Abstract
Present guidelines for classification of constitutional variants do not incorporate inferences from mutations seen in tumors, even when these are associated with a specific molecular phenotype. When somatic mutations and constitutional mutations lead to the same molecular phenotype, as for the mismatch repair genes, information from somatic mutations may enable interpretation of previously unclassified variants. To test this idea, we first estimated likelihoods that somatic variants in MLH1, MSH2, MSH6, and PMS2 drive microsatellite instability and characteristic IHC staining patterns by calculating likelihoods of high versus low normalized variant read fractions of 153 mutations known to be pathogenic versus those of 760 intronic passenger mutations from 174 paired tumor-normal samples. Mutations that explained the tumor mismatch repair phenotype had likelihood ratio for high variant read fraction of 1.56 (95% CI 1.42–1.71) at sites with no loss of heterozygosity and of 26.5 (95% CI 13.2–53.0) at sites with loss of heterozygosity. Next, we applied these ratios to 165 missense, synonymous, and splice variants observed in tumors, combining in a Bayesian analysis the likelihood ratio corresponding with the adjusted variant read fraction with pretest probabilities derived from published analyses and public databases. We suggest classifications for 86 of 165 variants: 7 benign, 31 likely benign, 22 likely pathogenic, and 26 pathogenic. These results illustrate that for mismatch repair genes, characterization of tumor mutations permits tumor mutation data to inform constitutional variant classification. We suggest modifications to incorporate molecular phenotype in future variant classification guidelines.
Keywords: variant classification guidelines, Lynch syndrome, colorectal cancer, Lynch-like, tumor mutations, likelihood-ratio, Bayesian analysis, variant of uncertain significance, molecular phenotype, loss of heterozygosity
Introduction
Established constitutional variant classification guidelines such as those proposed by the American College of Medical Genetics and Genomics (ACMGG) and Association for Molecular Pathology (AMP) do not incorporate inferences from tumor data.1 Inference from tumors can be difficult because clear correlations between pathogenic constitutional variants and tumor phenotype have not been established for many genes. In addition, tumors have much higher mutation rates than non-tumor tissue, so it can be difficult to ascribe clear functional effect to specific somatic mutations, as the majority of mutations observed in tumors are passenger mutations.2 In situations where tumor phenotype is highly correlated with specific genetic mutations, this phenotype has long been used to inform research on constitutional genetics.3, 4 We propose that establishing probability that an observed tumor mutation is a driver of that phenotype can inform systematic, quantitative variant classification.
Lynch syndrome is an inherited cancer susceptibility syndrome with high correlation of constitutional genotype to tumor phenotype. Families with Lynch syndrome have significantly higher risk than the general population for cancers of the colon, endometrium, stomach, ovary, and others. Loss-of-function mutations in the mismatch repair (MMR) genes MLH1 (MIM: 609310), MSH2 (MIM: 69309), MSH6 (MIM: 614350), and PMS2 (MIM: 614337) lead to microsatellite instability (MSI), the hallmark of Lynch syndrome. Characteristic immunohistochemical (IHC) staining patterns are also seen in Lynch syndrome-associated colorectal and endometrial tumors. MMR protein function influences IHC staining patterns and can indicate which gene is impaired. Loss of staining for MLH1 and PMS2 proteins suggests a pathogenic variant in MLH1; loss of MSH2 and MSH6 proteins suggests a pathogenic variant in MSH2; and isolated loss of MSH6 or PMS2 proteins suggests a pathogenic variant in the corresponding gene. Screening colorectal and endometrial tumors for these hallmark features has become standard practice in many healthcare systems.5, 6 Individuals with tumors that are MSI-high and/or show loss of IHC staining are referred for confirmatory constitutional testing following one of several testing algorithms that culminate with sequencing of one or several MMR genes.7, 8 Tumor phenotype is so strongly associated with constitutional mutations in Lynch syndrome that MSI findings have been used to determine a likelihood ratio that constitutional variants in MLH1 and MSH2 are associated with Lynch syndrome.9
Four recent independent studies have shown that two pathogenic somatic mutations (MLH1, MSH2, MSH6, and PMS2) or one such somatic mutation with loss of heterozygosity (LOH) results in the same tumor phenotype seen with constitutional MMR gene mutations.10, 11, 12, 13 Universal screening of colorectal tumors for MMR deficiency using MSI or IHC leads to genetic confirmation of Lynch syndrome causing mutations in 3% to 4% of individuals with colorectal cancer;14, 15 however, some tumors have MMR deficiency not explained by pathogenic constitutional variants or alternative causes such as MLH1 hypermethylation. Initially, it was thought that cases with unexplained mismatch repair deficiency were caused by constitutional mutations that could not be detected by current clinical sequencing tests. However, up to 69% of these cases can be attributed to two pathogenic somatic mutations or one mutation with LOH in the MMR genes.10, 11, 12, 13 Each independent study that has investigated double somatic mutations in these “Lynch-like” tumors has reported Lynch syndrome-associated mutations as somatic events and has shown that genetic “hits” have similar characteristics regardless of whether the first hit was inherited or somatic.10, 11, 12, 13
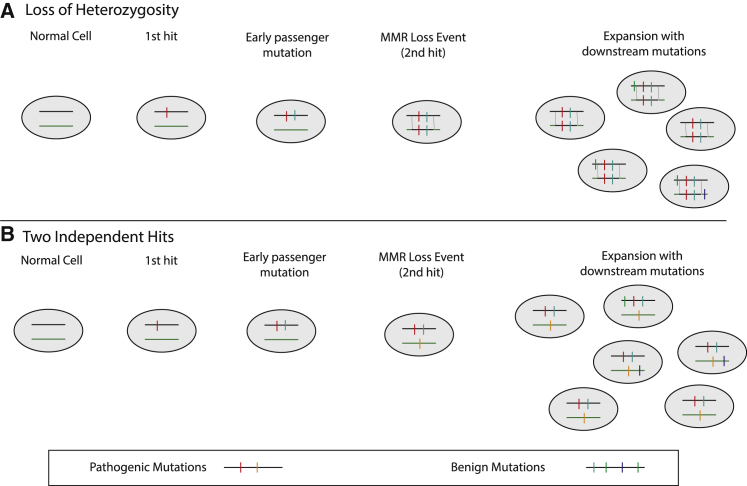
Determining the likelihood that a somatic mutation in an MMR gene is pathogenic in a tumor with mismatch repair deficiency requires differentiating “driver” mutations from the numerous “passenger” mutations that are a result of the increased mutability caused by defective DNA repair. After mismatch repair pathways are compromised by loss of both functional copies of an MMR gene, the cell becomes more mutable, resulting in many mutations across the genome.2 Some of these mutations alter other tumor pathways and eventually lead to cancer,16 while other mutations in the same genes may have no biochemical effect. This increased total mutational load, as opposed to specific mutations, may improve response to immune therapy.17, 18 However, when high mutational load is present, parsing out the functional consequence of specific mutations can be challenging (see Figure 1).
