Abstract
Background
Monogenean flatworms are the main ectoparasites of fishes. Representatives of the species-rich families Gyrodactylidae and Dactylogyridae, especially those infecting cichlid fishes and clariid catfishes, are important parasites in African aquaculture, even more so due to the massive anthropogenic translocation of their hosts worldwide. Several questions on their evolution, such as the phylogenetic position of Macrogyrodactylus and the highly speciose Gyrodactylus, remain unresolved with available molecular markers. Also, diagnostics and population-level research would benefit from the development of higher-resolution genetic markers. We aim to offer genetic resources for work on African monogeneans by providing mitogenomic data of four species (two belonging to Gyrodactylidae, two to Dactylogyridae), and analysing their gene sequences and gene order from a phylogenetic perspective.
Results
Using Illumina technology, the first four mitochondrial genomes of African monogeneans were assembled and annotated for the cichlid parasites Gyrodactylus nyanzae, Cichlidogyrus halli, Cichlidogyrus mbirizei (near-complete mitogenome) and the catfish parasite Macrogyrodactylus karibae (near-complete mitogenome). Complete nuclear ribosomal operons were also retrieved, as molecular vouchers. The start codon TTG is new for Gyrodactylus and for Dactylogyridae, as is the incomplete stop codon TA for Dactylogyridae. Especially the nad2 gene is promising for primer development. Gene order was identical for protein-coding genes and differed between the African representatives of these families only in a tRNA gene transposition. A mitochondrial phylogeny based on an alignment of nearly 12,500 bp including 12 protein-coding and two ribosomal RNA genes confirms that the Neotropical oviparous Aglaiogyrodactylus forficulatus takes a sister group position with respect to the other gyrodactylids, instead of the supposedly ‘primitive’ African Macrogyrodactylus. Inclusion of the African Gyrodactylus nyanzae confirms the paraphyly of Gyrodactylus. The position of the African dactylogyrid Cichlidogyrus is unresolved, although gene order suggests it is closely related to marine ancyrocephalines.
Conclusions
The amount of mitogenomic data available for gyrodactylids and dactylogyrids is increased by roughly one-third. Our study underscores the potential of mitochondrial genes and gene order in flatworm phylogenetics, and of next-generation sequencing for marker development for these non-model helminths for which few primers are available.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4893-5) contains supplementary material, which is available to authorized users.
Keywords: Cichlidae, Clariidae, Cichlidogyrus, Gene order, Gyrodactylus, Macrogyrodactylus, Mitogenome, Monogenea, Monopisthocotylea, Phylogenomics
Background
Ectoparasitic infections in bony fishes are dominated by monogeneans [1]. Among their most species-rich taxa are Gyrodactylidae and Dactylogyridae [2]. These include, respectively, the supergenera Gyrodactylus and Dactylogyrus, some of the most significant radiations of flatworm fish parasites [1]. Around 500 species of Gyrodactylus have been described at present ([3] and references therein), but the estimated species number is much higher [4]. These minute flatworms attach to their host by means of an opisthaptor, often used in monogenean taxonomy [5]. The resulting disruption of the epidermis may facilitate secondary infections by e.g. fungi or bacteria [6]. Some genera within these families, such as Gyrodactylus, Macrogyrodactylus, Dactylogyrus and Cichlidogyrus include fish pathogens, especially in captive-reared stocks and after anthropogenic co-introduction outside of their native range [2, 5, 7, 8]. In Africa, the most important aquaculture fishes are species of Cichlidae and Clariidae, including the Nile tilapia and the North African catfish, which have been introduced worldwide [9, 10]. These fish families are also relatively well-studied for monogenean parasites (e.g. [3, 11]). They harbour several originally African monogeneans that are widely distributed within and outside Africa, and that are important in the study of parasite ecology, evolution and invasion biology because of the economic and scientific importance of their hosts [12].
In view of the important threats that disease poses to the sustainable development of aquaculture in developing countries, a better monitoring and identification of aquatic pathogens is vital [13]. In Africa, better understanding of the diversity and ecology of fish parasites is needed to implement government policies on aquatic health management [14]. There is however a lack of monitoring, despite massive anthropogenic translocation of fishes that may lead to parasite co-introductions (e.g. [15]). Monogeneans, in particular, have been assessed as high-risk parasites in African aquaculture [16]. Since common procedures for the identification of these monogeneans are lethal to the host and require a high level of technical expertise, non-intrusive molecular diagnostics are called for (e.g. [17] for Cichlidogyrus). However, there is a lack of highly variable molecular markers for these animals [12].
In addition, the phylogenetic position of African monogenean lineages, including several endemic or recently discovered genera, is often poorly understood, also largely due to low phylogenetic coverage. For example, the currently most frequently used markers, situated in the nuclear ribosomal DNA region, have not fully resolved the position of the typically African Macrogyrodactylus. The representatives of this genus infect clariid catfishes, among other hosts [18, 19]. Malmberg [20] suggested, based on morphological data, that the genus comprises the earliest diverging lineage of gyrodactylids. This is a family of mainly viviparous monogeneans, although with some oviparous representatives [6]. However, mitogenomic phylogenetics recently suggested the Neotropical oviparous gyrodactylid Aglaiogyrodactylus forficulatus as sister to all other, viviparous, family members [21]. Also, Malmberg’s hypothesis was contradicted by nuclear phylogenetic data placing Macrogyrodactylus with other viviparous lineages [19]. Another long-standing issue in the phylogeny of this monogenean family, is the status of its most species-rich and well-studied genus, Gyrodactylus ([22] and references therein), first suggested to be paraphyletic by Kritsky & Boeger [23].
Recently, next-generation sequencing (NGS) approaches have facilitated marker development for non-model helminths [24]; this includes the assembly of mitogenomes for fish helminths [25, 26]. Here we want to apply this approach to the understudied, but highly diverse, African monogenean fauna. We targeted two common tilapia-infecting species of Cichlidogyrus (Dactylogyridae), the most speciose monogenean genus infecting African cichlid fishes [27]; one gyrodactylid parasite of cichlids; and a representative of Macrogyrodactylus. Through phylogenomic and gene order analysis, we address the following questions:
Are the Neotropical oviparous gyrodactylids still basal in a mitochondrial phylogeny when including the viviparous Macrogyrodactylus, which is supposedly the earliest divergent gyrodactylid lineage according to Malmberg [20]?
Does the phylogeny based on mitogenomic data confirm the paraphyly of Gyrodactylus?
Do the African representatives of Gyrodactylidae have the same gene order in their mitochondrial genome as the known Palearctic ones?
Do the African freshwater representatives of Dactylogyridae have the same gene order as seen in the only known dactylogyrid mitogenomes, from a Palearctic freshwater and an Indo-Pacific marine species?
Results
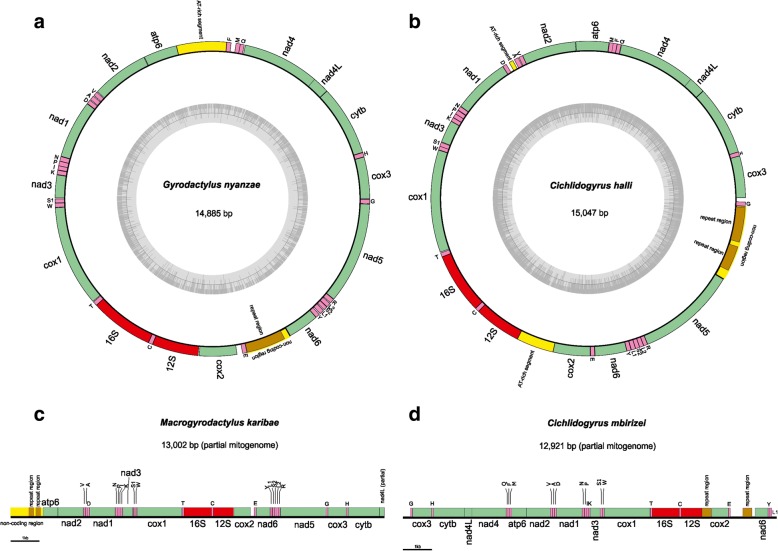
Genomic DNA sequencing on three quarters of a MiSeq v. 3 flowcell yielded 15,980,972 indexed paired-end 300 bp reads. Complete mitochondrial genomes were assembled for G. nyanzae (with a length of 14,885 base pairs (bp)) and C. halli (15,047 bp). A circular genome could not be assembled for C. mbirizei (12,921 bp) and M. karibae (13,002 bp) (Fig. 1). The annotated sequences were deposited in NCBI GenBank under accession numbers MG970255-8. The total number of reads mapped across all of the assembled mitochondrial genomes was 12,776, accounting for 0.8 % of the genomic readpool obtained, with an average coverage of 160, 31, 76 and 42 reads for G. nyanzae, C. halli, C. mbirizei and M. karibae, respectively. The coverage along the various protein-coding and ribosomal RNA (rRNA) genes is detailed in Table 1. All complete protein-coding genes (PCGs) were represented by a minimum of 15× coverage, with a minimum average coverage of 29× (Table 1). The ribosomal operons of G. nyanzae (6799 bp), M. karibae (6675 bp), C. halli (7496 bp) and C. mbirizei (7005 bp) were deposited as additional molecular vouchers for these species, under NCBI GenBank accession numbers MG973075-8; their annotation is provided in Additional file 1. We did not include these sequences in our phylogenetic analyses because of the lack of published complete ribosomal operons for other species represented.
Fig. 1.
Mitochondrial genomes of four African monogeneans, including two members of the Gyrodactylidae: (a) Gyrodactylus nyanzae, (c) Macrogyrodactylus karibae (partial genome) and two representatives of the Dactylogyridae: (b) Cichlidogyrus halli and (d) Cichlidogyrus mbirizei (partial genome). The GC content is displayed for complete mitogenomes
Table 1.
