Abstract
INTRODUCTION
Lung cancer continues to be the leading cause of cancer-related mortality worldwide. Early detection has proven essential to extend survival. Genomic and proteomic advances have provided impetus to the effort dedicated to detect and diagnose the disease at an earlier stage. Recently, the study of metabolites associated with tumor formation and progression has inaugurated the era of cancer metabolomics to aid in this effort.
OBJECTIVES
This review summarizes recent work regarding novel metabolites with the potential to serve as biomarkers for early lung tumor detection, evaluation of disease progression, and prediction of patient outcomes.
METHOD
We compare the metabolite profiling of cancer patients with that of healthy individuals, and the metabolites identified in tissue and biofluid samples and their usefulness as lung cancer biomarkers. We discuss metabolite alterations in tumor versus paired non-tumor lung tissues, as well as metabolite alterations in different stages of lung cancers and their usefulness as indicators of disease progression and overall survival. We evaluate metabolite dysregulation in different types of lung cancers, and those associated with lung cancer versus other lung diseases. We also examine metabolite differences between lung cancer patients and smokers/risk-factor individuals.
RESULT
Although an extensive list of metabolites has been evaluated to distinguish between these cases, refinement of methods is further required for adequate patient diagnosis.
CONCLUSION
We conclude that with technological advancement, metabolomics may be able to replace more invasive and costly diagnostic procedures while also providing the means to more effectively tailor treatment to patient-specific tumors.
Keywords: Cancer metabolomics, lung cancer, metabolism biomarkers, cancer diagnosis, cancer treatment
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with 1.8 million new cases being estimated in 2012 (Wong et al., 2017). In the United States alone, 154,050 deaths and 234,030 new cases are expected in 2018 (Siegel et al., 2018). Although the 5-year survival rate for lung and bronchial tumors is only 18% (15% for men and 21% for women), rates increase to 55% when the cancer is detected at an early, localized stage (Stage I and some Stage II) (ACS, 2017). The most common form of lung cancer (85%) is Non-Small Cell Lung Cancer (NSCLC) (ACS, 2017), which includes three subtypes, squamous cell carcinoma, adenocarcinoma and large cell carcinoma (Ambrosini et al., 2012). Approximately 10-15% of lung cancers are Small Cell Lung Cancers (SCLC), which are more aggressive than NSCLC and spread quickly (Jackman and Johnson, 2005). Lung carcinoid tumors represent less than 5% of lung cancers and these grow relatively slowly, do not metastasize and are not characterized by glandular or squamous differentiation (Ginsberg et al., 2007). Recent studies have focused on early diagnosis of the type and stage of disease in addition to treatment. In particular, gene and protein expression studies have provided information regarding mechanisms that direct tumor initiation and progression. However, such studies are insufficient to characterize the current physiological state of an individual, which may be influenced by disease-state and environmental factors (Carr et al., 2014, Kantae et al., 2017). The individual genetic variability between patients, along with the cost of performing genetic and proteomic analyses, preclude the monitoring of all the relevant processes in disease progression (Wishart, 2015).
Metabolomics has emerged as a valuable complement to genomics and proteomics to provide information regarding cellular metabolic processes that drive tumor formation and progression. Metabolomics enables evaluation of individual variability in the number and type of metabolites expressed in lung tumors. These metabolites can help differentiate the type, stage, and, potentially, the response to drug treatment. Most metabolomics studies specific to lung cancer are focused on developing cost-effective methods with high sensitivity and specificity for early tumor detection. The metabolic activity determines cell capacity for survival, and thus drives disease progression. Tumor cells employ various metabolic strategies to maintain the energy and biosynthetic precursor demands of proliferation, specifically to sustain an accelerated rate of aerobic glycolysis (Warburg, 1956, Fan et al., 2009). One of the basic strategies that characterizes the metabolic phenotype of tumor cells is the Warburg effect, which describes a high rate of glycolysis with subsequent lactic acid fermentation(Warburg, 1956, Vander Heiden et al., 2009).
The goal of this review is to summarize recent findings in identification of novel metabolites that could prove useful biomarkers in the early detection of lung tumors, determination of disease progression and prediction of patient survival. Figure 1 provides an overview of the types of metabolite comparisons evaluated in this review. Specific changes in metabolites in human subjects can be studied by: 1) extracting metabolites from tumor samples post-resection, 2) growing resected tumor cells in vitro followed by metabolite extraction, 3) implanting tumor cells resected from patients into a mouse model or 4) extracting metabolites from biofluids such as serum, sweat, urine and sputum samples. In addition to the alterations in levels of individual metabolites, stable-isotope labeling has been successfully used to determine pathways that are activated in different tumors. Fan et al. have successfully administered 13C glucose to mouse models and human subjects, which has enabled the detection of labeled metabolites involved in pathways such as glycolysis, Krebs cycle and anaplerotic pyruvate carboxylation, all of which are significantly altered in lung tumors (Lane et al., 2008, Lane and Fan, 2007, Lane et al., 2011, Fan et al., 2009, Fan et al., 2011, Bruntz et al., 2017, Fan et al., 2016, Higashi et al., 2014).
Figure 1.
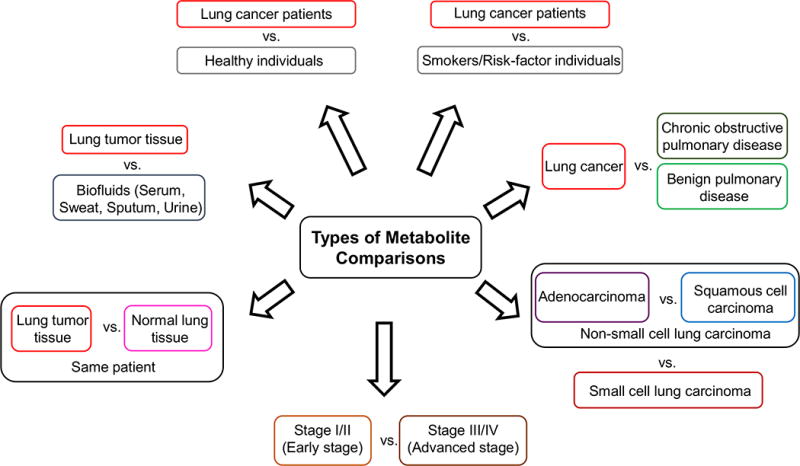
Overview of types of lung cancer metabolite comparisons evaluated in this review.
Two common metabolomics techniques used to analyze metabolites in lung cancer are mass spectrometry (MS) and nuclear magnetic resonance (NMR, which are of complementary value. The methods are described in detail elsewhere (Zhang et al., 2012a, Dunn et al., 2011, Lu et al., 2017), and are briefly discussed here. The techniques have been increasingly popular to obtain metabolic information from normal and tumor cells and from blood, sweat, urine and sputum samples (Emwas, 2015, Yu et al., 2017). While MS-based techniques offer a higher sensitivity, higher selectivity and broader metabolome coverage in any given sample, NMR-based techniques provide an easier method of sample preparation, reproducibility and extensive structural information, especially in stable-isotope enrichment measurements (Emwas, 2015, Lu et al., 2017). Typically, a combination of NMR and MS is utilized to obtain detailed profiles of relevant metabolites.
MS techniques acquire spectral data in the form of mass-to-charge (m/z) ratios and relative intensities of the measured compounds(Stringer et al., 2016, Alonso et al., 2015). The techniques allow for separation of metabolites with different chemical properties, which take varying amounts of time (retention time) to pass through a chromatographic column. LC-MS and GC-MS are two common techniques that are useful in detection of different types of analytes and have been used together in recent lung cancer metabolomics studies (Chen et al., 2015). In LC-MS, analytes undergo separation on a column, followed by ionization at an ion source, separation by a mass detector and detection (Lu et al., 2017). The proper selection of an ionization method is crucial to the detection of metabolites when using LC-MS. Most LC-MS methods utilize electrospray ionization, which does not ionize metabolites like cholesterol, for example. To overcome this, pressure and chemical ionization methods must be used. GC-MS, on the other hand does not require ionization of compounds, but instead relies on the use of a hot injection into the sample, followed by evaporation of molecules. GC-MS can measure a wide spectrum of water soluble metabolites and can measure volatile and low-molecular-weight metabolites that do not retain well on LC (Lu et al., 2017, Halket et al., 2005). However, GC-MS is not useful for measurement of thermolabile compounds such as ATP, NADPH or arginine due to possible decomposition after the hot injection (Kaspar et al., 2008, Lu et al., 2017).
NMR techniques, on the other hand, rely on the ability of protons to resonate in a high magnetic field. Samples are placed in a magnetic field and exposed to radiofrequency radiation. Each compound absorbs and releases electromagnetic radiation when subjected to a high power short duration radio frequency, based on the location of its associated protons (Stringer et al., 2016, James, 1998, Hornak). This leads to an NMR response, translated into a peak with parts per million (ppm) units to differentiate the positions of different atoms or chemical shifts (Stringer et al., 2016). This NMR response depends on the type of nucleus (1H or 13C), the chemical environment of the nucleus, and the spatial location of the nucleus in the magnetic field (Hornak, James, 1998). NMR can provide in-depth information on the structure, dynamics, chemical environment and reaction state of the molecules within the sample, which may be useful for studying the biology and determining therapeutic options for cancer.
The most sensitive and commonly used NMR technique in lung cancer metabolomics is 1D 1H-NMR (Fan et al., 2009, Puchades-Carrasco et al., 2016, Zhang et al., 2016, Rocha et al., 2015, Hao et al., 2016b, Deja et al., 2014).1D 1H-NMR has a small spectral width, with the resonances of most detected metabolites crowded within a narrow chemical shift range. Consequently, there is usually an overlap in the NMR resonances, leading to potentially ambiguous NMR assignments for different metabolites (Emwas et al., 2013). The 1D-carbon NMR spectroscopy offers the advantage of a wider spectral dispersion compared to the 1H-NMR, but is less sensitive and suffers from the low natural abundance of 13C nuclei, which negates its wide spectral width advantage (Emwas et al., 2013). The 2D-NMR offers a solution to the issues of overlapping resonances and low sensitivity, and offers higher resolution detection of metabolites than 1D-NMR (Lu et al., 2017). In metabolomics studies, heteronuclear (1H-13C) 2D-NMR experiments have been used extensively, including heteronuclear single quantum coherence (HSQC) and heteronuclear multiple bond correlation (HMBC), both of which take advantage of the high resolution and spectral width of the carbon for metabolite detection (Emwas, 2015, Emwas et al., 2013). Homonuclear (1H-1H) 2D-NMR has also been widely employed for metabolomic analysis, including total correlation spectroscopy (TOCSY), correlation spectroscopy (COSY) and diffusion ordered spectroscopy (DOSY) (Emwas, 2015, Emwas et al., 2013). The disadvantage of 2D-NMR compared to 1D-NMR is the requirement of a longer experimental time and extended period of analysis due to acquisition of large datasets.