Figure 1.
Illustration of Scenarios Explaining Different Variant Read Fractions for Tumor and Passenger Mutations in MMR Genes
(A) The first pathogenic mutation may or may not be followed by passenger mutations before the loss of heterozygosity (LOH) event. In the copy-neutral LOH event, a large segment is copied into the other chromosome after both a driver and an early passenger have happened on the same chromosome. Mutations in the segment with LOH are expected to be present in 100% of tumor alleles. Many additional downstream mutations may happen in daughter cells due to MMR deficiency, but these are mostly benign and expected to be seen in less than 100% of tumor alleles.
(B) The first pathogenic mutation may or may not be followed by passenger mutations before a second hit on the opposite allele eliminates MMR function. Many additional downstream mutations may happen in daughter cells due to MMR deficiency, but these are mostly benign and expected to be seen in less than 50% of tumor alleles.
Past work in tumors with unexplained MMR deficiency contains purely descriptive variant analysis.10, 11, 12, 13 The objective of our study is to establish quantitative metrics to relating to probability that mutations in MLH1, MSH2, MSH6, and PMS2 are driver or passenger using tumor variant allele fractions in the specific situations of two separate somatic hits or single somatic mutation with LOH. We estimate the likelihood that variants are responsible for the characteristic phenotype of MSI and loss of IHC patterns in MMR-deficient tumors. We then use Bayesian multifactorial analysis to combine this new data from paired tumor-normal samples with data from public databases to propose variant classification for 165 variants.
Material and Methods
Samples
Available samples consisted of 337 samples sent to the University of Washington, Department of Laboratory Medicine for ColoSeq Tumor testing between November 2014 and December 2017. All tumors were being evaluated for suspected somatic mutations causing the unexplained MMR deficiency. Tumors were predominantly colon and endometrial and had been tested using IHC staining for MMR genes. For most samples, initial sequencing had been performed and had not detected a constitutional variant explaining the phenotype. Study inclusion criteria also included available paired tumor and normal tissue, available IHC results, no other explanation for molecular phenotype (e.g., BRAF), and adequate tumor-sequencing quality. For this analysis we focused on data derived from somatic mutations, so we excluded samples with a constitutional pathogenic, likely pathogenic, or uncertain variants in MMR genes. This retrospective analysis of clinical data with waiver of consent was approved by the University of Washington IRB (00003358).
For each case, formalin-fixed paraffin-embedded (FFPE) tissue sections were evaluated by a board-certified anatomic pathologist to optimize the tumor content subject to DNA isolation and massively parallel sequencing. When appropriate, FFPE tissues were macro-dissected from unstained tissue sections to achieve the maximum possible tumor content. FFPE tissues were deparaffinized and DNA was extracted. Each tumor sample evaluated was submitted with a paired blood sample for constitutional variant analysis. Sequencing and variant calling were performed on tumor and normal samples as described previously.19, 20
Determination of Tumor Mutations and Sample Tumor Content
Constitutional and tumor variants were identified in paired samples using methods described previously.13, 19, 20 This analysis excluded calls with lower than 30× coverage, variant allele read fraction (VRF) < 0.04, or low mapping quality. We noted that VRF is not accurate for indels larger than 2 nucleotides and for changes located in or adjacent to homopolymer regions of greater than 5 repeats, so we excluded these mutations from our analysis. Extremely high homology for PMS2 exons 12–15 prevents accurate read mapping in this region, so this region was also excluded from our analysis. We used several methods to evaluate tumor fraction and LOH. We calculated tumor fraction using density plots of variant allele fractions to visualize separate subpopulations of tumor cells. We used B-allele frequency calculations determined using the sequenza (v.2.1.2)21 R package and change in variant allele fraction for heterozygous common polymorphisms between normal and tumor samples. We correlated calculated tumor fraction with variant allele fractions of known tumor driver mutations in APC, KRAS, ATM, and MMR genes. Estimates of tumor content were used to normalize variant read allele fraction (VRF) using the following formula:
Normalized VRF was then used to differentiate between putative driver mutations (i.e., those likely to be responsible for IHC patterns) and putative passenger mutations in subclones that happen to be in MMR genes.22
Likelihood that New and Uncertain Missense and Splice Variants Are Driver Mutations
Available samples that met inclusion criteria were used for LR calculations. Additional samples became available for multifactorial variant analysis after LR calculations were complete. LOH is a major predictor of expected normalized VRF. Because of this, we first separated tumors into those with and without evidence of LOH in the genes evaluated. We expected driver mutations responsible for MMR deficiency to appear clonal and be present at normalized VRF of approximately 0.5 if there were two (heterozygous) driver mutations in tumor and 1.0 if there was LOH in the tumor at the mutation site.12 On the other hand, passenger mutations in regions without LOH are likely to be present at VRF lower than 0.5, and normalized VRF lower than 1.0 in regions with LOH. Deviations from these ratios caused by stochastic variant read sampling and imprecision in tumor content estimates lead to distributions of normalized VRF for mutations causing the MMR phenotype. Passenger mutations may appear clonal if they are early events in tumor pathogenesis or if the tumor cell death-birth ratio is high and will have lower VRF if they appear late or are in slow growing tumors (see Figure 1).23 These factors necessitate empiric evaluation of VRF for many variants across multiple tumors to generate LR estimates.
We compared passenger (intronic) and driver (known pathogenic) VRF distributions with defined cutoffs for heterozygous tumor and tumor mutations with LOH. To rule out possible VRF bias based on gene location, we first showed that VRF of constitutional polymorphisms located in exons and introns did not significantly differ in the MMR genes tested (mean 0.473 and 0.474, respectively). VRF distribution for tumor passenger mutations in MMR genes were estimated using all intronic mutations from approximately 100 kb of captured intronic sequence in the MLH1, MSH2, MSH6, and PMS2 genes, which was included in the ColoSeq-Tumor capture design. We used the following reference sequences for variant annotations: MLH1 GenBank: NM_000249.3, MSH2 GenBank: NM_000251.2, MSH6 GenBank: NM_000179.2, and PMS2 GenBank: NM_000535.5. Only a very small proportion of known pathogenic MMR mutations are farther intronic than the 10 bp proximal to splice sites, so the possible few undescribed pathogenic intronic variants will have minimal effect on LR estimates. We tallied somatic intronic mutations seen in all four MMR genes to establish VRF distributions for benign variants. For driver mutation VRF distributions, we used those classified as InSiGHT class 5 combined with loss-of-function frameshift and nonsense mutations.