Minimum–maximum and average coverage (in number of reads) of the protein-coding and rRNA genes for the four assembled mitochondrial genomes
| Gyrodactylus nyanzae | Macrogyrodactylus karibae | Cichlidogyrus halli | Cichlidogyrus mbirizei | |||||
|---|---|---|---|---|---|---|---|---|
| Gene | Range | Average | Range | Average | Range | Average | Range | Average |
| cox3 | 151–184 | 165 | 21–48 | 39 | 41–59 | 51 | 101–131 | 118 |
| cytb | 136–174 | 155 | 16–44 | 29 | 51–74 | 61 | 115–165 | 139 |
| nad4L | 159–198 | 179 | – | – | 66–79 | 72 | 155–199 | 179 |
| nad4 | 125–217 | 177 | – | – | 42–75 | 56 | 112–187 | 140 |
| atp6 | 119–194 | 166 | 34–43 | 39 | 54–72 | 63 | 98–140 | 119 |
| nad2 | 130–195 | 167 | 36–56 | 47 | 49–68 | 56 | 121–177 | 149 |
| nad1 | 148–205 | 183 | 15–61 | 30 | 49–74 | 61 | 134–170 | 147 |
| nad3 | 151–175 | 163 | 27–44 | 35 | 48–72 | 59 | 152–180 | 166 |
| cox1 | 125–182 | 149 | 29–56 | 42 | 33–70 | 51 | 112–177 | 148 |
| 16S rRNA | 102–140 | 122 | 22–44 | 34 | 39–87 | 61 | 157–315 | 263 |
| 12S rRNA | 103–134 | 115 | 28–40 | 33 | 31–56 | 43 | 225–288 | 253 |
| cox2 | 129–151 | 139 | 37–50 | 44 | 48–69 | 57 | 199–349 | 252 |
| nad6 | 107–162 | 141 | 27–43 | 35 | 38–57 | 49 | 76–130 | 112 |
| nad5 | 127–183 | 160 | 24–49 | 38 | 27–70 | 49 | – | – |
Averages are rounded to the nearest integer. “-”indicates a partially characterized or missing gene
Mitogenome characterisation
The protein-coding, ribosomal RNA and tRNA genes are characterised in Table 2. The two complete mitogenomes were each comprised of 22 tRNA genes (including two for the amino acids serine and leucine each) and 12 intron-free PCGs and lack the atp8 gene. The genes coding for the large and small subunit of the mitochondrial rRNA were identified for all four species, as were most PCGs (Fig. 1). Only the nad5 gene of C. mbirizei and the nad4 gene and part of the nad4L gene of M. karibae were missing. Within the respective monogenean families, start and stop codons of most genes are conserved in these African species (Table 2). Within the two gyrodactylids, only the stop codons of the cytb, atp6, cox1 and nad6 genes differ; within dactylogyrids, this is only the case for the genes coding for cytb, nad3 and cox1. The only difference in start codon usage was found in the nad2 and nad6 gene in Cichlidogyrus. Abbreviated stop codons occur in the cox3 and nad2 genes of the two species of Cichlidogyrus.
Table 2.
Overview of the length of markers, the start and stop codons (protein-coding genes) and anticodons (tRNA genes) for the four assembled mitochondrial genomes
| Gyrodactylus nyanzae | Macrogyrodactylus karibae | Cichlidogyrus halli | Cichlidogyrus mbirizei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Length | Start/stop codon | Anticodon | Length | Start/stop codon | Anticodon | Position | Length | Start/stop codon | Anticodon | Length | Start/stop codon | Anticodon | |
| cox3 | 1-639 | 639 | ATG/TAA | 639 | ATG/TAA | 13-658 | 646 | ATG/T | 646 | ATG/T | ||||
| trnH | 642-709 | 68 | GTG | 63 | GTG | 659-722 | 64 | GTG | 63 | GTG | ||||
| cytb | 713-1789 | 1077 | ATG/TAA | 1080 | ATG/TAG | 723-1799 | 1077 | ATG/TAA | 1077 | ATG/TAG | ||||
| nad4L | 1792-2049 | 258 | ATG/TAA | 171 (partial) | ATG/- | 1803-2063 | 261 | GTG/TAG | 261 | GTG/TAG | ||||
| nad4 | 2022-3221 | 1200 | ATG/TAA | – | – | 2036-3250 | 1215 | ATG/TAG | 1212 | ATG/TAG | ||||
| trnQ | 3237-3299 | 63 | TTG | – | – | 3256-3317 | 62 | TTG | 62 | TTG | ||||
| trnM | 3298-3364 | 67 | CAT | – | – | 3374-3438 | 65 | CAT | 63 | CAT | ||||
| trnF | 3437-3504 | 68 | GAA | – | – | 3318-3381 | 64 | GAA | 64 | GAA | ||||
| AT-rich segment | 3505-4268 | 764 | – | – | – | – | ||||||||
| atp6 | 4269-4781 | 513 | ATG/TAA | 513 | ATG/TAG | 3443-3952 | 510 | ATG/TAG | 510 | ATG/TAG | ||||
| nad2 | 4788-5663 | 876 | ATG/TAA | 876 | ATG/TAA | 3960-4792 | 833 | ATG/TA | 833 | GTG/TA | ||||
| trnV | 5676-5741 | 66 | TAC | 63 | TAC | 4793-4855 | 63 | TAC | 64 | TAC | ||||
| trnA | 5754-5823 | 70 | TGC | 64 | TGC | 4866-4934 | 69 | TGC | 68 | TGC | ||||
| AT-rich region | – | – | – | – | – | – | – | 4958-5022 | 65 | |||||
| trnD | 5834-5899 | 66 | GTC | 65 | GTC | 5075-5136 | 62 | GTC | 64 | GTC | ||||
| nad1 | 5904-6791 | 888 | GTG/TAA | 888 | GTG/TAA | 5137-6024 | 888 | GTG/TAG | 888 | GTG/TAG | ||||
| trnN | 6796-6867 | 72 | GTT | 68 | GTT | 6027-6092 | 66 | GTT | 68 | GTT | ||||
| trnP | 6868-6939 | 72 | TGG | 64 | TGG | 6098-6162 | 65 | TGG | 64 | TGG | ||||
| trnI | 6938-7002 | 65 | GAT | 64 | GAT | 6162-6228 | 67 | GAT | 67 | GAT | ||||
| trnK | 7007-7073 | 67 | CTT | 62 | CTT | 6230-6292 | 63 | CTT | 64 | CTT | ||||
| nad3 | 7078-7428 | 351 | ATG/TAA | 351 | ATG/TAA | 6294-6641 | 348 | GTG/TAA | 348 | GTG/TAG | ||||
| trnS1 | 7441-7498 | 58 | GCT | 59 | GCT | 6648-6712 | 65 | GCT | 58 | GCT | ||||
| trnW | 7506-7570 | 65 | TCA | 65 | TCA | 6710-6769 | 60 | TCA | 63 | TCA | ||||
| cox1 | 7575-9122 | 1548 | ATG/TAA | 1548 | ATG/TAG | 6773-8356 | 1584 | ATG/TAG | 1581 | ATG/TAA | ||||
| trnT | 9132-9199 | 68 | TGT | 63 | TGT | 8369-8433 | 65 | TGT | 63 | TGT | ||||
| 16S rRNA | 9200-10,170 | 971 | 959 | 8434-9373 | 940 | 939 | ||||||||
| trnC | 10,172-10,237 | 66 | GCA | 60 | GCA | 9374-9436 | 63 | GCA | 61 | GCA | ||||
| 12S rRNA | 10,239-10,948 | 710 | 706 | 9437-10,166 | 730 | 722 | ||||||||
| AT-rich segment/repeat region | – | – | – | – | – | – | – | 10,167-10,743 | 577 | 320 | ||||
| cox2 | 10,963-11,538 | 576 | TTG/TAA | 579 | TTG/TAA | 10,744-11,316 | 573 | ATG/TAA | 576 | ATG/TAA | ||||
| trnE | 11,622-11,689 | 68 | TTC | 68 | TTC | 11,321-11,386 | 66 | TTC | 61 | TTC | ||||
| repeat region | 11,694-12,296 | 603 | – | – | – | – | – | 320 | ||||||
| nad6 | 12,391-12,867 | 477 | ATG/TAA | 477 | ATG/TAG | 11,390-11,881 | 492 | TTG/TAA | 447 | ATG/TAA | ||||
| trnY | 12,869-12,934 | 66 | GTA | 64 | GTA | 11,883-11,947 | 65 | GTA | 63 | GTA | ||||
| trnL1 | 12,951-13,021 | 71 | TAG | 66 | TAG | 11,951-12,018 | 68 | TAG | 63 | TAG | ||||
| trnS2 | 13,035-13,093 | 59 | TGA | 58 | TGA | 12,019-12,083 | 65 | TGA | – | – | ||||
| trnL2 | 13,097-13,163 | 67 | TAA | 68 | TAA | 12,084-12,148 | 65 | TAA | – | – | ||||
| trnR | 13,176-13,246 | 71 | TCG | 64 | TCG | 12,150-12,212 | 63 | TCG | – | – | ||||
| nad5 | 13,250-14,806 | 1557 | ATG/TAG | 1566 | ATG/TAG | 12,214-13,758 | 1545 | ATG/TAG | – | – | ||||
| repeat region | 174 | 13,911-14,302 | 392 | – | ||||||||||
| repeat region | 167 | 14,372-14,915 | 544 | – | ||||||||||
| trnG | 14,812-14,880 | 69 | TCC | 64 | TCC | 14,931-14,996 | 66 | TCC | 64 | TCC | ||||
Positions on the genome are only indicated for complete mitogenomes
Mitogenome gene arrangement differed between the African representatives of the dactylogyrids and gyrodactylids only in a single tRNA gene transposition. Protein-coding genes appeared in identical order (see below for pairwise gene order comparisons in a phylogenetic context). Several non-coding regions (NCRs) were observed in all four mitogenomes (Fig. 1). In G. nyanzae, one of them, a 603 bp stretch between the genes for nad6 and trnE, nearly perfectly repeats (except for one substitution) a fragment of 282 bp 2.1 times. The second one, an AT-rich segment (ca. 17% GC content) of 764 bp between the atp6 and trnF genes, was not identified as a repeat region. In contrast to this, and to the single repeat region of G. nyanzae, two consecutive repeat regions were identified adjacent to the atp6 gene in the partial mitogenome of M. karibae, one 174 bp long with a period of 87 bp (two repeats, 95% match) and the other one 167 bp long with a period of 73 bp (2.3 repeats, 99% match). It has to be noted however, that the possibility of a second, potentially longer non-coding region cannot be excluded due to the double amount of reads in this non-coding region. However, the annotation is incomplete and the exact location can only be inferred using conventional Sanger sequencing. Also the mitogenome of C. halli has two repeat regions, between the trnG and nad5 genes: a 392 bp fragment with repeats of 86 bp (4.6 repeats, 99% match), and a 544 bp fragment with repeats of 167 bp (3.3 repeats, 98% match). In addition, there are AT-rich segments between the cox2 and 12S rRNA genes (577 bp with a GC content of ca. 20%) and between the trnD and trnA genes (65 bp with a GC content of ca. 33%, displaying 58% sequence similarity with a motif in the former AT-rich segment). In the mitogenome of its congener C. mbirizei, a 320 bp stretch is duplicated (97% match) between the genes coding for cox2 and 12S rRNA on the one hand, and nad6 and trnE on the other hand.
