 10 µl diluted MitoT- Green for a final concentration of 100 nM. Samples were stained for 15 min at 37 °C in the dark, followed by two washes with pre-warmed exosome-depleted human serum RPMI media.
For transduction with CellLight Mitochondria GFP(Mito-GFP) a particle-per-cell value of 10 was used to determine the volume of BacMam 2.0 reagent for 1 × 105 cells/well in one well of a 96-well plate. 10 µl of CellLight Mitochondria GFP was used per 1 × 105 cells/well following the manufacturer's protocol. The transduction was performed in exosome-depleted human serum RPMI media for 24 h. Throughout the study, the levels of light energy used for excitation and the wavelength channel selections were kept constant, consistent with standard operating conditions of these instruments.
2.11. Mitochondrial DNA quantitation
Total DNA was harvested from 2.5 × 1010 exosomes from both study groups (2 sets of pooled exosomes from non-overlapping 5 healthy or 5 asthmatic subjects per pool). A third quantitation was performed on airway EV samples (2.5 ×1010 particles/subject) from a healthy individual and an asthmatic subject. Real time PCR for mtDNA was performed using mtDNA-F (5′-CACCCAAGAA CAGGGTTTGT-3′) and mtDNA-R (5′-TGGCCATGGG TATGTTGTTA A-3′). The quantity of DNA was calculated based on a standard curve with known amount of DNA. Data are represented as Mean+ /-SEM.
2.12. Labeling of T cells with MitoSox and MitoTracker red
Purified peripheral CD4+ T cells were labeled with MitoTracker Red or MitoSox (ThermoFisher, Waltham, MA). For MitoTracker Red staining, the stock dye (1 mM) was first diluted 1:1000 in PBS, and further diluted with 100 µl of cell suspension (1 ×106 cells) for a final concentration of 100 nM. Samples were stained for 15 min at 37 °C in the dark, followed by two washes with pre-warmed exosome-depleted human serum RPMI media. For MitoSox staining, the stock dye (5 mM) was diluted 1:100 in PBS, and further diluted with 100 µl of cell suspension (1 ×106 cells) for a final concentration of 5 µM. Samples were stained for 10 min at 37 °C in the dark, followed by three washes with pre-warmed exosome-depleted human serum RPMI media.
2.13. Co-culture of exosomes with T cells
Purified exosomes were cultured with T cells at a ratio of 10 exosome per T cell based on concentrations determined from NTA analysis. To normalize our conditions, we also used a ratio of exosome generating cells to CD4+ T cells. The ratio used was 2.5 × 105 MDRCs (exosome generating cells) to 1 × 106 CD4+ T cells. The exosome-T cell co-culture was performed for 24-h.
2.14. Statistical analysis
The statistical analysis was performed using GraphPad Prism software (La Jolla, CA, USA). Data are presented as mean ± standard deviation unless indicated otherwise in the figure legend. A p-value less than 0.05 was considered to be statistically significant. A two-tailed unpaired Student's t-test or ANOVA with Tukey post-hoc test for data with more than two groups was performed to determine statistical significance.
3. Results
3.1. EVs purified from airways of asthmatics are MitoT-Green+ and contain mitochondrial DNA
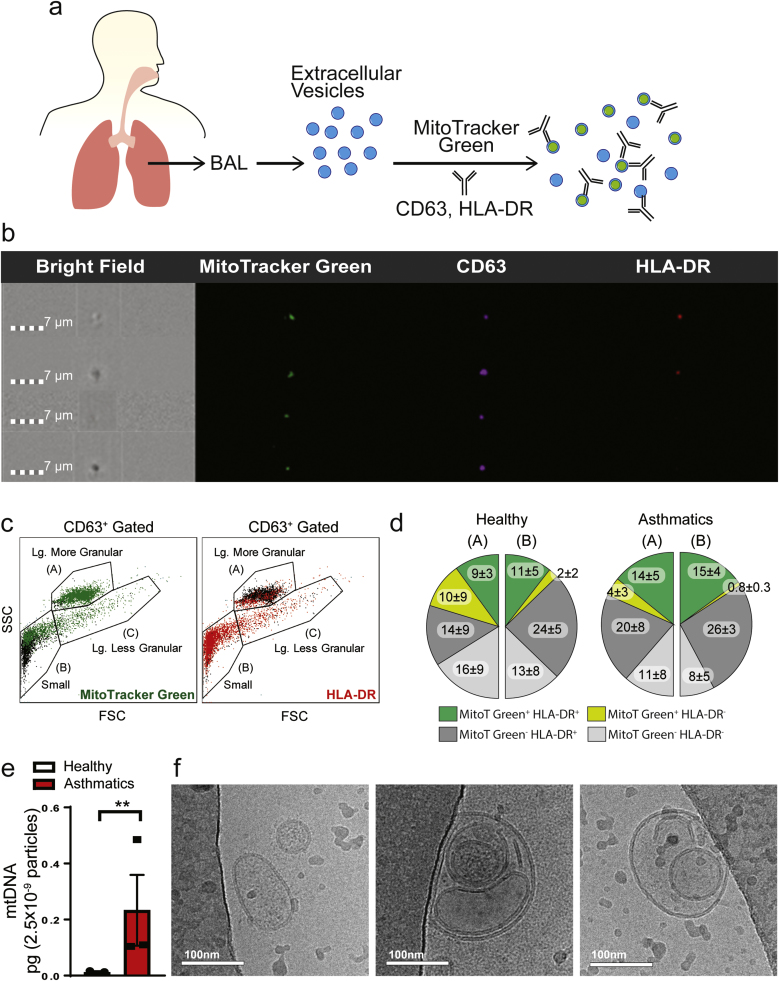
Complexity of exosome mediated cell-cell signaling has been attributed to the heterogeneity of their molecular cargo, which in turn may contribute to differences in their granularity, size and functions. Using an ImageStream imaging flow cytometer, we investigated this heterogeneity in EVs from BAL and identified three subpopulations of CD63+ EVs based on their forward and side scatter properties (larger more granular (A), larger less granular (C) and small (B)) (Fig. 1). In our efforts to determine if inclusion of membranous cellular components contributed to this enhanced granularity in specific subpopulations of EVs, we extensively characterized the EVs with MitoT-Green and additional exosome markers (Fig. 1a). The larger more granular CD63+ EVs stained positive for MitoT-Green with some overlap with the less granular populations, and with MitoT-Green+ CD63+ EVs segregating into both HLA-DR+ and HLA-DR- EVs (Fig. 1b & c). The total proportions of MitoT-Green+ HLA-DR+ within the larger granular (A) and the smaller population (B) of EVs were significantly higher in asthmatics compared to healthy controls (p = 0.0437) (Fig. 1d). Additionally, the proportions of MitoT-Green+HLA-DR+ EVs was significantly higher than MitoT-Green+HLA-DR- EVs in asthmatics (p = 0.0037). The proportions of large granular (A) MitoT-Green+HLA-DR- EVs of asthmatics were significantly (p = 0.0227) higher than the proportions of small (B) MitoT-Green+HLA-DR- EVs of asthmatics. Additionally, the proportions of HLA-DR+ EVs were significantly increased in the BAL of asthmatic subjects (Healthy = 26.4 ± 13.0% compared to Asthmatics= 39.5 ± 13.1%, p = 0.0174). The total MitoT-Green+ EVs were 33% ± 19.4 in healthy subjects and 34.3% ± 7.2 in asthmatics with no overall significant difference between the two groups. Quantitation of mitochondrial DNA (mtDNA) in the total EVs from BAL showed significantly higher levels in EVs from asthmatics compared to those from healthy controls (Fig. 1e), consistent with the presence of mitochondrial components. Cryo-electron microscopic analyses confirmed membrane bound vesicles of exosome size and larger EVs packaged with multiple vesicles, of which some were electron dense and consistent with potential inclusion of mitochondria but without recognizable cristae (Fig. 1f).