Metabolite alterations in cancer patients versus healthy individuals
The discovery of clinical biomarkers for early and specific detection of lung cancer has been a major focus of metabolomics research. Towards this end, many studies have focused on metabolite differences that can distinguish between healthy individuals and cancer patients with high specificity and sensitivity, in particular, for samples collected from serum. In terms of amino acids, Zhang et al. found that serum samples from lung cancer patients showed increased glutamine levels compared to healthy individuals (Zhang et al., 2016). Glutamine can be converted to glutamate, the deamination of which leads to the production of α-ketoglutarate, a TCA cycle intermediate. Glutamine can also be converted to aspartate, which forms oxaloacetate, malate and pyruvate through the TCA cycle. Thus, glutamine is essential for cancer cell growth and proliferation (Zhang et al., 2016). In contrast, as glutamine is involved in the synthesis of nucleotides and amino acids, Puchades-Carrasco et al. found decreases in glutamine levels in sera from lung cancer patients (Puchades-Carrasco et al., 2016). This difference in findings may be because the Zhang et al. study included only early stage (Ia and Ib) lung cancer patients, whereas the Puchades-Carrasco study included patients at all stages (I-IV) of lung cancer (Puchades-Carrasco et al., 2016, Zhang et al., 2016). This could indicate that as the disease progresses, the demand for glutamine increases and lower levels are observed in the sera from lung cancer patients. Lung tumors are also characterized by decreased serum levels of histidine and threonine, both of which are utilized in the glycine/serine/threonine and pyrimidine pathways that are upregulated in lung tumors (Puchades-Carrasco et al., 2016, Zhang et al., 2012b).
Alterations in serum levels of fatty acids and lipids between healthy individuals and lung cancer patients have also been observed, as dysregulation in fatty acid and lipid metabolism is very commonly observed in cancer (Table 1) (Reddy et al., 2016, Yeh et al., 2006, Hirsch et al., 2010, Zhang and Du, 2012, Currie et al., 2013). Serum levels of choline are commonly lowered in lung cancer patients as it is a precursor to membrane phospholipids, which are in high demand in proliferating tumors (Puchades-Carrasco et al., 2016). Subsequent increases in phosphatidylcholines, which are major components of cell membranes, are therefore commonly seen in lung tumors (Musharraf et al., 2015, Li et al., 2014, Chen et al., 2015, Yu et al., 2013, Guo et al., 2012, Dong et al., 2010). Lysophosphatidylcholines are membrane lipids that also have a proinflammatory function and these are also upregulated in lung cancer patients (Li et al., 2014). On the other hand, decreased levels of sphingosine, a membrane sphingolipid necessary for proliferation and apoptosis through activation of the sphingosine kinase pathway have been observed in lung cancer (Yu et al., 2013, Chen et al., 2015). Additionally, decreases in unsaturated lipids, low density lipoproteins and very low density lipoproteins have also been observed in sera from cancer patients, due to the requirement of excess lipids for growth, proliferation, invasion and metastasis in tumors (Zhang et al., 2016, Puchades-Carrasco et al., 2016). Other metabolite changes include decreases in prasterone sulfate and 2,3,4, -trihydroxybutyric acid and an increase in α-hydroxyisobutyric acid in cancer patients, both before and after surgical resection of the tumor as compared to healthy individuals, indicating that these may be useful biomarkers for lung cancer (Chen et al., 2015).
Table 1.
Metabolites in lung cancer (NA: Not applicable)
| Comparison | Upregulated Metabolites | Downregulated Metabolites | Metabolite Analysis Technique Used (Reference) |
|---|---|---|---|
| Tumor versus paired non-tumor tissue from the same patient | Lactate, alanine, citrate, glutamate, succinate, ribosyl moiety of nucleotides, aspartate, leucine, isoleucine, valine, oxidized glutathione, glutamine, creatine, phosphocholine, taurine, glycine, phenylalanine, tyrosine, myo-inositol, α – and β – glucose | NA | 1D – NMR, 2D – 1H TOCSY, GC-MS (Fan et al., 2009) |
| Lactic acid, propanoic acid, glycolic acid, L-alanine, oxalic acid, 2-hydroxybutyric acid, butanoic acid, ethanimidic acid, malonic acid, L-valine, L-leucin, phosphoric acid, L-isoleucine, L-proline, succinic acid, glycine, pyrimidine, fumaric acid, uracil, L-serine, aminomalonic acid, L-threonine, malic acid, aspartic acid, methionine, 5-oxoproline, L-cysteine, thiodiglycolic acid, 2-hydroxyglutaric acid, L-glutamic acid, L-phenylalanine, 4-hydroxyphenylaceticacid, N-acetylaspartic acid, D-ribose, α-aminoadipic acid, L-glutamine, 9H-purine, DL-ornithine, 4-hydroxyphenyllactic acid, L-tyrosine, D-gluconic acid, Stearic acid, L-tryptophan, Inosine, Adenosine, guanine | Lauric acid, myristic acid | GC/MS (Hori et al., 2011) | |
| All 19 amino acids, ATP, GTP, other adenosine and guanine phosphates, malate, fumarate, succinate, lactate, fructose 1,6-bisphosphate, pyruvate Phosphorylated TCA enzymes: phosphofructokinase, glyceraldehyde 3-phosphate dehydrogenase, phosphoglycerate kinase-1 and pyruvate kinase, pyruvate dehydrogenase |
Adenylate and guanylate charges, fructose 6-phosphate, phosphoenolpyruvate | CE – TOFMS, NanoLC – MS/MS (Kami et al., 2013) | |
| Lung cancer patients versus healthy individuals | Lactic acid, 2-hydroxyisobutyric acid, L-glycine, sarcosine, 3-hydroxybutyric acid, 2-hydroxyisovaleric acid, malonic acid, benzoic acid, octanoic acid, L-proline, fumaric acid, L-threonine, malic acid, 2- hexenedioic acid, 4-hydroxyproline, 2-propyl-glutaric acid, aconitic acid, uric acid | Glycerol, phosphoric acid, glyceric acid, aspartic acid, 5-oxoproline, L-glutamic acid, lauric acid | GC/MS (Hori et al., 2011) |
| Serine, Proline, Isoleucine, Ornithine | Asparagine, glutamine, citrulline, methionine, leucine, histidine, tryptophan | HPLC-ESI-MS (Miyagi et al., 2011) | |
| Lysophosphatidylcholines (LPC) – (18:1, 20:4, 20:3, 22:6) Sphingomyelins (SM) – SM (16:0/1) |
Oleamide | DI-ESI (+)-FTICR MS (Guo et al., 2012) | |
| Fatty acid amides (C20:1, C20:0, C22:2, C24:1, C22:1), lysophosphatidylcholines (LPC 16:0, 16:0 isomer, 18:0, 18:2, 20:4, 20:5, 22:3, C16:1, C20:3), diradylglycerolipids (DG 38:8), sphingomyelins (SM d18:1/14:0), | 1-arachidonyl glycerol, choline, carnitine (8:1, 10:1), palmitic acid, stearic acid, oleic acid, linoleic acid, arachidonic acid, palmitoleic acid | UPLC/Q-TOF MS (Li et al., 2014) | |
| Creatinine riboside, n-acetylneuraminic acid, cortisol sulfate, 561+ | NA | QTOF-MS (Mathe et al., 2014) | |
| Lactic acid, phosphoric acid, benzoic acid, naphthalene, d-glucose, altrose, palmitic acid, octadecanoic acid, stearic acid, cholesterol | 1-propene | GC-MS (Musharraf et al., 2015) | |
| Maltose, ethanolamine, glycerol, palmitic acid, lactic acid | Tryptophan, lysine, histidine, glutamic acid | GC-TOF MS (Miyamoto et al., 2015) | |
| Glycerophospho-N-arachidonoyl ethanolamine, phoshorylcholine, ϒ-linolenic acid, 9,12-octadecadienoic acid, oleic acid, α-hydroxyisobutyric acid | Sphingosine, prasterone sulfate, serine, 2,3,4-trihydroxybutyric acid | LC-Q-TOF-MS, GC-MS (Chen et al., 2015) | |
| Leucine, isoleucine, acetate, n-acetyl-cysteine, glutamate, methanol, glycerol, creatinine, lactate | HDL, LDL, VLDL, adipic acid, lipids (CH2-C=C and CH=CH), glutamine, choline, threonine, histidine | 1H-NMR (Puchades-Carrasco et al., 2016) | |
| B-hydroxybutyrate, acetoacetate, lactate, glutamate, glutamine, tyrosine, histidine, aspartate, asparagine, cysteine, isoleucine, leucine | LDL, VLDL, unsaturated lipids, α-glucose, β-glucose, choline, phosphocholine, glycerophosphocholine, betaine, TMAO, methionine, tryptophan | 1H-NMR (Zhang et al., 2016) | |
| Bisphenol A | Retinol, L-proline | LC-MS (Pamungkas et al., 2016) | |
| Putrescine/isobutyl decanoate, 189.09, diethyl glutarate, cysteamine, hexanal, cysteic acid, hydroxypyruvic acid | Phosphatidylcholine (PC 18:0/18:1 and 16:0/14:1), heptanoic acid, 3-methoxy-4-hydroxyphenylglycol glucuronide, aminopropylcadaverine, palmitaldehyde, 5-carboxy-gamma-chromanol | FIE-MS, GC-MS (Cameron et al., 2016) | |
| Stage I adenocarcinoma versus normal lung tissue | 5,6-dihydrouracil, xanthine, 5′-Deoxy-5′-methylthioadenosine, adenine-5-monophosphate, cysteine, glutamate, 4-hydroxybutyric acid, ribitol, fucose + rhamnose, arabidose, alpha-tocopherol, uridine diphosphate N-acetylglucosamine, uric acid | Citrulline, ornithine, lysine, spermidine, 1-monostearin, caprylic acid, glucose | GC-TOF (Wikoff et al., 2015) |
| Squamous cell carcinomas versus adenocarcinomas | Creatinine, glutathione | Myo-inositol, phosphatidylcholine, phosphoethanolamine, taurine | 1H-NMR, HRMAS-NMR (Rocha et al., 2015) |
| Pyruvate, lactate, valine, proline, isoleucine, histidine, 2-aminobutyrate, leucine and alloisoleucine | 2-oxoisocaproate, 4-hydroxybutyrate, lysine, arginine, dimethylamine, isobutyrate, 3-hydroxybutyrate, acetate, asparagine, phenylalanine | 1H-NMR, GC-MS (Hao et al., 2016) | |
| Small cell lung carcinomas versus non-small cell lung carcinomas | Phenylacetic acid, L-fucose, caprylic acid, acetic cid, propionic acid and glycine | NA | FIE-MS (O’Shea et al., 2016) |
| Early stage (I-II) lung carcinomas versus healthy individuals | Lactic acid, 2-hydroxybutyric acid, 3-hydroxybutyric acid, benzoic acid, octanoic acid, L-proline, fumaric acid, malic acid, aconitic acid, uric acid | Glyceric acid, aspartic acid, lauric acid | GC/MS (Hori et al., 2011) |
| Advanced stage lung cancer (III-IV) versus healthy individuals | Lactic acid, 2-hydroxybutyric acid, L-glycine, sarcosine, 2-hydroxyisovaleric acid, malonic acid, benzoic acid, octanoic acid, fumaric acid, malic acid, 2-hexenedioic acid, 2-propyl-glutaric acid, 4-hydroxyphenyllactic acid | Glyceric acid, 5-oxoproline, L-glutamic acid, indol-3-acetic acid | GC/MS (Hori et al., 2011) |
| Advanced stage (III-IV) versus Early stage (I-II) NSCLC carcinomas | Creatinine, N-acetylated glycoproteins (NAC1, NAC2), glycerol | Isoleucine, acetoacetate, lipid L8, creatinine, acetone, valine, isobutyrate, lactate | 1H-NMR (Deja et al., 2014) |
| Advanced Stage non-small cell carcinoma versus early stage non-small cell carcinoma | Leucine/isoleucine, lysine, N-acetyl-cysteine, glutamate, citrate, valine, glycerol, creatine, H α/β amino acids, lactate, phenylalanine | 3-hydroxybutyrate, adipic acid, glutamine, glucose, lipids (CH2-C=C) | 1H-NMR (Puchades-Carrasco et al., 2016) |
| Patients with non-small cell lung carcinoma versus patients with chronic obstructive pulmonary disease | N-acetylated glycoproteins 1 (NAC1), NAC2, leucine, lysine, mannose, choline, lipid (L3+L4), lipid (L6), isoleucine, valine, 3-hydroxybutyrate, acetoacetate, isobutyrate, α-glucose, glycine, glycerol | Acetate, citrate, methanol, trimethylamine | 1H-NMR (Deja et al., 2014) |
| Patients with non-small cell lung carcinoma versus patients with benign pulmonary disease | Very high density lipoproteins, low density lipoproteins, adipic acid, lipids (CH2-C=C, CH=CH-CH2-CH=CH and CH=CH), N-acetyl-cysteine, glutamine, choline, proline, methanol, creatinine, myo-inositol, threonine | Leucine/isoleucine, acetate, glutamate, creatine, lactate, glucose | 1H-NMR (Puchades-Carrasco et al., 2016) |
| Lung cancer patients versus smokers/risk-factor individuals | Trihexose, tetrahexose, urocanic acid, muconic acid | Suberic acid, nonanedioic acid, MG (22:2) | LC-QTOF MS/MS (Calderon-Santiago et al., 2015) |
Differential metabolite presence has also been detected in urine and sputum samples of cancer patients. In addition to the serum metabolites mentioned above, creatine riboside (CR) and N-acetylneuraminic acid (NANA) are significantly higher in the urine of these patients compared to healthy individuals (Mathe et al., 2014). Creatine riboside is a novel compound, and is predicted to be a product of high creatine levels and high phosphate flux within tumor cells (Mathe et al., 2014). NANA, is a sialic acid and plays a role in cell signalling processes and in protecting malignant cells from host cellular defense systems (Mathe et al., 2014, Schauer, 2004). High levels of hypersialylated structures are often expressed on the surface of tumor cells, which enhance tumor progression by facilitating an escape from apoptosis, metastatic formation and resistance to chemotherapy (Bull et al., 2014). Evaluating sputum samples, both isobutyl decanoate and diethyl glutarate, which are phospholipid fragments representing the products of lipid peroxidation in tumors, are increased in lung cancer patients (Cameron et al., 2016). As urine and sputum samples are relatively easy to collect, these metabolites could prove useful in early disease detection.