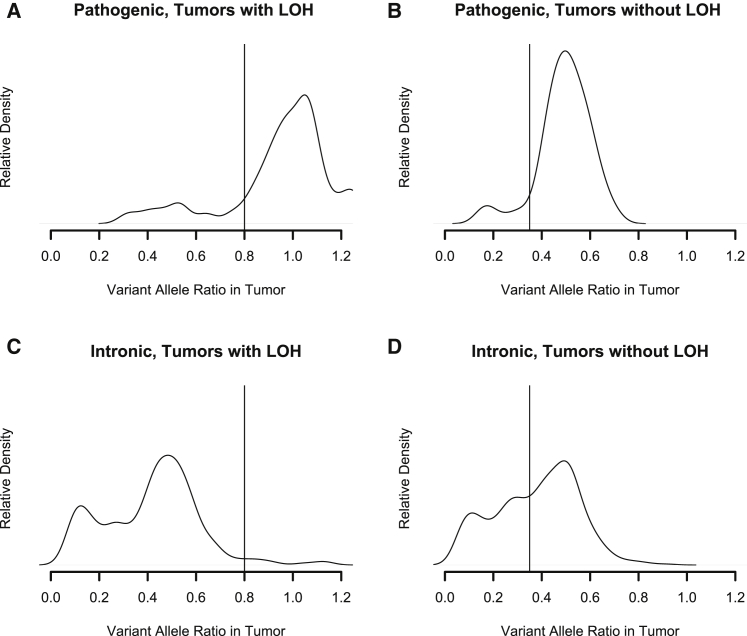
We set cutoffs for normalized variant read fraction at 0.8 for tumors with LOH, which was near the density plot inflection point below 1.0. We set the normalized VRF cutoff at 0.35 for tumors without LOH near the inflection point below 0.5 (Figure 2). We detected no significant difference between VRF distributions for colorectal and endometrial cancer, so we used the same VRF cutoffs and LR for all tumor types. Ratios of proportion of pathogenic and intronic (assumed benign) variants above and below these cutoffs defined likelihood ratios. Briefly, the LR that a variant is pathogenic is the ratio of two proportions: the proportion of known pathogenic driver mutations consistent with observed IHC above the cutoff and the proportion of intronic passenger mutations above the cutoff. The LR that a variant is benign is the ratio of similar proportions below the cutoff.24 Confidence intervals for LRs were calculated using standard methods. These LRs thus represent the likelihood that the observed VRF of a somatic change is that of a mutation causing MMR deficiency or that of an unimportant change often seen in MMR-deficient tumors.
Figure 2.
Distribution of Normalized Tumor Variant Read Fractions (VRFs) for Pathogenic Somatic Mutations in MLH1, MSH2, MSH6, and PMS2 in Tumors with and without Loss of Heterozygosity (LOH) in an MMR Gene
(A) VRF distribution of pathogenic mutations in tumors with LOH. We used 0.8 as a VRF cutoff in these tumors.
(B) VRF distribution of pathogenic mutations in tumors without LOH. We used 0.35 as a cutoff in these tumors.
(C) VRF distribution of intronic events observed in MMR tumors with LOH.
(D) VRF distribution of intronic events in MMR tumors without LOH.
Validation with Established Constitutional MMR Variant Data
For validation, we correlated evidence generated from missense, synonymous, and splice mutations observed in tumors with data on equivalent constitutional variants. We compared InSiGHT classifications of constitutional variants to identical somatic mutations seen in tumors.25 We also compared tumor data with published functional assay results26, 27, 28, 29, 30 as well as splice analysis31, 32, 33 and co-segregation data,32, 33 where available.
Evaluation in MMR-Proficient Tumors
Likelihood ratios were calculated using tumors that are MSI-high and have IHC patterns consistent with Lynch syndrome; however, false negative IHC and MSI results may occur. We evaluated data from 356 previously described microsatellite stable tumors without abnormal IHC staining ascertained as part of the Ohio Colorectal Cancer Prevention Initiative34 to determine whether normalized VRF cut-offs might identify MMR driver mutations where none are expected to be found.
Risk of Variant Misclassification Using Tumor Data
As tumor data have not previously been used extensively in variant classification, we sought to evaluate how often widespread use of mutation data obtained from tumor samples would be predicted to misclassify variants. Using observed frequencies of normalized VRF distributions for pathogenic missense, splice, and synonymous variants and observed VRF distributions for assumed benign intronic variants, we simulated this hypothetical situation. We assumed that a variant being observed is an independent event in each tumor, so the data from each sequential independent tumor can be combined until the variant is classified as definitively pathogenic or benign. Although true pathogenic and benign variants will have separate prior distributions, we used the null prior odds of 1 for simplicity. We repeated the simulation for 10,000 hypothetical benign and 10,000 hypothetical pathogenic variants.
Use of Tumor Data in Multifactorial Classification of MLH1, MSH2, MSH6, and PMS2
To illustrate how information from tumors can help classify constitutional variants, we performed multifactorial likelihood calculations for all missense, synonymous, and splice variants that potentially explained IHC staining patterns identified in available tumor-normal pairs that had not already been classified as definitively pathogenic (class 5) or benign (class 1) by the InSiGHT consortium. We did not perform multifactorial classification of frameshift and nonsense mutations, as these are already classified as pathogenic under current variant analysis standards. No calculations were performed for variants in exons 12–15 of PMS2, as very high homology with the PMS2CL pseudogene makes somatic variant calling challenging and may bias VRF.
We used quantitative probabilities of pathogenicity from InSiGHT as pretest probability, if available. If probabilities were not reported in InSiGHT, we used minimal implied InSiGHT or UMD probabilities of pathogenicity based on reported variant classification. Minimal probabilities of pathogenicity of 1%, 5%, 95%, and 99% corresponded to classifications of benign (class 1), likely benign (class 2), likely pathogenic (class 4), and pathogenic (class 5), respectively.35, 36 If no pretest probability or prior classification information was available, we calculated in silico priors from either MAPP+ PP2 or PP2 derived from Thompson et al. for missense mutations in MLH1, MSH2, and MSH6.9 For splice site variants, we calculated in silico priors by comparing z-scores above cutoffs defined by MaxEnt score distributions for all MMR genes with pretest probabilities defined by Vallee et al.37 and applied to MMR genes by Tricarico et al.38 (Table S1). Consistent with published multifactorial classification algorithms, we limited maximum prior probability to 0.9 and minimum prior probability to 0.1 for in silico predictions for missense variants and allowed splice predictions for variants disrupting canonical splice sites to define greater priors. No validated prior probabilities were available for synonymous variants not predicted to alter canonical splice sites and for missense variants in PMS2, so we used probabilities implied by ClinVar classification if there was consensus between laboratories. For the remaining variants we used the neutral pretest probability of 0.5. Prior probability was converted to prior-odds and multiplied by likelihood based on VRF described above to generate post-test odds and probability.
