Abstract
Backgrounds
Recent studies have identified the role of serologic markers in characterizing disease phenotype, location, complications, and severity among Northern Europeans (NE) with Crohn’s disease (CD). However, very little is known about the role of serology in CD among African Americans (AA). Our study explored the relationship between serology and disease phenotype in AA with CD, while controlling for genetic ancestry.
Methods
AAs with CD were enrolled as participants through multicenter collaborative efforts. Serological levels of IgA anti-Saccharomyces cervisiae antibody (ASCA), IgG ASCA, E. coli outermembrane porin C, anti-CBir1, and ANCA were measured using enzyme-linked immunosorbent assays. Genotyping was performed using Illumina immunochip technology; an admixture rate was calculated for each subject. Multiple imputation by chained equations was performed to account for data missing at random. Logistic regression was used to calculate adjusted odds ratio (OR) for associations between serological markers and both complicated disease and disease requiring surgery.
Results
A total of 358 patients were included in the analysis. The majority of our patients had inflammatory, noncomplicated disease (58.4%), perianal disease (55.7%), and documented colonic inflammation (86.8%). On multivariable analysis, both IgG ASCA and OmpC were associated with complicated disease (OR, 2.67; 95% CI, 1.67–4.28; OR, 2.23; 95% CI, 1.41–3.53, respectively) and disease requiring surgery (OR, 2.51; 95% CI, 1.49–4.22; OR, 3.57; 95% CI, 2.12–6.00). NE admixture to the African genome did not have any associations or interactions in relation to clinical outcome.
Conclusions
Our study comprises the largest cohort of AAs with CD. The utility of serological markers for the prognosis of CD in NE applies equally to AA populations.
Keywords: African Americans, Crohn’s disease, inflammatory bowel disease, serology
Crohn’s disease (CD) encompasses a spectrum of chronic, relapsing, and remitting inflammatory symptoms that affect pediatric and adult populations, with the peak incidence occurring in adolescents and young adults. Recent studies have explored the relationship between genetic, microbial, and immune responses in CD, including the role of antimicrobial serologies in characterizing disease phenotype, clinical course, and natural history.1–4 In particular, significant associations have been identified between the presence of antibodies to Saccharomyces cerevisiae (ASCA IgA and ASCA IgG), antibodies to Escherichia coli outer membrane porin C (anti-OmpC), antibodies to CBIR1 flagellin (anti-CBir1) and perinuclear anti-nuclear cytoplasmic antibodies (pANCA) with disease phenotype and clinical outcomes.5 ASCA, anti-CBir1, and anti-OmpC positivities have been associated with ileal disease, stricturing, and/or penetrating disease, a higher risk for surgery, and earlier disease onset, while ANCA positivity has been associated with colonic disease and a more “benign” disease course.4 The vast majority of these studies have originated from North America and Europe and have primarily focused on patient populations of Caucasians of Western European decent. Although the findings of these studies have contributed to our understanding of CD pathogenesis as well as to the development of tools both for differentiating types of inflammatory bowel disease (IBD) and predicting risk stratification, these findings need to be explored in non-Caucasian populations.1 An understanding of the relevance of these markers in demographically diverse groups is necessary if the potential clinical utility of IBD serology is to be extended to all populations.
Historically considered a disease of Northern European (NE) ancestries, it is now recognized that CD affects every race and ethnicity, with a rising incidence in non-European populations. Furthermore, CD characteristics, treatment responses, and disease course can vary in different populations.6–8 Specifically, the epidemiology, genetics, and natural history of IBD among African Americans (AAs) remains severely understudied.6–8 With regards to IBD-associated serology in AA, there has only been 1 study to date, which identified that ASCA has a similar sensitivity but lower specificity for CD, as well as an association with ileal involvement, complicated behavior, and surgery in AAs with CD.9 In addition to limiting investigations on ASCA alone, this study was further limited by its reliance on self-reported ethnicity. AAs are an admixed population, and the AA genome on average is comprised of 80% West African ancestry and 20% European ancestry.10 Due to differences in environmental and genetic influences among various ethnic populations, there is a need to further delineate the diagnostic value of these biomarkers among AA.4 Although genetic susceptibility among Caucasians and AAs is mostly similar, differences exist, including the association with NOD2 mutations, which is less influential in AAs, possibly due to Caucasian admixture among AAs. Hence, exploration of these serological markers in AAs among CD phenotypes may allow clinicians to risk stratify in an important and growing demographic of IBD patients. Our study was designed to explore the relationship between antibodies and disease phenotype in AAs with CD while controlling for genetic ancestry.
