Abstract
Metabolic labeling of glycans with bioorthogonal reporters has been widely used for glycan imaging and glycoproteomic profiling. One of the intrinsic limitations of metabolic glycan labeling is the lack of cell-type selectivity. The recently developed liposome-assisted bioorthogonal reporter (LABOR) strategy provides a promising means to overcome this limitation, but the mechanism of LABOR has not been investigated in detail. In this work, we performed a mechanistic study on LABOR and explored its multiplexing capability. Our studies support an endocytosis-salvage mechanism. The ligand-targeted liposomes encapsulating azidosugars are internalized into the endosome via the receptor-mediated endocytosis. Unlike the conventional drug delivery, LABOR does not rely on the endosomal escape pathways. Rather, the liposomes are allowed to enter the lysosome, inside which the azidosugars are released from the liposomes. The released azidosugars then intercept the salvage pathways of monosaccharides and get transported into the cytosol by lysosomal sugar transporters. Based on this mechanism, we expanded the scope of LABOR by evaluating a series of ligand-receptor pairs for targeting sialoglycans in various cell types. Different ligand types including small molecules, antibodies, aptamers, and peptides could be easily implemented into LABOR. Finally, we demonstrated that the sialoglycans in two distinct cell populations in a co-cultured system could be selectively labeled with two distinct chemical reporters by performing a multiplexed LABOR labeling.
TOC image
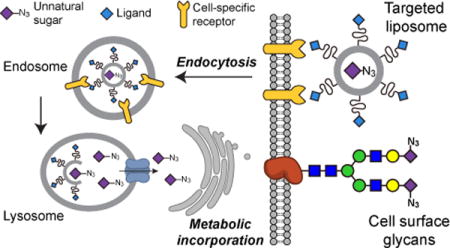
INTRODUCTION
Cells uptake monosaccharides, which serve as precursors for the biosynthesis of glycans in various glycoconjugates, including glycoproteins, glycolipids, and proteoglycans. Glycans play essential roles in regulating diverse biological and pathological processes.1 The metabolic machinery can be harnessed to incorporate exogenous monosaccharide analogs or unnatural sugars bearing a bioorthogonal functional group (e.g., the azide or alkyne) into cellular glycans.2–4 The incorporated azide or alkyne serves as a chemical handle for subsequent chemoselective conjugation with biophysical probes (e.g., fluorophores or affinity tags) functionalized with a complementary bioorthogonal group. Termed metabolic glycan labeling (MGL) or metabolic oligosaccharide engineering (MOE), this two-step chemical reporter strategy has emerged as a powerful tool for probing sialylation,5–9 mucin-type O-linked glycosylation,10–12 fucosylation,13–16 and O-GlcNAcyaltion17–21 in live cells and in living animals.
Unnatural sugars usually enter cells without discriminating cell types, resulting in global labeling of glycans in various cell types. The lack of cell-type selectivity hampers the utility of MGL in profiling glycosylation of specific cell types within a complex biological system.22 To overcome this limitation, we recently developed a liposome-assisted strategy for selective labeling of glycans in specific cell types.23 This strategy utilizes the ligand-targeted liposomes to encapsulate azidosugars and deliver them into the target cells via specific ligand-receptor recognition. The delivered azidosugars are metabolically incorporated into cellular glycans and the resulting azide-incorporated glycans are then labeled with desired probes using bioorthogonal chemistry. Furthermore, it was demonstrated that this strategy could be used to label and visualize tumor-associated sialoglycans and brain sialoglycans in living mice.24,25 Termed the LABOR (liposome-assisted bioorthogonal reporter) strategy, this technique has expanded the utility of MGL and enabled many potential applications. For example, it is of great interest to label glycans in a specific cell type when studying how glycans regulate cell-cell interactions, so that the origin of the labeled glycans is not obscured by surrounding cells sharing the common glycan structures. In addition, metabolic labeling of low-abundance glycans or glycans in low-abundance cells can tremendously benefit from cell-type selectivity.
Towards realizing those applications, we herein report the mechanistic investigation of LABOR and the exploration of its multiplexing capability. By using the ligand-targeted liposomes encapsulating 9AzSia, an azido analog of sialic acid (Sia), our studies support an endocytosis-salvage mechanism of LABOR (Figure 1). The liposomes are internalized into the endosome via the receptor-mediated endocytosis and then traffic to the lysosome, inside which 9AzSia is released out of the liposomes. The released 9AzSia intercepts the salvage pathway of Sia, and gets transported out of the lysosome to the cytoplasm by the lysosomal transporter sialin (SLC17A5), located on the lysosome membrane. Based on this mechanism, we performed a comprehensive evaluation of the scope of LABOR. A panel of ligand-receptor pairs were evaluated to target azidosugars to distinct cell types for cell-selective MGL. Finally, we demonstrated that LABOR could be used to simultaneously label two cell populations with two distinct chemical reporters.
Figure 1.
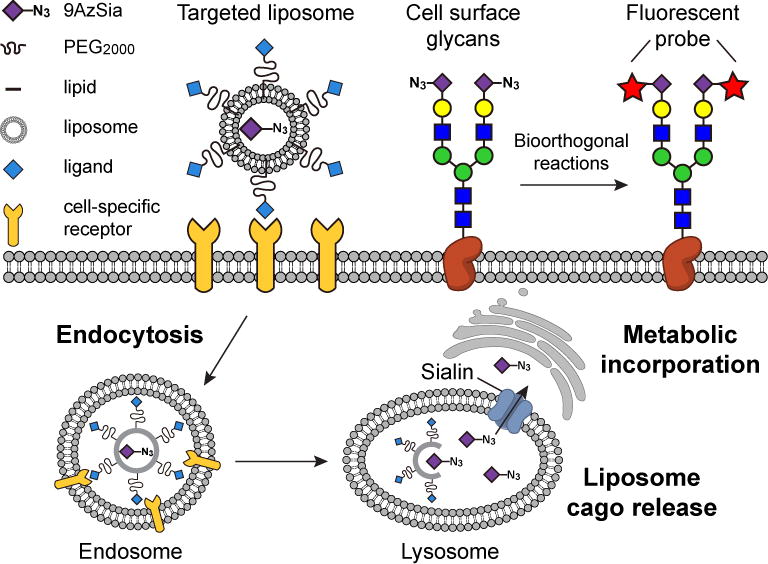
In LABOR, ligand-targeted liposomes encapsulating azidosugars are used as the chemical reporters. Through receptor-mediated endocytosis, the liposomes are internalized into the endosome and traffic to the lysosome. Unlike conventional drug delivery, which hinges on the endosomal escape, LABOR exploits the salvage pathways of monosaccharides. Using 9AzSia as an example, it is released into the lysosome after the liposomes translocate to the lysosome. The Sia transporter sialin on the lysosome then transports 9AzSia into the cytoplasm, where it enters the Sia biosynthetic pathway and gets incorporated into the cell-surface sialoglycans.
RESULTS AND DISCUSSION
Azidosugar-encapsulated ligand-targeted liposomes enter cells via receptor-mediated endocytosis
In MGL, azido or alkynyl analogs of ManNAc or Sia enter the de novo biosynthesis of Sia and are eventually incorporated into sialoglycans (Figure 1 and SI, Figure S1). Besides de novo biosynthesis, cells also recycle Sia from degraded sialoglycoconjugates (e.g., sialylated proteins and lipids) via the Sia salvage pathway, in which the lysosomal sialidases release Sia from sialoglycoconjugates (SI, Figure S1). The free Sia is then transported into the cytoplasm by the lysosomal Sia transporter sialin and reenters the Sia biosynthetic pathway.26 Based on these considerations, we postulated that LABOR intercepts the Sia salvage pathway by delivering 9AzSia into the lysosome. To elucidate the mechanism of LABOR, we used folic acid (FA)-conjugated liposomes encapsulating 9AzSia (f-LP-9AzSia)23 as a model system.