Suggested variant classifications followed IARC recommendations and InSiGHT classes (probabilities of pathogenicity of <0.1%, 0.1% to 5%, 5% to 95%, 95% to 99%, and >99% correspond to classifications of benign (class 1), likely benign (class 2), uncertain significance (class 3), likely pathogenic (class 4), and pathogenic (class 5), respectively.35, 36 This exercise was not intended to yield definitive classifications, as complete details of consortium information used for priors was not available to us; however, we do believe this is an effective illustration of the power of quantitative information from tumors.
Results
Samples
Of 337 samples, we excluded 16 for which no paired normal sample was available, 48 with constitutional findings that might explain molecular phenotype, and 26 with inadequate DNA quality or other explanation for molecular phenotype. Of the remaining 247 samples included in analysis, 142 samples were from colorectal tumors; 94 samples were from endometrial tumors; and 11 were from other tumor types including ovarian, cecal, small bowel, and bladder tumors. We detected a median of 5.5 somatic mutations per case subject in exons and introns of the MLH1, MSH2, MSH6, and PMS2 gene regions.
Likelihood Ratio Estimates
Likelihood ratio estimates were derived from 174 samples. In these samples we observed 153 pathogenic and 760 intronic variants (Figure 2). For tumors without evidence of LOH in MMR genes, normalized VRF above 0.35 had a LR for pathogenicity of 1.56 (95% CI 1.42–1.71). Normalized VRF below 0.35 were associated with a LR of 0.2 (95% CI 0.11–0.39) (Table S2). For tumors with evidence of LOH in MMR genes, normalized VRF above 0.80 had a LR for pathogenicity of 26.5 (95% CI 13.2–53.0). Normalized VRF below 0.80 was associated with a LR of 0.16 (95% CI 0.08–0.32) (Table S2).
Validation with Established Constitutional MMR Variants and Functional Data
We observed 36 mutations that had been classified as pathogenic or benign by the InSiGHT Consortium. Evidence from tumor mutations was consistent with classifications in 33 of 36 (92%) (see Table 1). Yeast assay, mammalian cell assay, RT-PCR, or splicing reporter assay data from published literature were available for eight additional variants.26, 27, 28, 29, 30, 31, 32, 33 Evidence from tumor mutations was consistent with assays findings for seven out of eight (88%) of these (see Table 1). Cosegregation analysis supporting pathogenicity has been reported for two families with unclassified constitutional variants where observed somatic mutations also support pathogenicity.
Table 1.
Validation with Established Constitutional MMR Variants and Functional Data
| Sample | Coordinates (hg19) | Ref | Var | Gene | Base Change | Protein Change | LOH | Adjusted VAF | LR | Prior Evidence Source | Prior Evidence Consistent with Tumor Observation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor Results Consistent with Published Information Supporting Benign Classification | |||||||||||
| 10113 | chr3:37090070 | G | T | MLH1 | c.1959G>T | p.Leu653(=) | 1 | 0.179 | 0.16 | InSiGHT Class 1 | yes |
| Tumor Results Supporting Benign Classification Inconsistent with Published Information Supporting Pathogenic Classification | |||||||||||
| 9618 | chr3:37038124 | C | T | MLH1 | c.131C>T | p.Ser44Phe | 1 | 0.254 | 0.16 | InSiGHT Class 5 | no |
| 10113 | chr3:37083822 | G | A | MLH1 | c.1731G>A | p.Ser577(=) | 1 | 0.751 | 0.16 | InSiGHT Class 5 | no |
| 13867 | chr3:37083823 | G | A | MLH1 | c.1731+1G>A | – | 1 | 0.381 | 0.16 | InSiGHT Class 5 | no |
| 11531 | chr2:47637384 | T | C | MSH2 | c.518T>C | p.Leu173Pro | 1 | 0.486 | 0.16 | cell assay: loss positive test of MMR activity29 | no |
| Tumor Results Consistent with Published Information Supporting Pathogenic Classification | |||||||||||
| 23459 | chr2:47643569 | G | A | MSH2 | c.1076+1G>A | – | 0 | 0.472 | 1.56 | InSiGHT Class 5 | yes |
| 23270 | chr2:47698103 | G | A | MSH2 | c.1662−1G>A | – | 0 | 0.618 | 1.56 | InSiGHT Class 5 | yes |
| 62_B02 | chr2:48033790 | G | A | MSH6 | c.4001G>A | p.Arg1334Gln | 0 | 0.450 | 1.56 | InSiGHT Class 5 | yes |
| 26732 | chr2:48033790 | G | A | MSH6 | c.4001G>A | p.Arg1334Gln | 0 | 0.708 | 1.56 | InSiGHT Class 5 | yes |
| 22846 | chr3:37035039 | A | G | MLH1 | c.1A>G | p.? | 1 | 1.198 | 26.5 | InSiGHT Class 5 | yes |
| 25027 | chr3:37038124 | C | T | MLH1 | c.131C>T | p.Ser44Phe | 0 | 0.404 | 1.56 | InSiGHT Class 5 | yes |
| 11775 | chr3:37038192 | G | A | MLH1 | c.199G>A | p.Gly67Arg | 0 | 0.482 | 1.56 | InSiGHT Class 5 | yes |
| 23274 | chr3:37038192 | G | A | MLH1 | c.199G>A | p.Gly67Arg | 1 | 1.065 | 26.5 | InSiGHT Class 5 | yes |
| 20039 | chr3:37038193 | G | A | MLH1 | c.200G>A | p.Gly67Glu | 1 | 0.983 | 26.5 | InSiGHT Class 5 | yes |
| 23526 | chr3:37042483 | C | T | MLH1 | c.245C>T | p.Thr82Ile | 0 | 0.519 | 1.56 | InSiGHT Class 5 | yes |
| 18534 | chr3:37045935 | C | T | MLH1 | c.350C>T | p.Thr117Met | 0 | 0.423 | 1.56 | InSiGHT Class 5 | yes |
