Abstract
Introduction
Current processing of renal biopsy samples provides limited information about immune mechanisms causing kidney injury and disease activity. We used flow cytometry with transplanted kidney biopsy samples to provide more information on the immune status of the kidney.
Methods
To enhance the information available from a biopsy, we developed a technique for reducing a fraction of a renal biopsy sample to single cells for multicolor flow cytometry and quantitation of secreted cytokines present within the biopsy sample. As proof of concept, we used our technique with transplant kidney biopsy samples to provide examples of clinically relevant immune information obtainable with cytometry.
Results
A ratio of CD8+ to CD4+ lymphocytes greater than or equal to 1.2 in transplanted allografts is associated with rejection, even before it is apparent by microscopy. Elevated numbers of CD45 leukocytes and higher levels of interleukin (IL)−6, IL-8, and IL-10 indicate more severe injury. Antibody binding to renal microvascular endothelial cells can be measured and corresponds to antibody-mediated forms of allograft rejection. Eculizumab binding to endothelial cells suggests complement activation, which may be independent of bound antibody. We compared intrarenal leukocyte subsets and activation states to those of peripheral blood from the same donor at the time of biopsy and found significant differences; thus the need for new techniques to evaluate immune responses within the kidney.
Conclusion
Assessment of leukocyte subsets, renal microvascular endothelial properties, and measurement of cytokines within a renal biopsy by flow cytometry enhance understanding of pathogenesis, indicate disease activity, and identify potential targets for therapy.
Keywords: cytokines, cytometry, immunology, kidney biopsy, rejection
Examination of renal biopsy samples with light, immunofluorescence, and electron microscopy is the mainstay of diagnosing renal disease. Pathology defined by these techniques is limited by the constraints of microscopy, the solid nature of the kidney, and timing of tissue procurement. Classic biopsy techniques may show an incomplete image of pathogenesis, especially in native kidney disease, in which biopsies are performed for an obvious clinical problem (proteinuria, hematuria, decline of glomerular filtration rate [GFR]), when the initiating event may no longer be present. Pathology is then a view of late response to injury, rather than the inciting event. Protocol transplant kidney biopsies used to identify early rejection improve the timely capture of injury, but information is still incomplete; no information is provided about circulating leukocytes migrating to the site of injury or the presence of soluble mediators that contribute to inflammation. Also, similar tissue architecture is associated with different conditions—for example, nonspecific interstitial inflammation in transplanted kidneys may represent rejection, viral infection, or fibrotic repair from prior injury. In addition, current renal pathology does not lend itself to quantitative assessment of inflammation and disease activity. Does the inflammation still present after treatment of rejection represent an improvement, or is it active rejection warranting additional therapy? Are fibrotic crescents always a sufficient sign that glomerular inflammation has resolved and that immunosuppressive therapy would not be useful?
Biopsy histopathology is often insufficient to direct therapy with specific agents, which becomes frustrating as new immune modulating agents are being developed. We have a limited understanding of the detailed immune processes in human kidneys; yet essentially all renal disease has an immune mechanism. Most of our understanding of the renal immune system is murine based; however, important differences exist between the immune systems of mice and humans.1, 2, 3, 4
Others have looked for ways to expand the information available from human renal biopsies. Halloran et al. and others have investigated whole-biopsy RNA in microarrays and identified natural killer (NK) cell and T-cell transcripts in transplanted kidneys with rejection and other inflammatory states.5, 6, 7 Flow cytometry, clinically important for hematopathology where easy access to single cells allows characterization and quantitation of cell phenotypes, has not been widely applied to solid organs such as the kidney. Cytometry has the advantage of detecting low levels of cell surface proteins that may be undetectable by immunofluorescent microscopy.8
We report a technique for reducing renal biopsy samples to single-cell suspensions for quantitative multicolor flow cytometry and measurement of secreted cytokines. Our data show how applying these methodologies can distinguish rejection from nonspecific inflammation earlier than apparent by traditional pathology. Data assessing endothelial cell antibody and complement binding suggest mechanism of injury, and intrarenal cytokine levels estimate inflammatory activity. All transplant biopsy samples studied, even those with normal histology, were performed for suspicion of pathology based on clinical parameters. Our purpose was not to compare our findings to Banff classifications but to develop a technique that provides more information on the immune status of the kidney. Although we have focused on biopsy samples from transplanted kidneys (because of their availability), the techniques are equally applicable to those from native kidneys.
Methods
Preparation of Renal Biopsy Samples and Peripheral Blood Leukocytes
Biopsy samples and peripheral blood were obtained after informed consent was provided and the study (STUDY00002529) was approved through our institution human subject review board. We adhered to the Declaration of Helsinki and the Declaration of Istanbul. At the time of biopsy, 3 to 5 mm of a 16- to 18-gauge tissue core was placed in 200 μl of Hanks Balanced Salt Solution. Transplant biopsies were performed for clinical indications as determined by the attending nephrologist. Biopsy samples designated as normal native were obtained from nontumor tissue of native kidneys immediately after nephrectomy for renal cell carcinoma or transitional cell carcinoma from donors who had normal renal function by laboratory testing and clinical history. Biopsy samples were stored at 4°C until processed. Cell viability was best when samples were processed the day of the biopsy, but tissue can be kept 24 to 36 hours with minimal effect on results. A 3- to 5-mm length of a 16-gauge biopsy sample yields ∼200,000 cells for cytometric analysis.
To the biopsy fragment was added 2 μl of 10% Collagenase P (Roche #11213857001, Roche Diagnostics Corporation, Indianapolis, IN) stock solution, and tissue was digested for 15 minutes on an orbital shaker at 37°C with mid- and end-digestion mechanical dissociation using Samco Scientific extended fine-tip transfer pipets (#231, ThermoScientific, Waltham, MA). Cells were centrifuged, and supernatant was collected and stored at −80°C for cytokine assay. A 0.2-ml quantity of 0.001% trypsin was added to the pelleted cells, and cells were incubated at 37°C for 7 minutes using mechanical dissociation with transfer pipet at the beginning and end of the incubation. Trypsin was neutralized with cell culture media containing calcium (Lonza’s endothelial medium or Roswell Park Memorial Institute [RPMI] medium). Cells were collected by centrifugation and stored overnight at 4°C in 150 μl of cell culture medium. Antibody labeling was done within 24 hours of tissue preparation.
Circulating leukocytes of tissue donors were obtained on the day of biopsy from a buffy coat of ethylenediamine tetraacetic acid (EDTA)−collected peripheral blood.
Antibody Labeling and Flow Cytometry
Antibodies were purchased from Biolegend (San Diego, CA) or were from hybridomas maintained in the laboratory. Eculizumab was obtained from the remains of a vial used for clinical treatment. Eculizumab and hybridomas were purified with Protein A or Protein A/G (Pierce, Waltham, MA) and conjugated to either fluorescein isothiocyanate (FITC) (F7250; Sigma, St. Louis, MO) or Alexa 680 (A-20172, Life Technologies, Waltham, MA). Each antibody was used as a direct conjugate to FITC, phycoerythrin (PE), allophycocyanin (APC), peridinin chlorphyll protein (PerCP), Alexa 680, or Pacific Blue. All work was done on ice. Cells were diluted in RPMI, aliquoted to a 96-well, round-bottom plate and centrifuged to pellet cells. Supernatant was removed and antibody cocktails diluted in RPMI were added in a final volume of 50 μl for kidney cells and 100 μl for leukocytes. Cells were incubated on ice for 60 minutes, washed in RPMI, and analyzed on a LSRII cytometer (BD Biosciences, San Jose, CA) adding 4',6-diamidino-2-phenylindole (DAPI) as a viability marker prior to analysis.
The LSR II is equipped with 355-, 405-, 488-, 561-, and 637-nm lasers. Data were analyzed with FCS Express (De Novo Software, Glendale, CA). Statistical analyses were done with Sigmaplot software (Systat Software, Inc, San Jose, CA).
Cytokines
Cytokines in the cell-free collagenase supernatant were measured in duplicate using Milliplex high-sensitivity assay (Millipore #HSCYMAG60SPMX13; EMD Millipore, Billerica, MA) with the Luminex platform per the manufacturer’s protocol. The high sensitivity kit measures interleukin (IL)−2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p, IL-13, tumor necrosis factor−α, IL-1β, interferon-γ, and granulocyte-macrophage colony-stimulating factor (GM-CSF). Concentration (pg/ml) was normalized to cubic millimeter of biopsy tissue.