The targeted drug delivery and gene delivery using ligand-targeted liposomes have been extensively studied and the drug-encapsulated liposomes are commonly reported to enter the cells via the receptor-mediated endocytosis.27 For example, it was shown that the drug-loaded liposomes functionalized with FA on the surface, upon binding to the cell-surface folate receptor (FR), were efficiently internalized into endosomes via FR-mediated endocytosis, and subsequently released the drug into the cytosol.28 To investigate whether f-LP-9AzSia enters the cells via the receptor-mediated endocytosis, we assayed its co-localization with the endosome and lysosome. The f-LP-9AzSia was fluorescently labeled with the lipophilic dye DiI, which did not significantly alter the liposome diameter or the 9AzSia encapsulation efficiency (SI, Table S1). The FR+ HeLa cells (i.e., HeLa cells with the expression of FR upregulated by culturing in FA-depleted media) were treated with DiI-labeled f-LP-9AzSia for 0.5 h, and confocal fluorescence microscopy showed binding of f/DiI-LP-9AzSia onto the cell surfaces (Figure 2A). Starting from 3 h, f/DiI-LP-9AzSia formed many intracellular puncta, which exhibited colocalization with the LysoTracker Deep Red, a live-cell indicator of lysosomes (Figure 2B, C). At 24 h, f/DiI-LP-9AzSia on the membrane had been completely internalized into the lysosomes (Figure 2D). By contrast, DiI-LP-9AzSia showed minimal binding to FR+ HeLa cells and no translocation to the lysosome, demonstrating that the FA-FR recognition was essential for liposome uptake (Figure 2E-H).
Figure 2.
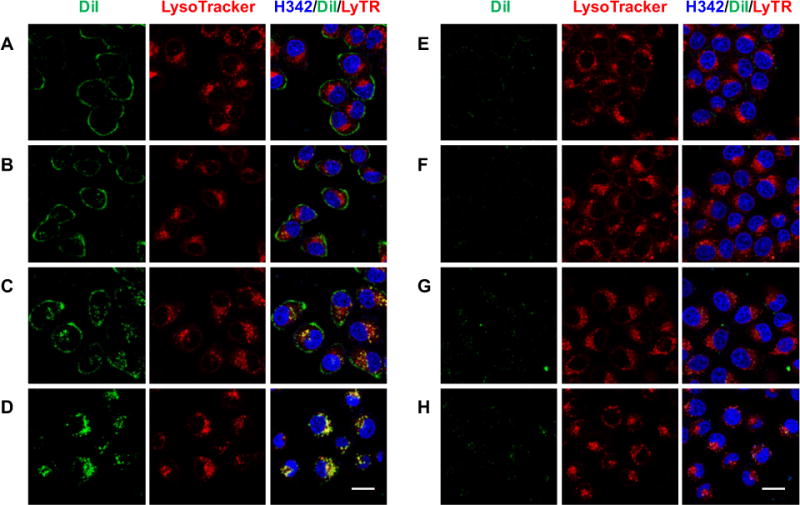
Live-cell tracking of DiI-labeled LP-9AzSia. FR+ HeLa cells were incubated with f/DiI-LP-9AzSia (A-D) or DiI-LP-9AzSia (E-H) at the 9AzSia-based concentration of 100 μM for 0.5 h. The cells were changed into fresh medium and imaged by confocal fluorescence microscoy at varied time points including 1.5 h (A, E), 3 h (B, F), 6 h (C, G) and 24 h (D, H). The liposomes were tracked with DiI (green), the lysosomes were stained with LysoTracker Deep Red (red), and the nuclei were stained with Hoechst 33342 (blue). The intensity of DiI at 24 h (D, H) was adjusted to 50% for the illustration purpose. Scale bar, 20 μm.
To evaluate the colocalization of f/DiI-LP-9AzSia with the endosome, the liposome-treated cells were fixed, permeabilized, and stained with an early endosome antigen 1 (EEA1) antibody (Figure 3). Partial cocolocalizaiton of f/DiI-LP-9AzSia with EEA1 was observed as early as 1.5 h. Under the fixed cell condition, we also observed similar colocalization of f/DiI-LP-9AzSia with the lysosome, as stained by a lysosomal associated membrane protein 1 (LAMP1) antibody (SI, Figure S2). These results indicate that f/DiI-LP-9AzSia traffic into the early endosome and lysosome successively after internalization via the FR-mediated endocytosis.
Figure 3.
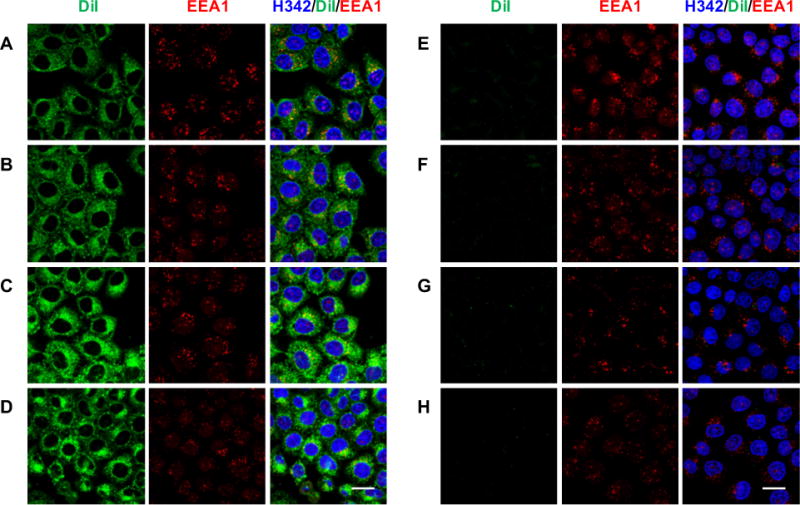
Colocalization of f/DiI-LP-9AzSia with the endosome. FR+ HeLa cells were incubated with 100 μM f/DiI-LP-9AzSia (A–D) or DiI-LP-9AzSia (E–H) for 0.5 h. The cells were changed into fresh medium and incubated to the time point of 1.5 h (A, E), 3 h (B, F), 6 h (C, G) or 24 h (D, H). The cells were then fixed with 4% PFA and permeabilized with 0.1% TritonX-100, followed by immunostaining with anti-EEA1 (red) and imaging by confocal fluorescence microscopy. The liposomes were tracked with DiI (green), and the nuclei were stained with Hoechst 33342 (blue). Scale bar, 20 μm.
After f/DiI-LP-9AzSia translocated into the lysosome, 9AzSia was eventually incorporated into cell-surface sialoglycans, as shown by reacting the cells with alkyne-biotin and staining with streptavidin-Alexa Fluor 647 (AF647) at 24 h (SI, Figure S3). Since Sia is de novo biosynthesized in the cytosol before being transported into the nucleus and converted to CMP-Sia (SI, Figure S1), 9AzSia needs to get out of the liposome and then the lysosome. To track the intracellular release of liposome-encapsulated molecules in the lysosomes, we loaded LPs with calcein at 300 mM, a concentration at which the fluorescence of calcein is self-quenched.29 Once being released out of the liposome, the calcein fluorescence is turned on due to dilution. In the aqueous solution, LP-calcein was relatively stable at pH 7.4 for up to 24 h (SI, Figure S4). Lowering the pH resulted in release of calcein. At pH 4.5 to 5.0, the pH of the interior of lysosomes, about 5%~10% release was observed at 24 h. Moreover, the enzymes inside the lysosome are capable of degrading the liposomal membrane.30 When the FR+ HeLa cells were treated with f/DiI-LP-calcein, only DiI fluorescence was observed on the cell surface (Figure 4A). From 3 h, the release of calcein into the lysosomes was observed (Figure 4B-D). As a comparison, only a minimal fluorescence was detected in cells treated with non-targeted liposomes (Figure 4E-H). These results indicate that the liposomes used in LABOR can be broken down in the lysosome, thus releasing the encapsulated unnatural sugars.
Figure 4.