Fig. 1.
Characterization of extracellular vesicles (EVs) from the bronchoalveolar lavage (BAL) fluid of healthy controls and asthmatic subjects. (a) Schematic diagram of characterization of BAL EVs by labeling with MitoTracker-Green (MitoT-Green) and anti-CD63 and HLA-DR antibodies (b) ImageStream flow cytometry images showing positive staining of MitoT-Green in airway EVs, along with CD63 and HLA-DR (n = 3 healthy, n = 4 asthmatics; 3 technical replicates each) (c) Representative plot of particle heterogeneity. Granularity is represented on the y-axis and the x-axis represents size (n = 3 healthy, n = 4 asthmatics; 3 technical replicates each) (d) Quantitation of the proportionality of EVs that are positive for MitoT-Green and HLA-DR. The majority of the MitoT-Green signal was found in small and large granular particles. Statistical significance was observed for the following comparison: MitoTGreen+HLA-DR+ Healthy (A+B) vs Asthmatics (A+B) p-value < 0.05; and MitoTGreen+HLA-DR- Asthmatics (A) vs Asthmatics (B) p-value < 0.05 (unpaired Student's T-test) (e) Quantitation of mitochondrial DNA in pooled BAL EVs (total n = 11/group, with 3 independent experiments (** p < 0.01; unpaired Student's T-test) (f) Representative Cryo-Electron Microscopy images of airway EVs (n = 3 independent experiments) showing heterogeneous morphology. Airway EVs were multi-vesicular with electron dense cargo.
3.2. MDRCs produce exosomes that contain polarized mitochondria
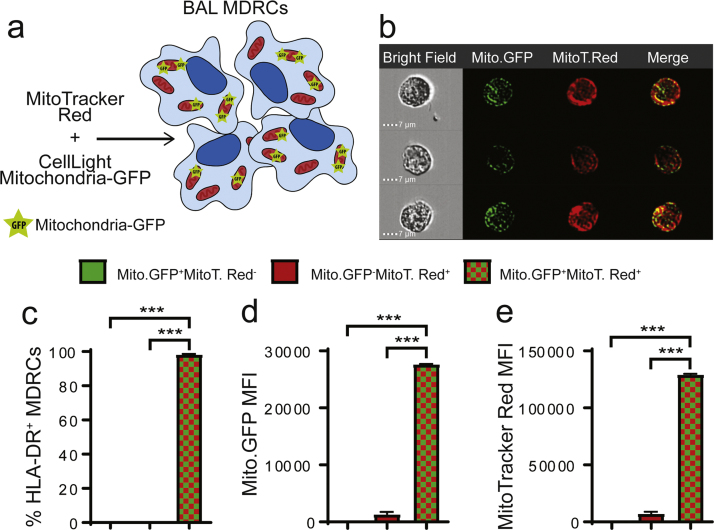
Recent studies have implicated ROS-producing MDRC subsets as critical regulators of T cell proliferation and airway responses [34], [35], [36], [37], [38], [48], with CD163+HLA-DR+human airway MDRCs increasing T cell proliferation in asthmatics in a ROS-dependent manner [34]. We investigated if these pro-inflammatory MDRC subsets produced EVs containing mitochondria. To confirm the presence of mitochondria, purified CD163+HLA-DR+ human airway MDRCs were transduced with CellLight Mitochondria-GFP (Mito-GFP) to mark the mitochondria present in EVs secreted by these cells (Fig. 2a). We then used MitoT-Red to label all MDRC mitochondria and to determine whether the mitochondrial network of MDRCs co-localized with the Mito-GFP signal (Fig. 2a). As shown in Fig. 2b & c, > 90% of the MitoT-Red overlapped with the Mito-GFP signal. Only a small percent of MDRCs (1.08% ± 0.007) were MitoT-Red+Mito-GFP- (Fig. 2. c-e) consistent with maintenance of membrane potential and a stable protein environment allowing expression of the Mito-GFP signal.
Fig. 2.
BAL MDRCs were transduced to express CellLight Mitochondria-GFP. Mitochondria-GFP (Mito-GFP) had proper subcellular localization as seen by the MitoTracker Red (MitoT-Red) overlay. (a) Schematic for validation of transduction and subcellular localization of Mito-GFP (b) ImageStream flow cytometry images illustrating Mito-GFP expression and co-localization with MitoT-Red. (c) Quantitation of % HLA-DR+ MDRCs that are positive for Mito-GFP, MitoT-Red, or both (*** p < 0.001; unpaired Student's T-test) (d-e) Quantitation of mean fluorescence intensity (MFI) of either Mito-GFP or MitoT-Red signal in HLA-DR+ MDRCs gated for either Mito-GFP+, MitoT-Red+, or Mito-GFP+MitoT-Red+ (*** p < 0.001; unpaired Student's T-test) (n = 3 subjects, 3 technical replicates).