MATERIALS AND METHODS
Study Design and Hypothesis
We conducted a cross-sectional study testing the hypothesis that serological levels of IgA ASCA, IgG ASCA, anti-OmpC, anti-CBir1, and ANCA are a risk factor for complicated disease and disease requiring surgery among AAs with CD. The Institutional Review Board of the participating sites (Emory University, Children’s Hospital of Atlanta, Atlanta VA Medical Center Children’s Hospital of Philadelphia, Cincinnati Children’s Hospital Medical Center, University Hospitals Case Western Medical Center, University of Maryland School of Medicine, Vanderbilt-Monroe Carell Jr. Children’s Hospital, UT Southwestern, UNC Chapel Hill, University of Chicago Children’s Hospital, LSU Health Science Center, Cooks Medical Center, and Willis-Knighton Physician Network) approved the study, and informed consent was obtained from all participants.
Study Population
The study population was recruited between August 2011 and March 2014 from 12 participating sites. Serum and genomic DNA along with clinical data were obtained on all the subjects and entered into an electronic database (RedCap). All clinical information was obtained either at the time of enrollment or by retrospective chart review. All cases had a diagnosis of CD, based on standard diagnostic criteria, readily available serological results, and clearly documented disease behavior. Related individuals were removed from the study.11
Clinical Characteristics of CD Patients
Patients’ demographics, date of diagnoses, disease location, disease behavior, surgical history, presence of extraintestinal manifestations (EIMs), smoking history, autoimmune history, family history, and history of biologic medication use were obtained either at the time of blood draw or via retrospective chart review. CD phenotype was classified in accordance with the Montreal Classification for adults and Paris Classification for children.12, 13 For disease location, patients were classified into 1 of 4 mutually exclusive groups: L1 (terminal ileal disease +/- limited cecal disease), L2 (colonic disease), L3 (ileocolonic disease), or L4 (isolated upper disease without evidence of ileal or colonic disease). The presence of upper gastrointestinal disease was categorized into 4 groups: 0 (no disease), L4a (upper disease proximal to the Ligament of Treitz), L4b (upper disease distal to the Ligament of Treitz and proximal to the distal 1/3 ileum), and L4ab.13 Each patient’s disease behavior was categorized into 1 of 4 groups: B1 (nonstricturing nonpenetrating disease), B2 (stricturing disease), B3 (penetrating disease), and B2B3 (both stricturing and penetrating disease, either at the same moment in time or separately over a period of time).13 Complicated disease was defined as B2, B3, or B2B3. Perianal disease was positive if the patient had any of the following: skin tag, anal fissure, perianal abscess, or perianal fistula. History of surgery was defined as any confirmed, documented surgical procedure necessary for the treatment or management of CD except surgery related to existing perianal disease.
Serological Analysis
Blood samples were collected at the time of enrollment. Sera were measured for expression of ASCA IgG, ASCA IgA, anti-OmpC, anti-CBir1, and ANCA antibodies in a blinded fashion by an enzyme-linked immunosorbent assay (ELISA). The tests were run at Cedars-Sinai using previously described protocols and standards.14 Antibody levels were measured relative to the Cedars-Sinai Laboratory standard and were expressed in ELISA units (EU/mL).
All serological values were treated as continuous variables except when used for bivariable analysis, where they were analyzed as categorical variables. All cutoff values were determined according to previous studies.14
Genotyping
DNA samples were derived from whole blood. All DNA samples were genotyped by immnochip using the Illumina Immunochip, and the genotyped cells were made by using GenomeStudio version 2011. We have then extracted the data for NOD2/CARD15 single nucleotide polymorphsims (SNPs) rs2066844, rs2066846, and rs5743293.1 and Genotyping Module version 1.9.4. Samples were genotyped at Cedars-Sinai Medical Center. For analysis purposes, patients were categorized by NOD2 genotype: NOD2-positive patients included those who carried at least 1 CD-associated NOD2 allele; NOD2-negative patients included those who carried only nonrisk alleles.
African Ancestry Estimation
Because AAs are well modeled as linear combinations of West African and European ancestries, we chose the WINPOP model in the LAMP package to estimate the locus-specific local ancestry. WINPOP takes allele frequencies from ancestral populations (YRI and CEU from HapMap) as input, and outputs the local ancestry estimate for each sample at each SNP as a proportion of YRI at values of 0, 0.5, or 1.15–17 The global YRI ancestry for each sample was estimated by using ADMIXTURE, which only requires sample genotypes and number of ancestral populations as input and outputs estimated proportion from each ancestral population with a numeric value ranging from 0 to 1.18
Statistical Analysis
Statistical analyses were performed using SAS 9.3 (SAS Institute Inc., Cary, NC) and R 3.1.1 (R Foundation for Statistical Computing, Vienna, Austria). Statistical tests were 2-sided, and a P value of less than 0.05 was considered statistically significant. The Shapiro-Wilk Test was used to test for normality on all continuous variables. Descriptive statistics are presented as median (interquartile range) and percent (95% confidence interval) for continuous and categorical outcomes, respectively. Univariate analyses were performed to determine associations between outcome, exposure, and predictor variables. Nonparametric Mann-Whitney tests were used for all continuous variables, and chi-square or Fisher exact tests were used for binomial variables. All independent terms were tested for linearity with the log odds of the outcome using 5-knot restricted cubic splines (RCS) according to the method of Harrell.19 One-knot linear splines were created based on review of the plot of the RCS and subject matter knowledge for those variables that did not meet this assumption. Multiple imputation by chained equations (MICE) was employed for multivariable analysis in order to account for data presumed to be missing at random.20 Imputed data sets were only used for multivariable modeling.
