Abstract
Background & Aims
Genetic factors are believed to affect risk for irritable bowel syndrome (IBS), but there have been no sufficiently powered and adequately sized studies. To identify DNA variants associated with IBS risk, we performed a genome-wide association study (GWAS) of the large UK Biobank population-based cohort, which includes genotype and health data from 500,000 participants.
Methods
We studied 7,287,191 high-quality single-nucleotide polymorphisms in individuals who self-reported a doctor’s diagnosis of IBS (cases; m=9576) compared to the remainder of the cohort (controls; n=336,499) (mean age of study subjects, 40–69 years). Genome-wide significant findings were further investigated in 2045 patients with IBS from tertiary centers and 7955 population controls from Europe and the United States, and a small general population sample from Sweden (n=249). Functional annotation of GWAS results was carried out by integrating data from multiple biorepositories, to obtain biological insights from the observed associations.
Results
We identified a genome-wide significant association on chromosome 9q31.2 (SNP rs10512344; P=3.57×10−8), in a region previously linked to age at menarche, and 13 additional loci of suggestive significance (P<5.0×10−6). Sex-stratified analyses revealed that the variants at 9q32.1 affect risk of IBS in only women (P=4.29×10−10 in UK Biobank) and also associate with constipation-predominant IBS in women (P=.015 in the tertiary cohort) and harder stools in women (P=.0012 in the population-based sample). Functional annotation of the 9q32.1 locus identified 8 candidate genes, including the elongator complex protein 1 gene (ELP1 or IKBKAP), which is mutated in patients with familial dysautonomia.
Conclusions
In a sufficiently powered GWAS of IBS, we associated variants at the locus 9q32.1 with risk of IBS in women. This observation may provide additional rationale for investigating the role of sex hormones and autonomic dysfunction in IBS.
Keywords: SNP, biobank research, genetics, bowel symptoms
INTRODUCTION
Irritable bowel syndrome (IBS) is the most common functional gastrointestinal disorder (FGID), affecting an average of 11% people worldwide with symptoms of abdominal pain and bloating, associated with constipation (IBS-C), diarrhea (IBS-D), or their combination (IBS-M).1 While expert consensus (Rome) criteria represent the current gold-standards to characterize IBS patients based on their symptoms, the underlying pathophysiology is still poorly understood, and this hampers the implementation of adequate therapeutic strategies, their efficacy and precision.2,3 IBS is a leading cause of work absenteeism, consumes 0.5% of annual healthcare budgets, and the identification of biomarkers suitable for patients stratification is therefore an active area of dedicated clinical research.3 In recent years, a number of factors have been proposed to contribute to IBS pathophysiology including miscommunication along the gut-brain axis, visceral hypersensitivity and altered motility, dysregulated immune activation, bile acids malabsorption, food intolerance and gut microbiota dysbiosis.3
Although with varying outcomes, a heritable component of IBS has long been demonstrated in independent twin and family studies,4 and more recently in a nationwide survey of first-, second- and third-degree relatives of Swedish IBS probands.5 Based on classical candidate-gene approaches, previous work from our group and others has highlighted the presence of rare pathogenic SI (sucrose-isomaltase) and SCN5A (voltage-gated sodium channel NaV1.5) variants in 2–4% of IBS patients, possibly linking their symptomatic manifestations to disaccharide intolerance and channelopathies, respectively.6,7 Additional associations with common single nucleotide polymorphisms (SNP) have been reported in the past (TNFSF15, TRPM8, CDC42, NPSR1, KDELR2 and other genes).8–13 Given the documented familial predisposition, and the evidence of diverse genetic associations with IBS, there is hope that genetic studies will identify actionable pathways that may allow enhanced therapy of IBS. To date, IBS genetic studies have been largely underpowered to detect modest genetic risk effects with genome-wide significance and no true unequivocal IBS risk locus has been thus far identified. Overall, large-scale gene-mapping efforts have been lacking, mainly due to the scarcity of adequately sized patient cohorts, and the heterogeneity of results observed across epidemiological studies. To try to overcome this issue, we recently proposed the use of large population-based cohorts for the study of IBS predisposition, leveraging large sample sizes and a harmonized (though less clinically informative) questionnaire-based definition of cases and controls.14
Here we report a genome-wide association study (GWAS) of IBS symptoms in the UK Biobank cohort, which includes health information and single nucleotide polymorphism (SNP) genotype data from 500,000 people aged 40–69 years, recruited across UK between 2006–2010. We used data on medical conditions recorded as doctor’s diagnoses, self-reported by UK Biobank participants via touchscreen questionnaires and later validated by a trained nurse during follow-up interviews. Genome-wide significant findings for a locus on chromosome 9q31.2 were further investigated in multi-national case-control cohorts from tertiary centers with expertise in IBS, and from a small general population sample from Sweden. The results point to female-specific genetic risk effects on IBS associated with constipation and harder stools.
MATERIALS AND METHODS
STUDY SUBJECTS
UK Biobank
UK Biobank (www.ukbiobank.ac.uk) is a population-based cohort study consisting of ~500,000 genotyped individuals (aged 40–69 years) recruited between 2006 and 2010 in the United Kingdom.15 Each participant underwent cognitive and physical assessment and, together with genotypes, a variety of phenotypic data have been collected, including demographics and health-related information. Health data (initially collected via a touchscreen questionnaire) included previous doctor’s diagnoses of non-cancer medical conditions as self-reported by UK Biobank participants (recorded at UK Biobank as data on “Non-cancer illness code, self-reported”). These were further investigated in a verbal interview by a trained nurse, who ratified and recorded the specific condition as self-reported doctor’s diagnosis. For the purpose of this study, individuals with a doctor’s diagnosis of IBS were selected as cases, while all other participants were regarded as controls. Prior to genotyping quality control and imputation, further selections criteria included (self-reported) British ancestry, and absence of independent diagnoses of inflammatory bowel disease (IBD) and celiac disease (CelD) (for both cases and controls) or IBS (for controls) in the data-fields “Non-cancer illness code, self-reported” and “Diagnoses – main ICD10” from hospital inpatient stay medical records. This resulted in 9,576 IBS cases and 336,499 controls included in downstream analyses (Table 1). UK Biobank received ethical approval from the competent Research Ethics Committee (REC reference for UK Biobank is 11/NW/0382)
Table 1.
