Abstract
The growing incidence of melanoma is a serious public health issue that merits a thorough understanding of potential causative risk factors, which includes exposure to ultraviolet radiation (UVR). Though UVR has been classified as a complete carcinogen and has long been recognized for its ability to damage genomic DNA through both direct and indirect means, the precise mechanisms by which the UVA and UVB components of UVR contribute to the pathogenesis of melanoma have not been clearly defined. In this review, we therefore highlight recent studies that have addressed roles for UVA radiation in the generation of DNA damage and in modulating the subsequent cellular responses to DNA damage in melanocytes, which are the cell type that gives rise to melanoma. Recent research suggests that UVA not only contributes to the direct formation of DNA lesions but also impairs the removal of UV photoproducts from genomic DNA through oxidation and damage to DNA repair proteins. Moreover, the melanocyte microenvironment within the epidermis of the skin is also expected to impact melanomagenesis, and we therefore discuss several paracrine signaling pathways that have been shown to impact the DNA damage response in UV-irradiated melanocytes. Lastly, we examine how alterations to the immune microenvironment by UVA-associated DNA damage responses may contribute to melanoma development. Thus, there appear to be multiple avenues by which UVA may elevate the risk of melanoma. Protective strategies against excess exposure to UVA wavelengths of light therefore have the potential to decrease the incidence of melanoma.
Keywords: Ultraviolet radiation, Ultraviolet A radiation, Melanoma, Melanocytes, DNA damage, DNA repair, Warburg effect, Nucleotide Excision repair, Immune suppression, DNA damage checkpoints, ATR kinase, DNA damage signaling, Skin Cancer
1. Introduction
1.1. Melanoma incidence
Cutaneous melanoma is a cancer that often arises from melanocytes located in regions of skin that are frequently exposed to sunlight, such as the face, neck, arms, and hands [Gilchrest et al., 1999]. Melanocytes reside within the basal layer of the epidermis and make contacts with many epidermal keratinocytes, which enables the transfer of the pigment melanin to these cells to protect them from the DNA damaging effects of ultraviolet radiation (UVR) from the sun [Nordlund, 2007; Gilchrest et al., 1996]. Though exposure to UVR from the sun or tanning beds is the most prominent risk factor for developing melanoma, several other factors, including eye color, fair skin, tendency to sunburn, inability to tan, numbers of moles, family history, age, and immunosuppression also contribute to disease risk [Gilchrest et al., 1999]. Melanoma is a major public health concern because of its rapidly growing incidence (at a rate of 3–7% per year for fair-skinned individuals), which is projected to double every 10–20 years [Lens and Dawes, 2004; Leiter and Garbe, 2008; MacKie et al., 2009]. It also has a relatively high rate of lethality in comparison to non-melanoma skin cancers (NMSCs). Indeed, more than 75% of all skin cancer-associated deaths are due to melanoma [Shenenberger, 2012]. In 2016, the National Institute of Health Surveillance, Epidemiology and End Results program (NIH-SEER) reported that there were an estimated 76,380 new cases (4.5% of all new cases of cancer) and 10,130 deaths (1.7% of all cancer deaths) from melanoma. NIH-SEER also reported that in 2013 there were approximately 1,034,460 people living with melanoma of the skin in the United States. The annual treatment cost for melanoma ranged between $44.9 million among Medicare patients with existing cases to $932.5 million among all newly diagnosed cases [Guy et al., 2012]. Understanding the factors that contribute to cutaneous melanomagenesis is therefore critical to reducing the burden of this growing disease in people worldwide.
1.2. Ultraviolet radiation
Ultraviolet radiation (UVR) is electromagnetic radiation with wavelengths in the range of 100 nm to 400 nm, which are shorter than visible light but longer than X-rays. The major source of UVR is sunlight, which makes sunlight the most prominent environmental carcinogen to which humans are routinely exposed. UVR can be subdivided into ultraviolet C (UVC; 100–280 nm), ultraviolet B (UVB; 280–320 nm) and ultraviolet A (UVA; 320–400nm) radiation based on the biological effects of the different wavelengths. Some properties of the different forms of UVR are provided in Table 1. Much of our understanding of UVR effects on DNA and cellular DNA damage responses have been derived from studies using a variety of model systems that have utilized UVC light sources. Though the germicidal properties of UVC are well known, the fact that UVC wavelengths of sunlight are effectively blocked by the ozone layer and do not penetrate the skin has led some to question the relevance of studies using UVC to human health. Nonetheless, analyses of mutagenesis in mammalian systems generally support the idea that all UV wavelengths generate similar patterns of mutations in DNA [Brash, 2015], and indeed deep sequencing of genomic DNA from melanoma tumors has revealed a plethora of UV signature mutations [Hodis et al., 2012; Berger et al., 2012; Krauthammer et al., 2012]. Thus, experimental data generated using UVC light sources provide a valuable resource for understanding fundamental aspects of DNA damage formation and the subsequent cellular responses in human cells.
Table 1. Properties of ultraviolet radiation (UVR).
The common properties of different wavelengths of UVR are provided.
| Properties | UVC | UVB | UVA |
|---|---|---|---|
| Wavelength range | 100–280 nm | 280–320 nm | 320–400 nm |
| % of UVR in terrestrial sunlight | completely blocked by ozone layer | 2–10% | 90–98% |
| Penetration through glass | Blocked | Blocked | Passes |
| Penetration through skin | Reaches top part of epidermis | Only penetrates epidermis | Epidermis and dermis |
However, because UVA and UVB wavelengths of sunlight can be absorbed by cells in the different layers of skin by various cellular biomolecules, UVA and UVB are therefore considered to be more relevant to human health. UVA and UVB have several unique properties that are relevant to understanding UVR-induced skin carcinogenesis. UVA comprises 90–95% of terrestrial sunlight UVR and can reach the dermal layer of human skin. In contrast, UVB only affects cells within the epidermal layer of the skin and comprises only a relatively small amount of the UVR from the sun at the surface of the Earth. Nevertheless, UVB is generally thought to be more carcinogenic than UVA because it is more efficiently absorbed by DNA. Although the sun is a natural source of UVA and UVB, it should be noted that there are other UV light sources that humans are sometimes exposed to, including tanning booths, curing lamps (fluorescent lamps, mercury vapor lamps), black light/UV-A lights/ Wood's lamps, high intensity discharge lamps, certain types of light-emitting diodes (LEDs), and lasers [Diffey, 2002]. Thus, within certain patient populations, exposures to these UVR sources may be relevant to understanding disease risk.
1.3. Epidemiological evidence linking UVA and melanoma
According to the International Agency of Research on Cancer of the World Health Organization (WHO), UVR including UVA is carcinogenic to humans. This classification is supported by several lines of evidence, including several epidemiological studies. For example, an association between tanning bed use and melanoma was used to explain an epidemic of melanoma that was observed in Iceland between 1990 and 2006 [Autier et al., 2011b]. Tanning beds primarily emit UVA, though some may emit a small amount of UVB. The UVA fluencies associated with tanning beds are 5 to 15 times greater than direct exposure from the sun. Several other studies have confirmed the association between tanning devices and elevated risk of melanoma [Buckel et al., 2006; Zhang et al., 2012; Le Clair and Cockburn, 2016; Dore and Chignol, 2012].
As will be described in greater detail below, UVA can act on various endogenous and therapeutic photosensitizers to generate reactive oxygen species that damage both DNA and the proteins that repair DNA damage [Brem et al., 2017; Karran and Brem, 2016; Brem and Karran, 2012; Attard and Karran, 2012], which may ultimately promote the mutagenic events that drive skin carcinogenesis [Karagas et al., 2007; Robinson et al., 2013]. For example, the immunosuppressive thiopurine prodrug azathioprine is widely used in organ transplant recipients to prevent rejection and in the management of inflammatory bowel conditions. However, patients treated with azathioprine show a significantly increased risk of skin cancers on parts of the body that are routinely exposed to the sun [Euvrard et al., 2003; Peyrin-Biroulet et al., 2011; Ramiscal and Brewer, 2013]. In addition to possibly preventing the efficient recognition and elimination of mutant pre-cancerous cells by the body’s immune system, some immunosuppressive drugs also act as UVA photosensitizers. For example, azathioprine treatment has been shown to sensitize human skin to UVA and to lower the minimal erythemal dose (MED) for UVA and solar simulating radiation [Perrett et al., 2008]. This UVA-dependent photosensitization also generates DNA damage and inhibits DNA repair protein function [Brem and Karran, 2012; Karran and Brem, 2016; Brem et al., 2017; Attard and Karran, 2012]. Epidemiological evidence has indicated that melanoma is a growing problem in organ transplant recipients receiving immunosuppressive drugs [Fattouh et al., 2017; Dahlke et al., 2014; Green and Olsen, 2015; Zwald et al., 2010], though the mechanism remains unclear. Furthermore, the observation that skin cancers tend to emerge to a greater extent on skin exposed to glass-penetrating UVA wavelengths of light may point to a role for UVA in skin carcinogenesis in organ transplant patients [Atkar et al., 2013].
1.4. UVA and melanoma in animal models
Though UVB wavelengths of light induce both melanoma and non-melanoma skin cancers in various experimental model systems, there has been some controversy regarding the role of UVA alone in causing melanoma. An early study using the Xiphophorus hybrid fish model suggested that UVA could cause melanoma [Setlow et al., 1993]. However, this finding was not supported by a follow-up study [Mitchell et al., 2010]. Similarly, though high dose exposure to UVA was thought to induce focal melanocytic hyperplasia in an opossum model of melanoma [Ley, 2001], the effect was found to be much weaker with UVA than with UVB [Robinson et al., 2000]. Moreover, additional studies in mice have shown that although UVA can induce squamous cell carcinoma (SCC), it was unable to directly induce melanoma [de Laat et al., 1997]. Thus, the general consensus from these model systems has been that UVA alone is not able to directly cause melanoma [Robinson et al., 2000; De Fabo et al., 2004; Mitchell and Fernandez, 2012].
However, there are significant differences between the skin of humans and these other model organisms that may impact melanoma susceptibility. In the skin of most animals, melanocytes are restricted to the hair follicle, which is not the case in human skin. The development and use of the hepatocyte growth factor/scatter factor (HGF/SF) transgenic mouse [Takayama et al., 1996; Noonan et al., 2003], which has extra-follicular melanocytes in skin and therefore more closely mimics human skin, has become a useful mouse model of UV-induced melanomagenesis [Recio et al., 2002; De Fabo et al., 2004; Noonan et al., 2001]. Interestingly, a single high dose UVA exposure of neonatal HGF/SF mice (in black C57BL/6 background) was shown to induce the formation of melanoma tumors [Noonan et al., 2012]. However, the same mice in an FVB albino background (which lack the pigment melanin) do not form melanoma after UVA exposure [De Fabo et al., 2004; Noonan et al., 2012]. These findings revealed that UVA may be promoting melanomagenesis in a melanin-dependent manner [Zaidi et al., 2012], which is surprising given that melanin has traditionally been thought to be protective for skin carcinogenesis [Tadokoro et al., 2005]. Furthermore, an elevation in staining for highly mutagenic 8-oxo-guanine (8-oxoG) DNA adducts, which are normally efficiently repaired, was reported in the skin of the UVA-irradiated C57BL/6-HGF mice in comparison to the FVB-HGF mice [Noonan et al., 2012]. Because these adducts represent the major lesion formed in DNA in response to oxidative stress, the authors suggested that these DNA lesions may be responsible for the mutagenesis that drives melanomagenesis. However, recent research showing a UVA- and melanin-dependent mechanism for the induction of non-oxidative, canonical bipyrimidine UV photoproducts [Premi et al., 2015] to be described below has called into question this interpretation. Nonetheless, these findings show that UVA alone can induce melanoma formation in an experimental mouse model.
1.5. Sunscreens, UVA, and melanoma
Previous studies have suggested that sunscreens provide little to no benefit against melanoma because the sunscreens may be primarily devised to prevent sunburns that are mainly caused by UVB [Autier et al., 1995; Autier et al., 2011a]. It has been proposed that the use of sunscreens that contain only UVB filters may therefore result in greater UVA exposure because the use of the sunscreen allows individuals to remain in the sun for longer periods of time without being burned. This hypothesis, and the current lack of effective UVA filters in many countries including the U.S. [Sargent and Travers, 2016], may provide a cogent explanation for why sunscreen users may experience an elevated risk of melanoma [Autier et al., 2011b; Autier et al., 2011a]. On the other hand, some studies suggest that broad spectrum sunscreens and their long-term monitoring may help to reduce the risk of melanoma [Gallagher et al., 2000]. For example, a long-term Australian study found that sunscreens that contain UVA filters appears to provide protection against malignant melanoma [Green et al., 2011]. Thus, in human patients, limiting the exposure to UVA may help to prevent the development of melanoma.
2. UVA radiation and the DNA damage response
2.1. UVA radiation and the formation of DNA damage
Nucleic acids, proteins, and lipids are all capable of absorbing UV wavelengths of light and thus likely contribute to varying degrees to cutaneous melanomagenesis [Pfeifer and Besaratinia, 2012]. Nonetheless, DNA damage by UVR is the most widely studied and best understood process and will therefore be the primary focus of this review. A variety of forms of DNA damage can be induced by UVR, including bipyrimidine dimers, oxidized bases, protein-DNA crosslinks, and cycloaddition reactions with breakdown products of lipid peroxides [Cadet et al., 2005; Cadet et al., 2015; Cadet et al., 2012]. Furthermore, hydroxyl radicals generated by photooxidative processes can attack DNA to generate single-strand breaks [Dedon, 2008], and double-strand breaks may form in response to aberrant DNA repair intermediates [Wakasugi et al., 2014; Kemp and Sancar, 2016] or as a consequence of replication fork breakage [Quinet et al., 2014; Iyer and Rhind, 2017; Toledo et al., 2013; Elvers et al., 2011; Kaufmann, 2010; Dungrawala et al., 2015]. Given their relative abundance and the degree to which the lesions have been studied in the context of melanoma, bipyrimidine dimers and oxidized purines will primarily be addressed here. A summary of the major lesions and their mechanisms of repair in human cells is provided in Table 2.
Table 2. Forms of DNA damage induced by UVR.
The major types of DNA damage induced by UVR are provided along with the principle mechanism of repair and common mutagenic effects.
| Type of damage | Repair Mechanism |
Mutagenic Effects |
|---|---|---|
|
| ||
| Bipyrimidine photoproducts | ||
| CPDs | NER | C-to-T transitions |
| (6–4)PPs | Cytosine deamination | |
| Dewar Isomers | Error-prone replication | |
|
| ||
| Oxidized purines | BER | G-to-T transversions |
| 8-oxoG | ||
|
| ||
| Single-strand breaks (SSBs) | SSBR | DSBs |
|
| ||
| Double-strand breaks (DSBs) | NHEJ | Chromosomal loss or translocations |
| HR | Insertions/deletions | |
| Error-prone replication | ||
The direct absorption of UV photons of light by DNA induces the formation of photoproducts in genomic DNA, which include cyclobutane pyrimidine dimers (CPDs), pyrimidine (6-4) pyrimidone photoproducts (6-4)PPs, and Dewar isomers [Cadet et al., 2005; Cadet et al., 2015; Cadet et al., 2012]. These photoproducts form between adjacent pyrimidine nucleotides in DNA and include the most commonly recognized “thymine dimers”. In CPDs, a cyclobutane ring forms between the 5,6 bonds of the two pyrimidine bases. In (6-4)PPs, a stable single bond forms between position 6 and 4 of the two adjacent bases, and upon further irradiation at wavelengths around and above 320 nm, these photoproducts can isomerize to form the Dewar photoproduct [Perdiz et al., 2000; Douki et al., 2003; Douki and Sage, 2016; Douki, 2016]. A variety of factors affect where these photoproducts form in the genome, including DNA sequence context, chromatin architecture, and transcription factor binding [Hu et al., 2017; Bryan et al., 2014; Mao et al., 2016; Sabarinathan et al., 2016; Perera et al., 2016; Poulos et al., 2016; Hu and Adar, 2017]. Thus, the biological consequences of UVR are dependent on not only the amount of UVR absorbed by DNA but also on the distribution and localization of lesions across the genome.