A multivariable logistic regression model was used to evaluate the primary associations among serologic levels with the presence of complicated disease and disease requiring surgery. All relevant demographic, phenotypic, genetic, and serological variables except history of surgery, history of biologic use, and outcome of interest were included in the initial model. Fast backward variable selection using Wald chi-square of individual factors at the 0.05 significance level was performed until only statistically significant variables remained.21 Odds ratios and 95% confidence intervals were calculated using the 75th percentile to the 25th percentile for all continuous variables as a way to assess which variables have the largest impact over the same range. All initial and final models were tested for interaction with percent African admixture.
RESULTS
Patient Characteristics
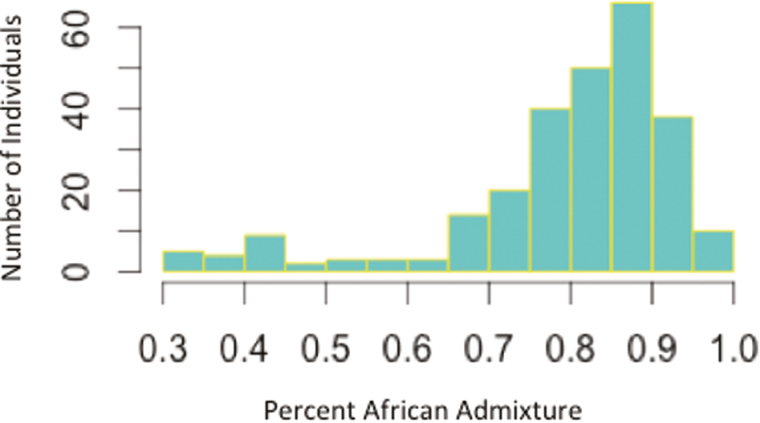
Data from 358 unrelated AA participants with CD were analyzed. The demographic and phenotypic characteristics of these participants are described in Table 1. The median age at diagnosis was 15 years (IQR, 12–22.5). Ileocolonic disease was observed in 53.6% of patients. Perianal disease was observed in 55.7%. Disease behavior was complicated in 41.6%. Biologic agents were used at some point in the disease course in 69.9% of patients. NOD2 positivity was observed in 5.9% of patients. More than 15% of data was missing for percent African admixture. The median serological levels and respective IQR for all serological markers are displayed in Table 1. The distribution of percent African admixture in the cohort is illustrated in Fig. 1: Percent admixture rates range from 0.31 to 1.0, with a median of 0.83 (IQR, 0.8–0.9).
Table 1.
Demographic and Phenotypic Characteristics of Cohort
| Variable | Median (IQR) or No. (%) |
|---|---|
| African admixturea | 0.83 (0.8–0.9) |
| Age (IGR), y | 20 (15–36) |
| Age at diagnosis (IQR), y | 15 (12–22.5) |
| Age category at diagnosis, y | |
| A1: younger than 16 | 180 (51.1%) |
| A2: between 17 and 40 | 147 (41.7%) |
| A3: older than 40 | 25 (7.1%) |
| Male sex, No. (%) | 181 (50.6%) |
| Family history of IBD, No. (%) | 27 (8.2%) |
| History of autoimmune disorder, No. (%) | 96 (27.7%) |
| Extraintestinal manifestations, No. (%) | 95 (27.7%) |
| Smoking (ever), No. (%) | 135 (42.4%) |
| Disease location, No. (%) | |
| L1: ileal | 42 (13.2%) |
| L2: colonic | 106 (33.2%) |
| L3: ileocolonic | 171 (53.6%) |
| L4: isolated upper | 0 (0.0%) |
| Upper disease modifier, No. (%) | |
| No disease | 198 (63.6%) |
| L4Ab | 73 (23.5%) |
| L4B | 20 (6.4%) |
| L4AB | 20 (6.4%) |
| Disease behavior, No. (%) | |
| B1: inflammatory | 209 (58.4%) |
| B2: stricturing | 6 (18.2%) |
| B3: penetrating | 37 (10.3%) |
| B2B3: stricturing and penetrating | 47 (13.1%) |
| Complicated disease, No. (%) | 149 (41.6%) |
| Perianal disease,a No. (%) | 156 (55.7%) |
| Surgical history, No. (%) | 128 (40.1%) |
| Biologic use (ever), No. (%) | 229 (69.9%) |
| NOD2 positivity, No. (%) | 20 (5.9%) |
| Serological responsec (level) | |
| ASCA IgA | 7.1 (2.3–26.7) |
| ASCA IgG | 21.1 (5.9–53.5) |
| Anti-OmpC | 15.7 (9.9–27.7) |
| Anti-CBir1 | 24.2 (14.4–46.1) |
| pANCA | 16.4 (10.3–23.4) |
| Serological response, No. (%) | |
| ASCA IgA | 125 (34.9%) |
| ASCA IgG | 117 (32.7%) |
| Anti-OmpC | 112 (31.3%) |
| Anti-CBir1 | 177 (49.4%) |
| pANCA | 61 (17.0%) |
aMissing Greater than 15% of the data.