Results
Tissue Preparation and Analysis
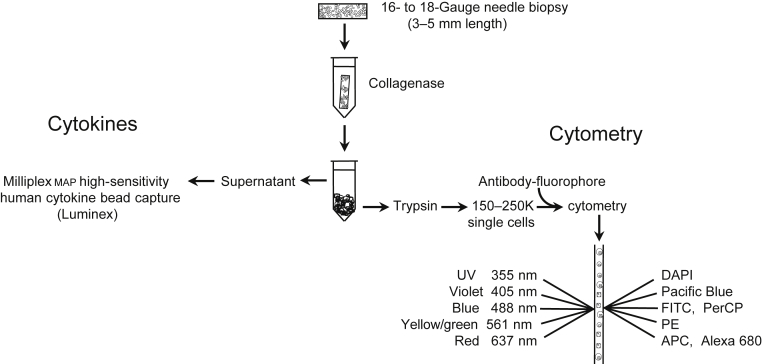
Figure 1 shows a flowchart of sample preparation technique. Complete digestion to single cells was confirmed by microscopic inspection and ensured that results were representative of the entire cell content of the biopsy sample. Digestion conditions were tested using peripheral blood and cultured cells that expressed epitopes of interest to verify that antigens were not lost during preparation.
Figure 1.
Flowchart outlining renal biopsy sample processing for multicolor flow cytometry and cytokine assay. Fluorophores used for cytometry were directly conjugated to antibodies that recognize cell surface antigens. APC, allophycocyanin; DAPI, 4’,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; PE, phycoerythrin; Per-CP, peridinin chlorophyll protein; UV, ultraviolet.
Cytometry samples include cells within the kidney parenchyma and those circulating through the biopsy core at the time of acquisition, such as leukocytes migrating to the tissue in response to inflammatory cytokines and erythrocytes. The supernatant resulting from collagenase digestion is used for measuring total biopsy sample cytokines. Biopsy cores used for analysis came from a mixture of cortex and medullary tissue, which did not affect results. Unlabeled cells were used to eliminate autofluorescence from analyses. Only viable cells, based on dye exclusion, were analyzed.
Intrarenal Leukocytes
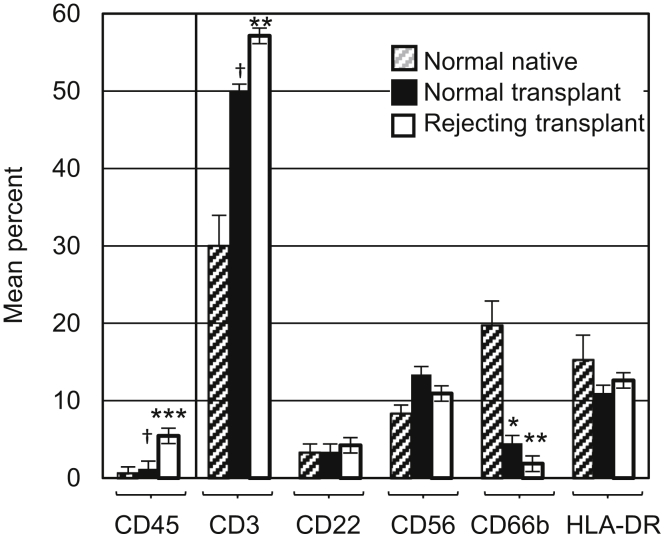
Intrarenal leukocytes were assessed by cytometry using antibodies to CD45 (leukocyte common antigen), CD66b (granulocytes), CD56 (natural killer cells), CD22 (B cells), and CD3 (T cells) (Figure 2). HLA-DR+ leukocytes are cells that were CD45+, CD66−, CD56−, CD22−, and CD3−, roughly equivalent to macrophage/dendritic cell populations. Normal native, normal transplant, and rejecting transplant biopsy samples were compared (Figure 2, Supplementary Table S1, Supplementary Figure S1). Tissue from normal native and normal transplant kidneys had distinct leukocyte differentials despite having similar histology. Normal native kidney had a significantly greater percentage of CD66+ granulocytes compared to transplanted kidney, whereas CD3+ T cells were increased in transplanted kidney with or without rejection. Rejection of any type or severity (specifically included in our analysis were Banff T-cell−mediated type IA, IB, IIA, IIB, and antibody-mediated9) was associated with a significant increase in CD45+ leukocytes.
Figure 2.
Leukocyte differentials of normal native, normal transplant, and rejecting transplanted kidneys, determined on biopsy samples with sufficient tissue to yield at least 100 CD45+ cells. Gating strategy: viable cells > CD45+ > markers for individual subsets (Supplementary Figure S1). CD45 data are the mean percentage of viable cells ± SE. CD3, CD22, CD56, CD66b, and human leukocyte antigen (HLA)−DR are mean percentage of viable CD45+ cells ± SE. HLA-DR includes cells that are HLA-DR+, CD45+, CD3−, CD22−, CD56−, and CD66b−. The Mann−Whitney rank sum test was used to test significant differences. *Significant difference between normal native and normal transplant (P = 0.007). **Significant difference between normal native and rejecting transplants (P < 0.001). ***Significant difference between normal transplant and rejecting transplant (P < 0.001). †Significant difference between normal native and normal transplant (P = 0.01). Normal native, n = 17; normal transplant, n = 7; rejecting transplant, n = 31.
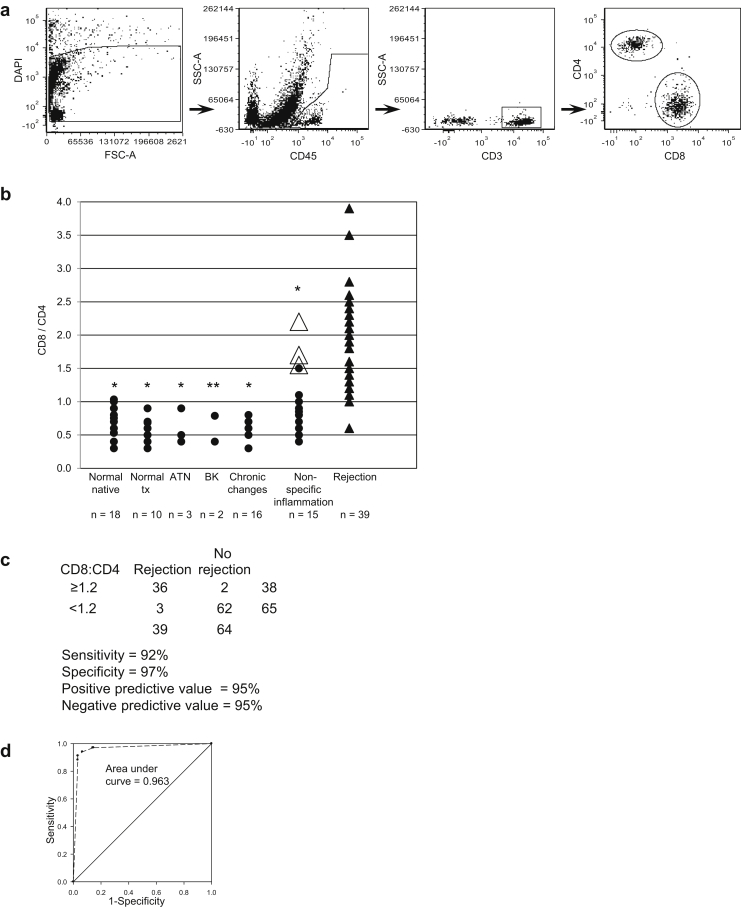
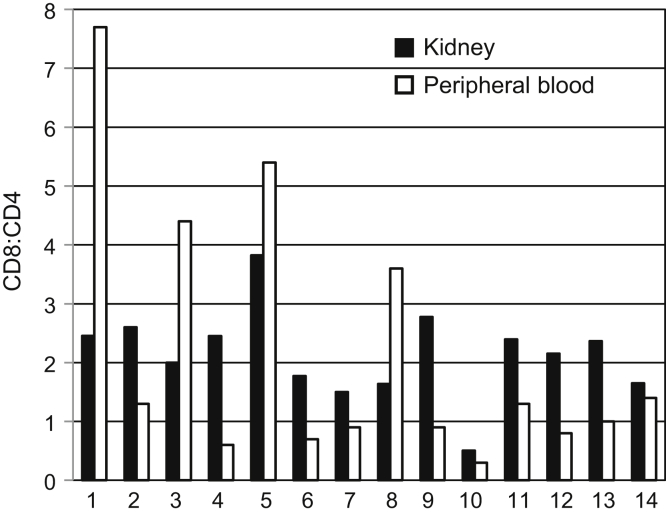
We next evaluated allograft CD4 and CD8 T-cell subsets (Figure 3). Gating strategy used is shown in Figure 3a. Rejection was associated with a higher ratio of CD8+ cells to CD4+ cells and distinguished rejection from other inflammatory conditions such as BK nephropathy, chronic scarring, and nonspecific inflammation (Figure 3b, Supplementary Table S2). Biopsy samples were classified as chronic changes when interstitial fibrosis, tubular atrophy, and glomerulosclerosis predominated, with leukocyte infiltrates localized to areas of fibrosis; as nonspecific inflammation when pathologic criteria for rejection were lacking (equivalent to Banff borderline,10 and chronic changes in which pathologists could not distinguish whether an acute process was also present, for example, leukocytes not entirely limited to areas of fibrosis but without significant tubulitis); or rejection according to pathologist interpretation and when the nephrologists judged pathologists’ interpretation of borderline to be sufficiently significant clinically to increase immunosuppression. Rejection included biopsy samples with both antibody-mediated and T-cell−mediated pathology of any degree of severity by Banff criteria. Ten of the 36 cases of rejection were antibody mediated; the others were cellular rejection of types IA, IB, IIA, and IIB. For biopsy samples in which at least 200 CD45+ cells were present in the sample, a ratio of CD8/CD4 cells greater than or equal to 1.2 was closely associated with rejection (Figure 3b, c, and d). In Figure 3b, open triangles indicate patients with high CD8/CD4 ratios who were judged not to have histologic rejection but who, on repeat biopsy weeks to 3 months later, in the setting of deteriorating renal function, were diagnosed with rejection. Thus, an elevated intrarenal CD8/CD4 ratio may indicate the presence of rejection earlier than apparent by current histologic criteria. Peripheral blood T-cell CD8/CD4 ratios from patients with rejection, obtained at the time of renal biopsy, did not correlate with biopsy sample CD8/CD4 ratios (Figure 4).