The formation of these UV photoproducts is dependent on the energy of the UV photons, and hence longer wavelengths of UVA are much less able to induce these photoproducts than UVC and UVB. Nonetheless, biologically relevant doses of UVA have been shown to induce CPD formation in both keratinocytes and melanocytes in human skin [Freeman et al., 1989; Young et al., 1998; Mouret et al., 2006]. Moreover, UVA can also facilitate the isomerization of UVB-induced (6-4)PPs into Dewar isomers [Cadet et al., 2005; Douki and Sage, 2016]. Thus, UVA has direct effects on DNA damage formation throughout the genome and may also modulate photoproducts that form in DNA from UVB photons. However, UVA has also long been recognized for its indirect effects on DNA damage formation through the excitation of various cellular photosensitizers, such as flavin, melanin, riboflavin, and porphyrins [Wondrak et al., 2006; Premi et al., 2015]. Along with UVA, these agents result in the production of reactive oxygen species (ROS) that can attack DNA to form single-strand breaks, oxidized pyrimidines, and oxidize purines, of which 7,8-dihydro-8-oxyguanine (8-oxoG) lesions are the most prevalent [Cadet et al., 2005; Cadet et al., 2012]. Though there has been debate about the relative contribution of oxidative stress-induced lesions versus bipyrimidine dimers by UVA on mutagenesis and carcinogenesis, a quantitative HPLC/mass spectrometry study showed that lesions associated with oxidative stress are not the major form of DNA damage induced by UVA in mammalian cells [Douki et al., 2003]. Rather, this study showed that UVA induces CPDs, 8-oxoG, oxidized pyrimidines, and single-strand breaks at a ratio of 10:3:1:1 [Douki et al., 2003; Courdavault et al., 2004]. Thus, CPDs likely represent the most abundant form of DNA damage induced in human skin by UVA wavelengths of light [Mouret et al., 2006]. Interestingly, this situation is slightly different in melanocytes, which exhibit a somewhat higher production of 8-oxoG upon UVA irradiation than keratinocytes [Mouret et al., 2012]. This elevation in 8-oxoG formation may be due in part to the additional ROS that melanocytes are exposed to during the biochemical synthesis of melanin [Denat et al., 2014]. Thus, UVA-induced oxidative stress may play a larger role in melanocyte cancers than keratinocyte cancers.
UVR rapidly induces CPD formation on a timescale of picoseconds, and for decades this process was the only known mechanism by which UVR generates CPDs in DNA. Interestingly, a recent report from Brash and colleagues described an unusual chemical process termed chemiexcitation in which UVA and UVB wavelengths of light can induce CPDs in DNA even hours after UV exposure [Premi et al., 2015; Premi and Brash, 2016] (Figure 1). This “dark CPD” process is particularly relevant to melanocytes and melanoma because it involves the pigment melanin. The exposure of cells to UVR causes the generation of nitric oxide and superoxide through the upregulation of the enzymes iNOS, NADPH oxidase, and enzymes involved in melanin synthesis. The nitric oxide and superoxide can react to form the oxidant peroxynitrite, which is able to degrade polymers of melanin into monomers capable of entering the nucleus. Peroxynitrite is also capable of exciting an electron to a triplet state, including within fragments of melanin, to produce an unstable and high energy dioxetane capable of reacting with DNA bases to produce CPDs. This report suggested that the production of dark CPDs is dependent on UVR-mediated ROS (reactive oxygen species) because the use of ROS scavengers, such as vitamin E, resulted in less dark CPD formation. Quantitative analyses of CPD formation in UVA-irradiated melanocytes revealed that more than half of all CPDs may arise after the UV exposure ends. Thus, this chemiexcitation mechanism for CPD formation may be responsible for most of the DNA damage and mutagenic events in UV-irradiated melanocytes and may therefore contribute to melanoma development. These findings may also explain the previous observation that the pigment melanin is apparently required for UVA- but not UVB-induced melanoma induction in the HGF/SF mouse [Noonan et al., 2012]. Interestingly, these dark CPDs were found to be more prominent in melanocytes containing pheomelanin, suggesting that pheomelanin is not only a poorer shield against initial direct CPD formation but is also more prone to form dark CPDs via chemiexcitation. Because much of the work on this novel mode of CPD induction was carried out using mouse skin cells [Premi et al., 2015], it will be important to confirm these findings in human skin.
Figure 1. Schematic for the chemiexcitation mechanism of CPD formation by UVA and melanin.
UVA exposure form the sun or other sources leads to the generation of nitric oxide and superoxide within mammalian cells. These reactive species combine to form peroxynitrate, which can then act to degrade melanin (induced by UVA) into monomers and then act on fragments of melanin to produce an unstable but high energy (triplet state) dioxetane that reacts with DNA bases to form CPDs. These CPDs can form hours after UVA exposure and hence are referred to as “Dark CPDs”.
2.2. DNA damage and mutagenesis
Both bipyrimidine dimers and oxidative photolesions induced by UVR are potentially mutagenic if not repaired, and therefore both forms of damage may be carcinogenic. Whereas cytosine bases are generally quite stable and only slowly undergo spontaneous deamination to form uracil (half-life of 30,000 years) [Frederico et al., 1990], cytosines within CPDs are much less stable (half-life of 2–100 hours) [Barak et al., 1995; Peng and Shaw, 1996; Burger et al., 2003; Tu et al., 1998]. When accurately copied by a DNA polymerase, the replication of these uracils leads to the fixation of C→T/G→A transitions in the DNA. These so-called “UV signature” mutations were first observed in 1964 [Howard and Tessman, 1964] in viral DNA and have since been observed in both naked DNA and DNA isolated from cultured cells and skin following UV exposure and in skin cancers [Brash, 2015; Brash et al., 1991; Sage et al., 2012]. Indeed, of relevance to melanoma, deep DNA sequencing of both gene promoters and exons has revealed a vast preponderance of C→T somatic mutations at bipyrimidines in sun-exposed melanomas [Hodis et al., 2012; Berger et al., 2012; Colebatch et al., 2016; Zhang et al., 2016]. These results have led to the identification of novel melanoma oncogenes and tumor suppressor genes [Krauthammer et al., 2012; Berger et al., 2012] and to the implication that alterations to gene regulatory elements, including in the telomerase (TERT) gene promoter [Lu et al., 2015; Griewank et al., 2014], likely contribute to melanomagenesis.
In addition to bipyrimidine dimers, oxidized bases, such as 8-oxoG, also form in DNA following UV exposure [Cadet et al., 2005; Cadet et al., 2012]. These ROS-dependent DNA lesions are known to generate G→T transversions through mispairing of the oxidized guanine with adenine instead of cytosine [Cooke et al., 2003; Shibutani et al., 1991]. Moreover, oxidation of dNTP pools can result in the misincorporation of 8-oxo-guanine nucleotide triphosphates into DNA opposite adenines during DNA synthesis to induce T→G mutations [Kamath-Loeb et al., 1997; Drobetsky et al., 1995] that may impact cell fate [Rai, 2010]. The importance of these mutagenic lesions to UVA-induced mutagenesis in skin is unclear however, as 8-oxoGs have been reported to contribute to only 6% of the total UVA mutation spectrum in mouse skin [Ikehata et al., 2008]. Thus, the relatively low level of G→T and T→G changes in melanoma genomic DNA [Hodis et al., 2012; Krauthammer et al., 2012] is consistent with a minor role of these oxidized bases in inducing skin tumors.
In addition to the base adducts described above, strand breaks can also be generated in genomic DNA following UV exposure (Table 2). Single-strand breaks involve the hydrolysis of the phosphodiester backbone in only one strand of DNA and hence are thought to be relatively easy to fix by simple re-ligation via single-strand break repair (SSBR). However, rare, unrepaired single-strand breaks (SSBs) are potentially problematic because an encountering RNA or DNA polymerase can lead to double-strand break (DSB) formation. DSBs breaks are particularly detrimental to cells, and when not lethal, may lead to chromosomal rearrangements that characterize cancer cell genomes [Hanahan and Weinberg, 2011; Bastian et al., 1998; Bauer, 2017]. Precisely how UVR induces double-strand breaks remains unclear. Bipyrimidine dimers are thought to be of greater concern than oxidized bases because they more severely block the progression of canonical replicative DNA polymerases. Thus, the stalling of replicative polymerases at such lesions can lead to the collapse of the replication fork and therefore to DSBs that must be repaired by homologous recombination [Dungrawala et al., 2015; Toledo et al., 2013; Iyer and Rhind, 2017; Quinet et al., 2014; Elvers et al., 2011; Kaufmann, 2010]. However, even in non-replicating cells, there are mechanisms that lead to DSB formation following UV exposure. For example, studies have shown that both DNA DSBs and the activation of DSB break-associated kinase signaling can be observed to take place in a manner dependent on the removal of the UV photoproduct by the nucleotide excision repair machinery [Wakasugi et al., 2014; Kemp and Sancar, 2016]. Depending on the mode of DSB repair, this repair may also introduce mutations or deletions into genomic DNA. Thus, in some circumstances, the repair of a canonical UV bipyrimidine lesion may lead to the aberrant formation of even more problematic DNA lesions.
2.3. Repair of UV-induced DNA damage
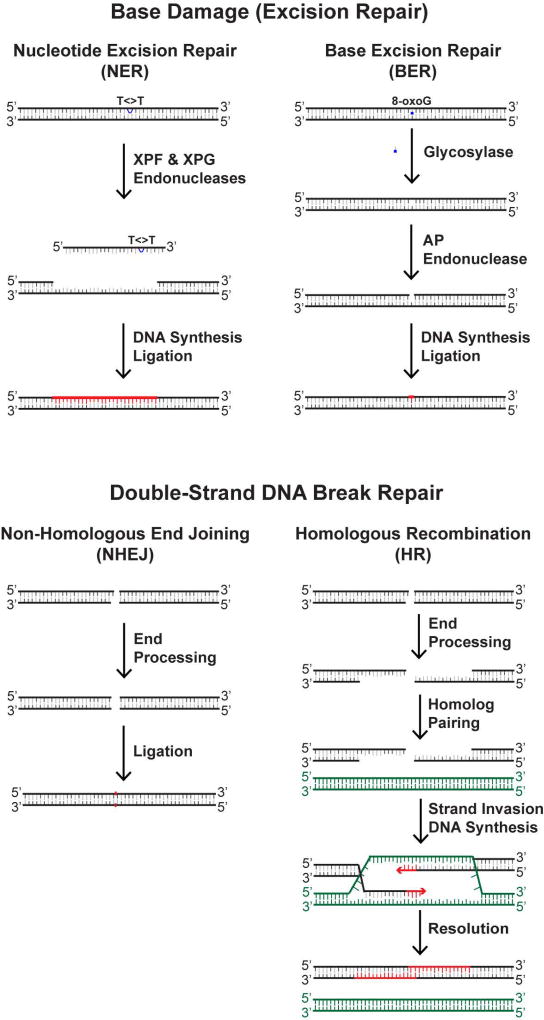
Several distinct DNA repair machineries (summarized in Figure 2) are responsible for fixing the DNA damage that is induced by UVR. The repair of bipyrimidine dimers and oxidized guanines in genomic DNA involves two different excision repair pathways that essentially involve the removal, or “excision”, of the damage from the DNA, by either nucleotide excision repair (NER) or base excision repair (BER). The major repair system of importance to UVR and melanoma is nucleotide excision repair (NER), which is a versatile system for removing a wide variety of lesions from DNA, including UV-induced CPDs and (6-4)PPs [Reardon and Sancar, 2005; Scharer, 2013]. In nucleotide excision repair, the lesion is recognized by one of two sub-pathways of NER. In the global genomic NER pathway (GG-NER), XPC (xeroderma pigmentosum group C) assists in the recognition of the lesion and then recruits additional proteins (RPA, replication protein A; TFIIH, transcription factor II-H; and XPA) to unwind the DNA around the lesion so that two nucleases (XPG and XPF) can make incisions at sites bracketing the lesion to release the damage in the form of a small (~30-nt-long), TFIIH-bound DNA oligonucleotide [Hu et al., 2013; Kemp and Sancar, 2012; Huang et al., 1992; Kemp et al., 2012; Kemp et al., 2014; Choi et al., 2014]. Though not essential for damage recognition or removal in vitro, the product of the XP-E gene, DDB2, may aid in the recognition of UV photoproducts along with XPC [Tang and Chu, 2002; Sugasawa, 2010; Ruthemann et al., 2016]. In transcription-coupled NER (TC-NER), the stalling of RNA polymerase II at the UV photoproduct is recognized by the Cockayne Syndrome (CS) proteins, which ultimate facilitates the recruitment of the other core repair proteins that unwind and incise the DNA. Regardless of the mode of damage recognition, once the damaged oligonucleotide has been removed, the gap that remains in the duplex is then filled in by a DNA polymerase and the remaining nick sealed. The importance of NER to human disease [Marteijn et al., 2014; Cleaver et al., 2009], including UV-induced melanoma, can be observed in patients with the disease xeroderma pigmentosum (XP) [Cleaver, 1968], who have a mutation in one of seven different “XP” genes that prevents NER and elevates melanoma and non-melanoma skin cancer risk by several thousand-fold [Kraemer et al., 1994; Lambert et al., 1995].
Figure 2. Schematics of major DNA repair systems that are utilized in UV-irradiated cells.
Nucleotide excision repair (NER) system is the major DNA repair pathway that is utilized in UV-irradiated cells because of the high number of bipyrimidine dimers induced by UVR and takes place via a dual incision mechanism (involving the XPF and XPG endonucleases) that removes bipyrimidine dimers (indicated by a thymine dimer T<>T) from DNA in form of a small (~30-nt-long) oligonucleotide. DNA synthesis and ligation complete the repair reaction to restore the duplex to its native state (indicated in red). Base excision repair (BER) is utilized to repair oxidized base damage (such as 8-oxo-guanine [8-oxoG] residues) in UV-irradiated cells. BER involves a glycosylase that first removes the damaged base from the DNA before the action of AP Endonuclease, which incises the phosphodiester backbone 5’ to the apurinic (AP) site. DNA synthesis and ligation then restore the duplex. DNA double-strand breaks also form in UV-irradiated cells and can be repaired by either non-homologous end-joining (NHEJ) or homologous recombination (HR). NHEJ simply involves re-ligating two broken ends to one another without significant end processing. In HR, more extensive end processing and end resection takes place, which allows for strand invasion and pairing to homologous DNA sequence elsewhere in the genome. DNA replication allow for missing sequence to be synthesized before resolution of the HR intermediate and restoration of the DNA duplex.
The BER pathway is initiated by the action of a glycosylase that removes the damaged nitrogenous base [Boiteux et al., 2017; Baute and Depicker, 2008; Liu and Wilson, 2012]. In the case of 8-oxoG, this is typically carried out by the action of the enzyme 8-oxo-guanine DNA glycosylase (OGG1) [Boiteux et al., 2017]. The loss of OGG1 in mice has been shown to increase the susceptibility to UVB-induced skin cancers [Kunisada et al., 2005]. Glycosylase action generates an apurinic (AP) site in the DNA that is cleaved at the 5’ side by the action of AP endonuclease 1 (APE1). To complete repair, DNA polymerase beta (Pol β) removes the 5’-deoxyribose and extends 1 nucleotide from the 3’-hydroxyl. In some cases, the replicative polymerases (pols δ and ε) extend 2–12 nucleotides using the intact strand as template. Regardless of the use of either the short or long patch repair mechanisms, a DNA ligase regenerates an intact phosphodiester backbone to restore the DNA duplex.
Because single-strand breaks (SSBs) are an intermediate of BER, the repair of SSBs generated via other means also utilizes some of the same cellular proteins [Abbotts and Wilson, 2017]. DSB repair is more complicated and is affected by several factors, including cell cycle status [Hustedt and Durocher, 2016], such that homologous recombination (HR) is favored during S phase when a homologous DNA template is available for repair. Non-homologous end joining (NHEJ) is the more dominant pathway for repairing double-strand breaks in human cells and involves essentially re-ligating the two broken ends of the DNA back together [Chang et al., 2017]. NHEJ is generally thought to be more mutagenic than HR because nucleolytic processing and error-prone polymerase function at DSB ends may lead to the loss of, or changes to, the genetic information. Nonetheless, the observation that melanomas frequently contain chromosomal translocations [Bastian et al., 1998; Kaufmann et al., 2014] suggests that DSB formation and aberrant repair take place during melanomagenesis.
2.4. UV DNA damage kinase signaling
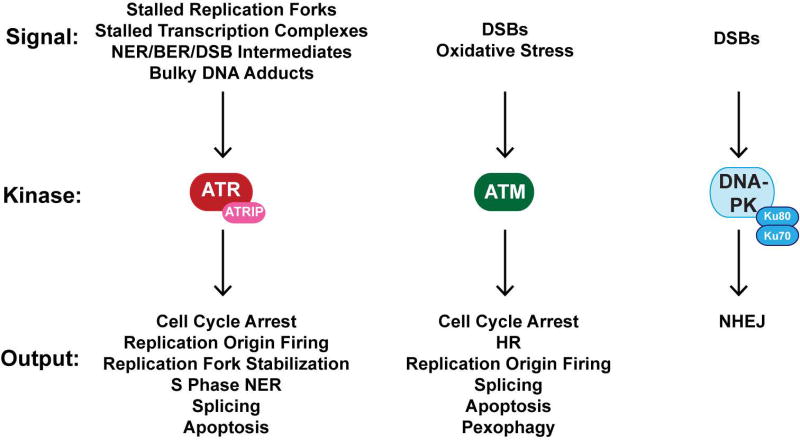
In addition to direct DNA repair, human cells also possess various kinase-based cell signaling pathways that regulate the cellular response to UV. Classically known as DNA damage checkpoints because of their role in transiently arresting cell cycle progression to allow more time for repair [Sancar et al., 2004; Ciccia and Elledge, 2010; Abraham, 2004], many studies have subsequently revealed numerous functions for these “checkpoint” pathways, including the direct regulation of DNA repair systems, DNA replication, and apoptosis. These pathways are governed by a family of phosphoinositide kinase-like protein kinases (PIKKs) known as ATR, ATM, and DNA-PK [Abraham, 2004; Blackford and Jackson, 2017] that initiate signaling in response to specific DNA damage signals. A schematic of these pathways and their stimuli are shown in Figure 3, and importantly, all three DNA damage kinases have been shown to be activated and to affect cell fate in cells irradiated with UV.