bL4A: disease proximal to the ligament of treitz; L4B: disease distall to the ligament of treitz but proximal to the distal 1/3 of the ileum.
cResults are classified as negative or positive according to the manufacturer’s determined cutoffs. A negative response was defined as: ASCA IgA < 20 EU/mL; ASCA IgG < 40 EU/mL; anti-OmpC < 23 EU/mL; anti-CBir1 < 25 EU/mL; pANCA < 17.46 EU/Ml.
anti-Cbir1 = flagellin-like bacterial antigen; Panca = perinuclear antinuclear cytoplasmic antibody.
FIGURE 1.
Percent African admixture. The distribution of African admixture among all study participants. The median percent admixture was 0.83 percent, indicating that the genetic make-up of the majority of AAs in our study are 83% African and 17% European.
Patient Demographic and Phenotypic Characteristics According to Serological Status
Among the 358 AA patients with CD, 155 (43.3%) were (IgG and/or IgA) ASCA positive, 112 (31.3%) were anti-OmpC positive, 177 (49.4%) were positive for anti-CBir1, and 61 (17.0%) were positive for ANCA. The demographic and phenotypic characteristics of participants according to serological status are displayed in Table 2. On univariate analysis, the following characteristics were associated with ASCA positivity: personal history of immune-mediated (P = 0.02), EIM (P = 0.02), complicated disease (P < 0.0001), ileal disease (P < 0.0001), and disease requiring surgery (P = 0.002). Anti-OmpC positivity was significantly associated with age at enrollment (P = 0.005), complicated disease (P = 0.02), and disease requiring surgery (P < 0.0001). Anti-CBir1 positivity was associated with age at enrollment (P = 0.001), age at diagnosis (P = 0.008), percent African admixture (P = 0.004), current smoke exposure (P = 0.004), and isolated ileal disease (P = 0.03). A negative ANCA was significantly associated with EIM (P = 0.01), complicated disease (P = 0.003), and ileal disease (P = 0.01).
Table 2.
Demographic and Phenotypic Characteristics According to Serologic Status
| ASCA a | anti-OmpC | anti-CBir1 | pANCA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Positive (n = 155) | Negative (n = 203) | P | Positive (n = 112) | Negative (n = 246) | P | Positive (n = 177) | Negative (n = 181) | P | Positive (n = 61) | Negative (n = 297) | P |
| Age (IGQ),b y | 21 (16–35) | 18 (15–37) | 0.11 | 25 (16–40) | 18 (15–33) | 0.005 | 18 (15–28) | 22 (16–44) | 0.001 | 18 (16–39) | 20 (15–36) | 0.61 |
| Age at diagnosis, y | 16 (13–22) | 15 (11–24) | 0.25 | 15 (12–23.5) | 15 (11.5– 21.5) | 0.44 | 15 (12–19) | 16 (12–25) | 0.008 | 16 (12–24) | 15 (12–22) | 0.31 |
| Male sex, No. (%) | 81 (52.3%) | 100 (49.2%) | 0.57 | 57 (50.9%) | 124 (50.4%) | 0.93 | 93 (53.5%) | 88 (48.6%) | 0.46 | 25 (41.0%) | 156 (53.5%) | 0.10 |
| NOD2 positive, No. (%) | 12 (8.7%) | 8 (4.7%) | 0.16 | 4 (4.2%) | 16 (7.5%) | 0.28d | 8 (5.4%) | 12 (7.5%) | 0.45 | 2 (3.9%) | 18 (7.0%) | 0.4d |
| African admixtureb,c | 0.80 (0.74– 0.88) | 0.84 (0.76– 0.89) | 0.41 | 0.84 (0.78– 0.90) | 0.82 (0.62– 0.88) | 0.06 | 0.84 (0.79– 0.90) | 0.82 (0.71– 0.93) | 0.004 | 0.83 (0.76– 0.89) | 9.83 (0.76– 0.89) | 0.80 |
| Family history of IBD, No. (%) | 11 (7.9%) | 16 (8.5%) | 0.83 | 7 (6.9%) | 20 (8.8%) | 0.56 | 12 (7.3%) | 15 (9.1%) | 0.55 | 4 (7.0%) | 23 (8.4%) | 0.72d |
| Autoimmune Disease, no. (%) | 33 (21.5%) | 63 (32.5%) | 0.02 | 31 (28.4%) | 65 (27.3%) | 0.83 | 45 (26.3%) | 51 (29.0%) | 0.58 | 14 (24.6%) | 82 (28.3%) | 0.57 |