Figure 3.
CD8/CD4 T cell ratios in renal biopsy samples with indicated diagnoses. Ratios were determined on biopsy samples with greater than or equal to 200 CD45+ cells in the cytometry sample. Gating strategy: viable cells > CD45+ > CD3+ > CD8+, CD4+. (a) Gating strategy: viable cells > CD45+ > CD3+ > CD8+, CD4+. DAPI, 4',6-diamidino-2-phenylindole; FSC-A, forward-scatter area; SSC-A, side-scatter area. (b) Scatter plot of CD8/CD4 ratios with indicated histologic classification as described in the text. Biopsy samples were from transplanted allografts except for the column normal native. Number of cases (n) in each category is indicated. The Mann−Whitney rank sum test was used for determining significance of CD8/CD4 ratio in cases of nonrejection compared to rejection (*P < 0.001, ** P = 0.004). Open triangles indicate transplants with high CD8/CD4 ratios that transitioned to rejection by histopathology criteria in a subsequent biopsy. ATN, acute tubular necrosis; BK, BK virus nephropathy; tx, transplant. (c) Predictive value of renal biopsy cytometry CD8/CD4 in determining rejection or no rejection. (d) Receiver operating characteristic curve and area under the curve.
Figure 4.
Representative CD8/CD4 ratios of renal CD3+ T cells compared to peripheral blood CD3+ T cells from the same patient sampled at time of biopsy in 14 representative cases of histologic rejection.
The activation state of intrarenal and peripheral T cells was assessed by evaluating the expression of CD69, an early marker of T-cell activation.11, 12, 13 Compared to peripheral blood, intrarenal CD4 and CD8 T cells had significantly increased expression of CD69 in normal native and transplanted kidneys (Figure 5a and c), and in transplanted kidneys with a range of rejection types and severity (including Banff T-cell−mediated type IA, IB, IIA, IIB, and antibody-mediated) (Figure 5b and d). This suggests that T cells within the kidney, even under conditions without obvious inflammation, are in a state of activation that differs from T cells in the peripheral circulation.
Figure 5.
CD69 expression on intrarenal and peripheral T cells from the same patient sampled at time of biopsy. Data are percentage of total (a,b) CD4+ or (c,d) CD8+ T cells that are CD69+. Gating strategy: viable cells > CD45+ > CD3+ > CD4+ or CD8+ > CD69+. Panels a and c compare normal native and normal transplanted kidney T cells (solid bars) to peripheral blood T cells (open bars). Panels b and d compare rejecting transplanted kidney T cells (solid bars) to peripheral blood T cells (open bars). Numbers below bar graph represent assigned specimen identifiers.
We also compared naive and memory markers, CD45RA and CD45RO, respectively on intrarenal and peripheral T cells from the same donor. CD4 and CD8 cells in native and transplanted kidneys were predominantly memory cells, based on the expression of CD45RO, whereas peripheral blood obtained at the time of tissue acquisition had relatively equal numbers of memory and naive cells (Supplementary Figure S2).
Renal Microvascular Endothelial Cells
Cytometry allows a detailed evaluation of renal microvascular endothelial cells (RMECs) beyond traditional histology. Prior studies have shown that human, but not rodent, RMECs co-express high levels of major histocompatibility complex class II proteins and the endothelial cell markers CD31 and CD34.14 RMEC are defined by excluding CD45+ leukocytes and CD324+ epithelial cells from gates of HLA-DR+/CD31+ or CD34+ cells.
We measured antibody bound to RMECs using antihuman lambda and kappa Ig light chain antibodies conjugated with the same dye so that total antibody binding to RMECs is assessed with a single fluorescence signal. Assay controls are shown in Supplementary Figure S3 demonstrating specificity using fluorescence minus 1 (FMO) and isotype controls, reproducibility, and the lack of RMEC Fc receptors.
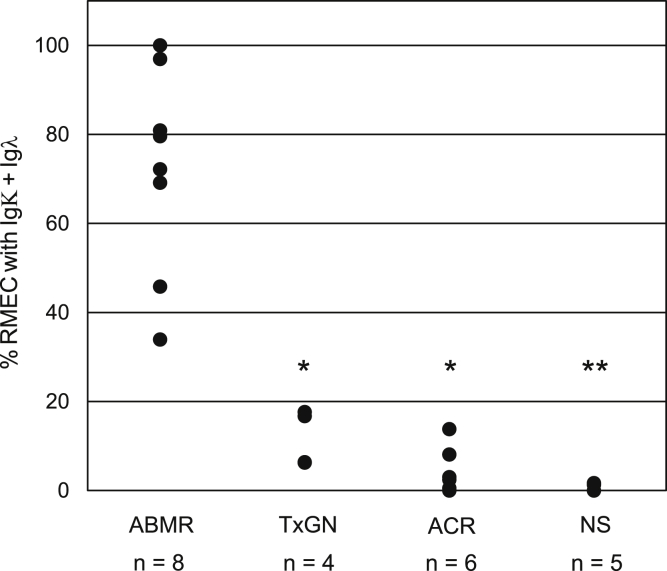
Measures of endothelial-bound antibodies in allograft biopsy samples with antibody-mediated rejection (ABMR) (active, acute, or chronic9), transplant glomerulopathy with advanced fibrosis but without inflammation (TxGN), acute cellular rejection (ACR), or nonspecific inflammation (equivalent to Banff borderline) (NS) are shown in Figure 6 and Supplementary Figure S4. A significantly higher percentage of light chain antibody bound to RMECs is associated with histologic antibody-mediated rejection, even when donor-specific antibody (DSA) or complement degradation product C4d are not detected (Supplementary Figure S4). Detailed clinical and cytometric data for antibody binding are available in Supplementary Figure S4 and Table S3. Clinical course of patients is also reflected in RMEC antibody binding (Supplementary Figure S4e).
Figure 6.
Percentage of renal microvascular endothelial cells (RMECs) with Igλ and Igκ antibody-bound plotted versus biopsy histopathology interpretation. ABMR, antibody-mediated rejection, active acute or chronic; ACR, acute cellular rejection; NS, nonspecific inflammation where histologic criteria for rejection were not met; TxGN, transplant glomerulopathy without inflammation. The Mann−Whitney rank sum test used to test significant difference between antibody-mediated rejection versus other diagnoses (*P < 0.001, **P = 0.002).
The presence of antibody bound to RMEC does not necessarily imply an injurious process, so we also assessed complement binding to RMECs using eculizumab, a clinically available monoclonal antibody that recognizes a site on C5 and prevents cleavage to C5a and C5b. Eculizumab was conjugated to fluorescein isothiocyanate (FITC). Human polyclonal IgG-FITC was used as an isotype control.