Figure 3. DNA damage response kinases activated in UV-irradiated cells.
The ATR, ATM, and DNA-PK protein kinases play pivotal roles in responding to various DNA damage signals in UV-irradiated cells. These kinases then initiate various signaling responses that may result in several different outcomes in the cells, including DNA repair, cell cycle arrest, modulation of DNA replication dynamics, altered gene expression, and cell death.
The ATR kinase is traditionally considered to be the major PIKK that is activated following exposure to UVR and primarily within the context of replication stress caused by DNA polymerase stalling at UV photoproducts [Sancar et al., 2004; Cimprich and Cortez, 2008; Nam and Cortez, 2011]. This polymerase stalling uncouples the actions of the replicative DNA helicase from that of the DNA polymerase [Byun et al., 2005] and generates long stretches of single-stranded DNA (ssDNA), which then becomes bound by RPA (the major ssDNA-binding protein in human cells) [Wold, 1997; Fanning et al., 2006; Zou et al., 2006]. RPA-coated ssDNA is thought to be the initial signal that then recruits ATR to the stalled replication fork via a specific interaction with the ATR-interacting protein ATRIP [Zou and Elledge, 2003; Cortez et al., 2001]. However, a number of studies have provided evidence for ATR activation in non-replicating cells as well, via ssDNA gaps generated by NER or APE1 [Vrouwe et al., 2011; Marini et al., 2006; Marti et al., 2006; Matsumoto et al., 2007; Hanasoge and Ljungman, 2007; O'Driscoll et al., 2003; Ray et al., 2016; Kemp et al., 2014] that may be enlarged by Exonuclease 1 [Lindsey-Boltz et al., 2014; Sertic et al., 2011; Giannattasio et al., 2010] or by stalled transcription complexes [Derheimer et al., 2007; Kemp and Sancar, 2016; Kemp, 2017]. Processing of BER intermediates by the enzyme APE2 has also been shown to lead to ATR activation [Willis et al., 2013]. Lastly, in vitro biochemical evidence support the notion that ATR and its activator protein TopBP1 may also directly sense UV photoproducts to activate ATR kinase activity independent of any further processing by DNA replication, transcription, or repair [Unsal-Kacmaz et al., 2002; Jiang and Sancar, 2006; Choi et al., 2007; Choi et al., 2009]. In addition to the diverse set of signals for ATR activation, the protein substrates that are targeted for phosphorylation by ATR are numerous and have generally been revealed to play important roles in regulating S phase progression [Cimprich and Cortez, 2008; Nam and Cortez, 2011], including stabilization of the replication fork [Dungrawala et al., 2015], repair of UV photoproducts during S phase events [Auclair et al., 2008], and control of translesion synthesis across UV photoproducts [Andrade-Lima et al., 2015; Gohler et al., 2011]. Nonetheless, recent reports also implicate ATR kinase signaling in the regulation of RNA splicing [Munoz et al., 2017] and in the induction of an apoptotic form of cell death in non-cycling UV-irradiated cells [Kemp and Sancar, 2016; Kemp, 2017].
The related kinase ATM is well-recognized for its activation in response to DNA DSBs, where it is recruited to DSBs by the Mre11-Nbs1-Rad50 (MRN) complex very soon after DSB formation to facilitate HR and delay cell cycle progression [Bakkenist and Kastan, 2004; Kitagawa and Kastan, 2005; Paull, 2015]. However, even in non-cycling cells where HR and cell cycle arrest are not relevant, ATM has been shown to be activated following UV irradiation via NER-dependent and -independent means [Kemp and Sancar, 2016; Tresini et al., 2015; Wakasugi et al., 2014] where it affects RNA splicing [Tresini et al., 2015] and proliferation after cell cycle re-entry [Wakasugi et al., 2014]. Moreover, ATM was shown to be directly activated by DNA containing bulky adducts [Kemp et al., 2011] and by oxidative stress through intermolecular disulfide formation [Paull, 2015; Guo et al., 2010]. Thus, ATM may be a direct sensor of not only DSBs but also reactive oxygen species and/or UV lesions in DNA, which is therefore important to consider in the context of UVA irradiation and melanoma. Proteomic studies have shown that ATM phosphorylates potentially hundreds of proteins in cells containing DNA damage [Matsuoka et al., 2007], including some that regulate mRNA splicing [Tresini et al., 2015]. Its activation in response to oxidative stress facilitates the autophagic degradation of peroxisomes [Zhang et al., 2015]. It should also be noted that there is extensive crosstalk and overlap between ATM and ATR function in cells containing DNA damage [Sancar et al., 2004; Ciccia and Elledge, 2010], and thus it can be difficult to define the particular kinase responsible for substrate phosphorylation following UV irradiation.
In contrast to ATR and ATM, DNA-PK is primarily thought to be activated in response to DSBs that are generated in DNA, and its functions are largely thought be limited to DSB repair. DNA-PK kinase activity plays an essential role in NHEJ, where it is recruited by its Ku subunits to the breaks to phosphorylate itself and other NHEJ factors that promote DSB repair [Jette and Lees-Miller, 2015; Davis et al., 2014]. However, like ATR [Choi et al., 2007; Choi et al., 2009] and ATM [Kemp et al., 2011], DNA-PK can also be directly activated by bulky DNA adducts [Kemp et al., 2011] that are present in UV-irradiated cells. NHEJ and DNA-PK function are particularly important in non-replicating cells exposed to UV, where the loss of DNA-PK kinase activity results in elevated lethality [Kemp and Sancar, 2016].
3. Regulation of the DNA damage response: UVA and melanocytes
3.1. DNA repair and DNA damage responses in melanocytes and melanoma cells
Because of the importance of DNA repair and DNA damage signaling in the response to UV-induced DNA damage, there has been great interest in determining whether defects in cellular DNA damage responses contribute to melanomagenesis. Using an immunoslot blot method to measure CPD and (6-4)PP removal from genomic DNA, an initial study using UVC-irradiated primary melanocytes and a panel of melanoma cell lines found that both the normal cells and 11 of 12 cancer cell lines repaired the UV photoproducts at similar rates [Gaddameedhi et al., 2010]. However, a more recent study that employed a flow cytometric methodology to study UV photoproduct repair at different cell cycle phases reported that a majority of melanoma cell lines were defective in NER during S phase [Belanger et al., 2014]. This repair deficiency was correlated with a reduced activation of the ATR kinase and may be explained by altered replicative stress responses caused by an insufficient supply of the protein RPA. Because of its numerous functions in DNA replication, NER, and ATR activation, RPA may become limiting for DNA metabolism under conditions of DNA damage and extensive replication stress [Belanger et al., 2016; Toledo et al., 2013; Guven et al., 2015; Kohler et al., 2016; Tsaalbi-Shtylik et al., 2014]. Interestingly, this finding was supported by additional studies using UVA or UVB light sources (instead of UVC), in which melanoma cells showed a reduced ability to remove UV-induced CPDs from genomic DNA in comparison to normal melanocytes [Murray et al., 2016; Budden et al., 2016]. These latter studies reported the defect to be specific for GG-NER and to involve the reduced expression of the damage sensor protein XPC. Thus, the initiation and/or progression of melanoma may be due in part to an inability to efficiently remove CPDs from genomic DNA.
Another possible factor that may affect how melanocytes recognize and response to UVR-induced DNA damage is the presence of melanin. Melanin can absorb sunlight and scavenge free radicals, and thus it is generally thought to shield cells from the damaging effects of UVR [Bustamante et al., 1993]. However, the level of reactive oxygen species in melanocytes is likely affected by both the ability of melanin to both scavenge ROS and to inadvertently promote ROS generation [Kipp and Young, 1999; Kvam and Tyrrell, 1999; Kvam and Dahle, 2004]. Using host-cell reactivation assays to monitor the repair of luciferase reporter plasmid DNAs damaged with either hydrogen peroxide or UVC, which yielded oxidative or canonical bipyrimidine UV photoproducts, respectively, one study found that the repair of both types of DNA damage was impaired in pigmented melanocytes in comparison to non-pigmented fibroblasts [Wang et al., 2010]. Using in vitro assays that measured repair protein incision or DNA repair synthesis on DNA templates damaged with UVC or H2O2 and incubated in cell-free extracts, the authors went on to show that melanocyte cell-free extracts were partially defective in DNA repair and repair synthesis and that addition of melanin to fibroblast cell-free extract inhibited the repair events. The inhibition of UV photoproduct removal by melanin was supported by another group that used a defined UV photoproduct-containing DNA substrate and cell-free extract in vitro [Gaddameedhi et al., 2010]. However, the authors noted that this inhibition of DNA repair by melanin was non-specific, as melanin also inhibited the ability of bacterial restriction endonucleases to cut DNA at their relevant target sequences. Thus, though melanin may be problematic under certain circumstances, the localization of melanin to cellular compartments outside the nucleus and away from DNA would be expected to limit this form of repair inhibition.
Effective arrest of the cell cycle following DNA damage induction is thought to be important for avoiding mutagenesis and genomic instability by preventing cells with damaged DNA from entering either S phase (where base lesions may be copied in an error-prone manner) or mitosis (where DNA breaks may lead to the loss of genetic information). Thus, G1 and G2 checkpoint responses, which are controlled by the ATR and ATM DNA damage response kinases, may serve as important barriers to tumorigenesis, including in melanoma. Interestingly, a majority of melanoma cell lines were found to display defects in one or both checkpoints following exposure to ionizing radiation [Kaufmann et al., 2008; Carson et al., 2012]. An additional study found a defective G2 checkpoint response to ssDNA gaps produced during the bypass of unrepaired UV photoproducts by the replication fork in melanoma cells [Wigan et al., 2012]. However, the S-phase checkpoint, which inhibits the firing of new replication origins following DNA damage and likely similarly functions to limit mutagenesis [Ciccia and Elledge, 2010; Sancar et al., 2004; Iyer and Rhind, 2017; Kaufmann, 2010], was found to be normal in UVC-irradiated melanoma cell lines [Cordeiro-Stone et al., 2016]. A recent report further identified a defect in the ATM-dependent cell cycle checkpoint in melanoma cells that was caused by overexpression of PLK1, which negatively regulates ATM signaling [Spoerri et al., 2016]. Because missense mutations in ATR and ATM have been reported in some melanomas [Dombernowsky et al., 2008; Chen et al., 2017], aberrant signaling by mutant kinases may further contribute to melanomagenesis.
3.2. UVA-dependent oxidation of DNA repair proteins
The observation that NER-deficient XP cells are extremely sensitive to UVC and UVB but are not hypersensitive to UVA [Keyse et al., 1983] has indicated that the biological effects of UVA may lay beyond its direct effects in inducing DNA damage. Moreover, CPDs induced by UVA in NER-proficient cells have been reported to take longer to be repaired than CPDs induced by UVB or UVC [Courdavault et al., 2004; Mouret et al., 2006], and indeed CPDs induced by UVB or UVC are moderately less toxic and mutagenic than CPDs induced by UVA [Enninga et al., 1986; Biverstal et al., 2008; Kappes et al., 2006; Runger et al., 2012]. Thus, the biological effects of UVA are likely influenced by it having additional cellular targets, potentially through the well-known induction of reactive oxygen species.
In both bacterial and human cells, exposure to UVA prior to irradiation with UVC or UVB results in increased cell lethality and a slower removal of canonical UV photoproducts [Tyrrell and Webb, 1973; Tyrrell et al., 1984; Smith and Paterson, 1982]. More recently, this phenomenon has been linked to UVA-dependent damage to DNA repair proteins, either alone or in conjunction with photosensitizing drugs [Brem et al., 2017; Karran and Brem, 2016]. A summary of DNA repair proteins that have been reported to undergo oxidation by either UVA or other mechanisms is provided in Table 3, which includes proteins involved in NER [Guven et al., 2015; Wang et al., 2001; Gueranger et al., 2014; Men et al., 2007; You et al., 2000; Montaner et al., 2007; Kelley et al., 2012; Xanthoudakis et al., 1992; Xanthoudakis and Curran, 1996; Xanthoudakis et al., 1994; Bennett et al., 2009; Andrews et al., 2006; Grosskopf et al., 2010; Zhou et al., 2015; Zhou et al., 2011; Wang et al., 2013; Bravard et al., 2006; Bravard et al., 2010; Morreall et al., 2015; Campalans et al., 2007; Girard et al., 2013; Fribourg et al., 2000; Ding et al., 2009], BER [Guven et al., 2015; Wang et al., 2001; Gueranger et al., 2014; Men et al., 2007; You et al., 2000; Montaner et al., 2007; Kelley et al., 2012; Xanthoudakis et al., 1992; Xanthoudakis and Curran, 1996; Xanthoudakis et al., 1994; Bennett et al., 2009; Andrews et al., 2006; Grosskopf et al., 2010; Zhou et al., 2015; Zhou et al., 2011; Wang et al., 2013; Bravard et al., 2006; Bravard et al., 2010; Morreall et al., 2015; Campalans et al., 2007; Girard et al., 2013; Fribourg et al., 2000; Ding et al., 2009], and DSB repair [Guven et al., 2015; Wang et al., 2001; Gueranger et al., 2014; Men et al., 2007; You et al., 2000; Montaner et al., 2007; Kelley et al., 2012; Xanthoudakis et al., 1992; Xanthoudakis and Curran, 1996; Xanthoudakis et al., 1994; Bennett et al., 2009; Andrews et al., 2006; Grosskopf et al., 2010; Zhou et al., 2015; Zhou et al., 2011; Wang et al., 2013; Bravard et al., 2006; Bravard et al., 2010; Morreall et al., 2015; Campalans et al., 2007; Girard et al., 2013; Fribourg et al., 2000; Ding et al., 2009].
Table 3. DNA repair proteins reported to undergo oxidation.
The indicated DNA repair proteins have been shown to undergo oxidation in mammalian cells. DNA metabolic processes and the functional effects of protein oxidation are provided along with references to the primary literature.
| Protein | DNA Metabolic Processes |
Effect of Protein Oxidation |
References |
|---|---|---|---|
|
| |||
| RPA | NER | Impairs ssDNA binding | Guven 2015; Wang 2001; Gueranger 2014; Men 2007; You 2000 |
| ATR signaling | Inhibits NER | ||
| Replication | |||
|
| |||
| PCNA | NER | Unknown | Montaner 2007 |
| Replication | |||
|
| |||
| APE1 | BER | May block reducing activity towards transcription factors | Kelley 2012; Xanthaudakis 1992, 1994, 1996 |
| Transcription | |||
|
| |||
| Ku | NHEJ | Alters DNA binding | Gueranger 2014; Bennett 2009; Andrews 2006 |
| Inhibits NHEJ | |||
|
| |||
| XPA | NER | Diminishes DNA binding | Grosskopf 2010; Zhou 2011, 2015 |
|
| |||
| XPE | NER | Reduced gene expression | Grosskop 2010 |
|
| |||
| OGG1 | BER | Decreases activity | Gueranger 2014; Bravard 2006; Bravard 2010; Morreall 2015; Camplans 2007 |
| Inhibits 8-oxoG removal | |||
| Localization to speckles | |||
|
| |||
| XRCC3 | HR | Chromatin re-localization | Girard 2013 |
| Sensitivity to camptothecin | |||
|
| |||
| TFIIH (p44) | NER | Unknown | Fribourg 2000 |
|
| |||
| PARP | BER | Inhibits activity | Zhou 2011, 2015; Wang 2013; Ding 2009 |
| SSBR | Interferes with BER | ||
|
| |||
| ATM | ATM signaling | Activates kinase activity | Guo 2010; Zhang 2015 |
| HR | Autophagy of peroxisomes | ||
The level of oxidative stress in cells may be exacerbated when UVA interacts with endogenous or exogenous photosensitizers, including 6-thioguanine or fluoroquinolone antibiotics such as ciprofloxacin. Treatment of cultured cells with UVA and photosensitizers has been shown to lead to oxidation of RPA, OGG1, and Ku, and to be associated with inhibition of NER, BER, and NHEJ, respectively [Gueranger et al., 2014; Guven et al., 2015]. Interestingly, RPA plays a particularly important role in multiple aspects of DNA metabolism, including DNA repair, replication, and ATR-dependent DNA damage checkpoint signaling [Wold, 1997; Fanning et al., 2006; Zou et al., 2006]. The depletion of the available pool of RPA protein in cells containing DNA damage and undergoing replication stress has been reported to be responsible for DSB formation [Toledo et al., 2013], and overexpression of RPA partly protects cells from UVA-mediated oxidative stress [Guven et al., 2015]. Thus, several aspects of the cellular response to UV-induced DNA damage may be affected by UVA-dependent oxidation of a single protein, RPA. These issues may be acutely relevant to melanocytes and melanomagenesis because the presence of melanin may exacerbate the effects of UVA on ROS production.