| EIM, No. (%) | 32 (21.5%) | 63 (32.5%) | 0.02 | 33 (31.4%) | 62 (26.1%) | 0.31 | 43 (25.2%) | 52 (30.2%) | 0.30 | 24 (40.7%) | 71 (29.1%) | 0.01 |
| Current smoke exposure, No. (%) | 35 (28.2%)b | 49 (29.2%) | 0.86 | 27 (29.0%) | 57 (28.6%) | 0.95 | 30 (20.6%) | 54 (37.0%) | 0.002 | 14 (27.5%) | 70 (29.1%) | 0.82 |
| Upper disease,e No. (%) | 50 (36.2%) | 63 (26.4%) | 0.97 | 32 (33.7%) | 81 (37.5%) | 0.52 | 56 (37.3%) | 57 (35.4%) | 0.72 | 18 (33.3%) | 95 (37.0%) | 0.61 |
| Perianal disease, No. (%) | 78 (61.4%) | 78 (51.0%) | 0.08 | 59 (64.8%) | 97 (51.3%) | 0.03 | 85 (57.4%) | 71 (53.8%) | 0.54 | 18 (42.9%) | 138 (58.0%) | 0.07 |
| Outcomes | ||||||||||||
| Complicated disease,f No. (%) | 85 (54.8%) | 64 (31.5%) | <0.0001 | 57 (50.9%) | 92 (37.4%) | 0.02 | 82 (46.3%) | 67 (37.0%) | 0.07 | 15 (24.6%) | 134 (45.1%) | 0.003 |
| Ileal disease,g No. (%) | 85 (54.8%) | 103 (56.0%) | <0.0001 | 68 (68.0%) | 145 (66.2%) | 0.75 | 109 (66.1%) | 104 (67.5%) | 0.78 | 28 (50.1%) | 185 (70.1%) | 0.01 |
| Isolated ileal disease, No. (%) | 35 (11.1%) | 46 (14.7%) | 0.35 | 24 (12.0%) | 57 (13.7%) | 0.68 | 27 (9.1%) | 54 (17.5%) | 0.03 | 12 (10.9%) | 69 (13.6%) | 0.59 |
| Surgery, No. (%) | 72 (49.3%) | 56 (32.4%) | 0.002 | 56 (56.0%) | 72 (32.9%) | <0.0001 | 62 (38.5%) | 66 (41.7%) | 0.55 | 16 (28.6%) | 112 (42.6%) | 0.05 |
IgA ASCA was classified as negative (0.0–20.0 EU/mL) and positive (≥20.0 EU/mL); IgG ASCA was classified as negative (0.0–40.0 EU/mL) and positive (≥40.0 EU/mL); Anti-OmpC was classified as negative (0.0–23.0 EU/mL) and positive (≥23.0 EU/mL); Anti-CBir1 was classified as negative (0.0–25.0 EU/mL) and positive (≥25.0 EU/mL); pANCA was classified as negative (0.0–30.0 EU/mL) and positive (≥30.0 EU/mL).
aASCA positivity is defined as being either IgA or IgG ASCA positive.
bMedian (IQR).
cMissing greater than 15% of the data.
dFisher exact test.
eUpper disease is defined as L4A, L4B, or L4AB positivity.
fComplicated disease is defined as B2 or B3 or B2B3 disease behavior.
gIleal disease is defined as the (L1) isolate ileal or (L3) ileocolonic disease.
anti-Cbir1 = flagellin-like bacterial antigen; pANCA = perinuclear antinuclear cytoplasmic antibody.
Patient Characteristics and Serological Levels Associated With Complicated Disease
One-knot linear splines were set at 15 EU/mL, 20 EU/mL, and 20 EU/mL for the variables ASCA IgG, anti-OmpC, and anti-CBir1, respectively (Supplemental Digital Content, Fig. S1). The final multivariable model (Table 3), after fast backwards selection of individual factors at the 0.05 significance level, showed that OmpC (OR, 2.23; 95% CI, 1.41–3.53), IgG ASCA (OR, 2.67; 95% CI, 1.67–4.28), and ileal disease (OR, 2.94; 95% CI, 5.26–8.77) were independently associated with complicated disease. There was no interaction between NOD2 positivity and any of the serological markers in the final model. Additionally, the interaction between percent African admixture and all covariates included in the initial and final model was assessed and found to be insignificant.
Table 3.
Adjusted Odds Ratio for Complicated Crohn’s Disease
| Odds Ratioa | 95% Confidence Interval | ||
|---|---|---|---|
| Anti-OmpCb | Comparing 27.7 EU/mL with 17.7 EU/mL | 2.23 | 1.41–3.53 |
| IgG ASCAb | Comparing 53.5 EU/mL with 5.9 EU/mL | 2.67 | 1.67–4.28 |
| Ileal disease | Positive to negative | 2.94 | 5.26–8.77 |
aORs were calculated by comparing the 75th percentile with the 25th percentile.
bOne-knot linear splines were created based on review of restricted cubic spline and subject matter knowledge.