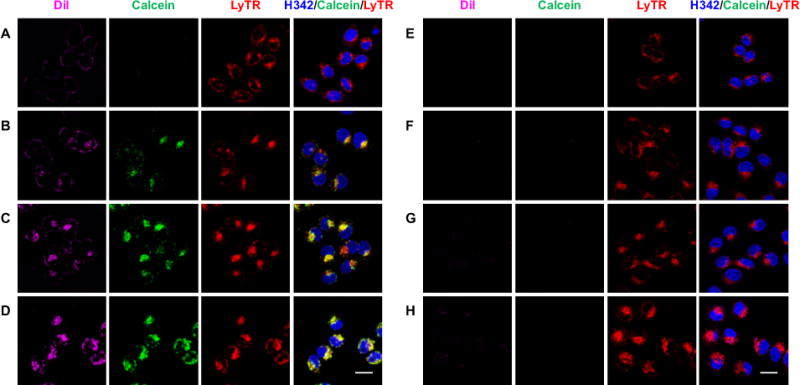
In the lysosome, f-LP releases the encapsulated molecules. FR+ HeLa cells were incubated with 40 μM f/DiI-LP-calcein (A–D) or DiI-LP-calcein (E–H) for 0.5 h, in which the local calcein concentration was 300 mM inside the liposome. The cells were changed into fresh medium and imaged by confocal fluorescence microscopy at varied time points including 1.5 h (A, E), 3 h (B, F), 6 h (C, G) and 24 h (D, H). The liposomes were tracked with DiI (magenta), the release of calcein from the liposome was monitored by the calcein fluorescence (green), the lysosomes were stained with LysoTracker Deep Red (red), and the nuclei were stained with Hoechst 33342 (blue). The intensity of DiI at 24 h (D, H) was adjusted to 50% for the illustration purpose. Scale bar, 20 μm.
Sialin can transport 9AzSia from the lysosome into the cytosol
The lysosomal Sia transporter sialin is responsible for the export of free Sia from the lysosome. Cells can uptake Sia via fluid phase pinocytosis, a clathrin-independent endocytic pathway.31 The negatively charged Sia is restricted from passive diffusion out of the endosome and reaches the lysosome, where it is then transported to the cytosol by sialin (SI, Figure S1). Sialin was also reported to transport N-(4-pentynoyl)neuramic acid (SiaNAl), an alkynyl analog of Sia.32 To test whether 9AzSia could be transported by sialin, we fed 9AzSia to FR+ HeLa cells overexpressing EGFP-sialin and the wild-type (WT) cells (SI, Figure S5). Presumably, 9AzSia was internalized into the lysosome via fluid phase pinocytosis. Flow cytometry analysis on cells reacted with DBCO-biotin and stained with streptavidin-AF647 showed increased labeling on sialin-overexpressed cells at various concentrations of 9AzSia, indicating that 9AzSia metabolism was dependent on sialin (Figure 5A). The sialin-overexpressed cells were then treated with f-LP-9AzSia at concentrations ranging from 10 μM to 500 μM. Increased 9AzSia incorporation was observed for all f-LP-9AzSia concentrations, comparing to the FR+ HeLa cells with no sialin overexpression (Figure 5B). Increased 9AzSia incorporation, to a less extend, was observed by treating the cells with LP-9AzSia (Figure 5C). Notably, overexpression of sialin should also increase the transport of recycled Sia into the cytosol and thus competing the incorporation of 9Azsia into sialoglycans. Nevertheless, these data demonstrate that sialin is capable of pumping 9AzSia into the cytosol from the lysosome and LABOR exploits the receptor-mediated endocytosis and the salvage pathway to metabolically label glycans with cell selectivity.
Figure 5.
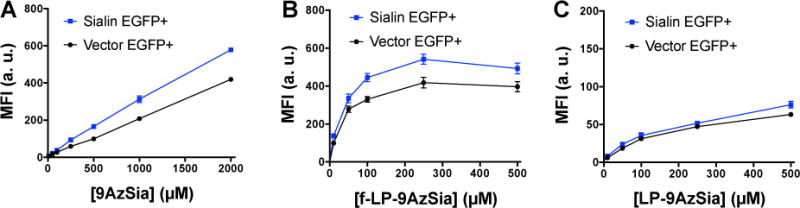
Sialin transports 9AzSia from the lysosome into the cytosol. FR+ HeLa cells were transfected with EGFP-sialin or the empty EGFP-C1 vector. The cells were incubated with 9AzSia (A), f-LP-9AzSia (B) or LP-9AzSia (C) at varied concentrations for 12 h. The cells were then reacted with DBCO-biotin and streptavidin-AF647, and analyzed by flow cytometry. MFI, mean fluorescence intensity. a. u., arbitrary unit. Error bars represent ± SD.
Evaluation of different ligand-receptor pairs for LABOR
The ligand-targeted liposomal delivery strategy permits versatile choices of ligands to target different cell-surface receptors and thus different cell types. We have previously used the small molecule ligand FA for targeting FR+ HeLa cells and the cyclic RGD peptide cRGDyK for the αVβ3-expressing B16-F10 cells.23,24 Aiming to demonstrate the broad applicability of the LABOR strategy, we evaluated several other types of ligands including monoclonal antibody trastuzumab, aptamer Sgc8, the linear RGD peptide, and glycan ligands BPCNeuAc and POBNeuAc (Table 1).
Table 1.
Various ligand-receptor pairs for LABOR
| LIGAND | LIGAND TYPE | RECEPTOR | DISEASE | CELL TARGETED |
|---|---|---|---|---|
| Trastuzumab | Antibody | Her2 | Breast cancer | SK-Br-3 |
| Sgc8 | Aptamer | PTK7 | Leukemia | CCRF-CEM |
|
BPCNeuAc POBNeuAc |
Glycan | human CD22 | B-cell lymphoma | hCD22_CHO BJAB K20 |
| cRGDyK24 RGD |
Peptide | αVβ3 integrin | Many cancers | B16-F10 MDA-MB-435 |
| Folate23 | Small molecule | Folate receptor | Many cancers | FR+ HeLa |
Trastuzumab (Herceptin) is a monoclonal antibody specific for the human epidermal growth factor receptor-2 (HER2), which is overexpressed in certain types of breast cancer.33 To prepare HER2-targeting liposomes encapsulating 9AzSia (tras-LP-9AzSia), trastuzumab was thiolated using 2-iminothiolane and subsequently conjugated to the maleimide-functionalized liposomes (SI, Table S1). Sgc8 is a 42-base aptamer that has high specificity and affinity for protein tyrosine kinase-7 (PTK7) on leukemia cell surfaces.34 Sgc8-LP-9AzSia was prepared by conjugating the 3′-thiol-modified Sgc8 aptamer with the maleimide-functionalized liposomes (SI, Table S1). BPCNeuAc (9-N-biphenylcarboxyl-NeuAcα2,6Galβ1,4GlcNAc; SI, Figure S6A) and POBNeuAc (9-N-phenoxybenzamido-NeuAcα2,6Galβ1,4GlcNAc; SI, Figure S6B) are two synthetic high affinity glycan ligands of human CD22 (siglec-2).35,36 As a B-lymphocyte-specific receptor, CD22 is a member of the sialic acid binding Ig-like lectin (Siglec) family and recognizes sialosides containing α2,6-linked sialic acids.37 Both BPCNeuAc and POBNeuAc were shown to bind hCD22 with high affinity and selectivity. Although BPCNeuAc showed cross-reactivity to macrophage-specific Siglec-1, in vivo targeting could be achieved by depleting of macrophages.38 POBNeuAc was reported to have no detectable crosstalk with other siglecs.36 BPCNeuAc and POBNeuAc were conjugated to the lipid polyethyleneglycol-distearoyl phosphoethanolamine (PEG-DSPE), and used to the 9AzSia-encapulated CD22-targeting liposomes (BPCNeuAc-LP-9AzSia and POBNeuAc-LP-9AzSia; SI, Table S1). The RGD peptide can serve as a ligand for αVβ3 integrin.39 RGD-LP-9AzSia was prepared using a procedure adapted from the previous report (SI, Table S1).24
With the ligand-modified liposomes encapsulating 9AzSia in hand, we evaluated their use for selective labeling of sialoglycans in the respective receptor-positive cells: CCRF-CEM, a human T-cell leukemia cell line expressing PTK7; SK-Br-3, a human breast cancer cell line expressing HER2; hCD22_CHO, the Chinese Hamster Ovary cell line overexpressed with hCD22; and MDA-MB-435 cells, a human breast cancer cell line expressing αVβ3 integrin. All these cell lines could be metabolically labeled with free 9AzSia on cell-surface sialylated glycans (SI, Figure S7). The cells were then incubated with sgc8-LP-9AzSia, tras-LP-9AzSia, BPCNeuAc-LP-9AzSia, POBNeuAc-LP-9AzSia and RGD-LP-9AzSia at varied concentrations ranging from 10 to 250 μM, respectively. As the comparison, the cells were treated with the naked liposome encapsulating 9AzSia, LP-9AzSia, at the same concentrations. After reacting with DBCO-biotin and staining with streptavidin-AF647, the cells were analyzed by flow cytometry, which showed that sgc8-LP-9AzSia, tras-LP-9AzSia, BPCNeuAc-LP-9AzSia, POBNeuAc-LP-9AzSia, and RGD-LP-9AzSia exhibited dose-dependent labeling of cell-surface sialoglycans (Figure 6). LP-9AzSia resulted in minimal or low levels of flurorescence labeling, presumably through non-specific endosytosis, in a cell type-dependent manner. More importantly, they all showed targeted labeling comparing to LP-9AzSia. The targeting efficiency of ligand-receptor pairs mediated glycan metabolic labeling was calculated by comparing mean fluorescence intensity of cells treated with targeted and non-targeted liposomes. As a result, the ligand-receptor pairs mediated an increase of cell-surface metabolic glycan labeling by 3.5 to 65 folds, respectively. One of the most effective targeted metabolic labeling was achieved in hCD22 targeting with BPCNeuAc-LP-9AzSia or POBNeuAc-LP-9AzSia at 25 to 50 μM (Figure 6C, D).