Common metabolites identified in tissue and biofluid samples and their usefulness as lung cancer biomarkers
Changes in metabolites that are similar in both resected tumors as well as biofluids from cancer patients may be important to consider. Such metabolites could then be detected in easy-to-collect biofluid samples and would serve as indicators of metabolic changes in the tumor itself. Additionally, changes in metabolites that differ in biofluids such as serum compared to tumor tissue are also useful. As normal tissue turns cancerous, the levels of certain metabolites increase in the tumor tissue and these are released into the blood, resulting in higher serum levels of the same metabolite. Conversely, certain metabolites move from the blood into the lung tumor to promote cell growth and proliferation, leading to decreased levels of these metabolites in the serum. Thus, metabolites that are common to both tumors and biofluids may not only serve as biomarkers for early detection, but also as indicators of disease progression. One of the most commonly upregulated metabolites in both lung tissue samples and serum samples from lung cancer patients is lactate (Fan et al., 2009, Kami et al., 2013, Rocha et al., 2015, Deja et al., 2014, Puchades-Carrasco et al., 2016, Zhang et al., 2016). This is not surprising as a higher glucose uptake and its subsequent conversion to lactate (the Warburg effect) is a common phenomenon observed in several different types of tumors, including NSCLC tumors, which are known to be highly glycolytic (Warburg, 1956, Vander Heiden et al., 2009). ATP is rapidly generated during aerobic glycolysis and the carbon from glucose is diverted into production of precursors for nucleotide, protein and lipid synthesis in lung tumor cells (Doherty and Cleveland, 2013). As a result, glucose is preferentially converted to lactate, instead of being metabolized into carbon dioxide through oxidative phosphorylation (Doherty and Cleveland, 2013). Research has shown that the production of lactate is higher in more aggressive lung tumors (Alderton, 2016), indicating that aggressive tumors are more glycolytic. Lactate dehydrogenase (LDH) catalyzes the conversion of pyruvate to lactate and vice versa and higher pre-treatment abundance of LDH is associated with aggressive tumor outcomes and lower patient survival (Xie et al., 2014, Hsieh et al., 2017). A specific subunit of LDH (LDH-A), is essential for cancer-initiating cell survival and proliferation in both K-RAS and EGFR-driven mouse models of NSCLC (Xie et al., 2014). The inactivation of LDH-A leads to decreased tumor proliferation and disease regression (Xie et al., 2014). Both lactate and LDH are potential biomarkers not just for detection, but also for determining the lung tumor stage. Increased creatine/creatinine levels have been found in both lung tumor tissues as well as in serum and urine samples from lung tumor patients (Fan et al., 2009, Rocha et al., 2015, Wikoff et al., 2015a, Puchades-Carrasco et al., 2016, Zhang et al., 2016, Mathe et al., 2014).
Creatine under normal conditions is produced in vital organs such as the liver, kidneys and pancreas and is transported to muscle tissues to provide energy. Increased creatine levels in tumor tissues leads to an increase in ATP production, which is associated with the highly energetic process of tumor growth and proliferation (Puchades-Carrasco et al., 2016). Creatine riboside is elevated in urine samples from NSCLC patients, and is associated with increased tumor size and poor prognosis, which makes it a potential biomarker (Mathe et al., 2014, Haznadar et al., 2016). Creatinine, on the other hand, is a waste product produced as cells utilize creatine, and is discarded from the body through the kidneys. Higher creatinine levels in the blood are an indicator of poor kidney function, which needs to be taken into consideration before patients are treated with nephrotoxic chemotherapy. In addition, glutamate is upregulated in lung tumor tissue as well as serum samples from lung tumor patients (Fan et al., 2009, Rocha et al., 2015, Wikoff et al., 2015a, Puchades-Carrasco et al., 2016, Zhang et al., 2016).
Glutamine, which is a non-essential amino acid involved in protein and lipid synthesis as well as a source of energy in normal cells, is known to act as a major source of nitrogen for synthesis of nucleotides and amino acids in conditions of hypermetabolism in tumor cells (Zhou et al., 2012). The increase in glutamate in lung tumor cells is likely due to increased hydrolysis of glutamine to produce glutamate and ammonia in the proliferative tumor microenvironment. Further evidence is that many, but not all types of lung tumors are glutamine dependent, leading to increased levels of glutamine seen in lung tumor tissues (Fan et al., 2009) and a subsequent decrease in serum glutamine levels in lung tumor patients (Deja et al., 2014, Puchades-Carrasco et al., 2016). The glutamine-dependency of certain lung tumor cell lines were associated with glutaminase 1 levels, an enzyme involved in glutamine metabolism (van den Heuvel et al., 2012). A study on liver and renal carcinoma cells found that hypoxic tumor environments lead to an increase in the production of hypoxia-inducible factors (HIFs), which in turn upregulates glutamate transporters and facilitates glutamate release (Hu et al., 2014). The same study also noted that HIFs increase the expression of glutamate receptors, which when bound by glutamate stimulates the proliferation, migration, invasion and resistance to apoptosis in both liver and renal tumor cells (Hu et al., 2014). Another possible explanation for increased glutamate levels may be due to the large number of hypoxic cells found in lung tumor tissues, especially in advanced stages. The upregulation of amino acids such as alanine/phenylalanine and leucine/isoleucine is also commonly seen in lung tumor tissue as well as serum samples from lung tumor patients and is also associated with the higher metabolic demands of tumor growth and proliferation (Fan et al., 2009, Rocha et al., 2015, Wikoff et al., 2015a, Deja et al., 2014, Puchades-Carrasco et al., 2016, Zhang et al., 2016).
In addition to metabolites that are upregulated in tumor samples, several metabolism-associated substances are downregulated in lung tumors. The most common is decreased glucose, associated with the accelerated rate of glycolysis of the Warburg Effect. This is true in both lung tumor tissue and biofluid samples, with glucose levels being significantly lower in lung tumor patients (Fan et al., 2009, Rocha et al., 2015, Deja et al., 2014, Zhang et al., 2016, Wikoff et al., 2015a). Levels of several amino acids, especially histidine, are lower in tumor cells, reflecting an increase in glycine/serine/threonine and pyrimidine metabolism in tumor cells (Miyamoto et al., 2015, Miyagi et al., 2011). Several studies (Table 1) found decreased histidine levels in lung cancer patients. This is true not only for lung tumor tissues but also for serum, plasma and sweat samples, making it an attractive biomarker for the detection of lung tumors (Wikoff et al., 2015a, Puchades-Carrasco et al., 2016, Miyamoto et al., 2015, Delgado-Povedano et al., 2016).
Noteworthy is the fact that the metabolites discussed above that were differentially regulated in lung tumor samples compared to controls, were also found in serum, plasma and sweat samples. Metabolites that are found in both biofluids and tumor tissue, especially those which increase in concentration in the bloodstream, as a result of secretion from the tumor or those that are taken up rapidly by the tumor from the blood may prove to be useful indicators of the lung tumor type, stage and progression of the disease.
Metabolite alterations in tumor versus paired non-tumor lung tissues
To understand the specific cellular and tissue level biochemical changes that occur in cancer, it is important to compare the metabolic changes between normal tissue and lung tumor tissue. This allows for the analysis metabolism without having to account for differences in genetic factors or environmental exposures, as the patient’s own non-cancerous lung tissue acts as a control. Metabolites that are upregulated or downregulated in tumor tissue compared to normal lung tissue are listed in Table 1. Lung cancer, like other cancers is also characterized by accelerated glycolysis with transformation of glucose to lactate, an increased activation of the pentose phosphate pathway, with increased production of nucleotides, and differences in TCA cycle intermediates (Fan et al., 2009) (Table 1).