The sliding window analysis showed concurring patterns and similar values of nucleotide diversity across the mitochondrial genes for the gyrodactylid and dactylogyrid comparisons (Fig. 2). The highest values were found in the genes coding for subunits of NADH dehydrogenase. The dN/dS ratios in the two pairwise comparisons vary, with the highest values in genes coding for subunits of NADH dehydrogenase (Fig. 3). Values remain around or below 0.1 and are higher for the comparison between the two dactylogyrids than between the two gyrodactylids.
Fig. 2.
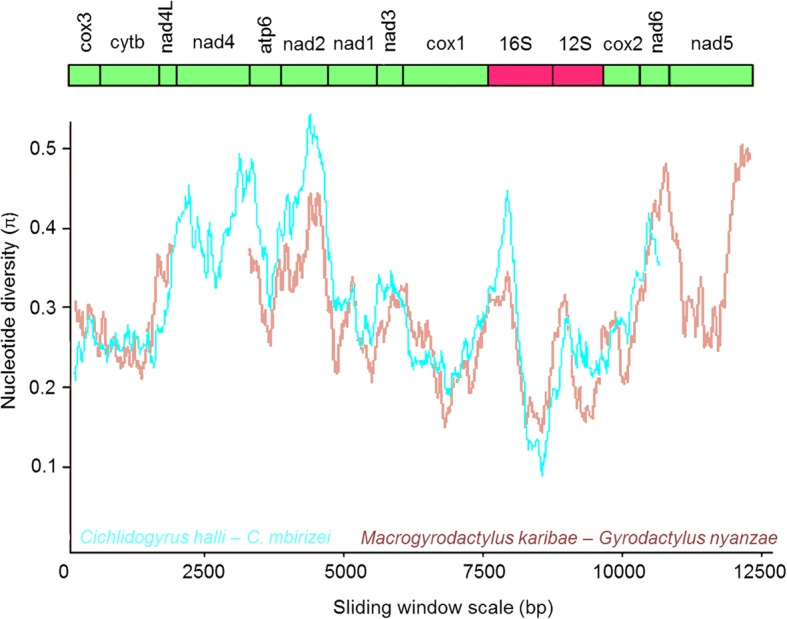
Sliding window analyses (window size 300 bp, step size 10 bp) of the alignment of mitochondrial protein-coding and ribosomal RNA genes used for the phylogenetic analyses of the four mitochondrial genomes of African monogeneans. The lines indicate the nucleotide diversity between two dactylogyrids (Cichlidogyrus halli and C. mbirizei, in blue) and two gyrodactylids (Gyrodactylus nyanzae and Macrogyrodactylus karibae, in red). Gene boundaries are indicated above the graph
Fig. 3.
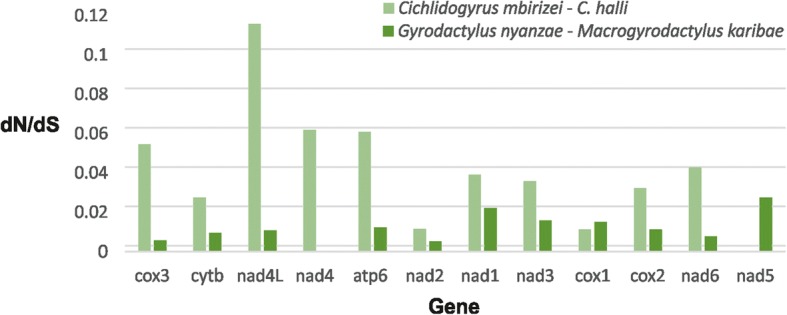
Ratio of non-synonymous to synonymous substitution rates for the protein-coding genes in two pairwise comparisons, between the mitogenomes of African dactylogyrid and gyrodactylid monogeneans, respectively. For Macrogyrodactylus karibae, no nad4 sequence was available, while the nad5 gene was lacking for Cichlidogyrus mbirizei
Phylogenetic and gene order analyses
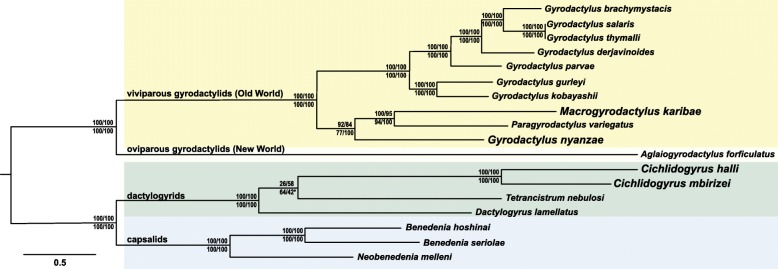
The concatenated alignment of 12 PCGs and two rRNA genes for 18 monogenean species contained 12,464 bp and 9184 variable sites, of which 8060 were parsimony-informative (although we do not analyse the data with parsimony). The topologies retrieved in ML and BI analyses were near-identical, except for the position of Tetrancistrum nebulosi; the resolution within Dactylogyridae is poor (Fig. 4). Capsalids and dactylogyrids firmly cluster together. Macrogyrodactylus karibae and Paragyrodactylus variegatus appear as sister taxa, albeit with long branches, presumably due to incomplete taxon coverage. Gyrodactylus nyanzae clusters with the clade of Macrogyrodactylus and Paragyrodactylus, rendering Gyrodactylus paraphyletic. Aglaiogyrodactylus is firmly positioned as basal to the other gyrodactylids.
Fig. 4.
Midpoint-rooted maximum likelihood phylogram of monopisthocotylean monogeneans based on 12 protein-coding and two ribosomal RNA genes. Support values displayed from (above branch): Shimodaira-Hasegawa-like approximate likelihood ratio test/ultrafast bootstrap, both implemented in IQ-TREE, (below branch) bootstrap in RAxML/Bayesian inference (posterior probability) in MrBayes. An asterisk (*) indicates that this partition was not withheld in the Bayesian consensus tree; the clade grouping Dactylogyrus lamellatus and Tetrancistrum nebulosi as sister to a monophyletic Cichlidogyrus was supported by a posterior probability of 58%. Branch lengths indicate the expected number of substitutions per site
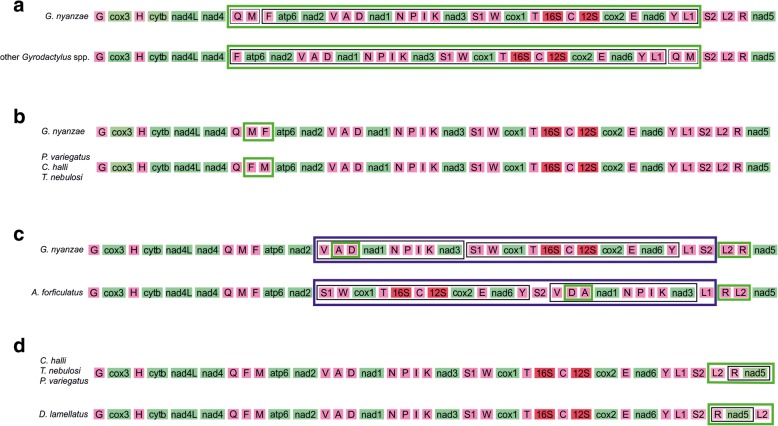
Within Gyrodactylidae, a transposition of two tRNA genes was the only difference in gene order between the African representatives and the Palearctic species of Gyrodactylus (Fig. 5a), while two adjacent tRNA genes were transposed between the African representatives and P. variegatus (Fig. 5b). The difference in mitochondrial gene order between the African gyrodactylids and the Neotropical Aglaiogyrodacylus forficulatus can be explained by a tandem duplication random loss (TDRL) event and two transpositions, or, alternatively, four transpositions (Fig. 5c). The gene order in the mitogenomes of both species of Cichlidogyrus was identical to that of their family member T. nebulosi, and the gyrodactylid P. variegatus. This gene order differed simply in one tRNA gene transposition from that of Gyrodactylus nyanzae (Fig. 5b) and from that of Dactylogyrus lamellatus (Fig. 5d).
Fig. 5.
Family diagram explaining gene order changes between (a) African Gyrodactylus nyanzae and its Palearctic congeners (a single transposition), (b) G. nyanzae and Paragyrodactylus variegatus (a single transposition), (c) G. nyanzae and the Neotropical oviparous gyrodactylid Aglaiogyrodactylus forficulatus (two transpositions and a tandem duplication random loss event (TDRL)) and (d) Dactylogyrus lamellatus and the other dactylogyrids (a single transposition). Green boxes indicate transpositions, a dark blue box a TDRL. Only protein-coding genes, tRNA genes and rRNA genes of species for which a complete mitogenome was assembled, are shown
Discussion
As the low number of available genetic markers imposes limitations on research on non-model flatworms [28], improved and cost-efficient NGS offers ever-more opportunities for genomic work on helminths [29]. Using Illumina technology we assembled, for African gyrodactylid and dactylogyrid monogeneans, one complete and one partial mitogenome each (Fig. 1).
So far only nine gyrodactylid [21, 30–36] and two dactylogyrid [37, 38] monogenean mitogenomes have been published. Our study substantially increases the quantity of available mitogenomic data on these two most diverse monogenean families, by one-third, and offers the first mitogenomes from African representatives. The mitochondrial nucleotide diversity of monogeneans is aptly illustrated by the fact that universal barcoding primers for these species-rich helminths are unavailable [28]. Hence utilising NGS technologies is promising for monogeneans and for other non-model organisms for which typically few or no PCR primers are available. Newly obtained mitogenomes can provide a relatively large set of (coding) molecular markers for molecular evolutionary research. These can be used to develop taxon-specific mitochondrial primers for phylogeographic or population genetic analyses. Challenges however remain, such as the characterisation of AT-rich and repeat regions, in view of the read length of only 300 bp (see also [39]). Also, it is questionable to what extent this NGS approach is workable for rare or opportunistically collected monogenean species, as it has been applied mostly on pools of a considerable number of individuals (this study) or on single larger worm specimens (e.g. [39]). Furthermore, in view of frequent mixed infections, ideally specimens are morphologically identified prior to DNA extraction. This renders the pooling of specimens labour-intensive and sensitive to contamination. Developing reliable NGS shotgun methodologies that can work with single monogeneans, often very small (< 500–1000 μm) in length, will be a worthwhile goal for future molecular ecological and evolutionary studies.