Figure 6.
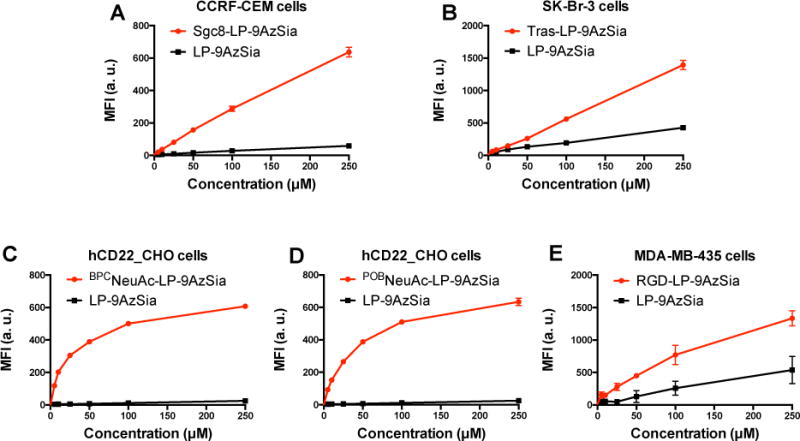
Targeted labeling of sialoglycans mediated by various ligand-receptor recognition. CCRF-CEM cells (A), SK-Br-3 cells (B), hCD22_CHO cells (C and D), and MDA-MB-435 cells (E) were treated sgc8-LP-9AzSia, tras-LP-9AzSia, BPCNeuAc-LP-9AzSia, POBNeuAc-LP-9AzSia and RGD-LP-9AzSia respectively, at varied concentrations for 24 h. For each ligand-targeted liposome, LP-9AzSia was used at the same concentrations for comparison. The cells were then reacted with DBCO-biotin, stained with streptavidin-AF647, and analyzed by flow cytometry. MFI, mean fluorescence intensity. a. u., arbitrary unit. Error bars represent ± SD.
To test the cell-type selectivity, we compared labeling of BPCNeuAc-LP-9Az and POBNeuAc-LP-9Az in hCD22_CHO cells with WT CHO cells (Figure 7). We first confirmed that these liposomes did not induce apparent cytotoxicity (SI, Figure S8). Both BPCNeuAc-LP-9Az and POBNeuAc-LP-9Az showed selective labeling of cell-surface sialoglycans in hCD22_CHO cells, while low labeling was observed in WT CHO cells (Figure 7A and B). The ratio of fluorescence intensity between two cells was ranging from 17 to 45 folds with the highest selectivity at 25 to 50 μM BPCNeuAc-LP-9AzSia or POBNeuAc-LP-9AzSia. The selective labeling of cell-surface sialoglycans by POBNeuAc-LP-9AzSia in hCD22_CHO cells was also confirmed by confocal fluorescence microscopy (Figure 8A). In contrast, no selective labeling was observed when treating hCD22_CHO and WT CHO cells with LP-9AzSia or 9AzSia (Figure 7C, D, and 8A). Furthermore, the co-culture of hCD22_CHO and WT CHO cells was treated with POBNeuAc-LP-9AzSia (Figure 8B). By differentiating hCD22_CHO and WT CHO cells by immunostaining of hCD22, selective labeling of cell-surface sialoglycans in hCD22_CHO cells was observed. In addition, cell-selective labeling of sialoglycans was demonstrated for Sgc8-LP-Sia9Az (SI, Figure S9) and RGD-LP-Sia9Az (SI, Figure S10). It is of note that the selectivity could dependent on the binding affinity between the ligand-receptor pairs and the cell types. Taken together, these results demonstrated that the CD22-targeted liposomes could selectively incorporate 9AzSia into sialoglycans on CD22-expressing cells in a receptor-dependent manner.
Figure 7.
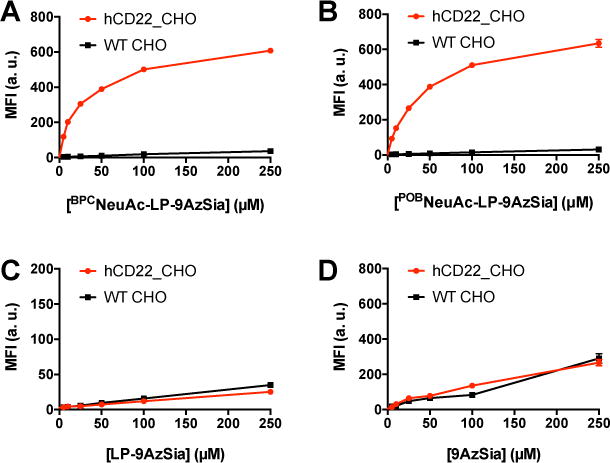
Cell-selective labeling of sialoglycans by CD22-targeting liposomes. hCD22_CHO and WT CHO cells were treated with BPCNeuAc-LP-9AzSia (A), POBNeuAc-LP-9AzSia (B), LP-9AzSia (C), and 9AzSia (D) at varied concentrations for 24 h. The cells were then reacted with DBCO-biotin, stained with streptavidin-AF647, and analyzed by flow cytometry. MFI, mean fluorescence intensity. a. u., arbitrary unit. Error bars represent ± SD.
Figure 8.
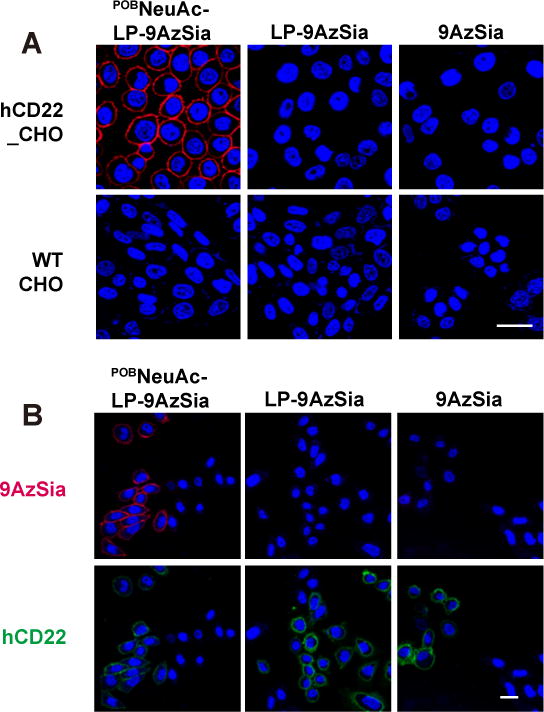
Cell-selective labeling in the co-culture of hCD22_CHO and WT CHO cells. (A) hCD22_CHO and WT CHO cells were treated with 100 μM POBNeuAc-LP-9AzSia, LP-9AzSia or 9AzSia for 24 h. The cells were then reacted with DBCO-biotin and stained with streptavidin-AF647, followed by imaging by confocal fluorescence microscopy. (B) The co-culture of hCD22_CHO and CHO cells was treated POBNeuAc-LP-9AzSia, LP-9AzSia or 9AzSia, followed with click-labeling and immunostaining with anti-hCD22 antibody-PE conjugate. The nuclei were stained with Hoechst 33342 (blue). Scale bars, 20 μm.