Increases in amino acid concentrations are commonly detected in lung tumor tissues (Fan et al., 2009, Hori et al., 2011, Kami et al., 2013). Alterations in amino acids and thereby the biosynthetic pathways that metabolize them are required for tumor growth and proliferation (Tsun and Possemato, 2015). Increased levels of alanine are observed in most tumors, including lung tumors, due to increased glycolysis. There is also an increased uptake of valine, leucine and isoleucine by lung tumors as these are required to fuel the production of Krebs cycle intermediates. Glutamine levels are usually found to be lower in tumor tissues, such as colon and stomach cancer as a large amount of glutamine is metabolized in tumor tissues (Hirayama et al., 2009). In contrast, Hori et al. found higher glutamine levels in lung tumor tissue compared to normal tissue (Hori et al., 2011). As explanation is that glutamine levels are usually higher in smokers and a large percentage of lung cancer patients in the above study were chronic smokers (Vulimiri et al., 2009). Although levels of most amino acids in lung tumors are found to be higher than in normal lung tissue, aspartate levels are similar (Kami et al., 2013). The reason could be that aspartate is a precursor for nucleic acids and several Krebs cycle intermediates, and is actively consumed in proliferating lung tumor tissues (Kami et al., 2013).
Fatty acid metabolism is greatly altered in rapidly proliferating tumor cells as these acids are required for biosynthesis of membrane lipids and signaling molecules (Currie et al., 2013). To meet this increased requirement, tumors exhibit a shift towards de novo fatty acid synthesis (Medes et al., 1953). This may explain the significant increases in the levels of several fatty acids that have been observed in lung tumor tissues compared to non-tumor tissue (Hori et al., 2011) (Table 1).
Metabolite alterations in different stages of lung cancers and their usefulness as indicators of disease progression and overall survival
Metabolites that can predict the progression of lung cancer and could potentially be used as biomarkers for overall patient survival have recently been identified in tumor tissues as well as in biofluids from cancer patients. Several serum metabolites were found to be differentially expressed in early stage (stages I-II) versus advanced stage (stages III-IV) lung tumors (Table 1). Increases in the levels of lactic acid, benzoic acid and fumaric acid were observed in advanced stage tumors, indicating their alteration during disease metastasis or increased inflammation (Hori et al., 2011). Glycerol, which is the backbone of all lipids, is also increased in sera from advanced-stage lung cancer patients, which is indicative of either lipid breakdown or cell membrane rearrangement (Deja et al., 2014). On the other hand, decreased levels of glyceric acid and L-glutamic acid were seen in advanced stage tumors, suggesting that these metabolites may be transferred from the blood into the lung tissue and then metabolized rapidly within the tissue (Hori et al., 2011). Early-stage patients were characterized by increased lactate levels (Warburg effect) and increased levels of all ketone bodies (2-hydroxybutyrate, 3-hydroxybutyrate, acetoacetate and acetone), which are common hallmarks of cancer metabolism. This increase in lactate and decrease in glucose concentration is more prominent in advanced-stage patients (Puchades-Carrasco et al., 2016). Similarly, increases in serum leucine/isoleucine, N-acetyl-cysteine and glutamate along with decreases in glutamine are seen in advanced stages of NSCLC compared to early stages (Puchades-Carrasco et al., 2016). The continued increase in levels of N-acetyl cysteine, involved in pathways that lead to the production of antioxidant species, are upregulated with disease progression in NSCLC patients (Puchades-Carrasco et al., 2016). These upregulated antioxidants are known to aid in lung cancer progression through the suppression of the well-known tumor suppressor, p53 (Sayin et al., 2014). Glutamine has been well-studied in NSCLC cells and glutaminolysis was found to be essential for tumor growth, thereby explaining the decrease in glutamine levels in advanced stages of the disease (van den Heuvel et al., 2012). Other metabolites that are increased in advanced stage lung cancer include adipic acid and phenylalanine. High adipic acid concentrations in advanced stages of lung cancer (Muntoni et al., 2009, Puchades-Carrasco et al., 2016) are observed due to changes in lipid metabolism in tumor cells. Phenylalanine is a precursor for tyrosine and neurotransmitter like dopamine, norepinephrine and epinephrine. High serum levels of phenylalanine is due to the downregulation of genes involved in its metabolism within NSCLC tumors (Long et al., 2015), suggesting a decrease in the ability of lung cancer cells to metabolize this amino acid in late stages of the disease (Puchades-Carrasco et al., 2016). A novel metabolite, N12-diacetylspermine, was found to be elevated in the serum of suspected lung cancer patients 0 to 6 months before the diagnosis of the disease and could serve as a good biomarker for early detection of lung tumors (Wikoff et al., 2015b).
Serum metabolites collected before, during and after chemotherapy are useful in predicting disease progression and overall survival in lung cancer patients (Hao et al., 2016b, Hao et al., 2016a). Elevated levels of metabolites such as hydroxylamine and octadecan-1-ol were observed in patients with advanced stage disease, who did not survive chemotherapy (Hao et al., 2016b, Hao et al., 2016a). On the other hand, amino acids glutamine, proline, valine, threonine and tyramine were abundant in patients in early stages of the disease, who had higher survival rates (Hao et al., 2016b, Hao et al., 2016a). Interestingly, metabolites such as glycine, phosphoric acid, pyroglutamic acid and octadecenoic acid are associated with advanced disease as well as higher survival and knowledge of these metabolites may prove useful in determination of an effective line of treatment for stage III-IV lung tumor patients (Hao et al., 2016b, Hao et al., 2016a). In addition to serum metabolites, higher levels of urine metabolites such as NANA, cortisol sulfate, creatine riboside and 561+ have also been associated with worse survival rates (Mathe et al., 2014). Creatine riboside was specifically associated with poor prognosis in early stage (I-II) lung tumor patients (Mathe et al., 2014). As mentioned earlier, creatine riboside and NANA can distinguish between lung cancer patients and healthy individuals, but high levels of these are also associated with poor survival. Thus, these are not only useful in early detection of the disease, but also in predicting patient survival after chemotherapy (Mathe et al., 2014). The infusion of 13C glucose in patients with early stage NSCLC allowed for the detection of enhanced pyruvate carboxylase (PC) activity in the tumor tissue, indicating that PC-mediated anaplerosis is essential for early stage NSCLC survival and growth (Sellers et al., 2015). Thus, high levels of PC in tissue sample resected from lung tumor patients may be indicative of early stage disease.
Metabolite alterations in different types of lung cancers
Recently, lung cancer metabolomics work has focused on identifying biomarkers that can help to detect specific lung cancer types. Metabolites that can specifically distinguish between stage I adenocarcinoma and normal lung tissue have been identified, as shown in Table 1. A significant elevation in 5,6-dihydrouracil, which is produced by the oxidation of the nucleotide uracil, was found to be the best discriminant of stage 1 adenocarcinoma compared with non-malignant tissue (Wikoff et al., 2015a). The catabolism of uracil to 5,6-dihydrouracil is catalyzed by the enzyme dihydropyrimidine dehydrogenase (DPD), the activity and expression of which is known to be increased in lung tumor patients (Miyoshi et al., 2005). Lowered levels of DPD are associated with higher survival in patients treated with DPD inhibitor 5-fluorouracil (Shintani et al., 2011). Thus, the knowledge of the ratio of uracil to 5,6-dihydrouracil levels and thereby DPD activity may help to identify patients that may benefit from treatment with DPD inhibitors (Wikoff et al., 2015b, Miyoshi et al., 2005, Shintani et al., 2011). In addition to alterations in nucleotide metabolism, the therapeutic potential of purinergic signaling has been of interest (Schmid et al., 2015). The levels of xanthine, a purine base have been found to be elevated in stage I adenocarcinomas (Wikoff et al., 2015a). Moreover, the conversion of xanthine to uric acid, the concentration of which is also elevated in adenocarcinoma, is mediated by xanthine oxidoreductase (XOR). Low XOR levels have been associated with poor prognosis and shortened survival times in NSCLC patients (Kim et al., 2011). Thus, levels of xanthine and uric acid could serve as indicators of XOR activity and prove to be useful biomarkers for lung tumors. On the other hand, metabolites ornithine and its product citrulline, involved in nitrogen disposal, were both downregulated in adenocarcinomas. As ornithine is involved in proline synthesis (Morris, 2004), the levels of proline were also higher in adenocarcinomas (Wikoff et al., 2015a). This indicates that in adenocarcinomas, ornithine is shunted from citrulline production and diverted to proline synthesis instead (Wikoff et al., 2015a) and levels of these metabolites may be useful biomarkers for stage I disease.
Serum metabolites that can aid in distinguishing between different histological types of lung tumors have been identified (Hori et al., 2011). A high percentage of patients in the adenocarcinoma group were characterized by the presence of glycine, 2-hydroxyisovaleric acid, methionine and phenylalanine; the squamous cell carcinoma group was typified by 3-hydroxybutyric acid and aconitic acid, and the small cell lung carcinoma group was characterized by pyruvic acid, benzoic acid, octanoic acid, isovalerylglycine, 2-hexenedioic acid and palmitoleic acid (Hori et al., 2011). The reason for the presence of these specific metabolites is unclear. Squamous cell carcinoma tissues were found to have relatively higher levels of ATP, GTP, adenosine phosphates, guanine phosphates and branched-chain amino acids compared to non-tumor tissues (Kami et al., 2013). This effect was more pronounced in squamous cell carcinoma compared to adenocarcinoma, pleomorphic carcinoma and large cell carcinoma tissue (Kami et al., 2013). Thus, relatively higher levels of these nucleotides and amino acids may prove useful in differentiating squamous cell carcinomas from other NSCLC tumors. Changes in metabolites between adenocarcinomas and squamous cell carcinomas have also been identified (Table 1) (Rocha et al., 2015). Adenocarcinomas were characterized by alterations in phospholipid metabolism, including marked differences in phosphocholine (PC), glycerophosphocholine (GPC) and phosphoethanolamine (PE) compared with squamous cell carcinomas (Rocha et al., 2015). The production of PC occurs through the phosphorylation of choline to PC by the enzyme choline kinase in the first step of the phoshaptidylcholine synthesis, the major cell membrane phospholipid. Choline kinase is known to be elevated in highly active tumors and has been proposed as a biomarker for early-stage lung cancer (de Molina et al., 2002, de Molina et al., 2007). Phospholipid breakdown is also associated with abnormal metabolism in tumors and proposed to be involved in promoting malignant transformation through mitogenic signal transduction (Rocha et al., 2015, Moestue et al., 2011). GPC is produced because of phosphatidylcholine degradation and therefore the increases in PC, GPC and PE, likely indicate both catabolic and anabolic processes, which are more prevalent in adenocarcinomas (Rocha et al., 2015).