Patient Characteristics and Serological Levels Associated With Surgery
One-knot linear splines were set at 25 years, 40 EU/mL, 20 EU/mL, and 20 EU/mL for the variables age at diagnosis, IgA ASCA, IgG ASCA, and anti-OmpC, respectively (Supplemental Digital Content, Fig. S2). The final multivariable model (Table 4), after fast backwards selection of individual factors at the 0.05 significance level, showed that disease requiring surgery was independently associated with NOD2 positivity (OR, 2.9; 95% CI, 1.01–8.33), IgG ASCA (OR, 2.51, 95% CI, 1.49–4.22), and anti-OmpC (OR, 3.57; 95% CI, 2.12–6.00). There was no interaction between NOD2 positivity and any of the serological markers in the final model. Additionally, interaction between percent African admixture and all covariates included in the initial models and the final remaining model was assessed and found to be insignificant.
Table 4.
Adjusted Odds Ratio for Disease Requiring Surgery
| Odds Ratioa | 95% Confidence Interval | ||
|---|---|---|---|
| NOD2 positivity | Positive to negative | 2.9 | 1.01–8.33 |
| IgG ASCAb | Comparing 53.3 EU/mL with 5.9 EU/mL | 2.51 | 1.49–4.22 |
| Anti-OmpCb | Comparing 27.7 EU/mL with 9.9 EU/mL | 3.57 | 2.12–6.00 |
aORs were calculated by comparing the 75th percentile with the 25th percentile.
bOne-knot linear splines were created based on review of restricted cubic spline and subject matter knowledge.
DISCUSSION
Our study was designed to describe the phenotypic behavior of CD in AAs and to explore the relationship between antimicrobial serology and disease outcomes, while controlling for genetic ancestry. Our series comprises the largest cohort of AAs with CD and is the first study to evaluate anti-OmpC, anti-CBir1, and ANCA and their association with phenotype and disease behavior.
We found that the majority of AAs with CD had inflammatory, noncomplicated disease type (58.4%), mostly the colon (86.8%). Fifty-three percent of patients with colonic disease also had disease affect the small bowel, and 55.7% of our patients had perianal disease at the time of enrollment. Our observations regarding primary disease location and disease behavior concur with the findings of both Mahid et al. and Hou et al., who published systematic reviews on the epidemiology and phenotypic presentation of IBD in AAs.7, 8 These findings are also consistent with those found in European ancestry populations, which is contrary to earlier literature reporting that AAs have a more severe course with different disease distribution.22, 23
However, we found a notably higher prevalence of perianal disease than previously reported. Mahid et al. reported the prevalence of perianal disease to be 26%, Hou et al. reported a prevalence of 25%, and Dassopoulos et al. reported a prevalence of 34%. In other reports, the prevalence of perianal disease ranged from 0% to 60%.24–31 The wide range appears to be a function of (i) the sample size, (ii) the disease severity of the patient population, and (iii) the definition of perianal disease. The perianal disease modifier, as described in the Montreal Classification, includes skin tags, anal fissures, perianal abscesses, or perianal fistulas, but many studies either did not clarify how perianal disease was defined or included only perianal abscesses and fistulas in their definition.12, 13
While numerous studies have reported the presence and magnitude of serological antibodies with complicated disease, ileal involvement, and earlier-onset disease in NE populations, less is known about these associations in AAs. Dassopoulos et al. conducted the first study to assess ASCA levels in AAs with CD. They reported that, similar to NEs, ASCA was independently associated with complicated behavior and surgery in AAs with CD.9 Our study expands upon this study by exploring the association between ASCA, anti-OmpC, anti-CBir1, and ANCA with complicated disease and disease requiring surgery. We not only confirmed the association between IgG ASCA with complicated disease and surgeries but also identified an association between anti-OmpC with complicated disease and disease requiring surgery in AAs. We also found that IgG ASCA remained statistically significant after controlling for ileal disease, NOD2 positivity, and percent African Admixture. In contrast to prior reports, in which ASCAs were known to be associated with white populations, ileal involvement, and NOD2 status, the association between ASCA and complicated disease and disease requiring surgery is not the result of European admixture.9, 32 Our findings do concur with prior literature regarding the association between anti-OmpC with complicated disease and the need for surgery in NE populations.5, 33, 34
Dubinsky et al. were the first to tabulate antibody sum scores in NEs and found that the frequency of internal penetrating disease, stricturing disease, and disease requiring surgery increased as the number of immune responses increased.5 In a subanalysis, we calculated antibody sum scores according to the method described by Dubinsky et al. and observed similar results (Supplemental Digital Content, Fig. S3).5 We saw an increase in complicated behavior types (B2, B3, and B2B3) with a higher sum score, and, conversely, inflammatory disease (B1) was least present in the highest-sum group.