Selective labeling of B lymphoma sialoglycans in a B-T cell co-culture
We next sought to demonstrate the application of LABOR for selective labeling of sialolgycans of B cells in the context of immunological systems. As a proof-of-concept experiment, we constricted a B-T cell mixture by co-culturing BJAB K20 cells, a human B-cell lymphoma cell line,40 and EL4 cells, a murine T-cell lymphoma cell line (Figure 9A). We first confirmed that K20 cells could be targeted by POBNeuAc-LP-9AzSia (Figure 9B). When treated with POBNeuAc-LP-9AzSia at concentrations ranging from 25 to 250 μM, K20 cells displayed 3 to 12-fold higher labeling of sialylated glycans than treated with LP-9AzSia, with the highest selectivity at 25 to 50 μM of 9AzSia. As a comparison, although 9AzSia was metabolically incorporated with a similar efficiency in EL4 cells and K20 cells, only background labeling was observed in EL4 cells treated with POBNeuAc-LP-9AzSia or LP-9AzSia (SI, Figure S11).
Figure 9.
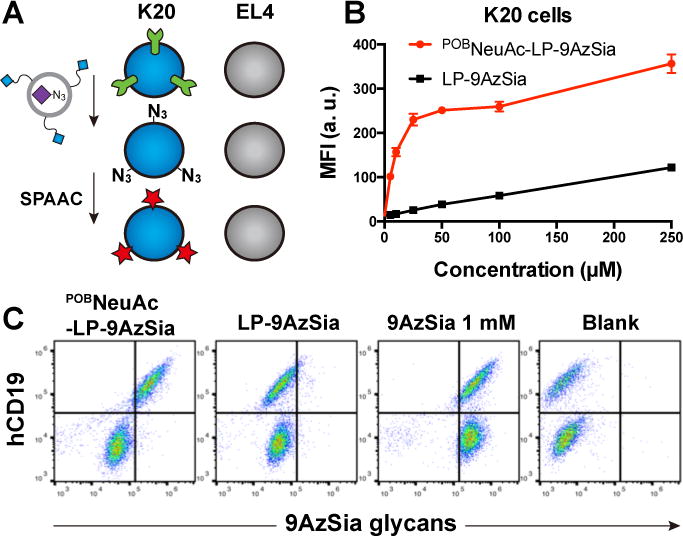
Selective labeling of sialoglycans on B-cell lymphoma cells in a B-T cell co-culture system. (A) Schematic of selective labeling of sialoglycans on K20 B cells by POBNeuAc-LP-9AzSia in the co-cultured mixture of K20 and EL4 cells. (B) Quantitative analysis of targeted labeling of sialoglycans in K20 cells with POBNeuAc-LP-9AzSia. The cells were treated with POBNeuAc-LP-9AzSia or LP-9AzSia at varied concentrations for 24 h. The cells were then reacted with DBCO-biotin, stained with streptavidin-AF647, and analyzed by flow cytometry. MFI, mean fluorescence intensity. a. u., arbitrary unit. Error bars represent ±SD. (C) Flow cytometry analysis of selective labeling of sialoglycans in the co-culture of K20 and EL4 cells. The co-cultured cells were incubated with 100 μM POBNeuAc-LP-9AzSia, 100 μM LP-9AzSia or 1 mM 9AzSia for 24 h, followed by reaction with DBCO-biotin and staining with streptavidin-AF647.
We then treated the co-culture of K20 and EL4 cells with POBNeuAc-LP-9AzSia, LP-9AzSia, or 9AzSia for 24 h. Cell-surface sialoglycans were labeled with DBCO-biotin and streptavidin-AF647. K20 cells were discriminated from EL4 cells by staining the cell surface receptor CD19, a B-cell marker, with anti-human CD19 AF488. In the cell mixture treated with POB-LP-9AzSia, only K20 cells showed a significant labeling of 9AzSia-incorported glycans, while the hCD19-negative EL4 cells exhibited low 9AzSia incorporation (Figure 9C). By contrast, similar labeling was observed in two cell populations when treated with LP-9AzSia or 9AzSia. These results indicate that POBNeuAc-LP-9AzSia can target B cells in a mixture of B and T cells, and selectively label the sialylated glycans on B cells, which possesses a promising application of this strategy for B-cell-specific glycan labeling in immune systems.
Multiplexed labeling of co-cultured cells with two distinct chemical reporters
Having established an arsenal of ligand-targeted liposomes for selective labeling of glycans in various cell types, we sought to demonstrate the multiplexing capability of the LABOR strategy. We established a co-culture of K20 cells with FR+ HeLa cells, as a model system of immune cell and cancer cell co-cultures (Figure 10A). We chose alkyne-functionalized sialic acid, SiaNAl, as a second reporter for sialoglycans. The K20 cells were incubated with SiaNAl, reacted with with azide-AF647 via copper(I)-catalyzed alkyne-azide cycloadditions (CuAAC) assisted with the BTTAA ligand.41 Flow cytometry analysis showed the incorporation of SiaNAl into K20 cell-surface glycans in a dose-dependent manner (SI, Figure S12). We then prepared BPCNeuAc-LP-SiaNAl using a similar procedure (SI, Table S1) and confirmed that BPCNeuAc-LP-SiaNAl could be used for targeted labeling of sialoglycans in K20 cells (SI, Figure S13).
Figure 10.
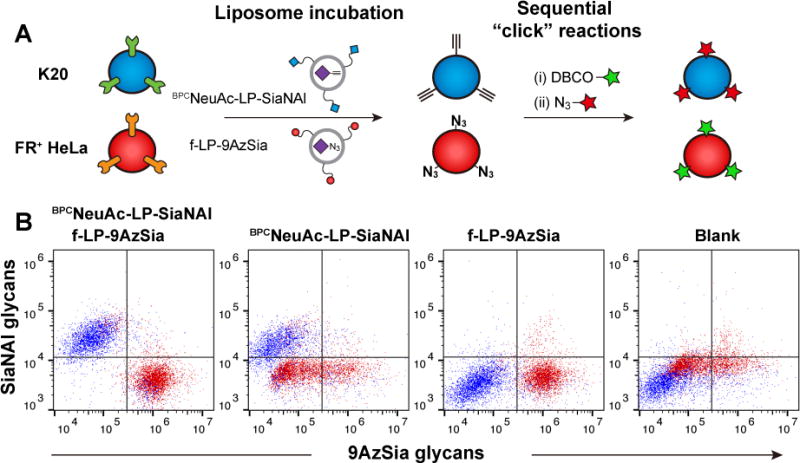
Multiplexed LABOR. (A) The co-culture of K20 and FR+ HeLa cells are treated simultaneously with BPCNeuAc-LP-SiaNAl and f-LP-9AzSia, which selectively target K20 and FR+ HeLa cells, respectively. The cells were then sequentially click-labeled for two-color detection. (B) The co-cultured cells were treated with 50 μM BPCNeuAc-LP-SiaNAl and 100 μM f-LP-9AzSia, 50 μM BPCNeuAc-LP-SiaNAl or 100 μM f-LP-9AzSia for 24 h. Cells were then collected and labeled sequentially with DBCO-Carboxyrhodamine 110 and azide-AF647, followed by flow cytometry analysis. Before reaciton with azide-AF647, the cells were treatd with TECP to reduce the un-reacted azides. K20 (blue) and FR+ HeLa (red) cells were distinguished based on their forward-scatter (FSC) and side-scatter (SSC) characteristics.
For multiplexed labeling, the co-culture of K20 and FR+ HeLa cells were treated simultaneously with BPCNeuAc-LP-SiaNAl and f-LP-9AzSia for 24 h. The co-cultured cells were then sequentially reacted with DBCO-carboxyrhodamine 110 for conjugating the azides, azide-AF647 for conjugating the alkynes (Figure 10A). The populations of K20 and FR+ HeLa cells were distinguished by gating their distinct forward-scatter (FSC) and side-scatter (SSC) signals in flow cytometry. K20 and FR+ HeLa cells were labeled with SiaNAl and 9AzSia, respectively (Figure 10B). By contrast, treating the co-culture with only BPCNeuAc-LP-SiaNAl or f-LP-9AzSia resulted in single-color labeling in the corresponding cells. These results demonstrate that by using two different ligand-receptor pairs, LABOR is able to label the glycans of different target cells with distinct chemical tags.