Demographics of cohorts included in the study
| DATASET | N | Mean age (SD) | F:M |
|---|---|---|---|
| UK Biobank | |||
| IBS | 9,576 | 56.4 (7.9) | 7,130:2,446 |
| Controls | 336,499 | 56.9 (8.0) | 178,076:158,423 |
|
| |||
| Case-control | |||
| IBS | 2,045 | 42.0 (14.8) | 1592:453 |
| IBS-C | 598 | 42.0 (14.1) | 536:62 |
| IBS-D | 876 | 42.3 (14.7) | 612:264 |
| IBS-M | 502 | 41.8 (15.8) | 401:101 |
| Controls | 7,955 | 56.5 (16.4) | 4334:3621 |
|
| |||
| PopCol | |||
| All | 249 | 53.6 (11.4) | 158:91 |
IBS cases and controls
Cases
We studied 2,045 unrelated IBS patients (598 IBS-C, 876 IBS-D and 502 IBS-M according to Rome Criteria) of European ancestry from centers in Sweden (multi-center study), The Netherlands (Maastricht University), Belgium (GSK multi-national cohort from TARGID Leuven), Italy (multi-center study) and USA (Mayo Clinic and UCLA) (Table 1), as part of a larger ongoing collaborative study on the genetics of IBS (www.mdalab.org/bellygenes). These cohorts have been extensively characterized and described in detail in previous studies.6,8–10,12,13 Population outliers were removed from the analysis based on principal component analysis of available genotype data (see below). Approval of genetic studies was obtained at respective centers from competent local ethics authorities. Karolinska Institutet’s Research Ethics Committee approved the global multi-center study and the bellygenes initiative (protocol nr 2016/1620-31/2).
Controls
A total of 7,955 ancestry-matched, healthy controls were randomly selected from previously published studies or general population cohorts with available genetic data. Their characteristics have been described previously (see below), and the demographics are reported in Table 1. Swedish controls were from the TwinGene subset of genotyped individuals from the Screening Across the Lifespan Twin (SALT) study.16 Italian blood donor controls were previously described in a study from the International IBD Genetics Consortium (IIBDGC).17 Healthy controls from the Stroke Genetics Network (SiGN) Study were included as Belgian controls.18 UK controls were from the population-based cohort Understanding Society, a longitudinal study which follows 40,000 UK households.19 The Health and Retirement Study HRS, which includes over 12,000 Americans with age above 50, was identified as control group for USA.20 Dutch study controls included healthy individuals from the Maastricht area,21 and participants in the 500 Functional Genomics Project.22 Informed consent was obtained from all IBS patients and controls at respective centers, and the global study protocol was approved by Karolinska Institutet’s Research Ethics Committee.
PopCol
The Population-based Colonoscopy study (PopCol) is a general population-based cohort with randomly selected participants from Stockholm, Sweden, also previously described in detail and included in genetic studies.7,9,23 The present study includes 249 subjects (63% females, mean age 56.5, Table 1) with available genotypes and recordings of defecation patterns (including stool consistency based on the Bristol Stool Form Scale - BSFS, daily average range 1.5–6.9), and excluding individuals with IBD and CelD. Informed consent was obtained from all participants and the PopCol study protocol was approved by Karolinska Institutet’s Research Ethics Committee (protocol nr 394/01).
GENOTYPING QUALITY CONTROL (QC) AND IMPUTATION
UK Biobank
UK Biobank full genetic data release (July 2017) was used in this study. This included genotype data for 488,377 UKB participants from two genotyping arrays (438,427 UK Biobank Axiom array and 49,950 UK BiLEVE Axiom array) sharing over 95% content. Details of the array design, genotyping and imputation procedures have been published elsewhere.15 Both the Haplotype Reference Consortium (HRC) and the UK10K + 1000 Genomes Phase 3 reference panels were used for imputation at UK Biobank, though only HRC-imputed markers have been used in this study following UK Biobank advice, because of inconsistencies in SNP marker annotation discovered for other panels. Before association testing, rigorous per-sample and per-marker quality control (QC) procedures were applied, as follows: we excluded individuals with call rate <95%, genotype-phenotype sex discrepancies, deviant heterozygosity (UK Biobank outlier flag), related individuals (KING kinship coefficient >0.0663) and population outliers identified via principal component analysis (PCA; absolute deviation from the median exceeding 6×interquartile range); we removed SNP markers with call rate <95% (or significant - P<0.05 - call rate difference between cases and controls), poor imputation accuracy (IMPUTE2 INFO score <0.8), minor allele frequency (MAF) <1% and markers out of Hardy Weinberg equilibrium (P<1.0×10−5). This yielded high-quality data for 346,075 individuals and 7,287,191 SNP markers, showing no population stratification (scaled genomic inflation factor λ1000=1.00). For the genome-wide significant locus at 9q31.2, individual cluster plots for all genotyped markers were visually inspected using Evoker 2.4.1 (www.sanger.ac.uk/science/tools/evoker) to ensure correct allele calling and absence of genotyping artifacts or batch effects.
Case-control and PopCol rs10512344 genotype data
For IBS cases and controls, rs10512344 genotypes were extracted from currently available imputed genotype data. IBS patients were genotyped on Illumina Infinium CoreExome24 as part of the bellygenes initiative project (www.mdalab.org/bellygenes), while controls genotypes were obtained from previous Illumina genotyping efforts at respective individual sites and data sources (study PIs or biobanks). rs10512344 data was extracted from imputed genotypes, obtained after QC and imputation (including harmonization of genotypes from different platforms using GenotypeHarmonizer; github.com/molgenis/systemsgenetics/wiki/Genotype-Harmonizer), following a pipeline similar to that applied to UK Biobank data. rs10512344 genotype data included in this study were all imputed with high-accuracy (IMPUTE2 INFO score range 0.977–0.992). PopCol rs10512344 genotypes were extracted from available QC’ed and imputed Illumina OmniExpressExome-8 v1 genotype data, which have already been used and described in previous publications.7,9 Imputation metrics ensured high imputation accuracy at this marker (IMPUTE2 INFO score=1.0).
GWAS AND FOLLOW-UP ANALYSES
UK Biobank GWAS
Data from 346,075 UK Biobank individuals (9,576 IBS cases and 336,499 controls) entered the regression model. Association tests were performed in PLINK2 (www.cog-genomics.org/plink/2.0/) using logistic regression under an additive genetic model, adjusting for sex, age, genotyping array and the first 10 principal components from PCA. Markers showing suggestive association (P<5×10−6) were further analyzed through functional genomics annotation in order to map and prioritize genes (see next section). UK Biobank GWAS results were reported on the Manhattan plot using the Bioconductor GWASTools package (bioconductor.org/packages/release/bioc/html/GWASTools.html).
Post GWAS functional genomics analyses
IBS risk loci were defined as non-overlapping genomic regions extending a linkage disequilibrium (LD) window (r2=0.6) from the association signals with P<5.0×10−6. Annotation of GWAS results including genes mapping to the identified risk loci was performed with Functional mapping and annotation of GWAS (FUMA, fuma.ctglab.nl/).24 Integrating data from multiple biorepositories, genes are prioritized by FUMA via positional mapping based on annotations obtained from ANNOVAR (annovar.openbioinformatics.org), potential regulatory functions (15-core chromatin state predicted by ChromHMM (compbio.mit.edu/ChromHMM) for 127 tissue/cell types) and effects on gene expression using expression quantitative trait loci (eQTLs) of several tissue types from BIOS QTL (molgenis58.target.rug.nl/biosqtlbrowser), BRAINEAC (www.braineac.org) and GTEx (www.gtexportal.org) databases. To test for overrepresentation of biological functions based on gene annotations (gene set enrichment analysis, GSEA), we screened the Molecular Signature Database (MsigDB, including hallmark gene sets, curated gene sets, motif gene sets, Gene Ontology (GO) gene sets, oncogenic signatures and immunologic signatures, software.broadinstitute.org/gsea/msigdb), using the list of FUMA mapped genes against a total of 19,264 protein coding genes in hypergeometric enrichment tests. Gene sets with an adjusted P<0.05 (FDR correction according to Benjamini-Hochberg) were considered significant evidence of enrichment.