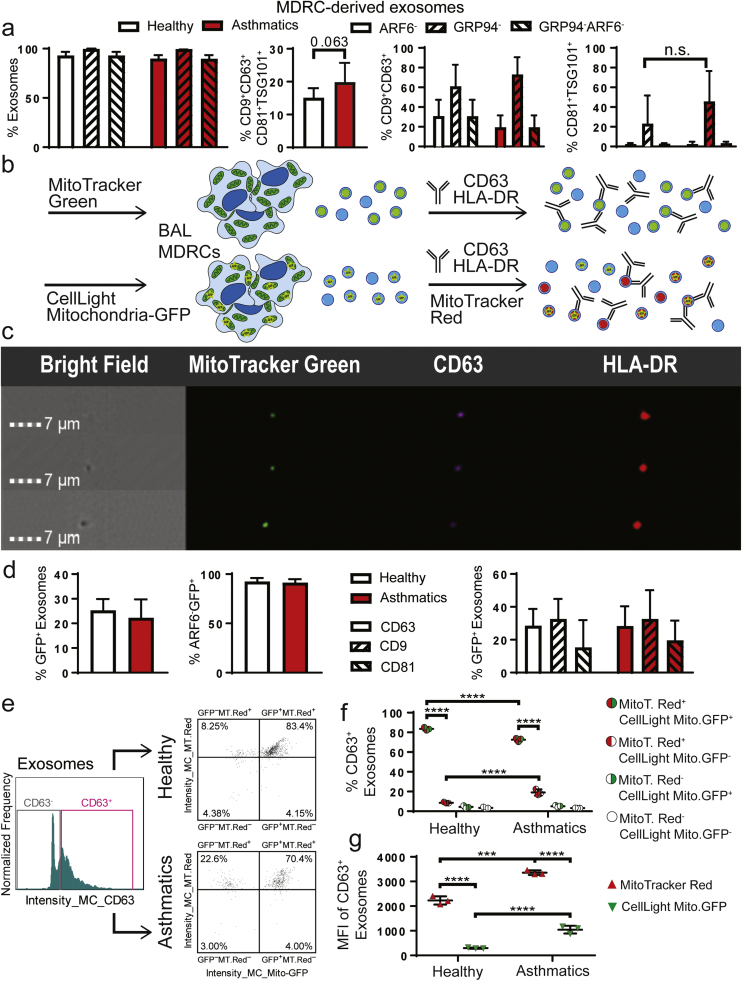
Further characterization of EVs produced by these MDRCs confirmed that they were almost 100% ARF6- exosomes and GRP94- devoid of contamination with endoplasmic reticulum components (Fig. 3a). We then used additional tetraspanin exosomal markers to phenotype MDRC-exosomes and determined the proportions of CD9+CD63+, CD81+TSG101+ as well as CD9+CD63+CD81+TSG101+ exosomes (Fig. 3a). Next, to determine if MDRCs contributed to exosomes MitoT-Green+ exosomes present in BAL, we stained MDRCs with MitoT-Green and purified MDRC exosomes (Fig. 3b). Additionally, we transduced MDRCs as in Fig. 2a and isolated MDRC-derived exosomes including those that were Mito-GFP+ (Fig. 3b). Similar to BAL EVs, MitoT-Green labeled MDRCs also produced MitoT-Green+ exosomes that were CD63+ HLA-DR+ (Fig. 3c). As shown in Fig. 3d, MDRCs transduced with Mito-GFP also produced GFP+ exosomes that were ARF6- and GRP94- with no significant difference between MDRCs isolated from healthy subjects or asthmatics. The proportions of Mito-GFP+ exosomes which were CD9+ and CD81+ were also not significantly different between healthy controls and asthmatics (Fig. 3d).
Fig. 3.
MDRC-derived exosomes contain polarized mitochondria. (a) Characterization of MDRC-derived exosomes after purification. Quantitation of MDRC-derived EVs that were negative for non-exosomal markers ARF6 and GRP94. Quantitation of percent MDRC-derived exosomes with Tetraspanins (CD63, CD9, CD81) and Tsg101, an ESCRT pathway molecule (no statistical significance observed; Student's T-test) (n = 3 healthy, n = 3 asthmatics; 3 technical replicates each) (b) Schematic for characterization and validation of Mito-GFP+exosomes derived from MDRCs. MDRC mitochondria were marked with MitoT-Green or Mito-GFP. MDRC-derived exosomes marked with Mito-GFP were labeled with MitoT-Red (c) ImageStream flow cytometry images of MDRC-derived exosomes positive for MitoT-Green, CD63, and HLA-DR (n = 3 healthy, n = 3 asthmatics; 3 technical replicates each) (d) Characterization of MDRC-derived Mito-GFP+ exosomes. BAL MDRCs were isolated by FACS sorting from n = 3 healthy and n = 3 asthmatic subjects. Purified MDRCs were transduced with Mito-GFP and exosomes isolated from cell culture supernatant at 48 h. Histogram showing quantitation of percentages of CD63+Mito-GFP+ MDRC-derived exosomes, ARF6-Mito-GFP+ MDRC-derived exosomes and individual percentages of CD63+, CD9+ and CD81+ MDRC-derived exosomes (no statistical significance observed; Student's T-test). (e) Histogram and scatter plots of exosomes isolated from MDRCs transduced with Mito-GFP and then labeled with MitoT-Red (n = 3 healthy, n = 3 asthmatics; 3 technical replicates each) (f) Quantitation of the MDRC-derived Mito-GFP+ exosomes labeled ex vivo with MitoT-Red. A significant fraction of exosomes generated were positive for both Mito-GFP and MitoT-Red. (**** p < 0.0001; Two-way ANOVA) (g) Quantitation of the mean fluorescent intensity (MFI) of the MDRC-derived Mito-GFP+ exosomes labeled with MitoT-Red ex vivo. MDRC-derived Mito-GFP+ exosomes from asthmatics had significantly higher MFI of MitoT- Red and Mito-GFP than those derived from MDRCs of healthy subjects (*** p < 0.001, **** p < 0.0001; Two-way ANOVA).
We then determined if MDRC-derived Mito-GFP+ exosomes had polarized mitochondria by evaluating their potential to take up MitoT-Red which requires a membrane potential. The percent of CD63+Mito-GFP+MitoT-Red+ exosomes were significantly higher than the CD63+ MitoT-Red+Mito-GFP- exosomes in both study groups (Fig. 3e). Although the percent of CD63+Mito-GFP+MitoT-Red+ MDRC-derived exosomes were significantly higher in healthy subjects compared to asthmatics (Fig. 3f), the MFI of MitoT-Red and Mito-GFP were both significantly higher in these exosomes from asthmatics compared to controls (Fig. 3g). This is mainly due to the fact that the MitoT-Red+Mito-GFP- exosomes were significantly increased in asthmatics (Fig. 3e). Taken together, these data confirms the presence of mitochondria in MDRC-derived exosomes which are polarized and suggest that they are competent in proton translocation and maintaining a membrane potential which also requires competency in electron transfer. Importantly, there are significant differences in the properties of exosomes between healthy subjects and asthmatics.