3.3. Regulation of melanocyte DNA damage responses by hormonal signaling
Given that human skin contains diverse cell types in addition to melanocytes, including keratinocytes, fibroblasts, and immune cells, melanocyte responses to UVR are therefore expected to be influenced by interactions with these other cell types through both contact-dependent and -independent means. Research over the past few years has uncovered two major hormonal signaling pathways that act in a paracrine manner to influence melanocyte responses to UV-induced DNA damage [Jarrett and D'Orazio, 2017; Jarrett et al., 2017]. Moreover, additional regulatory systems that have been shown to govern keratinocyte responses to UV and non-melanoma skin cancer risk, including the insulin-like growth factor-1 pathway and the circadian clock, may also be relevant to understanding melanomagenesis and are therefore briefly summarized here. These four systems are outlined in Table 4. It should be noted that most of the work to be described utilized UVB and/or UVC light sources, and thus the role of UVA remains to be fully explored. Moreover, the use of solar simulating light sources that include both UVA and UVB wavelengths will therefore be ideal in future validation studies.
Table 4. Paracrine regulatory systems that have been reported to affect the DNA damage response and/or skin carcinogenesis.
The indicated paracrine system is listed along with known regulatory targets in the DNA damage response.
| Regulatory System | Target(s) | References |
|---|---|---|
|
| ||
| Melanocortin/MC1R | NER (ATR and XPA) | Abdel-Malek 2009; Bohm 2005; Hauser 2006; Jarrett 2014, 2015; Kadekaro 2010; Song 2009; Swope 2014 |
|
| ||
| Endothelin/ETBR | NER (XPC) | Hosoda 1994; Kadekaro 2005; von Koschembahr 2015 |
|
| ||
| Insulin-like Growth Factor-1/IGF-1R | NER (XPC, XPB, XPF) | Lewis 2010; Loesch 2016; Fernandez 2016; Kemp 2017 |
| ATR kinase signaling | ||
|
| ||
| Circadian Clock | NER (XPA) | Gaddemeedhi 2011, 2015; Geyfman 2012; Wang 2017; Guo 2016; Manzella 2015 |
| BER (OGG1) | ||
| ATR kinase signaling | ||
The first known regulatory pathway in melanocytes involves the melanocortin 1 receptor (MC1R), which is a Gs protein-coupled receptor that is present in the melanocyte cell membrane. It is stimulated by α-melanocyte stimulating hormone (α-MSH), which is produced and released by keratinocytes in a p53-dependent manner following UV exposure [Cui et al., 2007; Fell et al., 2014]. The stimulation of MC1R by extracellular α-MSH leads to its cytoplasmic association with adenylyl cyclase and to cAMP production [Garcia-Borron et al., 2014; Garcia-Borron and Olivares, 2014; Swope et al., 2012]. Though a major effector of this signaling is the stimulation of melanin production [Abdel-Malek et al., 2000; D'Orazio et al., 2006], several other important processes are impacted in melanocytes, including a suppression of oxidative stress [Kadekaro et al., 2003; Kadekaro et al., 2012], the promotion of cell growth [Suzuki et al., 1999], and protection from apoptosis [McGill et al., 2002]. Of particular relevance to the DNA damage response, stimulation of the MC1R has also been shown in a number of studies to affect the efficiency of UV photoproduct removal [Abdel-Malek et al., 2009; Bohm et al., 2005; Hauser et al., 2006; Jarrett et al., 2014; Jarrett et al., 2015; Kadekaro et al., 2010; Song et al., 2009; Swope et al., 2014]. Moreover, recent work has uncovered a rapid response to MC1R signaling in which the activation of Protein Kinase A (PKA) by cAMP leads to the phosphorylation of the DNA damage response kinase ATR on Ser435, which facilitates the localization of XPA to sites of UV photodamage in the nucleus [Jarrett et al., 2014; Jarrett et al., 2016]. ATR is known to regulate NER through several mechanisms, including through the direct phosphorylation of XPA at a site that stabilizes the protein and limits its ubiquitin-mediated proteolysis [Park and Kang, 2016; Lee et al., 2014; Shell et al., 2009; Li et al., 2011; Li et al., 2013]. The MC1R-dependent and non-canonical ATR pathway was shown to facilitate an interaction of XPA with chromatin and to specifically promote the action of the NER endonuclease XPF to make the 5’ incision during UV photoproduct removal, which thus promotes NER and limits mutagenesis [Jarrett et al., 2016]. Interestingly, these responses were shown to be defective in cells expressing polymorphic versions of the MC1R that are common to fair-skinned individuals [Valverde et al., 1995; Wolf Horrell et al., 2016], which suggests that the enhanced melanoma risk in fair-skinned individuals may be due in part to this defective MC1R-cAMP-PKA-ATR-XPA signaling pathway [Jarrett et al., 2017; Jarrett et al., 2016].
A second paracrine system that has been shown to regulate melanocyte responses to UV DNA damage is the endothelin signaling pathway. Endothelins are known to affect melanocyte proliferation and pigment production [Yada et al., 1991], and similar to α-MSH, are secreted by UV-irradiated keratinocytes [Imokawa et al., 1992; Yohn et al., 1993]. Endothelins activate the endothelin B receptor (ETBR) on melanocytes, where they regulate melanocyte proliferation, development, and pigmentation [Baynash et al., 1994; Hosoda et al., 1994; Tada et al., 2002]. Of specific relevance to UV irradiation, ETBR signaling has been shown to affect NER and apoptosis in UV-irradiated melanocytes [Kadekaro et al., 2005; von Koschembahr et al., 2015]. Treatment of cultured melanocytes in vitro with endothelin 1 (ET1) reduced UV photoproduct levels and facilitated the activation of the JNK and p38 signaling pathways in a calcium-dependent manner [von Koschembahr et al., 2015]. Facilitation of NER was further correlated with an enhanced turnover of the UV photoproduct damage recognition protein XPC at sites of DNA damage. Thus, the mechanism by which ETBR signaling facilitated NER appears to be distinct from that of MC1R signaling.
Whereas the effect of the MC1R and ETBR pathways on UV responses have been specifically examined in melanocytes and are therefore directly relevant to melanomagenesis, an additional factor that has been investigated in keratinocytes in the context of non-melanoma skin cancer is the insulin-like growth factor-1 (IGF-1) system [Kemp et al., 2017b; Lewis et al., 2009]. In human skin, IGF-1 is primarily supplied to epidermal keratinocytes by dermal fibroblasts [Ando and Jensen, 1993; Barreca et al., 1992; Siddle, 2011], and this production is decreased in the skin of geriatric individuals and is correlated with increased fibroblast senescence [Lewis et al., 2008; Lewis et al., 2010]. In cultured keratinocytes in vitro and basal epidermal skin keratinocytes in vivo, the loss of IGF-1 receptor (IGF-1R) signaling slows CPD removal [Loesch et al., 2016; Fernandez et al., 2015] and partially abrogates canonical ATR signaling and the suppression of DNA synthesis following UVB exposure [Fernandez et al., 2015; Kemp et al., 2017a]. The association of these defective DNA damage responses with reduced IGF-1 signaling and advanced age may explain the observation that the majority of non-melanoma skin cancers occur in individuals over the age of 60 [Kraemer, 1997]. Though the correlation between patient age and melanoma is much weaker than for non-melanoma skin cancers [Fears et al., 1977; Armstrong and Cust, 2017], the fact that melanocytes express the IGF-1R [Hodak et al., 1996; Rodeck et al., 1987] and the observation that the number of mutations found in melanomas increases with advanced age [Krauthammer et al., 2012] would be consistent with a role for aging and/or the IGF-1 system in the regulation of the DNA damage response in melanocytes. Moreover, because MSH production by UV-irradiated keratinocytes is dependent on p53 [Cui et al., 2007; Fell et al., 2014], and p53 activation has been shown to be disrupted in UVB-irradiated keratinocytes with inactive IGF-1Rs [Lewis et al., 2008], it is possible that altered p53 signaling in IGF-1R-deficient keratinocytes negatively affects MC1R signaling in melanocytes. However, the reported reduction in melanocyte number in older people may serve to limit melanomagenesis in these patient populations by restricting the abundance of the cell type in the skin [Gilchrest et al., 1979]. Further work is therefore necessary to resolve this issue.
One final regulatory system that may influence melanomagenesis is the circadian clock, which controls diverse biochemical, physiological, and behavioral systems in humans and other organisms with a periodicity of approximately 24 h. At the organismal level in mammals, a master clock in the suprachiasmatic nucleus (SCN) in the brain receives external light cues via the optic nerve and then synchronizes peripheral clocks located in tissues throughout the body, including in the skin [Lin et al., 2009; Plikus et al., 2015; Matsui et al., 2016]. At the molecular level, the clock is regulated by a transcription-translation feedback loop that regulates gene expression throughout the course of a 24 h day [Takahashi, 2017] and in recent years has been shown to regulate the cellular response to DNA damage [Sancar et al., 2010; Sancar et al., 2015]. Using lymphocytes isolated from blood at different times of the day from human volunteers, the expression of the BER gene OGG1 was observed to be higher in the morning hours than in the evening hours, and thus lymphocytes accumulated less 8-oxoG at those time points when challenged with oxidative stress [Manzella et al., 2015]. Several studies in mice have investigated circadian regulation of the XPA gene, which is essential for the repair of UV-induced bipyrimidine dimers by the NER pathway. XPA mRNA and protein levels were observed to oscillate with a 24 h periodicity in mouse tissues (including skin), such that XPA mRNA and protein levels were found to be low in the morning and high in the evening [Kang et al., 2009; Kang et al., 2010; Kang et al., 2011; Gaddameedhi et al., 2011; Gaddameedhi et al., 2015; Geyfman et al., 2012; Wang et al., 2017]. This pattern of XPA expression was directly correlated with NER capacity throughout the day in these tissues [Kang et al., 2009; Gaddameedhi et al., 2011]. Moreover, UV skin carcinogenesis studies found that mice irradiated with UVB in the early morning were more susceptible to skin cancer formation, including invasive SCCs, than mice irradiated in the late afternoon/evening [Gaddameedhi et al., 2011]. Additional work also demonstrated that skin inflammation and erythema were similarly affected by the time of day of UV exposure in mice and were correlated with both low XPA levels and with elevated ATR kinase signaling to its downstream substrates CHK1 and p53 [Gaddameedhi et al., 2015]. Preliminary studies using human skin biopsies indicates that the clock also regulates XPA protein levels in human skin [Guan et al., 2016]. Thus, the interplay between the circadian clock and the cellular DNA damage response likely has important implications for human skin diseases [Dakup and Gaddameedhi, 2017; Sancar et al., 2015]. Because melanocytes also exhibit circadian properties [Hardman et al., 2015; Sandu et al., 2012; Slominski et al., 2015], it is expected that the time of day of UV exposure and the associated level of XPA protein and DNA damage signaling may also impact the risk of melanomagenesis.
3.4. UVA and immunosuppression
The recognition and elimination of mutant, pre-cancerous cells by the body’s immune system plays a major role in preventing cancers, including in the skin. UV has been known for decades to have immunosuppressive effects [Gonzalez Maglio et al., 2016; Prasad and Katiyar, 2017], and therefore immunosuppression is considered to be an important risk factor for melanoma. Indeed, organ transplant recipients taking immunosuppressive drugs have been reported to have higher rates of both melanoma and non-melanoma skin cancers [Kubica and Brewer, 2012]. Classical tumor studies in mice showed that whereas the transplantation of UV-induced skin tumors into syngeneic mice resulted in rejection of the tumors, the rejection did not take place if the recipient mice were exposed to UV light [Fisher and Kripke, 1977]. Later work went on to indicate that CPDs in DNA were specifically required to initiate system immunosuppression in the irradiated mice [Kripke et al., 1992]. For that reason, sunlight-induced immunosuppression may be a key factor that plays a role in the pathogenesis of the melanoma. Recent studies report that both UVA and UVB exert immunosuppressive effects [Damian et al., 1999; Damian et al., 2011; Matthews et al., 2010b; Matthews et al., 2010a; Hersey et al., 1987]. Sunscreen studies have confirmed that UVR-induced immune suppression can be prevented by using broad-spectrum sunscreens that provide UVA protection [Hersey et al., 1987]. Contact hypersensitivity studies provide the most compelling evidence of UVA-induced immunosuppression in humans. By using the nickel contact hypersensitivity test in human models, it was reported that there are immunosuppressive effects of UVA (370nm) at doses that corresponded to a 20 min exposure to mid-day summer sunlight [Damian et al., 2011].
The mechanism by which UVR induces immunosuppression remains unclear. Evidence has implicated UV-dependent damage to both lipids and DNA as contributing to the immunosuppression [Kripke et al., 1992; Damiani and Ullrich, 2016; Prasad and Katiyar, 2017]. Nonetheless, through the use of transgenic mice expressing bacterial photolyases that are capable of repairing UV-induced CPDs or (6-4)PPs in whole mouse skin or only the basal keratinocytes, one study showed that the presence of unrepaired CPDs in basal keratinocytes was responsible for systemic immunosuppression [Jans et al., 2006]. Moreover, complete loss of NER activity via mutation of XPA greatly enhances the likelihood of UV-induced systemic immunosuppression in mice [Garssen et al., 2000; Miyauchi-Hashimoto et al., 1996]. Additional studies using mice deficient in either GG-NER or TC-NER suggested that whereas a total NER defect is needed to increase systemic immunosuppression, a loss of TC-NER is sufficient to enhance local immunosuppression [Kolgen et al., 2003]. Though many studies of immunosuppression have utilized UVB light sources to maximize damage to genomic DNA in epidermal cells, Ngheim and colleagues concluded that UVA (60 kJ/m2 or higher) irradiation can also suppress the established immune response in mice [Nghiem et al., 2002]. Moreover, studies have also shown that the application of liposomes containing the bacterial T4N5 enzyme to target CPDs for repair blocked UVR-induced immune suppression [Nghiem et al., 2002; Ullrich et al., 2002; Yarosh, 2004], which further highlights the important role of canonical UV-induced bypyrimidine DNA photoproducts in immunosuppression.
Previous studies linking UV-induced DNA damage to immunosuppression have largely focused on DNA adduct levels and their repair. However, given the major roles for DNA damage kinase signaling in the cellular response to DNA damage described earlier, it is expected that there may be interplay between DNA damage signaling and immunosuppression [Neves-Costa and Moita, 2017]. Consistent with this hypothesis, a recent study uncovered a panel of human melanomas that contain mutations in the ATR kinase that abrogate its canonical signaling functions [Chen et al., 2017]. The loss of ATR function was found to be correlated with both the presence of multiple mutations and with alterations in inflammatory gene expression. A correlation of this phenotype with decreased T cell recruitment and elevated macrophage recruitment suggested that this loss of ATR signaling may facilitate tumor invasion. Thus, an altered immune microenvironment due to loss of UV-induced ATR kinase signaling in some developing melanomas may aid melanoma growth.
4. Summary and conclusions
Previously, there was a misconception that high intensity UVA from artificial tanning devices were safe and not associated with melanoma. However, more recent data from murine experimental studies, and human epidemiological and sunscreen use studies provide compelling evidence for a strong association between UVA and melanoma risk [Autier et al., 2011b; Autier et al., 2011a]. UVA is a complete carcinogen that may play key roles in both the initiation and promotion of melanoma. Consequently, reduced exposure to the sun and other UVA light sources should be employed to minimize the risk of melanomagenesis. Simple protective measures include the avoidance of direct sunlight during peak hours of the day by using hats, proper clothing and seeking shade. Moreover, because sunscreens have been shown to reduce DNA damage formation in UV-irradiated human skin [Olsen et al., 2017], the proper and long-term use of broad-spectrum sunscreens that contain both UVA and UVB filters should also be a part of the strategy to reduce the risk of the melanoma. There is also a clear need for the development of more effective UVA filters in sunscreens [Sargent and Travers, 2016]. Together, these approaches will be expected to limit the amount of unrepaired DNA damage induced by UVA and therefore the mutagenesis and immunosuppression that drive melanomagenesis.