Moreover, adenocarcinomas also showed significantly decreased acetate levels, which were not observed in squamous cell carcinomas (Rocha et al., 2015). Acetate can be converted to acetyl-CoA, which is the major driver of de novo lipid biosynthesis. Squamous cell carcinomas, on the other hand were characterized by greater glycolytic and glutaminolytic metabolic profiles when compared to adenocarcinomas (Rocha et al., 2015) (Table 1). Increases in the levels of lactate and decreases in glucose, which are commonly seen in cancer cells, were more pronounced in squamous cell carcinomas (Rocha et al., 2015). In addition, glutamate and alanine levels were also increased in squamous cell carcinomas compared to a much smaller glutamate increase and no increase in alanine in adenocarcinomas (Rocha et al., 2015). The transamination of glutamate and pyruvate, through the enzyme alanine transaminase, leads to the production of alanine and α-ketoglutarate, thereby indicating a higher glutaminolytic activity in squamous cell carcinomas (Rocha et al., 2015). Another important distinction between these histological types was glutathione (GSH) levels, which were significantly increased in squamous cell, but not in adenocarcinomas. High GSH levels and increased activity of GSH-related enzymes have been observed in lung tumors and are associated with resistance to chemotherapy and higher potential for metastasis (Blair et al., 1997, Ferruzzi et al., 2003, Ilonen et al., 2009, Traverso et al., 2013). Creatine levels were increased in both squamous cell and adenocarcinomas, but were significantly higher in squamous cell carcinomas compared to adenocarcinomas (Rocha et al., 2015). Thus, targeting lipid metabolism pathways may be useful for patients with adenocarcinomas whereas targeting of glutaminolytic pathways and glutathione metabolism may prove to be effective in treatment of squamous cell carcinomas.
Recently, sputum metabolites that can help distinguish patients with non-small cell lung cancer (NSCLC) and small cell lung cancer (SCC) have been identified (O’Shea et al., 2016) (Table 1). Glycine levels were found to be lower in NSCLC patients as compared to SCC patients (O’Shea et al., 2016). A possible explanation for this is the increase in the enzyme, glycine decarboxylase in NSCLC tumors, which degrades glycine (Zhang et al., 2012b). Decreases in L-fucose were observed in NSCLC patients, which is a deoxyhexose that is N- and O-linked to glycolipids and glycopeptides produced by mammalian cells (O’Shea et al., 2016, Becker and Lowe, 2003). Fucose is a precursor to GDP-fucose production, which is associated with transfer of fucose motifs on lipids and proteins. Thus, decreased L-fucose levels in NSCLC patients may be due to an increase in fucosylation in these tumors. In addition to glycine and fucose, fatty acids such as acetic acid, propionic acid and caprylic acid were significantly decreased in sputum from NSCLC patients compared to SCC, likely produced by lipid peroxidation (Wang et al., 2014, O’Shea et al., 2016).
Metabolite alterations in lung cancer versus other lung diseases
Patients diagnosed with lung cancer often exhibit the symptoms of chronic obstructive pulmonary disease (COPD) (Yao and Rahman, 2009). It has been suggested that COPD increases the risk of developing lung cancer (Potton et al., 2009, Wang et al., 2012). Metabolites that can distinguish between patients with COPD and patients with lung cancer have been recently identified (Table 1). Serum samples from patients with COPD were compared with those from patients with NSCLC to identify metabolic biomarkers that could differentiate between these different disease types (Deja et al., 2014). Decreased levels of acetate, citrate and methanol were observed in serum from NSCLC patients as compared to COPD patients (Deja et al., 2014). The decreased levels of citrate suggest that energetic demands of lung tumor cells are higher than those in lung diseases with chronic inflammation. Amino acids such as leucine and lysine were increased in NSCLC compared to COPD and both are ketogenic amino acids. Interestingly, leucine shows opposite trends with levels being higher in NSCLC patients and lower in COPD patients compared with healthy individuals and may be a good candidate to distinguish between these two types of lung diseases (Deja et al., 2014, Shingyoji et al., 2013, Miyagi et al., 2011, Ubhi et al., 2012, Wang et al., 2013). The major differences between COPD patients and early-stage NSCLC were associated with pathways related to protein biosynthesis, degradation of branched chain amino acids and ketone body metabolism (Deja et al., 2014). Overall metabolite expression was more similar in advanced stage NSCLC and COPD patients as compared to early stage NSCLC patients, which may be due to similar levels of lung tissue degradation and impairment (Deja et al., 2014). Elevated levels of ganglioside GM1 haves been observed in patients diagnosed with other pulmonary diseases (asthma, pneumonia, chronic obstructive pulmonary disorder, tuberculosis) compared to patients with lung cancer (Cameron et al., 2016). GM1, like other gangliosides, is involved in cell-cell recognition, cell-matrix attachment, cell growth and differentiation and is especially prominent in SCLC cell lines and tissues. Thus, GM1 can be a marker used to differentiate between SCLC patients and patients with other lung diseases (Cameron et al., 2016). Serum metabolites are also useful in discriminating between patients with benign pulmonary disease (BPD) and patients with NSCLC (Puchades-Carrasco et al., 2016) (Table 1). A combination of five metabolite markers, including glutamine, lactate, methanol, threonine and choline were described as being useful in discriminating between healthy individuals, BPD patients and NSCLC patients (Puchades-Carrasco et al., 2016). Of these, levels of threonine and choline were similar in BPD patients and healthy individuals and relatively higher than levels in NSCLC patients. This is because threonine is required for protein biosynthesis and choline is needed for production of membrane phospholipids in NSCLC tumors. Levels of lactate are usually higher in tumors and thus were found to be lower in BPD patients compared to NSCLC patients (Puchades-Carrasco et al., 2016).
Metabolite alterations in lung cancer patients versus smokers/risk-factor individuals
Cigarette smoke contains more than 70 established carcinogens, among which the most important causative agents are polycyclic aromatic hydrocarbons (PAHs) and tobacco-specific nitrosamines like 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) (Yuan et al., 2011). Urinary levels of the marker for PAH uptake, r-I,t-2,3,c-4-tetrahydroxy-1,2,3,4-tetrahydroxy phenanthrene (PheT) and metabolites of NNK such as 4-(methylnitosamino)-1-(3-pyridyl)-1-butanol and cotinine, and their glucoronides, were significantly higher in smokers who were subsequently diagnosed with lung cancer (Yuan et al., 2011). The levels of these markers also increase with corresponding increases in the number of cigarettes smoked per day (Yuan et al., 2011). Monitoring the levels of PheT and cotinine could therefore enhance the ability of clinicians to predict the risk of developing lung cancer in chronic smokers. In addition to these amrkers, a panel of metabolites was identified as suitable for distinguishing sweat samples of lung tumor patients from smokers/risk-factor individuals (Calderón-Santiago et al., 2015). This panel included short chain dicarboxylic acids such as nonanedioic acid and suberic acid which were downregulated in sweat from patients with lung cancer compared to smokers (Calderón-Santiago et al., 2015). These metabolites have also been observed in urine samples and identified as potential biomarkers to distinguish between patients with benign versus malignant tumors (Kim et al., 1998). The panel also included sugars such as trihexose, tetrahexose, which were upregulated in lung cancer patients compared to smokers (Calderón-Santiago et al., 2015). The presence of these is associated with the activity of the enzyme amylase, which is known to be altered in tumor tissues (Takeuchi et al., 1981). The levels of MG (22:2), a monoglyceride of an omega-6 fatty acid was also downregulated in lung cancer patients compared to smokers and is involved in the metabolism of essential fatty acids (Calderón-Santiago et al., 2015, Simopoulos, 2002, Simopoulos, 2008). A follow-up study revealed an increase in urocanic acid and muconic acid in cancer patients compared to smokers. Muconic acid has been previously linked to benzene exposure, which in turn is linked to smoking habits (Wiwanitkit et al., 2005) and cancer development (Snyder, 2012). Urocanic acid, produced through the degradation of histidine, is known to protect the skin from UV damage (Tuna et al., 2014) and modulates the production of immunosuppressive molecules from keratinocytes, nerves and mast cells (Hart et al., 2002, Kaneko et al., 2008, Khalil et al., 2001, Delgado-Povedano et al., 2016). Both urocanic acid and muconic acid were found to increase the specificity (from 80% to 96.9%) and sensitivity (from 79% to 83.8%) of lung cancer detection compared to the previous study (Delgado-Povedano et al., 2016). Low levels of bilirubin, an endogenous antioxidant, in male smokers was found to be associated with an increased risk of lung cancer incidence and mortality (Wen et al., 2015). Moreover, smoking cessation leads to increased bilirubin levels and is associated with lower risk of lung cancer and cardiovascular diseases (O’Malley et al., 2014). Retinol was found to be downregulated in patients who were current smokers as compared to non-smoker controls and was also downregulated in cancer patients compared to controls, indicating that lower levels of this metabolite in smokers may have increased their risk of developing lung cancer (Pamungkas et al., 2016). Retinol or Vitamin A has been found to be effective against breast, ovarian and prostate cancers in animal models, likely due to its ability to induce apoptosis (Bushue and Wan, 2010).
Conclusions and Future Directions
The goal of metabolomics research is to identify markers that can help in distinguishing between lung cancer and healthy patients, various lung cancer types and stages, and also aid in tumor detection. Previously, a list of genomic biomarkers that may be useful in the diagnosis of lung tumors was compiled (Hassanein et al., 2012). In this review, our aim was to highlight potential metabolism-associated biomarkers that have been identified as either upregulated or downregulated in lung cancer patients compared to healthy individuals, individuals with other lung diseases and risk-factor individual//smokers. We also summarized findings that identified metabolites which could be used to distinguish between different tumor types, stages and predict progression of the disease and overall patient survival.
Potential biomarkers belonging to glycolysis, Krebs cycle, amino acid and nucleotide biosynthesis, and membrane lipid biosynthesis and degradation could help distinguish between cancer patients and healthy individuals. Major alterations observed in cancerous tissue compared to non-cancerous tissue were accelerated glycolysis, alterations in levels of Krebs cycle intermediates, and activation of the pentose phosphate pathway. Increases in the levels of amino acids and fatty acids were also a common theme in lung tumor tissues. Metabolite changes may differ in magnitude or direction (upregulation or downregulation) in early stage versus advanced stage lung tumors, and these could be useful in diagnosis of the disease stage. Metabolic changes in biofluids can be easily tracked before, during and after chemotherapy, which could be useful for tracking tumor progression. Knowledge of metabolites that change specifically in different lung tumor types may not only aid in clinical diagnosis, but also provide appropriate molecular targets for the development of new anticancer drugs that could improve overall patient survival. Metabolites belonging to pathways associated with higher energy demands and increased growth of tumor cells such as amino acids, ketone bodies, lactate and methanol may help to distinguish lung tumors from benign or chronic lung diseases. These metabolites may also prove to be indicative of subsequent progression towards cancer in patients with lung diseases. Lastly, metabolites that distinguish between smokers and individuals with lung cancer may help to determine the risk associated with the development of cancer.
The knowledge that cancer cells have a distinct metabolic phenotype along with advances in specificity and sensitivity of techniques such as NMR and MS, have enabled the field of metabolomics in oncology to gain momentum over the past decade. Yet several challenges remain. Since metabolomics can vary greatly with age, sex, race, dietary habits, drugs and other environmental factors, findings must be carefully evaluated and confounding effects need to be disentangled. In addition to patient-related factors, it is necessary to consider differences in experimental methods (MS versus NMR) as well as analytical methods for determining metabolic biomarkers, when comparing the data obtained from various studies.