Mitogenome characterisation and potential for marker development
Throughout the PCGs in the four African mitogenomes, the typical start codons are mostly used: commonly ATG in gyrodactylids, and a combination of ATG and GTG in dactylogyrids. The same goes for the stop codons, typically TAA or TAG. Noteworthy exceptions are the cox2 gene of G. nyanzae and M. karibae and the nad6 gene of C. halli, with TTG as start codon. This has been reported in monogeneans before, e.g. in the cox2 gene of Paragyrodactylus variegatus [33]. However, it is reported for the first time here from a dactylogyrid monogenean [37, 38]; also, it is hitherto unique for a member of Gyrodactylus. It is somewhat unsurprising that the full breadth of codon usage diversity in this genus had not yet been captured, since existing mitogenomic data were limited to Palearctic species, all belonging to the subgenus Limnonephrotus, defined by Malmberg [40] on the basis of the excretory system. As regards abbreviated stop codons, the use of T had already been observed in a dactylogyrid monogenean, namely Dactylogyrus lamellatus [38]. The occurrence of TA as an incomplete stop codon, such as in the nad2 gene of both species of Cichlidogyrus, is newly reported for dactylogyrids. It has previously been reported in the same gene for Gyrodactylus brachymystacis [34].
Mitochondrial markers have a wide range of applications in micro-evolutionary and macro-evolutionary research on helminths. For most gyrodactylid and dactylogyrid monogeneans, a small set of established mitochondrial gene fragments (coding for cox1, cox2, nad2 and 16S rRNA) are the most variable markers available. These were applied in population genetics and demography [41, 42], in barcoding [43, 44], in phylogeography [45–48], to detect hybridisation [18] and to elucidate the phylogeny of closely related species [49] or genera [21, 50] or of higher-order taxa in monogeneans [51] and other flatworms (e.g. tapeworms [52]).
Within Palearctic gyrodactylids, nad2, nad4 and nad5 are the most variable genes in the mitochondrial genome and were therefore suggested as markers to study population-level processes [31, 34]. For African gyrodactylids and dactylogyrids, especially the nad2 gene seems promising for marker development as it is flanked by rather conservative stretches (Fig. 2). The dN/dS values for all mitochondrial PCGs fall well below 1 (Fig. 3), indicating purifying selection and confirming earlier mitogenomic work on monogeneans (e.g. [31, 38]). Overall purifying selection acting on mitochondrial genes has also been observed in a range of vertebrates [53, 54].
All hitherto known mitogenomes of species of Gyrodactylus, all representing the subgenus Limnonephrotus, contain two near-identical NCRs [34]. Conversely, such duplicated NCRs are absent in their congener G. nyanzae and, in our dataset, only found in C. mbirizei. Indeed, our results suggest substantial differences in the length, number and position of NCRs between African monogeneans even among gyrodactylids and within Cichlidogyrus (Fig. 1). There is no clear phylogenetic pattern, but a comparison with mitochondrial genomes of other gyrodactylid and dactylogyrid monogeneans indicates that non-coding (repeat) regions are commonly positioned in between certain pairs of genes: e.g. trnD and trnA in C. halli and Aglaiogyrodactylus forficulatus [21]; trnE and nad6 in G. nyanzae and C. mbirizei; nad5 and trnG in C. halli and Tetrancistrum nebulosi [37]; trnF and atp6 in G. nyanzae and its Palearctic congeners (e.g. [31, 34]); and 12S rRNA and cox2 in C. halli and C. mbirizei. Also a NCR containing two repeat regions in the vicinity of the nad5 gene, such as reported here for C. halli, has been reported before in dactylogyrids, namely by Zhang et al. [38] for Dactylogyrus lamellatus. Previous studies suggested the possibility of a functional role for certain NCRs [33] and the potential that NCRs offer for population-level research [38].
Ribosomal operons and utility
Characterising full nuclear ribosomal operons provides a wealth of information for established and prospective molecular markers. Ribosomal DNA codes for all the nuclear ribosomal genes (18S, 5.8S and 28S rRNA) and also includes the external and internal transcribed spacer regions (ETS, ITS1, ITS2). As tandemly repeated units, ribosomal operons occur in high number, and the remarkable variation in rate of molecular evolution within and between nuclear rRNA gene regions has driven their popularity as a source for molecular markers in Metazoa [55] and within the parasitic flatworms [56]. Within flatworms ITS regions are popular for discriminating between closely related species [57], and complete 18S and partial (D1-D3) regions of 28S rDNA were used for phylogenetics of Monogenea (e.g. [58]). In combination with mitochondrial genes, nuclear ribosomal RNA genes are invaluable for discriminating hybrid species, especially important when revealing the identity of disease-causing parasites (e.g. [59]). Many nuclear rRNA gene regions have been used to discriminate species, to resolve phylogenetic relations and as molecular ecological markers amongst monogeneans [49, 60]. Within the newly characterised mitogenomes of African monogeneans in this study, the full operon ranged in size between 6675 and 7496 bp largely reflecting differences in length of spacer regions. We consider this to be a rich resource for a diversity of future studies, especially in the emerging field of environmental (eDNA) metabarcoding and metagenomics where access to highly conserved, and high copy number markers will greatly benefit accurate species identification [61]. In addition, a pairwise or multiple alignment of full ribosomal operons will readily highlight regions of sequence variability and conservation suggesting potential marker regions and regions for PCR primer design. Future studies aimed at population genetics, hybridisation, biogeography, cryptic species recognition, and host-parasite interactions will benefit from access to the full rRNA operon and the full mitogenomes of these, and additional taxa. Certainly, characterisation of full ribosomal operons by means of NGS genome skimming is considerably easier, and cheaper than by long PCR and primer walking using Sanger technology.
Mitochondrial phylogeny, gene order and implications for the position of African gyrodactylid and dactylogyrid monogeneans
Our phylogenetic reconstruction based on 12 mitochondrial PCGs and 2 rRNA genes aimed to elucidate the position of African Macrogyrodactylus, Gyrodactylus and Cichlidogyrus (Fig. 4). All tree topologies firmly place the Neotropical oviparous Aglaiaogyrodactylus forficulatus as a sister lineage to all other representatives of Gyrodactylidae. This refutes Malmberg’s [20] hypothesis of Macrogyrodactylus being the most early divergent gyrodactylid. In addition, the inclusion of an African representative, G. nyanzae, renders Gyrodactylus paraphyletic. Hence, we provide the first mitochondrial data supporting the paraphyly of the genus, corroborating earlier phylogenetic hypotheses based on morphology [23] or nuclear rRNA genes [19, 22, 62].
The evolutionary distances and nucleotide diversity between the two representatives of Cichlidogyrus appear similar to, or even higher than, those between the two African gyrodactylids that are assigned to different genera, Macrogyrodactylus and Gyrodactylus (Figs. 2, 4). This corresponds to earlier work on these monogeneans that indicated the need for revision of Cichlidogyrus and Gyrodactylus. Vanhove et al. [62] and Přikrylová et al. [19] reported that genetic distances between gyrodactylid genera can reach the same order of magnitude as within the nominal genus Gyrodactylus, suggesting that a revision is necessary for several viviparous gyrodactylid genera including Gyrodactylus, although a monophyletic Macrogyrodactylus is strongly supported. Likewise, Pouyaud et al. [63] suggested that lineages within Cichlidogyrus sufficiently differ to be raised to generic status. In their analyses, the inclusion of Scutogyrus indeed rendered Cichlidogyrus paraphyletic, a finding confirmed in later analyses (e.g. [60]). The relationships between the only three dactylogyrid genera in the mitogenomic tree, all of them from the ‘Old World’, are not well resolved. Both Cichlidogyrus and Tetrancistrum have previously been mentioned as members of the Ancyrocephalinae (or Ancyrocephalidae). The monophyly of this (sub)family has often been challenged in earlier work (e.g. [50, 64, 65]). Two topologies (Tetrancistrum as a sister to Cichlidogyrus or, alternatively, to Dactylogyrus) have an equally low posterior probability under BI. Hence, our tree is not informative on the status of the Ancyrocephalinae versus the Dactylogyrinae, to which Dactylogyrus belongs. Although the polytomy makes it hard to favour either of the two alternative positions of Tetrancistrum, the gene order is identical between the representatives of Tetrancistrum and Cichlidogyrus in contrast to the representative of Dactylogyrus. We therefore consider the sister-group relation between the former two genera the biologically most likely hypothesis. This also corresponds to the nuclear rDNA-based results of Blasco-Costa et al. [66] suggesting that Tetrancistrum and Cichlidogyrus belong to the same clade of mostly marine ancyrocephalines. The affinity between Cichlidogyrus and marine genera, despite the likely sampling bias as many dactylogyrid genera have not yet been sequenced, is worth looking into because of the potential of cichlid parasites in elucidating the alleged role of marine dispersal in cichlid biogeography [67]. It would be worthwhile to consider mitochondrial gene order as a phylogenetic marker for further disentangling the relationships between purported dactylogyridean (sub)families.
While it is well-established that gene order is phylogenetically informative, it mainly seems to differ, certainly for PCGs, at the level of major flatworm lineages, such as between catenulids, triclads, polyclads and neodermatans [68]. Within the major flatworm clades, e.g. at order or family level, differences in mitogenome architecture mainly concern tRNA genes and NCRs (e.g. [69] for capsalids, [70] for triclads). This is confirmed in our results, where gene order differences within the dactylogyrids (Fig. 5d) and the viviparous gyrodactylids only concern tRNA genes (Fig. 5a, b). The transpositions seem to concur with evolutionary distance, e.g. simply two adjacent tRNA genes have swapped position within the Macrogyrodactylus-Paragyrodactylus-Gyrodactylus nyanzae clade (Fig. 5b). All viviparous gyrodactylids including Macrogyrodactylus have identical PCG orders. However, Aglaiogyrodactylus forficulatus displays a different PCG arrangement (Fig. 5c), which underscores its particular position within Gyrodactylidae, apart from the viviparous members of this family.