Low levels of eculizumab-FITC above control IgG-FITC were detected on RMEC from most biopsy samples analyzed (Figure 7), but the significance of this was unclear. Therefore we used clinical conditions to define relevant RMEC eculizumab−FITC binding. We assumed that RMEC-bound eculizumab−FITC from patients with normal native and transplanted kidneys without histologic signs of injury was unlikely to reflect pathology (Figure 7a). Using standardized cytometry settings, we set the level of clinically elevated eculizumab−FITC binding to RMECs at the upper limit of binding to normal kidneys, indicated by the vertical dotted lines in Figure 7. Assessment of transplant biopsy samples with nonspecific inflammation or chronic, noninflammatory changes (as previously defined) where complement is not thought to be causing injury did not show significant eculizumab−FITC binding (Figure 7b). Cases with acute mild cellular rejection (equivalent to Banff IA) had increased binding of eculizumab−FITC to RMECs (Figure 7c), without significant antibody binding (Supplementary Figure S4c). This raises the possibility that complement activation by the alternative pathway contributes to injury in mild acute-cellular rejection. Clinical course was also reflected in eculizumab−FITC binding, where we had a biopsy before and after successful treatment of acute mild cellular rejection with steroids; eculizumab−FITC binding to RMECs was reduced with resolution of rejection (Figure 7c; case 349a and 349b). Cases with more severe cellular rejection (equivalent to Banff scores greater than IA) did not show significant eculizumab−FITC binding (Figure 7d). Cases with active antibody-mediated rejection and transplant glomerulopathy (Figure 7e), where one might expect complement activation, had higher levels of eculizumab binding to RMECs. In Figure 7e, cases 343b and 386, which by histology had active inflammation, show more eculizumab bound to RMECs than cases 376 and 377, which received therapy for their transplant glomerulopathy before the cytometry-analyzed biopsy.
Figure 7.
Eculizumab binding to renal microvascular endothelial cells (RMECs). Solid black histograms are eculizumab bound to RMECs; gray line histograms are polyclonal IgG bound to RMECs (negative control). To the right of the dotted vertical line represents potentially clinically significant levels of eculizumab binding; the percentage of RMECs with this level of binding is indicated. (a) Representative biopsy samples from normal native and normal transplanted kidneys. (b−e) Representative transplant biopsy samples with indicated diagnoses as described in the text. The number in the upper right corner of the dot plots is the identifier code assigned to the biopsy sample. DSA, donor-specific antibody; MICA, major histocompatibility complex class–related chain A; ND, not determined.
Intrarenal Cytokines
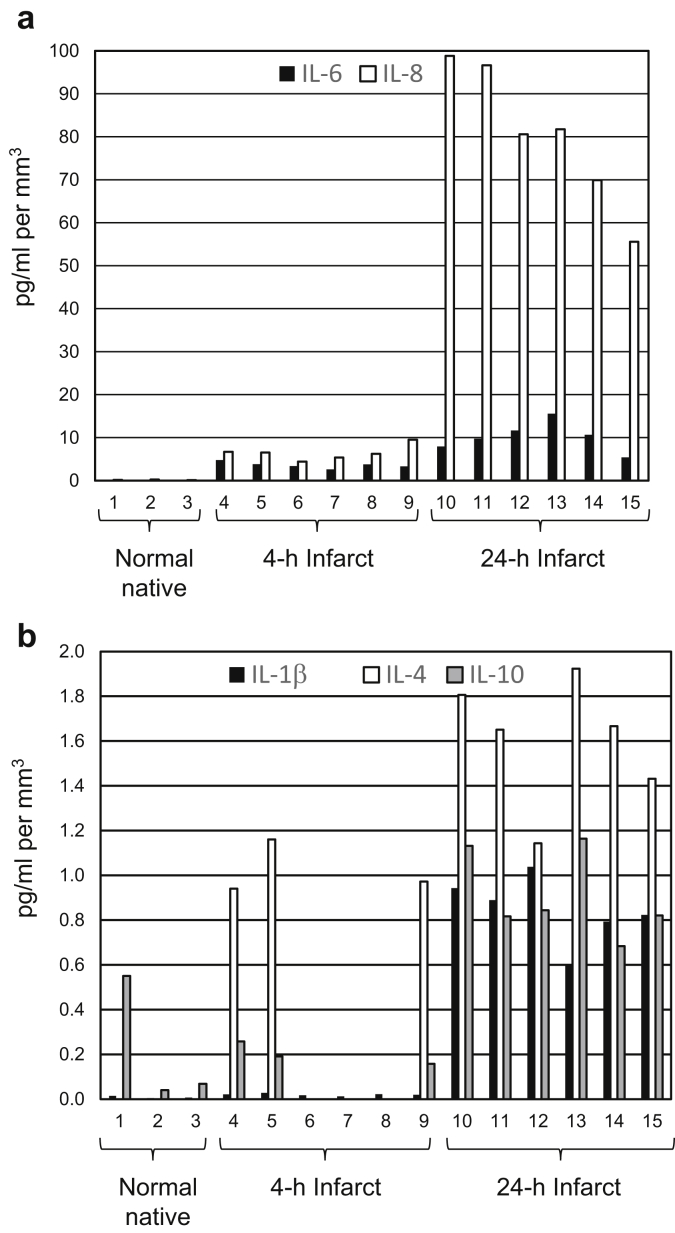
Intrarenal cytokines were measured in the supernatant from biopsy digestion using Milliplex high-sensitivity bead capture assay and Luminex technology. Biopsy supernatant contains glomerular filtrate, circulating plasma captured at the time of biopsy, and molecules secreted from cells within the biopsy sample during tissue processing. To validate clinical significance of intrarenal cytokine measurements, normal native kidneys removed for a renal mass or uroepithelial tumor served as negative controls, and native kidneys infarcted by interventional radiology procedures before nephrectomy served as positive controls. Native kidneys had measureable levels of IL-1β, IL-4, IL-6, IL-8, and IL-10 early after infarct, and all of the previously measured cytokines were elevated 24 hours after infarct (Figure 8 and Supplementary Table S4). Elevated levels of cytokines with infarction of native kidneys and higher levels with longer ischemia times indicate that cytokine measurements have clinical relevance.
Figure 8.
Comparison of cytokines (a) interleukin (IL)−6 and IL-8 and (b) IL-1β, IL-4, and IL-10 between normal native and infarcted native kidneys. Numbers 1 to 15 indicate 16-gauge, 4-mm biopsy sample fragments individually digested and assayed. Cytokine values are picograms per milliliter of supernatant from each of these digestions per cubic millimeter of tissue. Numbers 1 to 3 are from normal donors; 2 and 3 are from 2 different biopsy samples from the same donor; 4 to 9 are from a donor infarcted 4 hours before nephrectomy; 4 to 6 are different segments of the same biopsy sample; 7 to 9 are different segments of a second biopsy sample; 10 to 15 are from a donor infarcted 24 hours before nephrectomy; 10 to 12 are from different segments of the same biopsy sample; and 13 to 15 are from 3 other biopsy samples of the same kidney.
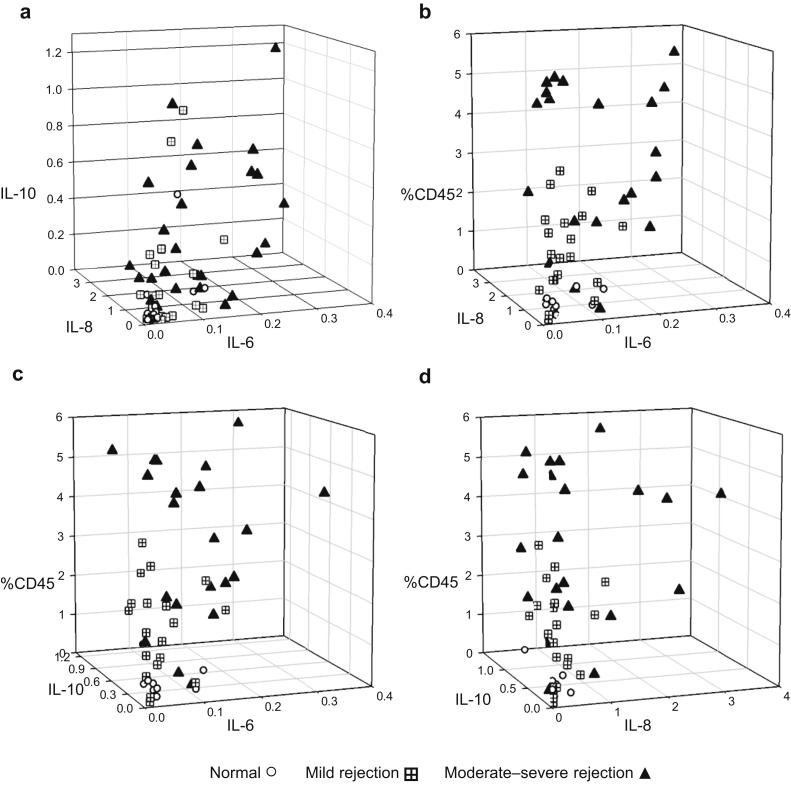
Intrarenal cytokines were measured in renal biopsy samples of native and transplanted kidneys and normalized to tissue volume. Intrarenal cytokine levels of IL-6, IL-8, and IL-10 were compared in transplanted kidneys with normal histology to those with rejection (Figure 9a). Rejection of any class or severity was associated with elevated levels of IL-6, IL-8, and IL-10.