3.3. Mitochondria that are transferred from MDRC-exosomes to T cells form a mitochondrial network
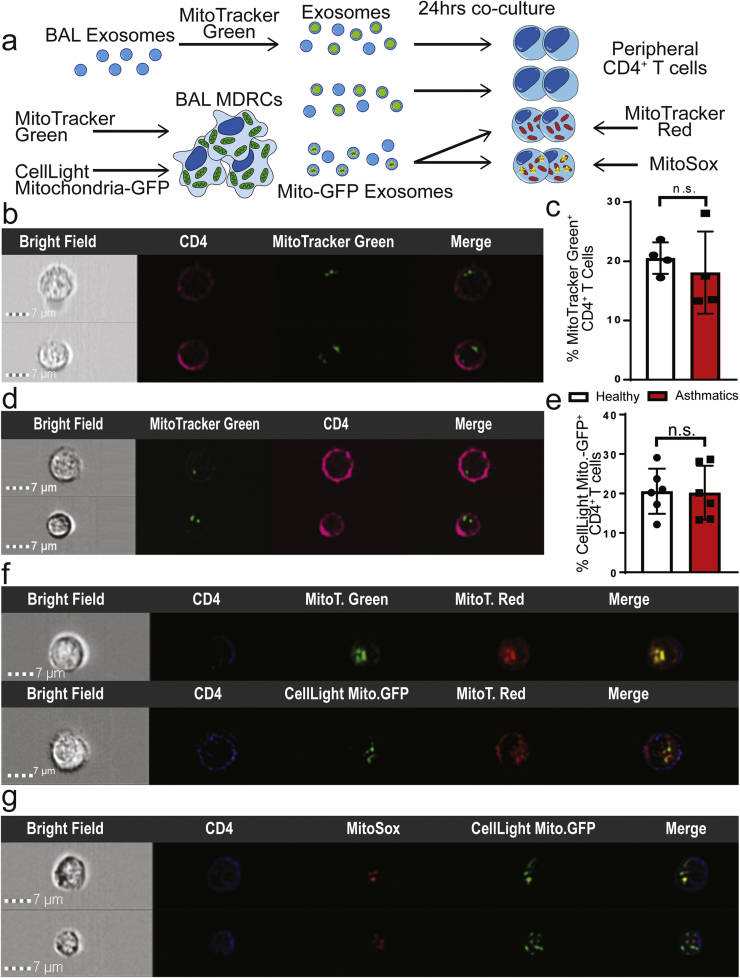
We then evaluated if mitochondria present in MDRC-derived exosomes could be transferred to autologous peripheral blood T cells (Fig. 4a). We first co-cultured MitoT-Green labeled BAL exosomes with peripheral T cells. As shown in Fig. 4b, the MItoT-Green+ BAL exosomes were transferred to T cells. Approximately, 20% of the T cells, in both healthy and asthmatics were MitoT-Green+ after exposure to exosomes (Fig. 4c). Additionally, we stained purified airway MDRCs with MitoT-Green and then co-cultured MDRC-derived exosomes with autologous peripheral T cells and observed MitoT-Green+ exosomes within the T cells (Fig. 4d) suggesting that these exosomes are capable of transferring the mitochondria to T cells. We then isolated exosomes from MDRCs transduced with Mito-GFP (as in Fig. 4a). In co-cultures of these exosomes with peripheral T cells, approximately 20% of the recipient T cells in both healthy and asthmatics contained transferred Mito-GFP+ exosomes. To determine if these exosomally transferred mitochondria merged with the T cell mitochondria and produced ROS, autologous peripheral T cells labeled with MitoSox or MitoT-Red were co-cultured with MDRC-derived Mito-GFP+ or MitoT-Green+ exosomes. As shown in Fig. 4f, both Mito-GFP+ and MitoT-Green+ exosomes co-localized with mitochondrial network of MitoT-Red labeled T cells. Additionally, MDRC-derived Mito-GFP+ exosomes co-localized partially with MitoSox labeled T cells (Fig. 4g) suggesting that the exosomally transferred mitochondria promote post-transfer ROS production in T cells Mito-GFP+ or MitoT-Green+ Mito-GFP+ or MitoT-Green+. Taken together, our studies demonstrate that airway pro-inflammatory regulatory myeloid cells transfer mitochondria that maintain a membrane potential via exosomes to T cells.
Fig. 4.
Exosomal transfer of mitochondria by MDRCs to T cells. Transferred exosomal mitochondria merge with T cell mitochondrial network. (a) Schematic for experimental approach to validate exosomal transfer of mitochondria in co-cultures of exosomes and peripheral CD4+ T cells. (b) ImageStream flow cytometry images showing MitoT-Green+ BAL-derived exosomes within peripheral autologous CD4+ T cells (c) Quantitation of % CD4+ T cells in which MitoT- Green+ airway exosomes were transferred following co-culture (no statistical significance observed; Student's T-test) (b & c: n = 4 healthy, n = 4asthmatics, 3 technical replicates each) (d) ImageStream flow cytometry images of autologous peripheral CD4+ T cells following transfer of MDRC-derived MitoT-Green+ exosomes (n = 4 healthy, n = 3 asthmatics; 3 technical replicates each) (e) Quantitation of % CD4+ T cells in co-cultures with transferred MDRC-derived Mito-GFP+ exosomes (no statistical significance observed; Student's T-test) (n = 6 healthy, n = 6 asthmatics; 3 technical replicates each) (f) ImageStream flow cytometry images showing overlap of host T cell MitoT-Red signal with transferred Mito-GFP from MDRC-derived exosomes, validating merging of these transferred exosomes with the mitochondrial network of the host cells (f & g: n = 3 healthy, n = 3 asthmatics; 3 technical replicates each). (g) ImageStream Flow cytometry images of MitoSox labeling indicating ROS in peripheral autologous CD4+ T cells that partially overlaps with transferred MDRC-derived Mito-GFP+ exosomes.