Conclusions
The first mitogenomic data for African monogeneans are provided, characterising two partial and two complete mitochondrial genomes. These confirm earlier results on the variability of and purifying selection on mitochondrial genes in monogeneans, and highlight some patterns in the location of NCRs. These mitogenomes increased the known diversity of start and stop codon usage in dactylogyrids and in species of Gyrodactylus. A phylogeny based on 14 mitochondrial markers firmly confirmed the Neotropical oviparous Aglaiogyrodactylus as ‘basal’ to the other gyrodactylids, rather than the allegedly ‘primitive’ Macrogyrodactylus. Furthermore, it provided additional evidence for the paraphyly of Gyrodactylus. While the gene order for PCGs remained constant throughout the species considered, the study suggested tRNA transpositions to be phylogenetically informative for relationships within the family level.
As highlighted above, (mitochondrial) gene sequences are established tools in the identification of monogeneans, including potentially pathogenic and invasive strains of fish parasites, but their availability for African species remains limited. We hope that this study will contribute to marker development and diagnostics, and hence to ecological and evolutionary studies of African monogeneans.
Methods
Sampling
Fish hosts were collected in the Haut-Katanga province of the D.R. Congo in 2014. Sampling was carried out under research permit no. 863/2014 from the Faculté des Sciences Agronomiques of the Université de Lubumbashi, D.R. Congo. Two individuals of North African catfish Clarias gariepinus (vouchers URA 2014-P-1-004 at the Université de Lubumbashi and MRAC 2015–06-P tag AB49120835 at the Royal Museum for Central Africa (RMCA), Belgium) were caught in the Kiswishi River at Futuka Farm (11°29’S 27°39’E) on August 30th-31st and a hybrid between Nile tilapia Oreochromis niloticus and Mweru tilapia Oreochromis mweruensis (voucher MRAC 2015–06-P tag 2655) at the Kipopo station of the Institut National pour l’Etude et la Recherche Agronomiques (11°34’S 27°21’E) on August 27th. Hosts were sacrificed using an overdose of tricaine methanesulfonate (MS222). Parasites isolated either in situ or later from preserved fish gills were fixed and preserved in analytical-grade ethanol. Individual monogenean specimens were temporarily water-mounted between slide and coverslip, and identified on the basis of their morphology using keys and features described in [3, 18, 27]. Identified specimens were pooled per species in absolute ethanol: four specimens of Macrogyrodactylus karibae (supplemented with two extracts from [18]), 43 of Cichlidogyrus mbirizei, 18 of Cichlidogyrus halli and 44 of Gyrodactylus nyanzae. While M. karibae is a typical gill parasite of Clarias gariepinus known from southern Africa ([18] and references therein), G. nyanzae and especially C. halli are known from a wide range of cichlids throughout Africa [3, 27]. The two latter species have previously been reported from tilapias in the Haut-Katanga province [71]. Cichlidogyrus mbirizei was only recently described from the Lake Tanganyika endemic Oreochromis tanganicae [72]. It was afterwards also found on Nile tilapia and its hybrid O. niloticus x mossambicus [73, 74] and is here for the first time reported from O. niloticus x mweruensis. Both species of Cichlidogyrus have been co-introduced outside Africa, in nature and in aquaculture settings (e.g. [73–75]).
DNA extraction and sequence assembly
Total genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instructions. The amount of double-stranded DNA isolated was measured with Qubit® 2.0 Fluorometer (Life Technologies, Paisley, UK) yielding 0.9 (M. karibae), 3.3 (C. halli), 3.2 (C. mbirizei) and 1.8 (G. nyanzae) ng/μl total DNA.
Samples for NGS were prepared and run at the DNA Sequencing Facility of the Natural History Museum, London, UK. Genomic DNA was indexed and libraries prepared using the TruSeq Nano DNA Sample Preparation Kit (Illumina, Inc., San Diego, USA), and run simultaneously on a MiSeq Illumina sequencer yielding 300 bp long paired-end reads. The new mitogenomes were directly assembled using Geneious v. 8.1.9 [76]. The sequences were first trimmed (error probability: 0.05, maximum ambiguity: 1) and then assembled. Partial cox1 sequences of Gyrodactylus salaris (NC008815 [30]) (for G. nyanzae), Macrogyrodactylus clarii (GU252718 [18]) (for M. karibae) and Cichlidogyrus zambezensis (KT037411 [49]) (for representatives of Cichlidogyrus) were used as reference sequence to extract cox1 reads from the Illumina genomic readpool to form the consensus sequence to subsequently map the reads on successive iterations. Trimmed reads were mapped back to the contigs in order to estimate the full mitochondrial genome coverage, trim the overlapping regions to create a circular molecule, and to inspect for potential mapping/assembly errors in problematic regions such as repetitive regions [77]. In instances where disagreements occurred between reads, the consensus sequence was generated by choosing the most frequently represented base.
Using nuclear ribosomal RNA gene sequences for Cichlidogyrus halli and Macrogyrodactylus congolensis from GenBank (accessions: HE792784 [60] and HF548680 [19] respectively), fragments of the ribosomal RNA operon were identified and assembled using the same iterative process as described for the mitochondrial genome. Exact coding positions of the 18S and 28S nuclear rDNAs, as well as the respective 5′ and 3′ boundaries of the external transcribed spacers, were determined using RNAmmer [78]. Subsequently the complete annotation was compared with the fully-annotated human rDNA repeating unit (GenBank accession: HSU13369).
Mitogenome annotation
The identity and boundaries of individual PCGs and rRNA genes were determined using the MITOS web server [79] in combination with the visualisation of open reading frames in Geneious and a comparison with alignments of available mitogenomes of closely related monopisthocotylean monogeneans. In addition to MITOS, the ARWEN v. 1.2 [80] and tRNAscan-SE v. 2.0 [81] web servers were used to identify the tRNA-coding regions. When results between applications conflicted, the solution proposing a 7 bp acceptor stem was chosen. We checked for repeat regions with Tandem Repeats Finder [82] and YASS [83]. The resulting mitogenomes were visualised in OGDRAW v. 1.1 [84].
Alignment, sequence analysis, phylogenetic reconstruction and gene order analysis
Ribosomal RNA genes were aligned by MAFFT v. 7 [85] using the Q-INS-i iterative refinement method, taking into account RNA secondary structure [86]. Codon-based alignment of all obtained PCGs was performed under the echinoderm and flatworm mitochondrial genetic code [87] using MUSCLE [88] implemented in SeaView v. 4.6.2 [89]. Since omitting unreliable portions of the alignment may increase resolution in phylogenomic reconstructions [90], an alternative alignment was obtained by trimming in Gblocks v. 0.91b [91], implemented for the PCGs in TranslatorX [92], carrying out codon-based MAFFT alignment followed by alignment cleaning in Gblocks. Options for a less stringent selection were selected, allowing smaller final blocks, gap positions within the final blocks, and less strict flanking positions. Especially for smaller datasets, trimming entails the risk of removing information contributing to phylogenetic signal [90]. Therefore, likelihood mapping [93] was performed in TREE-PUZZLE v. 5.3 [94] to compare the phylogenetic content of the complete and trimmed concatenated alignment. The percentage of fully, partially and unresolved quartets was 99.4, 0.5 and 0.1 in both cases, hence trimming did not increase phylogenetic content and the original alignment was preferred for downstream analyses (Fig. 6). Comparing, in DAMBE [95], the index of substitution saturation with its critical value at which sequences would start to fail to recover the true phylogeny, indicated little substitution saturation for this dataset [96].
Fig. 6.
Likelihood mapping (a) before and (b) after Gblocks trimming, demonstrating the high phylogenetic content and suggesting there is no need for alignment cleaning in the case of this dataset
Using the aligned sequences, two pairwise comparisons between members of the same monogenean family (C. halli versus C. mbirizei; G. nyanzae versus M. karibae) were made. Firstly, we visualised the nucleotide diversity by a sliding window analysis of nucleotide diversity (π) in DnaSP v. 5.10.01 [97], with a window size of 300 bp and a step size of 10 bp. To allow comparison between the dactylogyrid and gyrodactylid haplotypes, this approach was limited to the PCGs and rRNA genes. Secondly, for the PCGs of the same pairs of species, the proportion of non-synonymous versus synonymous substitutions (dN/dS ratio) was calculated in the codeml program of PAML [98] as implemented in PAL2NAL [99].
To situate the African monogeneans under study within their respective families, the PCGs and rRNA genes of all available dactylogyrid [37, 38] and gyrodactylid [21, 30–36] mitogenomes were included in phylogenetic analyses. The species of Capsalidae for which mitogenomes are available [69, 100, 101] were also included as they strongly cluster with the dactylogyrids [21, 38].
The best partition scheme and the optimal models of molecular evolution were determined based on the Bayesian Information Criterion using ModelFinder [102] with partition merging [103]. The selected partitions and models are shown in Table 3. These were used for Bayesian inference (BI) of phylogeny, whereby posterior probabilities were calculated in MrBayes v. 3.2 [104] over 10 million generations, sampling the Markov chain at a frequency of 100 generations. Chain stationarity was evidenced by a standard deviation of split frequencies of 8.10–4, absence of a trend in the probabilities plotted against the generations, and a potential scale reduction factor [105] converging towards 1. One-fourth of the samples were discarded as burn-in. The same partitions were used in a maximum likelihood (ML) search in IQ-TREE [106], using four gamma-rate categories and an edge-linked partition model. Nodal support was assessed through 10,000 ultrafast bootstrap [107] and 1000 Shimodaira-Hasegawa-like approximate likelihood ratio test [108] replicates. In addition, a ML tree was constructed in RAxML v. 8.1.21 [109] implemented in raxmlGUI v.1.3 [110], using codon-specific partitions under the GTR + Γ + I model with joint branch length optimization, and with 1000 bootstrap samples to calculate support values. ALTER [111] and GenBank 2 Sequin [112] were used for file conversion, and SequenceMatrix [113] to concatenate alignment files.
Table 3.