9q31.2 LOCUS-SPECIFIC ANALYSES
UK Biobank Sex-stratified analyses at the genome-wide significant locus 9q31.2 were performed as in the whole GWAS (excluding sex as covariate). Global and sex-stratified regional plots of 9q31.2 GWAS association signals were produced with ggplot2 (cran.r-project.org/web/packages/ggplot2/). Haplotype associations testing (SNP markers rs10512344 and rs10156597) for IBS in UK Biobank was performed using logistic regression and age, array and top ten PCs as covariates with haplo.stats (cran.r-project.org/web/packages/haplo.stats/). For the purpose of conducting colocalization and conditional analyses, we extracted 9q31.2 locus information from publicly available summary statistics data reported in the latest published age at menarche (AAM) GWAS meta-analysis,25 performed by the ReproGen consortium (www.reprogen.org). In order to investigate the interdependence of AAM and IBS association signals two summary statistics-based approaches were used: i) multi-trait-based conditional analysis using the Mendelian Randomization approach implemented in mtCOJO from the GCTA tools,26 and ii) Approximate Bayes Factor colocalization analysis to test whether the same or different causal variants are associated with multiple traits (IBS/AAM) using coloc (cran.r-project.org/web/packages/coloc/). The SNP marker rs10512344 was tested for association with IBS and stool consistency in additional cohorts (case-controls cohorts from tertiary centers and PopCol) in sex-stratified analyses: one-tailed association test with IBS in cases and controls was carried out on the combined cohorts using PLINK with logistic regression under an additive genetic model adjusted for age and country of origin; correlation between genotype and average BSFS scores was tested using linear regression methods implemented in R (cran.r-project.org) adjusting for age.
RESULTS
GWAS of IBS in UK Biobank
For GWAS purposes, we exploited the full release (July 2017) of UK Biobank data, including health and genotype information from 488,377 participants. IBS cases were identified based on doctor’s diagnoses self-reported by UK Biobank participants (collected via touchscreen questionnaire and ratified during a follow-up interview with a trained nurse; see Materials and Methods), while the remainder of the cohort was assigned to the control group. After exclusion of other relevant gastrointestinal (GI) diseases, people of non-British ancestry, related individuals, and individuals with missing data, we identified 9,576 IBS cases and 336,499 controls (Table 1). We applied stringent per-sample and per-marker quality controls (QC) to SNP data from UK Biobank (including both observed and imputed genotypes), in order to remove low-quality DNAs and rare or poor performing SNPs, and to identify and exclude population outliers based on principal components (PC) analysis of QC-filtered genotype data (Materials and Methods). This resulted in high-quality data for a total of 7,287,191 SNP markers, which were tested for association with IBS using age-, sex-, genotyping array- and principal component (PC)-adjusted logistic regression under an additive genetic model (Materials and Methods). The GWAS analysis yielded association signals of suggestive (P<5.0×10−6) significance for 14 independent loci, including a genome-wide significant (P<5.0×10−8) locus on chromosome 9q31.2 (rs10512344 [minor allele C] P=3.57×10−8, beta=0.190) (top panel in Figure 1, and Table 2).
Figure 1. Manhattan plot of GWAS results and regional plots for the 9q31.2 locus.
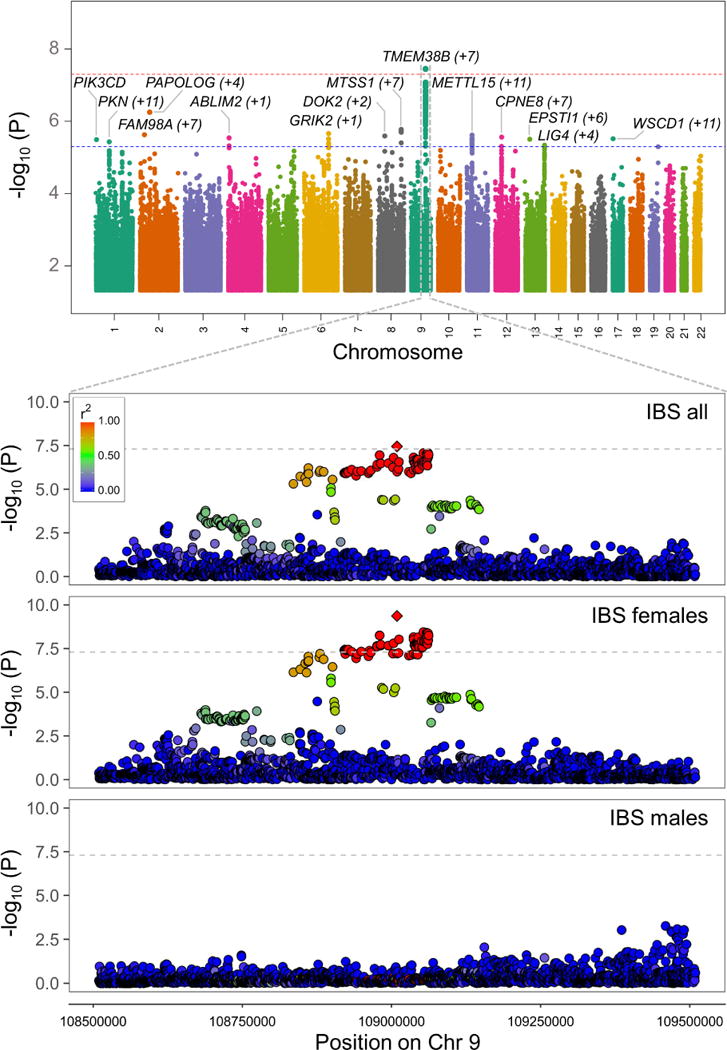
Top panel: GWAS association signals (−log10 P) are reported for SNP markers across all chromosomes, shown in alternate colors. Significance levels corresponding to genome wide (P=5.0×10−8) and suggestive (P=5.0×10−6) thresholds are indicated, respectively, with red and blue horizontal dashed lines. For each independent association signal, the nearest gene (mapping closest to the tag SNP) is reported, together with the number of additional genes from the same locus (in brackets). Bottom panel: 9q31.2 locus-specific regional plots of association signals in the entire UK Biobank cohort, and in sex-stratified analyses. SNPs are color-coded to reflect their linkage disequilibrium (r2) with the tag SNP rs10512344 (diamond symbol) according to the color-key in the upper left corner.
Table 2.