| 22162 | chr3:37045935 | C | T | MLH1 | c.350C>T | p.Thr117Met | 1 | 1.016 | 26.5 | InSiGHT Class 5 | yes |
| 25324 | chr3:37045935 | C | T | MLH1 | c.350C>T | p.Thr117Met | 1 | 0.885 | 26.5 | InSiGHT Class 5 | yes |
| 26434 | chr3:37045935 | C | T | MLH1 | c.350C>T | p.Thr117Met | 1 | 1.270 | 26.5 | InSiGHT Class 5 | yes |
| 23266 | chr3:37053358 | G | A | MLH1 | c.588+5G>A | – | 0 | 0.414 | 1.56 | InSiGHT Class 5 | yes |
| 15507 | chr3:37053590 | G | A | MLH1 | c.677G>A | p.Arg226Gln | 1 | 0.924 | 26.5 | InSiGHT Class 5 | yes |
| 24826 | chr3:37055984 | T | C | MLH1 | c.739T>C | p.Ser247Pro | 1 | 1.073 | 26.5 | InSiGHT Class 5 | yes |
| 60_C02 | chr3:37056036 | G | A | MLH1 | c.790+1G>A | – | 0 | 0.421 | 1.56 | InSiGHT Class 5 | yes |
| 13877 | chr3:37056036 | G | A | MLH1 | c.790+1G>A | – | 1 | 1.064 | 26.5 | InSiGHT Class 5 | yes |
| 22178 | chr3:37056036 | G | A | MLH1 | c.790+1G>A | – | 0 | 0.675 | 1.56 | InSiGHT Class 5 | yes |
| 22180 | chr3:37056036 | G | A | MLH1 | c.790+1G>A | – | 0 | 0.585 | 1.56 | InSiGHT Class 5 | yes |
| 25930 | chr3:37056036 | G | A | MLH1 | c.790+1G>A | – | 0 | 0.578 | 1.56 | InSiGHT Class 5 | yes |
| 21860 | chr3:37061954 | G | C | MLH1 | c.1038G>C | p.Gln346His | 0 | 0.509 | 1.56 | InSiGHT Class 5 | yes |
| 21860 | chr3:37061954 | G | C | MLH1 | c.1038G>C | p.Gln346His | 0 | 0.500 | 1.56 | InSiGHT Class 5 | yes |
| 16257 | chr3:37081676 | G | T | MLH1 | c.1559−1G>T | – | 1 | 1.041 | 26.5 | InSiGHT Class 5 | yes |
| 14421 | chr3:37083822 | G | A | MLH1 | c.1731G>A | p.Ser577(=) | 0 | 0.458 | 1.56 | InSiGHT Class 5 | yes |
| 12646 | chr3:37090053 | C | T | MLH1 | c.1942C>T | p.Pro648Ser | 1 | 1.064 | 26.5 | InSiGHT Class 5 | yes |
| 21546 | chr3:37090101 | G | A | MLH1 | c.1989+1G>A | – | 0 | 0.620 | 1.56 | InSiGHT Class 5 | yes |
| 23272 | chr3:37090101 | G | A | MLH1 | c.1989+1G>A | – | 0 | 0.556 | 1.56 | InSiGHT Class 5 | yes |
| 7243 | chr3:37090394 | G | A | MLH1 | c.1990−1G>A | – | 0 | 0.578 | 1.56 | InSiGHT Class 5 | yes |
| 25930 | chr3:37090446 | G | A | MLH1 | c.2041G>A | p.Ala681Thr | 0 | 0.614 | 1.56 | InSiGHT Class 5 | yes |
| 12036 | chr3:37090446 | G | A | MLH1 | c.2041G>A | p.Ala681Thr | 0 | 0.530 | 1.56 | InSiGHT Class 5 | yes |
| 19754 | chr3:37035147 | G | A | MLH1 | c.109G>A | p.Glu37Lys | 1 | 1.059 | 26.5 | cosegregation: LOD 2.1 UMDa | yes |
| 23461 | chr3:37038169 | T | A | MLH1 | c.176T>A | p.Ile59Asn | 0 | 0.667 | 1.56 | yeast assay: >67% loss of MMR activity26 | yes |
| 22182 | chr3:37055976 | G | T | MLH1 | c.731G>T | p.Gly244Val | 1 | 0.903 | 26.5 | yeast assay: partial loss of MMR function27 | yes |
| 21084 | chr3:37058999 | C | A | MLH1 | c.793C>A | p.Arg265Ser | 0 | 0.520 | 1.56 | yeast assay: loss of activity28 | yes |
| 23268 | chr3:37056038 | A | T | MLH1 | c.790+3A>T | – | 1 | 1.067 | 26.5 | splicing reporter assay and RT-PCR assay UMDb | yes |
| 21860 | chr3:37056039 | A | T | MLH1 | c.790+4A>T | – | 0 | 0.509 | 1.56 | splicing reporter assay31 | yes |
| 20046 | chr2:47702266 | G | A | MSH2 | c.1862G>A | p.Arg621Gln | 0 | 0.458 | 1.56 | cosegregation: LOD 0.6 UMDc | yes |
| 24631 | chr2:47672686 | G | A | MSH2 | c.1277−1G>A | – | 0 | 0.368 | 1.56 | splicing RT-PCR assay, UMDd | yes |
| 16529 | chr2:48026876 | T | C | MSH6 | c.1754T>C | p.Leu585Pro | 0 | 0.590 | 1.56 | MRR complementation assay: loss of activity30 | yes |
All four instances where tumor data were not consistent with clinical classifications or functional assay data were instances where pathogenic mutations were seen at VRF consistent with passenger status in tumors with clear evidence of LOH (Table 1). For each of the three instances with clinical classifications, InSiGHT contains substantial data that these variants are pathogenic. The one instance where tumor data were inconsistent with functional data, MSH2 (c.518T>C [p.Leu173Pro]), showed 14.5% repair efficiency in a mammalian MMR activity assay, was reported in an Amsterdam II family, and is classified as class 3 by InSiGHT.29
Evaluation in MMR-Proficient Tumors
In 356 microsatellite stable and IHC normal tumors we observed 8 potentially causative somatic mutations in MMR genes in 6 tumors (1.7%). Six mutations had normalized VRF over 0.35 in tumors without LOH and one had a normalized VRF over 0.8, consistent with LOH. Two separate tumors were observed with two heterozygous MMR gene mutations, each of which had a mutation profile consistent with the POLE-ultramutator tumor phenotype. In tumors without features of MMR deficiency, we were unable to distinguish whether somatic mutations are passengers that are the result of other mutational processes or if these are driver mutations seen in the context of false negative MSI and IHC results. Because likelihood ratios were developed in the context of a specific molecular phenotype (i.e., only in MSI-H tumors), we cannot extend likelihood findings to variants in tumors without the appropriate molecular phenotype.