Identifying tumor-tissue specific metabolites can help in studying various metabolic pathways affected in tumors and in developing effective treatment strategies. In particular, metabolites detected in biofluids, which can distinguish between cancer patients, risk factor individuals (smokers) and healthy individuals, could provide easy and cost-effective methods for first-line diagnosis. Special consideration should be given to metabolites that may be released by tumor cells and found in the sera of lung tumor patients (such as lactate) or those that may be taken up by tumor cells from the blood (such as glutamine and low-density lipoproteins). The use of biofluid metabolomics as an initial diagnostic tool would provide a means of early detection along with being an indicator of the status of the tumor itself. Substances such as lactate, creatinine/creatine, glutamate, alanine, leucine, glucose and histidine identified in serum samples as well as in tumor tissues could be useful in the development of a panel of potential metabolism-associated biomarkers, for detection and effective treatment.
Future studies focusing on the magnitude and direction of changes in common metabolites in tumor tissues and biofluids could help to identify metabolites specific to tumor stage with high sensitivity and specificity. Observation of changes in biofluid metabolites in patients, pre-therapy, during therapy and post-therapy could help to identify reliable markers for assessment of treatment responses without recourse to biopsies or multiple radiographic scans. Comparing metabolite profiles from different cancer types may also be useful in detection of biomarkers that may be exclusive to lung cancer. As long as it is feasible, studies should be conducted in a relatively consistent manner with comparable methods of sample collection, metabolite detection and analysis, and data interpretation. Ideally, the combination of genomic, proteomic, metabolomic and microbiome markers could provide an optimal panel of information to effectively diagnose and successfully treat lung cancer patients.
Acknowledgments
HBF acknowledges partial support by the National Institutes of Health/National Cancer Institute Grant R15CA203605.
Footnotes
Compliance with Ethical Requirements
Disclosure of potential conflicts of interest:
The authors declare no conflicts of interest.
Research involving Human Participants and/or Animals:
This work did not involve human participants or animals
Informed consent:
Is not applicable to this work
References
- ACS. Cancer Facts and Figures – 2017. 2017:1–76. [Google Scholar]
- ALDERTON GK. Tumour metabolism: Feeding the TCA cycle in vivo. Nat Rev Cancer. 2016;16:198. doi: 10.1038/nrc.2016.29. [DOI] [PubMed] [Google Scholar]
- ALONSO A, MARSAL S, JULIA A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3:23. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMBROSINI V, NICOLINI S, CAROLI P, NANNI C, MASSARO A, MARZOLA MC, RUBELLO D, FANTI S. PET/CT imaging in different types of lung cancer: an overview. Eur J Radiol. 2012;81:988–1001. doi: 10.1016/j.ejrad.2011.03.020. [DOI] [PubMed] [Google Scholar]
- BECKER DJ, LOWE JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- BLAIR SL, HEERDT P, SACHAR S, ABOLHODA A, HOCHWALD S, CHENG H, BURT M. Glutathione metabolism in patients with non-small cell lung cancers. Cancer research. 1997;57:152–155. [PubMed] [Google Scholar]
- BRUNTZ RC, LANE AN, HIGASHI RM, FAN TW. Exploring Cancer Metabolism using Stable Isotope Resolved Metabolomics (SIRM) Journal of Biological Chemistry. 2017 doi: 10.1074/jbc.R117.776054. jbc. R117. 776054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BULL C, STOEL MA, DEN BROK MH, ADEMA GJ. Sialic acids sweeten a tumor’s life. Cancer Res. 2014;74:3199–204. doi: 10.1158/0008-5472.CAN-14-0728. [DOI] [PubMed] [Google Scholar]
- BUSHUE N, WAN YJY. Retinoid pathway and cancer therapeutics. Advanced drug delivery reviews. 2010;62:1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALDERÓN-SANTIAGO M, PRIEGO-CAPOTE F, TURCK N, ROBIN X, JURADO-GÁMEZ B, SANCHEZ JC, DE CASTRO MDL. Human sweat metabolomics for lung cancer screening. Analytical and bioanalytical chemistry. 2015;407:5381–5392. doi: 10.1007/s00216-015-8700-8. [DOI] [PubMed] [Google Scholar]
- CAMERON SJ, LEWIS KE, BECKMANN M, ALLISON GG, GHOSAL R, LEWIS PD, MUR LA. The metabolomic detection of lung cancer biomarkers in sputum. Lung Cancer. 2016;94:88–95. doi: 10.1016/j.lungcan.2016.02.006. [DOI] [PubMed] [Google Scholar]
- CARR DF, ALFIREVIC A, PIRMOHAMED M. Pharmacogenomics: Current State-of-the-Art. Genes (Basel) 2014;5:430–43. doi: 10.3390/genes5020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN Y, MA Z, LI A, LI H, WANG B, ZHONG J, MIN L, DAI L. Metabolomic profiling of human serum in lung cancer patients using liquid chromatography/hybrid quadrupole time-of-flight mass spectrometry and gas chromatography/mass spectrometry. J Cancer Res Clin Oncol. 2015;141:705–18. doi: 10.1007/s00432-014-1846-5. [DOI] [PubMed] [Google Scholar]
- CURRIE E, SCHULZE A, ZECHNER R, WALTHER TC, FARESE RV., JR Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE MOLINA AR, RODRíGUEZ-GONZÁLEZ AN, GUTIÉRREZ R, MARTıNEZ-PINEIRO L, SÁNCHEZ JJ, BONILLA F, ROSELL R, LACAL JC. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochemical and biophysical research communications. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- DE MOLINA AR, SARMENTERO-ESTRADA J, BELDA-INIESTA C, TARÓN M, DE MOLINA VR, CEJAS P, SKRZYPSKI M, GALLEGO-ORTEGA D, DE CASTRO J, CASADO E. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. The lancet oncology. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- DEJA S, POREBSKA I, KOWAL A, ZABEK A, BARG W, PAWELCZYK K, STANIMIROVA I, DASZYKOWSKI M, KORZENIEWSKA A, JANKOWSKA R, MLYNARZ P. Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. J Pharm Biomed Anal. 2014;100:369–80. doi: 10.1016/j.jpba.2014.08.020. [DOI] [PubMed] [Google Scholar]
- DELGADO-POVEDANO MDM, CALDERÓN-SANTIAGO M, PRIEGO-CAPOTE F, JURADO-GÁMEZ B, LUQUE DE, CASTRO MD. Recent advances in human sweat metabolomics for lung cancer screening. Metabolomics. 2016;12 doi: 10.1007/s00216-015-8700-8. [DOI] [PubMed] [Google Scholar]
- DOHERTY JR, CLEVELAND JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–92. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONG J, CAI X, ZHAO L, XUE X, ZOU L, ZHANG X, LIANG X. Lysophosphatidylcholine profiling of plasma: discrimination of isomers and discovery of lung cancer biomarkers. Metabolomics. 2010;6:478–488. [Google Scholar]
- DUNN WB, BROADHURST DI, ATHERTON HJ, GOODACRE R, GRIFFIN JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chemical Society Reviews. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- EMWAS AHM, SALEK RM, GRIFFIN JL, MERZABAN J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics. 2013;9:1048–1072. [Google Scholar]
- EMWAS AH. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161–93. doi: 10.1007/978-1-4939-2377-9_13. [DOI] [PubMed] [Google Scholar]
- FAN TWM, LANE AN, HIGASHI RM, YAN J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics. 2011;7:257–269. doi: 10.1007/s11306-010-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN TW, LANE AN, HIGASHI RM. Stable isotope resolved metabolomics studies in ex vivo tissue slices. Bio-protocol. 2016;6 doi: 10.21769/bioprotoc.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN TW, LANE AN, HIGASHI RM, FARAG MA, GAO H, BOUSAMRA M, MILLER DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRUZZI E, FRANCESCHINI R, CAZZOLATO G, GERONI C, FOWST C, PASTORINO U, TRADATI N, TURSI J, DITTADI R, GION M. Blood glutathione as a surrogate marker of cancer tissue glutathione S-transferase activity in non-small cell lung cancer and squamous cell carcinoma of the head and neck. European Journal of Cancer. 2003;39:1019–1029. doi: 10.1016/s0959-8049(03)00122-9. [DOI] [PubMed] [Google Scholar]
- GINSBERG MS, GREWAL RK, HEELAN RT. Lung cancer. Radiol Clin North Am. 2007;45:21–43. doi: 10.1016/j.rcl.2006.10.004. [DOI] [PubMed] [Google Scholar]
- GUO Y, WANG X, QIU L, QIN X, LIU H, WANG Y, LI F, WANG X, CHEN G, SONG G, LI F, GUO S, LI Z. Probing gender-specific lipid metabolites and diagnostic biomarkers for lung cancer using Fourier transform ion cyclotron resonance mass spectrometry. Clin Chim Acta. 2012;414:135–41. doi: 10.1016/j.cca.2012.08.010. [DOI] [PubMed] [Google Scholar]
- HALKET JM, WATERMAN D, PRZYBOROWSKA AM, PATEL RK, FRASER PD, BRAMLEY PM. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J Exp Bot. 2005;56:219–43. doi: 10.1093/jxb/eri069. [DOI] [PubMed] [Google Scholar]
- HAO D, SARFARAZ MO, FARSHIDFAR F, BEBB DG, LEE CY, CARD CM, DAVID M, WELJIE AM. Erratum to: Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics. 2016a;12 doi: 10.1007/s11306-016-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAO D, SARFARAZ MO, FARSHIDFAR F, BEBB DG, LEE CY, CARD CM, DAVID M, WELJIE AM. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics. 2016b;12:58. doi: 10.1007/s11306-016-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HART PH, TOWNLEY SL, GRIMBALDESTON MA, KHALIL Z, FINLAY-JONES JJ. Mast cells, neuropeptides, histamine, and prostaglandins in UV-induced systemic immunosuppression. Methods. 2002;28:79–89. doi: 10.1016/s1046-2023(02)00201-3. [DOI] [PubMed] [Google Scholar]