Figure 9.
Three-dimensional scatter plots showing differences in cytokine and leukocyte profiles of representative normal transplant (n = 16), mild rejection, equivalent to Banff T-cell−mediated rejection IA (n = 25), and moderate to severe rejection, equivalent to Banff T-cell−mediated rejection >IA and antibody-mediated rejection (n = 31) biopsy samples. (a) IL-6 versus IL-8 versus IL-10; (b) IL-6 versus IL-8 versus CD45; (c) IL-6 versus IL-10 versus CD45; (d) IL-8 versus IL-10 versus CD45. Cytokine values are picograms per milliliter of digested supernatant per cubic millimeter of tissue. %CD45 represents the percentage of CD45+ leukocytes among the total number of viable cells in the biopsy sample. Normal versus moderate to severe rejection: P < 0.001 for IL-6, IL-8, IL-10, and %CD45. Mild versus moderate to severe rejection: P < 0.001 for IL-6 and %CD45; P = 0.01 for IL-8; and P = 0.007 for IL-10. P values were calculated using Mann−Whitney rank sum test.
Cytokine data were combined with cytometry results of percentage of CD45+ leukocytes per total viable cells in Figure 9b to d. The 3-dimensional scatter plots show distinct trends for different cytokine and leukocyte levels in kidneys with no rejection, mild (equivalent to Banff IA), or more severe forms of rejection (cellular rejection equivalent to Banff scores greater than IA and antibody-mediated rejection). Biopsy samples with more severe rejection have significantly greater numbers of leukocytes and higher levels of IL-6, IL-8, and IL-10 (Supplementary Table S5).
Discussion
The renal biopsy is the gold standard for assessing kidney injury. However, current techniques to evaluate tissue have limitations: pathogenesis is not always delineated, similar tissue architecture may result from different conditions, disease activity markers are often lacking, and insufficient information may be present to direct new therapies toward specific targets.
Our focus has been to develop methods that enhance the immunologic information available from a renal biopsy using standard amounts of tissue. The importance of evaluating renal tissue is demonstrated by our findings that leukocyte subsets and their activation states differ in kidneys and peripheral blood of the same individual (Figures 4 and 5 and Supplementary Figure S2), even when tissue has normal histology. Therefore, to know what is happening in the kidney, you need to look in the kidney. Leukocytes present in the kidney may not merely be idle passengers in the blood circulating through the organ. Their phenotype and activity may be altered within the kidney, creating a specific renal immune environment, consistent with reports of tissue-resident memory T cells in kidneys.15, 16, 17
Using techniques reported here, we now have the ability to study individual structural renal cells and leukocytes. In the same preparation used for cytometry, we are able to capture cytokines present in the biopsy sample. Although our techniques do not identify which cells secrete the cytokines, we show examples in which cytometry and cytokine levels provide relevant clinical information about the presence, severity, and pathogenesis of transplant rejection. Intrarenal leukocyte subsets, antibody and eculizumab binding to RMECs, and cytokine levels help to refine interpretations of biopsy results. Analyses of other leukocyte subsets with their activation markers, podocytes, tubular cells, and cell-costimulatory and inhibitory proteins on RMECs are underway.
Traditional renal histopathology focuses on the region of the kidney from which the biopsy was obtained, the cortex or the medulla. In our studies evaluating rejection, some samples contained cortex, others medulla, and some a mixture of medulla and cortex. Location of the tissue did not seem to affect leukocyte or cytokine results in assessing rejection. Others have made similar observations in their molecular studies of rejection using RNA extracts from biopsy samples.18 Our cytokine controls using normal and infarcted kidneys (Figure 8) also suggest that biopsy location is less important for measurements of inflammatory proteins, and supports the reproducibility of quantitating cytokines within an injured kidney. Different segments of biopsy cores and different biopsy samples from the same kidney provided similar levels of cytokines, particularly for the normal and 24-hour infarcted kidneys. The variability in IL-4 and IL-10 levels 4 hours after initiation of infarct may reflect the relatively short time period after injury, before the full effects of ischemia were operational.
Other methods used to obtain more information about kidney inflammation and rejection include gene profiling of renal tissue5, 6, 7 and blood,19 traditional immunocytochemistry to identify cells and cytokines,20 and cytometry of T-cell subsets.21 In our analysis, all cells and actual protein levels of cytokines within the entire biopsy sample fragment are assessed, rather than using transcripts or processed tissue, where components may be lost. We are able to identify microvascular endothelial cells and to assess relevant immune surface markers ex vivo. As we are able to provide this information within 18 hours of biopsy, our approach presents the opportunity to measure any cell surface protein and extracellular molecule where antibodies are available as a direct and timely adjunct to renal histopathology.
T-cell subsets have been used with variable results to distinguish rejection in transplanted kidneys using peripheral blood,22, 23, 24, 25, 26 tissue staining,27 fine-needle aspirates,28 and urine.29 However, the search for biomarkers of rejection has not yielded consistent candidates.30 Our focus has been to develop techniques to delineate more detailed and informative mechanisms of pathogenesis within renal tissue, namely, intrarenal indicators of inflammation. Our data suggest that the kidney has a localized immune response that may spill into urine and peripheral blood, but that both body fluids represent a dilution of the intrarenal immune process. This may explain inconsistencies in other studies.
The endothelium is the major site of antibody and complement binding associated with rejection. Current means of assessing antibody-mediated rejection include measuring the presence of donor-specific human leukocyte antigen (HLA), major histocompatibility complex class 1−related chain A (MICA), major histocompatibility complex class 1−related chain B (MICB), angiotensin receptor, and other antibodies31 by bead-bound antigen or enzyme-linked immunosorbent assay, and by looking for the deposition of C4d in regions of peritubular capillaries by microscopy.32 Others have focused on whether antibodies associated with rejection fix complement.33 Our cytometric assay for directly measuring antibody and complement binding to RMECs correlates with histopathology, even when donor-specific antibodies or C4d cannot be identified. It is also quantitative, as the numbers of endothelial cells binding antibody and complement are measured. Hence, the cytometric RMEC antibody binding assay may prove valuable for assessing clinically relevant antibody-mediated pathology. Our results suggest a role for activation of complement in mild acute-cellular rejection by non−antibody-dependent pathways. In cases in which eculizumab binds to RMECs, the possibility of using the antibody as a therapy might be considered, although in our series of mild acute rejection, steroids alone were sufficient to reduce complement activation. That RMEC−eculizumab binding was less in more severe cellular rejection may reflect the expected role of T-cell−mediated injury. RMEC−eculizumab binding in cases with transplant antibody-mediated rejection is consistent with an antibody-mediated mechanism for complement activation.
Low levels of eculizumab binding to normal RMECs raise the possibility that the complement component C5 may be more prevalent on microvascular endothelial cells than previously suspected, and that regulatory mechanisms have a role in defusing a potentially dangerous condition. What is unknown is whether the C5 that we detected with eculizumab was the intact C5 or the C5b fragment. Eculizumab binding sites could also be generated during tissue preparation.
Studies measuring cytokines to diagnose rejection have evaluated peripheral blood34, 35, 36, 37, 38 and urine39, 40, 41; where kidney tissue was evaluated, it was done by polymerase chain reaction amplification of cDNA.42, 43, 44, 45 To our knowledge, this is the first report of reliable cytokine protein measurements from human renal biopsy tissue without an interval of tissue amplification, cell culture, or stimulation. Our results suggest that IL-6, IL-8, and IL-10, particularly in combination with cytometry assessment of leukocytes in the sample, denote more severe tissue damage. That IL-10, considered an anti-inflammatory cytokine, is elevated in our study in more severe rejection has been reported by others.46 Additional studies with more biopsy samples are in progress, and may indicate clusters of cytokines that amend current classifications of rejection and inflammation.
In conclusion, just as hematopathology gained from cytometry by identifying offending cell types, nephrology can make similar advances by transforming small portions of kidney biopsy samples into nonsolid organs so that multicolor cytometry can precisely define and quantify cell populations of interest. We have shown examples in which cytometry and cytokine levels provide relevant quantitative clinical information regarding the presence, severity, and pathogenesis of transplant rejection. The techniques that we describe are also applicable to native kidney disease. They are not meant to replace histopathology of renal biopsy samples but, rather, add a higher resolution to the pathogenesis of renal disease.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This research was funded by the Institute of Translational Health Sciences and the Kidney Immunology Research Fund at the University of Washington, with generous donations from Mr. Robert Dolsen and the Dolsen Foundation, Mr. and Mrs. E. Kibble, Mr. John G. DuBois, and Dr. Alan and Susan Kendal. The authors gratefully acknowledge the support of these donors and the patients who provided tissue, which made the work possible.