Risk of Variant Misclassification Using Tumor Data
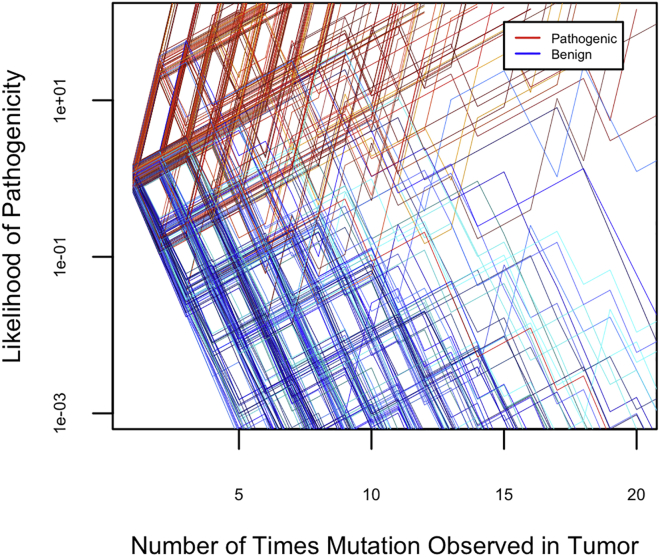
If data from sequential tumors with unexplained MMR deficiency are used widely in variant classification, the data from independent mutational events will accumulate over time (Figure 3). Longitudinal use of sequential tumor data to classify constitutional variants in MMR genes using only tumor data is predicted to result in misclassification of approximately 10 of every 10,000 pathogenic variants (0.1%) and 34 of every 10,000 benign variants (0.34%). Pathogenic variants are likely to be classified more quickly than benign variants because likelihood ratios supporting benign classification are modest compared to the likelihood ratios supporting pathogenic classification. This difference is because pathogenic mutations may be observed as passengers subsequent to other pathogenic driver mutations, while it is apparently more uncommon for a passenger to be observed at a VRF similar to those of driver mutations.
Figure 3.
Expected Variant Classification Pathways if Likelihood Data from Many Independent Tumors Is Combined over Time
A simulated sample of 200 pathogenic and 200 benign variants is shown.
Multifactorial Classification of Variants
To illustrate how data from tumors can be used in quantitative classification of constitutional variants, we generated multifactorial likelihood calculations for missense, synonymous, and splice variants consistent with IHC staining identified in 214 tumor-normal pairs (Table S3). Of the 247 tumor-normal pairs, 33 had only clearly pathogenic loss of function somatic mutations or had no identified somatic variants in MMR genes. We identified somatic events representing 165 missense, splice, and synonymous variants that had not previously been classified as definitively pathogenic or benign by the InSiGHT Consortium. ClinVar lacks entries for 90 (59%) of these variants. Twenty-two variants were represented by more than one somatic event (14%). Multifactorial classification calculation data are presented in Table S3.
Tumor evidence supported pathogenicity for 105 variants and was consistent with passenger status and benign classification for 73 variants (Table S3). For three variants—MLH1 (c.2040C>T [p.Cys680(=)]), MSH6 (c.1082G>A [p.Arg361His]), and MSH6 (c.3300G>A [p.Thr1100(=)])—somatic events contributed evidence both for and against pathogenicity. Based on our multifactorial likelihood calculation, we suggest classifying 7 variants as benign, 31 as likely benign, 22 as likely pathogenic, and 26 as pathogenic. Forty-four of these variants have yet to be reported as constitutional variants in individuals. If this tumor data were to be incorporated into InSiGHT classification, 32 variants might be updated (Table S3).
Discussion
Tumor data can support both pathogenic and benign variant classification in specific situations when the correlation between tumor phenotype and constitutional genotype is well defined. We use the specific example of observed somatic mutations in MMR genes in tumors with unexplained MMR deficiency (abnormal tumor screening without MLH1 methylation or constitutional MMR mutation). We illustrate this principle using multifactorial likelihood calculations for a large number of variants. This quantitative tumor data will add to the many sources of data used by expert panels focused on classifying variants in MMR genes and hasten variant classification efforts.
The high correlation between somatic findings and past classification of constitutional variants supports the observation that pathogenic changes are identical, whether somatic or constitutional, in their effect on tumor phenotype and MMR gene function.10, 11, 12, 13 The presence of passenger pathogenic hits in tumors is well established and indicates some risk of type 2 error when these data are used to classify a benign variant. Similarly, only modest evidence is obtained for heterozygous somatic mutations, as it is not unusual to see presumably benign intronic mutations at heterozygous read fractions. Nevertheless, quantification of misclassification risk illustrates that over time the overall risk of type 2 error from using tumor data will be low. Even without other forms of evidence, the large amount of data that tumors yield is expected to result in correct classification of the vast majority of variants in MMR genes (Figure 3).
In the past, in silico predictions of splice alterations have been considered more robust than computational predictions of missense mutations;36, 37 however, functional studies using RT-PCR or minigene constructs are considered important to confirm analysis.1 Even with RT-PCR and minigene data, in some situations evidence of clinical effect may be needed to confirm the pathogenicity of a splice-altering variant.39 Our data suggest that information obtained from tumor samples provides independent biological evidence of clinically meaningful splice disruptions. This evidence may allow a high degree of certainty about the pathogenicity of splice variants even in the absence of population data.
This analysis was performed on samples of predominantly colorectal or endometrial cancers with unexplained MMR deficiency. Finding double somatic mutations in persons with unexplained MMR deficiency can alleviate concern for Lynch syndrome. Paired tumor-normal analysis in this context does not necessarily prove that the individual does not have a constitutional MMR mutation, but it does dramatically reduce this probability.13 Another benefit of paired tumor-normal analysis is the ability to extract information about the functional effects of variants in MMR genes. Tumors present an ideal natural experiment to test the effect of genetic alterations in MMR genes.