4. Discussion
In this study, we report that EVs isolated from the airways of both healthy and asthmatic subjects contain mitochondria. However, these mitochondria containing EVs had increased mitochondrial DNA content in asthmatic subjects. Our investigations of potential airway cellular sources for these exosomal mitochondria suggest an important role for pro-inflammatory airway MDRCs. These MDRC-derived exosomes contain both a mitochondrial protein marker, Mito-GFP, and a chemical membrane potential based marker, MitoT-Green. Since the detection of these markers are based on different mechanisms, it is unlikely that selectivity has been introduced during the EV preparation. This provides convincing evidence for the presence of mitochondria in EVs. Furthermore, since the MitoT structural motif consists of a delocalized cation it accumulates across membranes with a net negative internal charge. This occurs if the membrane provides an intact compartment impermeable to ions particularly protons, although this does not imply specificity to mitochondria since other membranes are also capable of polarization. However, the combination of the presence of the mitochondrial marker protein Mito-GFP, mtDNA and contiguous membrane structures in EM studies, again strongly support our conclusion that airway MDRC-derived exosomes contain mitochondria. Furthermore, the MDRC-derived Mito-GFP+ exosomes can also be post-labeled with MitoT-Red, which is consistent with the capacity to maintain a polarized membrane and electron transfer. Importantly, these MDRC-derived exosomal mitochondria can be transferred to T cells ex vivo, following which they co-localize with the mitochondrial network of T cells and generate an increased MitoSox signal. Although the functional effects of donor mitochondria on recipient cells are yet to be determined, our studies support that sufficient elements of mitochondrial electron transport have been retained to allow maintenance of a membrane potential. Together, these studies provide the first evidence of MDRC to T cell transfer of mitochondria packaged within exosomes.
Mitochondria are known to not only regulate energy metabolism but also the survival and fate of eukaryotic cells. Mitochondrial dysfunction may contribute to diverse diseases, ranging from airway diseases [49], [50], [51], [52], neurological pathologies [53], [54], [55] to cancer [56], [57], [58]. There is emerging evidence that mitochondria translocate from one cell to the other both in vitro and in vivo, both in physiological and pathophysiological conditions that includes tissue injury [59], [60], [61] and cancer [62], [63], [64]. Intercellular trafficking of mitochondria has also been shown to exert potent biological effects such as the therapeutic effects of mesenchymal stem cells [65], [66]. There is emerging evidence that tunneling nanotubes are used by mesenchymal stem cells (MSCs) for intercellular transfer of mitochondria to macrophages that modifies their phagocytic function [62], [67], [68], [69]. Recent evidence shows that MSCs target depolarized mitochondria to microvesicles which are then fused and re-utilized by macrophages [70]. During the course of our studies, Morrisson et al. recently identified EVs as a vehicle for intercellular transfer of mitochondria between MSCs and macrophages in models of lung injury [71]. Although these studies evaluated the transfer of MitoT-Green labeled particles in the conditioned media of MSCs to macrophages, the identity of these particles and the functional capacity of mitochondria within exosomes were not clear.
Altered bioenergetics with increased mitochondrial ROS production and epithelial injury are key aspects of the pathogenesis of airway diseases such as asthma and chronic obstructive pulmonary disease [49]. Environmental exposures may contribute to mitochondrial dysfunction in the airways [51], [72]. Mitochondria may function as sensors of inflammation, infection and environmental insults and can respond to such stimuli through altered mitochondrial dynamics and dysfunction [51], [72]. Conversely, mitochondrial dysfunction has downstream influences on cytosolic and mitochondrial calcium regulation and metabolism with effects on airway contractility, responses to oxidative stress, proliferation, apoptosis, and fibrosis which are all hallmarks of airway disease pathophysiology [50], [51], [73], [74].
Oxidative stress has long been established as a driver of persistent inflammation in asthma [75], [76], [77]. We have reported previously that ROS-producing pro-inflammatory HLA-DR+ MDRCs enhance T cell proliferation in both mice and humans with asthma [34], [35], [36]. Our studies show that asthmatics have greater proportions of mitochondria containing EVs that express HLA-DR, a MHC-II molecule. The increased proportions of MitoT-Green+HLA-DR+ EVs suggest a potential role for these in antigen presentation. These data also suggest that the ROS-producing MDRCs may contribute significantly to the airway EVs containing mitochondria which is consistent with our studies with MDRCs. Although our studies reported here do not provide direct evidence, it is possible that the mitochondrial dysfunction in MDRCs is sensed by the T cells in asthmatics through the transfer of mitochondria. Several groups have isolated exosomes from BAL of healthy and asthmatic subjects [22], [30], [32]. The cargo of BAL exosomes from asthmatics include lipids that drive airway inflammation [24], [28] and microRNAs that have the potential to regulate T helper cell differentiation and macrophage functions [18], [22], [26]. None of these prior studies have identified polarized mitochondria or mitochondrial components that may alter the function of recipient T cells. Additionally, studies to-date have not identified MDRCs or any other immune cells as significant cellular sources for exosomes in the airways of asthmatics. Our findings support a potential novel role for exosome-mediated signaling in MDRC-T cell crosstalk. We propose that exosomal transfer of mitochondria from pro-inflammatory MDRCs results in its integration with the recipient cell mitochondrial network with the capacity to change pro-inflammatory function and signaling of these organelles. This modified retrograde signaling may regulate bioenergetic changes in target T cells and, ultimately, alter T cell differentiation and function in chronic inflammatory diseases such as asthma.
Acknowledgements
We acknowledge Marion Spell at the UAB Flow Cytometry Core Facilities, Shawn Williams at the High Resolution Imaging Facility and Cynthia M. Rodenberg at the Cryo-Electron Microscopy Core Facility for technical assistance in exosome flow cytometry, imaging and electron microscopy respectively.
Footnotes
This work was supported by FAMRI YCSA 2010, Parker B. Francis Fellowship and National Institutes of Health (NIH) grants, R01 HL128502 awarded to JSD, P01 HL114470 and R01 AG046210 awarded to VJT, Nathan Shock CenterP30 G050886 (JZ, VDU); and P30 AR048311 and P30 AI27667 for service and support provided by the UAB Comprehensive Flow Cytometry Core.