GWAS results including FUMA annotations.
| SNP | CHR | EA | OA | EAF CASES |
EAF CTRLS |
INFO | beta | SE | P | Positional map* | eQTL map** | Chromatin Interactions map*** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs140532807 | 1p36.22 | A | G | 0.015 | 0.011 | 0.89 | 0.289 | 0.062 | 3.22×10−6 | PIK3CD | ||
| rs10923043 | 1p22.2 | T | C | 0.565 | 0.548 | 0.99 | 0.069 | 0.015 | 3.70×10−6 | KYAT3 | COL24A1, ODF2L, CLCA2, CLCA1, CLCA4, SH3GLB1, SEP15, HS2ST1, LMO4, PKN2, BARHL2 | |
| rs116449111 | 2p22.3 | C | G | 0.974 | 0.979 | 0.97 | −0.218 | 0.046 | 2.36×10−6 | FAM98A | CAPN13, GALNT14, TTC27, LTBP1, RASGRP3, CRIM1, FEZ2, FAM98A | |
| rs116781238 | 2p16.1 | A | G | 0.056 | 0.048 | 0.95 | 0.161 | 0.032 | 5.61×10−7 | VRK2, PAPOLG, PUS10, PEX13, AHSA2 | ||
| rs1105615 | 4p16.1 | T | C | 0.173 | 0.160 | 0.99 | 0.091 | 0.02 | 2.88×10−6 | ABLIM2 | SORCS2 | |
| rs13212302 | 6q16.3 | T | C | 0.573 | 0.590 | 0.99 | −0.071 | 0.015 | 2.18×10−6 | ASCC3, GRIK2 | ||
| rs117107090 | 8p21.3 | A | G | 0.975 | 0.980 | 0.95 | −0.225 | 0.048 | 2.55×10−6 | LZTS1, DOK2, XPO7 | ||
| rs4871509 | 8q24.13 | A | G | 0.463 | 0.446 | 0.99 | 0.071 | 0.015 | 1.69×10−6 | MTSS1 | TRMT12, NDUFB9, MTSS1 | TATDN1, ZNF572, SQLE, KIAA0196, NSMCE2, MTSS1, TRMT12, NDUFB9 |
| rs10512344 | 9q31.2 | C | G | 0.048 | 0.040 | 0.98 | 0.190 | 0.034 | 3.57×10−8 | OR13C8, TAL2, TMEM38B, ZNF462, RAD23B, ELP1, FAM206A, TMEM245 | ||
| rs925870 | 11p14.1 | T | C | 0.860 | 0.848 | 0.99 | 0.100 | 0.021 | 2.41×10−6 | ANO3, MUC15, BBOX1, CCDC34, LGR4, LIN7C, BDNF, KIF18A, METTL15, KCNA4, FSHB, ARL14EP | ||
| rs61939612 | 12q12 | T | G | 0.390 | 0.373 | 0.97 | 0.072 | 0.015 | 2.74×10−6 | CPNE8 | CPNE8 | ALG10B, KIF21A, ABCD2, C12orf40, SLC2A13, MUC19, PDZRN4, CPNE8 |
| rs1540951 | 13q•14.11 | A | G | 0.318 | 0.334 | 0.98 | −0.075 | 0.016 | 3.14×10−6 | EPSTI1 | DNAJC15, EPSTI1 | AKAP11, FAM216B, ENOX1, CCDC122, LACC1, EPSTI1, DNAJC15 |
| rs147085754 | 13q33.3 | A | C | 0.030 | 0.025 | 0.99 | 0.198 | 0.043 | 4.57×10−6 | LIG4, FAM155A | DAOA, ARGLU1, ABHD13, LIG4, FAM155A | |
| rs146313367 | 17p13.2 | A | C | 0.084 | 0.075 | 0.97 | 0.125 | 0.027 | 3.04×10−6 | WSCD1 | NLRP1, AIPL1, FAM64A, PITPNM3, KIAA0753, TXNDC17, MED31, C17orf100, XAF1, FBXO39, TEKT1 |
CHR: chromosome and cytogenetic band; BP: genomic coordinates according to Hg19 build; EA: effect allele; OA: other allele; EAF CASES: effect allele frequency in cases; EAF CTRLS: effect allele frequency in controls; INFO: imputation quality score; beta: regression coefficient; SE: standard error.
genes physically mapped within the region;
genes mapped through eQTL effects;
genes mapped through chromatin interactions.
Functional annotation of GWAS results
To gain biological insight from the observed associations, we used Functional Mapping and Annotation of GWAS (FUMA GWAS)24 for computational annotation and interpretation of results (Materials and Methods). Based on genomic coordinates, regional linkage disequilibrium (LD), presence of expression quantitative trait loci (eQTL), and chromatin interactions, FUMA GWAS mapped a total of 93 genes to the 14 risk loci identified (Figure 2A and Table 2). These genes showed overall heterogeneity of mRNA expression across different human tissues from GTEx database (http://www.gtexportal.org), with a few transcripts showing higher expression in the brain (e.g. BARHL2, GRIK2) and intestine (e.g. CLCA1, CLCA4) (data not shown). A gene set enrichment analysis (see Materials and Methods) revealed, among others, that the IBS risk gene pool is significantly enriched for Intracellular calcium activated chloride channel activity, Ion gated channel activity and Anion channel activity among the Gene Ontology terms of the Molecular function domain, as well as for targets of microRNA species from the miR-15 family (Figure 2B).
Figure 2. FUMA analysis of GWAS results.
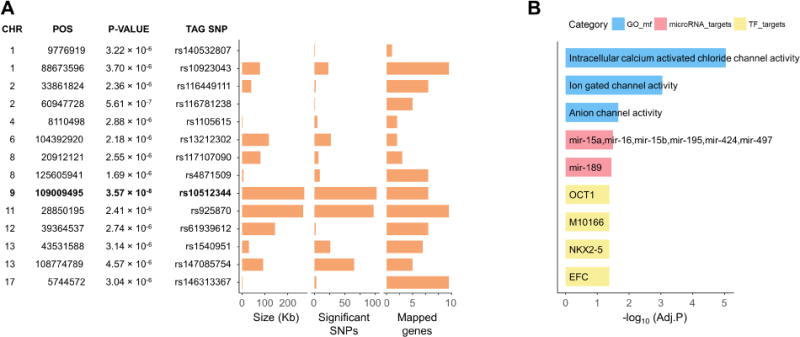
A) GWAS risk loci (P<5.0×10−6) with tag SNPs and FUMA annotations. The genome-wide significant locus 9q31.2 is highlighted in bold. B) GSEA significant findings (ranked based on FDR adjusted P-value) and color-coded categories Gene Ontology molecular function (GO_mf), miRNA targets (microRNA_targets), and transcription factor targets from the Molecular Signature Database (TF_targets).