The abundance of UV signature mutation in cutaneous melanomas highlights the relevance of UVR in melanomagenesis. However, the relative contributions of UVA and UVB wavelengths of light and the role of the melanocyte microenvironment in this process have been enigmatic. Recent research has therefore uncovered novel ways in which UVA may both induce CPD formation in melanocyte DNA and impair the rate of UV photoproduct repair via oxidation of NER proteins. Moreover, alterations to the melanocyte microenvironment by paracrine factors and the immune system may further exacerbate these effects. Defects in DNA repair and the cellular DNA damage response to UVA are expected to ultimately give rise to gene mutations that drive the initiation and promotion of melanomas. Because many of the effects of UVA are thought to be due in great part to its ability to produce ROS, novel strategies should be considered to protect the skin against excessive UVA exposure as well and to ameliorate the effects of ROS generation. Nonetheless, further research is therefore required to provide a more complete understanding behind the pathogenesis of UVA-induced melanoma.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions: AQK, JBT, and MGK all contributed to the writing of this manuscript.
References
- Abbotts R, Wilson DM., 3rd Coordination of DNA single strand break repair. Free Radic Biol Med. 2017;107:228–244. doi: 10.1016/j.freeradbiomed.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Malek Z, Scott MC, Suzuki I, Tada A, Im S, Lamoreux L, Ito S, Barsh G, Hearing VJ. The melanocortin-1 receptor is a key regulator of human cutaneous pigmentation. Pigment Cell Res. 2000;13(Suppl 8):156–162. doi: 10.1034/j.1600-0749.13.s8.28.x. [DOI] [PubMed] [Google Scholar]
- Abdel-Malek ZA, Ruwe A, Kavanagh-Starner R, Kadekaro AL, Swope V, Haskell-Luevano C, Koikov L, Knittel JJ. alpha-MSH tripeptide analogs activate the melanocortin 1 receptor and reduce UV-induced DNA damage in human melanocytes. Pigment Cell Melanoma Res. 2009;22:635–644. doi: 10.1111/j.1755-148X.2009.00598.x. [DOI] [PubMed] [Google Scholar]
- Abraham RT. PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Ando Y, Jensen PJ. Epidermal growth factor and insulin-like growth factor I enhance keratinocyte migration. J Invest Dermatol. 1993;100:633–639. doi: 10.1111/1523-1747.ep12472297. [DOI] [PubMed] [Google Scholar]
- Andrade-Lima LC, Andrade LN, Menck CF. ATR suppresses apoptosis after UVB irradiation by controlling both translesion synthesis and alternative tolerance pathways. J Cell Sci. 2015;128:150–159. doi: 10.1242/jcs.161596. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Lehman JA, Turchi JJ. Kinetic analysis of the Ku-DNA binding activity reveals a redox-dependent alteration in protein structure that stimulates dissociation of the Ku-DNA complex. J Biol Chem. 2006;281:13596–13603. doi: 10.1074/jbc.M512787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong BK, Cust AE. Sun exposure and skin cancer, and the puzzle of cutaneous melanoma: A perspective on Fears et al. Mathematical models of age and ultraviolet effects on the incidence of skin cancer among whites in the United States. American Journal of Epidemiology 1977; 105: 420–427. Cancer Epidemiol. 2017;48:147–156. doi: 10.1016/j.canep.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Atkar R, Ocampo M, Euvrard S, McGregor J, Kanitakis J, Harwood C. Ultraviolet radiation exposure through window glass may be associated with localization of nonmelanoma skin cancer in organ transplant recipients: a study in France and the UK. Br J Dermatol. 2013;169:484–485. doi: 10.1111/bjd.12379. [DOI] [PubMed] [Google Scholar]
- Attard NR, Karran P. UVA photosensitization of thiopurines and skin cancer in organ transplant recipients. Photochem Photobiol Sci. 2012;11:62–68. doi: 10.1039/c1pp05194f. [DOI] [PubMed] [Google Scholar]
- Auclair Y, Rouget R, Affar el B, Drobetsky EA. ATR kinase is required for global genomic nucleotide excision repair exclusively during S phase in human cells. Proc Natl Acad Sci U S A. 2008;105:17896–17901. doi: 10.1073/pnas.0801585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autier P, Boniol M, Dore JF. Is sunscreen use for melanoma prevention valid for all sun exposure circumstances? J Clin Oncol. 2011a;29:e425–6. doi: 10.1200/JCO.2010.34.4275. author reply e427. [DOI] [PubMed] [Google Scholar]
- Autier P, Dore JF, Eggermont AM, Coebergh JW. Epidemiological evidence that UVA radiation is involved in the genesis of cutaneous melanoma. Curr Opin Oncol. 2011b;23:189–196. doi: 10.1097/CCO.0b013e3283436e5d. [DOI] [PubMed] [Google Scholar]
- Autier P, Dore JF, Schifflers E, Cesarini JP, Bollaerts A, Koelmel KF, Gefeller O, Liabeuf A, Lejeune F, Lienard D. Melanoma and use of sunscreens: an Eortc case-control study in Germany, Belgium and France. The EORTC Melanoma Cooperative Group. Int J Cancer. 1995;61:749–755. doi: 10.1002/ijc.2910610602. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Barak Y, Cohen-Fix O, Livneh Z. Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA. Significance for UV light mutagenesis. J Biol Chem. 1995;270:24174–24179. doi: 10.1074/jbc.270.41.24174. [DOI] [PubMed] [Google Scholar]
- Barreca A, De Luca M, Del Monte P, Bondanza S, Damonte G, Cariola G, Di Marco E, Giordano G, Cancedda R, Minuto F. In vitro paracrine regulation of human keratinocyte growth by fibroblast-derived insulin-like growth factors. J Cell Physiol. 1992;151:262–268. doi: 10.1002/jcp.1041510207. [DOI] [PubMed] [Google Scholar]
- Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–2175. [PubMed] [Google Scholar]
- Bauer J. The Molecular Revolution in Cutaneous Biology: Era of Cytogenetics and Copy Number Analysis. J Invest Dermatol. 2017;137:e57–e59. doi: 10.1016/j.jid.2016.11.043. [DOI] [PubMed] [Google Scholar]
- Baute J, Depicker A. Base excision repair and its role in maintaining genome stability. Crit Rev Biochem Mol Biol. 2008;43:239–276. doi: 10.1080/10409230802309905. [DOI] [PubMed] [Google Scholar]
- Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Belanger F, Angers JP, Fortier E, Hammond-Martel I, Costantino S, Drobetsky E, Wurtele H. Mutations in Replicative Stress Response Pathways Are Associated with S Phase-specific Defects in Nucleotide Excision Repair. J Biol Chem. 2016;291:522–537. doi: 10.1074/jbc.M115.685883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger F, Rajotte V, Drobetsky EA. A majority of human melanoma cell lines exhibits an S phase-specific defect in excision of UV-induced DNA photoproducts. PLoS One. 2014;9:e85294. doi: 10.1371/journal.pone.0085294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SM, Neher TM, Shatilla A, Turchi JJ. Molecular analysis of Ku redox regulation. BMC Mol Biol. 2009;10 doi: 10.1186/1471-2199-10-86. 86-2199-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, Zhang H, Zeid R, Ren X, Cibulskis K, Sivachenko AY, Wagle N, Sucker A, Sougnez C, Onofrio R, Ambrogio L, Auclair D, Fennell T, Carter SL, Drier Y, Stojanov P, Singer MA, Voet D, Jing R, Saksena G, Barretina J, Ramos AH, Pugh TJ, Stransky N, Parkin M, Winckler W, Mahan S, Ardlie K, Baldwin J, Wargo J, Schadendorf D, Meyerson M, Gabriel SB, Golub TR, Wagner SN, Lander ES, Getz G, Chin L, Garraway LA. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biverstal A, Johansson F, Jenssen D, Erixon K. Cyclobutane pyrimidine dimers do not fully explain the mutagenicity induced by UVA in Chinese hamster cells. Mutat Res. 2008;648:32–39. doi: 10.1016/j.mrfmmm.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Bohm M, Wolff I, Scholzen TE, Robinson SJ, Healy E, Luger TA, Schwarz T, Schwarz A. alpha-Melanocyte-stimulating hormone protects from ultraviolet radiation-induced apoptosis and DNA damage. J Biol Chem. 2005;280:5795–5802. doi: 10.1074/jbc.M406334200. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Coste F, Castaing B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic Biol Med. 2017;107:179–201. doi: 10.1016/j.freeradbiomed.2016.11.042. [DOI] [PubMed] [Google Scholar]
- Brash DE. UV signature mutations. Photochem Photobiol. 2015;91:15–26. doi: 10.1111/php.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci U S A. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravard A, Campalans A, Vacher M, Gouget B, Levalois C, Chevillard S, Radicella JP. Inactivation by oxidation and recruitment into stress granules of hOGG1 but not APE1 in human cells exposed to sub-lethal concentrations of cadmium. Mutat Res. 2010;685:61–69. doi: 10.1016/j.mrfmmm.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Bravard A, Vacher M, Gouget B, Coutant A, de Boisferon FH, Marsin S, Chevillard S, Radicella JP. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol Cell Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R, Guven M, Karran P. Oxidatively-generated damage to DNA and proteins mediated by photosensitized UVA. Free Radic Biol Med. 2017;107:101–109. doi: 10.1016/j.freeradbiomed.2016.10.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R, Karran P. Multiple forms of DNA damage caused by UVA photoactivation of DNA 6-thioguanine. Photochem Photobiol. 2012;88:5–13. doi: 10.1111/j.1751-1097.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- Bryan DS, Ransom M, Adane B, York K, Hesselberth JR. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 2014;24:1534–1542. doi: 10.1101/gr.174052.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel TB, Goldstein AM, Fraser MC, Rogers B, Tucker MA. Recent tanning bed use: a risk factor for melanoma. Arch Dermatol. 2006;142:485–488. doi: 10.1001/archderm.142.4.485. [DOI] [PubMed] [Google Scholar]
- Budden T, Davey RJ, Vilain RE, Ashton KA, Braye SG, Beveridge NJ, Bowden NA. Repair of UVB-induced DNA damage is reduced in melanoma due to low XPC and global genome repair. Oncotarget. 2016;7:60940–60953. doi: 10.18632/oncotarget.10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Fix D, Liu H, Hays J, Bockrath R. In vivo deamination of cytosine-containing cyclobutane pyrimidine dimers in E. coli: a feasible part of UV-mutagenesis. Mutat Res. 2003;522:145–156. doi: 10.1016/s0027-5107(02)00310-x. [DOI] [PubMed] [Google Scholar]
- Bustamante J, Bredeston L, Malanga G, Mordoh J. Role of melanin as a scavenger of active oxygen species. Pigment Cell Res. 1993;6:348–353. doi: 10.1111/j.1600-0749.1993.tb00612.x. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Grand A, Douki T. Solar UV radiation-induced DNA Bipyrimidine photoproducts: formation and mechanistic insights. Top Curr Chem. 2015;356:249–275. doi: 10.1007/128_2014_553. [DOI] [PubMed] [Google Scholar]
- Cadet J, Mouret S, Ravanat JL, Douki T. Photoinduced damage to cellular DNA: direct and photosensitized reactions. Photochem Photobiol. 2012;88:1048–1065. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Campalans A, Amouroux R, Bravard A, Epe B, Radicella JP. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci. 2007;120:23–32. doi: 10.1242/jcs.03312. [DOI] [PubMed] [Google Scholar]
- Carson C, Omolo B, Chu H, Zhou Y, Sambade MJ, Peters EC, Tompkins P, Simpson DA, Thomas NE, Fan C, Sarasin A, Dessen P, Shields JM, Ibrahim JG, Kaufmann WK. A prognostic signature of defective p53-dependent G1 checkpoint function in melanoma cell lines. Pigment Cell Melanoma Res. 2012;25:514–526. doi: 10.1111/j.1755-148X.2012.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Ruiz-Vega R, Vasudeva P, Espitia F, Krasieva TB, de Feraudy S, Tromberg BJ, Huang S, Garner CP, Wu J, Hoon DS, Ganesan AK. ATR Mutations Promote the Growth of Melanoma Tumors by Modulating the Immune Microenvironment. Cell Rep. 2017;18:2331–2342. doi: 10.1016/j.celrep.2017.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Gaddameedhi S, Kim SY, Hu J, Kemp MG, Sancar A. Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Res. 2014;42:e29. doi: 10.1093/nar/gkt1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Lindsey-Boltz LA, Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci U S A. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- Colebatch AJ, Di Stefano L, Wong SQ, Hannan RD, Waring PM, Dobrovic A, McArthur GA, Papenfuss AT. Clustered somatic mutations are frequent in transcription factor binding motifs within proximal promoter regions in melanoma and other cutaneous malignancies. Oncotarget. 2016;7:66569–66585. doi: 10.18632/oncotarget.11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Cordeiro-Stone M, McNulty JJ, Sproul CD, Chastain PD, Gibbs-Flournoy E, Zhou Y, Carson C, Rao S, Mitchell DL, Simpson DA, Thomas NE, Ibrahim JG, Kaufmann WK. Effective intra-S checkpoint responses to UVC in primary human melanocytes and melanoma cell lines. Pigment Cell Melanoma Res. 2016;29:68–80. doi: 10.1111/pcmr.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Courdavault S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutat Res. 2004;556:135–142. doi: 10.1016/j.mrfmmm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter SR, Fisher DE. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- Dahlke E, Murray CA, Kitchen J, Chan AW. Systematic review of melanoma incidence and prognosis in solid organ transplant recipients. Transplant Res. 2014;3 doi: 10.1186/2047-1440-3-10. 10-1440-3-10. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakup P, Gaddameedhi S. Impact of the Circadian Clock on UV-Induced DNA Damage Response and Photocarcinogenesis. Photochem Photobiol. 2017;93:296–303. doi: 10.1111/php.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damian DL, Barnetson RS, Halliday GM. Low-dose UVA and UVB have different time courses for suppression of contact hypersensitivity to a recall antigen in humans. J Invest Dermatol. 1999;112:939–944. doi: 10.1046/j.1523-1747.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- Damian DL, Matthews YJ, Phan TA, Halliday GM. An action spectrum for ultraviolet radiation-induced immunosuppression in humans. Br J Dermatol. 2011;164:657–659. doi: 10.1111/j.1365-2133.2010.10161.x. [DOI] [PubMed] [Google Scholar]
- Damiani E, Ullrich SE. Understanding the connection between platelet-activating factor, a UV-induced lipid mediator of inflammation, immune suppression and skin cancer. Prog Lipid Res. 2016;63:14–27. doi: 10.1016/j.plipres.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fabo EC, Noonan FP, Fears T, Merlino G. Ultraviolet B but not ultraviolet A radiation initiates melanoma. Cancer Res. 2004;64:6372–6376. doi: 10.1158/0008-5472.CAN-04-1454. [DOI] [PubMed] [Google Scholar]
- de Laat A, van der Leun JC, de Gruijl FR. Carcinogenesis induced by UVA (365-nm) radiation: the dose-time dependence of tumor formation in hairless mice. Carcinogenesis. 1997;18:1013–1020. doi: 10.1093/carcin/18.5.1013. [DOI] [PubMed] [Google Scholar]
- Dedon PC. The chemical toxicology of 2-deoxyribose oxidation in DNA. Chem Res Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- Denat L, Kadekaro AL, Marrot L, Leachman SA, Abdel-Malek ZA. Melanocytes as instigators and victims of oxidative stress. J Invest Dermatol. 2014;134:1512–1518. doi: 10.1038/jid.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer FA, O'Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, Ljungman M. RPA and ATR link transcriptional stress to p53. Proc Natl Acad Sci U S A. 2007;104:12778–12783. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffey BL. Sources and measurement of ultraviolet radiation. Methods. 2002;28:4–13. doi: 10.1016/s1046-2023(02)00204-9. [DOI] [PubMed] [Google Scholar]
- Ding W, Liu W, Cooper KL, Qin XJ, de Souza Bergo PL, Hudson LG, Liu KJ. Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J Biol Chem. 2009;284:6809–6817. doi: 10.1074/jbc.M805566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombernowsky SL, Weischer M, Allin KH, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Risk of cancer by ATM missense mutations in the general population. J Clin Oncol. 2008;26:3057–3062. doi: 10.1200/JCO.2007.14.6613. [DOI] [PubMed] [Google Scholar]
- D'Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, Ito S, Fisher DE. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- Dore JF, Chignol MC. Tanning salons and skin cancer. Photochem Photobiol Sci. 2012;11:30–37. doi: 10.1039/c1pp05186e. [DOI] [PubMed] [Google Scholar]