Best partition scheme for the dataset of two ribosomal RNA genes and 12 protein-coding genes in the mitochondrial genomes of 14 monopisthocotylean monogenean flatworms
| Partition | Model of molecular evolution |
|---|---|
| 12S + 16S + first codon positions of atp6, nad2, nad3, nad4, nad4L, nad5, nad6 | TIM2 + I + G |
| second codon positions of atp6, nad1, cox1, cox2, cox3, cytb | TVM + I + G |
| third codon positions of atp6, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, cox1, cox2, cox3, cytb | TIM2 + I + G |
| first codon positions of nad1, cox1, cox2, cox3, cytb | TIM2 + I + G |
| second codon positions of nad2, nad3, nad4, nad4L, nad5, nad6 | TVM + I + G |
The number of gamma rate categories was set to four
Gene orders were compared, and family diagrams of gene orders constructed, using CREx [114]. For those genes that were available from the partial mitogenomes, gene order was identical between M. karibae and G. nyanzae, and between C. mbirizei and C. halli, respectively. Therefore, only the complete mitogenomes could be included in gene order analyses. For the same reason of comparability, non-coding regions (NCRs) were omitted in gene order analysis.
Additional file
Table S1. Annotation of the ribosomal operons of the four African monogenean species. (TXT 1 kb)
Acknowledgements
The authors cordially thank W. Fannes, E.J. Vreven, A. Chocha Manda, E. Abwe, B. Katemo Manda, G. Kapepula Kasembele, M. Kasongo Ilunga Kayaba, C. Kalombo Kabalika, C. Mukweze Mulelenu, M. Katumbi Chapwe and J. Snoeks for their help with sampling and/or planning this research, I. Přikrylová and A. Pariselle for advice on monogenean identification, R. Väinölä and M. Monnens for helpful input regarding the analyses, and two anonymous reviewers for their suggestions which greatly improved the manuscript.
Funding
This research was supported by the Belgian Federal Science Policy Office (BRAIN-be Pioneer Project BR/132/PI/TILAPIA), Czech Science Foundation project no. P505/12/G112 (ECIP) and the SYNTHESYS Project (http://www.synthesys.info/) (GB-TAF-2984 and GB-TAF-4940) which is financed by European Community Research Infrastructure Action under the FP7 Integrating Activities Programme. Fieldwork was supported by the University Development Cooperation of the Flemish Interuniversity Council (VLIR-UOS) (South Initiative Renforcement des capacités locales pour une meilleure évaluation biologique des impacts miniers au Katanga (RDC) sur les poissons et leurs milieux aquatiques, ZRDC2014MP084), the Mbisa Congo project, a framework agreement project of the RMCA with the Belgian Development Cooperation and travel grant K220314 N (to MPMV) from the Research Foundation – Flanders (FWO-Vlaanderen). MWPJ is currently supported by a BOF Reserve Fellowship from Hasselt University. The funding bodies had no role in the design of the study, in the collection, analysis, and interpretation of the data, or in writing the manuscript.
Availability of data and materials
The sequence data produced and analysed during the current study were deposited in NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and are freely available under accession numbers MG970255-8 and MG973075-8. Voucher specimens are available in the invertebrate collection of the Royal Museum for Central Africa (RMCA), Tervuren, Belgium. The posterior ends (with opisthaptor) of four of the specimens of Macrogyrodactylus karibae used were deposited under accession numbers M.T.38263–6; for the other monogenean species, entire animals were used for DNA extraction, and conspecifics from the same host specimen are available under accession numbers MRAC M.T.38239–62.
Abbreviations
- BI
Bayesian inference
- bp
Base pairs
- MITObim
Mitochondrial baiting and iterative mapping
- ML
Maximum likelihood
- NCR
Non-coding region
- NGS
Next-generation sequencing
- PCG
Protein-coding gene
- rDNA
Ribosomal DNA
- rRNA
Ribosomal RNA
- TDRL
Tandem duplication random loss event
Authors’ contributions
MPMV conceived the study, collected and identified specimens, analysed data and drafted the manuscript. AGB carried out experiments and analysed data. MWPJ prepared and identified specimens. DTJL analysed data, oversaw the study and provided lab facilities. TH conceived and oversaw the study, carried out experiments, analysed data and provided lab facilities. All authors contributed to drafting the manuscript and read and approved the final version of the manuscript.
Ethics approval and consent to participate
In the absence of relevant animal welfare regulations in the D.R. Congo, the same strict codes of practice enforced within the European Union were applied. Sampling was carried out under research permit no. 863/2014 from the Faculté des Sciences Agronomiques of the Université de Lubumbashi. Since this research did not involve human subjects, human material, or human data, consent to participate did not apply.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4893-5) contains supplementary material, which is available to authorized users.
Contributor Information
Maarten P. M. Vanhove, Email: maarten.vanhove@uhasselt.be
Andrew G. Briscoe, Email: A.Briscoe@nhm.ac.uk
Michiel W. P. Jorissen, Email: michiel.jorissen@uhasselt.be
D. Tim J. Littlewood, Email: t.littlewood@nhm.ac.uk
Tine Huyse, Email: tine.huyse@africamuseum.be.
References
- 1.Cribb TH, Chisholm LA, Bray RA. Diversity in the Monogenea and Digenea: does lifestyle matter? Int J Parasitol. 2002;32:321–328. doi: 10.1016/S0020-7519(01)00333-2. [DOI] [PubMed] [Google Scholar]
- 2.Pugachev ON, Gerasev PI, Gussev AV, Ergens R, Khotenowsky I. Guide to Monogenoidea of freshwater fish of Palaearctic and Amur regions. Milan: Ledizione-LediPublishing; 2009. [Google Scholar]
- 3.Zahradníčková P, Barson M, Luus-Powell WJ, Přikrylová I. Species of Gyrodactylus Von Nordmann, 1832 (Platyhelminthes: Monogenea) from cichlids from Zambezi and Limpopo river basins in Zimbabwe and South Africa: evidence for unexplored species richness. Syst Parasitol. 2016;93:679–700. doi: 10.1007/s11230-016-9652-x. [DOI] [PubMed] [Google Scholar]
- 4.Bakke TA, Harris PD, Cable J. Host specificity dynamics: observations on gyrodactylid monogeneans. Int J Parasitol. 2002;32:281–308. doi: 10.1016/S0020-7519(01)00331-9. [DOI] [PubMed] [Google Scholar]
- 5.Paladini G, Longshaw M, Gustinelli A, Shinn AP. Parasitic diseases in aquaculture: their biology, diagnosis and control. In: Austin B, Newaj-Fyzul A, editors. Diagnosis and control of diseases of fish and shellfish. Chichester: John Wiley & Sons, Ltd; 2017. pp. 37–107. [Google Scholar]
- 6.Bakke TA, Cable J, Harris PD. The biology of gyrodactylid monogeneans: the “Russian doll-killers”. Adv Parasit. 2007;64:161–376. doi: 10.1016/S0065-308X(06)64003-7. [DOI] [PubMed] [Google Scholar]
- 7.Paperna I. Parasites, infections and diseases of fishes in Africa – An Update CIFA Technical Paper 31. Rome: Food and Agriculture Organization of the United Nations; 1996. [Google Scholar]
- 8.Lio-Po GD, Lim LHS. Infectious diseases of warmwater fish in fresh water. In: Woo PTK, Bruno DW, editors. Diseases and disorders of finfish in cage culture. 2nd edition. Wallingford and. Boston: CAB International; 2014. pp. 193–253. [Google Scholar]
- 9.Pouomogne V. Clarias gariepinus. In: Cultured aquatic species information Programme. FAO fisheries and aquaculture department. 2010. http://www.fao.org/fishery/culturedspecies/Clarias_gariepinus/en. Accessed 3 Jan 2017.