9q31.2 locus-specific analyses
The genome-wide significant 9q31.2 locus (see Materials and Methods for additional QC of genotype data from this region) has also been consistently associated with age at menarche (AAM) in several GWAS studies,25,27 and it is known that IBS prevalence in women is higher than in men.1 To test for potential sex-specific genetic risk effects, we therefore repeated our association analysis upon stratification of UK Biobank participants in male (2,446 cases and 158,423 controls) and female (7,130 cases and 178,076 controls) groups. As shown in Figure 1 (bottom panel), this revealed the association signal to be absent in males (rs10512344 P=0.79), while it is entirely accounted for by the female group, where both the strength of association and the magnitude of risk effects increased despite the reduction in sample size (rs10512344 P=4.29×10−10; beta=0.245; Table 3). The 9q31.2 SNP with strongest genetic risk effects in AAM (rs10156597; P=3.70×10−107 in the most recent meta-analysis of AAM GWAS25) is not in linkage disequilibrium (r2=0.04) with the top IBS-risk marker rs10512344, and the analysis of their haplotypic combinations in UK Biobank data suggests the association with IBS is not dependent on the AAM-effect allele (Figure 3A). An AAM-conditioned analysis of the entire 9q31.2 region, using the latest AAM GWAS meta-analysis summary statistics with mtCOJO (Materials and Methods), did not disclose a significant reduction of the IBS signal of association based upon AAM-conditioning (Figure 3B). Finally, approximate Bayes factor colocalization analysis (Materials and Methods) provided further evidence for AAM-independence of the IBS association signal (9q31.2 locus associated to both traits but with different causal variants, posterior probability for H3=99.98%).
Table 3.
Sex-stratified association results for the SNP rs10512344.
| females | males | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Dataset Tested trait |
EAF | Beta | SE | P | Beta | SE | P |
| UK Biobank | |||||||
| IBS | 0.0404 | 0.245 | 0.039 | 4.29×10−10 | 0.019 | 0.073 | 0.789 |
|
| |||||||
| Case-control | |||||||
| IBS | 0.0362 | 0.228 | 0.127 | 0.036 | 0.192 | 0.183 | 0.147 |
| IBS-C | 0.0357 | 0.383 | 0.177 | 0.015 | 0.256 | 0.428 | 0.274 |
| IBS-D | 0.0348 | 0.008 | 0.190 | 0.483 | 0.166 | 0.224 | 0.229 |
| IBS-M | 0.0342 | 0.232 | 0.204 | 0.255 | −0.120 | 0.388 | 0.757 |
|
| |||||||
| PopCol | |||||||
| Avg BSFS score | 0.0241 | −1.105 | 0.335 | 0.0012 | 0.291 | 0.387 | 0.455 |
EAF=Effect allele frequency; SE=standard error; Avg BSFS=Average daily Bristol Stool Form Scale score.
Figure 3. Haplotype association and conditional analysis.
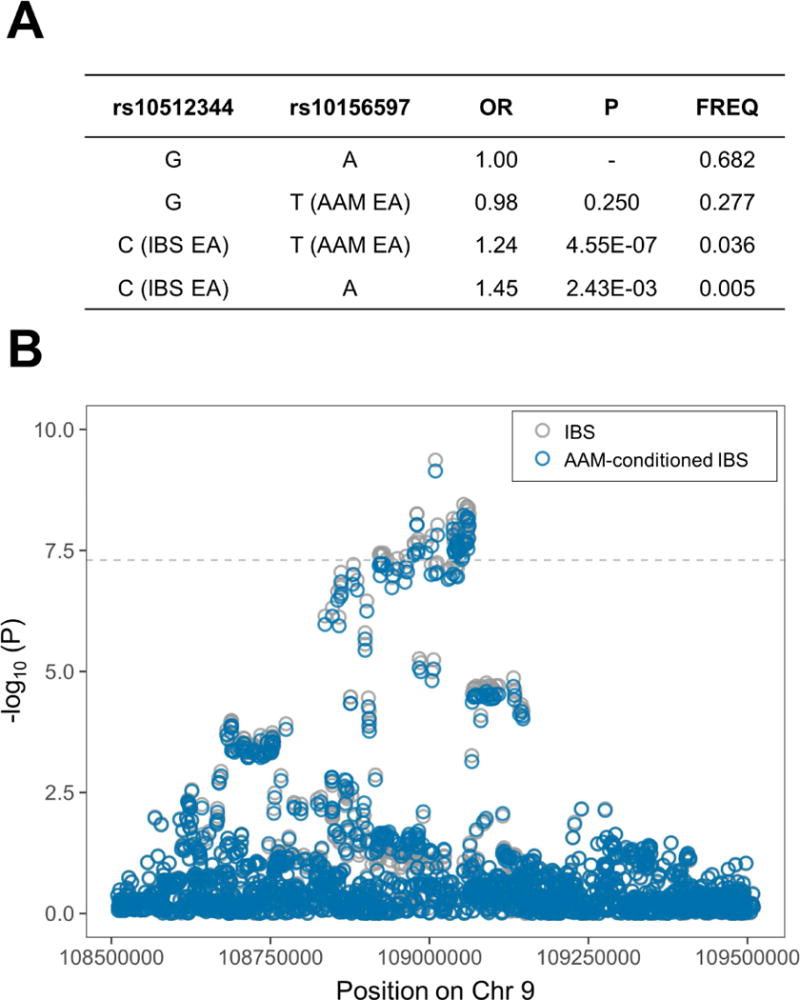
A) Summary statistics from association tests between IBS/AAM tag SNPs haplotypes and IBS in UK Biobank (females only); OR=odds ratio, P=P-value, FREQ=haplotype frequency. B) regional plot of the 9q31.2 locus before and after conditional analysis on AAM. The horizontal dashed line shows the genome-wide significance P-value threshold (5.0×10−8). Grey and blue circles correspond to un-conditioned and AAM-conditioned P-values, respectively.
9q31.2 follow-up analyses
We further characterized the 9q31.2 association through additional analyses in independent datasets, including Rome Criteria-defined IBS patients recruited at expert neurogastroenterology tertiary centres: we extracted genotype data for 2,045 cases and 7,955 population-based controls of European descent (Materials and Methods) currently available from an ongoing large-scale IBS genetics project (www.mdalab.org/bellygenes), and tested the rs10512344 marker again in sex-stratified analyses. As reported in Table 3, we observed genetic risk effects in the same direction and of the same magnitude as in the GWAS and, despite the relatively small sample size (>4000 IBS cases would be needed to replicate 9q31.2 GWAS findings with adequate statistical power >80%), we detected a significant female-specific association in the constipation-predominant IBS group (P=0.015; beta=0.383). Finally, analysis of genotype and bowel diary data available for 249 individuals from the PopCol population-based cohort (Materials and Methods) revealed that the IBS/constipation risk allele rs10512344 C also strongly correlates with harder stools in females (P=0.0012, beta=−1.105; Table 3).
DISCUSSION
We report here the first powered GWAS of IBS, implemented by taking advantage of available health and genotype data from the large UK Biobank resource, which includes approximately half a million participants. Through the analysis of 7,287,191 SNP markers in 9,576 cases and 336,499 controls, we identify 1 genome-wide significant risk locus on chromosome 9q31.2, as well 13 additional loci of suggestive significance that represent ideal candidates for future targeted studies in other large independent cohorts. The large sample size, the nested case-control design and the rigorous per individual/per genotype data QC pipeline are among the major strengths of this study, while a limitation results from the selection of cases and controls based on indirect evidence of a clinical diagnosis (patients report that a doctor has diagnosed them with IBS via touchscreen questionnaire and during a follow-up interview with a trained nurse). However, because of the remarkable opportunity deriving from increased sample size (questionnaire data are available for several large population-based cohorts), the use of similar self-reported traits and conditions has been increasingly and successfully applied in many recent GWAS, including GWAS of common diseases in UK Biobank (www.ukbiobank.ac.uk/genetic-publications). In addition, evidence of replication is provided for the 9q31.2 locus in a multi-national cohort of 2,045 Rome Criteria-based IBS cases from tertiary centres with expertise in IBS; this corroborates initial GWAS findings and highlights the need to assemble large cohorts suitable for clinical validation of genetic findings.