These findings suggest that the phenotypes of CD in Caucasians and AAs are not as dissimilar as initially believed.22, 23 We hypothesized that the genetic differences between AAs and NEs would result in statistically significant, clinically meaningful differences between antimicrobial antibodies and their relationship with disease phenotype in CD. We also hypothesized that there would be an interaction between African admixture and serological levels, which was also not observed. Contrary to our initial hypotheses, we found that antimicrobial antibodies and their association with disease phenotype and behavior can be interpreted similarly between AAs and NEs. We postulate that these similarities are more of a function of similarities among the intestinal microbiota than differences in genetic make-up, emphasizing the role of the environment and the limitation of genetics to diagnose IBD or predict disease behavior.1 These findings are further corroborated by twin concordance studies that have demonstrated that only a small percentage of the risk of developing inflammatory bowel disease is related to genetics.35
Limitations of our study include the substantial missing data from our participant population and the predominance of participants diagnosed younger than age 16 years. While there was no missing data related to serological levels, we were missing more than 15% of data in variables measuring the presence of perianal disease and percent African admixture. Eleven additional confounding variables were missing values for anywhere between 1.4% and 14.0% of the participants. As a result, our sample size decreased by more than 50% when we performed the multivariable analysis using our original model. We addressed this limitation by performing multiple imputations by chained equations, a principled method of dealing with missing data.20 We were also limited by the absence of demographic or phenotypic information on those patients who chose not to participate. Additionally, our study population was based on a population with a pediatric majority. The majority of our patients were diagnosed younger than age 16 years, which is not reflective of the general population and likely suggests a population with more severe disease.
In contrast to the hypothesis that the diagnostic value of antimicrobial and autoimmune antibodies would vary among different ethnic and geographic populations due to differences in environmental and genetic influences, we found that with respect to ASCA and anti-OmpC, they behave similarly in AA and NE populations recruited from similar geographic areas.4 Our study also provides reassurance that the utility of serological markers for prognosis of CD applies equally to AA populations.
Supplementary Material
ACKNOWLEDGMENTS
Author contributions: Madeline Bertha helped conceptualize the paper, contributed to data acquisition, performed statistical analysis, wrote the manuscript, and reviewed and approved the final manuscript. Arthi Vasantharoopan contributed to statistical analysis, contributed to the manuscript, and reviewed and approved the final manuscript. Archana Kumar helped conceptualize the paper, contributed to data acquisition, contributed to statistical analysis, contributed to the manuscript, and reviewed and approved the final manuscript. Beau Bruce helped conceptualize the paper, performed statistical analysis, contributed to the manuscript, and reviewed and approved the final manuscript. Tatyana Hofmekler contributed to the manuscript and reviewed and approved the final manuscript. Jarod Prince contributed to data acquisition, contributed to the manuscript, and reviewed and approved the final manuscript. Cary Sauer helped contribute to the manuscript and reviewed and approved the final manuscript. Dermot McGovern conceptualized the manuscript, performed the assays, helped contributed to the manuscript, and reviewed and approved the final manuscript. Subra Kugathasan is the principle investigator of this project and responsible for the overall conduct, results, and conclusions of the paper. He conceptualized the paper, contributed to the manuscript, and reviewed and approved the final manuscript. All other authors contributed to the acquisition of data.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.ibdjournal.org).
Conflicts of interest: The authors have no financial relationships relevant to this article to disclose. The authors have no conflicts of interest to disclose.
Supported by NIH/NIDDK R01 DK087694 (S.K.) and National Center for Advancing Translational Sciences of National Institute of Health Award Number UL1TR000454 (M.B.).
REFERENCES
- 1. Gerich ME, McGovern DP. Towards personalized care in ibd. Nat Rev Gastroenterol Hepatol. 2014;11:287–299. [DOI] [PubMed] [Google Scholar]
- 2. Mow WS, Landers CJ, Steinhart AH et al. High-level serum antibodies to bacterial antigens are associated with antibiotic-induced clinical remission in Crohn’s disease: a pilot study. Dig Dis Sci. 2004;49:1280–1286. [DOI] [PubMed] [Google Scholar]
- 3. Papp M, Altorjay I, Dotan N et al. ; Hungarian IBD Study Group New serological markers for inflammatory bowel disease are associated with earlier age at onset, complicated disease behavior, risk for surgery, and nod2/card15 genotype in a hungarian ibd cohort. Am j Gastroenterol. 2008;103:665–681. [DOI] [PubMed] [Google Scholar]
- 4. Prideaux L, De Cruz P, Ng SC et al. Serological antibodies in inflammatory bowel disease: a systematic review. Inflamm Bowel Dis. 2012;18:1340–1355. [DOI] [PubMed] [Google Scholar]