Footnotes
Figure S1. Gating for differentials. Each dot in the plot represents a cell. Gates are set using peripheral blood (a−e) where leukocyte subsets are more abundant and hence easier to identify. Viable cells are selected based on exclusion of DAPI (DAPI−, A and F). Within the viable cells, leukocytes are selected based on the expression of CD45, the leukocyte common antigen (CD45+, B and G). Subsets of viable CD45+ leukocytes are determined based on the gates shown in C to E and H to J. The number of cells within the subset gates compared to the total CD45+ cells are used to determine the percentage of each leukocyte subpopulation for the differentials. CD56+ cells indicate NK cells, CD22 are B cells, CD66b are granulocytes. As described in the text, CD45+ cells that are CD3−, CD56−, CD22−, or CD66b− but that express HLA-DR approximate a macrophage/dendritic cell population.
Figure S2. Comparison of CD45RO/CD45RA ratios in T cells from renal biopsy samples and matched peripheral blood lymphocytes. Gating: viable cells > CD45 > CD3 > CD4 (a) or CD8 (b) > CD45RO and CD45RA. In all but 1 case (*), the ratio of CD45RO+ cells to CD45RA+ cells is higher in the kidney than in peripheral blood for both CD4 (P = 0.015) and CD8 (P = 0.045) T cells. There is no significant difference in the CD45RO/CD45RA ratios between native and transplant samples. The CD45RO/CD45RA ratio is significantly higher in CD4+ T cells in both renal (P = 0.014) and peripheral blood (P = 0.011) when CD4+ T cells are compared to CD8+ T cells. P values calculated using the Student t test.
Figure S3. Cytometry assay controls for RMEC antibody binding using representative biopsy samples. Gating: viable cells > CD45−/CD324−/HLA-DR+/CD31+ or CD34+. (A) Fluorescence minus 1 (FMO) and isotype controls for anti-human λ and κ (B) Reproducibility of anti-human λ and κ binding to RMECs when the same biopsy sample is labeled with 4 different antibody cocktails. (C) FcR expression on RMECs isolated from normal native kidney. Above the horizontal line on each dotplot indicates positive expression of FcRs based on kidney and peripheral blood leukocytes expression of the FcRs.
Figure S4. RMEC antibody binding in transplant biopsy samples. Gating: viable cells > CD45−/CD324−/HLA-DR+/CD31+ or CD34+. Representative dot plots of anti- λ + κ bound to RMECs in cases of (A) antibody-mediated rejection (ABMR), including active acute and chronic; and (B) transplant glomerulopathy without acute inflammation. Most of these kidneys had been transplanted more than 20 years before biopsy, with the exception of kidney 377. For case 377, the biopsy was performed after treatment 5 months earlier for major histocompatibility complex class 1−related chain A (MICA) antibody-mediated rejection. (C) Acute cellular rejection. (D) Nonspecific inflammation where histologic conditions for rejection were not met (equivalent to Banff borderline). DSA and C4d information appear under each dot plot where results were available. Where a percent sign is noted for C4d, it reflects the pathologist’s estimate of the amount of peritubular capillaries with C4d. In some cases, DSA were not determined (ND) because either there was no clinical indication to do so or donor HLA type was unknown. In cases 341 and 374, DSA were detected but below the level generally interpreted as positive by our HLA laboratory. In case 377, HLA DSA was negative but MICA antibody was detected. (E) Dot plots of anti- λ + κ bound to RMEC from cases with serial biopsy samples. Clinical course for each case indicated by arrows and text below dot plot. In case 1, the biopsy sample initially showed nonspecific inflammation with no antibody binding; subsequently, when renal function worsened, antibodies bound to RMECs were detected. In case 2, a patient with persistent rejection 6 weeks after treatment showed continued RMEC antibody binding. In case 3, a patient with acute DSA+, C4d+ antibody-mediated rejection had slightly decreased levels of RMEC antibody binding after treatment, with resolution of DSA but persistent C4d+ tissue staining. Number in the lower left corner of the dot plots is the identifier code assigned to the biopsy sample. The positive level for combined λ and κ binding to RMEC, indicated by the vertical line on the dot plots, was set using the level of light chains detected on peripheral blood leukocytes from the same donor.
Table S1. Analysis details of leukocyte differentials as shown in Figure 2.
Table S2. Analysis details of CD8/CD4 ratio data as shown in Figure 3.
Table S3. Analysis details of percent Igκ− plus Igλ-positive renal microvascular endothelial cells (RMECs) as shown in Figure 6 and Figure S4 .
Table S4. Analysis details of cytokines in normal and infarcted kidneys as shown in Figure 8.
Table S5. Analysis details of 3-dimensional cytokine and leukocyte data from Figure 9.
Supplementary material is linked to the online version of the paper at www.kireports.org.
Supplementary Material
Gating for differentials. Each dot in the plot represents a cell. Gates are set using peripheral blood (a−e) where leukocyte subsets are more abundant and hence easier to identify. Viable cells are selected based on exclusion of DAPI (DAPI−, A and F). Within the viable cells, leukocytes are selected based on the expression of CD45, the leukocyte common antigen (CD45+, B and G). Subsets of viable CD45+ leukocytes are determined based on the gates shown in C to E and H to J. The number of cells within the subset gates compared to the total CD45+ cells are used to determine the percentage of each leukocyte subpopulation for the differentials. CD56+ cells indicate NK cells, CD22 are B cells, CD66b are granulocytes. As described in the text, CD45+ cells that are CD3−, CD56−, CD22−, or CD66b− but that express HLA-DR approximate a macrophage/dendritic cell population.
Comparison of CD45RO/CD45RA ratios in T cells from renal biopsy samples and matched peripheral blood lymphocytes. Gating: viable cells > CD45 > CD3 > CD4 (a) or CD8 (b) > CD45RO and CD45RA. In all but 1 case (*), the ratio of CD45RO+ cells to CD45RA+ cells is higher in the kidney than in peripheral blood for both CD4 (P = 0.015) and CD8 (P = 0.045) T cells. There is no significant difference in the CD45RO/CD45RA ratios between native and transplant samples. The CD45RO/CD45RA ratio is significantly higher in CD4+ T cells in both renal (P = 0.014) and peripheral blood (P = 0.011) when CD4+ T cells are compared to CD8+ T cells. P values calculated using the Student t test.
Cytometry assay controls for RMEC antibody binding using representative biopsy samples. Gating: viable cells > CD45−/CD324−/HLA-DR+/CD31+ or CD34+. (A) Fluorescence minus 1 (FMO) and isotype controls for anti-human λ and κ (B) Reproducibility of anti-human λ and κ binding to RMECs when the same biopsy sample is labeled with 4 different antibody cocktails. (C) FcR expression on RMECs isolated from normal native kidney. Above the horizontal line on each dotplot indicates positive expression of FcRs based on kidney and peripheral blood leukocytes expression of the FcRs.
RMEC antibody binding in transplant biopsy samples. Gating: viable cells > CD45−/CD324−/HLA-DR+/CD31+ or CD34+. Representative dot plots of anti- λ + κ bound to RMECs in cases of (A) antibody-mediated rejection (ABMR), including active acute and chronic; and (B) transplant glomerulopathy without acute inflammation. Most of these kidneys had been transplanted more than 20 years before biopsy, with the exception of kidney 377. For case 377, the biopsy was performed after treatment 5 months earlier for major histocompatibility complex class 1−related chain A (MICA) antibody-mediated rejection. (C) Acute cellular rejection. (D) Nonspecific inflammation where histologic conditions for rejection were not met (equivalent to Banff borderline). DSA and C4d information appear under each dot plot where results were available. Where a percent sign is noted for C4d, it reflects the pathologist’s estimate of the amount of peritubular capillaries with C4d. In some cases, DSA were not determined (ND) because either there was no clinical indication to do so or donor HLA type was unknown. In cases 341 and 374, DSA were detected but below the level generally interpreted as positive by our HLA laboratory. In case 377, HLA DSA was negative but MICA antibody was detected. (E) Dot plots of anti- λ + κ bound to RMEC from cases with serial biopsy samples. Clinical course for each case indicated by arrows and text below dot plot. In case 1, the biopsy sample initially showed nonspecific inflammation with no antibody binding; subsequently, when renal function worsened, antibodies bound to RMECs were detected. In case 2, a patient with persistent rejection 6 weeks after treatment showed continued RMEC antibody binding. In case 3, a patient with acute DSA+, C4d+ antibody-mediated rejection had slightly decreased levels of RMEC antibody binding after treatment, with resolution of DSA but persistent C4d+ tissue staining. Number in the lower left corner of the dot plots is the identifier code assigned to the biopsy sample. The positive level for combined λ and κ binding to RMEC, indicated by the vertical line on the dot plots, was set using the level of light chains detected on peripheral blood leukocytes from the same donor.