- HASSANEIN M, CALLISON JC, CALLAWAY-LANE C, ALDRICH MC, GROGAN EL, MASSION PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5:992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAZNADAR M, CAI Q, KRAUSZ KW, BOWMAN ED, MARGONO E, NORO R, THOMPSON MD, MATHE EA, MUNRO HM, STEINWANDEL MD, GONZALEZ FJ, BLOT WJ, HARRIS CC. Urinary Metabolite Risk Biomarkers of Lung Cancer: A Prospective Cohort Study. Cancer Epidemiol Biomarkers Prev. 2016;25:978–86. doi: 10.1158/1055-9965.EPI-15-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIGASHI RM, FAN TW-M, LORKIEWICZ PK, MOSELEY HNB, LANE AN. Stable Isotope-Labeled Tracers for Metabolic Pathway Elucidation by GC-MS and FT-MS. In: RAFTERY D, editor. Mass Spectrometry in Metabolomics: Methods and Protocols. New York, NY: Springer New York; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRAYAMA A, KAMI K, SUGIMOTO M, SUGAWARA M, TOKI N, ONOZUKA H, KINOSHITA T, SAITO N, OCHIAI A, TOMITA M, ESUMI H, SOGA T. Quantitative Metabolome Profiling of Colon and Stomach Cancer Microenvironment by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Cancer Research. 2009;69:4918. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- HIRSCH HA, ILIOPOULOS D, JOSHI A, ZHANG Y, JAEGER SA, BULYK M, TSICHLIS PN, SHIRLEY LIU X, STRUHL K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–61. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORI S, NISHIUMI S, KOBAYASHI K, SHINOHARA M, HATAKEYAMA Y, KOTANI Y, HATANO N, MANIWA Y, NISHIO W, BAMBA T, FUKUSAKI E, AZUMA T, TAKENAWA T, NISHIMURA Y, YOSHIDA M. A metabolomic approach to lung cancer. Lung Cancer. 2011;74:284–92. doi: 10.1016/j.lungcan.2011.02.008. [DOI] [PubMed] [Google Scholar]
- HORNAK JP. The Basics of NMR, 1996±2002 [Google Scholar]
- HSIEH AH, TAHKAR H, KOCZWARA B, KICHENADASSE G, BECKMANN K, KARAPETIS C, SUKUMARAN S. Pre-treatment serum lactate dehydrogenase as a biomarker in small cell lung cancer. Asia Pac J Clin Oncol. 2017 doi: 10.1111/ajco.12674. [DOI] [PubMed] [Google Scholar]
- HU H, TAKANO N, XIANG L, GILKES DM, LUO W, SEMENZA GL. Hypoxia-inducible factors enhance glutamate signaling in cancer cells. Oncotarget. 2014;5:8853. doi: 10.18632/oncotarget.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ILONEN IK, RÄSÄNEN JV, SIHVO EI, KNUUTTILA A, SALMENKIVI KM, AHOTUPA MO, KINNULA VL, SALO JA. Oxidative stress in non-small cell lung cancer: role of nicotinamide adenine dinucleotide phosphate oxidase and glutathione. Acta Oncologica. 2009;48:1054–1061. doi: 10.1080/02841860902824909. [DOI] [PubMed] [Google Scholar]
- JACKMAN DM, JOHNSON BE. Small-cell lung cancer. The Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- JAMES TL. Fundamentals of NMR. Online Textbook: Department of Pharmaceutical Chemistry, University of California, San Francisco. 1998:1–31. [Google Scholar]
- KAMI K, FUJIMORI T, SATO H, SATO M, YAMAMOTO H, OHASHI Y, SUGIYAMA N, ISHIHAMA Y, ONOZUKA H, OCHIAI A. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics. 2013;9:444–453. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANEKO K, SMETANA-JUST U, MATSUI M, YOUNG AR, JOHN S, NORVAL M, WALKER SL. cis-Urocanic acid initiates gene transcription in primary human keratinocytes. The Journal of Immunology. 2008;181:217–224. doi: 10.4049/jimmunol.181.1.217. [DOI] [PubMed] [Google Scholar]
- KANTAE V, KREKELS EH, ESDONK MJ, LINDENBURG P, HARMS AC, KNIBBE CA, VAN DER GRAAF PH, HANKEMEIER T. Integration of pharmacometabolomics with pharmacokinetics and pharmacodynamics: towards personalized drug therapy. Metabolomics. 2017;13:9. doi: 10.1007/s11306-016-1143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KASPAR H, DETTMER K, GRONWALD W, OEFNER PJ. Automated GC–MS analysis of free amino acids in biological fluids. Journal of Chromatography B. 2008;870:222–232. doi: 10.1016/j.jchromb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- KHALIL Z, TOWNLEY SL, GRIMBALDESTON MA, FINLAY-JONES JJ, HART PH. cis-Urocanic acid stimulates neuropeptide release from peripheral sensory nerves. Journal of investigative dermatology. 2001;117:886–891. doi: 10.1046/j.0022-202x.2001.01466.x. [DOI] [PubMed] [Google Scholar]
- KIM AW, BATUS M, MYINT R, FIDLER MJ, BASU S, BONOMI P, FABER LP, WIGHTMAN SC, WARREN WH, MCINTIRE M. Prognostic value of xanthine oxidoreductase expression in patients with non-small cell lung cancer. Lung Cancer. 2011;71:186–190. doi: 10.1016/j.lungcan.2010.05.006. [DOI] [PubMed] [Google Scholar]
- KIM KR, PARK HG, PAIK MJ, RYU HS, OH KS, MYUNG SW, LIEBICH HM. Gas chromatographic profiling and pattern recognition analysis of urinary organic acids from uterine myoma patients and cervical cancer patients. Journal of Chromatography B: Biomedical Sciences and Applications. 1998;712:11–22. doi: 10.1016/s0378-4347(98)00155-8. [DOI] [PubMed] [Google Scholar]
- LANE AN, FAN TW. NMR-based Stable Isotope Resolved Metabolomics in systems biochemistry. Arch Biochem Biophys. 2017 doi: 10.1016/j.abb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANE AN, FAN TW, BOUSAMRA M, 2ND, HIGASHI RM, YAN J, MILLER DM. Stable isotope-resolved metabolomics (SIRM) in cancer research with clinical application to nonsmall cell lung cancer. OMICS. 2011;15:173–82. doi: 10.1089/omi.2010.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANE AN, FAN TWM. Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY. Metabolomics. 2007;3:79–86. [Google Scholar]
- LANE AN, FAN TWM, HIGASHI RM. Isotopomer‐Based Metabolomic Analysis by NMR and Mass Spectrometry. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- LI Y, SONG X, ZHAO X, ZOU L, XU G. Serum metabolic profiling study of lung cancer using ultra high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;966:147–53. doi: 10.1016/j.jchromb.2014.04.047. [DOI] [PubMed] [Google Scholar]
- LONG F, SU JH, LIANG B, SU LL, JIANG SJ. Identification of gene biomarkers for distinguishing small-cell lung cancer from non-small-cell lung cancer using a network-based approach. BioMed research international. 2015 doi: 10.1155/2015/685303. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU W, SU X, KLEIN MS, LEWIS IA, FIEHN O, RABINOWITZ JD. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu Rev Biochem. 2017;86:277–304. doi: 10.1146/annurev-biochem-061516-044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATHE EA, PATTERSON AD, HAZNADAR M, MANNA SK, KRAUSZ KW, BOWMAN ED, SHIELDS PG, IDLE JR, SMITH PB, ANAMI K, KAZANDJIAN DG, HATZAKIS E, GONZALEZ FJ, HARRIS CC. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 2014;74:3259–70. doi: 10.1158/0008-5472.CAN-14-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEDES G, THOMAS A, WEINHOUSE S. Metabolism of Neoplastic Tissue. IV. A Study of Lipid Synthesis in Neoplastic Tissue Slices in Vitro. Cancer Research. 1953;13:27. [PubMed] [Google Scholar]
- MIYAGI Y, HIGASHIYAMA M, GOCHI A, AKAIKE M, ISHIKAWA T, MIURA T, SARUKI N, BANDO E, KIMURA H, IMAMURA F, MORIYAMA M, IKEDA I, CHIBA A, OSHITA F, IMAIZUMI A, YAMAMOTO H, MIYANO H, HORIMOTO K, TOCHIKUBO O, MITSUSHIMA T, YAMAKADO M, OKAMOTO N. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS One. 2011;6:e24143. doi: 10.1371/journal.pone.0024143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIYAMOTO S, TAYLOR SL, BARUPAL DK, TAGUCHI A, WOHLGEMUTH G, WIKOFF WR, YONEDA KY, GANDARA DR, HANASH SM, KIM K, FIEHN O. Systemic Metabolomic Changes in Blood Samples of Lung Cancer Patients Identified by Gas Chromatography Time-of-Flight Mass Spectrometry. Metabolites. 2015;5:192–210. doi: 10.3390/metabo5020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIYOSHI T, KONDO K, FUJINO H, TAKAHASHI Y, SAWADA N, SAKIYAMA S, TSUYUGUCHI M, KIMURA S, SUMITOMO M, MONDEN Y. Thymidylate synthase and dihydropyrimidine dehydrogenase in non-small cell lung cancer: relationship between mRNA expression and activity. Anticancer research. 2005;25:923–930. [PubMed] [Google Scholar]
- MOESTUE S, SITTER B, FROST BATHEN T, TESSEM MB, SUSANN GRIBBESTAD I. HR MAS MR spectroscopy in metabolic characterization of cancer. Current topics in medicinal chemistry. 2011;11:2–26. doi: 10.2174/156802611793611869. [DOI] [PubMed] [Google Scholar]
- MORRIS SM. Enzymes of arginine metabolism. The Journal of nutrition. 2004;134:2743S–2747S. doi: 10.1093/jn/134.10.2743S. [DOI] [PubMed] [Google Scholar]
- MUNTONI S, ATZORI L, MEREU R, SATTA G, MACIS MD, CONGIA M, TEDDE A, DESOGUS A. Serum lipoproteins and cancer. Nutrition, Metabolism and Cardiovascular Diseases. 2009;19:218–225. doi: 10.1016/j.numecd.2008.06.002. [DOI] [PubMed] [Google Scholar]
- MUSHARRAF SG, MAZHAR S, CHOUDHARY MI, RIZI N, ATTA UR R. Plasma metabolite profiling and chemometric analyses of lung cancer along with three controls through gas chromatography-mass spectrometry. Sci Rep. 2015;5:8607. doi: 10.1038/srep08607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’SHEA K, CAMERON SJ, LEWIS KE, LU C, MUR LA. Metabolomic-based biomarker discovery for non-invasive lung cancer screening: A case study. Biochim Biophys Acta. 2016;1860:2682–7. doi: 10.1016/j.bbagen.2016.07.007. [DOI] [PubMed] [Google Scholar]
- O’MALLEY SS, WU R, MAYNE ST, JATLOW PI. Smoking Cessation Is Followed by Increases in Serum Bilirubin, an Endogenous Antioxidant Associated With Lower Risk of Lung Cancer and Cardiovascular Disease. Nicotine & Tobacco Research. 2014;16:1145–1149. doi: 10.1093/ntr/ntu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAMUNGKAS AD, PARK C, LEE S, JEE SH, PARK YH. High resolution metabolomics to discriminate compounds in serum of male lung cancer patients in South Korea. Respir Res. 2016;17:100. doi: 10.1186/s12931-016-0419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POTTON E, MCCAUGHAN F, JANES S. Chronic obstructive pulmonary disease and lung cancer. Respiratory Medicine: COPD Update. 2009;5:34–37. [Google Scholar]
- PUCHADES-CARRASCO L, JANTUS-LEWINTRE E, PÉREZ-RAMBLA C, GARCÍA-GARCÍA F, LUCAS R, CALABUIG S, BLASCO A, DOPAZO J, CAMPS C, PINEDA-LUCENA A. Serum metabolomic profiling facilitates the non-invasive identification of metabolic biomarkers associated with the onset and progression of non-small cell lung cancer. Oncotarget. 2016;7:12904. doi: 10.18632/oncotarget.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REDDY AV, KILLAMPALLI LK, PRAKASH AR, NAAG S, SREENATH G, BIRAGGARI SK. Analysis of lipid profile in cancer patients, smokers, and nonsmokers. Dental Research Journal. 2016;13:494–499. doi: 10.4103/1735-3327.197036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROCHA CM, BARROS AS, GOODFELLOW BJ, CARREIRA IM, GOMES A, SOUSA V, BERNARDO J, CARVALHO L, GIL AM, DUARTE IF. NMR metabolomics of human lung tumours reveals distinct metabolic signatures for adenocarcinoma and squamous cell carcinoma. Carcinogenesis. 2015;36:68–75. doi: 10.1093/carcin/bgu226. [DOI] [PubMed] [Google Scholar]