- 10.Deines AM, Wittmann ME, Deines JM, Lodge DM. Tradeoffs among ecosystem services associated with global tilapia introductions. Rev Fish Sci Aquac. 2016;24:178–191. doi: 10.1080/23308249.2015.1115466. [DOI] [Google Scholar]
- 11.Beletew M, Getahun A, Vanhove MPM. First report of monogenean flatworms from Lake Tana, Ethiopia: gill parasites of the commercially important Clarias gariepinus (Teleostei: Clariidae) and Oreochromis niloticus tana (Teleostei: Cichlidae) Parasite Vector. 2016;9:410. doi: 10.1186/s13071-016-1691-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanhove MPM, Hablützel PI, Pariselle A, Šimková A, Huyse T, Raeymaekers JAM. Cichlids: a host of opportunities for evolutionary parasitology. Trends Parasitol. 2016;32:820–832. doi: 10.1016/j.pt.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Bondad-Reantaso MG, Subasinghe RP, Arthur JR, Ogawa K, Chinabut S, Adlard R, et al. Disease and health management in Asian aquaculture. Vet Parasitol. 2005;132:249–272. doi: 10.1016/j.vetpar.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Akoll P, Konecny R, Mwanja WW, Nattabi JK, Agoe C, Schiemer F. Parasite fauna of farmed Nile tilapia (Oreochromis niloticus) and African catfish (Clarias gariepinus) in Uganda. Parasitol Res. 2012;110:315–323. doi: 10.1007/s00436-011-2491-4. [DOI] [PubMed] [Google Scholar]
- 15.Smit NJ, Malherbe W, Hadfield KA. Alien freshwater fish parasites from South Africa: diversity, distribution, status and the way forward. Int J Parasitol Parasites Wildl. 2017;6:386–401. doi: 10.1016/j.ijppaw.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akoll P, Konecny R, Mwanja WW, Schiemer F. Risk assessment of parasitic helminths on cultured Nile tilapia (Oreochromis niloticus, L.) Aquaculture. 2012;356:123–127. doi: 10.1016/j.aquaculture.2012.05.027. [DOI] [Google Scholar]
- 17.Ek-Huchim JP, Jimenez-Garcia I, Pérez-Vega JA, Rodríguez-Canul R. Non-lethal detection of DNA from Cichlidogyrus spp. (Monogenea, Ancyrocephalinae) in gill mucus of the Nile tilapia Oreochromis niloticus. Dis Aquat Org. 2012;98:155–162. doi: 10.3354/dao02435. [DOI] [PubMed] [Google Scholar]
- 18.Barson M, Přikrylová I, Vanhove MPM, Huyse T. Parasite hybridization in African Macrogyrodactylus spp. (Monogenea, Platyhelminthes) signals historical host distribution. Parasitology. 2010;137:1585–1595. doi: 10.1017/S0031182010000302. [DOI] [PubMed] [Google Scholar]
- 19.Přikrylová I, Vanhove MPM, Janssens SB, Billeter PA, Huyse T. Tiny worms from a mighty continent: high diversity and new phylogenetic lineages of African monogeneans. Mol Phylogenet Evol. 2013;67:43–52. doi: 10.1016/j.ympev.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Malmberg G. On the evolution within the family Gyrodactylidae (Monogenea) Int J Parasitol. 1998;28:1625–1635. doi: 10.1016/S0020-7519(98)00058-7. [DOI] [PubMed] [Google Scholar]
- 21.Bachmann L, Fromm B, Patella de Azambuja L, Boeger WA. The mitochondrial genome of the egg-laying flatworm Aglaiogyrodactylus forficulatus (Platyhelminthes: Monogenoidea) Parasite Vector. 2016;9:285. doi: 10.1186/s13071-016-1586-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmore SR, Cone DK, Lowe G, King SK, Jones SRM, Abbott CL. Molecular phylogeny of Gyrodactylus (Monogenea) parasitizing fishes in fresh water, estuarine, and marine habitats in Canada. Can J Zool. 2012;90:776–786. doi: 10.1139/z2012-040. [DOI] [Google Scholar]
- 23.Kritsky DC, Boeger WA. Phylogeny of the Gyrodactylidae and the phylogenetic status of Gyrodactylus Nordmann, 1832 (Platyhelminthes: Monogenoidea) In: Combes C, Jourdane J, editors. Taxonomie, Ecologie et Evolution des Métazoaires Parasites. Taxonomy, Ecology and Evolution of Metazoan Parasites (Livre hommage à Louis Euzet) Perpignan: Presses Universitaires de Perpignan; 2003. pp. 37–58. [Google Scholar]
- 24.Minárik G, Bazsalovicsová E, Zvijáková Ľ, Štefka J, Pálková L, Králová-Hromadová I. Development and characterization of multiplex panels of polymorphic microsatellite loci in giant liver fluke Fascioloides magna (Trematoda: Fasciolidae), using next-generation sequencing approach. Mol Biochem Parasit. 2014;195:30–33. doi: 10.1016/j.molbiopara.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brabec J, Kostadinova A, Scholz T, Littlewood DTJ. Complete mitochondrial genomes and nuclear ribosomal RNA operons of two species of Diplostomum (Platyhelminthes: Trematoda): a molecular resource for taxonomy and molecular epidemiology of important fish pathogens. Parasite Vector. 2015;8:336. doi: 10.1186/s13071-015-0949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pariselle A, Euzet L. Systematic revision of dactylogyridean parasites (Monogenea) from cichlid fishes in Africa, the Levant and Madagascar. Zoosystema. 2009;31:849–898. doi: 10.5252/z2009n4a6. [DOI] [Google Scholar]
- 28.Vanhove MPM, Tessens B, Schoelinck C, Jondelius U, Littlewood DTJ, Artois T, et al. Problematic barcoding in flatworms: a case-study on monogeneans and rhabdocoels (Platyhelminthes) ZooKeys. 2013;365:355–379. doi: 10.3897/zookeys.365.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wit J, Gilleard JS. Resequencing helminth genomes for population and genetic studies. Trends Parasitol. 2017;33:388–399. doi: 10.1016/j.pt.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Huyse T, Plaisance L, Webster BL, Mo TA, Bakke TA, Bachmann L, et al. The mitochondrial genome of Gyrodactylus salaris (Platyhelminthes: Monogenea), a pathogen of Atlantic salmon (Salmo salar) Parasitology. 2007;134:739–747. doi: 10.1017/S0031182006002010. [DOI] [PubMed] [Google Scholar]
- 31.Huyse T, Buchmann K, Littlewood DTJ. The mitochondrial genome of Gyrodactylus derjavinoides (Platyhelminthes: Monogenea) – a mitogenomic approach for Gyrodactylus species and strain identification. Gene. 2008;417:27–34. doi: 10.1016/j.gene.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Plaisance L, Huyse T, Littlewood DTJ, Bakke TA, Bachmann L. The complete mitochondrial DNA sequence of the monogenean Gyrodactylus thymalli (Platyhelminthes: Monogenea), a parasite of grayling (Thymallus thymallus) Mol Biochem Parasitol. 2007;154:190–194. doi: 10.1016/j.molbiopara.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Ye F, King SD, Cone DK, You P. The mitochondrial genome of Paragyrodactylus variegatus (Platyhelminthes: Monogenea): differences in major non-coding region and gene order compared to Gyrodactylus. Parasite Vector. 2014;7:377. doi: 10.1186/1756-3305-7-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye F, Easy RH, King SD, Cone DK, You P. Comparative analyses within Gyrodactylus (Platyhelminthes: Monogenea) mitochondrial genomes and conserved polymerase chain reaction primers for gyrodactylid mitochondrial DNA. J Fish Dis. 2017;40:541–555. doi: 10.1111/jfd.12539. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Zou H, Zhou S, Wu SG, Li WX, Wang GT. The complete mitochondrial genome of Gyrodactylus kobayashii (Platyhelminthes: Monogenea) Mitochondr DNA Part B. 2016;1:154–155. doi: 10.1080/23802359.2016.1144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou H, Zhang D, Li W, Zhou S, Wu S, Wang G. The complete mitochondrial genome of Gyrodactylus gurleyi (Platyhelminthes: Monogenea) Mitochondr DNA Part B. 2016;1:383–385. doi: 10.1080/23802359.2016.1172042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Wu X, Li Y, Xie M, Li A. The complete mitochondrial genome of Tetrancistrum nebulosi (Monogenea: Ancyrocephalidae) Mitochondr DNA Part A. 2014;27:22–23. doi: 10.3109/19401736.2013.867441. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Zou H, Wu SG, Li M, Jakovlić I, Zhang J, et al. Sequencing, characterization and phylogenomics of the complete mitochondrial genome of Dactylogyrus lamellatus (Monogenea: Dactylogyridae). J Helminthol. 2017; 10.1017/S0022149X17000578. [DOI] [PubMed]
- 39.Briscoe AG, Bray RA, Brabec J, Littlewood DTJ. The mitochondrial genome and ribosomal operon of Brachycladium goliath (Digenea: Brachycladiidae) recovered from a stranded minke whale. Parasitol Int. 2016;65:271–275. doi: 10.1016/j.parint.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Malmberg G. The excretory systems and the marginal hooks as a basis for the systematics of Gyrodactylus (Trematoda, Monogenea) Ark Zool. 1970;23:1–235. [Google Scholar]
- 41.Bueno-Silva M, Boeger WA, Pie MR. Choice matters: incipient speciation in Gyrodactylus corydori (Monogenoidea: Gyrodactylidae) Int J Parasitol. 2011;41:657–667. doi: 10.1016/j.ijpara.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Kmentová N, Gelnar M, Mendlová M, Van Steenberge M, Koblmüller S, Vanhove MPM. Reduced host-specificity in a parasite infecting non-littoral Lake Tanganyika cichlids evidenced by intraspecific morphological and genetic diversity. Sci Rep. 2016;6:39605. doi: 10.1038/srep39605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen H, Bakke TA, Bachmann L. DNA taxonomy and barcoding of monogenean parasites: lessons from Gyrodactylus. Trends Parasitol. 2007;23:363–367. doi: 10.1016/j.pt.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Bueno-Silva M, Boeger WA. Neotropical Monogenoidea. 58. Three new species of Gyrodactylus (Gyrodactylidae) from Scleromystax spp. (Callichthyidae) and the proposal of COII gene as an additional fragment for barcoding gyrodactylids. Folia Parasit. 2014;61:213. doi: 10.14411/fp.2014.028. [DOI] [PubMed] [Google Scholar]
- 45.Meinilä M, Kuusela J, Ziętara MS, Lumme J. Initial steps of speciation by geographic isolation and host switch in salmonid pathogen Gyrodactylus salaris (Monogenea: Gyrodactylidae) Int J Parasitol. 2004;34:515–526. doi: 10.1016/j.ijpara.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Plaisance L, Rousset V, Morand S, Littlewood DTJ. Colonization of pacific islands by parasites of low dispersal abilities: phylogeography of two monogenean species parasitizing butterflyfishes in the indo-West Pacific Ocean. J Biogeogr. 2008;35:76–87. [Google Scholar]
- 47.Wang M, Yan S, Brown CL, Shaharom-Harrison F, Shi S-F, Yang T-B. Phylogeography of Tetrancistrum nebulosi (Monogenea, Dactylogyridae) on the host of mottled spinefoot (Siganus fuscescens) in the South China Sea, inferred from mitochondrial COI and ND2 genes. Mitochondr DNA Part A. 2016;27:3865–3875. doi: 10.3109/19401736.2014.971240. [DOI] [PubMed] [Google Scholar]
- 48.Huyse T, Oeyen M, Larmuseau MHD, Volckaert FAM. Co-phylogeographic study of the flatworm Gyrodactylus gondae and its goby host Pomatoschistus minutus. Parasitol Int. 2017;66:119–125. doi: 10.1016/j.parint.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 49.Vanhove MPM, Pariselle A, Van Steenberge M, Raeymaekers JAM, Hablützel PI, Gillardin C, et al. Hidden biodiversity in an ancient Lake: phylogenetic congruence between lake Tanganyika tropheine cichlids and their monogenean flatworm parasites. Sci Rep. 2015;5:13669. doi: 10.1038/srep13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plaisance L, Littlewood DTJ, Olson PD, Morand S. Molecular phylogeny of gill monogeneans (Platyhelminthes, Monogenea, Dactylogyridae) and colonization of indo-West Pacific butterflyfish hosts (Perciformes, Chaetodontidae) Zool Scr. 2005;34:425–436. doi: 10.1111/j.1463-6409.2005.00191.x. [DOI] [Google Scholar]