Analysis details of leukocyte differentials as shown in Figure 2.
Analysis details of CD8/CD4 ratio data as shown in Figure 3.
Analysis details of percent Igκ− plus Igλ-positive renal microvascular endothelial cells (RMECs) as shown in Figure 6 and Figure S4 .
Analysis details of cytokines in normal and infarcted kidneys as shown in Figure 8.
Analysis details of 3-dimensional cytokine and leukocyte data from Figure 9.
References
- 1.Mestas J., Hughes C.C. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 2.Payne K.J., Crooks G.M. Immune-cell lineage commitment: translation from mice to humans. Immunity. 2007;26:674–677. doi: 10.1016/j.immuni.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 3.von Willebrand E., Lautenschlager I., Inkinen K. Distribution of the major histocompatibility complex antigens in human and rat kidney. Kidney Int. 1985;27:616–621. doi: 10.1038/ki.1985.55. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler S.F. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 5.Halloran P.F., Reeve J.P., Pereira A.B. Antibody-mediated rejection, T cell-mediated rejection, and the injury-repair response: new insights from the Genome Canada studies of kidney transplant biopsies. Kidney Int. 2014;85:258–264. doi: 10.1038/ki.2013.300. [DOI] [PubMed] [Google Scholar]
- 6.Hidalgo L.G., Sellares J., Sis B. Interpreting NK cell transcripts versus T cell transcripts in renal transplant biopsies. Am J Transplant. 2012;12:1180–1191. doi: 10.1111/j.1600-6143.2011.03970.x. [DOI] [PubMed] [Google Scholar]
- 7.Hidalgo L.G., Sis B., Sellares J. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant. 2010;10:1812–1822. doi: 10.1111/j.1600-6143.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- 8.Muczynski K.A., Ekle D.M., Anderson S.K. Normal human HLA-DR-expressing microvascular endothelial cells: characterization, isolation and regulation of MHC class II expression. J Am Soc Nephrol. 2003;14:1336–1348. doi: 10.1097/01.asn.0000061778.08085.9f. [DOI] [PubMed] [Google Scholar]
- 9.Haas M., Sis B., Racusen L.C. Banff 2013 meeting report: inclusion of C4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant. 2014;14:272–283. doi: 10.1111/ajt.12590. [DOI] [PubMed] [Google Scholar]
- 10.Becker J.U., Chang A., Nickeleit V. Banff borderline changes suspicious for acute t cell-mediated rejection: where do we stand? Am J Transplant. 2016;16:2654–2660. doi: 10.1111/ajt.13784. [DOI] [PubMed] [Google Scholar]
- 11.Marzio R., Mauel J., Betz-Corradin S. CD69 and regulation of the immune function. Immunopharmacol Immunotoxicol. 1999;21:565–582. doi: 10.3109/08923979909007126. [DOI] [PubMed] [Google Scholar]
- 12.Testi R., Phillips J.H., Lanier L.L. T cell activation via Leu-23 (CD69) J Immunol. 1989;143:1123–1128. [PubMed] [Google Scholar]
- 13.Wieland E., Shipkova M. Lymphocyte surface molecules as immune activation biomarkers. Clin Biochem. 2016;49:347–354. doi: 10.1016/j.clinbiochem.2015.07.099. [DOI] [PubMed] [Google Scholar]
- 14.Muczynski K.A., Cotner T., Anderson S.K. Unusual expression of human lymphocyte antigen class II in normal renal microvascular endothelium. Kidney Int. 2001;59:488–497. doi: 10.1046/j.1523-1755.2001.059002488.x. [DOI] [PubMed] [Google Scholar]
- 15.Ma C., Mishra S., Demel E.L. TGF-beta controls the formation of kidney-resident t cells via promoting effector t cell extravasation. J Immunol. 2017;198:749–756. doi: 10.4049/jimmunol.1601500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay L.K., Minnich M., Kragten N.A. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459–463. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- 17.van Aalderen M.C., Remmerswaal E.B., Heutinck K.M. Clinically relevant reactivation of polyomavirus BK (BKPyV) in HLA-A02-positive renal transplant recipients is associated with impaired effector-memory differentiation of BKPyV-specific CD8+ T Cells. PLoS Pathog. 2016;12:e1005903. doi: 10.1371/journal.ppat.1005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madill-Thomsen K.S., Wiggins R.C., Eskandary F. The effect of cortex/medulla proportions on molecular diagnoses in kidney transplant biopsies: rejection and injury can be assessed in medulla. Am J Transplant. 2017;17:2117–2128. doi: 10.1111/ajt.14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarwal M., Chua M.S., Kambham N. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 20.Sementilli A., Franco M. Renal acute cellular rejection: correlation between the immunophenotype and cytokine expression of the inflammatory cells in acute glomerulitis, arterial intimitis, and tubulointerstitial nephritis. Transplant Proc. 2010;42:1671–1676. doi: 10.1016/j.transproceed.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 21.Totterman T.H., Hanas E., Bergstrom R. Immunologic diagnosis of kidney rejection using FACS analysis of graft-infiltrating functional and activated T and NK cell subsets. Transplantation. 1989;47:817–823. doi: 10.1097/00007890-198905000-00014. [DOI] [PubMed] [Google Scholar]
- 22.von Willebrand E. OKT4/8 ratio in the blood and in the graft during episodes of human renal allograft rejection. Cell Immunol. 1983;77:196–201. doi: 10.1016/0008-8749(83)90019-9. [DOI] [PubMed] [Google Scholar]
- 23.Henny F.C., van Es A., Oljans P.J. Prognostic value of T lymphocyte subset ratios for renal transplant survival in patients on different immunosuppressive regimens. Clin Exp Immunol. 1986;65:373–380. [PMC free article] [PubMed] [Google Scholar]
- 24.Sheikh I.A., Al-Menawy L., Shaheen F.A.M. The diagnosis of acute renal allograft rejection using T-lymphocyte subsets in the peripheral blood: a better test now? Saudi J Kidney Dis Transplant. 1995;6:15–21. [PubMed] [Google Scholar]
- 25.Vallotton L., Hadaya K., Venetz J.P. Monitoring of CD4+CD25highIL-7Ralphahigh activated T cells in kidney transplant recipients. Clin J Am Soc Nephrol. 2011;6:2025–2033. doi: 10.2215/CJN.09611010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ordonez L., Bernard I., Chabod M. A higher risk of acute rejection of human kidney allografts can be predicted from the level of CD45RC expressed by the recipients' CD8 T cells. PLoS One. 2013;8:e69791. doi: 10.1371/journal.pone.0069791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Divella C., Rossini M., Loverre A. Immunohistochemical characterization of glomerular and tubulointerstitial infiltrates in renal transplant patients with chronic allograft dysfunction. Nephrol Dial Transplant. 2010;25:4071–4077. doi: 10.1093/ndt/gfq377. [DOI] [PubMed] [Google Scholar]
- 28.Xavier P.D., Lema G.L., Magalhaes M.C. Flow cytometry assessment of graft-infiltrating lymphocytes can accurately identify acute rejection in kidney transplants. Clin Transplant. 2014;28:177–183. doi: 10.1111/ctr.12293. [DOI] [PubMed] [Google Scholar]
- 29.van Doesum W.B., Abdulahad W.H., van Dijk M.C. Characterization of urinary CD4(+) and CD8(+) T cells in kidney transplantation patients with polyomavirus BK infection and allograft rejection. Transpl Infect Dis. 2014;16:733–743. doi: 10.1111/tid.12273. [DOI] [PubMed] [Google Scholar]
- 30.Lo D.J., Kaplan B., Kirk A.D. Biomarkers for kidney transplant rejection. Nat Rev Nephrol. 2014;10:215–225. doi: 10.1038/nrneph.2013.281. [DOI] [PubMed] [Google Scholar]
- 31.Jackson A.M., Sigdel T.K., Delville M. Endothelial cell antibodies associated with novel targets and increased rejection. J Am Soc Nephrol. 2015;26:1161–1171. doi: 10.1681/ASN.2013121277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regele H., Bohmig G.A., Habicht A. Capillary deposition of complement split product C4d in renal allografts is associated with basement membrane injury in peritubular and glomerular capillaries: a contribution of humoral immunity to chronic allograft rejection. J Am Soc Nephrol. 2002;13:2371–2380. doi: 10.1097/01.asn.0000025780.03790.0f. [DOI] [PubMed] [Google Scholar]
- 33.Al Mahri A., Holgersson J., Alheim M. Detection of complement-fixing and non-fixing antibodies specific for endothelial precursor cells and lymphocytes using flow cytometry. Tissue Antigens. 2012;80:404–415. doi: 10.1111/j.1399-0039.2012.01954.x. [DOI] [PubMed] [Google Scholar]