- Douki T. Relative Contributions of UVB and UVA to the Photoconversion of (6-4) Photoproducts into their Dewar Valence Isomers. Photochem Photobiol. 2016;92:587–594. doi: 10.1111/php.12605. [DOI] [PubMed] [Google Scholar]
- Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- Douki T, Sage E. Dewar valence isomers, the third type of environmentally relevant DNA photoproducts induced by solar radiation. Photochem Photobiol Sci. 2016;15:24–30. doi: 10.1039/c5pp00382b. [DOI] [PubMed] [Google Scholar]
- Drobetsky EA, Turcotte J, Chateauneuf A. A role for ultraviolet A in solar mutagenesis. Proc Natl Acad Sci U S A. 1995;92:2350–2354. doi: 10.1073/pnas.92.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvers I, Johansson F, Groth P, Erixon K, Helleday T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 2011;39:7049–7057. doi: 10.1093/nar/gkr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga IC, Groenendijk RT, Filon AR, van Zeeland AA, Simons JW. The wavelength dependence of u.v.-induced pyrimidine dimer formation, cell killing and mutation induction in human diploid skin fibroblasts. Carcinogenesis. 1986;7:1829–1836. doi: 10.1093/carcin/7.11.1829. [DOI] [PubMed] [Google Scholar]
- Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348:1681–1691. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattouh K, Ducroux E, Decullier E, Kanitakis J, Morelon E, Boissonnat P, Sebbag L, Jullien D, Euvrard S. Increasing incidence of melanoma after solid organ transplantation: a retrospective epidemiological study. Transpl Int. 2017;30:1172–1180. doi: 10.1111/tri.13011. [DOI] [PubMed] [Google Scholar]
- Fears TR, Scotto J, Schneiderman MA. Mathematical models of age and ultraviolet effects on the incidence of skin cancer among whites in the United States. Am J Epidemiol. 1977;105:420–427. doi: 10.1093/oxfordjournals.aje.a112400. [DOI] [PubMed] [Google Scholar]
- Fell GL, Robinson KC, Mao J, Woolf CJ, Fisher DE. Skin beta-endorphin mediates addiction to UV light. Cell. 2014;157:1527–1534. doi: 10.1016/j.cell.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez TL, Van Lonkhuyzen DR, Dawson RA, Kimlin MG, Upton Z. Insulin-like growth factor-I and UVB photoprotection in human keratinocytes. Exp Dermatol. 2015;24:235–238. doi: 10.1111/exd.12637. [DOI] [PubMed] [Google Scholar]
- Fisher MS, Kripke ML. Systemic alteration induced in mice by ultraviolet light irradiation and its relationship to ultraviolet carcinogenesis. Proc Natl Acad Sci U S A. 1977;74:1688–1692. doi: 10.1073/pnas.74.4.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Freeman SE, Hacham H, Gange RW, Maytum DJ, Sutherland JC, Sutherland BM. Wavelength dependence of pyrimidine dimer formation in DNA of human skin irradiated in situ with ultraviolet light. Proc Natl Acad Sci U S A. 1989;86:5605–5609. doi: 10.1073/pnas.86.14.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Kellenberger E, Rogniaux H, Poterszman A, Van Dorsselaer A, Thierry JC, Egly JM, Moras D, Kieffer B. Structural characterization of the cysteine-rich domain of TFIIH p44 subunit. J Biol Chem. 2000;275:31963–31971. doi: 10.1074/jbc.M004960200. [DOI] [PubMed] [Google Scholar]
- Gaddameedhi S, Kemp MG, Reardon JT, Shields JM, Smith-Roe SL, Kaufmann WK, Sancar A. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 2010;70:4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddameedhi S, Selby CP, Kemp MG, Ye R, Sancar A. The circadian clock controls sunburn apoptosis and erythema in mouse skin. J Invest Dermatol. 2015;135:1119–1127. doi: 10.1038/jid.2014.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RP, Rivers JK, Lee TK, Bajdik CD, McLean DI, Coldman AJ. Broad-spectrum sunscreen use and the development of new nevi in white children: A randomized controlled trial. JAMA. 2000;283:2955–2960. doi: 10.1001/jama.283.22.2955. [DOI] [PubMed] [Google Scholar]
- Garcia-Borron JC, Abdel-Malek Z, Jimenez-Cervantes C. MC1R, the cAMP pathway, and the response to solar UV: extending the horizon beyond pigmentation. Pigment Cell Melanoma Res. 2014;27:699–720. doi: 10.1111/pcmr.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Borron JC, Olivares C. Melanocortin 1 receptor and skin pathophysiology: beyond colour, much more than meets the eye. Exp Dermatol. 2014;23:387–388. doi: 10.1111/exd.12310. [DOI] [PubMed] [Google Scholar]
- Garssen J, van Steeg H, de Gruijl F, de Boer J, van der Horst GT, van Kranen H, van Loveren H, van Dijk M, Fluitman A, Weeda G, Hoeijmakers JH. Transcription-coupled and global genome repair differentially influence UV-B-induced acute skin effects and systemic immunosuppression. J Immunol. 2000;164:6199–6205. doi: 10.4049/jimmunol.164.12.6199. [DOI] [PubMed] [Google Scholar]
- Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, Cam E, Millar SE, Smyth P, Ihler A, Takahashi JS, Andersen B. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A. 2012;109:11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Gilchrest BA, Blog FB, Szabo G. Effects of aging and chronic sun exposure on melanocytes in human skin. J Invest Dermatol. 1979;73:141–143. doi: 10.1111/1523-1747.ep12581580. [DOI] [PubMed] [Google Scholar]
- Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N Engl J Med. 1999;340:1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- Gilchrest BA, Park HY, Eller MS, Yaar M. Mechanisms of ultraviolet light-induced pigmentation. Photochem Photobiol. 1996;63:1–10. doi: 10.1111/j.1751-1097.1996.tb02988.x. [DOI] [PubMed] [Google Scholar]
- Girard PM, Graindorge D, Smirnova V, Rigolet P, Francesconi S, Scanlon S, Sage E. Oxidative stress in mammalian cells impinges on the cysteines redox state of human XRCC3 protein and on its cellular localization. PLoS One. 2013;8:e75751. doi: 10.1371/journal.pone.0075751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler T, Sabbioneda S, Green CM, Lehmann AR. ATR-mediated phosphorylation of DNA polymerase eta is needed for efficient recovery from UV damage. J Cell Biol. 2011;192:219–227. doi: 10.1083/jcb.201008076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Maglio DH, Paz ML, Leoni J. Sunlight Effects on Immune System: Is There Something Else in addition to UV-Induced Immunosuppression? Biomed Res Int. 2016;2016:1934518. doi: 10.1155/2016/1934518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AC, Olsen CM. Increased risk of melanoma in organ transplant recipients: systematic review and meta-analysis of cohort studies. Acta Derm Venereol. 2015;95:923–927. doi: 10.2340/00015555-2148. [DOI] [PubMed] [Google Scholar]
- Green AC, Williams GM, Logan V, Strutton GM. Reduced melanoma after regular sunscreen use: randomized trial follow-up. J Clin Oncol. 2011;29:257–263. doi: 10.1200/JCO.2010.28.7078. [DOI] [PubMed] [Google Scholar]
- Griewank KG, Murali R, Puig-Butille JA, Schilling B, Livingstone E, Potrony M, Carrera C, Schimming T, Moller I, Schwamborn M, Sucker A, Hillen U, Badenas C, Malvehy J, Zimmer L, Scherag A, Puig S, Schadendorf D. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju246. Print 2014 Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf C, Schwerdtle T, Mullenders LH, Hartwig A. Antimony impairs nucleotide excision repair: XPA and XPE as potential molecular targets. Chem Res Toxicol. 2010;23:1175–1183. doi: 10.1021/tx100106x. [DOI] [PubMed] [Google Scholar]
- Guan L, Suggs A, Ahsanuddin S, Tarrillion M, Selph J, Lam M, Baron E. 2016 Arte Poster Competition First Place Winner: Circadian Rhythm and UV-Induced Skin Damage: An In Vivo Study. J Drugs Dermatol. 2016;15:1124–1130. [PubMed] [Google Scholar]
- Gueranger Q, Li F, Peacock M, Larnicol-Fery A, Brem R, Macpherson P, Egly JM, Karran P. Protein oxidation and DNA repair inhibition by 6-thioguanine and UVA radiation. J Invest Dermatol. 2014;134:1408–1417. doi: 10.1038/jid.2013.509. [DOI] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- Guven M, Brem R, Macpherson P, Peacock M, Karran P. Oxidative Damage to RPA Limits the Nucleotide Excision Repair Capacity of Human Cells. J Invest Dermatol. 2015;135:2834–2841. doi: 10.1038/jid.2015.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy GP, Jr, Ekwueme DU, Tangka FK, Richardson LC. Melanoma treatment costs: a systematic review of the literature, 1990–2011. Am J Prev Med. 2012;43:537–545. doi: 10.1016/j.amepre.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hanasoge S, Ljungman M. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 2007;28:2298–2304. doi: 10.1093/carcin/bgm157. [DOI] [PubMed] [Google Scholar]
- Hardman JA, Tobin DJ, Haslam IS, Farjo N, Farjo B, Al-Nuaimi Y, Grimaldi B, Paus R. The peripheral clock regulates human pigmentation. J Invest Dermatol. 2015;135:1053–1064. doi: 10.1038/jid.2014.442. [DOI] [PubMed] [Google Scholar]
- Hauser JE, Kadekaro AL, Kavanagh RJ, Wakamatsu K, Terzieva S, Schwemberger S, Babcock G, Rao MB, Ito S, Abdel-Malek ZA. Melanin content and MC1R function independently affect UVR-induced DNA damage in cultured human melanocytes. Pigment Cell Res. 2006;19:303–314. doi: 10.1111/j.1600-0749.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- Hersey P, MacDonald M, Burns C, Schibeci S, Matthews H, Wilkinson FJ. Analysis of the effect of a sunscreen agent on the suppression of natural killer cell activity induced in human subjects by radiation from solarium lamps. J Invest Dermatol. 1987;88:271–276. doi: 10.1111/1523-1747.ep12466123. [DOI] [PubMed] [Google Scholar]
- Hodak E, Gottlieb AB, Colen S, Anzilotti M, Krueger JG. In vivo expression of the insulin-like growth factor-I (IGF-I) receptor in congenital pigmented nevi. J Cutan Pathol. 1996;23:19–24. doi: 10.1111/j.1600-0560.1996.tb00772.x. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, Hammer RE, Richardson JA, Baynash AG, Cheung JC, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Howard BD, Tessman I. Identification of the Altered Bases in Mutated Single-Stranded Dna. 3. Mutagenesis by Ultraviolet Light. J Mol Biol. 1964;9:372–375. doi: 10.1016/s0022-2836(64)80214-x. [DOI] [PubMed] [Google Scholar]
- Hu J, Adar S. The Cartography of UV-induced DNA Damage Formation and DNA Repair. Photochem Photobiol. 2017;93:199–206. doi: 10.1111/php.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Adebali O, Adar S, Sancar A. Dynamic maps of UV damage formation and repair for the human genome. Proc Natl Acad Sci U S A. 2017;114:6758–6763. doi: 10.1073/pnas.1706522114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Choi JH, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A. Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5' and the 6th phosphodiester bond 3' to the photodimer. Proc Natl Acad Sci U S A. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- Ikehata H, Kawai K, Komura J, Sakatsume K, Wang L, Imai M, Higashi S, Nikaido O, Yamamoto K, Hieda K, Watanabe M, Kasai H, Ono T. UVA1 genotoxicity is mediated not by oxidative damage but by cyclobutane pyrimidine dimers in normal mouse skin. J Invest Dermatol. 2008;128:2289–2296. doi: 10.1038/jid.2008.61. [DOI] [PubMed] [Google Scholar]
- Imokawa G, Yada Y, Miyagishi M. Endothelins secreted from human keratinocytes are intrinsic mitogens for human melanocytes. J Biol Chem. 1992;267:24675–24680. [PubMed] [Google Scholar]
- Iyer DR, Rhind N. The Intra-S Checkpoint Responses to DNA Damage. Genes (Basel) 2017;8 doi: 10.3390/genes8020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Garinis GA, Schul W, van Oudenaren A, Moorhouse M, Smid M, Sert YG, van der Velde A, Rijksen Y, de Gruijl FR, van der Spek PJ, Yasui A, Hoeijmakers JH, Leenen PJ, van der Horst GT. Differential role of basal keratinocytes in UV-induced immunosuppression and skin cancer. Mol Cell Biol. 2006;26:8515–8526. doi: 10.1128/MCB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, Carter KM, D'Orazio JA. Paracrine regulation of melanocyte genomic stability: a focus on nucleotide excision repair. Pigment Cell Melanoma Res. 2017;30:284–293. doi: 10.1111/pcmr.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, D'Orazio JA. Hormonal Regulation of the Repair of UV Photoproducts in Melanocytes by the Melanocortin Signaling Axis. Photochem Photobiol. 2017;93:245–258. doi: 10.1111/php.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, Wolf Horrell EM, Boulanger MC, D'Orazio JA. Defining the Contribution of MC1R Physiological Ligands to ATR Phosphorylation at Ser435, a Predictor of DNA Repair in Melanocytes. J Invest Dermatol. 2015;135:3086–3095. doi: 10.1038/jid.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, Wolf Horrell EM, Christian PA, Vanover JC, Boulanger MC, Zou Y, D'Orazio JA. PKA-mediated phosphorylation of ATR promotes recruitment of XPA to UV-induced DNA damage. Mol Cell. 2014;54:999–1011. doi: 10.1016/j.molcel.2014.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jarrett SG, Wolf Horrell EM, D'Orazio JA. AKAP12 mediates PKA-induced phosphorylation of ATR to enhance nucleotide excision repair. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw871. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol Cell Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadekaro AL, Chen J, Yang J, Chen S, Jameson J, Swope VB, Cheng T, Kadakia M, Abdel-Malek Z. Alpha-melanocyte-stimulating hormone suppresses oxidative stress through a p53-mediated signaling pathway in human melanocytes. Mol Cancer Res. 2012;10:778–786. doi: 10.1158/1541-7786.MCR-11-0436. [DOI] [PubMed] [Google Scholar]
- Kadekaro AL, Kanto H, Kavanagh R, Abdel-Malek Z. Significance of the melanocortin 1 receptor in regulating human melanocyte pigmentation, proliferation, and survival. Ann N Y Acad Sci. 2003;994:359–365. doi: 10.1111/j.1749-6632.2003.tb03200.x. [DOI] [PubMed] [Google Scholar]
- Kadekaro AL, Kavanagh R, Kanto H, Terzieva S, Hauser J, Kobayashi N, Schwemberger S, Cornelius J, Babcock G, Shertzer HG, Scott G, Abdel-Malek ZA. alpha-Melanocortin and endothelin-1 activate antiapoptotic pathways and reduce DNA damage in human melanocytes. Cancer Res. 2005;65:4292–4299. doi: 10.1158/0008-5472.CAN-04-4535. [DOI] [PubMed] [Google Scholar]
- Kadekaro AL, Leachman S, Kavanagh RJ, Swope V, Cassidy P, Supp D, Sartor M, Schwemberger S, Babcock G, Wakamatsu K, Ito S, Koshoffer A, Boissy RE, Manga P, Sturm RA, Abdel-Malek ZA. Melanocortin 1 receptor genotype: an important determinant of the damage response of melanocytes to ultraviolet radiation. FASEB J. 2010;24:3850–3860. doi: 10.1096/fj.10-158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Hizi A, Kasai H, Loeb LA. Incorporation of the guanosine triphosphate analogs 8-oxo-dGTP and 8-NH2-dGTP by reverse transcriptases and mammalian DNA polymerases. J Biol Chem. 1997;272:5892–5898. doi: 10.1074/jbc.272.9.5892. [DOI] [PubMed] [Google Scholar]
- Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci U S A. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Reardon JT, Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011;39:3176–3187. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes UP, Luo D, Potter M, Schulmeister K, Runger TM. Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J Invest Dermatol. 2006;126:667–675. doi: 10.1038/sj.jid.5700093. [DOI] [PubMed] [Google Scholar]
- Karagas MR, Stukel TA, Umland V, Tsoukas MM, Mott LA, Sorensen HT, Jensen AO, Nelson HH, Spencer SK, Perry AE, Stern RS. Reported use of photosensitizing medications and basal cell and squamous cell carcinoma of the skin: results of a population-based case-control study. J Invest Dermatol. 2007;127:2901–2903. doi: 10.1038/sj.jid.5700934. [DOI] [PubMed] [Google Scholar]