- 51.Park J-K, Kim K-H, Kang S, Kim W, Eom KS. Littlewood DTJ. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes) BMC Evol Biol. 2007;7:11. doi: 10.1186/1471-2148-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waeschenbach A, Webster BL, Littlewood DTJ. Adding resolution to ordinal level relationships of tapeworms (Platyhelminthes: Cestoda) with large fragments of mtDNA. Mol Phylogenet Evol. 2012;63:834–847. doi: 10.1016/j.ympev.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 53.Castellana S, Vicario S, Saccone C. Evolutionary patterns of the mitochondrial genome in Metazoa: exploring the role of mutation and selection in mitochondrial protein-coding genes. Genome Biol Evol. 2011;3:1067–1079. doi: 10.1093/gbe/evr040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobsen MW, Da Fonseca RR, Bernatchez L, Hansen MM. Comparative analysis of complete mitochondrial genomes suggests that relaxed purifying selection is driving high nonsynonymous evolutionary rate of the NADH2 gene in whitefish (Coregonus ssp.) Mol Phylogenet Evol. 2016;95:161–170. doi: 10.1016/j.ympev.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Mallatt J, Craig CW, Yoder MJ. Nearly complete rRNA genes assembled from across the metazoan animals: effects of more taxa, a structure-based alignment, and paired-sites evolutionary models on phylogeny reconstruction. Mol Phylogenet Evol. 2010;55:1–17. doi: 10.1016/j.ympev.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 56.Lockyer AE, Olson PD, Littlewood DTJ. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): implications and a review of the cercomer theory. Biol J Linn Soc. 2003;78:155–171. doi: 10.1046/j.1095-8312.2003.00141.x. [DOI] [Google Scholar]
- 57.Nolan MJ, Cribb TH. The use and implications of ribosomal DNA sequencing for the discrimination of digenean species. Adv Parasit. 2005;60:101–163. doi: 10.1016/S0065-308X(05)60002-4. [DOI] [PubMed] [Google Scholar]
- 58.Olson PD, Littlewood DTJ. Phylogenetics of the Monogenea – evidence from a medley of molecules. Int J Parasitol. 2002;32:233–244. doi: 10.1016/S0020-7519(01)00328-9. [DOI] [PubMed] [Google Scholar]
- 59.Huyse T, Webster BL, Geldof S, Stothard JR, Diaw OT, Polman K, et al. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathog. 2009;5:e1000571. doi: 10.1371/journal.ppat.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendlová M, Desdevises Y, Civáňová K, Pariselle A, Šimková A. Monogeneans of west African cichlid fish: evolution and cophylogenetic interactions. PLoS One. 2012;7:e37268. doi: 10.1371/journal.pone.0037268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang CQ, Leasi F, Obertegger U, Kieneke A, Barraclough TG, Fontaneto D. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. P Natl Acad Sci USA. 2012;109:16208–16212. doi: 10.1073/pnas.1209160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanhove MPM, Snoeks J, Volckaert FAM, Huyse T. First description of monogenean parasites in Lake Tanganyika: the cichlid Simochromis diagramma (Teleostei, Cichlidae) harbours a high diversity of Gyrodactylus species (Platyhelminthes, Monogenea) Parasitology. 2011;138:364–380. doi: 10.1017/S0031182010001356. [DOI] [PubMed] [Google Scholar]
- 63.Pouyaud L, Desmarais E, Deveney M, Pariselle A. Phylogenetic relationships among monogenean gill parasites (Dactylogyridea, Ancyrocephalidae) infesting tilapiine hosts (Cichlidae): systematic and evolutionary implications. Mol Phylogenet Evol. 2006;38:241–249. doi: 10.1016/j.ympev.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 64.Šimková A, Plaisance L, Matějusová I, Morand S, Verneau O. Phylogenetic relationships of the Dactylogyridae Bychowsky, 1933 (Monogenea: Dactylogyridea): the need for the systematic revision of the Ancyrocephalinae Bychowsky, 1937. Syst Parasitol. 2003;54:1–11. doi: 10.1023/A:1022133608662. [DOI] [PubMed] [Google Scholar]
- 65.Šimková A, Matějusová I, Cunningham CO. A molecular phylogeny of the Dactylogyridae sensu Kritsky and Boeger (1989) (Monogenea) based on the D1-D3 domains of large subunit rDNA. Parasitology. 2006;133:43–54. doi: 10.1017/S0031182006009942. [DOI] [PubMed] [Google Scholar]
- 66.Blasco-Costa I, Míguez-Lozano R, Sarabeev V, Balbuena JA. Molecular phylogeny of species of Ligophorus (Monogenea: Dactylogyridae) and their affinities within the Dactylogyridae. Parasitol Int. 2012;61:619–627. doi: 10.1016/j.parint.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Pariselle A, Boeger WA, Snoeks J, Bilong Bilong CF, Morand S, Vanhove MPM. The monogenean parasite fauna of cichlids: a potential tool for host biogeography. Int J Evol Biol 2011;2011:471480. [DOI] [PMC free article] [PubMed]
- 68.Rosa MT, Oliveira DS, Loreto EL. Characterization of the first mitochondrial genome of a catenulid flatworm: Stenostomum leucops (Platyhelminthes) J Zool Syst Evol Res. 2017;55:98–105. doi: 10.1111/jzs.12164. [DOI] [Google Scholar]
- 69.Zhang J, Wu X, Li Y, Zhao M, Xie M, Li A. The complete mitochondrial genome of Neobenedenia melleni (Platyhelminthes: Monogenea): mitochondrial gene content, arrangement and composition compared with two Benedenia species. Mol Biol Rep. 2014;41:6583–6589. doi: 10.1007/s11033-014-3542-6. [DOI] [PubMed] [Google Scholar]
- 70.Solà E, Álvarez-Presas M, Frías-López C, Littlewood DTJ, Rozas J, Riutort M. Evolutionary analysis of mitogenomes from parasitic and free-living flatworms. PLoS One. 2015;10:e0120081. doi: 10.1371/journal.pone.0120081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jorissen MWP, Pariselle A, Huyse T, Vreven EJ, Snoeks J, Volckaert FAM, et al. Diversity, endemicity and host-specificity of monogenean gill parasites (Platyhelminthes) of cichlids in the Bangweulu-Mweru ecoregion. J Helminthol. 2017; 10.1017/S0022149X17000712. [DOI] [PubMed]
- 72.Muterezi Bukinga F, Vanhove MPM, Van Steenberge M, Pariselle A. Ancyrocephalidae (Monogenea) of Lake Tanganyika: III: Cichlidogyrus infecting the world’s biggest cichlid and the non-endemic tribes Haplochromini, Oreochromini and Tylochromini (Teleostei, Cichlidae) Parasitol Res. 2012;111:2049–2061. doi: 10.1007/s00436-012-3052-1. [DOI] [PubMed] [Google Scholar]
- 73.Lerssutthichawal T, Maneepitaksanti W, Purivirojkul W. Gill monogeneans of potentially cultured tilapias and first record of Cichlidogyrus mbirizei Bukinga et al., 2012, in Thailand. Walailak J Sci & Tech. 2016;13:543–553. [Google Scholar]
- 74.Lim S-Y, Ooi A-L, Wong W-L. Gill monogeneans of Nile tilapia (Oreochromis niloticus) and red hybrid tilapia (Oreochromis spp.) from the wild and fish farms in Perak, Malaysia: infection dynamics and spatial distribution. Springerplus. 2016;5:1609. doi: 10.1186/s40064-016-3266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maneepitaksanti W, Nagasawa K. Monogeneans of Cichlidogyrus Paperna, 1960 (Dactylogyridae), gill parasites of tilapias, from Okinawa prefecture. Japan Biogeography. 2012;14:111–119. [Google Scholar]
- 76.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Briscoe AG, Hopkins KP, Waeschenbach A. High-throughput sequencing of complete mitochondrial genomes. In: Bourlat SJ, editor. Marine genomics. New York, NY: Humana Press; 2016. pp. 45–64. [DOI] [PubMed] [Google Scholar]
- 78.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 80.Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 81.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noe L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33:W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRA - a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212. doi: 10.1186/1471-2105-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Telford MJ, Herniou EA, Russell RB, Littlewood DTJ. Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. P Natl Acad Sci USA. 2000;97:11359–11364. doi: 10.1073/pnas.97.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 90.Philippe H, de Vienne D, Ranwez V, Roure B, Baurain D, Delsuc F. Pitfalls in supermatrix phylogenomics. Eur J Taxon. 2017;(283):1–25.
- 91.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 92.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. P Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 95.Xia X. DAMBE6: new tools for microbial genomics, phylogenetics and molecular evolution. J Hered. 2017;108:431–437. doi: 10.1093/jhered/esx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xia X. Assessing substitution saturation with DAMBE. Theory. In: Lemey P, Salemi M, Vandamme A-M, editors. The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing. Cambridge: Cambridge University Press; 2009. pp. 615–623. [Google Scholar]
- 97.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 98.Yang ZPAML. 4: a program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 99.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Perkins EM, Donnellan SC, Bertozzi T, Whittington ID. Closing the mitochondrial circle on paraphyly of the Monogenea (Platyhelminthes) infers evolution in the diet of parasitic flatworms. Int J Parasitol. 2010;40:1237–1245. doi: 10.1016/j.ijpara.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 101.Kang S, Kim J, Lee J, Kim S, Min GS, Park JK. The complete mitochondrial genome of an ectoparasitic monopisthocotylean fluke Benedenia hoshinai (Monogenea: Platyhelminthes) Mitochondr DNA. 2012;23:176–178. doi: 10.3109/19401736.2012.668900. [DOI] [PubMed] [Google Scholar]
- 102.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Meth. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–472. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- 106.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 109.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 110.Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 111.Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada DALTER. Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:W14–W18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Massouh A, Schubert J, Yaneva-Roder L, Ulbricht-Jones ES, Zupok A, Johnson MTJ, et al. Spontaneous chloroplast mutants mostly occur by replication slippage and show a biased pattern in the plastome of Oenothera. Plant Cell. 2016;28:911–929. doi: 10.1105/tpc.15.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 114.Bernt M, Merkle D, Ramsch K, Fritzsch G, Perseke M, Bernhard D, et al. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics. 2007;23:2957–2958. doi: 10.1093/bioinformatics/btm468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Annotation of the ribosomal operons of the four African monogenean species. (TXT 1 kb)
Data Availability Statement
The sequence data produced and analysed during the current study were deposited in NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and are freely available under accession numbers MG970255-8 and MG973075-8. Voucher specimens are available in the invertebrate collection of the Royal Museum for Central Africa (RMCA), Tervuren, Belgium. The posterior ends (with opisthaptor) of four of the specimens of Macrogyrodactylus karibae used were deposited under accession numbers M.T.38263–6; for the other monogenean species, entire animals were used for DNA extraction, and conspecifics from the same host specimen are available under accession numbers MRAC M.T.38239–62.