Current ACMGG-AMP variant classification guidelines do not include use of data from tumor variants. However, evidence of de novo somatic driver mutations that define a highly characteristic tumor phenotype might be considered to be similar to a de novo constitutional mutation in a gene explaining a highly characteristic phenotype. We have shown that concerns about the high frequency of other tumor mutations potentially explaining the characteristic phenotype can be mitigated by quantifying this frequency. Gene-specific expert panels, like the ClinGen panel for MMR genes, will undoubtedly be equipped to incorporate quantitative likelihood data into variant classifications by quantifying ACMGG-AMP criteria40 or using custom criteria. Although we believe quantitative gene-specific methods are superior to general qualitative rules, we propose a modification to ACMGG-AMP variant classification guidelines to incorporate general molecular phenotype data, such as tumor data (Table 2). Molecular phenotype data differ from functional assay data because phenotypes are observed in nature rather than being engineered in the laboratory setting. Any evaluation of molecular phenotype will necessarily need to be validated to demonstrate the strength of correlation of molecular phenotype with known pathogenic genotypes. We have given an example of how this might be done for MMR variants seen in tumors. Although some molecular phenotype data may be acceptable as very strong evidence in the future, we believe that combining data from other sources with molecular data will help avoid rare false positives (see Figure 3). We hope that this work will facilitate systematic analysis of tumor variants in clinical laboratories and future research on the use of information from tumors or other molecular phenotypes to classify constitutional variants responsible for other genetic syndromes.
Table 2.
Proposed Addition to ACMG-AMP Guidelines for Variant Classification to Incorporate Molecular Phenotype with Examples from Specific Rules for Somatic MMR Gene Mutations
|
Benign |
Pathogenic |
||||
|---|---|---|---|---|---|
| Strong | Supporting | Supporting | Moderate | Strong | Very Strong |
| ACMG General Rule | |||||
| variant observed in a context that has been proven to be incompatible with molecular phenotype of pathogenic variants | variant observed in the context where other findings entirely explain molecular phenotype | variant observed in tissue contributes to phenotype associated with established pathogenic variants | variant likely to explain phenotype associated with pathogenic variants | variant observed in tissue explains phenotype associated with established pathogenic variants | |
| MMR-Specific Rule | |||||
| observed multiple times as a passenger somatic mutation in MMR gene where other mutations explain MSI and IHC patterns | somatic mutation at a VAF consistent with a passenger mutation in MMR gene where MSI and IHC are explained by other mutations | one of two somatic mutations in MMR gene that together explain MSI and IHC status without observed LOH | – | somatic mutation with clear LOH in MMR gene entirely explains MSI and IHC status | |
Declaration of Interests
T.W. consults for Color Genomics. H.H. is on the scientific advisory boards for InVitae and Genome Medical and owns stock in Genome Medical. All other authors declare no competing interests.
Acknowledgments
This research supported by the Damon Runyon Cancer Research Foundation (DRR-33-15), the NHGRI (R21HG008513), and the Fred Hutch/University of Washington Cancer Consortium (NCI 5P30 CA015704-39). Research was also partially supported by Ohio Colorectal Cancer Prevention Initiative research grant (Pelotonia, philanthropic funds).
Published: June 7, 2018
Footnotes
Supplemental Data include three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.05.001.
Web Resources
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Supek F., Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teare M.D., Rohde K., Santibáñez Koref M.F. The use of loss of constitutional heterozygosity data to ascertain the location of predisposing genes in cancer families. J. Med. Genet. 1994;31:448–452. doi: 10.1136/jmg.31.6.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemminki A., Peltomäki P., Mecklin J.P., Järvinen H., Salovaara R., Nyström-Lahti M., de la Chapelle A., Aaltonen L.A. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat. Genet. 1994;8:405–410. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S.A. Current Lynch syndrome tumor screening practices: a survey of genetic counselors. J. Genet. Couns. 2014;23:38–47. doi: 10.1007/s10897-013-9603-5. [DOI] [PubMed] [Google Scholar]
- 6.Beamer L.C., Grant M.L., Espenschied C.R., Blazer K.R., Hampel H.L., Weitzel J.N., MacDonald D.J. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J. Clin. Oncol. 2012;30:1058–1063. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giardiello F.M., Allen J.I., Axilbund J.E., Boland C.R., Burke C.A., Burt R.W., Church J.M., Dominitz J.A., Johnson D.A., Kaltenbach T., US Multi-Society Task Force on Colorectal Cancer Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology. 2014;147:502–526. doi: 10.1053/j.gastro.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Hegde M., Ferber M., Mao R., Samowitz W., Ganguly A. ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis) Genet. Med. 2014;16:101–116. doi: 10.1038/gim.2013.166. [DOI] [PubMed] [Google Scholar]
- 9.Thompson B.A., Goldgar D.E., Paterson C., Clendenning M., Walters R., Arnold S., Parsons M.T., Michael D W., Gallinger S., Haile R.W., Colon Cancer Family Registry A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Hum. Mutat. 2013;34:200–209. doi: 10.1002/humu.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sourrouille I., Coulet F., Lefevre J.H., Colas C., Eyries M., Svrcek M., Bardier-Dupas A., Parc Y., Soubrier F. Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam. Cancer. 2013;12:27–33. doi: 10.1007/s10689-012-9568-9. [DOI] [PubMed] [Google Scholar]
- 11.Mensenkamp A.R., Vogelaar I.P., van Zelst-Stams W.A., Goossens M., Ouchene H., Hendriks-Cornelissen S.J., Kwint M.P., Hoogerbrugge N., Nagtegaal I.D., Ligtenberg M.J. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146:643–646 e8. doi: 10.1053/j.gastro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Geurts-Giele W.R., Leenen C.H., Dubbink H.J., Meijssen I.C., Post E., Sleddens H.F., Kuipers E.J., Goverde A., van den Ouweland A.M., van Lier M.G. Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J. Pathol. 2014;234:548–559. doi: 10.1002/path.4419. [DOI] [PubMed] [Google Scholar]