- SAYIN VI, IBRAHIM MX, LARSSON E, NILSSON JA, LINDAHL P, BERGO MO. Antioxidants accelerate lung cancer progression in mice. Science translational medicine. 2014;6:221ra15–221ra15. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- SCHAUER R. Sialic acids: fascinating sugars in higher animals and man. Zoology. 2004;107:49–64. doi: 10.1016/j.zool.2003.10.002. [DOI] [PubMed] [Google Scholar]
- SCHMID S, KÜBLER M, AYATA CK, LAZAR Z, HAAGER B, HOßFELD M, MEYER A, CICKO S, ELZE M, WIESEMANN S. Altered purinergic signaling in the tumor associated immunologic microenvironment in metastasized non-small-cell lung cancer. Lung Cancer. 2015;90:516–521. doi: 10.1016/j.lungcan.2015.10.005. [DOI] [PubMed] [Google Scholar]
- SELLERS K, FOX MP, BOUSAMRA M, 2ND, SLONE SP, HIGASHI RM, MILLER DM, WANG Y, YAN J, YUNEVA MO, DESHPANDE R, LANE AN, FAN TW. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest. 2015;125:687–98. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHINGYOJI M, II, ZASA T, HIGASHIYAMA M, IMAMURA F, SARUKI N, IMAIZUMI A, YAMAMOTO H, DAIMON T, TOCHIKUBO O, MITSUSHIMA T. The significance and robustness of a plasma free amino acid (PFAA) profile-based multiplex function for detecting lung cancer. BMC cancer. 2013;13:77. doi: 10.1186/1471-2407-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHINTANI Y, INOUE M, FUNAKOSHI Y, MATSUMURA A, OHTA M, MAEDA H, OKUMURA M. Low dihydropyrimidine dehydrogenase correlates with prolonged survival in patients with lung adenocarcinoma treated with 5-fluorouracil. Anticancer research. 2011;31:4665–4671. [PubMed] [Google Scholar]
- SIEGEL RL, MILLER KD, JEMAL A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- SIMOPOULOS AP. The importance of the ratio of omega-6/omega-3 essential fatty acids. Biomedicine & pharmacotherapy. 2002;56:365–379. doi: 10.1016/s0753-3322(02)00253-6. [DOI] [PubMed] [Google Scholar]
- SIMOPOULOS AP. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Experimental biology and medicine. 2008;233:674–688. doi: 10.3181/0711-MR-311. [DOI] [PubMed] [Google Scholar]
- SNYDER R. Leukemia and benzene. International journal of environmental research and public health. 2012;9:2875–2893. doi: 10.3390/ijerph9082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRINGER KA, MCKAY RT, KARNOVSKY A, QUEMERAIS B, LACY P. Metabolomics and Its Application to Acute Lung Diseases. Front Immunol. 2016;7:44. doi: 10.3389/fimmu.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKEUCHI T, FUJIKI H, KAMEYA T. Characterization of amylases produced by tumors. Clinical chemistry. 1981;27:556–559. [PubMed] [Google Scholar]
- THEODORIDIS G, GIKA HG, WILSON ID. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom Rev. 2011;30:884–906. doi: 10.1002/mas.20306. [DOI] [PubMed] [Google Scholar]
- TRAVERSO N, RICCIARELLI R, NITTI M, MARENGO B, FURFARO AL, PRONZATO MA, MARINARI UM, DOMENICOTTI C. Role of glutathione in cancer progression and chemoresistance. Oxidative medicine and cellular longevity. 2013 doi: 10.1155/2013/972913. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSUN ZY, POSSEMATO R. Amino acid management in cancer. Semin Cell Dev Biol. 2015;43:22–32. doi: 10.1016/j.semcdb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUNA D, SOBOLEWSKI AL, DOMCKE W. Photochemical mechanisms of radiationless deactivation processes in urocanic acid. The Journal of Physical Chemistry B. 2014;118:976–985. doi: 10.1021/jp411818j. [DOI] [PubMed] [Google Scholar]
- UBHI BK, RILEY JH, SHAW PA, LOMAS DA, TAL-SINGER R, MACNEE W, GRIFFIN JL, CONNOR SC. Metabolic profiling detects biomarkers of protein degradation in COPD patients. European Respiratory Journal. 2012;40:345–355. doi: 10.1183/09031936.00112411. [DOI] [PubMed] [Google Scholar]
- VAN DEN HEUVEL AP, JING J, WOOSTER RF, BACHMAN KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012;13:1185–94. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANDER HEIDEN MG, CANTLEY LC, THOMPSON CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science (New York, NY) 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VULIMIRI SV, MISRA M, HAMM JT, MITCHELL M, BERGER A. Effects of Mainstream Cigarette Smoke on the Global Metabolome of Human Lung Epithelial Cells. Chemical Research in Toxicology. 2009;22:492–503. doi: 10.1021/tx8003246. [DOI] [PubMed] [Google Scholar]
- WANG C, DONG R, WANG X, LIAN A, CHI C, KE C, GUO L, LIU S, ZHAO W, XU G. Exhaled volatile organic compounds as lung cancer biomarkers during one-lung ventilation. Scientific reports. 2014;4:7312. doi: 10.1038/srep07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG H, YANG L, ZOU L, HUANG D, GUO Y, PAN M, TAN Y, ZHONG H, JI W, RAN P, ZHONG N, LU J. Association between chronic obstructive pulmonary disease and lung cancer: a case-control study in Southern Chinese and a meta-analysis. PLoS One. 2012;7:e46144. doi: 10.1371/journal.pone.0046144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG L, TANG Y, LIU S, MAO S, LING Y, LIU D, HE X, WANG X. Metabonomic profiling of serum and urine by 1H NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PloS one. 2013;8:e65675. doi: 10.1371/journal.pone.0065675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARBURG O. On the Origin of Cancer Cells. Science. 1956;123:309. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- WEN CP, ZHANG F, LIANG D, WEN C, GU J, SKINNER H, CHOW WH, YE Y, PU X, HILDEBRANDT MA, HUANG M, CHEN CH, HSIUNG CA, TSAI MK, TSAO CK, LIPPMAN SM, WU X. The ability of bilirubin in identifying smokers with higher risk of lung cancer: a large cohort study in conjunction with global metabolomic profiling. Clin Cancer Res. 2015;21:193–200. doi: 10.1158/1078-0432.CCR-14-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIKOFF WR, GRAPOV D, FAHRMANN JF, DEFELICE B, ROM WN, PASS HI, KIM K, NGUYEN U, TAYLOR SL, GANDARA DR, KELLY K, FIEHN O, MIYAMOTO S. Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev Res (Phila) 2015a;8:410–8. doi: 10.1158/1940-6207.CAPR-14-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIKOFF WR, HANASH S, DEFELICE B, MIYAMOTO S, BARNETT M, ZHAO Y, GOODMAN G, FENG Z, GANDARA D, FIEHN O, TAGUCHI A. Diacetylspermine Is a Novel Prediagnostic Serum Biomarker for Non-Small-Cell Lung Cancer and Has Additive Performance With Pro-Surfactant Protein B. J Clin Oncol. 2015b;33:3880–6. doi: 10.1200/JCO.2015.61.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WISHART DS. Is Cancer a Genetic Disease or a Metabolic Disease? EBioMedicine. 2015;2:478–9. doi: 10.1016/j.ebiom.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIWANITKIT V, SUWANSAKSRI J, SOOGARUN S. Monitoring of urine trans, trans-muconic acid level among smokers and non-smokers. Respiratory medicine. 2005;99:788–791. doi: 10.1016/j.rmed.2004.10.017. [DOI] [PubMed] [Google Scholar]
- WONG MCS, LAO XQ, HO KF, GOGGINS WB, TSE SLA. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7:14300. doi: 10.1038/s41598-017-14513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIE H, HANAI J, REN JG, KATS L, BURGESS K, BHARGAVA P, SIGNORETTI S, BILLIARD J, DUFFY KJ, GRANT A, WANG X, LORKIEWICZ PK, SCHATZMAN S, BOUSAMRA M, 2ND, LANE AN, HIGASHI RM, FAN TW, PANDOLFI PP, SUKHATME VP, SETH P. Targeting lactate dehydrogenase–a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014;19:795–809. doi: 10.1016/j.cmet.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO H, RAHMAN I. Current concepts on the role of inflammation in COPD and lung cancer. Curr Opin Pharmacol. 2009;9:375–83. doi: 10.1016/j.coph.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YEH CS, WANG JY, CHENG TL, JUAN CH, WU CH, LIN SR. Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by Microarray-Bioinformatics analysis. Cancer letters. 2006;233:297–308. doi: 10.1016/j.canlet.2005.03.050. [DOI] [PubMed] [Google Scholar]
- YU L, LI K, ZHANG X. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: mini review. Oncotarget. 2017;8:115774–115786. doi: 10.18632/oncotarget.22404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU X, WU Q, LU W, WANG Y, MA X, CHEN Z, YAN C. Metabonomics study of lung cancer cells based on liquid l chromatography-mass spectrometry. Chinese journal of chromatography. 2013;31:691–696. doi: 10.3724/sp.j.1123.2012.12039. [DOI] [PubMed] [Google Scholar]
- YUAN JM, GAO YT, MURPHY SE, CARMELLA SG, WANG R, ZHONG Y, MOY KA, DAVIS AB, TAO L, CHEN M, HAN S, NELSON HH, YU MC, HECHT SS. Urinary levels of cigarette smoke constituent metabolites are prospectively associated with lung cancer development in smokers. Cancer Res. 2011;71:6749–57. doi: 10.1158/0008-5472.CAN-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG A, SUN H, WANG P, HAN Y, WANG X. Modern analytical techniques in metabolomics analysis. Analyst. 2012a;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- ZHANG F, DU G. Dysregulated lipid metabolism in cancer. World J Biol Chem. 2012;3:167–74. doi: 10.4331/wjbc.v3.i8.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG WC, SHYH-CHANG N, YANG H, RAI A, UMASHANKAR S, MA S, SOH BS, SUN LL, TAI BC, NGA ME, BHAKOO KK, JAYAPAL SR, NICHANE M, YU Q, AHMED DA, TAN C, SING WP, TAM J, THIRUGANANAM A, NOGHABI MS, PANG YH, ANG HS, MITCHELL W, ROBSON P, KALDIS P, SOO RA, SWARUP S, LIM EH, LIM B. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012b;148:259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- ZHANG X, ZHU X, WANG C, ZHANG H, CAI Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget. 2016;7:63437. doi: 10.18632/oncotarget.11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU W, CAPELLO M, FREDOLINI C, RACANICCHI L, PIEMONTI L, LIOTTA LA, NOVELLI F, PETRICOIN EF. Proteomic Analysis Reveals Warburg Effect and Anomalous Metabolism of Glutamine in Pancreatic Cancer Cells. Journal of Proteome Research. 2012;11:554–563. doi: 10.1021/pr2009274. [DOI] [PMC free article] [PubMed] [Google Scholar]