- 5. Dubinsky MC, Kugathasan S, Mei L et al. ; Western Regional Pediatric IBD Research Alliance; Pediatric IBD Collaborative Research Group; Wisconsin Pediatric IBD Alliance Increased immune reactivity predicts aggressive complicating Crohn’s disease in children. Clin Gastroenterol Hepatol. 2008;6:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Straus WL, Eisen GM, Sandler RS et al. Crohn’s disease: does race matter? The Mid-Atlantic Crohn’s Disease Study Group. Am j Gastroenterol. 2000;95:479–483. [DOI] [PubMed] [Google Scholar]
- 7. Hou JK, El-Serag H, Thirumurthi S. Distribution and manifestations of inflammatory bowel disease in Asians, Hispanics, and African Americans: a systematic review. Am j Gastroenterol. 2009;104:2100–2109. [DOI] [PubMed] [Google Scholar]
- 8. Mahid SS, Mulhall AM, Gholson RD et al. Inflammatory bowel disease and African Americans: a systematic review. Inflamm Bowel Dis. 2008;14:960–967. [DOI] [PubMed] [Google Scholar]
- 9. Dassopoulos T, Nguyen GC, Talor MV et al. ; NIDDK IBD Genetics Consortium nod2 mutations and anti-saccharomyces cerevisiae antibodies are risk factors for Crohn’s disease in African Americans. Am j Gastroenterol. 2010;105:378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tishkoff SA, Reed FA, Friedlaender FR et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernstein CN, Fried M, Krabshuis JH et al. World Gastroenterology Organization practice guidelines for the diagnosis and management of ibd in 2010. Inflamm Bowel Dis. 2010;16:112–124. [DOI] [PubMed] [Google Scholar]
- 12. Satsangi J, Silverberg MS, Vermeire S et al. The montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levine A, Griffiths A, Markowitz J et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–1321. [DOI] [PubMed] [Google Scholar]
- 14. Landers CJ, Cohavy O, Misra R et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. [DOI] [PubMed] [Google Scholar]
- 15. Price AL, Helgason A, Palsson S et al. The impact of divergence time on the nature of population structure: an example from Iceland. Plos Genet. 2009;5:e1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pasaniuc B, Sankararaman S, Kimmel G et al. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25:i213–i221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12:523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrell F. Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression, and Survival Analysis. New York: Springer-Verlag, Inc; 2001. [Google Scholar]
- 20. Azur MJ, Stuart EA, Frangakis C, Leaf PJ. Multiple imputation by chained equations: what is it and how does it work?Int j Methods Psychiatr Res. 2011;20:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lawless JF, Singhal K. Efficient screening of non-normal regression models. Biometrics. 1978;34:318–327. [Google Scholar]
- 22. Agrez MV, Valente RM, Pierce W et al. Surgical history of Crohn’s disease in a well-defined population. Mayo Clin Proc. 1982;57:747–752. [PubMed] [Google Scholar]
- 23. Sonnenberg A, McCarty DJ, Jacobsen SJ. Geographic variation of inflammatory bowel disease within the United States. Gastroenterology. 1991;100:143–149. [DOI] [PubMed] [Google Scholar]
- 24. Basu D, Lopez I, Kulkarni A et al. Impact of race and ethnicity on inflammatory bowel disease. Am j Gastroenterol. 2005;100:2254–2261. [DOI] [PubMed] [Google Scholar]
- 25. Cross RK, Jung C, Wasan S et al. Racial differences in disease phenotypes in patients with Crohn’s disease. Inflamm Bowel Dis. 2006;12:192–198. [DOI] [PubMed] [Google Scholar]
- 26. Deveaux PG, Kimberling J, Galandiuk S. Crohn’s disease: presentation and severity compared between black patients and white patients. Dis Colon Rectum. 2005;48:1404–1409. [DOI] [PubMed] [Google Scholar]
- 27. Goldman CD, Kodner IJ, Fry RD et al. Clinical and operative experience with non-Caucasian patients with Crohn’s disease. Dis Colon Rectum. 1986;29:317–321. [DOI] [PubMed] [Google Scholar]
- 28. Mahid SS, Minor KS, Stromberg AJ et al. Active and passive smoking in childhood is related to the development of inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:431–438. [DOI] [PubMed] [Google Scholar]
- 29. Nguyen GC, Torres EA, Regueiro M et al. Inflammatory bowel disease characteristics among African Americans, Hispanics, and non-Hispanic whites: characterization of a large North American cohort. Am j Gastroenterol. 2006;101:1012–1023. [DOI] [PubMed] [Google Scholar]
- 30. Ogunbi SO, Ransom JA, Sullivan K et al. Inflammatory bowel disease in African-American children living in Georgia. j Pediatr. 1998;133:103–107. [DOI] [PubMed] [Google Scholar]
- 31. Simsek H, Schuman BM. Inflammatory bowel disease in 64 black patients: analysis of course, complications, and surgery. j Clin Gastroenterol. 1989;11:294–298. [DOI] [PubMed] [Google Scholar]
- 32. Adeyanju O, Okou DT, Huang C et al. Common nod2 risk variants in African Americans with Crohn’s disease are due exclusively to recent Caucasian admixture. Inflamm Bowel Dis. 2012;18:2357–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferrante M, Henckaerts L, Joossens M et al. New serological markers in inflammatory bowel disease are associated with complicated disease behaviour. Gut. 2007;56:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mow WS, Vasiliauskas EA, Lin YC et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. [DOI] [PubMed] [Google Scholar]
- 35. Halfvarson J, Bodin L, Tysk C et al. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–1773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.