- 13.Haraldsdottir S., Hampel H., Tomsic J., Frankel W.L., Pearlman R., de la Chapelle A., Pritchard C.C. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147:1308–1316.e1. doi: 10.1053/j.gastro.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira L., Balaguer F., Lindor N., de la Chapelle A., Hampel H., Aaltonen L.A., Hopper J.L., Le Marchand L., Gallinger S., Newcomb P.A., EPICOLON Consortium Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearlman R., Frankel W.L., Swanson B., Zhao W., Yilmaz A., Miller K., Bacher J., Bigley C., Nelsen L., Goodfellow P.J., Ohio Colorectal Cancer Prevention Initiative Study Group Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3:464–471. doi: 10.1001/jamaoncol.2016.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofstra R.M., Spurdle A.B., Eccles D., Foulkes W.D., de Wind N., Hoogerbrugge N., Hogervorst F.B., IARC Unclassified Genetic Variants Working Group Tumor characteristics as an analytic tool for classifying genetic variants of uncertain clinical significance. Hum. Mutat. 2008;29:1292–1303. doi: 10.1002/humu.20894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguiar P.N., Jr., Tadokoro H., Forones N.M., de Mello R.A. MMR deficiency may lead to a high immunogenicity and then an improvement in anti-PD-1 efficacy for metastatic colorectal cancer. Immunotherapy. 2015;7:1133–1134. doi: 10.2217/imt.15.84. [DOI] [PubMed] [Google Scholar]
- 18.Westdorp H., Fennemann F.L., Weren R.D., Bisseling T.M., Ligtenberg M.J., Figdor C.G., Schreibelt G., Hoogerbrugge N., Wimmers F., de Vries I.J. Opportunities for immunotherapy in microsatellite instable colorectal cancer. Cancer Immunol. Immunother. 2016;65:1249–1259. doi: 10.1007/s00262-016-1832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard C.C., Smith C., Salipante S.J., Lee M.K., Thornton A.M., Nord A.S., Gulden C., Kupfer S.S., Swisher E.M., Bennett R.L. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J. Mol. Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pritchard C.C., Salipante S.J., Koehler K., Smith C., Scroggins S., Wood B., Wu D., Lee M.K., Dintzis S., Adey A. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J. Mol. Diagn. 2014;16:56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favero F., Joshi T., Marquard A.M., Birkbak N.J., Krzystanek M., Li Q., Szallasi Z., Eklund A.C. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niknafs N., Beleva-Guthrie V., Naiman D.Q., Karchin R. SubClonal hierarchy inference from somatic mutations: automatic reconstruction of cancer evolutionary trees from multi-region next generation sequencing. PLoS Comput. Biol. 2015;11:e1004416. doi: 10.1371/journal.pcbi.1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bozic I., Gerold J.M., Nowak M.A. Quantifying clonal and subclonal passenger mutations in cancer evolution. PLoS Comput. Biol. 2016;12:e1004731. doi: 10.1371/journal.pcbi.1004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Easton D.F., Deffenbaugh A.M., Pruss D., Frye C., Wenstrup R.J., Allen-Brady K., Tavtigian S.V., Monteiro A.N., Iversen E.S., Couch F.J., Goldgar D.E. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plazzer J.P., Sijmons R.H., Woods M.O., Peltomäki P., Thompson B., Den Dunnen J.T., Macrae F. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam. Cancer. 2013;12:175–180. doi: 10.1007/s10689-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 26.Ellison A.R., Lofing J., Bitter G.A. Human MutL homolog (MLH1) function in DNA mismatch repair: a prospective screen for missense mutations in the ATPase domain. Nucleic Acids Res. 2004;32:5321–5338. doi: 10.1093/nar/gkh855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi M., Shimodaira H., Andreutti-Zaugg C., Iggo R., Kolodner R.D., Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–4604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- 28.Wanat J.J., Singh N., Alani E. The effect of genetic background on the function of Saccharomyces cerevisiae mlh1 alleles that correspond to HNPCC missense mutations. Hum. Mol. Genet. 2007;16:445–452. doi: 10.1093/hmg/ddl479. [DOI] [PubMed] [Google Scholar]
- 29.Ollila S., Sarantaus L., Kariola R., Chan P., Hampel H., Holinski-Feder E., Macrae F., Kohonen-Corish M., Gerdes A.-M., Peltomäki P. Pathogenicity of MSH2 missense mutations is typically associated with impaired repair capability of the mutated protein. Gastroenterology. 2006;131:1408–1417. doi: 10.1053/j.gastro.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 30.Kantelinen J., Kansikas M., Candelin S., Hampel H., Smith B., Holm L., Kariola R., Nyström M. Mismatch repair analysis of inherited MSH2 and/or MSH6 variation pairs found in cancer patients. Hum. Mutat. 2012;33:1294–1301. doi: 10.1002/humu.22119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianchi F., Raponi M., Piva F., Viel A., Bearzi I., Galizia E., Bracci R., Belvederesi L., Loretelli C., Brugiati C. An intronic mutation in MLH1 associated with familial colon and breast cancer. Fam. Cancer. 2011;10:27–35. doi: 10.1007/s10689-010-9371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandval P., Fabre A.J., Gaildrat P., Baert-Desurmont S., Buisine M.-P., Ferrari A., Wang Q., Béroud C., Olschwang S. UMD-MLH1/MSH2/MSH6 databases: description and analysis of genetic variations in French Lynch syndrome families. Database. 2013;2013 doi: 10.1093/database/bat036. bat036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Béroud C., Collod-Béroud G., Boileau C., Soussi T., Junien C. UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum. Mutat. 2000;15:86–94. doi: 10.1002/(SICI)1098-1004(200001)15:1<86::AID-HUMU16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Hampel H., Pearlman R., Beightol M., Zhao W., Jones D., Frankel W.L., Goodfellow P.J., Yilmaz A., Miller K., Bacher J., Ohio Colorectal Cancer Prevention Initiative Study Group Assessment of tumor sequencing as a replacement for Lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol. 2018 doi: 10.1001/jamaoncol.2018.0104. Published online March 29, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V., IARC Unclassified Genetic Variants Working Group Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson B.A., Spurdle A.B., Plazzer J.P., Greenblatt M.S., Akagi K., Al-Mulla F., Bapat B., Bernstein I., Capellá G., den Dunnen J.T. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vallée M.P., Di Sera T.L., Nix D.A., Paquette A.M., Parsons M.T., Bell R., Hoffman A., Hogervorst F.B., Goldgar D.E., Spurdle A.B., Tavtigian S.V. Adding in silico assessment of potential splice aberration to the integrated evaluation of BRCA gene unclassified variants. Hum. Mutat. 2016;37:627–639. doi: 10.1002/humu.22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tricarico R., Kasela M., Mareni C., Thompson B.A., Drouet A., Staderini L., Gorelli G., Crucianelli F., Ingrosso V., Kantelinen J., InSiGHT Variant Interpretation Committee Assessment of the InSiGHT interpretation criteria for the clinical classification of 24 MLH1 and MSH2 gene variants. Hum. Mutat. 2017;38:64–77. doi: 10.1002/humu.23117. [DOI] [PubMed] [Google Scholar]
- 39.de la Hoya M., Soukarieh O., López-Perolio I., Vega A., Walker L.C., van Ierland Y., Baralle D., Santamariña M., Lattimore V., Wijnen J. Combined genetic and splicing analysis of BRCA1 c.[594-2A>C; 641A>G] highlights the relevance of naturally occurring in-frame transcripts for developing disease gene variant classification algorithms. Hum. Mol. Genet. 2016;25:2256–2268. doi: 10.1093/hmg/ddw094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavtigian S.V., Greenblatt M.S., Harrison S.M., Nussbaum R.L., Prabhu S.A., Boucher K.M., Biesecker L.G. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018 doi: 10.1038/gim.2017.210. Published online January 4, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.