CONCLUSION
Unlike proteins, glycans are challenging to label in a cell-selective manner. Most genetically encoded protein tags can be readily confined in specific cell types by exploiting the cell- or tissue-specific promoters.42 Because glycans are not genetically encoded, chemical labeling methods like MGL are often employed for glycan imaging and glycoproteomic profiling.2 However, the chemical reporters for glycans cannot be controlled by cell-specific promoters.22 Alternatively, LABOR exploits ligand-targeted liposomes to enable cell-selective metabolic labeling of glycans with chemical reporters. In this work, we investigated the mechanism of LABOR. Our results revealed that LABOR exploits the receptor-mediated endocytosis and the salvage pathways of monosaccharides for cell-selective labeling. By targeting specific cell-surface receptors, it shuttles azidosugars into the endosome and then the lysosome via endocytosis. The conventional use of ligand-targeted liposomes for drug delivery hinges on the endosomal escape pathways to release drugs into the cytosol, because the drugs are usually degraded once inside the lysosome.43 By contrast, LABOR does not rely on the endosomal escape. The liposomes are allowed to enter the lysosome, in which the encapsulated azidosugars are released due to liposomal membrane degradation by lysosomal enzymes and acidic environment. The azidosugars then enter the monosaccharide salvage pathway and get transported into the cytosol by the lysosomal sugar transporters. Notably, pH-sensitive liposomes could be exploited to facilitate the lysosomal release of azidosugars.30
The endocytosis-salvage mechanism of LABOR provides guidelines for future development and applications of LABOR. Regarding the compatibility of unnatural sugars with LABOR, one should probably explore those that can be transported by the lysosomal transporters.44 For example, GlcNAc and GalNAc could be transported to the cytosol after glycoprotein degradation in lysosomes,45 indicating the possibility of cell-selective metabolic labeling of O-linked glycosylation and O-GlcNAcylation using LABOR. On the other hand, even though ManNAc analogs have been widely used for sialic acid metabolic labeling, they might not be proper for LABOR because no lysosomal ManNAc transporter has been reported. A possible solution to this limitation is to explore liposomes that can fuse with the cell membrane and thus deliver the cargo directly into the cytosol.46–48
LABOR is a versatile strategy. Various ligand-receptor pairs can be exploited to targeted glycans in respective cells and tissues. We demonstrated that different ligand types including small molecules, aptamers, peptides, antibodies, and glycans can be easily implemented into LABOR. Furthermore, the multiplexing capability of LABOR demonstrated in this work should be of great use facilitating studies on glycan-mediated cell-cell interactions. For example, cell-surface sialoglycans on cancer cells play an important roles in immune evasion.49,50 Thus, multiplexed labeling of sialoglycans on immune cells and tumor cells would facilitate the study of tumor–immune interactions.
EXPERIMENTAL SECTION
Chemical Compounds
All chemical reagents utilized in the study were of analytical grade, purchased from commercial suppliers and used without further purification unless noted. DBCO-Carboxyrhodamine 110, alkynyl-PEG4-biotin (alkyne-biotin), DBCO-Cy5 and sulfo-DBCO-PEG4-biotin (DBCO-biotin) were obtained from Click Chemistry Tools (Scottsdale, AZ, USA). Streptavidin-AF647, azide-AF647 and LysoTracker Deep Red were purchased from Invitrogen (San Diego, CA, USA). 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), cholesterol (Chol), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethyleneglycoll)-2000] (MeO-PEG-DSPE) and maleimide-activated PEG2000-DSPE (Mal-PEG-DSPE) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). N-hydroxysuccinimide-activated PEG2000-DSPE (NHS-PEG-DSPE) was purchased from NOF America Corporation (White Plains, NY, USA). DiI (1,1′-Dioctadecyl-3,3,3′,3′-Tetramethylindocarbocyanine Perchlorate) was purchased from Sigma-Aldrich (St. Louis, MO, USA). 9AzSia51, SiaNAl52 and BTTAA41 were synthesized as previously described.
Targeted Ligands and Ligand-Conjugated Lipids
The 9-N-biphenylcarboxamido-N-acetyl-neuraminic acid trisaccharide (9-N-BPC-NeuAcα2-6Galβ1-4GlcNAcβ-O-ethylamine; abbreviated herein as BPCNeuAc) and 9-N-phenoxybenzamido-N-acetyl-neuraminic acid trisaccharide (9-N-POB-NeuAcα2-6Galβ1-4GlcNAcβ-O-ethylamine; abbreviated herein as POBNeuAc) were prepared as previously described.38 Argine-glycine-aspartic acid tripeptide (RGD) was purchased from Scilight-Peptide (Beijing, China). Trastuzumab (Herceptin®) was purchased from Roche Ltd. (Basel, Switzerland). Sgc8 aptamer was synthesized on a DNA synthesizer using standard bases with a thiol (S-S)-modified 5′ end. The Sgc8 sequence: 5′-thiol-ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GA-3′. RGD, BPCNeuAc and POBNeuAc were conjugated with NHS-PEG-DSPE to give RGD-PEG-DSPE, BPCNeuAc-PEG-DSPE, and POBNeuAc-PEG-DSPE using the previously reported procedures.24,38 FA-PEG-DSPE was synthesized as previously described.53
Liposome Preparation and Characterization
FA- and RGD-functionalized liposomes were composed of DOPC, Chol, FA-PEG-DSPE or RGD-PEG-DSPE in a 50:50:0.5 molar ratio. CD22-targeted liposomes were composed of DOPC, Chol, and BPCNeuAc-PEG-DSPE (or POBNeuAc-PEG-DSPE) in a 60:35:5 molar ratio. The maleimide-modified liposomes were composed of DOPC, Chol, MeO-PEG-DSPE and Mal-PEG-DSPE in a 50:50:5:1 molar ratio. For the non-targeted liposomes, the same molar amount of MeO-PEG-DSPE was used instead of the ligand-containing lipids. The liposomes were prepared by the film hydration and extrusion method as reported in our previously work.23 Briefly, lipids dissolved in chloroform or methanol were mixed and dried into a lipid film and kept in vacuo overnight. To prepare fluorescently labeled liposomes, 0.1 mol % of DiI was added into the lipid mixture. For the liposomal encapsulation of sugars or calcein, the resulting lipid film was hydrated by PBS (PH7.4), or aqueous solutions containing 9AzSia (300 mM), SiaNAl (300 mM) or calcein (300 mM), respectively. After sonication for 5 to 10 min, the mixture was frozen in liquid nitrogen and thawed in 37°C water bath for 10 times. The suspension was then extruded with a mini-extruder (Avanti Polar Lipids) at room temperature (RT) using a polycarbonate membrane (Whatman, GE Healthcare) with a pore size of 0.2 μm. After 15-20 cycles of extrusion, the non-encapsulated sugar or calcein was removed by gel filtration through Sephadex G-50 (Sigma-Aldrich).
For preparation of trastuzumab-functionalized liposomes, trastuzumab (1 eq) was thiolated by 2-iminothiolane (100 eq) at 4°C for 1 h. The thiolated trastuzumab (1 eq) was mixed with the purified liposomes containing Mal-PEG-DSPE (10 eq), and incubated at 4°C overnight, followed by gel filtration through Sepharose CL-4B (GE Healthcare) to remove the unconjugated trastuzumab. For preparation of Sgc8-functionalized liposomes, the 5′-thiol-modified Sgc8 aptamer was first activated by 100 mM TCEP at 4°C for 30 min and the resulting aptamer (5 eq) was incubated with liposomes containing Mal-PEG-DSPE (1 eq) overnight, followed by removing the unconjugated Sgc8 by Sephadex-G50.
The diameters of the prepared liposomes were measured using dynamic light scattering (DLS) on a Brookhaven ZetaPALS instrument. The concentration of encapsulated calcein was determined using a microplate reader (Synergy™ 4, Bio-Tek, USA) at λex = 490 and λem = 520 nm after complete lysis of liposomes by 0.1% Triton X-100. The concentrations of encapsulated 9AzSia, SiaNAl and the lipid:sugar ratio were determined using reversed-phase HPLC after complete lysis of liposomes by 1% Triton X-100. Analytical HPLC was carried out on an Agilent 1260 Infinity Quaternary HPLC System equipped with a VWD UV-Vis detector, and a ZORBAX Eclipse XDB-C18 column (Analytical, 4.6 × 150 mm).