- 34.Diller R., Palmes D., Dietl K.H. Interleukin-6, interleukin-8, and interleukin-10 in kidney transplantation: improved risk strategy? Transplant Proc. 2003;35:1333–1337. doi: 10.1016/s0041-1345(03)00529-3. [DOI] [PubMed] [Google Scholar]
- 35.Karczewski M., Karczewski J., Poniedzialek B. Distinct cytokine patterns in different states of kidney allograft function. Transplant Proc. 2009;41:4147–4149. doi: 10.1016/j.transproceed.2009.08.067. [DOI] [PubMed] [Google Scholar]
- 36.Wu D.J., Qian M.J., Rong R.M. [Expression of inflammation cytokines and network analysis in acute rejection of renal transplantation] Zhonghua Yi Xue Za Zhi. 2012;92:2976–2979. doi: 10.3760/cma.j.issn.0376-2491.2012.42.007. [DOI] [PubMed] [Google Scholar]
- 37.De Serres S.A., Mfarrej B.G., Grafals M. Derivation and validation of a cytokine-based assay to screen for acute rejection in renal transplant recipients. Clin J Am Soc Nephrol. 2012;7:1018–1025. doi: 10.2215/CJN.11051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mota A.P., Vilaca S.S., das Merces F.L., Jr. Cytokines signatures in short and long-term stable renal transplanted patients. Cytokine. 2013;62:302–309. doi: 10.1016/j.cyto.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Kaden J., Priesterjahn R. Increasing urinary IL-6 levels announce kidney graft rejection. Transpl Int. 2000;13(suppl 1):S34–S41. doi: 10.1007/s001470050271. [DOI] [PubMed] [Google Scholar]
- 40.Karczewski M., Karczewski J., Poniedzialek B. Cytometric analysis of TH1/TH2 cytokines in the urine of patients undergoing kidney transplantation. Ann Transplant. 2009;14:25–28. [PubMed] [Google Scholar]
- 41.Reinhold S.W., Staub R.H., Kruger B. Elevated urinary sVCAM-1 IL6 and TNFR1 concentrations indicate acute kidney transplant rejection in the first 2 weeks after transplantation. Cytokines. 2013;57:379–388. doi: 10.1016/j.cyto.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 42.Hribova P., Kotsch K., Brabcova I. Cytokines and chemokine gene expression in human kidney transplantation. Transplant Proc. 2005;37:760–763. doi: 10.1016/j.transproceed.2004.12.177. [DOI] [PubMed] [Google Scholar]
- 43.Hueso M., Navarro E., Moreso F. Intragraft expression of the IL-10 gene is up-regulated in renal protocol biopsies with early interstitial fibrosis, tubular atrophy, and subclinical rejection. Am J Pathol. 2010;176:1696–1704. doi: 10.2353/ajpath.2010.090411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rekers N.V., Bajema I.M., Mallat M.J. Quantitative polymerase chain reaction profiling of immunomarkers in rejecting kidney allografts for predicting response to steroid treatment. Transplantation. 2012;94:596–602. doi: 10.1097/TP.0b013e31825db651. [DOI] [PubMed] [Google Scholar]
- 45.Sharma V.K., Ding R., Li B. Molecular correlates of human renal allograft rejection. Transplant Proc. 1998;30:2364–2366. doi: 10.1016/s0041-1345(98)00656-3. [DOI] [PubMed] [Google Scholar]
- 46.Spivey T.L., Uccellini L., Ascierto M.L. Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J Transl Med. 2011;9:174. doi: 10.1186/1479-5876-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gating for differentials. Each dot in the plot represents a cell. Gates are set using peripheral blood (a−e) where leukocyte subsets are more abundant and hence easier to identify. Viable cells are selected based on exclusion of DAPI (DAPI−, A and F). Within the viable cells, leukocytes are selected based on the expression of CD45, the leukocyte common antigen (CD45+, B and G). Subsets of viable CD45+ leukocytes are determined based on the gates shown in C to E and H to J. The number of cells within the subset gates compared to the total CD45+ cells are used to determine the percentage of each leukocyte subpopulation for the differentials. CD56+ cells indicate NK cells, CD22 are B cells, CD66b are granulocytes. As described in the text, CD45+ cells that are CD3−, CD56−, CD22−, or CD66b− but that express HLA-DR approximate a macrophage/dendritic cell population.
Comparison of CD45RO/CD45RA ratios in T cells from renal biopsy samples and matched peripheral blood lymphocytes. Gating: viable cells > CD45 > CD3 > CD4 (a) or CD8 (b) > CD45RO and CD45RA. In all but 1 case (*), the ratio of CD45RO+ cells to CD45RA+ cells is higher in the kidney than in peripheral blood for both CD4 (P = 0.015) and CD8 (P = 0.045) T cells. There is no significant difference in the CD45RO/CD45RA ratios between native and transplant samples. The CD45RO/CD45RA ratio is significantly higher in CD4+ T cells in both renal (P = 0.014) and peripheral blood (P = 0.011) when CD4+ T cells are compared to CD8+ T cells. P values calculated using the Student t test.
Cytometry assay controls for RMEC antibody binding using representative biopsy samples. Gating: viable cells > CD45−/CD324−/HLA-DR+/CD31+ or CD34+. (A) Fluorescence minus 1 (FMO) and isotype controls for anti-human λ and κ (B) Reproducibility of anti-human λ and κ binding to RMECs when the same biopsy sample is labeled with 4 different antibody cocktails. (C) FcR expression on RMECs isolated from normal native kidney. Above the horizontal line on each dotplot indicates positive expression of FcRs based on kidney and peripheral blood leukocytes expression of the FcRs.
RMEC antibody binding in transplant biopsy samples. Gating: viable cells > CD45−/CD324−/HLA-DR+/CD31+ or CD34+. Representative dot plots of anti- λ + κ bound to RMECs in cases of (A) antibody-mediated rejection (ABMR), including active acute and chronic; and (B) transplant glomerulopathy without acute inflammation. Most of these kidneys had been transplanted more than 20 years before biopsy, with the exception of kidney 377. For case 377, the biopsy was performed after treatment 5 months earlier for major histocompatibility complex class 1−related chain A (MICA) antibody-mediated rejection. (C) Acute cellular rejection. (D) Nonspecific inflammation where histologic conditions for rejection were not met (equivalent to Banff borderline). DSA and C4d information appear under each dot plot where results were available. Where a percent sign is noted for C4d, it reflects the pathologist’s estimate of the amount of peritubular capillaries with C4d. In some cases, DSA were not determined (ND) because either there was no clinical indication to do so or donor HLA type was unknown. In cases 341 and 374, DSA were detected but below the level generally interpreted as positive by our HLA laboratory. In case 377, HLA DSA was negative but MICA antibody was detected. (E) Dot plots of anti- λ + κ bound to RMEC from cases with serial biopsy samples. Clinical course for each case indicated by arrows and text below dot plot. In case 1, the biopsy sample initially showed nonspecific inflammation with no antibody binding; subsequently, when renal function worsened, antibodies bound to RMECs were detected. In case 2, a patient with persistent rejection 6 weeks after treatment showed continued RMEC antibody binding. In case 3, a patient with acute DSA+, C4d+ antibody-mediated rejection had slightly decreased levels of RMEC antibody binding after treatment, with resolution of DSA but persistent C4d+ tissue staining. Number in the lower left corner of the dot plots is the identifier code assigned to the biopsy sample. The positive level for combined λ and κ binding to RMEC, indicated by the vertical line on the dot plots, was set using the level of light chains detected on peripheral blood leukocytes from the same donor.
Analysis details of leukocyte differentials as shown in Figure 2.
Analysis details of CD8/CD4 ratio data as shown in Figure 3.
Analysis details of percent Igκ− plus Igλ-positive renal microvascular endothelial cells (RMECs) as shown in Figure 6 and Figure S4 .
Analysis details of cytokines in normal and infarcted kidneys as shown in Figure 8.
Analysis details of 3-dimensional cytokine and leukocyte data from Figure 9.