References
- 1.Anderson G.P. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 2.Boushey H.A., Fahy J.V. Basic mechanisms of asthma. Environ. Health Perspect. 1995;103(Suppl 6):229–233. doi: 10.1289/ehp.95103s6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes P.J. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008;8:183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 4.Hammad H., Lambrecht B.N. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat. Rev. Immunol. 2008;8:193–204. doi: 10.1038/nri2275. [DOI] [PubMed] [Google Scholar]
- 5.Ray A., Khare A., Krishnamoorthy N., Qi Z., Ray P. Regulatory T cells in many flavors control asthma. Mucosal Immunol. 2010;3:216–229. doi: 10.1038/mi.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gennai S., Monsel A., Hao Q., Park J., Matthay M.A., Lee J.W. Microvesicles derived from human mesenchymal stem cells restore alveolar fluid clearance in human lungs rejected for transplantation. Am. J. Transplant. 2015;15:2404–2412. doi: 10.1111/ajt.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monsel A., Zhu Y.G., Gennai S., Hao Q., Hu S., Rouby J.J., Rosenzwajg M., Matthay M.A., Lee J.W. Therapeutic effects of human mesenchymal stem cell-derived microvesicles in severe pneumonia in mice. Am. J. Respir. Crit. Care Med. 2015;192:324–336. doi: 10.1164/rccm.201410-1765OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H., Abston E., Zhang D., Rai A., Jin Y. Extracellular vesicle: an emerging mediator of intercellular crosstalk in lung inflammation and injury. Front Immunol. 2018;9:924. doi: 10.3389/fimmu.2018.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H., Zhang D., Zhu Z., Dela Cruz C.S., Jin Y. Epithelial cell-derived microvesicles activate macrophages and promote inflammation via microvesicle-containing microRNAs. Sci. Rep. 2016;6:35250. doi: 10.1038/srep35250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone M.L., Zhao Y., Robert Smith J., Weiss M.L., Kron I.L., Laubach V.E., Sharma A.K. Mesenchymal stromal cell-derived extracellular vesicles attenuate lung ischemia-reperfusion injury and enhance reconditioning of donor lungs after circulatory death. Respir. Res. 2017;18:212. doi: 10.1186/s12931-017-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbins P.D., Morelli A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller L., Mitsuhashi M., Simms P., Gooding W.E., Whiteside T.L. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci. Rep. 2016;6:20254. doi: 10.1038/srep20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins P.D., Dorronsoro A., Booker C.N. Vol. 126. 2016. Regulation of chronic inflammatory and immune processes by extracellular vesicles; pp. 1173–1180. (J Clin Invest). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiltbrunner S., Larssen P., Eldh M., Martinez-Bravo M.J., Wagner A.K., Karlsson M.C., Gabrielsson S. Exosomal cancer immunotherapy is independent of MHC molecules on exosomes. Oncotarget. 2016 doi: 10.18632/oncotarget.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell B.M., Kirk I.D., Hiltbrunner S., Gabrielsson S., Bultema J.J. Designer exosomes as next-generation cancer immunotherapy. Nanomedicine. 2016;12:163–169. doi: 10.1016/j.nano.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Shinohara H., Kuranaga Y., Kumazaki M., Sugito N., Yoshikawa Y., Takai T., Taniguchi K., Ito Y., Akao Y. Regulated polarization of tumor-associated macrophages by miR-145 via colorectal cancer-derived extracellular vesicles. J. Immunol. 2017;199:1505–1515. doi: 10.4049/jimmunol.1700167. [DOI] [PubMed] [Google Scholar]
- 17.Salimu J., Webber J., Gurney M., Al-Taei S., Clayton A., Tabi Z. Dominant immunosuppression of dendritic cell function by prostate-cancer-derived exosomes. J. Extracell. Vesicles. 2017;6:1368823. doi: 10.1080/20013078.2017.1368823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sastre B., Canas J.A., Rodrigo-Munoz J.M., Del Pozo V. Novel modulators of asthma and allergy: exosomes and MicroRNAs. Front Immunol. 2017;8:826. doi: 10.3389/fimmu.2017.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hough K.P., Chanda D., Duncan S.R., Thannickal V.J., Deshane J.S. Exosomes in immunoregulation of chronic lung diseases. Allergy. 2016 doi: 10.1111/all.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Admyre C., Telemo E., Almqvist N., Lotvall J., Lahesmaa R., Scheynius A., Gabrielsson S. Exosomes - nanovesicles with possible roles in allergic inflammation. Allergy. 2008;63:404–408. doi: 10.1111/j.1398-9995.2007.01600.x. [DOI] [PubMed] [Google Scholar]
- 21.Cazzoli R., Buttitta F., Di Nicola M., Malatesta S., Marchetti A., Rom W.N., Pass H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013;8:1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levanen B., Bhakta N.R., Torregrosa Paredes P., Barbeau R., Hiltbrunner S., Pollack J.L., Skold C.M., Svartengren M., Grunewald J., Gabrielsson S. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 2013;131:894–903. doi: 10.1016/j.jaci.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thery C., Boussac M., Veron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 24.Subra C., Grand D., Laulagnier K., Stella A., Lambeau G., Paillasse M., De Medina P., Monsarrat B., Perret B., Silvente-Poirot S. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puhka M., Takatalo M., Nordberg M.E., Valkonen S., Nandania J., Aatonen M., Yliperttula M., Laitinen S., Velagapudi V., Mirtti T. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics. 2017;7:3824–3841. doi: 10.7150/thno.19890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Ying X., Wang X., Wu X., Zhu Q., Wang X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017;38:522–528. doi: 10.3892/or.2017.5697. [DOI] [PubMed] [Google Scholar]
- 27.Boelens M.C., Wu T.J., Nabet B.Y., Xu B., Qiu Y., Yoon T., Azzam D.J., Twyman-Saint Victor C., Wiemann B.Z., Ishwaran H. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulshreshtha A., Ahmad T., Agrawal A., Ghosh B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J. Allergy Clin. Immunol. 2013;131:1194–1203. doi: 10.1016/j.jaci.2012.12.1565. (1203 e1191-1114) [DOI] [PubMed] [Google Scholar]