Liposome Stability Analysis
Calcein was encapsulated in liposomes at a concentration of 300 mM (LP-calcein), at which concentration its fluorescence is self-quenched.29 Leakage of calcein from the liposomes and its dilution in the buffer results in an increase of fluorescence. The 200 μL PBS solutions of 40 μM LP-calcein, in which the local concentration of calcein inside the liposome was 300 mM, were incubated in a 37°C water bath at varied pH values for up to 24 h. At a series of time points, a 50-μL aliquot of LP-calcein was diluted into the same volume of ice-old PBS (pH7.4) and was kept on ice until measurement. Calcein fluorescence was measured at λex = 490 and λem = 520 nm on a microplate reader. To calibrate the assay, 0% leakage of each sample was measured before incubation at 37°C (I0), 100% leakage was achieved by the addition of Triton X-100 with final concentration of 0.05% (I100). The percentage of calcein leakage was calculated as follow: (IpH – I0)/(I100 – I0) × 100%, where I0 is the fluorescence of pre-disrupted liposomes, IpH is the intensity measured at various pH, and I100 is the totally dequenched calcein fluorescence.
Cell Culture
The cells were maintained at 37°C and 5% CO2 in a water-saturated incubator. SK-BR-3 and MCF-7, both human breast cell lines, were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin (p/s). MDA-MB-435 cells were cultured in L15 medium supplemented with 10% FBS and p/s. Chinese hamster ovary (CHO) cells and CHO cells expressing human CD22 (hCD22_CHO)38 were cultured in DMEM/F12 supplemented with 10% FBS and p/s or 500 μg/mL Hygromycin-B (Roche), respectively. K20 (human B cell line), Ramos (human B cell line), EL4 (murine T cell line), CCRF-CEM (human T cell line) and HeLa were cultured in RPMI-1640 supplemented with 10% FBS and p/s. FR+ HeLa was induced from wild type HeLa cells as previously described,23 and was maintained in folic acid-depleted RPMI-1640 supplemented with 10% FBS and p/s.
Cell-Surface Glycan Labeling
Cells were seeded at 4.0 × 105 cells/mL for flow cytometry experiments or 1.5 × 105 cells/mL for fluorescence microscopy analysis, cultured for 18 h, and incubated for 24 h with the corresponding liposomes or unnatural sugars at concentrations indicated. For flow cytometry analysis, the cells were transferred into a V-bottom 96-well tissue culture plate (Corning), pelleted (800 × g, 5 min), and rinsed with PBS containing 1% FBS (1% FBS-PBS). Cell-surface azides were labeled with 50 μM DBCO-biotin in 0.5% FBS-PBS 30 min at RT. The cells were rinsed with PBS three times with 1% FBS-PBS, then incubated with the same buffer containing 2 μg/mL streptavidin-AF647 on ice for 30 min in darkness. For human CD19 staining, cells were incubated with anti-human CD19 AF488 (BD Pharmingen) according to the manufacturer’s protocol. Flow cytometry analysis was performed on a BD C6 Accuri flow cytometer.
For confocal fluorescence imaging, cells were rinsed with PBS. Cell-surface azides were labeled via CuAAC in PBS containing 50 μM alkyne-biotin, 2.5 mM sodium ascorbate, and BTTAA-CuSO4 complex (50 μM CuSO4, BTTAA:CuSO4 at 6:1 molar ratio) at RT for 5 min. The cells were rinsed three times with 1% FBS-PBS, then incubated with the same buffer containing 2 μg/mL streptavidin-AF647 on ice for 30 min in darkness. For hCD22 staining, the biotin and hCD22 were simultaneously labeled with 1%FBS-PBS containing 2 μg/mL streptavidin-AF647 and 5% (v/v) PE anti-human CD22 (BD Pharmingen) for 30 min at RT. The cells were fixed on ice with 4% paraformaldehyde (PFA, Sigma-Aldrich) for 15 min. The nuclei were then stained with 5 μg/mL Hoechst 33342 (Thermo Fisher Scientific) on ice for 20 min before fluorescence imaging. Fluorescence microscopy was performed on a Zeiss LSM 700 laser scanning confocal microscope.
For multiplexed LABOR, FR+ HeLa and K20 cells were transferred into a V-bottom 96-well plate and rinsed with 1% FBS-PBS. First, the azides were labeled with 50 μM DBCO-Carboxyrhodamine 110 in 0.5% FBS-PBS for 15 min at RT. After washing with 1% FBS-PBS for three times, the cells were treated with 50 mM TCEP for 10 min to reduce the unreacted azides. Next, the alkynes were labeled via CuAAC in 0.5% FBS-PBS containing 50 μM azido-AF647, 2.5 mM sodium ascorbate, and BTTAA-CuSO4 complex (50 μM CuSO4, BTTAA:CuSO4 at 6:1 molar ratio) at RT for 5 min. The reaction was quenched by adding 2 μL of 50 mM copper chelator bathocuproine disulphonate (BCS). The cells were rinsed with 1% FBS-PBS and analyzed by flow cytometry.
Liposome Tracking
Cells were seeded on Lab-Tek™ 8-well chamber slides (Thermo Scientific) and cultured for 24 to 36 h. For live-cell imaging, the cells were stained with 5 μg/mL Hoechst 33342 and 50 nM LysoTracker Deep Red at 37°C for 15 min. The cells were then incubated with DiI-labeled liposomes (100 μM lipid) for 30 min, changed into fresh medium containing 50 nM LysoTracker Deep Red, incubated at 37°C, and imaged by confocal fluorescence microscopy at indicated time points.
For immunofluorescence imaging, cells were incubated with DiI-labeled liposomes (100 μM lipid) for 30 min, changed into fresh medium, and incubated for varied durations of time up to 24 h. After fixing with 4% PFA for 10 min and permeabilizing with 0.1% TritonX-100 for 10 min, the cells were blocked with 5% BSA for 30 min and stained with 4 μg/mL rabbit anti-EEA1 (Abcam) or 10 μg/mL mouse anti-LAMP1 (Abcam) in 1% BSA at RT for 1 h. The cells were then stained with 4 μg/mL anti-rabbit IgG-AF633 (Invitrogen) or 10 μg/mL anti-mouse IgG-AF488 (Invitrogen) at RT for 1 h. After staining the nuclei with 5 μg/mL Hoechst 33342 for 10 min, the cells were subjected to confocal fluorescence imaging.
Sialin Expression Regulation
The amino-terminus of sialin was fused to GFP by subcloning at the EcoRI and XmaI sites of the pEGFP-C1 vector (Clontech). FR+ HeLa cells were transfected by electroporation using the Gene Pulser Xcell electroporation system (Bio-Rad). Typically, 3 × 106 cells were distributed in 100 μL OPTI-MEM (Gibico) and mixed with 3 μg plasmid. The cells were then subjected to ten square pulses (160 V, 3 mSec) delivered at 1 Hz by 2-mm-spaced electrodes. The cells were diluted with 9 mL of culture medium and distributed into 18 wells (15-mm diameter) of a 24-well cultural plate and cultured for two days. The cells were then incubated with free 9AzSia or liposomes for 12 h, labeled with DBCO-biotin and Streptavidin-AF647, and analyzed by flow cytometry.
MTS Cell Viability Assay
Cells were seeded at a density of 1 × 104 cells/well in a 96-well plate containing 100 μL medium and cultured for 24 h before treated with 9AzSia or liposomes at varied concentrations. After 24 h, the cell viability was measured using the MTS (3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay (Promega, cat. no. G3582) on a microplate reader. Briefly, the cultural medium was replaced with 90 μL PBS containing 5% FBS, and MTS (10 μL) was then added to each well After incubation for 2 h at 37°C, the absorbance at 490 nm was measured for each well, and normalized to that of control cells with no 9AzSia or liposomes. The experiments were carried out in triplicate.