GWAS-downstream analyses and functional annotation of risk loci with FUMA show that the IBS risk pool is significantly enriched for targets of microRNAs from the miR-15 family, as well as for genes involved in cellular activities mediated by ion channels. Members of the miR-15 family have been reported to affect the expression of serotonin receptors involved in the regulation of motor function (miR-16) and epithelial barrier integrity (miR-16 and miR-125b), and their dysregulated expression has been recently associated with clinical symptoms in IBS patients.28 The link to ion channel biology adds to emerging evidence for a role of channelopathies in IBS pathophysiology.29 Previous studies have identified IBS-risk effects due to rare and common variants in the ion channel genes SCN5A (the voltage-gated sodium channel NaV1.5 expressed on the myenteric plexus interstitial cells of Cajal)6,30 and TRPM8 (the “cold” receptor target of menthol and other intestinal smooth muscle relaxants),9 and have also highlighted ion channel activity among the GO biological pathways most relevant to the control of stool frequency.31 In particular, the specific GO term with strongest evidence of enrichment in the current study (intracellular calcium activated chloride channel activity, FDR-adjusted P=9.10×10−6) includes four calcium-dependent chloride channels (CaCC) at two risk loci on chromosome 1p22.2 (CLCA1, CLCA2 and CLCA4) and 11p14.1 (ANO3). Among other functions, these channels play important roles in intestinal fluid secretion and secretory diarrhoea (CLC1/2/4), as well as in the control of gut peristalsis mediated by the interstitial cells of Cajal (ANO3). These are interesting observations that may provide rationale for future translational opportunities, since ion channels are attractive drug targets accessible on the cell surface and represent potential gateways to therapeutically actionable pathways. Nevertheless, the computational annotation of IBS GWAS results needs to be interpreted with caution because i) it mostly includes regions of suggestive significance (13/14 loci) that may reveal false positives in replication analyses and ii) it may be driven by fewer enriched loci with many genes from the same GO term category (while possibly only 1 gene/locus ultimately mediates specific risk effects). At least the latter does not appear to vitiate our FUMA results for ion channel enrichment (still highly significant under a conservative assumption of only 1 causative gene per locus, not shown). The identification of a genome-wide significant risk locus for IBS is the main finding from our study. The association at 9q31.2 is tagged by the SNP marker rs10512344, whose risk allele C is not common in UK Biobank and other populations of European ancestry (4% in UK Biobank, and 2–5.1% in other EU reference cohorts, www.ensembl.org), hence this variant will only affect IBS risk in a small proportion of individuals. However, while genetic risk scores (including rs10512344 C) may be later exploited for stratifying patients into different treatment groups when more GWAS data are accumulated, the essential value of these early findings is in their interpretation for pathway discovery. The observation of female-specific IBS risk effects, together with its known association with AAM, make the 9q31.2 locus particularly interesting. Although not previously detected in independent cohort studies of similarly large sample size,32 Day et al. recently reported an association between IBS and early AAM in UK Biobank,33 which raises the question as to whether our genetic findings may reflect this overlap (that is, AAM genetic risk effects also drive, at least in part, the association with IBS). We have shown this is not the case by applying three alternative methodological approaches, demonstrating that disease association signals do not colocalise at the 9q31.2 locus, where IBS risk variants differ from AAM risk variants. It is noteworthy that the association between early AAM and IBS is of borderline significance in UK Biobank, a similar trend was also observed for late AAM, and significant early/late AAM associations were detected for other 48 adverse outcomes across a range of cancer, cardio-metabolic, gynaecological/obstetric, gastrointestinal, musculoskeletal, and neuro-cognitive categories.33 Hence, while the association with young age at menarche and IBS may not be unequivocal, it does not appear to be specific.
Nevertheless, irrespective of specific associations with individual risk variants, the 9q31.2 region has been linked to a series of human conditions and traits (voice breaking in males, body mass index, waist circumference, breast and prostate cancer; GWAS catalog www.ebi.ac.uk/gwas/) that, like AAM, are under the influence of hormonal stimuli, especially those involving mechanisms of action of sex hormones. This is particularly interesting in the context of IBS, because of the known sex differences in its prevalence, symptomatic manifestations, response to treatment and, possibly, pathophysiology. While IBS symptoms are twice more common in women at the level of the general population, the female:male ratio increases to 5:1 in patients from specialized tertiary centers,34 where better treatment outcomes have also been observed, for instance, in the colonic transit response of women treated with alosetron (5-HT3 receptor antagonist) compared to men.35 In addition, colonic transit appears to be generally slower in women compared to men,36 and female IBS patients most often report symptoms of constipation and feeling of incomplete evacuation (opposite to men who more frequently suffer from diarrhoea).37 Our targeted 9q31.2 follow-up analyses are in line with these observations, as the rs10512344 risk genotype primarily associated with constipation-predominant IBS in female cases from tertiary centers, and correlated with harder stools in women but not men from a population-based cohort.