- 29.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torregrosa Paredes P., Esser J., Admyre C., Nord M., Rahman Q.K., Lukic A., Radmark O., Gronneberg R., Grunewald J., Eklund A. Bronchoalveolar lavage fluid exosomes contribute to cytokine and leukotriene production in allergic asthma. Allergy. 2012;67:911–919. doi: 10.1111/j.1398-9995.2012.02835.x. [DOI] [PubMed] [Google Scholar]
- 31.Admyre C., Bohle B., Johansson S.M., Focke-Tejkl M., Valenta R., Scheynius A., Gabrielsson S. B cell-derived exosomes can present allergen peptides and activate allergen-specific T cells to proliferate and produce TH2-like cytokines. J. Allergy Clin. Immunol. 2007;120:1418–1424. doi: 10.1016/j.jaci.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 32.Admyre C., Grunewald J., Thyberg J., Gripenback S., Tornling G., Eklund A., Scheynius A., Gabrielsson S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur. Respir. J. 2003;22:578–583. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 33.Admyre C., Johansson S.M., Paulie S., Gabrielsson S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur. J. Immunol. 2006;36:1772–1781. doi: 10.1002/eji.200535615. [DOI] [PubMed] [Google Scholar]
- 34.Deshane J.S., Redden D.T., Zeng M., Spell M.L., Zmijewski J.W., Anderson J.T., Deshane R.J., Gaggar A., Siegal G.P., Abraham E. Subsets of airway myeloid-derived regulatory cells distinguish mild asthma from chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2015;135:413–424. doi: 10.1016/j.jaci.2014.08.040. (e415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshane J., Zmijewski J.W., Luther R., Gaggar A., Deshane R., Lai J.F., Xu X., Spell M., Estell K., Weaver C.T. Free radical-producing myeloid-derived regulatory cells: potent activators and suppressors of lung inflammation and airway hyperresponsiveness. Mucosal Immunol. 2011;4:503–518. doi: 10.1038/mi.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Jin T.H., Farhana A., Freeman J., Estell K., Zmijewski J.W., Gaggar A., Thannickal V.J., Schwiebert L.M., Steyn A.J., Deshane J.S. Exposure to cigarette smoke impacts myeloid-derived regulatory cell function and exacerbates airway hyper-responsiveness. Lab Invest. 2014;94:1312–1325. doi: 10.1038/labinvest.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray P., Arora M., Poe S.L., Ray A. Lung myeloid-derived suppressor cells and regulation of inflammation. Immunol. Res. 2011;50:153–158. doi: 10.1007/s12026-011-8230-1. [DOI] [PubMed] [Google Scholar]
- 38.Arora M., Poe S.L., Oriss T.B., Krishnamoorthy N., Yarlagadda M., Wenzel S.E., Billiar T.R., Ray A., Ray P. TLR4/MyD88-induced CD11b+Gr-1 int F4/80+ non-migratory myeloid cells suppress Th2 effector function in the lung. Mucosal Immunol. 2010;3:578–593. doi: 10.1038/mi.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bateman E.D., Hurd S.S., Barnes P.J., Bousquet J., Drazen J.M., FitzGerald M., Gibson P., Ohta K., O'Byrne P., Pedersen S.E. Global strategy for asthma management and prevention: gina executive summary. Eur. Respir. J. 2008;31:143–178. doi: 10.1183/09031936.00138707. [DOI] [PubMed] [Google Scholar]
- 40.Anderson J.T., Zeng M., Li Q., Stapley R., Moore D.R., 2nd, Chenna B., Fineberg N., Zmijewski J., Eltoum I.E., Siegal G.P. Elevated levels of NO are localized to distal airways in asthma. Free Radic. Biol. Med. 2011;50:1679–1688. doi: 10.1016/j.freeradbiomed.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. (Chapter 3: Unit3 22) [DOI] [PubMed] [Google Scholar]
- 42.Dragovic R.A., Gardiner C., Brooks A.S., Tannetta D.S., Ferguson D.J., Hole P., Carr B., Redman C.W., Harris A.L., Dobson P.J. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prado N., Marazuela E.G., Segura E., Fernandez-Garcia H., Villalba M., Thery C., Rodriguez R., Batanero E. Exosomes from bronchoalveolar fluid of tolerized mice prevent allergic reaction. J. Immunol. 2008;181:1519–1525. doi: 10.4049/jimmunol.181.2.1519. [DOI] [PubMed] [Google Scholar]
- 45.Hosseini-Beheshti E., Pham S., Adomat H., Li N., Tomlinson Guns E.S. Exosomes as biomarker enriched microvesicles: characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes. Mol. Cell Proteom. 2012;11:863–885. doi: 10.1074/mcp.M111.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caby M.P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 47.Dearborn A.D., Wall E.A., Kizziah J.L., Klenow L., Parker L.K., Manning K.A., Spilman M.S., Spear J.M., Christie G.E., Dokland T. Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. Elife. 2017:6. doi: 10.7554/eLife.30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Upham J.W., Denburg J.A., O'Byrne P.M. Rapid response of circulating myeloid dendritic cells to inhaled allergen in asthmatic subjects. Clin. Exp. Allergy. 2002;32:818–823. doi: 10.1046/j.1365-2222.2002.01375.x. [DOI] [PubMed] [Google Scholar]
- 49.Agarwal A.R., Yin F., Cadenas E. Metabolic shift in lung alveolar cell mitochondria following acrolein exposure. Am. J. Physiol. Lung Cell Mol. Physiol. 2013;305:L764–L773. doi: 10.1152/ajplung.00165.2013. [DOI] [PubMed] [Google Scholar]
- 50.Delmotte P., Yang B., Thompson M.A., Pabelick C.M., Prakash Y.S., Sieck G.C. Inflammation alters regional mitochondrial Ca(2)+ in human airway smooth muscle cells. Am. J. Physiol. Cell Physiol. 2012;303:C244–C256. doi: 10.1152/ajpcell.00414.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aravamudan B., Thompson M.A., Pabelick C.M., Prakash Y.S. Mitochondria in lung diseases. Expert Rev. Respir. Med. 2013;7:631–646. doi: 10.1586/17476348.2013.834252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trian T., Allard B., Ozier A., Maurat E., Dupin I., Thumerel M., Ousova O., Gillibert-Duplantier J., Le Morvan V., Begueret H. Selective dysfunction of p53 for mitochondrial biogenesis induces cellular proliferation in bronchial smooth muscle from asthmatic patients. J. Allergy Clin. Immunol. 2016;137:1717–1726. doi: 10.1016/j.jaci.2015.10.031. (e1713) [DOI] [PubMed] [Google Scholar]
- 53.Mattson M.P., Gleichmann M., Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner M., Jurcoane A., Hildebrand C., Guresir E., Vatter H., Zanella F.E., Berkefeld J., Pilatus U., Hattingen E. Metabolic changes in patients with aneurysmal subarachnoid hemorrhage apart from perfusion deficits: neuronal mitochondrial injury? AJNR Am. J. Neuroradiol. 2013;34:1535–1541. doi: 10.3174/ajnr.A3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ben-Shachar D., Karry R. Neuroanatomical pattern of mitochondrial complex I pathology varies between schizophrenia, bipolar disorder and major depression. PLoS One. 2008;3:e3676. doi: 10.1371/journal.pone.0003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamp D.W., Shacter E., Weitzman S.A. Chronic inflammation and cancer: the role of the mitochondria. Oncol. (Williston Park) 2011;25:400–410. (413) [PubMed] [Google Scholar]