- Karran P, Brem R. Protein oxidation, UVA and human DNA repair. DNA Repair (Amst) 2016;44:178–185. doi: 10.1016/j.dnarep.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK. The human intra-S checkpoint response to UVC-induced DNA damage. Carcinogenesis. 2010;31:751–765. doi: 10.1093/carcin/bgp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK, Carson CC, Omolo B, Filgo AJ, Sambade MJ, Simpson DA, Shields JM, Ibrahim JG, Thomas NE. Mechanisms of chromosomal instability in melanoma. Environ Mol Mutagen. 2014;55:457–471. doi: 10.1002/em.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK, Nevis KR, Qu P, Ibrahim JG, Zhou T, Zhou Y, Simpson DA, Helms-Deaton J, Cordeiro-Stone M, Moore DT, Thomas NE, Hao H, Liu Z, Shields JM, Scott GA, Sharpless NE. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128:175–187. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr Mol Pharmacol. 2012;5:36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG. DNA damage-induced ATM- and Rad-3-related (ATR) kinase activation in non-replicating cells is regulated by the XPB subunit of transcription factor IIH (TFIIH) J Biol Chem. 2017;292:12424–12435. doi: 10.1074/jbc.M117.788406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Gaddameedhi S, Choi JH, Hu J, Sancar A. DNA repair synthesis and ligation affect the processing of excised oligonucleotides generated by human nucleotide excision repair. J Biol Chem. 2014;289:26574–26583. doi: 10.1074/jbc.M114.597088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Lindsey-Boltz LA, Sancar A. The DNA damage response kinases DNA-dependent protein kinase (DNA-PK) and ataxia telangiectasia mutated (ATM) Are stimulated by bulky adduct-containing DNA. J Biol Chem. 2011;286:19237–19246. doi: 10.1074/jbc.M111.235036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem. 2012;287:22889–22899. doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Sancar A. ATR Kinase Inhibition Protects Non-cycling Cells from the Lethal Effects of DNA Damage and Transcription Stress. J Biol Chem. 2016;291:9330–9342. doi: 10.1074/jbc.M116.719740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Sancar A. DNA excision repair: where do all the dimers go? Cell Cycle. 2012;11:2997–3002. doi: 10.4161/cc.21126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Spandau DF, Simman R, Travers JB. Insulin-like Growth Factor 1 Receptor Signaling Is Required for Optimal ATR-CHK1 Kinase Signaling in Ultraviolet B (UVB)-irradiated Human Keratinocytes. J Biol Chem. 2017a;292:1231–1239. doi: 10.1074/jbc.M116.765883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Spandau DF, Travers JB. Impact of Age and Insulin-Like Growth Factor-1 on DNA Damage Responses in UV-Irradiated Human Skin. Molecules. 2017b;22 doi: 10.3390/molecules22030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse SM, Moss SH, Davies DJ. Action spectra for inactivation of normal and xeroderma pigmentosum human skin fibroblasts by ultraviolet radiations. Photochem Photobiol. 1983;37:307–312. doi: 10.1111/j.1751-1097.1983.tb04478.x. [DOI] [PubMed] [Google Scholar]
- Kipp C, Young AR. The soluble eumelanin precursor 5,6-dihydroxyindole-2-carboxylic acid enhances oxidative damage in human keratinocyte DNA after UVA irradiation. Photochem Photobiol. 1999;70:191–198. [PubMed] [Google Scholar]
- Kitagawa R, Kastan MB. The ATM-dependent DNA damage signaling pathway. Cold Spring Harb Symp Quant Biol. 2005;70:99–109. doi: 10.1101/sqb.2005.70.002. [DOI] [PubMed] [Google Scholar]
- Kohler C, Koalick D, Fabricius A, Parplys AC, Borgmann K, Pospiech H, Grosse F. Cdc45 is limiting for replication initiation in humans. Cell Cycle. 2016;15:974–985. doi: 10.1080/15384101.2016.1152424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolgen W, van Steeg H, van der Horst GT, Hoeijmakers JH, van Vloten WA, de Gruijl FR, Garssen J. Association of transcription-coupled repair but not global genome repair with ultraviolet-B-induced Langerhans cell depletion and local immunosuppression. J Invest Dermatol. 2003;121:751–756. doi: 10.1046/j.1523-1747.2003.12476.x. [DOI] [PubMed] [Google Scholar]
- Kraemer KH. Sunlight and skin cancer: another link revealed. Proc Natl Acad Sci U S A. 1997;94:11–14. doi: 10.1073/pnas.94.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kripke ML, Cox PA, Alas LG, Yarosh DB. Pyrimidine dimers in DNA initiate systemic immunosuppression in UV-irradiated mice. Proc Natl Acad Sci U S A. 1992;89:7516–7520. doi: 10.1073/pnas.89.16.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica AW, Brewer JD. Melanoma in immunosuppressed patients. Mayo Clin Proc. 2012;87:991–1003. doi: 10.1016/j.mayocp.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisada M, Sakumi K, Tominaga Y, Budiyanto A, Ueda M, Ichihashi M, Nakabeppu Y, Nishigori C. 8-Oxoguanine formation induced by chronic UVB exposure makes Ogg1 knockout mice susceptible to skin carcinogenesis. Cancer Res. 2005;65:6006–6010. doi: 10.1158/0008-5472.CAN-05-0724. [DOI] [PubMed] [Google Scholar]
- Kvam E, Dahle J. Melanin synthesis may sensitize melanocytes to oxidative DNA damage by ultraviolet A radiation and protect melanocytes from direct DNA damage by ultraviolet B radiation. Pigment Cell Res. 2004;17:549–550. doi: 10.1111/j.1600-0749.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Kvam E, Tyrrell RM. The role of melanin in the induction of oxidative DNA base damage by ultraviolet A irradiation of DNA or melanoma cells. J Invest Dermatol. 1999;113:209–213. doi: 10.1046/j.1523-1747.1999.00653.x. [DOI] [PubMed] [Google Scholar]
- Lambert WC, Kuo HR, Lambert MW. Xeroderma pigmentosum. Dermatol Clin. 1995;13:169–209. [PubMed] [Google Scholar]
- Le Clair MZ, Cockburn MG. Tanning bed use and melanoma: Establishing risk and improving prevention interventions. Prev Med Rep. 2016;3:139–144. doi: 10.1016/j.pmedr.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Park JM, Leem SH, Kang TH. Coordinated regulation of XPA stability by ATR and HERC2 during nucleotide excision repair. Oncogene. 2014;33:19–25. doi: 10.1038/onc.2012.539. [DOI] [PubMed] [Google Scholar]
- Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancer--the role of sunlight. Adv Exp Med Biol. 2008;624:89–103. doi: 10.1007/978-0-387-77574-6_8. [DOI] [PubMed] [Google Scholar]
- Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol. 2004;150:179–185. doi: 10.1111/j.1365-2133.2004.05708.x. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Travers JB, Somani AK, Spandau DF. The IGF-1/IGF-1R signaling axis in the skin: a new role for the dermis in aging-associated skin cancer. Oncogene. 2010;29:1475–1485. doi: 10.1038/onc.2009.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Travers JB, Spandau DF. A new paradigm for the role of aging in the development of skin cancer. J Invest Dermatol. 2009;129:787–791. doi: 10.1038/jid.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Yi Q, Travers JB, Spandau DF. UVB-induced senescence in human keratinocytes requires a functional insulin-like growth factor-1 receptor and p53. Mol Biol Cell. 2008;19:1346–1353. doi: 10.1091/mbc.E07-10-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RD. Dose response for ultraviolet radiation A-induced focal melanocytic hyperplasia and nonmelanoma skin tumors in Monodelphis domestica. Photochem Photobiol. 2001;73:20–23. doi: 10.1562/0031-8655(2001)073<0020:drfura>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Li Z, Musich PR, Cartwright BM, Wang H, Zou Y. UV-induced nuclear import of XPA is mediated by importin-alpha4 in an ATR-dependent manner. PLoS One. 2013;8:e68297. doi: 10.1371/journal.pone.0068297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Musich PR, Serrano MA, Dong Z, Zou Y. XPA-mediated regulation of global nucleotide excision repair by ATR Is p53-dependent and occurs primarily in S-phase. PLoS One. 2011;6:e28326. doi: 10.1371/journal.pone.0028326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P, Paus R, Takahashi JS, Andersen B. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009;5:e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Kemp MG, Reardon JT, DeRocco V, Iyer RR, Modrich P, Sancar A. Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J Biol Chem. 2014;289:5074–5082. doi: 10.1074/jbc.M113.542787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem Sci. 2012;37:162–172. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesch MM, Collier AE, Southern DH, Ward RE, Tholpady SS, Lewis DA, Travers JB, Spandau DF. Insulin-like growth factor-1 receptor regulates repair of ultraviolet B-induced DNA damage in human keratinocytes in vivo. Mol Oncol. 2016;10:1245–1254. doi: 10.1016/j.molonc.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zhang J, Nagahawatte P, Easton J, Lee S, Liu Z, Ding L, Wyczalkowski MA, Valentine M, Navid F, Mulder H, Tatevossian RG, Dalton J, Davenport J, Yin Z, Edmonson M, Rusch M, Wu G, Li Y, Parker M, Hedlund E, Shurtleff S, Raimondi S, Bhavin V, Donald Y, Mardis ER, Wilson RK, Evans WE, Ellison DW, Pounds S, Dyer M, Downing JR, Pappo A, Bahrami A. The genomic landscape of childhood and adolescent melanoma. J Invest Dermatol. 2015;135:816–823. doi: 10.1038/jid.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20(Suppl 6):vi1–7. doi: 10.1093/annonc/mdp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzella N, Bracci M, Strafella E, Staffolani S, Ciarapica V, Copertaro A, Rapisarda V, Ledda C, Amati M, Valentino M, Tomasetti M, Stevens RG, Santarelli L. Circadian Modulation of 8-Oxoguanine DNA Damage Repair. Sci Rep. 2015;5:13752. doi: 10.1038/srep13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2016;113:9057–9062. doi: 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Falconi MM. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci U S A. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci U S A. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui MS, Pelle E, Dong K, Pernodet N. Biological Rhythms in the Skin. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17060801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, Inobe M, Matsunaga T. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Matthews YJ, Halliday GM, Phan TA, Damian DL. A UVB wavelength dependency for local suppression of recall immunity in humans demonstrates a peak at 300 nm. J Invest Dermatol. 2010a;130:1680–1684. doi: 10.1038/jid.2010.27. [DOI] [PubMed] [Google Scholar]
- Matthews YJ, Halliday GM, Phan TA, Damian DL. Wavelength dependency for UVA-induced suppression of recall immunity in humans. J Dermatol Sci. 2010b;59:192–197. doi: 10.1016/j.jdermsci.2010.07.005. [DOI] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, Jordan SA, Jackson IJ, Korsmeyer SJ, Golub TR, Fisher DE. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Men L, Roginskaya M, Zou Y, Wang Y. Redox-dependent formation of disulfide bonds in human replication protein A. Rapid Commun Mass Spectrom. 2007;21:2743–2749. doi: 10.1002/rcm.3144. [DOI] [PubMed] [Google Scholar]
- Mitchell D, Fernandez A. The photobiology of melanocytes modulates the impact of UVA on sunlight-induced melanoma. Photochem Photobiol Sci. 2012;11:69–73. doi: 10.1039/c1pp05146f. [DOI] [PubMed] [Google Scholar]
- Mitchell DL, Fernandez AA, Nairn RS, Garcia R, Paniker L, Trono D, Thames HD, Gimenez-Conti I. Ultraviolet A does not induce melanomas in a Xiphophorus hybrid fish model. Proc Natl Acad Sci U S A. 2010;107:9329–9334. doi: 10.1073/pnas.1000324107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi-Hashimoto H, Tanaka K, Horio T. Enhanced inflammation and immunosuppression by ultraviolet radiation in xeroderma pigmentosum group A (XPA) model mice. J Invest Dermatol. 1996;107:343–348. doi: 10.1111/1523-1747.ep12363295. [DOI] [PubMed] [Google Scholar]
- Montaner B, O'Donovan P, Reelfs O, Perrett CM, Zhang X, Xu YZ, Ren X, Macpherson P, Frith D, Karran P. Reactive oxygen-mediated damage to a human DNA replication and repair protein. EMBO Rep. 2007;8:1074–1079. doi: 10.1038/sj.embor.7401084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreall J, Limpose K, Sheppard C, Kow YW, Werner E, Doetsch PW. Inactivation of a common OGG1 variant by TNF-alpha in mammalian cells. DNA Repair (Amst) 2015;26:15–22. doi: 10.1016/j.dnarep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci U S A. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouret S, Forestier A, Douki T. The specificity of UVA-induced DNA damage in human melanocytes. Photochem Photobiol Sci. 2012;11:155–162. doi: 10.1039/c1pp05185g. [DOI] [PubMed] [Google Scholar]
- Munoz MJ, Nieto Moreno N, Giono LE, Cambindo Botto AE, Dujardin G, Bastianello G, Lavore S, Torres-Mendez A, Menck CF, Blencowe BJ, Irimia M, Foiani M, Kornblihtt AR. Major Roles for Pyrimidine Dimers, Nucleotide Excision Repair, and ATR in the Alternative Splicing Response to UV Irradiation. Cell Rep. 2017;18:2868–2879. doi: 10.1016/j.celrep.2017.02.066. [DOI] [PubMed] [Google Scholar]
- Murray HC, Maltby VE, Smith DW, Bowden NA. Nucleotide excision repair deficiency in melanoma in response to UVA. Exp Hematol Oncol. 2016;5 doi: 10.1186/s40164-016-0035-4. 6-016-0035-4. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Costa A, Moita LF. Modulation of inflammation and disease tolerance by DNA damage response pathways. FEBS J. 2017;284:680–698. doi: 10.1111/febs.13910. [DOI] [PubMed] [Google Scholar]
- Nghiem DX, Kazimi N, Mitchell DL, Vink AA, Ananthaswamy HN, Kripke ML, Ullrich SE. Mechanisms underlying the suppression of established immune responses by ultraviolet radiation. J Invest Dermatol. 2002;119:600–608. doi: 10.1046/j.1523-1747.2002.01845.x. [DOI] [PubMed] [Google Scholar]
- Noonan FP, Dudek J, Merlino G, De Fabo EC. Animal models of melanoma: an HGF/SF transgenic mouse model may facilitate experimental access to UV initiating events. Pigment Cell Res. 2003;16:16–25. doi: 10.1034/j.1600-0749.2003.00014.x. [DOI] [PubMed] [Google Scholar]
- Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, De Fabo EC, Merlino G. Neonatal sunburn and melanoma in mice. Nature. 2001;413:271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- Noonan FP, Zaidi MR, Wolnicka-Glubisz A, Anver MR, Bahn J, Wielgus A, Cadet J, Douki T, Mouret S, Tucker MA, Popratiloff A, Merlino G, De Fabo EC. Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat Commun. 2012;3:884. doi: 10.1038/ncomms1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund JJ. The melanocyte and the epidermal melanin unit: an expanded concept. Dermatol Clin. 2007;25:271–81. vii. doi: 10.1016/j.det.2007.04.001. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Olsen CM, Wilson LF, Green AC, Biswas N, Loyalka J, Whiteman DC. Prevention of DNA damage in human skin by topical sunscreens. Photodermatol Photoimmunol Photomed. 2017;33:135–142. doi: 10.1111/phpp.12298. [DOI] [PubMed] [Google Scholar]
- Park JM, Kang TH. Transcriptional and Posttranslational Regulation of Nucleotide Excision Repair: The Guardian of the Genome against Ultraviolet Radiation. Int J Mol Sci. 2016;17:E1840. doi: 10.3390/ijms17111840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015;84:711–738. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- Peng W, Shaw BR. Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC-->TT transitions. Biochemistry. 1996;35:10172–10181. doi: 10.1021/bi960001x. [DOI] [PubMed] [Google Scholar]
- Perdiz D, Grof P, Mezzina M, Nikaido O, Moustacchi E, Sage E. Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J Biol Chem. 2000;275:26732–26742. doi: 10.1074/jbc.M001450200. [DOI] [PubMed] [Google Scholar]
- Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, Wong JW. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532:259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- Perrett CM, Walker SL, O'Donovan P, Warwick J, Harwood CA, Karran P, McGregor JM. Azathioprine treatment photosensitizes human skin to ultraviolet A radiation. Br J Dermatol. 2008;159:198–204. doi: 10.1111/j.1365-2133.2008.08610.x. [DOI] [PubMed] [Google Scholar]
- Peyrin-Biroulet L, Khosrotehrani K, Carrat F, Bouvier AM, Chevaux JB, Simon T, Carbonnel F, Colombel JF, Dupas JL, Godeberge P, Hugot JP, Lemann M, Nahon S, Sabate JM, Tucat G, Beaugerie L Cesame Study Group. Increased risk for nonmelanoma skin cancers in patients who receive thiopurines for inflammatory bowel disease. Gastroenterology. 2011;141:1621–28. e1–5. doi: 10.1053/j.gastro.2011.06.050. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Besaratinia A. UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem Photobiol Sci. 2012;11:90–97. doi: 10.1039/c1pp05144j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Van Spyk EN, Pham K, Geyfman M, Kumar V, Takahashi JS, Andersen B. The circadian clock in skin: implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J Biol Rhythms. 2015;30:163–182. doi: 10.1177/0748730414563537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos RC, Thoms JA, Guan YF, Unnikrishnan A, Pimanda JE, Wong JW. Functional Mutations Form at CTCF-Cohesin Binding Sites in Melanoma Due to Uneven Nucleotide Excision Repair across the Motif. Cell Rep. 2016;17:2865–2872. doi: 10.1016/j.celrep.2016.11.055. [DOI] [PubMed] [Google Scholar]