Increasing female preponderance of IBS is detected from the time of puberty to adult age, hence a pathogenetic role of sex hormones has been postulated and actively investigated in IBS. In addition, several chronic pain disorders often overlapping with IBS (chronic fatigue syndrome, fibromyalgia, migraine, chronic pelvic pain and others) show increased prevalence in women and correlation of symptoms with hormonal status.38,39 Sex hormones contribute important peripheral and central regulatory signals along the gut-brain axis, directly and indirectly (via other hormones) affecting gut motor and sensory functions, permeability, and mucosal immune activation.39,40 In particular, estrogens and their receptors (which contribute to induce menarche) show wide and abundant expression in the central and enteric nervous systems, and interact with other hormones such as serotonin and corticotropin-releasing factor, which are known to mediate visceral responses to stress among other functions.41 The effect of sex hormones on GI function is probably best typified by the fluctuations of bowel behaviour with the different phases of the menstrual cycle,39,42 and the appearance of symptoms perimenstrually in otherwise healthy women.43 Ovarian hormone variations have been linked not only to changes in sensorimotor gastrointestinal function, but also to hyperresponsiveness to stress with potential repercussions on gut immune activation and barrier function, and the emotional system responsible for visceral pain perception.43,44 Of note, female IBS patients have been shown to be affected by menstruation to a greater degree than healthy women,39,45 with exacerbation of bowel symptoms possibly due to their reported reduced rectal sensitivity at the time of menses.46 While the 9q31.2 association signal overlaps with an intergenic region, FUMA GWAS analysis mapped eight genes to this risk locus based on functional annotation of chromatin interactions (Table 2). Most of these genes are either poorly characterized (ORF13C8, ZNF462, FAM206A, TMEM245), or they have a potential role in cancer (TAL2, RAD23B) and osteogenesis imperfecta (TMEM38B). Hence, the gene ELP1 (elongator complex protein 1 gene, also known as IKBKAP) may represent the current best candidate to impact IBS risk at 9q31.2. ELP1 codes for a protein that is part of the RNA polymerase II elongator complex involved in the control of RNA transcription via chromatin remodelling and histone acetylation.47 It is expressed primarily in the adrenal and pituitary glands, the ovary, and the cerebellum (www.gtexportal.org/home/gene/IKBKAP), which may suggest hormonal regulation (including distal effects on chromatin rearrangements as described for estrogen receptors and other sex hormones).48 Most notably, ELP1 (IKBKAP) mutations cause familial dysautonomia (FD), a recessively inherited disease due to impaired development of sensory, sympathetic and parasympathetic neurons.49 FD patients thus typically suffer from sensory neuropathy characterized by pain insensitivity, GI dysmotility, altered temperature sensation, and cardiovascular instability. Of interest in view of the findings reported here, female FD patients often also show delayed age at menarche and a very high prevalence of premenstrual symptoms.50 Autonomic response to visceral stressors is dysregulated in IBS patients,51 and gender differences in autonomic nervous system reactivity to colorectal distention have been reported.52 At the same time, the increased prevalence of comorbidities also characterized by autonomic dysfunction (such as the join hypermobility syndrome), is increasingly recognized in IBS patients. Hence, should ELP1 be the causative gene at the 9q31.2 locus, it is possible to speculate that genotype-driven alterations of its function or expression may contribute also to IBS pathogenesis, possibly involving biological mechanisms under the control of sex hormones. Overall, the findings reported here open up for new lines of research in IBS and constipation, and warrant further investigation at the clinical and experimental levels.
Acknowledgments
This research has been conducted using the UK Biobank Resource under Application Number 17435. The UK Household Longitudinal Study (Understanding Society) is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website www.understandingsociety.ac.uk/.
Grant Support: This study was supported by grants from the Swedish Research Council (VR project nrs 2013-03862 and 2017-02403), the Health Department of the Basque Government (grant 2015111133), and the Spanish Ministry of Economy and Competitiveness (ISCIII grant FIS PI17/00308) to MDA, and NIH grants 1P50 DK64539, R01 DK048351, P30 DK 41301 to EAM and LC. The research leading to these results has received funding from the EU FP7 under grant nr 313010 (BBMRI-LPC). MGN was supported by an ERC Consolidator Grant (#310372) and a Spinoza Grant of the Netherlands Organization for Scientific Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests: The authors declare no conflict of interest
Author Contributions: MDA and AZ study concept and design; LB, FBr, LA, AA, AD, GL, PTS, PK, BO, MS, SW, GN, RC, PUS, FG, MN, PP, MB, GBa, AL, VT, MGN, DJ, LC, EAM, MMW, GBo, MC patients characterization, data acquisition; FBo, TZ, KGE, FH statistical analyses; FBo, TZ, MDA, MH, AF, data analysis and interpretation; MDA obtained funding, administrative and technical support, and study supervision; FBo and MDA drafted the manuscript, with input and critical revision from all other authors. All authors approved the final draft of the manuscript.
References
- 1.Sperber AD, Dumitrascu D, Fukudo S, et al. The global prevalence of IBS in adults remains elusive due to the heterogeneity of studies: A Rome Foundation working team literature review. Gut. 2017;66:1075–1082. doi: 10.1136/gutjnl-2015-311240. [DOI] [PubMed] [Google Scholar]
- 2.Drossman DA, Hasler WL. Rome IV - Functional GI disorders: Disorders of gut-brain interaction. Gastroenterology. 2016;150:1257–1261. doi: 10.1053/j.gastro.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 3.Enck P, Aziz Q, Barbara G, et al. Irritable bowel syndrome. Nat Rev Dis Prim. 2016;2:1–24. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito YA. The Role of Genetics in IBS. Gastroenterol Clin North Am. 2011;40:45–67. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waehrens R, Ohlsson H, Sundquist J, et al. Risk of irritable bowel syndrome in first-degree, second-degree and third-degree relatives of affected individuals: a nationwide family study in Sweden. Gut. 2015;64:215–221. doi: 10.1136/gutjnl-2013-305705. [DOI] [PubMed] [Google Scholar]
- 6.Beyder A, Mazzone A, Strege PR, et al. Loss-of-function of the voltage-gated sodium channel NaV1.5 (Channelopathies) in patients with irritable bowel syndrome. Gastroenterology. 2014;146:1659–1668. doi: 10.1053/j.gastro.2014.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henström M, Diekmann L, Bonfiglio F, et al. Functional variants in the sucrase-isomaltase gene associate with increased risk of irritable bowel syndrome. Gut. 2016 doi: 10.1136/gutjnl-2016-312456. gutjnl-2016-312456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zucchelli M, Camilleri M, Andreasson AN, et al. Association of TNFSF15 polymorphism with irritable bowel syndrome. Gut. 2011;60:1671–1677. doi: 10.1136/gut.2011.241877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henström M, Hadizadeh F, Beyder A, et al. TRPM8 polymorphisms associated with increased risk of IBS-C and IBS-M. Gut. 2017;66:1725–1727. doi: 10.1136/gutjnl-2016-313346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wouters MM, Lambrechts D, Knapp M, et al. Genetic variants in CDC42 and NXPH1 as susceptibility factors for constipation and diarrhoea predominant irritable bowel syndrome. Gut. 2014;63:1103–11. doi: 10.1136/gutjnl-2013-304570. [DOI] [PubMed] [Google Scholar]
- 11.Henström M, Zucchelli M, Söderhäll C, et al. NPSR1 polymorphisms influence recurrent abdominal pain in children: A population-based study. Neurogastroenterol Motil. 2014;26:1417–1425. doi: 10.1111/nmo.12401. [DOI] [PubMed] [Google Scholar]