Supplementary Material
Acknowledgments
We thank Prof. Hai Qi at Tsinghua University for providing EL4 cells, and Prof. Songlin Wang at Capital Medical University School of Basic Medicine for providing the sialin plasmid. This work was supported by the National Natural Science Foundation of China (No. 21425204, No. 21521003, and No. 21672013), the National Key Research and Development Projects (No. 2016YFA0501500), and National Institutes of Health (AI050143).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website: http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Hart GW, Copeland RJ. Cell. 2010;143(5):672. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laughlin ST, Bertozzi CR. Proc Natl Acad Sci USA. 2009;106(1):12. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng B, Xie R, Dong L, Chen X. Chembiochem. 2016;17(1):11. doi: 10.1002/cbic.201500344. [DOI] [PubMed] [Google Scholar]
- 4.Wratil PR, Horstkorte R, Reutter W. Angew Chem Int Ed. 2016;55(33):9482. doi: 10.1002/anie.201601123. [DOI] [PubMed] [Google Scholar]
- 5.Saxon E, Bertozzi CR. Science. 2000;287(5460):2007. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 6.Feng L, Hong S, Rong J, You Q, Dai P, Huang R, Tan Y, Hong W, Xie C, Zhao J, Chen X. J Am Chem Soc. 2013;135(25):9244. doi: 10.1021/ja402326z. [DOI] [PubMed] [Google Scholar]
- 7.Dehnert KW, Baskin JM, Laughlin ST, Beahm BJ, Naidu NN, Amacher SL, Bertozzi CR. Chembiochem. 2012;13(3):353. doi: 10.1002/cbic.201100649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rong J, Han J, Dong L, Tan Y, Yang H, Feng L, Wang Q-W, Meng R, Zhao J, Wang S-Q, Chen X. J Am Chem Soc. 2014;136(50):17468. doi: 10.1021/ja508484c. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Cheng B, Li J, Zhang Z, Hong W, Chen X, Chen PR. Angew Chem Int Ed. 2015;54(18):5364. doi: 10.1002/anie.201409145. [DOI] [PubMed] [Google Scholar]
- 10.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100(25):14846. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Proc Natl Acad Sci USA. 2006;103(13):4819. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320(5876):664. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawa M, Hsu T-L, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong C-H. Proc Natl Acad Sci USA. 2006;103(33):12371. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. J Am Chem Soc. 2006;128(37):12078. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soriano Del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J Am Chem Soc. 2010;132(47):16893. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehnert KW, Beahm BJ, Huynh TT, Baskin JM, Laughlin ST, Wang W, Wu P, Amacher SL, Bertozzi CR. ACS Chem Biol. 2011;6(6):547. doi: 10.1021/cb100284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vocadlo DJ, Hang HC, Kim E-J, Hanover JA, Bertozzi CR. Proc Natl Acad Sci USA. 2003;100(16):9116. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaro BW, Yang Y-Y, Hang HC, Pratt MR. Proc Natl Acad Sci USA. 2011;108(20):8146. doi: 10.1073/pnas.1102458108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. J Am Chem Soc. 2014;136(35):12283. doi: 10.1021/ja504063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin W, Gao L, Chen X. Chembiochem. 2015;16(18):2571. doi: 10.1002/cbic.201500544. [DOI] [PubMed] [Google Scholar]
- 21.Liu T-W, Myschyshyn M, Sinclair DA, Cecioni S, Beja K, Honda BM, Morin RD, Vocadlo DJ. Nat Chem Biol. 2017;13(2):161. doi: 10.1038/nchembio.2247. [DOI] [PubMed] [Google Scholar]
- 22.Xie R, Hong S, Chen X. Curr Opin Chem Biol. 2013;17(5):747. doi: 10.1016/j.cbpa.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Xie R, Hong S, Feng L, Rong J, Chen X. J Am Chem Soc. 2012;134(24):9914. doi: 10.1021/ja303853y. [DOI] [PubMed] [Google Scholar]
- 24.Xie R, Dong L, Huang R, Hong S, Lei R, Chen X. Angew Chem Int Ed. 2014;53(51):14082. doi: 10.1002/anie.201408442. [DOI] [PubMed] [Google Scholar]
- 25.Xie R, Dong L, Du Y, Zhu Y, Hua R, Zhang C, Chen X. Proc Natl Acad Sci USA. 2016;113(19):5173. doi: 10.1073/pnas.1516524113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verheijen FW, Verbeek E, Aula N, Beerens CE, Havelaar AC, Joosse M, Peltonen L, Aula P, Galjaard H, van der Spek PJ, Mancini GM. Nature Genetics. 1999;23(4):462. doi: 10.1038/70585. [DOI] [PubMed] [Google Scholar]
- 27.Allen TM. Nat Rev Cancer. 2002;2(10):750. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 28.Lee RJ, Low PS. Biochim Biophys Acta. 1995;1233(2):134. doi: 10.1016/0005-2736(94)00235-h. [DOI] [PubMed] [Google Scholar]
- 29.Slepushkin VA, Simões S, Dazin P, Newman MS, Guo LS, Pedroso de Lima MC, Düzgünes N. J Biol Chem. 1997;272(4):2382. doi: 10.1074/jbc.272.4.2382. [DOI] [PubMed] [Google Scholar]
- 30.Harding CV, Collins DS, Slot JW, Geuze HJ, Unanue ER. Cell. 1991;64(2):393. doi: 10.1016/0092-8674(91)90647-h. [DOI] [PubMed] [Google Scholar]
- 31.Bardor M, Nguyen DH, Diaz S, Varki A. J Biol Chem. 2005;280(6):4228. doi: 10.1074/jbc.M412040200. [DOI] [PubMed] [Google Scholar]
- 32.Gilormini PA, Lion C, Vicogne D, Levade T, Potelle S, Mariller C, Guérardel Y, Biot C, Foulquier F. Chem Commun. 2016;52(11):2318. doi: 10.1039/c5cc08838k. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, Zhong W, He J, Li H, Zhang H, Zhou G, Li B, Lu Y, Zou H, Kou G, Zhang D, Wang H, Guo Y, Zhong Y. Int J Pharm. 2009;374(1–2):145. doi: 10.1016/j.ijpharm.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Chemistry. 2008;14(6):1769. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 35.Blixt O, Han S, Liao L, Zeng Y, Hoffmann J, Futakawa S, Paulson JC. J Am Chem Soc. 2008;130(21):6680. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rillahan CD, Macauley MS, Schwartz E, He Y, McBride R, Arlian BM, Rangarajan J, Fokin VV, Paulson JC. Chem Sci. 2014;5(6):2398. doi: 10.1039/C4SC00451E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macauley MS, Crocker PR, Paulson JC. Nat Rev Immunol. 2014;14(10):653. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WC, Completo GC, Sigal DS, Crocker PR, Saven A, Paulson JC. Blood. 2010;115(23):4778. doi: 10.1182/blood-2009-12-257386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danhier F, Le Breton A, Préat V. Mol Pharm. 2012;9(11):2961. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- 40.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. Science. 1999;284(5418):1372. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 41.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano Del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew Chem Int Ed. 2011;50(35):8051. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engström PG, Frith MC, Forrest ARR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Nature Genetics. 2006;38(6):626. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 43.Varkouhi AK, Scholte M, Storm G, Haisma HJ. J Control Release. 2011;151(3):220. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Pisoni RL, Thoene JG. Biochim Biophys Acta. 1991;1071(4):351. doi: 10.1016/0304-4157(91)90002-e. [DOI] [PubMed] [Google Scholar]
- 45.Rome LH, Hill DF. Biochem J. 1986;235(3):707. doi: 10.1042/bj2350707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dutta D, Pulsipher A, Luo W, Yousaf MN. J Am Chem Soc. 2011;133(22):8704. doi: 10.1021/ja2022569. [DOI] [PubMed] [Google Scholar]
- 47.Pulsipher A, Griffin ME, Stone SE, Brown JM, Hsieh-Wilson LC. J Am Chem Soc. 2014;136(19):6794. doi: 10.1021/ja5005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kube S, Hersch N, Naumovska E, Gensch T, Hendriks J, Franzen A, Landvogt L, Siebrasse J-P, Kubitscheck U, Hoffmann B, Merkel R, Csiszár A. Langmuir. 2017;33(4):1051. doi: 10.1021/acs.langmuir.6b04304. [DOI] [PubMed] [Google Scholar]
- 49.Crocker PR, Paulson JC, Varki A. Nat Rev Immunol. 2007;7(4):255. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 50.Hudak JE, Canham SM, Bertozzi CR. Nat Chem Biol. 2014;10(1):69. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han S, Collins BE, Bengtson P, Paulson JC. Nat Chem Biol. 2005;1(2):93. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 52.Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Angew Chem Int Ed. 2009;48(22):4030. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabizon A, Horowitz AT, Goren D, Tzemach D, Mandelbaum-Shavit F, Qazen MM, Zalipsky S. Bioconjug Chem. 1999;10(2):289. doi: 10.1021/bc9801124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