- Prasad R, Katiyar SK. Crosstalk Among UV-Induced Inflammatory Mediators, DNA Damage and Epigenetic Regulators Facilitates Suppression of the Immune System. Photochem Photobiol. 2017;93:930–936. doi: 10.1111/php.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premi S, Brash DE. Chemical excitation of electrons: A dark path to melanoma. DNA Repair (Amst) 2016;44:169–177. doi: 10.1016/j.dnarep.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premi S, Wallisch S, Mano CM, Weiner AB, Bacchiocchi A, Wakamatsu K, Bechara EJ, Halaban R, Douki T, Brash DE. Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science. 2015;347:842–847. doi: 10.1126/science.1256022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet A, Vessoni AT, Rocha CR, Gottifredi V, Biard D, Sarasin A, Menck CF, Stary A. Gap-filling and bypass at the replication fork are both active mechanisms for tolerance of low-dose ultraviolet-induced DNA damage in the human genome. DNA Repair (Amst) 2014;14:27–38. doi: 10.1016/j.dnarep.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Rai P. Oxidation in the nucleotide pool, the DNA damage response and cellular senescence: Defective bricks build a defective house. Mutat Res. 2010;703:71–81. doi: 10.1016/j.mrgentox.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Ramiscal JA, Brewer JD. Thiopurines and risk of nonmelanoma skin cancer in inflammatory bowel disease. JAMA Dermatol. 2013;149:92–94. doi: 10.1001/2013.jamadermatol.616. [DOI] [PubMed] [Google Scholar]
- Ray A, Blevins C, Wani G, Wani AA. ATR- and ATM-Mediated DNA Damage Response Is Dependent on Excision Repair Assembly during G1 but Not in S Phase of Cell Cycle. PLoS One. 2016;11:e0159344. doi: 10.1371/journal.pone.0159344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- Recio JA, Noonan FP, Takayama H, Anver MR, Duray P, Rush WL, Lindner G, De Fabo EC, DePinho RA, Merlino G. Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis. Cancer Res. 2002;62:6724–6730. [PubMed] [Google Scholar]
- Robinson ES, Hill RH, Jr, Kripke ML, Setlow RB. The Monodelphis melanoma model: initial report on large ultraviolet A exposures of suckling young. Photochem Photobiol. 2000;71:743–746. doi: 10.1562/0031-8655(2000)071<0743:tmmmir>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Robinson SN, Zens MS, Perry AE, Spencer SK, Duell EJ, Karagas MR. Photosensitizing agents and the risk of non-melanoma skin cancer: a population-based case-control study. J Invest Dermatol. 2013;133:1950–1955. doi: 10.1038/jid.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeck U, Herlyn M, Menssen HD, Furlanetto RW, Koprowsk H. Metastatic but not primary melanoma cell lines grow in vitro independently of exogenous growth factors. Int J Cancer. 1987;40:687–690. doi: 10.1002/ijc.2910400520. [DOI] [PubMed] [Google Scholar]
- Runger TM, Farahvash B, Hatvani Z, Rees A. Comparison of DNA damage responses following equimutagenic doses of UVA and UVB: a less effective cell cycle arrest with UVA may render UVA-induced pyrimidine dimers more mutagenic than UVB-induced ones. Photochem Photobiol Sci. 2012;11:207–215. doi: 10.1039/c1pp05232b. [DOI] [PubMed] [Google Scholar]
- Ruthemann P, Balbo Pogliano C, Naegeli H. Global-genome Nucleotide Excision Repair Controlled by Ubiquitin/Sumo Modifiers. Front Genet. 2016;7:68. doi: 10.3389/fgene.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- Sage E, Girard PM, Francesconi S. Unravelling UVA-induced mutagenesis. Photochem Photobiol Sci. 2012;11:74–80. doi: 10.1039/c1pp05219e. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Gaddameedhi S, Selby CP, Ye R, Chiou YY, Kemp MG, Hu J, Lee JH, Ozturk N. Circadian clock, cancer, and chemotherapy. Biochemistry. 2015;54:110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Kang TH, Reardon JT, Lee JH, Ozturk N. Circadian clock control of the cellular response to DNA damage. FEBS Lett. 2010;584:2618–2625. doi: 10.1016/j.febslet.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sandu C, Dumas M, Malan A, Sambakhe D, Marteau C, Nizard C, Schnebert S, Perrier E, Challet E, Pevet P, Felder-Schmittbuhl MP. Human skin keratinocytes, melanocytes, and fibroblasts contain distinct circadian clock machineries. Cell Mol Life Sci. 2012;69:3329–3339. doi: 10.1007/s00018-012-1026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent EV, Travers JB. Examining the differences in current regulatory processes for sunscreens and proposed safety assessment paradigm. Regul Toxicol Pharmacol. 2016;79:125–141. doi: 10.1016/j.yrtph.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci U S A. 2011;108:13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Grist E, Thompson K, Woodhead AD. Wavelengths effective in induction of malignant melanoma. Proc Natl Acad Sci U S A. 1993;90:6666–6670. doi: 10.1073/pnas.90.14.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SM, Li Z, Shkriabai N, Kvaratskhelia M, Brosey C, Serrano MA, Chazin WJ, Musich PR, Zou Y. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J Biol Chem. 2009;284:24213–24222. doi: 10.1074/jbc.M109.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenenberger DW. Cutaneous malignant melanoma: a primary care perspective. Am Fam Physician. 2012;85:161–168. [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Siddle K. Signalling by insulin and IGF receptors: supporting acts and new players. J Mol Endocrinol. 2011;47:R1–10. doi: 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- Slominski AT, Hardeland R, Reiter RJ. When the circadian clock meets the melanin pigmentary system. J Invest Dermatol. 2015;135:943–945. doi: 10.1038/jid.2014.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Paterson MC. Lethality and the induction and repair of DNA damage in far, mid or near UV-irradiated human fibroblasts: comparison of effects in normal, xeroderma pigmentosum and Bloom's syndrome cells. Photochem Photobiol. 1982;36:333–343. doi: 10.1111/j.1751-1097.1982.tb04383.x. [DOI] [PubMed] [Google Scholar]
- Song X, Mosby N, Yang J, Xu A, Abdel-Malek Z, Kadekaro AL. alpha-MSH activates immediate defense responses to UV-induced oxidative stress in human melanocytes. Pigment Cell Melanoma Res. 2009;22:809–818. doi: 10.1111/j.1755-148X.2009.00615.x. [DOI] [PubMed] [Google Scholar]
- Spoerri L, Brooks K, Chia K, Grossman G, Ellis JJ, Dahmer-Heath M, Skalamera D, Pavey S, Burmeister B, Gabrielli B. A novel ATM-dependent checkpoint defect distinct from loss of function mutation promotes genomic instability in melanoma. Pigment Cell Melanoma Res. 2016;29:329–339. doi: 10.1111/pcmr.12466. [DOI] [PubMed] [Google Scholar]
- Sugasawa K. Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat Res. 2010;685:29–37. doi: 10.1016/j.mrfmmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Suzuki I, Im S, Tada A, Scott C, Akcali C, Davis MB, Barsh G, Hearing V, Abdel-Malek Z. Participation of the melanocortin-1 receptor in the UV control of pigmentation. J Investig Dermatol Symp Proc. 1999;4:29–34. doi: 10.1038/sj.jidsp.5640177. [DOI] [PubMed] [Google Scholar]
- Swope V, Alexander C, Starner R, Schwemberger S, Babcock G, Abdel-Malek ZA. Significance of the melanocortin 1 receptor in the DNA damage response of human melanocytes to ultraviolet radiation. Pigment Cell Melanoma Res. 2014;27:601–610. doi: 10.1111/pcmr.12252. [DOI] [PubMed] [Google Scholar]
- Swope VB, Jameson JA, McFarland KL, Supp DM, Miller WE, McGraw DW, Patel MA, Nix MA, Millhauser GL, Babcock GF, Abdel-Malek ZA. Defining MC1R regulation in human melanocytes by its agonist alpha-melanocortin and antagonists agouti signaling protein and beta-defensin 3. J Invest Dermatol. 2012;132:2255–2262. doi: 10.1038/jid.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada A, Pereira E, Beitner-Johnson D, Kavanagh R, Abdel-Malek ZA. Mitogen- and ultraviolet-B-induced signaling pathways in normal human melanocytes. J Invest Dermatol. 2002;118:316–322. doi: 10.1046/j.0022-202x.2001.01694.x. [DOI] [PubMed] [Google Scholar]
- Tadokoro T, Yamaguchi Y, Batzer J, Coelho SG, Zmudzka BZ, Miller SA, Wolber R, Beer JZ, Hearing VJ. Mechanisms of skin tanning in different racial/ethnic groups in response to ultraviolet radiation. J Invest Dermatol. 2005;124:1326–1332. doi: 10.1111/j.0022-202X.2005.23760.x. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama H, La Rochelle WJ, Anver M, Bockman DE, Merlino G. Scatter factor/hepatocyte growth factor as a regulator of skeletal muscle and neural crest development. Proc Natl Acad Sci U S A. 1996;93:5866–5871. doi: 10.1073/pnas.93.12.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst) 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- Tresini M, Warmerdam DO, Kolovos P, Snijder L, Vrouwe MG, Demmers JA, van IJcken WF, Grosveld FG, Medema RH, Hoeijmakers JH, Mullenders LH, Vermeulen W, Marteijn JA. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523:53–58. doi: 10.1038/nature14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaalbi-Shtylik A, Moser J, Mullenders LH, Jansen JG, de Wind N. Persistently stalled replication forks inhibit nucleotide excision repair in trans by sequestering Replication protein A. Nucleic Acids Res. 2014;42:4406–4413. doi: 10.1093/nar/gkt1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Dammann R, Pfeifer GP. Sequence and time-dependent deamination of cytosine bases in UVB-induced cyclobutane pyrimidine dimers in vivo. J Mol Biol. 1998;284:297–311. doi: 10.1006/jmbi.1998.2176. [DOI] [PubMed] [Google Scholar]
- Tyrrell RM, Webb RB. Reduced dimer excision in bacteria following near ultraviolet (365 nm) radiation. Mutat Res. 1973;19:361–364. doi: 10.1016/0027-5107(73)90238-8. [DOI] [PubMed] [Google Scholar]
- Tyrrell RM, Werfelli P, Moraes EC. Lethal action of ultraviolet and visible (blue-violet) radiations at defined wavelengths on human lymphoblastoid cells: action spectra and interaction sites. Photochem Photobiol. 1984;39:183–189. doi: 10.1111/j.1751-1097.1984.tb03426.x. [DOI] [PubMed] [Google Scholar]
- Ullrich SE, Kripke ML, Ananthaswamy HN. Mechanisms underlying UV-induced immune suppression: implications for sunscreen design. Exp Dermatol. 2002;11(Suppl 1):13–16. doi: 10.1034/j.1600-0625.11.s.1.4.x. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Makhov AM, Griffith JD, Sancar A. Preferential binding of ATR protein to UV-damaged DNA. Proc Natl Acad Sci U S A. 2002;99:6673–6678. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- von Koschembahr AM, Swope VB, Starner RJ, Abdel-Malek ZA. Endothelin-1 protects human melanocytes from UV-induced DNA damage by activating JNK and p38 signalling pathways. Exp Dermatol. 2015;24:269–274. doi: 10.1111/exd.12638. [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Pines A, Overmeer RM, Hanada K, Mullenders LH. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J Cell Sci. 2011;124:435–446. doi: 10.1242/jcs.075325. [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Sasaki T, Matsumoto M, Nagaoka M, Inoue K, Inobe M, Horibata K, Tanaka K, Matsunaga T. Nucleotide excision repair-dependent DNA double-strand break formation and ATM signaling activation in mammalian quiescent cells. J Biol Chem. 2014;289:28730–28737. doi: 10.1074/jbc.M114.589747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhou X, Liu W, Sun X, Chen C, Hudson LG, Jian Liu K. Arsenite-induced ROS/RNS generation causes zinc loss and inhibits the activity of poly(ADP-ribose) polymerase-1. Free Radic Biol Med. 2013;61:249–256. doi: 10.1016/j.freeradbiomed.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, van Spyk E, Liu Q, Geyfman M, Salmans ML, Kumar V, Ihler A, Li N, Takahashi JS, Andersen B. Time-Restricted Feeding Shifts the Skin Circadian Clock and Alters UVB-Induced DNA Damage. Cell Rep. 2017;20:1061–1072. doi: 10.1016/j.celrep.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HT, Choi B, Tang MS. Melanocytes are deficient in repair of oxidative DNA damage and UV-induced photoproducts. Proc Natl Acad Sci U S A. 2010;107:12180–12185. doi: 10.1073/pnas.1005244107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, You JS, Lee SH. Role of zinc-finger motif in redox regulation of human replication protein A. Antioxid Redox Signal. 2001;3:657–669. doi: 10.1089/15230860152543005. [DOI] [PubMed] [Google Scholar]
- Wigan M, Pinder A, Giles N, Pavey S, Burgess A, Wong S, Sturm RA, Gabrielli B. A UVR-induced G2-phase checkpoint response to ssDNA gaps produced by replication fork bypass of unrepaired lesions is defective in melanoma. J Invest Dermatol. 2012;132:1681–1688. doi: 10.1038/jid.2012.41. [DOI] [PubMed] [Google Scholar]
- Willis J, Patel Y, Lentz BL, Yan S. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc Natl Acad Sci U S A. 2013;110:10592–10597. doi: 10.1073/pnas.1301445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wolf Horrell EM, Boulanger MC, D'Orazio JA. Melanocortin 1 Receptor: Structure, Function, and Regulation. Front Genet. 2016;7:95. doi: 10.3389/fgene.2016.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondrak GT, Jacobson MK, Jacobson EL. Endogenous UVA-photosensitizers: mediators of skin photodamage and novel targets for skin photoprotection. Photochem Photobiol Sci. 2006;5:215–237. doi: 10.1039/b504573h. [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, Curran T. Redox regulation of AP-1: a link between transcription factor signaling and DNA repair. Adv Exp Med Biol. 1996;387:69–75. [PubMed] [Google Scholar]
- Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao GG, Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci U S A. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada Y, Higuchi K, Imokawa G. Effects of endothelins on signal transduction and proliferation in human melanocytes. J Biol Chem. 1991;266:18352–18357. [PubMed] [Google Scholar]
- Yarosh DB. DNA repair, immunosuppression, and skin cancer. Cutis. 2004;74:10–13. [PubMed] [Google Scholar]
- Yohn JJ, Morelli JG, Walchak SJ, Rundell KB, Norris DA, Zamora MR. Cultured human keratinocytes synthesize and secrete endothelin-1. J Invest Dermatol. 1993;100:23–26. doi: 10.1111/1523-1747.ep12349932. [DOI] [PubMed] [Google Scholar]
- You JS, Wang M, Lee SH. Functional characterization of zinc-finger motif in redox regulation of RPA-ssDNA interaction. Biochemistry. 2000;39:12953–12958. doi: 10.1021/bi001206f. [DOI] [PubMed] [Google Scholar]
- Young AR, Potten CS, Nikaido O, Parsons PG, Boenders J, Ramsden JM, Chadwick CA. Human melanocytes and keratinocytes exposed to UVB or UVA in vivo show comparable levels of thymine dimers. J Invest Dermatol. 1998;111:936–940. doi: 10.1046/j.1523-1747.1998.00435.x. [DOI] [PubMed] [Google Scholar]
- Zaidi MR, De Fabo EC, Noonan FP, Merlino G. Shedding light on melanocyte pathobiology in vivo. Cancer Res. 2012;72:1591–1595. doi: 10.1158/0008-5472.CAN-11-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, Pandita RK, Charaka VK, Pandita TK, Kastan MB, Walker CL. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Qureshi AA, Geller AC, Frazier L, Hunter DJ, Han J. Use of tanning beds and incidence of skin cancer. J Clin Oncol. 2012;30:1588–1593. doi: 10.1200/JCO.2011.39.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Dutton-Regester K, Brown KM, Hayward NK. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res. 2016;29:266–283. doi: 10.1111/pcmr.12459. [DOI] [PubMed] [Google Scholar]
- Zhou X, Cooper KL, Sun X, Liu KJ, Hudson LG. Selective Sensitization of Zinc Finger Protein Oxidation by Reactive Oxygen Species through Arsenic Binding. J Biol Chem. 2015;290:18361–18369. doi: 10.1074/jbc.M115.663906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, Hudson LG. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J Biol Chem. 2011;286:22855–22863. doi: 10.1074/jbc.M111.232926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwald FO, Christenson LJ, Billingsley EM, Zeitouni NC, Ratner D, Bordeaux J, Patel MJ, Brown MD, Proby CM, Euvrard S, Otley CC, Stasko T Melanoma Working Group of The International Transplant Skin Cancer Collaborative and Skin Care in Organ Transplant Patients, Europe. Melanoma in solid organ transplant recipients. Am J Transplant. 2010;10:1297–1304. doi: 10.1111/j.1600-6143.2010.03078.x. [DOI] [PubMed] [Google Scholar]