- 57.Ralph S.J., Rodriguez-Enriquez S., Neuzil J., Saavedra E., Moreno-Sanchez R. The causes of cancer revisited: "mitochondrial malignancy" and ROS-induced oncogenic transformation - why mitochondria are targets for cancer therapy. Mol. Asp. Med. 2010;31:145–170. doi: 10.1016/j.mam.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Bonnet S., Archer S.L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R., Lee C.T., Lopaschuk G.D., Puttagunta L., Bonnet S. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Mahrouf-Yorgov M., Augeul L., Da Silva C.C., Jourdan M., Rigolet M., Manin S., Ferrera R., Ovize M., Henry A., Guguin A. Mesenchymal stem cells sense mitochondria released from damaged cells as danger signals to activate their rescue properties. Cell Death Differ. 2017;24:1224–1238. doi: 10.1038/cdd.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Zhang Y., Yeung S.C., Liang Y., Liang X., Ding Y., Ip M.S., Tse H.F., Mak J.C., Lian Q. Mitochondrial transfer of induced pluripotent stem cell-derived mesenchymal stem cells to airway epithelial cells attenuates cigarette smoke-induced damage. Am. J. Respir. Cell Mol. Biol. 2014;51:455–465. doi: 10.1165/rcmb.2013-0529OC. [DOI] [PubMed] [Google Scholar]
- 61.Spees J.L., Olson S.D., Whitney M.J., Prockop D.J. Mitochondrial transfer between cells can rescue aerobic respiration. Proc. Natl. Acad. Sci. USA. 2006;103:1283–1288. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu J., Zheng X., Li F., Yu Y., Chen Z., Liu Z., Wang Z., Xu H., Yang W. Tunneling nanotubes promote intercellular mitochondria transfer followed by increased invasiveness in bladder cancer cells. Oncotarget. 2017;8:15539–15552. doi: 10.18632/oncotarget.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Cui J., Sun X., Zhang Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2011;18:732–742. doi: 10.1038/cdd.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasquier J., Guerrouahen B.S., Al Thawadi H., Ghiabi P., Maleki M., Abu-Kaoud N., Jacob A., Mirshahi M., Galas L., Rafii S. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J. Transl. Med. 2013;11:94. doi: 10.1186/1479-5876-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmad T., Mukherjee S., Pattnaik B., Kumar M., Singh S., Kumar M., Rehman R., Tiwari B.K., Jha K.A., Barhanpurkar A.P. Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J. 2014;33:994–1010. doi: 10.1002/embj.201386030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinha P., Islam M.N., Bhattacharya S., Bhattacharya J. Intercellular mitochondrial transfer: bioenergetic crosstalk between cells. Curr. Opin. Genet Dev. 2016;38:97–101. doi: 10.1016/j.gde.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X., Gerdes H.H. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 2015;22:1181–1191. doi: 10.1038/cdd.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vignais M.L., Caicedo A., Brondello J.M., Jorgensen C. Cell connections by tunneling nanotubes: effects of mitochondrial trafficking on target cell metabolism, homeostasis, and response to therapy. Stem Cells Int. 2017;2017:6917941. doi: 10.1155/2017/6917941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson M.V., Morrison T.J., Doherty D.F., McAuley D.F., Matthay M.A., Kissenpfennig A., O'Kane C.M., Krasnodembskaya A.D. Mitochondrial transfer via tunneling nanotubes is an important mechanism by which mesenchymal stem cells enhance macrophage phagocytosis in the in vitro and in vivo models of ARDS. Stem Cells. 2016;34:2210–2223. doi: 10.1002/stem.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phinney D.G., Di Giuseppe M., Njah J., Sala E., Shiva S., St Croix C.M., Stolz D.B., Watkins S.C., Di Y.P., Leikauf G.D. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat. Commun. 2015;6:8472. doi: 10.1038/ncomms9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morrison T.J., Jackson M.V., Cunningham E.K., Kissenpfennig A., McAuley D.F., O'Kane C.M., Krasnodembskaya A.D. Mesenchymal stromal cells modulate macrophages in clinically relevant lung injury models by extracellular vesicle mitochondrial transfer. Am. J. Respir. Crit. Care Med. 2017;196:1275–1286. doi: 10.1164/rccm.201701-0170OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aravamudan B., Kiel A., Freeman M., Delmotte P., Thompson M., Vassallo R., Sieck G.C., Pabelick C.M., Prakash Y.S. Cigarette smoke-induced mitochondrial fragmentation and dysfunction in human airway smooth muscle. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L840–L854. doi: 10.1152/ajplung.00155.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prakash Y.S. Airway smooth muscle in airway reactivity and remodeling: what have we learned? Am. J. Physiol. Lung Cell Mol. Physiol. 2013;305:L912–L933. doi: 10.1152/ajplung.00259.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiegman C.H., Michaeloudes C., Haji G., Narang P., Clarke C.J., Russell K.E., Bao W., Pavlidis S., Barnes P.J., Kanerva J. Oxidative stress-induced mitochondrial dysfunction drives inflammation and airway smooth muscle remodeling in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2015;136:769–780. doi: 10.1016/j.jaci.2015.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calhoun W.J., Reed H.E., Moest D.R., Stevens C.A. Enhanced superoxide production by alveolar macrophages and air-space cells, airway inflammation, and alveolar macrophage density changes after segmental antigen bronchoprovocation in allergic subjects. Am. Rev. Respir. Dis. 1992;145:317–325. doi: 10.1164/ajrccm/145.2_Pt_1.317. [DOI] [PubMed] [Google Scholar]
- 76.Barnes P.J. Reactive oxygen species and airway inflammation. Free Radic. Biol. Med. 1990;9:235–243. doi: 10.1016/0891-5849(90)90034-g. [DOI] [PubMed] [Google Scholar]
- 77.Sahiner U.M., Birben E., Erzurum S., Sackesen C., Kalayci O. Oxidative stress in asthma. World Allergy Organ J. 2011;4:151–158. doi: 10.1097/WOX.0b013e318232389e. [DOI] [PMC free article] [PubMed] [Google Scholar]