- 12.Ek WE, Reznichenko A, Ripke S, et al. Exploring the genetics of irritable bowel syndrome: a GWA study in the general population and replication in multinational case-control cohorts. Gut. 2015;64:1774–1782. doi: 10.1136/gutjnl-2014-307997. [DOI] [PubMed] [Google Scholar]
- 13.Camilleri M, Carlson P, Zinsmeister AR, et al. Neuropeptide S Receptor Induces Neuropeptide Expression and Associates With Intermediate Phenotypes of Functional Gastrointestinal Disorders. Gastroenterology. 2010;138:98–107.e4. doi: 10.1053/j.gastro.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Amato M. Genes and functional GI disorders: from casual to causal relationship. Neurogastroenterol Motil. 2013;25:638–649. doi: 10.1111/nmo.12173. [DOI] [PubMed] [Google Scholar]
- 15.Bycroft C, Freeman C, Petkova D, et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv. 2017:166298. [Google Scholar]
- 16.Lichtenstein P, Sullivan PF, Cnattingius S, et al. The Swedish Twin Registry in the third millennium: an update. Twin Res Hum Genet. 2006;9:875–82. doi: 10.1375/183242706779462444. [DOI] [PubMed] [Google Scholar]
- 17.Franke A, McGovern DPB, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meschia JF, Arnett DK, Ay H, et al. Stroke Genetics Network (SiGN) study: design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke. 2013;44:2694–702. doi: 10.1161/STROKEAHA.113.001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benzeval M, Davillas A, Kumari M, et al. Understanding Society: The UK Household Longitudinal Study Biomarker User Guide and GLossary. Inst Soc Econ Res Univ Essex. 2014 [Google Scholar]
- 20.Sonnega A, Faul JD, Ofstedal MB, et al. Cohort Profile: the Health and Retirement Study (HRS) Int J Epidemiol. 2014;43:576–585. doi: 10.1093/ije/dyu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mujagic Z, Tigchelaar EF, Zhernakova A, et al. A novel biomarker panel for irritable bowel syndrome and the application in the general population. Sci Rep. 2016;6:26420. doi: 10.1038/srep26420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Oosting M, Smeekens SP, et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016;167:1099–1110.e14. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Walter SA, Kjellström L, Nyhlin H, et al. Assessment of normal bowel habits in the general adult population: the Popcol study. Scand J Gastroenterol. 2010;45:556–566. doi: 10.3109/00365520903551332. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, Van Bochoven A, et al. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day FR, Thompson DJ, Helgason H, et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–841. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elks CE, Perry JRB, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez C, Rodiño-Janeiro BK, Lobo B, et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut. 2017;66:1537–1538. doi: 10.1136/gutjnl-2016-311477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyder A, Farrugia G. Ion channelopathies in functional GI disorders. Am J Physiol Gastrointest Liver Physiol. 2016;311:G581–G586. doi: 10.1152/ajpgi.00237.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strege PR, Mazzone A, Bernard CE, et al. Irritable bowel syndrome (IBS) patients have SCN5A channelopathies that lead to decreased Na V 1.5 current and mechanosensitivity. Am J Physiol Liver Physiol. 2017 doi: 10.1152/ajpgi.00016.2017. ajpgi.00016.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jankipersadsing SA, Hadizadeh F, Bonder MJ, et al. A GWAS meta-analysis suggests roles for xenobiotic metabolism and ion channel activity in the biology of stool frequency. Gut. 2017;66:756–758. doi: 10.1136/gutjnl-2016-312398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palacios N, Chan AT, Ascherio A, et al. Hormonal Factors and Risk of Irritable Bowel Syndrome in a Large Prospective Study of Women. Gastroenterology. 2012;142:S-833. [Google Scholar]
- 33.Day FR, Elks CE, Murray A, et al. Puberty timing associated with diabetes, cardiovascular disease and also diverse health outcomes in men and women: The UK Biobank study. Sci Rep. 2015;5:1–12. doi: 10.1038/srep11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heitkemper M, Jarrett M, Bond EF, et al. Impact of sex and gender on irritable bowel syndrome. Biol Res Nurs. 2003;5:56–65. doi: 10.1177/1099800403005001006. [DOI] [PubMed] [Google Scholar]
- 35.Viramontes BE, Camilleri M, McKinzie S, et al. Gender-related differences in slowing colonic transit by a 5-HT3 antagonist in subjects with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol. 2001;96:2671–2676. doi: 10.1111/j.1572-0241.2001.04138.x. [DOI] [PubMed] [Google Scholar]
- 36.Meier R, Beglinger C, Dederding JP, et al. Influence of age, gender, hormonal status and smoking habits on colonic transit time. Neurogastroenterol Motil. 1995;7:235–8. doi: 10.1111/j.1365-2982.1995.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 37.Lovell RM, Ford AC. Effect of Gender on Prevalence of Irritable Bowel Syndrome in the Community: Systematic Review and Meta-Analysis. Am J Gastroenterol. 2012;107:991–1000. doi: 10.1038/ajg.2012.131. [DOI] [PubMed] [Google Scholar]
- 38.Houghton LA, Heitkemper M, Crowell MD, et al. Age, gender, and women’s health and the patient. Gastroenterology. 2016;150:1332–1343e4. doi: 10.1053/j.gastro.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Heitkemper MM, Chang L. Do fluctuations in ovarian hormones affect gastrointestinal symptoms in women with irritable bowel syndrome? Gend Med. 2009;6:152–167. doi: 10.1016/j.genm.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hogan AM, Collins D, Baird AW, et al. Estrogen and its role in gastrointestinal health and disease. Int J Colorectal Dis. 2009;24:1367–1375. doi: 10.1007/s00384-009-0785-0. [DOI] [PubMed] [Google Scholar]
- 41.Taché Y, Bonaz B. Corticotropin-releasing factor receptors and stress-related alterations of gut motor function. J Clin Invest. 2007;117:33–40. doi: 10.1172/JCI30085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longstreth GF, Thompson WG, Chey WD, et al. Functional Bowel Disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein MT, Graff LA, Avery L, et al. Gastrointestinal symptoms before and during menses in healthy women. BMC Womens Health. 2014;14:14. doi: 10.1186/1472-6874-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meleine M, Matricon J. Gender-related differences in irritable bowel syndrome: Potential mechanisms of sex hormones. World J Gastroenterol. 2014;20:6725–6743. doi: 10.3748/wjg.v20.i22.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitehead WE, Cheskin LJ, Heller BR, et al. Evidence for exacerbation of irritable bowel syndrome during menses. Gastroenterology. 1990;98:1485–1489. doi: 10.1016/0016-5085(90)91079-l. [DOI] [PubMed] [Google Scholar]
- 46.Houghton LA, Lea R, Jackson N, et al. The menstrual cycle affects rectal sensitivity in patients with irritable bowel syndrome but not healthy volunteers. Gut. 2002;50:471–474. doi: 10.1136/gut.50.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kojic M, Wainwright B. The Many Faces of Elongator in Neurodevelopment and Disease. Front Mol Neurosci. 2016;9:115. doi: 10.3389/fnmol.2016.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Dily F, Beato M. Signaling by steroid hormones in the 3D nuclear space. Int J Mol Sci. 2018;19:306. doi: 10.3390/ijms19020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norcliffe-Kaufmann L, Slaugenhaupt SA, Kaufmann H. Familial dysautonomia: History, genotype, phenotype and translational research. Prog Neurobiol. 2017;152:131–148. doi: 10.1016/j.pneurobio.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Maayan C, Sela O, Axelrod F, et al. Gynecological aspects of female familial dysautonomia. Isr Med Assoc J. 2000;2:679–683. [PubMed] [Google Scholar]
- 51.Camilleri M, McKinzie S, Busciglio I, et al. Prospective Study of Motor, Sensory, Psychologic, and Autonomic Functions in Patients With Irritable Bowel Syndrome. Clin Gastroenterol Hepatol. 2008;6:772–81. doi: 10.1016/j.cgh.2008.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng P, Shih W, Alberto M, et al. Autonomic response to a visceral stressor is dysregulated in irritable bowel syndrome and correlates with duration of disease. Neurogastroenterol Motil. 2013;25:e650–659. doi: 10.1111/nmo.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]


